Abstract
The E3 ubiquitin ligase ring finger protein 115 (RNF115) is overexpressed in more than half of human breast tumors and is implicated in the pathogenesis and progression of breast cancer. However, the mechanism behind RNF115 overexpression in breast tumors remains largely unknown. Here we report that ubiquitin‐specific protease 9X (USP9X), a substrate‐specific deubiquitinating enzyme, stabilizes RNF115 and thereby regulates its biological functions in breast cancer cells. Immunoprecipitation and GST pull‐down assays showed that USP9X interacted with RNF115. Depletion of RNF115 by siRNAs or overexpression of RNF115 did not significantly affect USP9X expression. In contrast, knockdown of USP9X in breast cancer cells by siRNAs reduced RNF115 protein abundance, which was partially restored following treatment with proteasome inhibitor MG‐132. Moreover, depletion of USP9X reduced the half‐life of RNF115 and increased its ubiquitination. Conversely, overexpression of USP9X resulted in an accumulation of RNF115 protein, accompanied by a decrease in its ubiquitination. RNF115 mRNA levels were unaffected by overexpression or knockdown of USP9X. Furthermore, USP9X protein expression levels correlated positively with RNF115 in breast cancer cell lines and breast tumor samples. Importantly, reintroduction of RNF115 in USP9X‐depleted cells partially rescued the reduced proliferation, migration, and invasion of breast cancer cells by USP9X knockdown. Collectively, these findings indicate that USP9X is a stabilizer of RNF115 protein and that the USP9X‐RNF115 signaling axis is implicated in the breast cancer malignant phenotype.
Keywords: breast cancer, deubiquitinating enzyme, E3 ubiquitin ligase, ring finger protein 115, ubiquitin‐specific protease 9X
Abbreviations
- BCA2
breast cancer‐associated gene 2
- CHX
cycloheximide
- DUB
deubiquitinating enzyme
- ERα
estrogen receptor α
- IP
immunoprecipitation
- qPCR
quantitative PCR
- RNF115
ring finger protein 115
- shNC
negative control shRNA
- shUSP9X
shRNA targeting USP9X
- siNC
negative control siRNA
- SMURF1
SMAD‐specific E3 ubiquitin protein ligase 1
- USP
ubiquitin‐specific protease
- YAP
Yes‐associated protein
1. INTRODUCTION
Breast cancer is the most commonly diagnosed cancer and the leading cause of cancer death among women.1 Emerging evidence suggests that dysregulation of the ubiquitin‐proteasome system is intimately implicated in breast cancer pathogenesis and progression through regulating protein homeostasis, protein‐protein interaction, and signal transduction.2, 3 Specificity within this system is dictated by E3 ubiquitin ligases, which catalyze the final step of ubiquitin conjugation by transferring ubiquitin from ubiquitin‐conjugating enzymes to their substrates.4 There are 2 major classes of mechanistically distinct E3s, characterized by the RING (or RING‐like) and HECT domains.5 Of these, the majority are RING finger and RING finger‐related E3s.6, 7 Ring finger protein 115, also known as BCA2, is a poorly characterized RING finger‐type E3 ubiquitin ligase.8, 9 The RNF115 gene encodes a protein of 305 amino acids, comprising an N‐terminal BCA2 zinc‐finger domain, a central AKT phosphorylation domain, and a C‐terminal RING H2 domain.10, 11, 12 The BCA2 zinc‐finger domain specifically binds to ubiquitin and is susceptible to becoming ubiquitinated, whereas the RING domain is implicated in catalyzing the ubiquitination of RNF115‐interacting proteins and/or autoubiquitination.8, 11 RNF115 was originally isolated through subtractive hybridization cloning from breast cancer cells.8, 9 Subsequent studies reported that RNF115 is overexpressed in more than 50% of invasive breast tumors and is important for regulating breast cancer cell proliferation, migration, and invasion.8, 11, 13 Moreover, its high expression is associated with regional recurrence, lymph node metastasis, and unfavorable prognosis of patients with breast cancer.8, 14 Mechanistic investigations reveal that RNF115 promotes breast cancer cell proliferation through targeting the cyclin‐dependent kinase inhibitor p21Waf/Cip1 for ubiquitin‐dependent degradation.13 Given the functional importance of RNF115 in driving breast cancer, understanding the mechanism underlying its overexpression in breast tumors should facilitate the development of new therapeutic agents. A recent study revealed that estrogen enables transcriptional activation of RNF115 in breast cancer cells through enhancing the binding of ERα to its promoter.15 In addition to gene transcription, RNF115 possesses an intrinsic autoubiquitination activity and is thought to be regulated by the ubiquitin‐proteasome pathway.8, 11 However, the mechanisms for regulating its protein stability remain undefined.
Protein ubiquitination is counterbalanced by DUBs, which remove ubiquitin chains from target proteins to regulate their functions.16, 17 To date, approximately 100 DUBs have been identified in the human genome.17, 18 Among these DUBs, the largest family is the USPs.16, 18 Notably, USP9X,19 one of the USP family of DUBs, has been shown to be upregulated in breast tumors3, 20, 21 and to promote breast cancer cell survival, migration, tumorigenesis, and chemoresistance by deubiquitinating and stabilizing its substrates, such as transcription factor FOXO3a,22 SMURF1,23 YAP1,21 centriolar satellite protein CEP131,20 and pseudokinase Tribbles homolog 3.24 Consequently, inhibition or knockdown of USP9X enhances the sensitivity of breast cancer cells to chemotherapeutic drugs.21, 25 Interestingly, a recent quantitative proteomic study identified RNF115 as one of significantly downregulated proteins in USP9X‐depleted human lung cancer A549 cells,26 indicating that RNF115 could be a potential substrate of USP9X. However, the functional and mechanistic insights into regulation of RNF115 by USP9X in breast cancer cells remain unexplored.
In this study, we report that USP9X interacts with and stabilizes RNF115 by antagonizing its ubiquitination and proteasomal degradation. Functional rescue experiments further indicate that the USP9X‐RNF115 signaling axis is linked with breast cancer cell proliferation, migration, and invasion.
2. MATERIALS AND METHODS
2.1. Cell culture and reagents
Human breast cancer cell lines (MCF‐7, T47D, ZR‐75‐1, SK‐BR‐3, MDA‐MB‐231, MDA‐MB‐468, Hs578T, BT20, and BT549), human cervical cancer HeLa cell line, human mammary epithelial HMEC cell line, and human embryonic kidney 293T (HEK293T) cell line were obtained from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China), where cell lines have been authenticated by short tandem repeat profiling and monitoring cell morphology, biologic behavior, and mycoplasma contamination. HMEC cells were cultured in high‐glucose DMEM containing 5% FBS (ExCell Bio, Shanghai, China), 1% penicillin/streptomycin, 20 ng/μL human epidermal growth factor, 0.5 μg/mL hydrocortisone, and 10 μg/mL insulin. Other cell lines were grown in high‐glucose DMEM medium containing 10% FBS and 1% penicillin/streptomycin. All cells were cultured in a humidified incubator at 37°C with 5% CO2. Culture media and supplements were obtained from BasalMedia (Shanghai, China). Proteasome inhibitor MG‐132 and protein synthesis inhibitor CHX were obtained from Selleck (Shanghai, China) and Cell Signaling Technology (Danvers, MA, USA), respectively. All chemicals and reagents were from Sigma‐Aldrich (St. Louis, MO, USA) unless otherwise noted.
