Abstract
JARID1B/KDM5B histone demethylase's mRNA is markedly overexpressed in breast cancer tissues and cell lines and the protein has been shown to have a prominent role in cancer cell proliferation and DNA repair. However, the mechanism of its post‐transcriptional regulation in cancer cells remains elusive. We performed a computational analysis of transcriptomic data from a set of 103 breast cancer patients, which, along with JARID1B upregulation, showed a strong downregulation of 2 microRNAs (miRNAs), mir‐381 and mir‐486, potentially targeting its mRNA. We showed that both miRNAs can target JARID1B 3′UTR and reduce luciferase's activity in a complementarity‐driven repression assay. Moreover, MCF7 breast cancer cells overexpressing JARID1B showed a strong protein reduction when transfected with mir‐486. This protein's decrease is accompanied by accumulation of DNA damage, enhanced radiosensitivity and increase of BRCA1 mRNA, 3 features previously correlated with JARID1B silencing. These results enlighten an important role of a miRNA’s circuit in regulating JARID1B's activity and suggest new perspectives for epigenetic therapies.
Keywords: breast cancer, DNA damage, Ionizing radiation, KDM5 histone demethylase, microRNA
1. INTRODUCTION
Chromatin structure dynamics regulate many aspects of cellular processes.1, 2 Histone tail modifications have a major role in determining structural and functional changes through a sophisticated signaling code, which is interpreted by a complex network of interacting proteins.3 Histone methylation in particular has been shown to be a key factor in transcription regulation and DDR.4 Mono‐methylation, di‐methylation and tri‐methylation can be catalyzed by specialized histone methyl transferases at specific lysine (K) and arginine (R) residues on histone tails and erased by specific histone demethylases (HDM). Two families of HDM have been identified in eukaryotes: the LSD1 family and the Jumonji C‐domain‐containing family. Jumonji C‐domain‐containing HDM (JHDM) are Fe2+ and α‐ketoglutarate‐dependent hydroxylases. They are divided into 7 phylogenetically distinct subfamilies (JHDM1‐3, JARID, PHF2/PHF8, UTX/UTY and JmjC domain only), with each group sharing a common set of substrates.5, 6
Jumonji C‐domain‐containing HDM have an important role in cancer epigenetics7 and, among them, those capable of demethylating specifically H3K4 (JARID 1A‐1D) look particularly interesting as potential therapeutic targets.8, 9
Indeed, JARID1A was found to be upregulated in gastric10 and cervical cancer.11 In particular, in gastric cancer, Zeng and colleagues observed that JARID1A is responsible for the repression of p16, p21 and p27.10
In parallel, JARID1B (previously known as PLU‐1), was initially identified as a markedly upregulated gene in breast cancer12 and later found overexpressed in prostate,13 bladder and lung cancer14 and melanomas.15 More importantly, Yamane and colleagues found that downregulation of JARID1B in MCF7 breast cancer cell line decreased cell growth.16 This was confirmed in an in vivo mouse model, in which shJARID1B led to suppression of mammary tumor growth.16, 17 Analogous results were obtained in bladder and lung cancer cell lines.14 Taken together, these observations lead to the conclusion that JARID1B could be a crucial player in regulating the development and growth of certain types of cancer. Nonetheless, JARID1B is downregulated in triple negative breast cancer subtype cell lines as MDA‐MB‐231. This negative modulation was proposed to be functional to cancer cell migration, invasiveness and metastatic potential, which characterize this subtype of aggressive breast cancer cell line.18 Thus, depending on the cellular environment, JARID1B might be considered as an oncogene or a tumor suppressor.
Moreover, the JARID1 subfamily has recently been confirmed to be deeply involved in the mechanism of DNA double strand break (DSB) repair (DDR). Although H3K4me3 was previously suggested to have a positive role in DNA repair,19 more recent observations by Li and colleagues20 suggest that it is H3K4me3 removal, rather than its presence, that facilitates DDR protein recruitment: JARID1B is, in fact, specifically enriched in DNA damage sites after irradiation, in a PARP1‐dependent and macroH2A1.1‐dependent mechanism. Interestingly, recruitment and catalytic activity of JARID1B are both required for local H3K4me3 demethylation, which allows for Ku70/80 or BRCA1 association to the site of damage, respectively, in non‐homologous end joining and homologous recombination (HR) pathways.20 In parallel, JARID1B‐depleted irradiated cells displayed an enhanced γ‐H2AX signal, along with a G1 arrest and a phosphorylated‐p53 (a downstream effector of ATM/ATR‐mediated CHK1/CHK2 activation). More interestingly, p53 activation and H2AX phosphorylation were also present in non‐irradiated cells, suggesting that JARID1B depleted cells are more prone to spontaneous DNA breaks.20 In addition to this, Gong and colleagues also described a similar role for JARID1A: this protein is recruited to the site of damage in a PARP1‐dependent manner and its demethylating activity on pre‐existing H3K4me3 is required for ZMYND8 recruitment to the site of damage,21 which, in turn, allows NuRD complex association to initiate HR‐related remodeling events.22, 23 In partial contrast with these observations, Penterling and colleagues suggested that JARID1A's role in DDR is not dependent on its catalytic activity. However, depletion of this protein enhances cells’ sensitivity to irradiation‐induced death. The authors proposed that this is probably because demethylating activity at the sites of damage is mostly carried out by JARID1B, as observed in Li et al (2014).20 Instead, sensitivity to ionizing radiation observed upon JARID1A depletion might be due to concurrent histone hyperacetylation.24
MicroRNAs (miRNAs) are endogenous small non‐coding RNAs that regulate gene expression by binding to the 3′ UTR of the mRNA targets.25 Increasing evidence shows a deregulation of miRNA expression in human cancer, indicating that they are potential biomarkers for cancer diagnosis and prognosis, as well as therapeutic targets or tools.26, 27 In this work, we demonstrate that one of the mostly downregulated miRNAs in human breast cancer, miR‐486, regulates JARID1B expression in human cell lines. In addition, we show that miR‐486 induced JARID1B downregulation is accompanied by accumulation of DNA damage and enhanced radiosensitivity.
2. MATERIALS AND METHODS
2.1. Computational analysis of breast cancer patients’ transcriptome
Gene expression data was obtained for the TCGA public database (cancergenome.nih.gov). For this research, datasets of mRNAs and small RNAs (RNA‐Seq) from breast cancer patients were used. Computational analysis was performed using R version 3.4.3. Only the subset of data related to samples that had both normal and cancer counterparts was taken into consideration, to assess differences in gene expression related to single patients. For this reason, all data related to “cancer‐only” or “normal‐only” samples (without a counterpart) were discarded from the analysis. The same principle was used to compare mRNA and miRNA data; therefore, the actual subset of patients used was composed of those who had all 4 kinds of data (mRNA normal, mRNA cancer, miRNA normal and miRNA cancer). The lists of the significant modulated mRNAs/miRNAs was obtained by applying the following criteria: (i) mRNAs/miRNAs whose expression profiles were characterized by an interquartile range (IQR) value that stood under the 10th percentile of the IQR value distribution were discarded; (ii) mRNAs/miRNAs with a foldchange <2 were discarded; and (iii) mRNAs/miRNAs with a false discovery rate (FDR) >.01 were discarded. These lists were cross‐checked to identify differentially expressed miRNAs that could be targeting differentially expressed mRNAs of interest. Data for miRNA‐mRNA targeting prediction was downloaded from TargetScan.org.
2.2. Kaplan‐Meier
We performed Kaplan‐Meier analyses28 by using clinical and expression data provided by the TCGA portal relating to 1062 patients affected by breast cancer. The patient samples are split into 2 groups (low‐expression and high‐expression) according to the expression level of a given transcript with respect to distribution of the expression values of all other transcripts. In particular, low‐expression and high‐expression groups refer to patients with expression levels lower than or greater than the 50th percentile, respectively. A log‐rank test was performed to evaluate the P‐value associated with each switch gene: the lower the P‐value, the better the separation between the prognoses of the 2 groups.
2.3. DNA constructs
miRNA overexpression plasmids (pSP65/U1‐miR‐381‐3p, pSP65/U1‐miR‐486‐5p miRNA OE plasmids) were obtained by amplifying miR‐381‐3p and miR‐486‐5p genomic regions, containing approximately 100 nt upstream and 100 nt downstream of the miRNA stem‐loop sequence (http://www.mirbase.org/). The PCR fragments were cloned downstream of the U1 promoter of a pSP65/U1vector obtained, as previously described.29
Transfected MCF7 cells displayed a high fold‐increase in miRNA expression (data not shown). Jarid1B 3′ UTR from human genomic DNA was amplified and cloned into the Xho1 and EcoR1 unique sites downstream of the Renilla coding sequence of Psi‐CHECK‐2 vector (Promega, Madison, WI, USA) (Psi‐CHECK/Jarid1B). To generate Jarid1B 3′ UTR mutant constructs, mutations at miR‐381‐3p (Psi‐CHECK/Jarid1B 381 mut) or miR‐486‐5p (Psi‐CHECK/Jarid1B 486 mut) seed binding sites were introduced into the Jarid1B 3′ UTR using synthetic oligonucleotides by generating partially complementary PCR fragments. These fragments were used as templates for PCR to generate complete mutated Jarid1B 3′ UTR fragments, and further cloned as described for wild‐type 3′ UTR. All constructs have been checked by sequencing. See Table S1 for primer sequences.
