Abstract
MicroRNAs (miRs) are widely involved in regulating tumor development and progression. miR-758-3p has been reported to suppress the progression of various cancer types, including hepatocellular carcinoma. However, whether miR-758-3p has a role in bladder cancer (BC) has not been previously reported, and was thus investigated in the present study. It was revealed that miR-758-3p was downregulated in BC tissues and cell lines. Transfection with miR-758-3p mimics suppressed the proliferation, migration and invasion of BC cells, and inhibition of miR-758-3p had the opposite effect. In terms of the underlying mechanisms, a luciferase reporter assay revealed that Notch receptor 2 (NOTCH2) is a direct target gene of miR-758-3p in BC cells. Transfection with miR-758-3p mimics decreased the mRNA and protein levels of NOTCH2. Furthermore, an inverse correlation between miR-758-3p and NOTCH2 levels was identified. Finally, overexpression of NOTCH2 significantly rescued the proliferation, migration and invasion of BC cells transfected with miR-758-3p mimics. Taken together, the present study indicated that miR-758-3p suppresses BC cell proliferation, migration and invasion by targeting NOTCH2.
Keywords: microRNA-758-3p, bladder cancer, proliferation, invasion, notch ligand 2
Introduction
Bladder cancer (BC) is the fourth most prevalent solid tumor type in males and the seventh most prevalent in females worldwide (1). It is accountable for ~3% of cancer-associated deaths (2). Although certain therapeutic methods, including radiotherapy, surgery and chemotherapy, have been developed for BC treatment, the recurrence rate remains high (2,3) and the prognosis of BC patients is poor (4). Therefore, it is urgently required to explore the regulatory mechanisms underlying the occurrence of BC, which will contribute to the identification of therapeutic targets and the development of novel treatments.
MicroRNAs (miRNAs/miRs) are a class of non-coding RNAs and post-transcriptionally regulate gene expression by recognizing the complementary sequence in the 3′ untranslated region (3′-UTR) of their target mRNAs (5,6). miRNAs exert vital functions in a broad variety of biological processes, including development, cell proliferation and apoptosis (7). Dysregulated expression of miRNAs is usually observed in almost all cancer types, including colorectal (8), liver (9) and bladder cancer (4). Increasing evidence indicates that certain miRNAs may serve as oncogenes or tumor suppressors to regulate BC development and progression (4,10). For instance, Yuan et al (11) reported that miR-124-3p inhibits the growth and metastasis of BC by degrading the mRNA of aurora kinase A. Furthermore, Feng et al (12) indicated that miR-556-3p contributes to BC cell proliferation and invasiveness through inhibiting DAB2 interacting protein expression. Another previous study indicated that miR-758-3p inhibits hepatocellular carcinoma progression (13). miR-758-3p is also implicated in cervical cancer (14). However, the biological functions of miR-758-3p in BC have not been previously reported. Due to the significance of miR-758-3p in the abovementioned cancer types, the present study sought to investigate the function and potential mechanisms of miR-758-3p in BC.
In the present study, it was demonstrated that miR-758-3p expression was downregulated in BC tissues and cell lines. Furthermore, transfection with miR-758-3p mimics markedly repressed the proliferation, migration and invasion of BC cells. It was also revealed that Notch receptor 2 (NOTCH2) was a direct target of miR-758-3p. In summary, the present study illustrated that miR-758-3p inhibits BC progression via targeting NOTCH2, suggesting that miR-758-3p may be a promising therapeutic target for BC treatment.
Materials and methods
Human tissues
A total of 33 BC tissues (age range, 61±8.1 years; female, n=4; male, n=29) and matched normal tissues (at least 3 cm away from the tumor border and with no microscopic evidence of tumor cells) were collected from patients diagnosed with BC at the Xiangyang Central Hospital (Xiangyang, China) from January 2014 to September 2016. All patients provided written informed consent. Samples from patients who received radiotherapy or chemotherapy prior to surgery were excluded. The tissues were stored in liquid nitrogen at −80°C until use. The clinicopathological characteristics of the 33 patients with BC were also recorded. The present study was approved by the Ethics Committee of Xiangyang Central Hospital (Xiangyang, China).