2.2. Expression plasmids, siRNAs, and transfection
pDEST51 and pDEST51‐V5‐USP9X expression vectors were kindly provided by Professor Stephen A. Wood (Eskitis Institute for Drug Discovery, Griffith University, Brisbane, Australia). Full‐length human RNF115 cDNA was purchased from Vigene Biosciences (Shandong, China) and subcloned into pCDH‐CMV‐MCS‐EF1‐Puro (System Biosciences, Mountain View, CA, USA), pLVX‐IRES‐neo (Clontech, Mountain View, CA, USA), and pGEX‐6p‐1 (kindly provided by Yanhui Xu, Fudan University, Shanghai, China) expression vectors to generate pCDH‐Flag‐RNF115, pLVX‐Flag‐RNF115, and GST‐RNF115, respectively. The primers used for molecular cloning are provided in Table S1. Hemagglutinin‐ubiquitin expression vector has been described previously.27, 28 Small hairpin RNAs targeting USP9X and shNC lentiviral expression vectors were kindly provided by Hu Zhou (Shanghai Institute of Materia Medica, Chinese Academy of Sciences, Shanghai, China). Short interfering RNAs were synthesized by GenePharma (Shanghai, China). The shRNA and siRNA targeting sequences are provided in Table S2. Transfection of plasmids and siRNAs was carried out using Neofect DNA transfection reagent (TengyiBio, Shanghai, China) and Lipofectamine 2000 (Invitrogen, Waltham, MA, USA), respectively, according to the manufacturer's protocol. Protein and mRNA levels were estimated after 48 hours of transfection by immunoblotting and qPCR, respectively. To establish USP9X‐depleted and RNF115‐overexpressing stable cell lines, packaging plasmid psPAX2, envelope plasmid pMD2.G, shUSPX lentiviral vectors, and pLVX‐IRES‐neo‐RNF115 vector were cotransfected into HEK293T cells. Viral supernatant was collected 48 hours after transfection and filtered through a 0.45‐μm filter. Target cells were infected twice with the viral supernatant in the presence of 8 μg/mL polybrene (Sigma‐Aldrich) and selected with 1‐4 μg/mL puromycin (Cayman, Ann Arbor, MI, USA) or 0.5 mg/mL Geneticin (Invitrogen).
2.3. Quantitative PCR
Total RNAs were isolated using TRIzol reagent (Invitrogen) and converted into cDNAs using PrimeScript Reverse Transcription Master Mix (Takara, Dalian, China). Quantitative PCR was undertaken using SYBR Premix Ex Taq (Tli RNaseH Plus; Takara). The primers used for qPCR analysis of RNF115 and USP9X were designed by PrimerBank (https://pga.mgh.harvard.edu/primerbank/) (Table S3); GAPDH primers have been described previously.29 Results normalized to GAPDH are presented as fold induction relative to control.
2.4. Antibodies, immunoblotting, immunoprecipitation, and immunofluorescence
The detailed information for primary Abs used in this study is provided in Table S4. Secondary Abs for immunoblotting and immunofluorescence were from Cell Signaling Technology. Immunoblotting, IP, and immunofluorescence have previously been described.27, 30 Briefly, for immunoblotting analyses, cells were lysed in modified RIPA buffer (50 mmol/L Tris‐HCl, pH 7.4, 150 mmol/L NaCl, 1% NP‐40, 0.25% sodium deoxycholate, 0.1% SDS, and 1 mmol/L EDTA) containing 1× protease inhibitor cocktail and 1× phosphatase inhibitor cocktail (Bimake, Shanghai, China). Proteins were quantified using the bicinchoninic acid assay (Yeasen, Shanghai, China), resolved by SDS‐PAGE, and transferred onto PVDF membrane (Millipore, Billerica, MA, USA). Antibody detection was carried out using an enhanced chemiluminescent substrate kit (Yeasen). For IP assays, cells were lysed in NP‐40 lysis buffer (50 mmol/L Tris‐HCl, pH 8, 150 mmol/L NaCl, 0.5% NP‐40, 10% glycerol, 2 mmol/L MgCl2, and 1 mmol/L EDTA) supplemented with protease and phosphatase inhibitors and centrifuged at 18 000 g at 4°C for 30 minutes. Then the supernatant was collected and immunoprecipitated with the indicated primary Ab at 4°C overnight, followed by protein A/G magnetic beads (Bimake) for another 2 hours at 4°C. The beads were washed with NP‐40 buffer 3 times. The proteins released from the beads were boiled for 5 minutes and subjected to immunoblotting analysis. For immunofluorescent staining, HEK293T cells were seeded on 12‐mm coverslips and cotransfected with Flag‐RNF115 and V5‐USP9X. After 48 hours of transfection, cells were fixed with 4% paraformaldehyde for 10 minutes, pre‐extracted in 0.1% Triton X‐100 for 5 minutes, blocked in 5% normal goat serum in PBST for 1 hour, and incubated with primary rabbit anti‐V5 and mouse anti‐Flag Abs overnight at 4°C. After washing with PBST buffer 3 times, cells were incubated with secondary Abs conjugated with mouse 488‐Alexa (green) and rabbit 555‐Alexa (red). DNA was counterstained with DAPI (Abcam, Shanghai, China). Microscopic analyses were carried out using a Leica SP5 confocal microscope (Leica Microsystems, Buffalo Grove, IL, USA).
2.5. GST pull‐down assay
The GST pull‐down assays were carried out as described previously.27 In brief, pGEX‐6p‐1 and pGEX‐6p‐1‐RNF115 constructs were transformed in Escherichia coli strain BL21 (DE3; Tiangen Biotech, Beijing, China), induced with 0.2 mmol/L isopropyl‐β‐D‐thiogalactoside (Invitrogen), and purified using Glutathione Sepharose 4B bath beads (GE Healthcare, Beijing, China). The GST and GST‐RNF115 proteins were added to 800 μL cellular lysates from MCF‐7 cells and rotated for 3 hours at 4°C. After washing with NP‐40 buffer 3 times, proteins were eluted and subjected to SDS‐PAGE for immunoblotting analysis.
2.6. Ubiquitination assay
HEK293T cells were transfected with siRNAs targeting USP9X or siNC using Lipofectamine 2000 (Invitrogen). After 6 hours of transfection, HA‐ubiquitin and Flag‐RNF115 were cotransfected using Neofect DNA transfection reagent (TengyiBio). To address the effect of USP9X overexpression on RNF115 ubiquitination, Flag‐RNF115, HA‐ubiquitin, and V5‐USP9X were cotransfected into HEK293T cells. After 48 hours of transfection, cells were treated with 10 μmol/L MG‐132 for 4 hours, washed with PBS, and lysed with 200 μL denaturing buffer (4% SDS, 300 mmol/L NaCl, 10 mmol/L DTT, 20 mmol/L N‐ethylmaleimide, 10 mmol/L iodoacetamide, and 10 mmol/L HEPES, pH 7.5) supplemented with protease and phosphatase inhibitors. Lysates were collected in a 1.5‐mL Eppendorf tube and immediately boiled for 10 minutes. Lysates were then diluted in 1.8 mL ice‐cold dilution buffer (1.7% Thesit [Sigma‐Aldrich], 150 mmol/L NaCl, 20 mmol/L N‐ethylmaleimide, and 50 mmol/L HEPES, pH 7.5) containing protease and phosphatase inhibitors and incubated with mouse anti‐DYKDDDDK (Flag) affinity gel (Bimake) for 3 hours. The ubiquitination of RNF115 was assessed by immunoblotting analysis with an anti‐HA Ab.
