Abstract
Collective invasion is an important strategy of cancers of epithelial origin, including colorectal cancer (CRC), to infiltrate efficiently into local tissues as collective cell groups. Within the groups, cells at the invasive front, called leader cells, are highly polarized and motile, thereby providing the migratory traction that guides the follower cells. However, its underlying mechanisms remain unclear. We have previously shown that signaling emanating from the receptor tyrosine kinase Ror2 can promote invasion of human osteosarcoma cells and that intraflagellar transport 20 (IFT20) mediates its signaling to regulate Golgi structure and transport. Herein, we investigated the role of Ror2 and IFT20 in collective invasion of CRC cells, where Ror2 expression is either silenced or nonsilenced. We show by cell biological analyses that IFT20 promotes collective invasion of CRC cells, irrespective of expression and function of Ror2. Intraflagellar transport 20 is required for organization of Golgi‐associated, stabilized microtubules, oriented toward the direction of invasion in leader cells. Our results also indicate that IFT20 promotes reorientation of the Golgi apparatus toward the front side of leader cells. Live cell imaging of the microtubule plus‐end binding protein EB1 revealed that IFT20 is required for continuous polarized microtubule growth in leader cells. These results indicate that IFT20 plays an important role in collective invasion of CRC cells by regulating organization of Golgi‐associated, stabilized microtubules and Golgi polarity in leader cells.
Keywords: collective invasion, colorectal cancer, IFT20, noncentrosomal microtubule, Ror2
Abbreviations
- γ‐TuRC
γ‐tubulin‐ring complex
- AKAP450
A‐kinase anchor protein 450
- CAMSAP
calmodulin‐regulated spectrin‐associated protein
- CRC
colorectal cancer
- IFT20
intraflagellar transport 20
- LEF
lymphoid enhancer transcription factors
- MT
microtubule
- NZ
nocodazole
- TCF
T‐cell factor
1. INTRODUCTION
Ror2 is a member of the Ror‐family of receptor tyrosine kinases, which plays critical roles during developmental morphogenesis, tissue repair, and cancer progression.1 Ror2 acts with Frizzled proteins as a cognate receptor for Wnt5a to mediate the noncanonical Wnt pathway, which activates signaling molecules, including JNK and c‐Src.2, 3 Ror2 is expressed highly in various types of cancer cells, including osteosarcoma, melanoma, and breast cancer cells, resulting in constitutive activation of Ror2‐mediated signaling (Ror2 signaling).1 We have recently shown that constitutively activated Ror2 signaling in osteosarcoma SaOS2 cells induces expression of a gene encoding IFT20, a Golgi‐localized protein, which normally mediates the assembly and maintenance of the primary cilia, even though SaOS2 cells lack the primary cilia.4 Our findings also revealed that Ror2‐mediated expression of IFT20 in cancer cells is a critical event to drive cancer cell invasion by promoting MT nucleation at the Golgi apparatus, but not the centrosome.4 Golgi‐derived MTs are highly stable and organized in an asymmetrical manner, which allows Golgi ministacks to link together for properly oriented Golgi ribbon formation, thereby facilitating polarized post‐Golgi transport and cell polarization.5, 6, 7 However, the centrosome organizes radial symmetric array of MTs, responsible for pericentrosomal localization of the Golgi ministacks without supporting Golgi ribbon formation.5 In fact, Ror2 and IFT20 are required for polarized positioning of the Golgi during invasion of SaOS2 cells.4
Besides its role in activating the noncanonical Wnt pathway, Wnt5a‐Ror2 signaling can inhibit the canonical Wnt pathway,8 which is mediated by Lrp5/6 and Frizzled receptors to induce stabilization and nuclear translocation of ß‐catenin. Nuclear ß‐catenin binds to TCF/LEF, thereby activating transcription of their target genes, including Cyclin‐D1 and c‐Myc. The canonical Wnt pathway is activated aberrantly in various human cancers, including CRC, and is a primary mechanism of CRC development.9 Wnt5a‐Ror2 signaling inhibits the canonical Wnt pathway at the level of TCF/LEF‐mediated transcription.8 In fact, Wnt5a and Ror2 genes are silenced frequently in CRC cells, and reactivation of either Wnt5a or Ror2 inhibits TCF/LEF‐mediated transcription in and proliferation of CRC cells,10, 11, 12 indicating that Wnt5a‐Ror2 signaling can show a suppressive function for CRC. Interestingly, it has been reported that high expression of Ror2 is associated with poor prognosis in patients with CRC,13 suggesting that Ror2 might also have a role in promoting CRC progression, at least under particular conditions.
Cancer cells, retaining epithelial characteristics, such as differentiated CRC cells, invade predominantly as groups (ie, strands, sheets, and/or clusters), termed collective invasion, by maintaining their cell‐to‐cell adhesion.14, 15 Within the groups, cells at the invasive front (leader cells) are highly polarized and motile, thereby providing the migratory traction, and through cell‐to‐cell junctions, they pull the trailing cells (follower cells) at their rear.14, 15 Engagement of integrins occurs at anterior protrusions of leader cells towards the ECMs16, 17 with concomitant increased expression and activity of MMPs, resulting in polarized ECM degradation.18
Here we investigated the role of IFT20 as well as Ror2 in invasive cell migration using several CRC cell lines, in which Ror2 expression is either silenced (DLD1) or nonsilenced (HCT116 and SW480). We show that knockdown of Ror2 in HCT116 cells resulted in decreased IFT20 levels and impaired collective invasion, which are not observed in Ror2‐knockdown SW480 cells. Knockdown of IFT20 inhibited collective invasion of all the 3 cell lines. We further show that IFT20 can promote organization of Golgi‐associated, stabilized MTs and reorientation of the Golgi toward the direction of invasion in leader cells, probably by regulating growth dynamics, but not nucleation of MTs. Taken together, our present study unravels a novel function of IFT20 in collective invasion of CRC cells through the MT‐mediated regulation of the Golgi.