2.4. Cell culture and transfection
Human breast cancer HEK293, MCF7, T47D and MDA‐MB‐231 cells were a gift from Dr Lucia Gabriele (Istituto Superiore di Sanità, Rome, Italy), who purchased them from ATCC and maintained them in culture. Cells were cultured for, at most, 10‐12 passages, then replaced by cells from early passages, kept in liquid nitrogen since collection. The presence of mycoplasma was checked every 6 months using a Mycoplasma PCR Detection Kit (Sigma‐Aldrich, St. Louis, MO, USA).
Cells were grown in high‐glucose DMEM medium plus FBS, 1 mmol L−1 L‐glutamine, 100 U/mL penicillin and 100 μg/mL streptomycin (Thermo Fisher Scientific, Waltham, MA, USA). For RNA and protein analysis, transfections were performed at 70% confluency using Lipofectamine 2000 Reagent, according to the manufacturer's protocol (Thermo Fisher Scientific). Cells were transfected in 6‐well plates with 4 μg of miRNA overexpression plasmids and cell lysates were prepared at 48 and 72 hours after plasmid transfections for subsequent analysis as specified. Transfected MCF7 cells displayed a high fold‐increase in miRNA expression (data not shown).
2.5. RNA extraction and quantitative RT‐PCR
Total RNA extraction from transfected cells was performed using an miRNeasy Mini Kit, according to the manufacturer's guidelines (Qiagen, Germantown, MD, USA). Quantitative RT‐PCR analysis of miRNAs was done by miScript II RT‐PCR kit (Qiagen) with HiFlex buffer allowing retrotranscription of both mRNA and miRNA species. Relative quantification of gene expression was conducted with the Applied Biosystems 7500 Real‐Time PCR System and data analysis was performed using the comparative ΔΔCT method. Endogenous genes used as internal controls for miRNA and mRNA were U6 snRNA and GAPDH mRNA, respectively.
2.6. Luciferase assay
Psi‐CHECK‐2 vector (Promega) containing both Renilla and firefly luciferase genes was used to clone JARID1B 3′UTR downstream of the Renilla gene (psi‐CHECK/Jarid1B). For transfection efficiency, the Renilla luciferase signal was normalized upon firefly signal. To perform luciferase assays, cells were seeded on 96‐well plates, co‐transfected with both luciferase containing plasmid (psi‐CHECK/Jarid1B and empty psi‐CHECK) and miR‐OE plasmids (pSP65/U1‐miR‐381‐3p, pSP65/U1‐miR‐486‐5p). Luciferase assays were performed 24 and 48 hours after transfection, using the Dual‐Luciferase Reporter Assay System (Promega), according to protocol guidelines. To evaluate firefly (reference) activity, 100 μL of Luciferase Assay Reagent II (LAR II) was added to each well and luminescence was measured using a Wallac VICTOR2 1420 (Perkin‐Elmer, Waltham, MA, USA) plate reader. Renilla luciferase activity was expressed relatively to firefly activity.
miRNA effect on psi‐CHECK/Jarid1B reporter expression was evaluated as a percentage of luciferase activity from pSP65/U1‐miR‐381‐3p or pSP65/U1‐miR‐486‐5p transfected cells compared to control vector (pSP65/U1) transfected cells. Data shown is the result of at least 4 independent experiments.
2.7. Protein expression analysis
Transfected cells were harvested after 48 hours, DMEM was removed, cells were washed with Dulbecco's PBS (Carlo Erba Reagents) and then resuspended in 60 μL RIPA buffer (Sigma‐Aldrich) with protease inhibitor cocktail (Complete, EDTA‐free Protease Inhibitor Cocktail, Roche, Basel, Switzerland). Cell lysates were obtained according to the manufacturer's guidelines and stored at −20°C.
Western blot analysis was used to evaluate the JARID1B amount. For each sample, 40 μg of proteins were run on an 8% denaturing PAGE to resolve both 180 kDa JARID1B and 43 kDa actin that was used as loading control. Histones were resolved on a 12% PAGE, using 20 μg of RIPA extracted total proteins. Transfer efficiency was verified via ponceau staining (Sigma Diagnostics). Filters were hybridized with rabbit anti‐JARID1B (1:200, Cell Signaling Technology, Cat#3273 RRID: AB_1264191), rabbit anti‐actin (1:1000, Sigma‐Aldrich, Cat#A2066 RRID: AB_476693), rabbit anti‐H3K4me3 (1:1000. Active Motif, Cat#39915 RRID: AB_2687512), rabbit anti‐H2AX (1:1000, Cell Signaling Technology, Cat#7631, RRID: AB_10860771) and rabbit anti‐Phospho‐H2AX (1:1000, Cell Signaling Technology, Cat#9718, RRID: AB_2118009) at 37°C overnight. Goat anti‐rabbit HRP conjugate (1:10 000, Thermo Fisher Scientific Cat#31460, RRID: AB_228341) was used as a secondary antibody. For quantitative analysis, western blots were analyzed with a ChemiDoc XRS+ Imaging System (Bio‐Rad, Hercules, CA, USA). The signal for each protein was normalized to actin detected on the same blot, and ratios with average control values were determined.
2.8. Chromatin immunoprecipitation assay
ChIP assay was performed using the MAGnify Chromatin Immunoprecipitation System (Thermo Fisher Scientific). Then, 24‐hours post‐transfection, MCF7 cells were prepared according to the manufacturer's instructions. DNA‐bound protein was immune‐precipitated using the following antibodies: anti‐JARID1B (1:50, Cell Signaling Technology, Cat#3273 RRID: AB_1264191), anti‐H3K4me3 (1:50. Cell Signaling Technology, Cat# #9751 RRID: AB_2616028) or anti‐IgG (as negative control provided by kit). PCR amplification (35 cycles) was performed on 1 μL of input or 5 μL of DNA immuno‐precipitated with anti‐JARID1B, anti‐H3K4me3 or anti‐IgG, using DreamTaq DNA Polymerase (Thermo Scientific) and the following primers:
BRCA1 Fw 5′‐TAAGCCGCAACTGGAAGAGT‐3′
BRCA1 Rv 5′‐CAGAAAGAGCCAAGCGTCTC‐3′
ACTB Fw 5′‐GCCGGGACCTGACTGACTA‐3′
ACTB Rv 5′‐TGGTGATGACCTGGCCGT‐3′
2.9. Flow‐cytometry
Flow‐cytometry analysis of DNA content was performed using an EPICS xl flow‐cytometer (Beckman‐Coulter). At the indicated times from transfection, MCF7 cells were trypsinized, pelleted, washed with PBS and, finally, resuspended in PBS containing .1% Triton‐X‐100 (AppliChem) and 40 μg/mL propidium iodide (Sigma‐Aldrich). The samples were incubated for 20 min at 37°C and then analyzed, acquiring 10 000 events for each sample. Acquired data were analyzed using the WinMDI software by Joe Trotter, available at http://facs.scripps.edu
2.10. Irradiation and clonogenic assay
Cells were X‐ray irradiated at 200 V and 6 mA (Gilardoni MLG 300/6‐D) 24 hours post‐transfection to produce an equivalent absorbed dose of 1 cGy/s. X‐ray and sham‐irradiated cells were then incubated for 4 hours. Cells were harvested, seeded on 60‐mm wells at a density of 103 cells/well and incubated for 14 days. Fixing and staining of cell colonies were performed in .3% Methylene Blue, 80% ethanol solution for 30 minutes and extensively washed with ddH2O. Colorimetric images were taken using the ChemiDoc XRS+ Imaging System (Bio‐Rad). Colonies were counted using Fiji software,30 applying the “Find maxima” function and a background noise threshold of 3000. Proliferative capacities of each experimental group (pSP65/U1, pSP65/U1‐miR‐381‐3p and pSP65/U1‐miR‐486‐5p) was then calculated as the relative number of colonies for each radiation dose (1, 3 and 10 Gy), relativel to sham‐irradiated samples, to normalize any radiation‐independent effects of miRNAs on cells.