2.7. Cycloheximide chase assays
To determine the half‐life of RNF115 protein, CHX chase assays were carried out as described previously.27, 30 After 48 hours of transfection, cells were treated with 100 μg/mL CHX for the indicated times and harvested for immunoblotting analysis.
2.8. Colony formation, cell migration, and invasion assays
For colony formation assays, MCF‐7 and MDA‐MB‐231 cells stably expressing shNC, shUSP9X, and shUSP9X together with pLVX or pLVX‐Flag‐RNF115 were plated onto 12‐well plates (1000 and 500 cells per well for MCF‐7 and MDA‐MB‐231 cell lines, respectively) in triplicate for 16 days. Media were replaced every 3 days. Cells were fixed with methanol for 30 minutes and stained with 0.5% crystal violet solution for 1 hour. The colonies were counted. For wound‐healing assays, MDA‐MB‐231 cells were plated onto 6‐well plates. When cells were grown to confluency, the wound was created by 200 μL tips, the floated cells were removed through PBS washing, and the culture medium replaced by serum‐free medium. Images of the wound were taken at the indicated time points using an Olympus IX51 inverted microscope (Tokyo, Japan), and the percentage of the wound closure was quantified from 3 independent replicates as described previously.29 For migration and invasion assays, an 8‐μm pore‐size polycarbonate membrane Transwell chamber and BioCoat Matrigel invasion chamber (Corning, Shanghai, China) were used, respectively.29 MDA‐MB‐231 cells in serum‐free medium were plated in the top chamber (3 × 104 cells for migration assays and 6 × 104 cells for invasion assays). Growth medium containing 10% FBS was used as a chemoattractant in the lower chamber. After 18 hours of incubation, cells were fixed with methanol for 30 minutes and stained with 0.5% crystal violet solution for 1 hour. The migrated and invaded cells were photographed using an Olympus IX51 inverted microscope at 200× magnification.
2.9. Statistical analysis
All data are presented as the mean ± SE from at least 3 independent experiments. Two‐tailed unpaired Student's t test was used for comparing 2 groups of data. The correlation between USP9X and RNF115 expression in cell lines and tumor samples was analyzed using the χ2 test. R is Pearson's correlation coefficient. P values of less than 0.05 were considered statistically significant.
3. RESULTS
3.1. Ubiquitin‐specific protease 9X and RNF115 are correlatively upregulated in breast cancer cell lines and breast tumor samples
To investigate the relationship between USP9X and RNF115 in human breast cancer, we analyzed the expression levels of USP9X and RNF115 in 31 pairs of primary breast cancer specimens and matched adjacent noncancerous breast tissues by immunoblotting. As shown in Figure 1A,B, USP9X was overexpressed in 87% (27/31) of breast cancer samples as compared with adjacent noncancerous tissues. Ring finger protein 115 protein levels were increased in 96% (26/27) of USP9X‐overexpressing cancer samples. In addition, among 13% (4/31) of cancer tissues with USP9X low expression, 50% (2/4) of cancer tissues showed lower RNF115 expression compared to adjacent normal tissues. The correlation between USP9X and RNF115 protein expression was also observed in multiple breast cancer cell lines (Figure 1C,D). These findings suggest that USP9X and RNF115 are concomitantly overexpressed in human breast cancer cell lines and tumor samples from patients.
Figure 1.
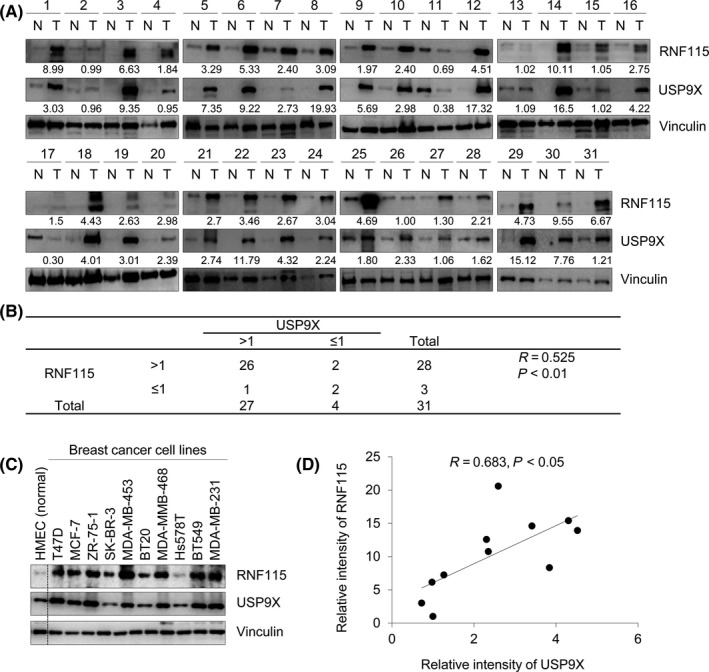
Ring finger protein 115 (RNF115) is positively correlated with ubiquitin‐specific protease 9X (USP9X) expression in breast cancer cell lines and tumor samples. A, Immunoblotting analysis of USP9X and RNF115 in 31 pairs of breast cancer specimens (T) and matched adjacent noncancerous breast tissues (N). Western blots were quantified using ImageJ software (https://imagej.nih.gov/ij/). Protein levels of USP9X and RNF115 were normalized to those of internal control vinculin. Relative expression levels of USP9X and RNF115 (T/N) are shown under corresponding lanes in these western blots. B, The correlation between RNF115 and USP9X protein levels in these samples was analyzed using the χ2 test. R is Pearson's correlation coefficient. C, Immunoblotting analysis of USP9X and RNF115 proteins in 10 breast cancer cell lines and normal HMEC cell line. D, A correlation between the protein expression levels of RNF115 and USP9X in these cell lines was analyzed as described in B
3.2. Ring finger protein 115 does not modulate USP9X expression
Ring finger protein 115 is an E3 ubiquitin ligase that catalyzes ubiquitination of its substrates to trigger their proteasomal degradation.8, 13 To address whether USP9X is a potential substrate of RNF115, we first examined whether treatment of cells with proteasomal inhibitor MG‐132 could result in an accumulation of USP9X protein. As shown in Figure S1, we found that MG‐132 treatment had no significant effects on USP9X protein levels. As a positive control, MG‐132 treatment resulted in a significant increase in the protein levels of p53, which is known to be degraded through the proteasome pathway.31 These results suggest that the ubiquitin‐proteasome pathway might not be involved in USP9X degradation. To confirm these results, MCF‐7 and MDA‐MB‐231 cells were transfected with siNC and 3 independent siRNAs targeting RNF115 (siRNF115 #1‐3). As shown in Figure 2A, 3 RNF115 siRNAs effectively reduced RNF115 protein levels after 48 hours of transfection. However, siRNA‐mediated knockdown of RNF115 had no remarkable effect on USP9X levels. Consistently, overexpression of RNF115 in HEK293T cells did not significantly affect protein abundance of endogenous (Figure 2B) or exogenously expressed USP9X (Figure 2C). These results suggest that RNF115 might not modulate USP9X expression.