2. MATERIALS AND METHODS
2.1. Cells and transfection
DLD1, HCT116, and SW480 cells were obtained from JCRB cell bank (Osaka, Japan), RIKEN BioResource Center (Tsukuba, Japan), and ATCC (Manassas, VA, USA), respectively, and maintained in RPMI‐1640 (Nacalai Tesque, Kyoto, Japan) containing 10% (v/v) FBS at 37°C in a humidified atmosphere of 5% (v/v) CO2. Cells (1 × 106/mL) suspended in 10% (v/v) DMSO in FBS were frozen and stored in liquid nitrogen. Cells were used for experiments within 7 passages after thawing the frozen stocks in general. Cells were transfected with the respective siRNAs and plasmids by using Lipofectamine RNAiMAX (Thermo Fisher Scientific, Waltham, MA, USA) and ViaFect (Promega, Madison, WI, USA) transfection reagents, respectively, according to the manufacturers’ instructions. Briefly, siRNAs (20 nmol/L) or plasmids (1 μg/mL) were mixed with the transfection reagents diluted in Opti‐MEM (Thermo Fisher Scientific), incubated for 20 minutes at room temperature, and added to cells. For rescue experiments, siRNA‐transfected cells were incubated for 24 hours and further transfected with siRNA‐resistant plasmids. At 48 hours post‐siRNA transfection, the resultant cells were replated for 2‐D invasion assay. The sequences of si‐IFT20#1, si‐IFT20#2, and si‐Ror2 were described previously.3, 4 Negative control siRNA (si‐Ctrl) was purchased from Sigma (St. Louis, MO, USA). The plasmid containing the siRNA‐resistant (sr)‐IFT20 gene in pIRES2‐ZsGreen1 vector (Clontech, Mountain View, CA, USA) was described previously.4 To establish DLD1 cells stably expressing EB1‐GFP, DLD1 cells were transfected with the plasmid encoding EB1‐GFP (a gift from Y. Mimori‐Kiyosue)19 by using a square wave electroporator (CUY21Edit; Nepagene, Chiba, Japan), followed by selection with G418 at a final concentration of 500 μg/mL. We confirmed that there were no obvious differences in velocities of EB1‐GFP movement among 6 independent clones, including the clone used in the present study (data not shown).
2.2. Antibodies
Rabbit anti‐Ror2 Ab was prepared as described previously.20 The following Abs were purchased commercially: mouse anti‐GM130 Ab (Medical and Biological Laboratories [MBL], Nagoya, Japan), anti‐γ‐tubulin Ab (GTU‐88; Sigma), anti‐acetylated tubulin Ab (6‐11B‐1; Sigma), and anti‐AKAP450 Ab (15; BD Biosciences, San Jose, CA, USA); rabbit anti‐IFT20 Ab (13615‐1‐AP; Proteintech, Chicago, IL, USA), anti‐GM130 Ab (PM061; MBL), and anti‐α‐tubulin Ab (PM054; MBL).
2.3. Western blot analyses
Western blotting was carried out as described previously.21 Briefly, cells were solubilized in ice‐cold lysis buffer (50 mmol/L Tris‐HCl [pH 7.5], 150 mmol/L NaCl, 1% [v/v] Nonidet P‐40 [NP‐40], 1 mmol/L EDTA, 10 mmol/L NaF, 1 mmol/L Na3VO4, 10 μg/mL aprotinin, 10 μg/mL leupeptin, and 1 mmol/L p‐Amidinophenylmethanesulfonyl fluoride), and the lysates were clarified by centrifugation at 15 000 g for 15 minutes. Protein concentration was determined using the BCA protein assay (Thermo Fisher Scientific). Proteins (10 μg) were separated by SDS‐PAGE and transferred onto Immobilon‐P membranes (Merck Millipore, Darmstadt, Germany) using the blotting device (ATTO, Tokyo, Japan). Membranes were blocked with 5% (w/v) dried skim milk and immunoblotted with the respective primary Abs, followed by HRP‐conjugated secondary Abs. Immunoreactive bands were visualized with Western Lightning Plus‐ECL (Perkin Elmer, Waltham, MA, USA) and detected using the ECL detection system (LAS‐1000; Fujifilm, Tokyo, Japan).
2.4. Two‐dimensional invasion assay and immunofluorescence
The 2‐D invasion assay was carried out as described previously.4 In brief, the 2‐well culture insert with 0.5 mm gap between wells (ibidi, Munich, Germany) was placed on a fibronectin‐coated coverslip. Cells transfected with the respective siRNAs were plated onto the culture insert and grown to confluent monolayers. After removal of the inserts, the monolayers were overlaid with Matrigel (BD Biosciences), followed by incubation for 4 hours before an addition of growth medium. Cells were then cultured for up to 18 hours to allow invasion toward the space between the monolayers. The invasion ratio was calculated by dividing the width at each time point by the width at 0 hour. Cells were fixed with 3.7% (w/v) paraformaldehyde in PBS at room temperature, followed by methanol at −20°C. After blocking with 5% (w/v) BSA in PBS, cells were stained with the respective Abs and DAPI (Sigma). Fluorescence images were obtained using a laser scanning confocal imaging system (LSM700; Carl Zeiss, Jena, Germany) with 40×/1.4 NA and 63×/1.4 NA Plan‐Apochromat oil immersion objective lenses (Carl Zeiss). Super‐resolution imaging was undertaken using a spinning disk super‐resolution microscope (SpinSR10; Olympus, Tokyo, Japan) with a 100×/1.49 NA UAPON 100×OTIRF oil immersion objective lens (Olympus). Fluorescence images were processed using ImageJ software (Bethesda, MD, USA). Cells located at the edge and approximately 30 μm away from the edge were defined as leader and follower cells, respectively. Fluorescence intensities of α‐tubulin and acetylated tubulin in leader cells, normalized by those in follower cells, were quantified using Image J. To analyze the Golgi polarization, the percentages of the leader and follower cells in which their Golgi apparatus were within the 120° sector emerging from the center of the nucleus and facing toward the leading edge were measured.
2.5. Live cell imaging
To allow individual cell imaging, DLD1 cells stably expressing EB1‐GFP were plated for 2‐D invasion assay as described above, except a glass‐bottom dish was used. After cells were allowed to invade for 16 hours, fluorescence images were collected from living cells every 0.3 second using a laser scanning confocal imaging system (LSM700) with 63×/1.4 NA Plan‐Apochromat oil immersion objective lenses. Detection and tracking of EB1‐GFP in individual cells were undertaken using MatLab software (Mathworks, Natick, MA, USA) with the U‐Track algorism22, 23 and the MTrackJ plugin in ImageJ.24
2.6. Cell viability assay
Cells transfected with the respective siRNAs were analyzed by 2‐D invasion assay as described above. After cells were allowed to invade for 0 and 18 hours, medium was replaced with fresh medium containing 10% (v/v) WST‐8 (CCK‐8: Dojindo, Kumamoto, Japan). Cells were then incubated for 150 minutes at 37°C, and the absorbance of the culture supernatant was measured at 450 nm using an EnSpire multimode plate reader (Perkin Elmer).
2.7. Transwell invasion assay
Transwell invasion assay was carried out as described previously.3 Briefly, cells were loaded onto the upper well of the Transwell chamber with 8‐μm φ pore membrane (Coster, Cambridge, MA, USA), precoated with Matrigel, on an upper side of the chamber. The lower well was filled with 600 μL RPMI‐1640 containing 20% (v/v) FBS. After incubation for 24 hours, cells invaded to lower surface of the membrane were counted.
2.8. Microtubule nucleation assay
Microtubule nucleation assay was carried out as described previously.4 In brief, cells plated on fibronectin‐coated coverslips were treated with 3 μg/mL NZ in culture medium for 2 hours to depolymerize MTs. Cells were washed with ice‐cold RPMI‐1640 containing 20 mmol/L HEPES (pH 7.5) 6 times on ice to remove NZ and then incubated for 0‐10 minutes in RPMI‐1640 containing 20 mmol/L HEPES (pH 7.5) and 1% (v/v) FBS at 25°C. Cells were fixed with methanol at −20°C. Fixed cells were stained with Abs against α‐tubulin, GM130, and γ‐tubulin. Serial optical confocal z sections spanning the entire cell were obtained and stacked using a maximal intensity projection.