3. RESULTS
3.1. JARID1B/KDM5B upregulation in breast cancer patients correlates with a downregulation of predicted miRNAs targeting it
We performed a computational analysis of transcriptomic data from a set of 103 breast cancer patients (Cancer Genome Atlas public database). The analysis revealed that in tumor tissues, almost 4000 genes are modulated at least 2‐fold. Among these, several epigenetic players were found to be differentially expressed (summarized in Table 1). As expected from previous observations, JARID1B/KDM5B is upregulated in breast cancer patients, showing a 2‐fold increase, with highly statistical significance (FDR = 1.48 × 10−25). All the epigenetic modifiers reported in Table 1 are genes previously known to be modulated in breast cancer (DNMT3B,31 EZH2,32 HDAC4,33 HOTAIR,34 KAT2B,33 KDM5B,12 MEF2C and35 SMYD334), except for SMYD1/KMT3D, which was never reported to be modulated in this type of tumor. Interestingly, this latter gene is among the most repressed ones (−97.4% of expression with respect to normal tissue, FDR = 4.46 × 10−28). Next, we analyzed the list of significantly downregulated miRNAs to identify those potentially targeting JARID1B. According to TargetScan (http://www.targetscan.org), 12 miRNAs (reported in Table S2) showed the required sequence complementarity with JARID1B 3′UTR region. We focused on the 2 most repressed ones, as they are, in principle, the best candidates that could account for a JARID1B modulation in breast cancer tissue: miR‐486‐5p is found to be almost completely silenced (log2‐foldchange = −3.97, FDR = 3.27 × 10−31) and it is actually the most repressed miRNA of the whole analysis, while miR‐381‐3p shows a 3‐fold decrease (log2‐foldchange = −1.554, FDR = 2.91 × 10−32). miR‐486‐5p displays a higher sequence complementarity with the JARID1B 3′UTR with respect to miR‐381‐3p, that extends outside the seed region, suggesting a stronger targeting efficacy (Figure S1). JARID1B is also a bona fide target of both miRNAs according to microRNA.org (http://www.microrna.org) prediction. The box plot in Figure 1A summarizes JARID1B, miR‐381‐3p and miR‐486‐5p modulation in breast cancer samples with respect to normal tissues: in absolute figures, miR‐381‐3p is generally less expressed than miR‐486‐5p in normal conditions, while their expression is almost equalized in cancer cells. In particular, miR‐486‐5p shows a wide variability in the level of expression in normal conditions, that is sensibly reduced in cancer conditions, suggesting the emergence of a regulative mechanism that is not operating in normal conditions.
Table 1.
Modulated genes coding for epigenetic players in breast cancer tissues
| Gene | FDR | log2 fold change | Molecular function |
|---|---|---|---|
| EZH2 | 5.25E‐32 | 2.084353478 | H3K27 methylation |
| HOTAIR | 1.24E‐08 | 1.94264031 | PRC2 interactor |
| SMYD3 | 7.12E‐21 | 1.213517762 | H3K4 methylation |
| DNMT3B | 3.86E‐15 | 1.084809605 | DNA de novo methylation |
| KDM5B | 1.48E‐25 | 1.019012074 | H3K4me2/3 demethylation |
| HDAC4 | 2.57E‐26 | −1.134060683 | Histone deacetylation |
| MEF2C | 1.38E‐20 | −1.162203015 | HDAC4 interactor |
| KAT2B | 3.77E‐25 | −1.324736764 | Histone acetylation |
| SMYD1 | 4.46E‐28 | −5.279628314 | H3K4 methylation |
FDR, false discovery rate.
Figure 1.
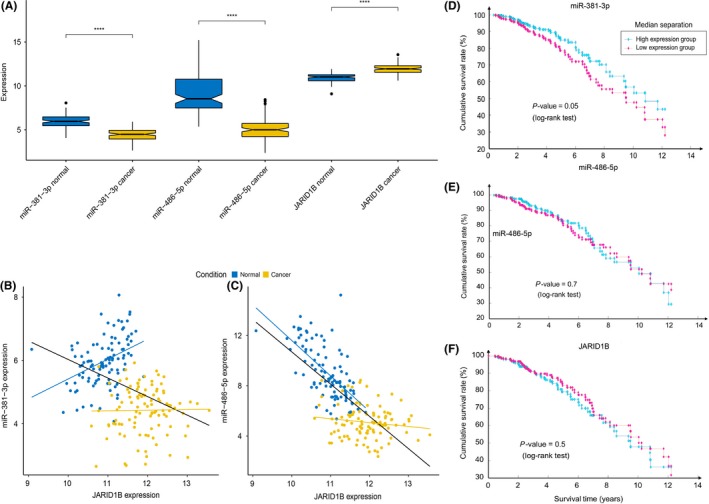
JARID1B/KDM5B, miR‐381‐3p and miR‐486‐5p expression level in breast cancer patients and association of their expression level with patients’ survival. A, Box plot showing miR‐381‐3p, miR‐486‐5p and JARID1B/KDM5B expression in normal (blue) and cancer (yellow) subsets. Statistical significance was assessed according to 2‐tailed paired Student's t test, ****P < .0001. B, Scatter plot of miR‐381‐3p vs JARID1B/KDM5B expression in both normal (blue) and cancer (yellow) samples. Pearson coefficients are calculated for total (ρ = −.40), normal‐only (ρN = .40) and cancer‐only (ρC = .02) datasets and respective correlation lines (black, blue, yellow) are reported in the plot. C, Scatter plot for miR‐486‐5p vs JARID1B/KDM5B expression in both normal (blue) and cancer (yellow) samples. Pearson coefficients are calculated for total (ρ = −.72), normal‐only (ρN = −.65) and cancer‐only (ρC = −.14) datasets and respective correlation lines (black, blue and yellow) are reported in the plot. D, Kaplan‐Meier diagram showing survival for patients with higher (>50th percentile, cyan) or lower (<50th percentile, magenta) expression of miR‐381. E, Kaplan‐Meier diagram showing survival for patients with higher (>50th percentile, cyan) or lower (<50th percentile, magenta) expression of miR‐486. F, Kaplan‐Meier diagram showing survival for patients with higher (>50th percentile, cyan) or lower (<50th percentile, magenta) expression of JARID1B
A scatter plot of miR‐381‐3p vs JARID1B expression (Figure 1B) reveals that normal samples form a separated cluster with respect to cancer samples. Correlation between miRNAs and their target protein levels does not always affect mRNA levels, as the canonical miRNA mode of action is limited to the repression of translation rather than leading to destabilization of mRNAs of their targets. However, it is possible that, in cancer cells, both phenotypes (miRNA downregulation and JARID1B mRNA upregulation) might arise independently. Concerning miR‐381‐3p, the Pearson coefficient calculated on the whole dataset suggests that its expression is anti‐correlated with that of JARID1B (Figure 1B, ρ = −.4); however, this anti‐correlation is not conserved in the 2 separated subsets of normal‐only and cancer‐only samples, meaning that, even though breast cancer cells display a concomitant JARID1B upregulation and miR‐381‐3p downregulation, these 2 alterations might arise independently from each other.
In contrast, the miR‐486‐5p scatter plot reveals a similar sample clustering (Figure 1C). However, in this case, its anti‐correlation might effectively account for a regulative mechanism, as it is preserved in the normal‐only subset, while it is lost in the cancer‐only subset, suggesting that in physiological conditions, the miRNA could be engaged in downregulating JARID1B mRNA accumulation but if its concentration is lower than a threshold level (as it might be in breast cancer), this mechanism is no longer relevant. Once again, anti‐correlation between a miRNA and its target mRNA does not always mean that there is a functional link between the 2 levels of expression: it is more likely an additive effect rather than a causative one. Therefore, it is possible that miR‐486‐5p contributes to the post‐transcriptional downregulation of JARID1B mRNA in normal conditions and that, in contrast, in cancer cells, this cooperative repression is lost, allowing higher levels of JARID1B mRNA to produce higher levels of JARID1B protein.
We next analyzed the association of the 2 miRNAs’ expression level with patient survival. The Kaplan‐Meier diagram in Figure 1D shows that patients with higher expression (>50th percentile) of miR‐381 have a significantly longer survival (P = .05) than those with a lower expression, while no significant effect is observed for miR‐486 (Fig. 1E). This could be due to the fact that while a consistent part of the cancer samples shows expression levels of miR‐381 comparable to the control population, most of the tumors have a very low expression level of miR‐486 compared to the normal population, making internal differences irrelevant for regulating the JARID1B protein level. The JARID 1B mRNA level itself does not show an impact on survival (Fig. 1F), contrary to what has recently been observed for its protein level, which generally negatively correlates with survival,36 confirming that post‐transcriptional regulation is crucial.