Figure 2.
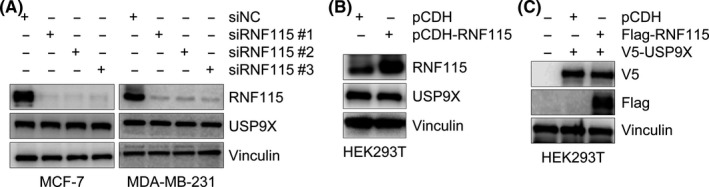
Ring finger protein 115 (RNF115) does not affect ubiquitin‐specific protease 9X (USP9X) expression. A, MCF‐7 and MDA‐MB‐231 cells were transfected with negative control siRNA (siNC) or 3 different siRNAs targeting RNF115 (siRNF115 #1‐3). After 48 h of transfection, cells were harvested and analyzed by immunoblotting with the indicated Abs. B,C, HEK293T cells were transfected with expression vector encoding empty vector pCDH or pCDH‐RNF115 with or without V5‐USP9X. After 48 h of transfection, total cellular lysates were subjected to immunoblotting with the indicated Abs
3.3. Ubiquitin‐specific protease 9X regulates RNF115 at the protein level, but not mRNA level
To test whether USP9X regulates RNF115 expression, we knocked down endogenous USP9X using 3 independent siUSP9Xs in MCF‐7, T47D, SK‐BR‐3, and BT474 cells and then examined RNF115 protein levels by immunoblotting analysis. Results showed that the levels of RNF115 were markedly decreased following USP9X depletion by siRNAs (Figure 3A). Similarly, stable knockdown of USP9X in MDA‐MB‐231, BT549, and HeLa cells using 2 different shUSP9Xs also resulted in a downregulation of RNF115 protein (Figure S2). In contrast, the mRNA levels of RNF115 were not significantly altered following USP9X knockdown (Figure 3B). In addition, overexpression of USP9X increased the protein abundance of endogenous (Figure 3C) and exogenously expressed RNF115 (Figure 3D). Quantitative PCR analysis showed that USP9X overexpression did not significantly affect the mRNA levels of RNF115 (Figure 3E,F). These results suggest that USP9X regulates RNF115 at the protein level.
Figure 3.
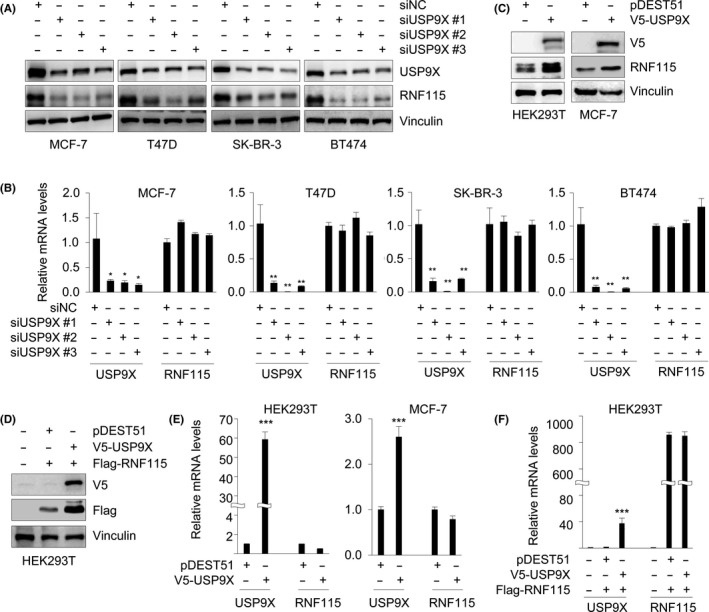
Ubiquitin‐specific protease 9X (USP9X) regulates ring finger protein 115 (RNF115) at the protein but not mRNA level. A,B, MCF‐7, T47D, SK‐BR‐3, and BT474 cells were transfected with negative control siRNA (siNC) or 3 different siRNAs targeting USP9X (siUSP9X #1‐3). After 48 h of transfection, cells were subjected to immunoblotting (A) or quantitative PCR (B) analysis of USP9X and RNF115 protein and mRNA levels, respectively. C,D, HEK293T and MCF‐7 cells were transfected with the indicated expression vectors. After 48 h of transfection, cells were subjected to immunoblotting analysis with the indicated Abs. E,F, HEK293T and MCF‐7 cells were transfected with the indicated expression vectors. After 48 h of transfection, cells were subjected to quantitative PCR analysis of USP9X and RNF115 mRNA levels. In B, E, and F, *P < .05; **P < .01; ***P < .001
3.4. Ubiquitin‐specific protease 9X enhances the stability of RNF115 protein
To determine the effect of USP9X on protein stability of RNF115, MCF‐7 and T47D cells stably expressing vectors encoding shNC or 3 shUSP9Xs were treated with DMSO or 10 μmol/L MG‐132 for 6 hours. Immunoblotting analysis showed that depletion of USP9X resulted in a decrease in RNF115 protein levels, which were partially restored after MG‐132 treatment (Figure 4A). To further determine the impact of USP9X on RNF115 protein turnover, MCF‐7 and T47D cells were transfected with siNC or two siUSP9Xs. After 48 hours of transfection, cells were treated with 100 μg/mL CHX for the indicated times and then harvested for immunoblotting. As expected, the half‐life of RNF115 was reduced following depletion of USP9X (Figure 4B,C). These results suggest that USP9X controls the stability of RNF115.
Figure 4.