Cell culture and transfection
The J82, UMUC3, T24 and 5637 BC cell lines as well as the SV-HUC-1 normal bladder cell line were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). Cells were maintained in RPMI-1640 medium (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) containing 10% fetal bovine serum (FBS; Invitrogen; Thermo Fisher Scientific, Inc.) and 1% penicillin/streptomycin.
For cell transfection, the miR-758-3p mimics (5′-UUUGUGACCUGGUCCACUAACC-3′), miR-758-3p inhibitor (5′-GGUUAGUGGACCAGGUCACAAA-3′), inhibitor control (5-GCGUAACUAAUACAUCGGAUUCGU-3) and mimic control (5′-ACAUCUGCGUAAGAUUCGAGUCUA-3′) were purchased from GenePharma (Shanghai, China). Cells were transfected with miR-758-3p mimics or controls using Lipofectamine 2000™ (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. For NOTCH2 overexpression, the sequence encoding the NOTCH2 intracellular segment was inserted into the pcDNA3 vector to generate pcDNA3-NOTCH2. Then pcDNA3-NOTCH2 vector (1 µg) was transfected into BCa cell lines using Lipofectamine 2000™ (Invitrogen; Thermo Fisher Scientific, Inc.). After 48 h, the overexpression efficiency was evaluated and gene expression was determined using reverse transcription-quantitative polymerase chain reaction.
RT-qPCR
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) was used to extract total RNA from cells. Total RNA (1 µg) was reverse transcribed into cDNA using the PrimeScript™ RT reagent kit (Takara Biotechnology Co., Ltd., Dalian, China), according to the manufacturer's protocol. qPCR was subsequently performed using the SYBR Green I Supermix (Takara Biotechnology Co., Ltd.), according to the manufacturer's protocol using an iCycler IQ multicolor Detection System (Bio-Rad Laboratories, Hercules, CA, USA). The following thermocycling conditions were used for the qPCR: Initial denaturation at 95°C for 10 min; 40 cycles of 95°C for 15 sec and 60°C for 1 min. The primer pairs used were as follows: miR-758-3p forward, 5′-ACACTCCAGCTGGGTTTGTGACCTGGTCCA-3′ and reverse, 5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGGGTTAGTG-3′; U6 forward, 5′-CTCGCTTCGGCAGCACA-3′ and reverse, 5′-AACGCTTCACGAATTTGCGT-3′; NOTCH2 forward, 5′-CAAGGAACCTGCTTTGATGACA-3′ and reverse, 5′-GGGGAACAGGGAGCCAATAC-3′; and GAPDH forward, 5′-GCACCGTCAAGGCTGAGAAC-3′ and reverse, 5′-TGGTGAAGACGCCAGTGGA-3′. The mRNA levels were quantified using the 2−∆∆Cq method and U6 was used as a normalization control (15).
Cell Counting Kit (CCK)-8 proliferation assay
Cell proliferation was measured using a CCK-8 proliferation assay (Dojindo Molecular Technologies, Inc., Kumamoto, Japan). Cells were seeded into 96-well plates at a density of 2×103 cells/well) and cultured for the indicated durations. Following the addition of 10 µl CCK-8 reagent, the plates were incubated for 1 h at 37°C. Subsequently, the absorbance at 450 nm was determined using a microplate reader (Berthold Technologies GmbH, Bad Wildbad, Germany).
Colony formation assay
Cells were seeded into 6-well plates at 1×103 cells/well and cultured for 12 days. The colonies were fixed using methanol for 15 min at room temperature, stained using 0.5% crystal violet for 20 min at room temperature. The total number of visible colonies was examined under an optical light microscope (magnification, ×40; Olympus Corporation, Tokyo, Japan).