2.9. RNA isolation and real‐time PCR
RNA isolation, reverse transcription, and real‐time PCR analysis were described previously.4 Briefly, total RNAs were isolated and reverse‐transcribed using Isogen (Nippon Gene, Tokyo, Japan) and PrimeScript RT reagent kit (Takara Bio, Shiga, Japan), respectively. Real‐time PCR was undertaken using LightCycler 480 SYBR Green I Master mix (Roche Diagnostics, Mannheim, Germany) on the LightCycler 480 system (Roche Diagnostics). The following primers were used: IFT20, 5′‐ CAGAACTCCTCTAGGGAACCTG ‐3′ (forward) and 5′‐GCTCTATGGTCTGCTGGGTAA‐3′ (reverse); Ror2, 5′‐CAATTCCACTGGTCATCGCT‐3′ (forward) and 5′‐TGAGGGGCATTTCCATGTC‐3′ (reverse); and 18S ribosomal RNA, 5′‐ATGGCCGTTCTTAGTTGGTG‐3′ (forward) and 5′‐CGCTGAGCCAGTCAGTGTAG‐3′ (reverse).
2.10. Statistical analysis
Statistical significance was analyzed using the Student's t test. P < .05 was considered statistically significant.
3. RESULTS
3.1. Intraflagellar transport 20 promotes invasive migration of CRC cells, irrespective of expression status and function of Ror2
We have recently shown that constitutively activated Ror2 signaling induces expression of IFT20 in osteosarcoma and breast cancer cells, thereby promoting their invasion.4 In contrast, Ror2 signaling is often inactivated in CRC cells due to silencing of the Ror2 gene.11, 12 To examine the relationship between expression levels of Ror2 and IFT20 in CRC cells, we used 3 CRC cell lines, DLD1, HCT116 and SW480. Expression of Ror2 was detectable in HCT116 and SW480, but not DLD1 cells, but these 3 cell lines express readily detectable levels of IFT20 (Figure 1A). As reported for both ciliated and non‐ciliated cells,4, 25, 26 a significant pool of IFT20 was detected at the Golgi apparatus in these 3 cell lines, as assessed by co‐immunostaining of IFT20 with GM130, a marker for the cis‐Golgi (Figure S1). Consistent with our previous findings in osteosarcoma and breast cancer cells, siRNA‐mediated knockdown of Ror2 resulted in decreased mRNA and protein levels of IFT20 in HCT116 cells (Figure 1B,C). In contrast, Ror2 knockdown failed to affect mRNA and protein levels of IFT20 in SW480 cells (Figure 1D,E), indicating that Ror2 might regulate expression of IFT20, depending on cell lines.
Figure 1.
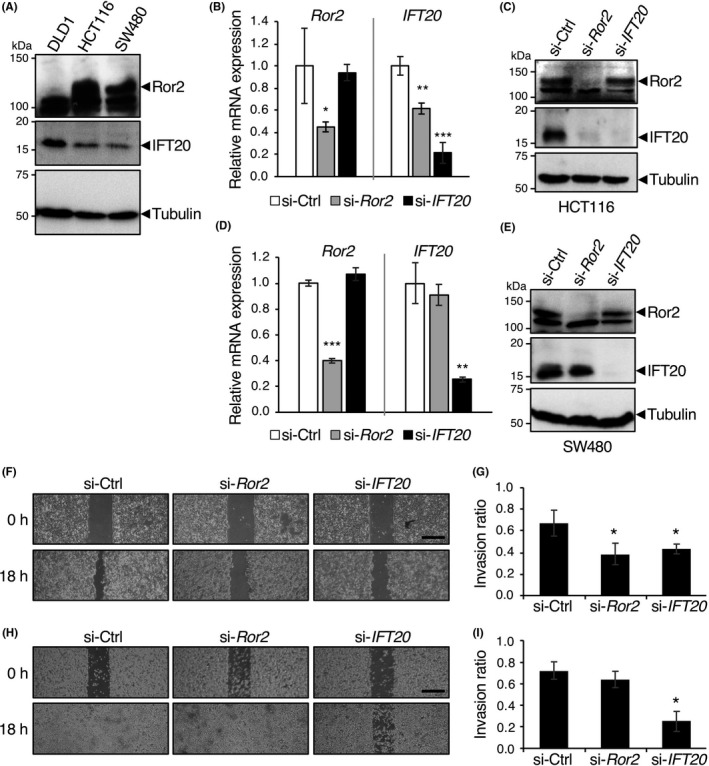
Suppressed expression of intraflagellar transport 20 (IFT20) impairs invasive migration of Ror2‐expressing colorectal cancer (CRC) cells. A, Expression levels of Ror2 and IFT20 in the CRC cell lines DLD1, HCT116, and SW480. Whole cell lysates from the respective CRC cells were analyzed by western blotting with Abs against the indicated proteins. B‐E, Decreased RNA (B,D) and protein (C,E) levels of IFT20 in HCT116 (B,C), but not SW480 cells (D,E), transfected with Ror2 siRNA. Cells were transfected with control (Ctrl), Ror2, or IFT20 siRNA and analyzed by real‐time PCR (B,D) and western blotting (C,E) to monitor amounts of the indicated RNAs and proteins, respectively. F‐I, HCT116 (F,G) and SW480 (H,I) cells were transfected with the indicated siRNAs and subjected to 2‐D invasion assay. Representative phase contrast images before (0 h) and after incubation for 18 h are shown (F,H). Scale bar, 500 μm. Invasion ratios of the respective cells after incubation for 18 h were determined (G,I). Data are expressed as mean ± SD (n = 3). *P < .05, t test
We next examined whether Ror2 and IFT20 play a role in invasion of HCT116 and SW480 cells. Transfection of HCT116 cells with siRNA against either Ror2 or IFT20 significantly inhibited their invasive migration (Figure 1F,G). However, transfection of SW480 cells with siRNA against IFT20, but not Ror2, inhibited their invasive migration (Figure 1H,I). These findings suggest that IFT20 is critically important for promoting CRC cell invasion, irrespective of Ror2 expression.
To further investigate a Ror2‐independent function of IFT20 in invasion of CRC cells, we next used DLD1 cells, in which expression of Ror2 was undetectable (Figure 1A). In agreement with the findings in HCT116 and SW480 cells, 2‐D invasion assay revealed that knockdown of IFT20 significantly inhibited the invasive migration of DLD1 cells (Figure 2A‐D). Any apparent differences in cell viability were not seen in DLD1 cells transfected with si‐Ctrl or si‐IFT20 during their 2‐D invasion assay (Figure 2E), indicating that the effect of si‐IFT20 on cell proliferation and viability might be negligible. Suppressed expression of IFT20 also inhibited invasive migration of DLD1 cells in 3‐D matrices as assessed by the Transwell invasion assay (Figure 2F). These results suggest that IFT20 plays an important role in promoting invasion of CRC cells, irrespective of their expression status of Ror2.