3.2. Validation of the inhibitory effect of miR‐381‐3p and miR‐486‐5p on JARID1B translation
Until very recently,37 no miRNAs showing sequence complementarity with JARID1B 3′UTR had been experimentally proven to be able to target its mRNA in vivo and reduce luciferase's activity in a complementarity‐driven repression assay.
To address whether miR‐381 and miR‐486 could effectively exploit their function by targeting complementary regions on JARID1B mRNA, the 3′UTR region of this gene was used as a recombinant 3′UTR for Renilla luciferase gene expressed by a reporter vector. The resulting construct was introduced by transfection in HEK 293 cells. We therefore transfected HEK 293 cells with plasmids overexpressing miR‐381‐3p and miR‐486‐5p, pSP65/U1‐miR‐381‐3p, pSP65/U1‐miR‐486‐5p, respectively. This assay only accounts for a qualitative evaluation of the molecular complementarity‐driven targeting mechanism performed by miRNAs, as the amount of ectopically‐expressed miRNAs obtained via U1 promoter does not correspond to any physiological or cancerous conditions reported. However, the results obtained (Figure 2A) clearly demonstrate that upon overexpression of both miRNAs, Renilla relative activity is decreased with respect to pSP65/U1 empty plasmid‐transfected cells. The statistical significance of this effect is high and reproducible, both at 24 and 48 hours, with the exception of the 24‐hour post‐transfection effects of miR‐381, whose modulation is only barely significant (P = .053). As a control, we produced constructs carrying the JARID1B mRNA 3′UTR region with mutated seeds for miR‐381‐3p and miR‐486‐5p, respectively. Transfection of HEK 293 cells carrying these constructs with the corresponding miRNAs did not cause any significant reduction of luciferase activity (Figure 2B). These data provide molecular evidence that both miR‐381‐3p and miR‐486‐5p are able to target JARID1B 3′UTR and, presumably, impair its translation into protein.
Figure 2.
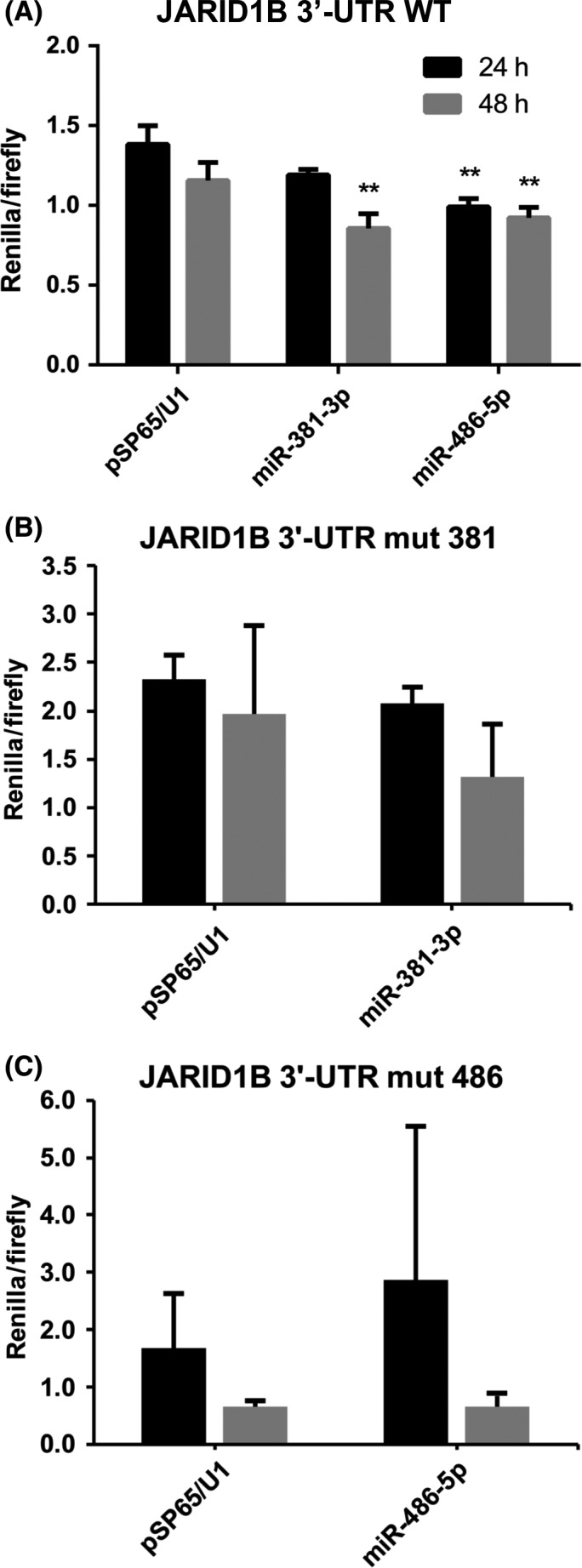
miR‐381‐3p and miR‐486‐5p target JARID1B 3′UTR. Renilla activity (normalized on firefly luciferase activity) measured from psiCHECK‐2 and pSP65/U1‐miR co‐transfected HEK 293 cells at 24 and 48 h post‐transfection, n = 6. Data are represented as the mean ± SD of Renilla/Firefly activity. Statistical significance was assessed according to a 2‐tailed Student's t test, **P < .01
3.3. MiR‐381‐3p and miR‐486‐5p overexpression impairs JARIDB protein accumulation in MCF7 breast cancer cells
To prove the role of miRNAs in regulating JARID1B translation in breast cancer, we transfected them in the breast cancer cell line MCF7 that overexpresses JARID1B.38, 39
Protein samples from 48‐hour post‐transfection cells were used to assess JARID1B protein levels by western blot analysis (Figure 3A). The results (summarized in Figure 3B) show that both miRNAs could account for the suggested repressive mechanism: overexpression of miR‐381‐3p reduces JARID1B's protein level by almost 40%, while miR‐486‐5p seems to be even more effective, as it reduces the protein level by around 60%. Nonetheless, H3K4me3 (JARID1B substrate) levels in bulk chromatin, are not significantly affected by JARID1B knockdown (Figure 3C), suggesting that this epigenetic regulator is probably not involved in a continuous global histone demethylation in breast cancer cells but rather in a more specific activity. However, we cannot rule out the possibility that a residual amount of the protein could still be able to carry out bulk chromatin demethylation to the same extent.
Figure 3.
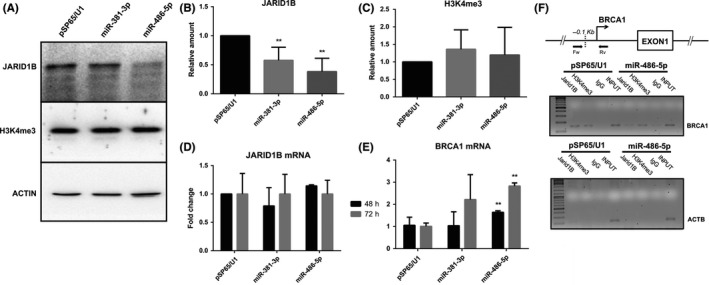
miR‐381‐3p and miR‐486‐5p overexpression impairs JARID1B mRNA translation without affecting JARID1B mRNA levels and upregulates the JARID1B target gene BRCA1. A, Representative western blotting image of JARID1B protein and H3K4me3 levels in cell lysate from MCF7 cells 24 h after transfection with control pSP65/U1, pSP65/U1‐miR‐381‐3p or pSP65/U1‐miR‐486‐5p. B and C, Relative quantification of JARID1B protein (B) and H3K4me3 (C) levels in western blotting analysis, n = 7. Data are represented as the mean ± SD of JARID1B or H3K4me3 levels relative to actin. Statistical significance was assessed according to 2‐tailed paired Student's t test, **P < .01. D and E, Quantitative RT‐PCR analysis of JARID1B (D) and BRCA1 (E) mRNA levels from MCF7 cells 48 and 72 hours after transfection with control pSP65/U1, pSP65/U1‐miR‐381‐3p or pSP65/U1‐miR‐486‐5p, n = 4. JARID1B and BRCA1 raw data (Ct) were normalized on GAPDH expression levels (ΔCt) as endogenous control and ΔΔCt was calculated with respect to control transfection (pSP65/U1). Data are represented as the mean ± SD. Statistical significance was assessed according to 2‐tailed Student's t test, **P < .01. F, ChIP assay (see Materials and Methods) to test JARID1B association and H3K4 trimethylation level at the BRCA1 promoter. The location of the primers used for the PCR amplification of the BRCA1 proximal promoter segment is reported in the map at the top. The ACTB exon V region is used as a negative control for JARID1B binding and H3K4 tri‐methylation
3.4. Overexpression of miR‐381‐3p and miR‐486‐5p does not affect the levels of JARID1B mRNA
Using quantitative RT‐PCR, the levels of JARID1B mRNA were measured in MCF7 cells transfected with pSP65/U1‐miR‐381‐3p, pSP65/U1‐miR‐486‐5p miRNA overexpressing plasmids. The results (Figure 3D) revealed that miRNAs do not induce any significant effect on JARID1B mRNA accumulation, as its levels appear unchanged in miRNA overexpressing cells with respect to control conditions.