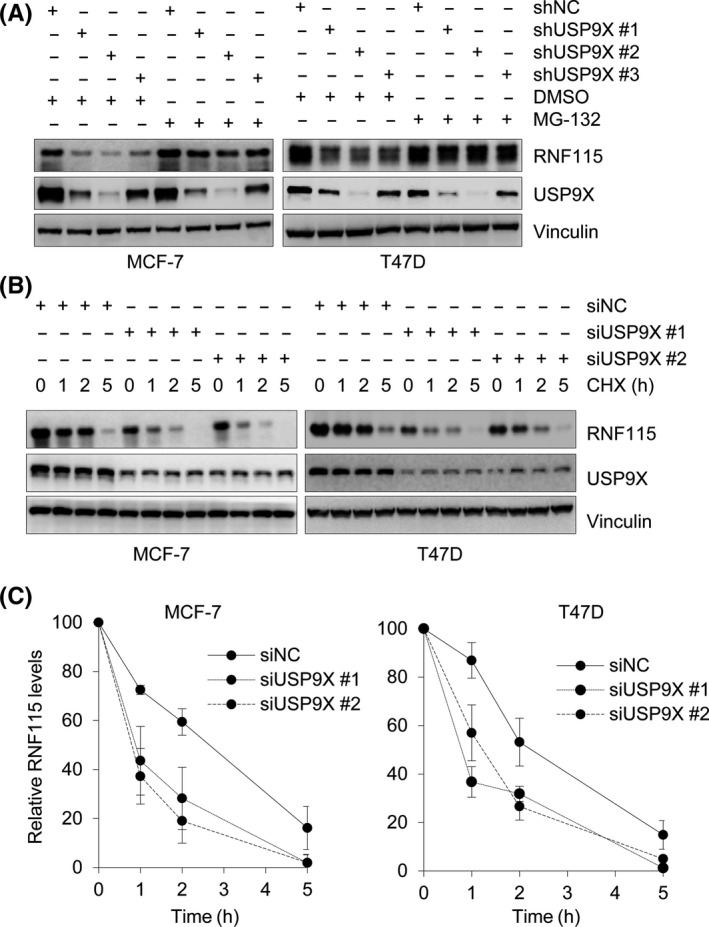
Ubiquitin‐specific protease 9X (USP9X) enhances ring finger protein 115 (RNF115) stability. A, MCF‐7 and T47D cells stably expressing negative control shRNA (shNC) or shRNAs targeting USP9X were treated with DMSO or 10 μmol/L MG‐132 for 6 h, followed by immunoblotting analysis with the indicated Abs. B,C, MCF‐7 and T47D cells were transfected with negative control siRNA (siNC) or 2 different siUSP9Xs. After 48 h of transfection, cells were treated with 100 μg/mL cycloheximide for the indicated times. Total cellular lysates were subjected to immunoblotting analysis with the indicated Abs (B). Quantitative results of relative RNF115 protein levels (RNF115/vinculin) from 3 independent experiments are shown (C)
3.5. Ubiquitin‐specific protease 9X interacts with RNF115 and counteracts its ubiquitination
To investigate the mechanisms regulating RNF115 protein stability by USP9X, we next examined whether USP9X interacts with RNF115. To do this, HEK293T cells were transfected with expression vectors encoding V5‐USP9X and Flag‐RNF115 alone or in combination. After 48 hours of transfection, total cellular lysates were subjected to IP assays using either an anti‐V5 or an anti‐Flag Ab. As shown in Figure 5A, V5‐USP9X was immunoprecipitated with Flag‐RNF115 when coexpressed. Immunofluorescent staining showed that V5‐USP9X and Flag‐RNF115 colocalized in cotransfected HEK293T cells (Figure S3). This interaction was also validated with transfected Flag‐RNF115 and endogenous USP9X in HEK293T cells (Figure 5B). To examine whether endogenous USP9X interacts with endogenous RNF115, endogenous RNF115 protein was immunoprecipitated using an anti‐RNF115 Ab in MCF‐7 and T47D cells. Results showed that RNF115 was efficiently coimmunoprecipitated with USP9X in both cell lines (Figure 5C). To validate the direct interaction between USP9X and RNF115 in vitro, GST pull‐down assays were carried out using purified GST‐RNF115 from bacteria and total cellular lysates of MCF‐7 cells. Results showed that GST‐RNF115 fusion protein, but not GST negative control, was able to pull down USP9X (Figure 5D). Together, these results suggest that USP9X interacts with RNF115.
Figure 5.
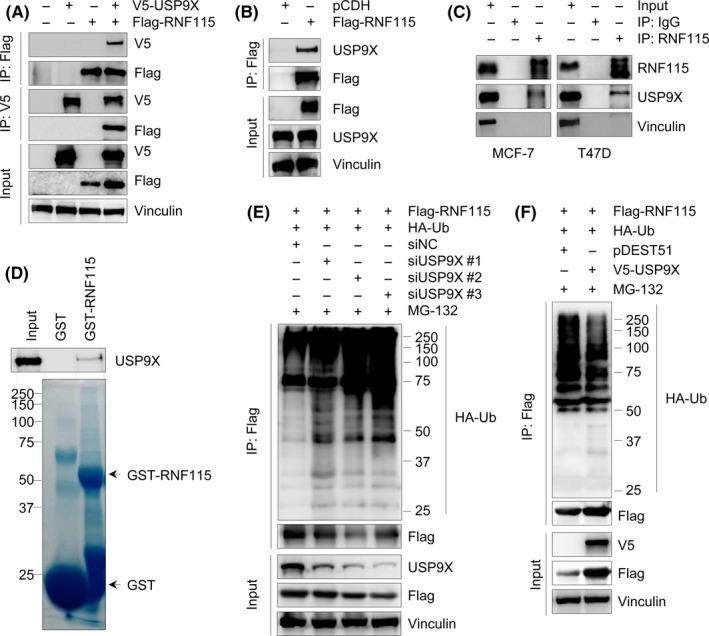
Ubiquitin‐specific protease 9X (USP9X) interacts with and promotes ring finger protein 115 (RNF115) deubiquitination. A,B, HEK293T cells were transfected with the indicated expression vectors. After 48 h of transfection, total cellular lysates were subjected to immunoprecipitation (IP) and immunoblotting analysis with the indicated Abs. C, Lysates from MCF‐7 or T47D cells were immunoprecipitated with control IgG or an anti‐RNF115 Ab, followed by immunoblotting analysis with the indicated Abs. D, Purified GST or GST‐RNF115 fusion protein was incubated with total cellular lysates of MCF‐7 cells for 3 h, followed by immunoblotting analysis with an anti‐USP9X Ab. GST or GST‐RNF115 protein was stained with Coomassie blue solution as loading controls. E, HEK293T cells were cotransfected with Flag‐RNF115, HA‐Ubiquitin (Ub), negative control siRNA (siNC), or siUSP9Xs (siUSP9X #1‐3) for 48 h. Lysates were subjected to IP assays using Flag M2 affinity gel, followed by immunoblotting analysis with the indicated Abs. F, HEK293T cells were cotransfected with Flag‐RNF115, HA‐Ub, V5‐USP9X, or pDEST51 vector. After 48 h of transfection, lysates were subjected to IP and immunoblotting analysis as described in E
To test whether USP9X regulates RNF115 through deubiquitination, HEK293T cells were co‐transfected with Flag‐RNF115, HA‐ubiquitin, siNC, or siUSP9X. After 48 hours of transfection, cells were treated with 10 μmol/L MG‐132 for 4 hours and subjected to IP assays with Flag M2 affinity gel. Immunoblotting analysis showed that depletion of USP9X increased the ubiquitination of RNF115 protein (Figure 5E). Conversely, overexpression of USP9X decreased RNF115 ubiquitination (Figure 5F). These results indicate that USP9X stabilizes RNF115 by deubiquitination and prevention of its proteasome‐mediated degradation.