Figure 2.
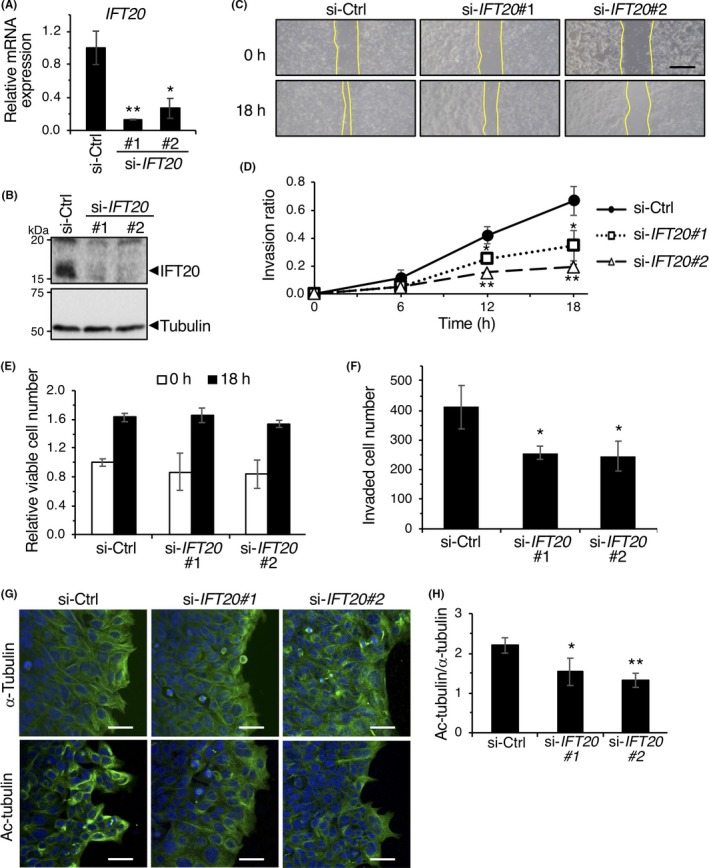
Intraflagellar transport 20 (IFT20) is required for accumulation of acetylated microtubules during collective invasion. A,B, Suppressed mRNA (A) and protein (B) expression of IFT20 in DLD1 cells transfected with siRNAs against IFT20. C,D, siRNA‐transfected DLD1 cells were analyzed by 2‐D invasion assay. Representative phase contrast images of cells before (0 h) and after incubation for 18 h are shown (C). Yellow lines indicate edges of cell layers. Scale bar, 500 μm. Invasion ratios of the respective cells after incubation for 0, 6, 12, and 18 h were determined 2(D). Data are expressed as mean ± SD (n = 3). *P < .05; **P < .005, t test. E, Relative viable cell number of siRNA‐transfected DLD1 cells at 0 and 18 h in 2‐D invasion assay was measured using WST8. Data are expressed as mean ± SD (n = 3). F, siRNA‐transfected DLD1 cells were analyzed by Transwell invasion assay. Cells invaded to the lower surface of the Transwell membranes were counted. Data are expressed as mean ± SD (n = 3). *P < .05, t test. G,H, siRNA‐transfected DLD1 cells were subjected to 2‐D invasion assay for 18 h. Cells were stained with Abs against α‐tubulin or acetylated (Ac)‐tubulin (green) and counterstained with DAPI (blue). G, Representative confocal images of cells. Scale bar, 20 μm. H, Fluorescence intensities of α‐tubulin and Ac‐tubulin in leader cells, normalized by those in follower cells, were quantified, and the ratio of fluorescence intensities of Ac‐tubulin and α‐tubulin were determined. Data are expressed as mean ± SD (n = 3, >30 cells/experiment). *P < .05; **P < .005, t test
3.2. Intraflagellar transport 20 regulates organization of Golgi‐associated MTs during collective invasion
In directionally migrating cells, a subset of MTs is stabilized to form and maintain polarized arrays of MTs.27 We detected prominent accumulation of acetylated MTs in si‐Ctrl‐transfected leader cells (Figure 2G), indicating that these leader cells organized stabilized MT network during collective invasion. In contrast, si‐IFT20 transfected leader cells showed less accumulation of acetylated tubulin (Figure 2G,H). Similar results were obtained when HCT116 cells were transfected with siRNA against either Ror2 or IFT20 (Figure S2A,B). These results suggest that IFT20 is required for organization of stabilized MT networks in leader cells during collective invasion.
We also noticed that neither leader nor follower cells of CRC cell layers showed typical astral arrays of MTs emanating from the centrosome (Figure S3A), consistent with previous reports showing that MTs are mainly noncentrosomal ones in confluent epithelial cells.28, 29 Instead, MTs were tightly associated with the Golgi apparatus (Figure S3B), indicating that the noncentrosomal MT network in DLD1 cells consists of, at least partly, Golgi‐associated MTs during collective invasion. It has also been known that noncentrosomal, Golgi‐associated MTs are highly stabilized.30, 31 In fact, co‐immunostaining analysis with anti‐GM130 and anti‐acetylated tubulin Abs revealed that leader cells, transfected with si‐Ctrl, showed highly organized and polarized MTs, closely associating with the Golgi apparatus (Figure 3A). In contrast, in leader cells transfected with si‐IFT20, acetylated MTs associated with the Golgi apparatus were quite poor and disorganized, although their Golgi structures were almost unaffected (Figure 3A,B). These results indicate that IFT20 is required for proper organization of Golgi‐associated MTs that are polarized and stabilized during collective migration. It is also worth noting that impaired organization of Golgi‐associated MTs was seen in HCT116 cells transfected with siRNA against either Ror2 or IFT20, but could be restored by the ectopic expression of sr‐IFT20 (Figure S2C‐E), indicating that Ror2 signaling acts through IFT20 to regulate organization of Golgi‐associated MTs in HCT116 cells.
Figure 3.