While an effect of miRNAs at protein level but not at mRNA level is generally observed in mammalian cells,40 these results suggest that anti‐correlation between miRNAs and JARID1B transcript levels discussed above cannot be read as a causal relationship: in other words, JARID1B mRNA upregulation in cancer is not connected to a lack of these specific miRNAs. However, one possible speculation is that these two phenotypes arise independently in cancer cells as a reinforcing mechanism that, by repressing JARID1B expression at both transcriptional and post‐transcriptional levels, results in a proliferative advantage for them.
3.5. JARID1B target gene BRCA1 is upregulated upon miR‐486‐5p overexpression
JARID1B has been shown to be involved in regulating the mRNA accumulation of important regulatory genes. In particular, its repressing activity on BRCA1 with important functional implications has been demonstrated in several breast cancer cell lines, including MCF7.18
We therefore tested whether the effects of miR‐381‐3p and miR‐486‐5p overexpression on JARID1B abundance in MCF7 cells could influence BRCA1 mRNA accumulation.
The results shown in Figure 3 confirm that both miR‐381‐3p and miR‐486‐5p overexpression increase the amount of BRCA1 mRNA (1.6‐fold and 2.8‐fold, respectively) 48 hours from transfection.
We also tested other genes that were previously shown to be modulated when JARID1B was downregulated by siRNA.38, 39 In particular, MT1F is significantly repressed, as previously shown,37 while CAV1 is only slightly and not significantly induced (Figure S2).
To test the effects of miR‐486‐5p overexpression on JARID1B association in BRCA1 promoter and H3H4 tri‐methylation, we performed ChIP experiments (see Materials and Methods). Figure 3F shows that JARID1B is binding to BRCA1 promoter in MCF7 transfected with pSP65/U1 empty plasmid, as previously shown,16 while no association with the control amplicon (ACTB exon5) is detected. When the cells are transfected with the pSP65/U1‐miR‐486‐5p miRNA overexpressing plasmid, a reduction in the JARID1B association is evident. In the same ChIP experiments, we did not observe an evident increase of H3K4me3 upon miR‐486‐5p overexpression. This is not surprising because JARID1B binds to the BRAC1 promoter in tight association with the NURD complex and cooperates with it in transcriptional repression.18 Recovery of H3K4 trimethylation could be dispensable for the observed transcriptional de‐repression or delayed in time.
3.6. Overexpression of miR‐381‐3p and miR‐486‐5p affect cell cycle dynamics
BRCA1 repression is a crucial event in breast cancer to escape cell cycle control. The protein is involved in promoting G1/S arrest through both P53‐dependent or P53‐independent mechanisms.41 We therefore tested the effects of miR‐381‐3p and miR‐486‐5p on cell cycle dynamics by cytofluorimetric analysis. The results reported in Figure S3 show that transfection with miR‐381‐3p and miR‐486‐5p causes a significant increase in G1/G0 cell population and a decrease in S population at 24 hours, which are still detectable at 48 hours (statistically significant only for miR‐381‐3p). These results are coherent with the increase in BRCA1 expression caused by miRNA transfection, although additional regulators could be involved. On the other hand we do not observe any significant increase in the subG1 population, which could be diagnostic of apoptotic effects. This is not surprising because MCF7 cells are apoptosis‐resistant due to lack of Caspase‐3.42 Therefore, we did not perform more specific apoptosis assays.
3.7. miR‐381‐3p and miR‐486‐5p overexpression increases cells sensitivity to radiation and accumulation of DBS in MCF7 cells
Given the emerging role of JARID1B as a key player of DDR, we hypothesize that lowering its expression could impair, to some extent, the ability of the cell to overcome DNA double‐strand breaks. Therefore, JARID1B targeting by miRNAs should, in principle, sensitize cells to X‐ray irradiation‐induced DNA damage and result in a greater proportion of cells unable to proliferate.
This rationale was used to test the miRNA effects and the results are summarized in Figure 4.
Figure 4.
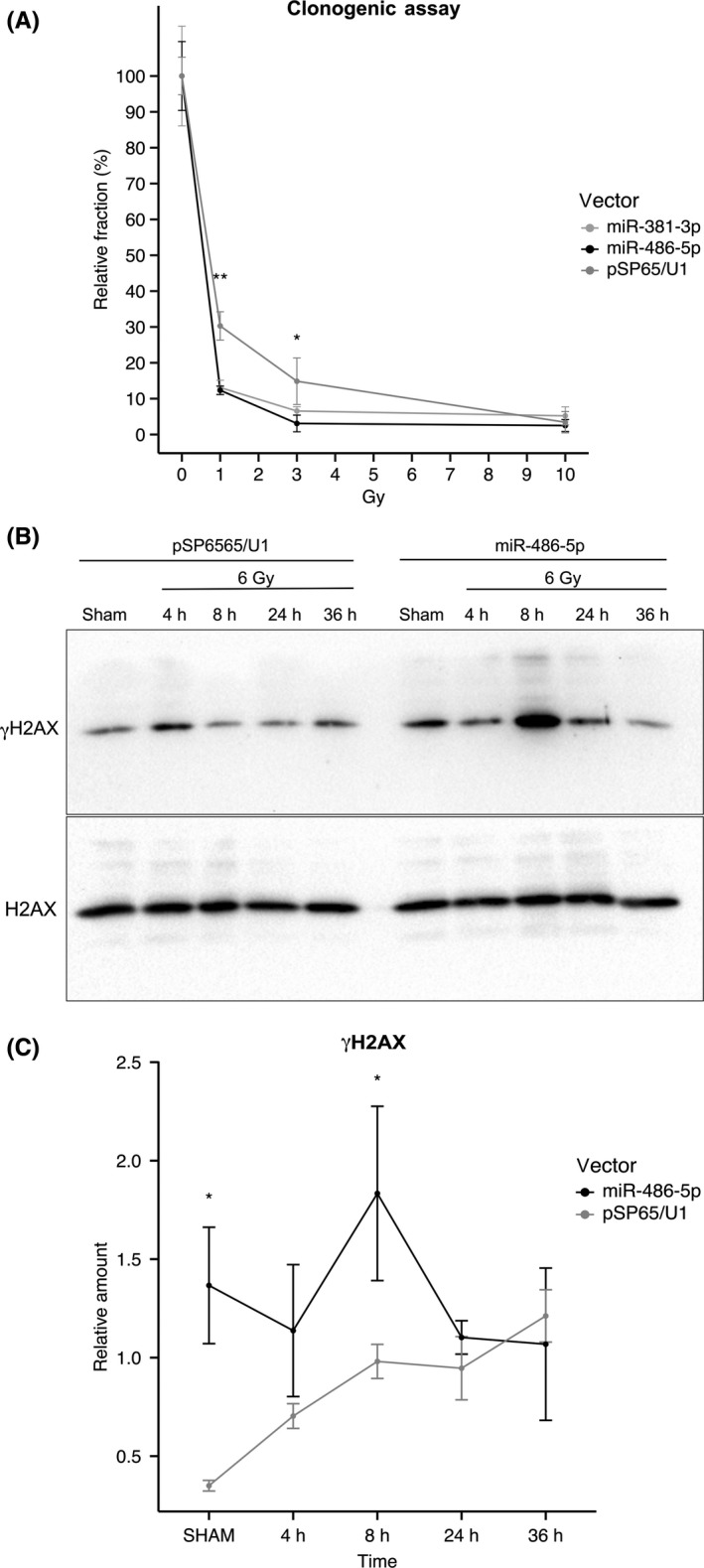
MiR‐381‐3p and miR‐486‐5p overexpression increases cells’ sensitivity to radiation and accumulation of DBS in MCF7 cells. A, Clonogenic assay on MCF7 irradiated with 0, 1, 3 or 10 Gy 24‐h post‐transfection with control pSP65/U1, pSP65/U1‐miR‐381‐3p or pSP65/U1‐miR‐486‐5p, n = 3. The relative surviving fraction of each experimental group was calculated as the plating efficiency for each radiation dose (1, 3 and 10 Gy), with respect to the sham‐irradiated sample. For every experimental group it is indicated as the mean value of the relative surviving fraction ± SD. Statistical significance was assessed according to a 2‐tailed Student's t test, *P < .05; **P < .01. B, Representative western blotting image of γH2AX levels of cell lysate of transfected and irradiated MCF7 cells; 24 h after transfection with pSP65/U1 or pSP65/U1‐miR‐486‐5p MCF7 cells are exposed to 0 or 6 Gy of X‐rays and irradiated samples are collected after 4, 8, 24 or 36 h. C, Relative quantification of γH2AX levels in western blotting analysis, n = 3. Data are represented as the mean ± SD of γH2AX levels relative to total H2AX. Statistical significance was determined using 2‐way ANOVA, *P < .05
In Figure 4A, both miR‐381‐3p and miR‐486‐5p were observed to decrease the fraction of surviving cells able to proliferate: for 1 and 3 Gy irradiation doses, proliferative capacity, measured as the fraction of plated cells able to proliferate and give rise to colonies with respect to sham‐irradiated controls, was decreased by almost half with respect to the empty vector transfection group (pSP65/U1). This variation is statistically significant for both miRNAs at 1‐Gy and 3‐Gy doses (P < .01 and P < .05 respectively). A 10‐Gy radiation dose neutralizes every effect, because the number of cells able to proliferate after this treatment is too low to appreciate any differential sensitivity. To further characterize these observations, we tested whether DNA damage was preferentially accumulated in miR‐486‐transfected MCF7 cells by analyzing kinetics of γ‐H2AX phosphorylation. Figure 4B and C shows that γ‐H2AX phosphorylation is significantly increased in miR‐486‐transfected sham‐irradiated MCF7 cells, as compared with cells transfected with empty vector. This shows that damage accumulates in miR‐486‐transfected even in the absence of a genotoxic treatment. Irradiation with 6 Gy of X‐rays, as expected, induced γ‐H2AX phosphorylation in both cell lines, although accumulation was faster in the miR‐486‐transfected cell line, which shows a significantly higher phosphorylation level at the 8‐hour time‐point. At later time points, γ‐H2AX phosphorylation in the miR‐486‐transfected line tends to level up with the cells transfected with the empty vector.