3.6. Ubiquitin‐specific protease 9X regulates breast cancer cell proliferation, migration, and invasion partially through RNF115
As both USP9X and RNF115 have been reported to promote breast cancer cell proliferation, migration, and invasion,13, 20, 21, 23 we next determined whether the biological function of USP9X in breast cancer cells depends on RNF115. USP9X‐depleted MCF‐7 and MDA‐MB‐231 cells were infected with lentiviral expression vector encoding either pLVX empty vector or pLVX‐Flag‐RNF115. Immunoblotting analysis showed that shUSP9X significantly reduced USP9X protein levels, accompanied by a decrease in RNF115 protein levels. When infected with lentivirus vector encoding RNF115, RNF115 protein levels were restored, but USP9X protein levels remained unchanged (Figure 6A). Colony formation assays showed that depletion of USP9X impeded the colony formation of MCF‐7 and MDA‐MB‐231 cells, and overexpression of RNF115 rescued colony formation in USP9X‐depleted cells (Figure 6B,C). To investigate the impact of the USP9X‐RNF115 axis on breast cancer cell migration and invasion, highly invasive and metastatic MDA‐MB‐231 cell line was used.23 Wound‐healing assays showed that USP9X knockdown delayed the migration of MDA‐MB‐231 cells, while reintroduction of RNF115 partially restored the motility of USP9X‐depleted cells (Figure 6D,E). Consistently, migration and invasion assays showed that knockdown of USP9X reduced the number of migrated and invaded cells, and RNF115 overexpression rescued USP9X depletion‐induced cell migration and invasion inhibition (Figure 6F,G). These data suggests USP9X promotes breast carcinogenesis partially through stabilizing RNF115.
Figure 6.
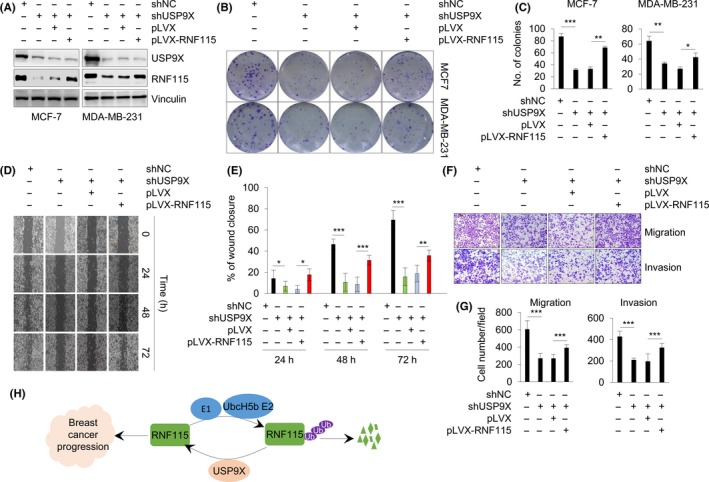
Ubiquitin‐specific protease 9X (USP9X) regulates cell proliferation, migration, and invasion partially through ring finger protein 115 (RNF115). A, MCF‐7 and MDA‐MB‐231 cells stably expressing shUSP9X were infected with lentiviral expression vector encoding empty vector pLVX or pLVX‐RNF115. After antibiotic selection, expression status of USP9X and RNF115 in established cell lines was validated by immunoblotting analysis. B,C, Resultant stable cell lines were used for colony formation assays. Representative images (B) and corresponding quantitative results (C) of survival colonies are shown. D,E, Wound‐healing assays were carried out using the above established stable MDA‐MB‐231 cell lines (A). Representative images (D) and corresponding quantitative results (E) of wound‐healing assay are shown. F,G, Resultant stable MDA‐MB‐231 cell lines were subjected to Transwell migration and Matrigel invasion assays. Representative images of cell migration (F, upper panel) and invasion (F, bottom panel) and corresponding quantitative results (G) are shown. H, The proposed working model. In C, E, and G, *P < .05; **P < .01; ***P < .001
4. DISCUSSION
In this study, we discovered that USP9X is a deubiquitinase of RNF115, which binds to RNF115 and antagonizes its autoubiquitination and subsequent proteasomal degradation, ultimately resulting in the promotion of cancer cell proliferation, migration, and invasion (Figure 6H).
First, USP9X is a novel binding partner but not a ubiquitinated substrate of E3 ubiquitin‐protein ligase RNF115. Although RNF115 possesses ubiquitin ligase activity,8 only a few of its substrates have been identified to date. For example, it was reported that RNF115 targets p21 for ubiquitin‐mediated degradation to promote breast cancer cell proliferation.13 In addition, RNF115 has been shown to stimulate ubiquitination and degradation of c‐Myc and epidermal growth factor receptor in other model systems.32, 33 Interestingly, we found that depletion or overexpression of RNF115 does not affect USP9X protein abundance (Figure 2). In support of our findings, a recent study showed that SMURF1, a member of the Nedd4 family of HECT ubiquitin ligases, interacts with USP9X but this association does not lead to USP9X degradation.23
Second, USP9X dictates the stability of RNF115. Overexpression of RNF115 has been documented in breast cancer cells,8, 10, 15 but the underlying mechanism for its aberrant expression remains elusive. Recently, Kona et al15 reported that ERα enables transcriptional activation of RNF115 in ERα‐positive breast tumors. In addition, just like many other E3s, RNF15 undergoes autoubiquitination and self‐degradation.8, 11 USP9X is a highly conserved member of the USP family of deubiquitinating enzymes and regulates multiple cellular functions through deubiquitinating and stabilizing its substrates.25 In this study, we showed that siRNA‐mediated silencing of USP9X resulted in a downregulation of RNF115 protein without affecting its mRNA expression (Figure 3). Intriguingly, high levels of USP9X correlated with elevated RNF115 expression in breast cancer cell lines and clinical breast tumor tissues. In line with our findings, a recent quantitative proteomics analysis showed that depletion of USP9X is accompanied by a decrease in RNF115 expression in human lung cancer A549 cells.26 Moreover, reduced RNF115 expression in USP9X‐depleted cells was restored following treatment with proteasome inhibitor MG‐132. The CHX chase assays further confirmed that USP9X depletion remarkably shortened the half‐life of RNF115 (Figure 4). These results suggest that USP9X enhances RNF115 protein stability. Biochemical evidence further indicated that depletion of USP9X increased, whereas overexpression of USP9X decreased, RNF115 ubiquitination (Figure 5). These results suggest that USP9X as a deubiquitinase stabilizes RNF115 through inhibiting its autoubiquitination and proteasomal degradation. In support of our findings, USP9X has been shown to regulate the stability of several E3 ubiquitin ligases, such as Itch,34 MARCH7,35 and SMURF1,23 through antagonizing their autoubiquitination and self‐inflicted degradation.