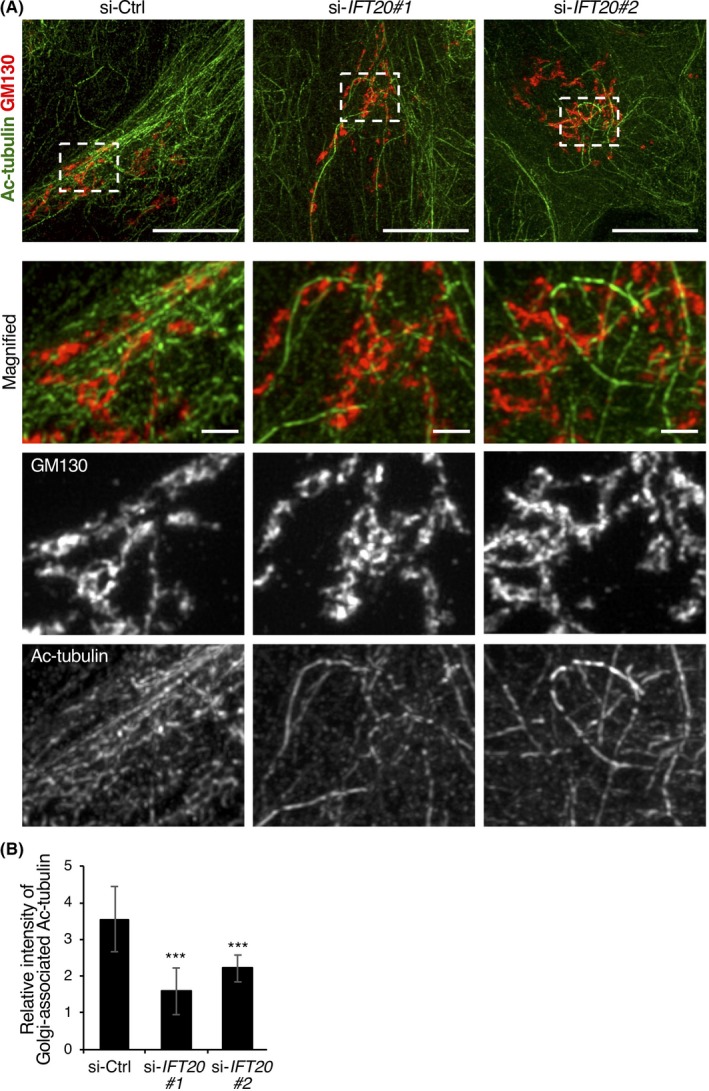
Intraflagellar transport 20 (IFT20) is required for organization of Golgi‐associated microtubules during collective invasion. A, DLD1 cells transfected with the indicated siRNAs were subjected to 2‐D invasion assay for 18 h. Cells were stained with Abs against acetylated (Ac)‐tubulin (green) and GM130 (red) and analyzed by super‐resolution microscopy. Representative leader cells are shown with the leading edge to the right. Lower panels show enlarged views of the boxed region in the upper panels. Scale bar, 10 μm (upper panels) and 1 μm (lower panels). Note that acetylated microtubules are highly organized and associated with the Golgi apparatus in control (si‐Ctrl) cells, but poorly organized in si‐IFT20 cells. B, Fluorescence intensities of Ac‐tubulin surrounding the Golgi in leader cells were quantified. Data are expressed as mean ± SD of 12 cells. ***P < .001, t test
We have recently shown that IFT20 promotes nucleation of MTs at the Golgi apparatus, but not the centrosome, in SaOS2 cells.4 Thus, we examined whether IFT20 knockdown can affect the MT nucleation at the Golgi apparatus in DLD1 cells. Detection of repolymerized MTs following removal of NZ indicated that, unlike SaOS2 cells, DLD1 cells showed MT nucleation primarily at the centrosome, but not the Golgi apparatus (Figure S4A). Thus, it is likely that Golgi‐associated MTs in DLD1 cells are originally nucleated at and released from the centrosome, as proposed in epithelial cells.32 As siRNAs against IFT20 failed to affect MT nucleation at the centrosome (Figure S4B), IFT20 might regulate attachment of the released MTs to the Golgi apparatus and/or stabilization of the attached MTs.
3.3. Intraflagellar transport 20 regulates polarization of the Golgi apparatus during collective invasion
Polarized cell migration requires reorientation of the Golgi apparatus toward the leading edge, a process depending on MTs, to facilitate directed secretion.7 In fact, in si‐Ctrl‐transfected DLD1 cells, there was an approximately 2.5‐fold increase in the number of leader cells in which the Golgi apparatus was facing toward the leading edge within 18 hours of collective invasion (Figure 4A,B). However, such Golgi reorientation in leader cells was inhibited remarkably by si‐IFT20 transfection (Figure 4A,B). In contrast to leader cells, any apparent changes in the number of follower cells with reoriented Golgi apparatus were not observed in follower cells from DLD1 cells transfected with either si‐Ctrl or si‐IFT20 (Figure 4B). These results indicate that IFT20 might promote reorientation of the Golgi apparatus in leader cells probably by regulating organization of Golgi‐associated MTs during collective invasion.
Figure 4.
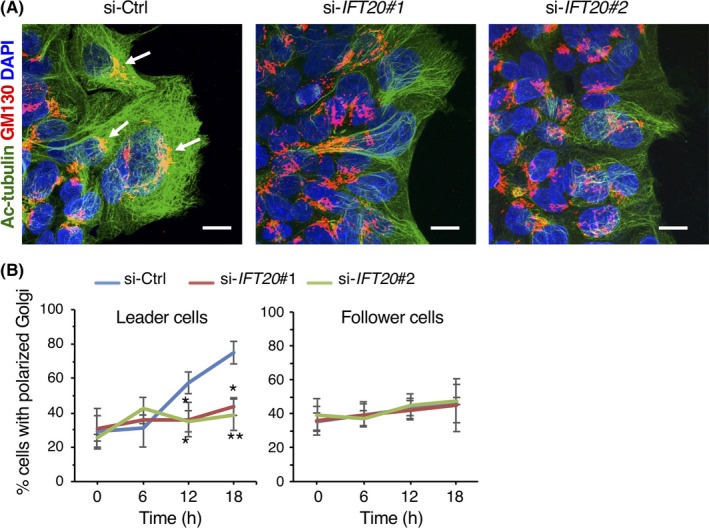
Intraflagellar transport 20 (IFT20) is required for polarization of the Golgi toward the leading edge during collective invasion of colorectal cancer cells. A, DLD1 cells transfected with the indicated siRNAs were subjected to 2‐D invasion assay for 18 h. Cells were stained with Abs against acetylated (Ac)‐tubulin (green) and GM130 (red), and counterstained with DAPI (blue). Arrows indicate the Golgi facing to the leading edge in leader cells. Scale bar, 10 μm. B, Percentages of leader and follower cells, in which the Golgi apparatus was detected within the 120° sector emerging from the center of the nucleus and facing toward the leading edge, were measured. Data are expressed as mean ± SD (n = 3). *P < .05; **P < .005, t test. si‐Ctrl, negative control siRNA
3.4. Intraflagellar transport 20 regulates direction of MT growth
As it has been established that MT dynamics play a central role in regulating the positioning of the Golgi apparatus, we next undertook live cell imaging of DLD1 cells stably expressing the MT plus‐end tracking protein EB1 fused to GFP as a marker for MT plus‐end growth.33 In leader cells undergoing collective invasion, we could detect comet‐like signals of EB1‐GFP, which were moving in a nonradial pattern, irrespective of IFT20 knockdown (Movie S1), consistent with their non‐radial MT arrays (Figures 3A and S3). Tracking analysis of the comets revealed that the number of EB1‐GFP comets moving toward the leading edge was substantially higher in si‐Ctrl‐transfected cells than in si‐IFT20‐transfected cells (Figure 5A). Furthermore, pause events of the comets were more frequently observed in si‐IFT20‐transfected cells than those in si‐Ctrl‐transfected ones, although their overall moving speeds were comparable between these groups (Figure 5B,C). These results indicate that IFT20 is required for polarized and continuous MT growth during collective invasion.