3.8. Analysis of the effects of miR‐381‐3p and miR‐486‐5p on JARID1B expression in other breast cancer cell lines
To understand to what degree the effects of miR‐381‐3p and miR‐486‐5p can be extended to other breast cancer cell lines, we repeated some of the experiments using T47D, another luminal breast cancer line, which, as for MCF7, should overproduce JARID1B and MDA‐MB‐231, a metastatic ER‐negative breast cancer cell line, which instead should express JARID1B at a lower level because the protein seems to reduce its metastatic potential.18
The western blot in Figure 5A shows that, indeed, T47D expresses JARID1B protein at a level very similar to MCF7, while the band is barely detectable in the MDA‐MB‐231 lane. Next, we looked at the expression of the 2 miRNAs. MiR‐381‐3p was undetectable in all 3 cell lines (not shown). Interestingly, MDA‐MB‐231 cells express approximately 4‐fold higher levels of miR‐486‐5p as compared with MCF7 (Figure 5B), suggesting that the miRNA might be involved in downregulation of JARID1B in this cell line. In support of this hypothesis, MDA‐MB‐231 cells accumulate JARID1B mRNA at a level comparable to MCF7, while it is at least 2‐fold higher in T47D (Figure 5C).
Figure 5.
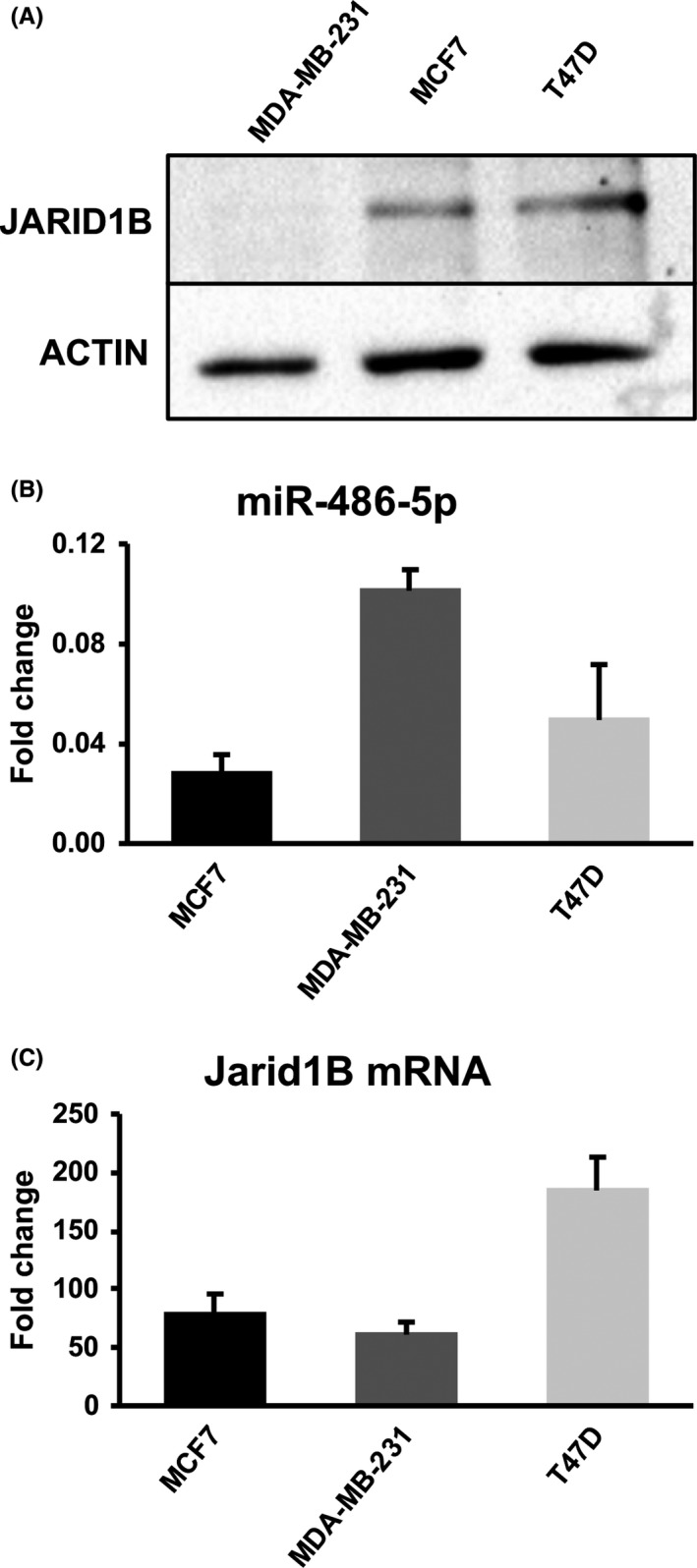
Expression of JARID1B protein and mRNA and of miR‐486‐5p in different human breast cancer cell lines. A, Representative western blotting image of JARID1B protein levels in the indicated breast cancer cell lines. B, Real time RT‐PCR quantitation of miR‐486‐5p in the indicated breast cancer cell lines. The histogram reports the amounts relative to the U6 endogenous calibrator. Data are the average of 3 experiments; SD is indicated. C, Real‐time RT‐PCR quantitation of JARID1B mRNA in the indicated breast cancer cell lines. The histogram reports the amounts relative to the GADPH endogenous calibrator. Data are the average of 3 experiments; SD is indicated
Next, we monitored the effects of transfecting T47D cells with the 2 miRNAs on the levels of JARID1B protein. As shown in Figure S4, transfection with miR‐381‐3p and miR‐486‐5p leads to a significant reduction of the protein but only at 48 hours from transfection. This delay in the decrease of the protein could be due to the higher amount of JARID1B mRNA present in the T47D cells compared to the MCF7. We also tested the effects of miRNAs on T47D radiosensitivity using a clonogenic assay (Figure S5). We detected a mild effect for miR‐486‐5p, while miR‐381‐3p did not show any effect. This limited action could be explained by the delayed decline of JARID1B abundance. The same reason could explain the absence of significant effects of both miRNAs on the cell cycle kinetics (Figure S6).
4. DISCUSSION
The JARID1 subfamily of HDM is the only group of enzymes able to remove the H3K4me3 mark, which was proposed to have an essential role in regulating gene transcription. However, their expression in adult tissues is quite low and differentiated among members: JARID1A is probably the only member that is broadly expressed. The other members show instead limited and tissue‐specific expression, suggesting precise roles in gene regulation. However, it is now clear that JARID1B is highly expressed during development, especially in mammalian embryonic stem cells and zygotes. Emerging evidence is supporting a role for JARID1 enzymes in regulating H3K4 methylation dynamics at the level of broad non‐canonical H3K4me3 domains during zygote genome activation,39, 44 and cell fate and self‐renewal genes in embryonic stem cell differentiation and reprogramming.45, 46, 47, 48 These observations suggest that active reshaping the of H3K4me3 pattern of the human epigenetic landscape is mostly performed in highly dynamic epigenomes, such as those of early developmental stages, rather than in somatic differentiated cells, providing principles for the explanation of such low levels of JARID1 enzymes found in adult tissues. However, their role in regulating processes as proliferation, pluripotency and differentiation are still to be fully addressed, as we are now beginning to understand that their transcriptional role is far more complex than previously thought.