Third, the USP9X‐RNF115 axis contributes to the malignant phenotype of breast cancer cells. USP9X has been implicated as both an oncogene and a tumor suppressor, depending on the type and stage of cancer.19, 36, 37 For example, elevated expression of USP9X has been documented in several types of human cancers, including non‐small‐cell lung cancer, esophageal squamous cell carcinoma, B‐cell lymphoma, cervical carcinoma, and melanoma, and its high expression is closely linked with tumor growth, invasion, metastasis, and chemoresistance.26, 36, 38, 39, 40, 41, 42, 43 Conversely, USP9X has tumor suppressor functions in pancreatic ductal adenocarcinoma37, 44, 45 and colorectal cancer.46 Interestingly, 2 recent studies showed that USP9X stabilizes large tumor suppressor kinase 2, an upstream component of the Hippo pathway, to negatively regulate YAP and its target genes to suppress tumor growth in pancreatic44, 45 and breast44 cancer. In contrast, another study reported that USP9X deubiquitinates and stabilizes YAP1, a major downstream effector of the Hippo pathway, thus promoting breast cancer cell survival and chemoresistance.21 Moreover, several other studies have shown that USP9X is overexpressed in breast tumors and promotes breast tumorigenesis, progression, and chemoresistance through distinct mechanisms.20, 21, 23, 25, 47 For example, it was shown that depletion of USP9X hinders motility of metastatic MDA‐MB‐231 breast cancer cells,23 which could be an indication for the role of this protein in invasion and metastasis of breast cancer cells. Moreover, depletion or inhibition of USP9X enhances the sensitivity of breast cancer cells to DNA‐damaging chemotherapeutic drugs, such as mitomycin C, cisplatin, and etoposide.21, 25 In addition, USP9X contributes to obesity‐induced breast cancer metastasis.47 Here, we showed that shRNA‐mediated knockdown of USP9X resulted in a decrease in breast cancer cell proliferation, migration, and invasion. However, re‐expression of RNF115 in USP9X‐depleted cells partially rescued cell proliferation, migration, and invasion induced by USP9X knockdown (Figure 6). These results highlight the functional importance of the USP9X‐RNF115 axis in the development and/or progression of breast cancer. Clearly, future studies are warranted to investigate whether RNF115 overexpression could rescue breast cancer progression induced by USP9X knockdown in vivo.
In conclusion, findings presented here establish USP9X as a positive regulator of RNF115 stability and the USP9X‐RNF115 signaling axis as an important regulatory mechanism of breast cancer. Given that the inhibitors of USP9X and RNF115 have been identified,25, 48, 49 these findings provide a rationale for potential therapeutic interventions in the treatment of breast cancer.
CONFLICTS OF INTEREST
The authors declare no potential conflicts of interest.
Supporting information
ACKNOWLEDGEMENTS
The authors thank Professors Stephen A. Wood (Eskitis Institute for Drug Discovery, Griffith University, Australia), Hu Zhou (Shanghai Institute of Materia Medica, Chinese Academy of Sciences, China), and Yanhui Xu (Fudan University, China) for kindly providing V5‐USP9X, shUSP9X, and pGEX‐6p‐1 expression vectors, respectively. This work is supported, in whole or in part, by the National Natural Science Foundation of China (Nos. 81572584 and 81772805), the Program for Professor of Special Appointment (Eastern Scholar) at Shanghai Institutions of Higher Learning (No. 2013‐06), the Science and Technology Innovation Action Plan of Shanghai Municipal Science and Technology Commission (No. 16JC1405400), and a start‐up fund for new investigators from Fudan University (to DQL).
Lu Q, Lu D, Shao Z‐M, Li D‐Q. Deubiquitinase ubiquitin‐specific protease 9X regulates the stability and function of E3 ubiquitin ligase ring finger protein 115 in breast cancer cells. Cancer Sci. 2019;110:1268–1278. 10.1111/cas.13953
Funding information
National Natural Science Foundation of China, Grant/Award Numbers 81572584, 81772805; Program for Professor of Special Appointment (Eastern Scholar) at Shanghai Institutions of Higher Learning, Grant/Award Number 2013‐06; Science and Technology Innovation Action Plan of Shanghai Municipal Science and Technology Commission, Grant/Award Number 16JC1405400; Fudan University.
Contributor Information
Zhi‐Ming Shao, Email: zhimingshao@fudan.edu.cn.
Da‐Qiang Li, Email: daqiangli1974@fudan.edu.cn.
REFERENCES
- 1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394‐424. [DOI] [PubMed] [Google Scholar]
- 2. Jin C, Yang YA, Anver MR, Morris N, Wang X, Zhang YE. Smad ubiquitination regulatory factor 2 promotes metastasis of breast cancer cells by enhancing migration and invasiveness. Cancer Res. 2009;69:735‐740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Deng S, Zhou H, Xiong R, et al. Over‐expression of genes and proteins of ubiquitin specific peptidases (USPs) and proteasome subunits (PSs) in breast cancer tissue observed by the methods of RFDD‐PCR and proteomics. Breast Cancer Res Treat. 2007;104:21‐30. [DOI] [PubMed] [Google Scholar]
- 4. Buetow L, Huang DT. Structural insights into the catalysis and regulation of E3 ubiquitin ligases. Nat Rev Mol Cell Biol. 2016;17:626‐642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ravid T, Hochstrasser M. Diversity of degradation signals in the ubiquitin‐ proteasome system. Nat Rev Mol Cell Biol. 2008;9:679‐690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lipkowitz S, Weissman AM. RINGs of good and evil: RING finger ubiquitin ligases at the crossroads of tumour suppression and oncogenesis. Nat Rev Cancer. 2011;11:629‐643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399‐434. [DOI] [PubMed] [Google Scholar]
- 8. Burger AM, Gao Y, Amemiya Y, et al. A novel RING‐type ubiquitin ligase breast cancer‐associated gene 2 correlates with outcome in invasive breast cancer. Cancer Res. 2005;65:10401‐10412. [DOI] [PubMed] [Google Scholar]
- 9. Burger AM, Zhang X, Li H, et al. Down‐regulation of T1A12/mac25, a novel insulin‐like growth factor binding protein related gene, is associated with disease progression in breast carcinomas. Oncogene. 1998;16:2459‐2467. [DOI] [PubMed] [Google Scholar]
- 10. Burger A, Amemiya Y, Kitching R, Seth AK. Novel RING E3 ubiquitin ligases in breast cancer. Neoplasia. 2006;8:689‐695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Amemiya Y, Azmi P, Seth A. Autoubiquitination of BCA2 RING E3 ligase regulates its own stability and affects cell migration. Mol Cancer Res. 2008;6:1385‐1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bacopulos S, Amemiya Y, Yang W, et al. Effects of partner proteins on BCA2 RING ligase activity. BMC Cancer. 2012;12:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang Z, Nie Z, Chen W, et al. RNF115/BCA2 E3 ubiquitin ligase promotes breast cancer cell proliferation through targeting p21Waf1/Cip1 for ubiquitin‐mediated degradation. Neoplasia. 2013;15:1028‐1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wymant JM, Hiscox S, Westwell AD, Urbe S, Clague MJ, Jones AT. The role of BCA2 in the endocytic trafficking of EGFR and significance as a prognostic biomarker in cancer. J Cancer. 2016;7:2388‐2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kona FR, Stark K, Bisoski L, Buac D, Cui Q, Dou QP. Transcriptional activation of breast cancer‐associated gene 2 by estrogen receptor. Breast Cancer Res Treat. 2012;135:495‐503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Komander D, Clague MJ, Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10:550‐563. [DOI] [PubMed] [Google Scholar]