Figure 5.
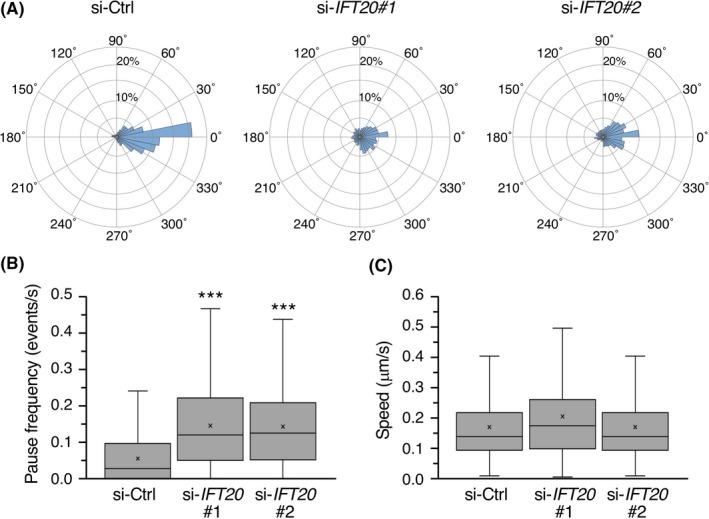
Suppressed expression of intraflagellar transport 20 (IFT20) impairs polarized and continuous microtubule growth during collective invasion of colorectal cancer cells. DLD1 cells expressing EB1‐GFP were transfected with the indicated siRNAs and subjected to 2‐D invasion assay. After 16 h of incubation, movement of EB1‐GFP in leader cells was analyzed by time‐lapse fluorescence imaging [Link]and tracking programs. A, Circular histograms showing the percentages of EB1‐GFP comets whose moving direction was within each of 36 equal sectors (10°). 0° indicates the direction perpendicular to the leading edge of the original cell sheet. The analysis included 2300‐2500 comets from 15 cells per each condition. B,C, Quantification of pause frequency (B) and moving speed (C) of EB1‐GFP comets. We defined a pause as an absence of movement in either direction for at least 0.9 s. Data are presented as box‐and‐whisker plots. n = 300‐520 comets; ***P < .001, t test. si‐Ctrl, negative control siRNA
4. DISCUSSION
Colorectal cancer is the third most commonly diagnosed cancer and the fourth leading cause of cancer‐related mortality worldwide.34 Cancer invasion and metastasis are the main causes of mortality in these patients.35 In fact, despite advancements in therapeutic strategies, many patients with CRC die due to development of metastases even after surgery.36 Therefore, elucidating the mechanisms of invasion and metastasis of CRC will help to improve the prognosis of the disease. Cancers of epithelial origin, including CRC, take advantage of collective invasion for local tissue infiltration.37, 38 During this process, leader cells engage to activate integrins in anterior protrusions toward the ECM16, 17 and show an increased expression and activity of MMPs, leading to polarized ECM degradation.18 Therefore, it has been postulated that leader cells possess the polarized Golgi apparatus toward the direction of invasion, coupled with polarized MT organization, to transport such membrane and secretory proteins, as well as membrane lipids, efficiently toward their anterior protrusions. However, the mechanisms underlying coordinated organization of MTs and Golgi apparatus in leader cells remain to be elucidated. In the present study, we show that IFT20 regulates the organization of polarized MTs, associated with the Golgi apparatus, to promote collective invasion.
Ror2 is expressed highly in various types of cancer cells, contributing to aberrant activation of Ror2 signaling that affects their proliferation and/or invasion, depending on cellular contexts.1 We have recently shown that constitutively activated Ror2 signaling induces expression of IFT20 to regulate polarized positioning of the Golgi apparatus during invasion of osteosarcoma cells.4 In this study, we found that HCT116 cells use this Ror2‐IFT20 axis to promote their invasion. However, we have shown that IFT20 also promotes invasion of DLD1 cells, in which the Ror2 gene is silenced. Furthermore, IFT20 promotes invasion of SW480 cells (expressing Ror2), where Ror2 itself fails to mediate expression of IFT20 and to promote their invasion. Thus, although IFT20 seems to be critically important for invasion of CRC cells, the role of Ror2 in IFT20 expression and invasion might be varied among CRC cell lines. At present, the molecular basis of Ror2‐dependent and ‐independent mechanisms of IFT20 expression remains unclear. As the Ror2 gene is often silenced in CRC cells,11, 12 it would be of interest to investigate the relationship between the epigenetic status of the Ror2 gene and Ror2‐dependency of IFT20 expression and invasiveness of CRC cells.
Noncentrosomal MTs are observed predominantly in differentiated cell types, such as polarized epithelial cells, mature neurons, and muscle cells, although their arrangement and localization appear to be varied among different cell types.39, 40, 41, 42 In osteosarcoma cells, IFT20 promotes nucleation of Golgi‐derived MTs.4 Accumulating evidence indicates that γ‐TuRC is involved in the nucleation of noncentrosomal MTs and/or anchoring their minus ends.43, 44 A‐kinase anchor protein 450, a scaffolding protein localized at the centrosome and Golgi apparatus,45 can recruit γ‐TuRC to the cis‐Golgi by interacting with a cis‐Golgi matrix protein, GM130.46, 47, 48, 49, 50 Intraflagellar transport 20 binds to both AKAP450 and GM130 at the cis‐Golgi, thereby regulating proper interaction between these 2 proteins for MT nucleation.4 We found that DLD1 cells show MT nucleation primarily at the centrosome, but not the Golgi apparatus (Figure S4A), suggesting that the γ‐TuRC‐AKAP450 complex might not be present at the cis‐Golgi in DLD1 cells. In fact, we failed to detect obvious expression of AKAP450 at the cis‐Golgi, as assessed by co‐immunostaining with anti‐AKAP450 and anti‐GM130 Abs in DLD1 cells, but it was detected clearly at the centrosome (Figure S5), indicating that IFT20 also has a nucleation‐independent function for organizing Golgi‐associated MTs. It has been shown that centrosomal MTs are released and attached through their minus ends to the apical cell membrane during epithelial cell differentiation.51 Thus, IFT20 might regulate attachments of MTs released from the centrosome to the Golgi and/or stabilization of the attached MTs. In this regard, it is worth noting that CAMSAP2 recognizes MT minus ends and tethers them to the cis‐Golgi membrane to protect them from depolymerization, independent of γ‐TuRC‐mediated MT nucleation at the cis‐Golgi.52 Additionally, CAMSAP2 is known to cooperate with EB1 and EB3 at the Golgi apparatus to regulate MT growth dynamics and Golgi reorientation.53, 54 Our findings indicate that IFT20 is required for polarized and continuous MT growth and for Golgi reorientation during collective invasion. Thus, there might be a link between IFT20 and CAMSAP2 in regulating MT dynamics at the Golgi apparatus, required for proper organization of Golgi‐associated MTs and Golgi polarity during collective invasion. Further studies will be required to clarify this issue.