Moreover, these speculations are not in contrast with what is observed in the context of cancer cells: like other epigenetic modifiers such as EZH2 and DNMT3B, JARID1B is found to be mainly expressed in developmental stages or in cancer (as was confirmed by the bioinformatic analysis presented in this work, see Table 1). This is probably due to their role in shaping epigenetic features that regulate the global cell transcriptome: alterations in the expression of epigenetic modifiers in somatic cells might lead to an unnecessary epigenetic plasticity that could eventually facilitate de‐differentiation, which, in turn, is a necessary step towards neoplastic transformation.
In this work, we confirmed that JARID1B is transcriptionally generally upregulated in breast cancer. We show a general downregulation of miR‐381‐3p and miR‐486‐5p in breast tumors, proving that they are both able to target JARID1B 3′UTR region and to efficiently repress its translation into protein. These observations assume even more importance if we consider that miR‐486‐5p is the most downregulated miRNA of the whole breast cancer transcriptome, its expression being almost completely shut down. Moreover, miR‐381‐3p expression level, which is more variable in cancer samples, appears to be positively correlated to patients’ survival. Another miRNA targeting JARID1B (mir‐137, Ref. 37) was recently found downregulated in breast cancer cells, confirming that shutdown of post‐transcriptional repression by miRNAs is crucial in order to allow overexpression of JARID1B. The strong downregulation of SMYD1 histone methylase which we observed in cancer tissues (Table 1) also points to the relevance of the H3K4me3 levels control for breast cancer cells (Figure 6).
Figure 6.
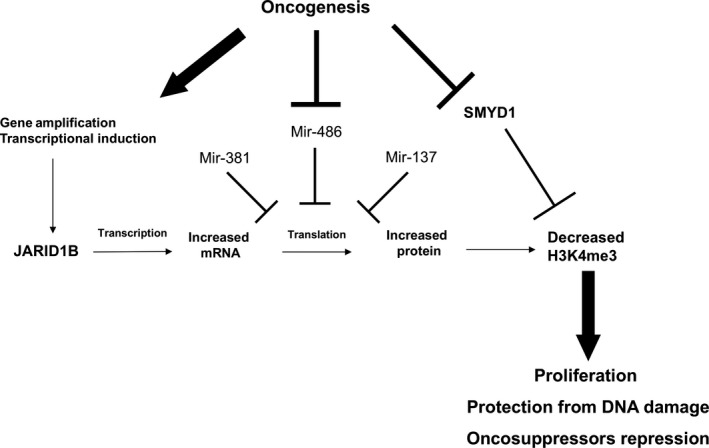
Graphic summary of the dysregulation of H3K4 tri‐methylation observed in breast cancer cells and of the putative biological effects
Indeed, here we show that miRNA‐mediated repression of JARID1B translation induces DNA damage accumulation, as revealed by monitoring the level of phosphorylated γ‐H2AX, and causes an enhanced sensitivity to radiation‐induced cell death, supporting emerging evidence depicting this HDM as a key regulator of cancer cells resistance to genotoxic damage. Very similar effects were recently shown by inhibiting the catalytic activity of HDM UTX, specific for H3K27 in other cancer cell lines, including the MDA‐MB‐231 breast cancer line.49 Obviously, these observations suggest a straightforward biomedical application in cancer therapy: targeting JARID1B by microRNAs could, in principle, sensitize cells to radiation‐induced cell death.
Moreover, we observed that this JARID1B post‐transcriptional downregulation determines alteration of the expression of its regulatory target gene BRCA1 and cell cycle alterations (G1/G0 population increase and S population decrease) potentially related with it. This gene represents an emblematic trait d'union that ties together the emerging role of JARID enzymes in DDR and their already known role in transcription regulation. JARID1B seems to act as both a negative regulator of a DDR factor (BRCA1) and as a positive factor itself, for DNA repair. Thus, its role cannot be limited to that of an oncogene or a tumor suppressor but probably depends on the context in which it is found. Certainly, molecular insights on chromatin modifications required for efficient DSB repair will be key to understand the role of JARID1B in cancer.
Finally, it is worth noting that the almost 3‐fold reduction in JARID1B protein abundance that we showed in MCF7 miR‐486‐transfected cells does not cause an evident increase of H3K4 tri‐methylation in bulk chromatin. This observation suggests that in these cells JARID1B overexpression is not strictly required to maintain a general low level of histone tri‐methylation in the genome but is rather important for regulating the expression of specific genes (ie BRCA1) and for efficient DNA repair.
The expression of both miRNAs appears to be downregulated not only in tumors but also in luminal breast cancer cell lines. MiR‐381‐3p expression is not detectable not only in MCF7 but also in T47D breast cancer lines, while miR‐486‐5p is much more expressed in the metastatic ER‐negative MDA‐MB‐231 breast cancer cell line, which poorly expresses JARID1B compared to the 2 luminal breast cancer lines. This suggests a possible involvement of miR‐486‐5p in the downregulation of JARID1B in this cell line which shows levels of mRNA similar to MCF7 but much lower levels of protein. In T47D, JARID1B appears to be less responsive than MCF7 to transfection with the 2 miRNAs, showing a delayed reduction of the protein abundance. This could be due to a less favorable ratio between miRNAs and JARID1B mRNA, which in these cells is at least 2‐fold more abundant. Stoichiometry between miRNAs and mRNAs has often been shown to be crucial for effectiveness of repression.
In conclusion, JARID1B overexpression in human breast cancer tissues correlates with downregulation of miRNAs targeting its mRNA. In human MCF7 breast cancer cell line a causal relationship between the 2 phenomena is demonstrated. Moreover, in this cancer cell line, miRNAsinduced JARID1B downregulation, although not changing the level of bulk chromatin H3K4 tri‐methylation, causes hypersensitivity to genotoxic damage, BRCA1 de‐repression and cell cycle alterations. Although the case of T47D cell line shows that responsiveness to miRNA action could be dependent on the strength of transcriptional regulation, and its balance with miRNA abundance, these results shed light on JARID1B post‐transcriptional regulation and open the way for possible therapeutic use of these miRNAs and/or chemical inhibitors of JARID enzymes in those cancers in which they have a primary role.
CONFLICT OF INTEREST
The authors have no conflict of interest.
Supporting information
ACKNOWLEDGMENT
We thank Elena Di Nisio, Luca Bombardi and Simone Fabozzi for technical help.
Mocavini I, Pippa S, Licursi V, et al. JARID1B expression and its function in DNA damage repair are tightly regulated by miRNAs in breast cancer. Cancer Sci. 2019;110:1232–1243. 10.1111/cas.13925
Contributor Information
Carlo Presutti, Email: carlo.presutti@uniroma1.it.
Rodolfo Negri, Email: rodolfo.negri@uniroma1.it.