Migration and invasion assays
Transwell chambers (BD Biosciences, Franklin Lakes, NJ, USA) were used for Transwell migration and invasion assays. Cells (5×104) in 100 µl serum-free medium were seeded into the upper chamber [pre-coated with Matrigel® (1:6 dilution; BD Biosciences) for the invasion assay]. The lower chamber was filled with 600 µl medium containing 10% FBS. Following 24-h incubation, cells that had migrated to the lower side of the membrane were fixed with polyoxymethylene at room temperature for 30 min and stained with 0.5% crystal violet at room temperature for 30 min. Images of the cells were captured under an optical microscope.
Western blot analysis
Total protein was extracted from cells using radioimmunoprecipitation assay buffer (Thermo Fisher Scientific, Inc.). Total protein was quantified using a bicinchoninic acid assay and 40 µg protein/lane was separated via SDS-PAGE on a 12% gel. The separated proteins were transferred onto polyvinylidene fluoride membranes (Thermo Fisher Scientific, Inc.) and blocked for 3 h at room temperature with 5% non-fat milk in PBS (Thermo Fisher Scientific, Inc.) containing 0.1% Tween-20 (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). The membranes were incubated with the following primary antibodies: Anti-NOTCH2 (1:1,500; cat. no. 5732) and mouse anti-GAPDH (1:5,000; cat. no. 5174; both Cell Signaling Technology, Danvers, MA, USA) overnight at 4°C. Subsequently, the membranes were incubated with horseradish peroxidase-conjugated secondary antibodies (1:5,000; cat. no. ab7090; Abcam, Cambridge, MA, USA) for 1 h at room temperature. Protein bands were visualized using the Pierce™ ECL Western Blotting Substrate (Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. Protein expression was quantified using ImageJ software (version 1.41; National Institutes of Health, Bethesda, MD, USA).
Luciferase assay
The potential binding site of NOTCH2 3′-UTR for miR-758-3p was predicted using the TargetScan7 tool (http://www.targetscan.org/vert_71/). The sequences containing the wild-type (WT) or site-mutated (Mut) region of NOTCH2 were synthesized by Sangon (Shanghai, China) and inserted into the pGL3 vector (Promega Corporation, Madison, WI, USA). For the luciferase reporter assay, miR-758-3p or NC mimics and the respective reporter plasmids were transfected into BC cells using Lipofectamine 2000™ (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. After 24 h, the Renilla and firefly luciferase activity was determined using the Dual-Luciferase Reporter Assay System (Promega Corp.) according to the manufacturer's protocols and a luminometer (Infinite 200 PRO NanoQuant; Tecan Group Ltd., Männedorf, Switzerland).
Statistical analysis
Statistical analysis was performed using SPSS 20.0 (IBM Corp., Armonk, NY, USA) or GraphPad Prism version 5 (GraphPad Software, Inc., LA Jolla, CA, USA). The assays were performed as three independent replicates. Values are expressed as the mean ± standard deviation. P-values were calculated using Student's t-test or one-way analysis of variance followed by Tukey's post hoc test. The association between miR-758-3p expression and the clinicopathological characteristics of patients with BC was analyzed using the Chi-square test. Spearman's rank correlation analysis was performed to analyze the correlation between miR-758-3p and NOTCH2 expression levels. P<0.05 was considered to indicate statistical significance.
Results
miR-758-3p is downregulated in BC tissues and cell lines
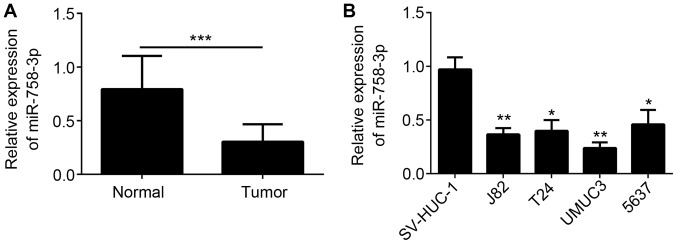
To investigate the function of miR-758-3p in BC, its expression was analyzed in tumor tissues and adjacent normal tissues of 33 BC patients. As presented Fig. 1A, miR-758-3p was downregulated in BC tissues compared with that in the matched normal tissues. In addition, miR-758-3p expression was downregulated in BC cell lines compared with that in the SV-HUC-1 normal bladder cell line (Fig. 1B). The association between miR-758-3p expression and the clinicopathological characteristics of patients with BC was examined (Table I).