In summary, our results reveal that IFT20 promotes collective invasion of CRC cells, irrespective of expression status and function of Ror2. Intraflagellar transport 20 promotes reorientation of the Golgi apparatus toward the front side of leader cells by regulating organization of Golgi‐associated MTs, which are stabilized and reoriented toward the direction of invasion in leader cells. Our results also indicate that IFT20 regulates polarized and continuous growth of Golgi‐associated MTs in leader cells. These findings unravel a novel function of IFT20 in regulating collective invasion of CRC cells through organization of Golgi‐associated MTs and Golgi polarity in leader cells. It can also be envisaged that IFT20 would be a novel diagnostic and therapeutic target molecule for collective invasion of CRC cells.
CONFLICTS OF INTEREST
The authors declare that they have no conflicts of interest.
Supporting information
ACKNOWLEDGMENTS
We thank Y. Mimori‐Kiyosue (RIKEN) for the plasmid encoding EB1‐GFP. This work was supported in part by a grant‐in‐aid for Scientific Research (B) (16H05152 [Y.M.]) from MEXT, a grant from the Japan Agency of Medical Research and Development (AMED) (18gm5010001s0901 [Y.M.]), a grant from the Japan Society for the Promotion of Science Bilateral Open Partnership Joint Research Projects (M.N.), and a grant from the Mitsubishi Foundation (ID 29145 [Y.M.]).
Aoki T, Nishita M, Sonoda J, Ikeda T, Kakeji Y, Minami Y. Intraflagellar transport 20 promotes collective cancer cell invasion by regulating polarized organization of Golgi‐associated microtubules. Cancer Sci. 2019;110:1306–1316. 10.1111/cas.13970
Contributor Information
Michiru Nishita, Email: nishita@med.kobe-u.ac.jp.
Yasuhiro Minami, Email: minami@kobe-u.ac.jp.
REFERENCES
- 1. Nishita M, Enomoto M, Yamagata K, Minami Y. Cell/tissue‐tropic functions of Wnt5a signaling in normal and cancer cells. Trends Cell Biol. 2010;20:346‐354. [DOI] [PubMed] [Google Scholar]
- 2. Oishi I, Suzuki H, Onishi N, et al. The receptor tyrosine kinase Ror2 is involved in non‐canonical Wnt5a/JNK signalling pathway. Genes Cells. 2003;8:645‐654. [DOI] [PubMed] [Google Scholar]
- 3. Enomoto M, Hayakawa S, Itsukushima S, et al. Autonomous regulation of osteosarcoma cell invasiveness by Wnt5a/Ror2 signaling. Oncogene. 2009;28:3197‐3208. [DOI] [PubMed] [Google Scholar]
- 4. Nishita M, Park SY, Nishio T, et al. Ror2 signaling regulates Golgi structure and transport through IFT20 for tumor invasiveness. Sci Rep. 2017;7:1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Miller PM, Folkmann AW, Maia AR, Efimova N, Efimov A, Kaverina I. Golgi‐derived CLASP‐dependent microtubules control Golgi organization and polarized trafficking in motile cells. Nat Cell Biol. 2009;11:1069‐1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vinogradova T, Paul R, Grimaldi AD, et al. Concerted effort of centrosomal and Golgi‐derived microtubules is required for proper Golgi complex assembly but not for maintenance. Mol Biol Cell. 2012;23:820‐833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yadav S, Puri S, Linstedt AD. A primary role for Golgi positioning in directed secretion, cell polarity, and wound healing. Mol Biol Cell. 2009;20:1728‐1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mikels AJ, Nusse R. Purified Wnt5a protein activates or inhibits beta‐catenin‐TCF signaling depending on receptor context. PLoS Biol. 2006;4:e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bienz M, Clevers H. Linking colorectal cancer to Wnt signaling. Cell. 2000;103:311‐320. [DOI] [PubMed] [Google Scholar]
- 10. Ying J, Li H, Yu J, et al. WNT5A exhibits tumor‐suppressive activity through antagonizing the Wnt/beta‐catenin signaling, and is frequently methylated in colorectal cancer. Clin Cancer Res. 2008;14:55‐61. [DOI] [PubMed] [Google Scholar]
- 11. Lara E, Calvanese V, Huidobro C, et al. Epigenetic repression of ROR2 has a Wnt‐mediated, pro‐tumourigenic role in colon cancer. Mol Cancer. 2010;9:170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ma SS, Srivastava S, Llamosas E, et al. ROR2 is epigenetically inactivated in the early stages of colorectal neoplasia and is associated with proliferation and migration. BMC Cancer. 2016;16:508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mei H, Lian S, Zhang S, Wang W, Mao Q, Wang H. High expression of ROR2 in cancer cell correlates with unfavorable prognosis in colorectal cancer. Biochem Biophys Res Commun. 2014;453:703‐709. [DOI] [PubMed] [Google Scholar]
- 14. Friedl P, Gilmour D. Collective cell migration in morphogenesis, regeneration and cancer. Nat Rev Mol Cell Biol. 2009;10:445‐457. [DOI] [PubMed] [Google Scholar]
- 15. Clark AG, Vignjevic DM. Modes of cancer cell invasion and the role of the microenvironment. Curr Opin Cell Biol. 2015;36:13‐22. [DOI] [PubMed] [Google Scholar]
- 16. Hegerfeldt Y, Tusch M, Brocker EB, Friedl P. Collective cell movement in primary melanoma explants: plasticity of cell‐cell interaction, beta1‐integrin function, and migration strategies. Cancer Res. 2002;62:2125‐2130. [PubMed] [Google Scholar]
- 17. Klinowska TC, Soriano JV, Edwards GM, et al. Laminin and beta1 integrins are crucial for normal mammary gland development in the mouse. Dev Biol. 1999;215:13‐32. [DOI] [PubMed] [Google Scholar]
- 18. Nabeshima K, Inoue T, Shimao Y, et al. Front‐cell‐specific expression of membrane‐type 1 matrix metalloproteinase and gelatinase A during cohort migration of colon carcinoma cells induced by hepatocyte growth factor/scatter factor. Cancer Res. 2000;60:3364‐3369. [PubMed] [Google Scholar]
- 19. Mimori‐Kiyosue Y, Shiina N, Tsukita S. The dynamic behavior of the APC‐binding protein EB1 on the distal ends of microtubules. Curr Biol. 2000;10:865‐868. [DOI] [PubMed] [Google Scholar]
- 20. Kani S, Oishi I, Yamamoto H, et al. The receptor tyrosine kinase Ror2 associates with and is activated by casein kinase Iepsilon. J Biol Chem. 2004;279:50102‐50109. [DOI] [PubMed] [Google Scholar]