- 17. Harrigan JA, Jacq X, Martin NM, Jackson SP. Deubiquitylating enzymes and drug discovery: emerging opportunities. Nat Rev Drug Discov. 2018;17:57‐78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nijman SM, Luna‐Vargas MP, Velds A, et al. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773‐786. [DOI] [PubMed] [Google Scholar]
- 19. Murtaza M, Jolly LA, Gecz J, Wood SA. La FAM fatale: USP9X in development and disease. Cell Mol Life Sci. 2015;72:2075‐2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li X, Song N, Liu L, et al. USP9X regulates centrosome duplication and promotes breast carcinogenesis. Nat Commun. 2017;8:14866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li L, Liu T, Li Y, et al. The deubiquitinase USP9X promotes tumor cell survival and confers chemoresistance through YAP1 stabilization. Oncogene. 2018;37:2422‐2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zheng X, Zhai B, Koivunen P, et al. Prolyl hydroxylation by EglN2 destabilizes FOXO3a by blocking its interaction with the USP9x deubiquitinase. Genes Dev. 2014;28:1429‐1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xie Y, Avello M, Schirle M, et al. Deubiquitinase FAM/USP9X interacts with the E3 ubiquitin ligase SMURF1 protein and protects it from ligase activity‐dependent self‐degradation. J Biol Chem. 2013;288:2976‐2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Izrailit J, Jaiswal A, Zheng W, Moran MF, Reedijk M. Cellular stress induces TRB3/USP9x‐dependent Notch activation in cancer. Oncogene. 2017;36:1048‐1057. [DOI] [PubMed] [Google Scholar]
- 25. Fu P, Du F, Liu Y, et al. WP1130 increases cisplatin sensitivity through inhibition of usp9x in estrogen receptor‐negative breast cancer cells. Am J Transl Res. 2017;9:1783‐1791. [PMC free article] [PubMed] [Google Scholar]
- 26. Chen X, Yu C, Gao J, et al. A novel USP9X substrate TTK contributes to tumorigenesis in non‐small‐cell lung cancer. Theranostics. 2018;8:2348‐2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sun R, Xie HY, Qian JX, et al. FBXO22 possesses both protumorigenic and antimetastatic roles in breast cancer progression. Cancer Res. 2018;78:5274‐5286. [DOI] [PubMed] [Google Scholar]
- 28. Zhang Y, Liao XH, Xie HY, Shao ZM, Li DQ. RBR‐type E3 ubiquitin ligase RNF144A targets PARP1 for ubiquitin‐dependent degradation and regulates PARP inhibitor sensitivity in breast cancer cells. Oncotarget. 2017;8:94505‐94518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liao XH, Zhang Y, Dong WJ, Shao ZM, Li DQ. Chromatin remodeling protein MORC2 promotes breast cancer invasion and metastasis through a PRD domain‐mediated interaction with CTNND1. Oncotarget. 2017;8:97941‐97954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang FL, Cao JL, Xie HY, et al. Cancer‐associated MORC2‐mutant M276I regulates an hnRNPM‐mediated CD44 splicing switch to promote invasion and metastasis in triple‐negative breast cancer. Cancer Res. 2018;78:5780‐5792. [DOI] [PubMed] [Google Scholar]
- 31. Brooks CL, Gu W. p53 ubiquitination: Mdm2 and beyond. Mol Cell. 2006;21:307‐315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Narita R, Kitaura H, Torii A, et al. Rabring7 degrades c‐Myc through complex formation with MM‐1. PLoS One. 2012;7:e41891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Smith CJ, Berry DM, McGlade CJ. The E3 ubiquitin ligases RNF126 and Rabring7 regulate endosomal sorting of the epidermal growth factor receptor. J Cell Sci. 2013;126:1366‐1380. [DOI] [PubMed] [Google Scholar]
- 34. Mouchantaf R, Azakir BA, McPherson PS, Millard SM, Wood SA, Angers A. The ubiquitin ligase itch is auto‐ubiquitylated in vivo and in vitro but is protected from degradation by interacting with the deubiquitylating enzyme FAM/USP9X. J Biol Chem. 2006;281:38738‐38747. [DOI] [PubMed] [Google Scholar]
- 35. Nathan JA, Sengupta S, Wood SA, et al. The ubiquitin E3 ligase MARCH7 is differentially regulated by the deubiquitylating enzymes USP7 and USP9X. Traffic. 2008;9:1130‐1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schwickart M, Huang X, Lill JR, et al. Deubiquitinase USP9X stabilizes MCL1 and promotes tumour cell survival. Nature. 2010;463:103‐107. [DOI] [PubMed] [Google Scholar]
- 37. Perez‐Mancera PA, Rust AG, van der Weyden L, et al. The deubiquitinase USP9X suppresses pancreatic ductal adenocarcinoma. Nature. 2012;486:266‐270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Guo H, Zhang X, Chen Q, Bao Y, Dong C, Wang X. miR‐132 suppresses the migration and invasion of lung cancer cells by blocking USP9X‐induced epithelial‐mesenchymal transition. Am J Transl Res. 2018;10:224‐234. [PMC free article] [PubMed] [Google Scholar]
- 39. Wang Y, Liu Y, Yang B, et al. Elevated expression of USP9X correlates with poor prognosis in human non‐small cell lung cancer. J Thorac Dis. 2015;7:672‐679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Engel K, Rudelius M, Slawska J, et al. USP9X stabilizes XIAP to regulate mitotic cell death and chemoresistance in aggressive B‐cell lymphoma. EMBO Mol Med. 2016;8:851‐862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Peng J, Hu Q, Liu W, et al. USP9X expression correlates with tumor progression and poor prognosis in esophageal squamous cell carcinoma. Diagn Pathol. 2013;8:177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rolen U, Kobzeva V, Gasparjan N, et al. Activity profiling of deubiquitinating enzymes in cervical carcinoma biopsies and cell lines. Mol Carcinog. 2006;45:260‐269. [DOI] [PubMed] [Google Scholar]
- 43. Potu H, Peterson LF, Kandarpa M, et al. Usp9x regulates Ets‐1 ubiquitination and stability to control NRAS expression and tumorigenicity in melanoma. Nat Commun. 2017;8:14449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Toloczko A, Guo F, Yuen HF, et al. Deubiquitinating enzyme USP9X suppresses tumor growth via LATS kinase and core components of the hippo pathway. Cancer Res. 2017;77:4921‐4933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhu C, Ji X, Zhang H, et al. Deubiquitylase USP9X suppresses tumorigenesis by stabilizing large tumor suppressor kinase 2 (LATS2) in the Hippo pathway. J Biol Chem. 2018;293:1178‐1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Khan OM, Carvalho J, Spencer‐Dene B, et al. The deubiquitinase USP9X regulates FBW7 stability and suppresses colorectal cancer. J Clin Invest. 2018;128:1326‐1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wu Y, Yu X, Yi X, et al. Aberrant phosphorylation of SMAD4 Thr277‐mediated USP9x‐SMAD4 interaction by free fatty acids promotes breast cancer metastasis. Cancer Res. 2017;77:1383‐1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Brahemi G, Kona FR, Fiasella A, et al. Exploring the structural requirements for inhibition of the ubiquitin E3 ligase breast cancer associated protein 2 (BCA2) as a treatment for breast cancer. J Med Chem. 2010;53:2757‐2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kapuria V, Peterson LF, Fang D, Bornmann WG, Talpaz M, Donato NJ. Deubiquitinase inhibition by small‐molecule WP1130 triggers aggresome formation and tumor cell apoptosis. Cancer Res. 2010;70:9265‐9276. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