Figure 1.
miR-758-3p is downregulated in BC tissues and cell lines. (A) The expression of miR-758-3p in 33 BC and matched normal bladder tissues was measured by RT-qPCR. (B) RT-qPCR analysis indicated that miR-758-3p was downregulated in BC cell lines (J82, T24, UMUC3 and 5637 cells) compared with that in SV-HUC-1 cells. *P<0.05, **P<0.01 and ***P<0.001 as indicated or vs. SV-HUC-1. BC, bladder cancer; miR, microRNA; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
Table I.
Association between miR-758-3p expression and clinicopathological characteristics of patients with bladder cancer (n=33).
| miR-758-3p expression | |||
|---|---|---|---|
| Clinicopathological characteristic | Low (n=18) | High (n=15) | P-value |
| Age (years) | 0.169 | ||
| <60 | 5 | 8 | |
| ≥60 | 13 | 7 | |
| Sex | 0.607 | ||
| Female | 3 | 1 | |
| Male | 15 | 14 | |
| TNM stage | 0.038 | ||
| I/II | 4 | 9 | |
| III/IV | 14 | 6 | |
| Lymph node metastasis | 0.034 | ||
| Yes | 9 | 13 | |
| No | 9 | 2 | |
miR, microRNA; TMN, tumor, node, metastasis.
miR-758-3p suppresses the proliferation, migration and invasion of BC cells
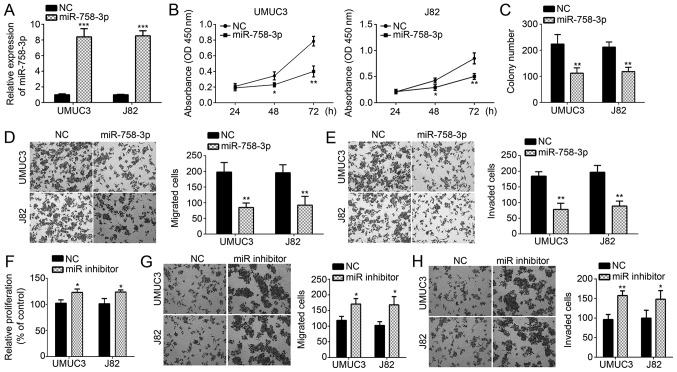
To explore the role of miR-758-3p in BC, miR-758-3p mimics were transfected into UMUC3 and J82 cells. RT-qPCR analysis confirmed that miR-758-3p levels were markedly increased in UMUC3 and J82 cells after transfection (Fig. 2A). CCK-8 and colony formation assays were then performed to evaluate the cell proliferation ability. The results indicated that miR-758-3p overexpression inhibited the proliferation and colony formation of UMUC3 and J82 cells (Fig. 2B and C). Furthermore, as indicated by the Transwell assay, transfection of miR-758-3p mimics into UMUC3 and J82 cells markedly inhibited migration and invasion (Fig. 2D and E). In addition, to further validate the function of miR-758-3p, UMUC3 and J82 cells were transduced with miR-758-3p inhibitor. Through CCK-8 and Transwell assays, it was revealed that miR-758-3p inhibition significantly promoted proliferation, migration and invasion (Fig. 2F-H). Taken together, miR-758-3p suppresses BC proliferation and progression.
Figure 2.