- 21. Nishita M, Yoo SK, Nomachi A, et al. Filopodia formation mediated by receptor tyrosine kinase Ror2 is required for Wnt5a‐induced cell migration. J Cell Biol. 2006;175:555‐562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jaqaman K, Loerke D, Mettlen M, et al. Robust single‐particle tracking in live‐cell time‐lapse sequences. Nat Methods. 2008;5:695‐702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Myers KA, Applegate KT, Danuser G, Fischer RS, Waterman CM. Distinct ECM mechanosensing pathways regulate microtubule dynamics to control endothelial cell branching morphogenesis. J Cell Biol. 2011;192:321‐334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Meijering E, Dzyubachyk O, Smal I. Methods for cell and particle tracking. Methods Enzymol. 2012;504:183‐200. [DOI] [PubMed] [Google Scholar]
- 25. Finetti F, Paccani SR, Riparbelli MG, et al. Intraflagellar transport is required for polarized recycling of the TCR/CD3 complex to the immune synapse. Nat Cell Biol. 2009;11:1332‐1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Follit JA, Tuft RA, Fogarty KE, Pazour GJ. The intraflagellar transport protein IFT20 is associated with the Golgi complex and is required for cilia assembly. Mol Biol Cell. 2006;17:3781‐3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gundersen GG, Bulinski JC. Selective stabilization of microtubules oriented toward the direction of cell migration. Proc Natl Acad Sci U S A. 1988;85:5946‐5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Waterman‐Storer CM, Salmon ED. Actomyosin‐based retrograde flow of microtubules in the lamella of migrating epithelial cells influences microtubule dynamic instability and turnover and is associated with microtubule breakage and treadmilling. J Cell Biol. 1997;139:417‐434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dong C, Xu H, Zhang R, Tanaka N, Takeichi M, Meng W. CAMSAP3 accumulates in the pericentrosomal area and accompanies microtubule release from the centrosome via katanin. J Cell Sci. 2017;130:1709‐1715. [DOI] [PubMed] [Google Scholar]
- 30. Skoufias DA, Burgess TL, Wilson L. Spatial and temporal colocalization of the Golgi apparatus and microtubules rich in detyrosinated tubulin. J Cell Biol. 1990;111:1929‐1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Thyberg J, Moskalewski S. Relationship between the Golgi complex and microtubules enriched in detyrosinated or acetylated alpha‐tubulin: studies on cells recovering from nocodazole and cells in the terminal phase of cytokinesis. Cell Tissue Res. 1993;273:457‐466. [DOI] [PubMed] [Google Scholar]
- 32. Bartolini F, Gundersen GG. Generation of noncentrosomal microtubule arrays. J Cell Sci. 2006;119:4155‐4163. [DOI] [PubMed] [Google Scholar]
- 33. Matov A, Applegate K, Kumar P, et al. Analysis of microtubule dynamic instability using a plus‐end growth marker. Nat Methods. 2010;7:761‐768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359‐E386. [DOI] [PubMed] [Google Scholar]
- 35. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646‐674. [DOI] [PubMed] [Google Scholar]
- 36. Dekker E, Sanduleanu S. Colorectal cancer: strategies to minimize interval CRC in screening programmes. Nat Rev Gastroenterol Hepatol. 2016;13:10‐12. [DOI] [PubMed] [Google Scholar]
- 37. Cheung KJ, Ewald AJ. A collective route to metastasis: seeding by tumor cell clusters. Science. 2016;352:167‐169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ilina O, Friedl P. Mechanisms of collective cell migration at a glance. J Cell Sci. 2009;122:3203‐3208. [DOI] [PubMed] [Google Scholar]
- 39. Nishita M, Satake T, Minami Y, Suzuki A. Regulatory mechanisms and cellular functions of non‐centrosomal microtubules. J Biochem. 2017;162:1306‐10. [DOI] [PubMed] [Google Scholar]
- 40. Conde C, Caceres A. Microtubule assembly, organization and dynamics in axons and dendrites. Nat Rev Neurosci. 2009;10:319‐332. [DOI] [PubMed] [Google Scholar]
- 41. Guerin CM, Kramer SG. Cytoskeletal remodeling during myotube assembly and guidance: coordinating the actin and microtubule networks. Commun Integr Biol. 2009;2:452‐457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Oddoux S, Zaal KJ, Tate V, et al. Microtubules that form the stationary lattice of muscle fibers are dynamic and nucleated at Golgi elements. J Cell Biol. 2013;203:205‐213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Akhmanova A, Hoogenraad CC. Microtubule minus‐end‐targeting proteins. Curr Biol. 2015;25:R162‐R171. [DOI] [PubMed] [Google Scholar]
- 44. Sanchez AD, Feldman JL. Microtubule‐organizing centers: from the centrosome to non‐centrosomal sites. Curr Opin Cell Biol. 2017;44:93‐101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Takahashi M, Shibata H, Shimakawa M, Miyamoto M, Mukai H, Ono Y. Characterization of a novel giant scaffolding protein, CG‐NAP, that anchors multiple signaling enzymes to centrosome and the golgi apparatus. J Biol Chem. 1999;274:17267‐17274. [DOI] [PubMed] [Google Scholar]
- 46. Choi YK, Liu P, Sze SK, Dai C, Qi RZ. CDK5RAP2 stimulates microtubule nucleation by the gamma‐tubulin ring complex. J Cell Biol. 2010;191:1089‐1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Roubin R, Acquaviva C, Chevrier V, et al. Myomegalin is necessary for the formation of centrosomal and Golgi‐derived microtubules. Biol Open. 2013;2:238‐250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang Z, Wu T, Shi L, et al. Conserved motif of CDK5RAP2 mediates its localization to centrosomes and the Golgi complex. J Biol Chem. 2010;285:22658‐22665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wang Z, Zhang C, Qi RZ. A newly identified myomegalin isoform functions in Golgi microtubule organization and ER‐Golgi transport. J Cell Sci. 2014;127:4904‐4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rivero S, Cardenas J, Bornens M, Rios RM. Microtubule nucleation at the cis‐side of the Golgi apparatus requires AKAP450 and GM130. EMBO J. 2009;28:1016‐1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Moss DK, Bellett G, Carter JM, et al. Ninein is released from the centrosome and moves bi‐directionally along microtubules. J Cell Sci. 2007;120:3064‐3074. [DOI] [PubMed] [Google Scholar]
- 52. Wu J, de Heus C, Liu Q, et al. Molecular pathway of microtubule organization at the Golgi apparatus. Dev Cell. 2016;39:44‐60. [DOI] [PubMed] [Google Scholar]
- 53. Yang C, Wu J, de Heus C, et al. EB1 and EB3 regulate microtubule minus end organization and Golgi morphology. J Cell Biol. 2017;216:3179‐3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wei J, Xu H, Meng W. Noncentrosomal microtubules regulate autophagosome transport through CAMSAP2‐EB1 cross‐talk. FEBS Lett. 2017;591:2379‐2393. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