REFERENCES
- 1. Zhou VW, Goren A, Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nat Rev Genet. 2011;12:7‐18. [DOI] [PubMed] [Google Scholar]
- 2. Nair N, Shoaib M, Sørensen CS. Chromatin dynamics in genome stability: roles in suppressing endogenous DNA damage and facilitating DNA repair. Int J Mol Sci. 2017;18:1486‐1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhang T, Cooper S, Brockdorff N. The interplay of histone modifications – writers that read. EMBO Rep. 2015;16:1467‐1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol. 2005;6:838‐849. [DOI] [PubMed] [Google Scholar]
- 5. Klose RJ, Kallin EM, Zhang Y. JmjC‐domain‐containing proteins and histone demethylation. Nat Rev Genet. 2006;7:715‐727. [DOI] [PubMed] [Google Scholar]
- 6. Dimitrova E, Turberfield AH, Klose RJ. Histone demethylases in chromatin biology and beyond. EMBO Rep. 2015;16:1620‐1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. D'Oto A, Tian Q‐W, Davidoff AM, Yang J. Histone demethylases and their roles in cancer epigenetics. J Med Oncol Ther. 2016;1:34‐40. [PMC free article] [PubMed] [Google Scholar]
- 8. Mannironi C, Proietto M, Bufalieri F, et al. An high‐throughput in vivo screening system to select H3K4‐specific histone demethylase inhibitors. PLoS ONE. 2014;9:e86002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Taylor‐Papadimitriou J, Burchell J. JARID1/KDM5 demethylases as cancer targets? Expert Opin Ther Targets. 2017;21:5‐7. [DOI] [PubMed] [Google Scholar]
- 10. Zeng J, Ge Z, Wang L, et al. The histone demethylase RBP2 Is overexpressed in gastric cancer and its inhibition triggers senescence of cancer cells. Gastroenterology. 2010;138:981‐992. [DOI] [PubMed] [Google Scholar]
- 11. Hidalgo A, Baudis M, Petersen I, et al. Microarray comparative genomic hybridization detection of chromosomal imbalances in uterine cervix carcinoma. BMC Cancer. 2005;5:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lu PJ, Sundquist K, Baeckstrom D, et al. A novel gene (PLU‐1) containing highly conserved putative DNA/chromatin binding motifs is specifically up‐regulated in breast cancer. J Biol Chem. 1999;274:15633‐15645. [DOI] [PubMed] [Google Scholar]
- 13. Xiang Y, Zhu Z, Han G, et al. JARID1B is a histone H3 lysine 4 demethylase up‐regulated in prostate cancer. Proc Natl Acad Sci USA. 2007;104:19226‐19231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hayami S, Yoshimatsu M, Veerakumarasivam A, et al. Overexpression of the JmjC histone demethylase KDM5B in human carcinogenesis: involvement in the proliferation of cancer cells through the E2F/RB pathway. Mol Cancer. 2010;9:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Roesch A, Fukunaga‐Kalabis M, Schmidt EC, et al. A temporarily distinct subpopulation of slow‐cycling melanoma cells is required for continuous tumor growth. Cell. 2010;141:583‐594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yamane K, Tateishi K, Klose RJ, et al. PLU‐1 is an H3K4 demethylase involved in transcriptional repression and breast cancer cell proliferation. Mol Cell. 2007;25:801‐812. [DOI] [PubMed] [Google Scholar]
- 17. Catchpole S, Spencer‐Dene B, Hall D, et al. PLU‐1/JARID1B/KDM5B is required for embryonic survival and contributes to cell proliferation in the mammary gland and in ER+ breast cancer cells. Int J Oncol. 2011;38:1267‐1277. [DOI] [PubMed] [Google Scholar]
- 18. Klein BJ, Piao L, Xi Y, et al. The histone‐H3K4‐specific demethylase KDM5B binds to its substrate and product through distinct PHD fingers. Cell Rep. 2014;6:325‐335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Faucher D, Wellinger RJ. Methylated H3K4, a transcription‐associated histone modification, is involved in the DNA damage response pathway. PLoS Genet. 2010;6:e1001082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li X, Liu L, Yang S, et al. Histone demethylase KDM5B is a key regulator of genome stability. Proc Natl Acad Sci USA. 2014;111:7096‐7101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gong F, Clouaire T, Aguirrebengoa M, Legube G, Miller KM. Histone demethylase KDM5A regulates the ZMYND8‐NuRD chromatin remodeler to promote DNA repair. J Cell Biol. 2017;216:1959‐1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gong F, Chiu L‐Y, Cox B, et al. Screen identifies bromodomain protein ZMYND8 in chromatin recognition of transcription‐associated DNA damage that promotes homologous recombination. Genes Dev. 2015;29:197‐211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Savitsky P, Krojer T, Fujisawa T, et al. Multivalent histone and DNA engagement by a PHD/BRD/PWWP triple reader cassette recruits ZMYND8 to K14ac‐rich chromatin. Cell Rep. 2016;17:2724‐2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Penterling C, Drexler GA, Böhland C, et al. Depletion of histone demethylase jarid1a resulting in histone hyperacetylation and radiation sensitivity does not affect DNA double‐strand break repair. PLoS ONE. 2016;11:e0156599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281‐297. [DOI] [PubMed] [Google Scholar]
- 26. Iorio MV, Croce CM. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med. 2017;9:852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med. 2014;20:460‐469. [DOI] [PubMed] [Google Scholar]
- 28. Rich JT, Neely JG, Paniello RC, Voelker CCJ, Nussenbaum B, Wang EW. A practical guide to understanding Kaplan‐Meier curves. Otolaryngol Head Neck Surg. 2010;143:331‐336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Denti MA, Rosa A, Sthandier O, De Angelis FG, Bozzoni I. A new vector, based on the PolII promoter of the U1 snRNA gene, for the expression of siRNAs in mammalian cells. Mol Ther. 2004;10:191‐199. [DOI] [PubMed] [Google Scholar]
- 30. Schindelin J, Arganda‐Carreras I, Frise E, et al. Fiji: an open‐source platform for biological‐image analysis. Nat Methods. 2012;9:676‐682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Patani N, Jiang WG, Newbold RF, Mokbel K. Histone‐modifier gene expression profiles are associated with pathological and clinical outcomes in human breast cancer. Anticancer Res. 2011;31:4115‐4125. [PubMed] [Google Scholar]
- 32. Tsang DPF, Cheng ASL. Epigenetic regulation of signaling pathways in cancer: role of the histone methyltransferase EZH2. J Gastroenterol Hepatol. 2011;26:19‐27. [DOI] [PubMed] [Google Scholar]
- 33. Girault I, Tozlu S, Lidereau R, Bièche I. Expression analysis of DNA methyltransferases 1, 3A, and 3B in sporadic breast carcinomas. Clin Cancer Res. 2003;9:4415‐4422. [PubMed] [Google Scholar]
- 34. Gupta RA, Shah N, Wang KC, et al. Long non‐coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071‐1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ozdağ H, Teschendorff AE, Ahmed AA, et al. Differential expression of selected histone modifier genes in human solid cancers. BMC Genom. 2006;7:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bamodu OA, Huang W‐C, Lee W‐H, et al. Aberrant KDM5B expression promotes aggressive breast cancer through MALAT1 overexpression and downregulation of hsa‐miR‐448. BMC Cancer. 2016;16:160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Denis H, Van Grembergen O, Delatte B, et al. MicroRNAs regulate KDM5 histone demethylases in breast cancer cells. Mol BioSyst. 2016;12:404‐413. [DOI] [PubMed] [Google Scholar]
- 38. Scibetta AG, Santangelo S, Coleman J, et al. Functional analysis of the transcription repressor PLU‐1/JARID1B. Mol Cell Biol. 2007;27:7220‐7235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dahl JA, Jung I, Aanes H, et al. Broad histone H3K4me3 domains in mouse oocytes modulate maternal‐to‐zygotic transition. Nature. 2016;537:548‐552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Iwakawa H, Tomari Y. The functions of MicroRNAs: mRNA decay and translational repression. Trends Cell Biol. 2015;25:651‐665. [DOI] [PubMed] [Google Scholar]
- 41. Deng C. BRCA1: cell cycle checkpoint, genetic instability, DNA damage response and cancer evolution. Nucleic Acid Res. 2006;34:1416‐1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Essmann F, Engels IH, Totzke G, et al. Apoptosis resistance of MCF‐7 breast carcinoma cells to ionizing radiation is independent of P53 cell cycle control bur caused by the lack of Caspase‐3 and a caffeine‐inhibitable event. Cancer Res. 2004;64:7065‐7072. [DOI] [PubMed] [Google Scholar]
- 43. Vaquerizas JM, Torres‐Padilla M‐E. Developmental biology: panoramic views of the early epigenome. Nature. 2016;537:494‐496. [DOI] [PubMed] [Google Scholar]
- 44. Zhang B, Zheng H, Huang B, et al. Allelic reprogramming of the histone modification H3K4me3 in early mammalian development. Nature. 2016;537:553‐557. [DOI] [PubMed] [Google Scholar]
- 45. Dey BK, Stalker L, Schnerch A, Bhatia M, Taylor‐Papidimitriou J, Wynder C. The histone demethylase KDM5b/JARID1b plays a role in cell fate decisions by blocking terminal differentiation. Mol Cell Biol. 2008;28:5312‐5327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kidder BL, Hu G, Zhao K. KDM5B focuses H3K4 methylation near promoters and enhancers during embryonic stem cell self‐renewal and differentiation. Genome Biol. 2014;15:R32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Xie L, Pelz C, Wang W, et al. KDM5B regulates embryonic stem cell self‐renewal and represses cryptic intragenic transcription. EMBO J. 2011;30:1473‐1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cacci E, Negri R, Biagioni S, Lupo G. Histone methylation and microRNA‐dependent regulation of epigenetic activities in neural progenitor self‐renewal and differentiation. Curr Top Med Chem. 2017;17:794‐807. [DOI] [PubMed] [Google Scholar]
- 49. Rath BH, Waung I, Camphausen K, Tofilon PJ. Inhibition of the histone H3K27 demethylase UTX enhances tumor cell radiosensitivity. Mol Cancer Ther. 2018;17:1070‐1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