Overexpression of miR-758-3p suppresses the proliferation, migration and invasion of BC cells. (A) Reverse transcription-quantitative polymerase chain reaction analysis of miR-758-3p expression in UMUC3 and J82 cells transfected with miR-758-3p mimics or NC. (B) A CCK-8 assay was used to assess the proliferation ability and (C) a clonogenic assay was used to assess the colony formation ability of UMUC3 and J82 cells transfected with miR-758-3p mimics or NC. (D and E) Transwell assays were used to determine the migration and invasion of UMUC3 and J82 cells transfected with miR-758-3p mimics or NC (magnification, ×100). (F) A CCK-8 assay was used to assess the proliferation ability of UMUC3 and J82 cells transfected with miR-758-3p inhibitor or NC. (G and H) Transwell assays were used to determine the migration and invasion of UMUC3 and J82 cells transfected with miR-758-3p inhibitor or NC (magnification, ×100). *P<0.05, **P<0.01 and ***P<0.001 vs. NC. BC, bladder cancer; miR, microRNA; OD, optical density; NC, negative control; CCK-8, Cell Counting Kit-8.
NOTCH2 is a target of miR-758-3p in BC cells
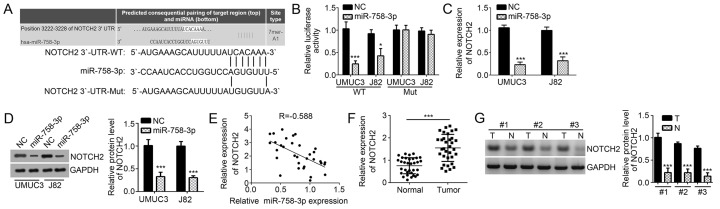
To further determine the mechanisms of miR-758-3p in BC, the downstream target genes of miR-758-3p were searched with TargetScan software. Among all candidates of predicted potential targets of miR-758-3p, NOTCH2 ranked high and was previously reported to promote BC progression (16). Thus, NOTCH2 was selected for further investigation. There was a complementary sequence of miR-758-3p in the 3′-UTR region of NOTCH2 mRNA (Fig. 3A). To confirm the direct binding interaction in vitro, WT and Mut luciferase reporter plasmids were constructed and used in a luciferase reporter assay. The results demonstrated that miR-758-3p mimics inhibited the luciferase intensity of the NOTCH2-WT reporter plasmid in UMUC3 and J82 cells, while mutation of the complementary binding site abrogated this effect (Fig. 3B). In a further experiment, miR-758-3p mimics markedly decreased NOTCH2 expression in UMUC3 and J82 cells (Fig. 3C and D). In addition, the expression of NOTCH2 was examined in BC tissues, revealing an inverse association between the expression of miR-758-3p and NOTCH2 (Fig. 3E). Furthermore, NOTCH2 levels were determined in BC tissues by RT-qPCR and western blot analysis, demonstrating that NOTCH2 expression was significantly upregulated in BC tissues compared with that in adjacent normal tissues (Fig. 3F and G).
Figure 3.
NOTCH2 is a target of miR-758-3p in BC cells. (A) The predicted complementary site in the 3′-UTR of NOTCH2 with miR-758-3p. (B) A luciferase reporter assay in UMUC3 and J82 cells indicated that miR-758-3p mimics inhibited the luciferase activity of the reporter plasmid carrying the WT fragment from the 3′-UTR of NOTCH2 but not the mutant 3′-UTR fragment. (C and D) miR-758-3p mimics reduced the mRNA and protein levels of NOTCH2 in UMUC3 and J82 cells, as indicated by RT-qPCR and western blot analysis, respectively. (E) The correlation between NOTCH2 mRNA and miR-758-3p expression in BC tissues from 33 cases was determined by Spearman's correlation analysis. (F and G) mRNA and protein levels of NOTCH2 in BC tissues and adjacent normal tissues were measured by RT-qPCR and western blot analysis, respectively. *P<0.05; ***P<0.001 vs. control group. UTR, untranslated region; WT, wild-type; miR, microRNA; Mut, mutated; NC, negative control; Hsa, Homo sapiens; T, tumor tissue; N, normal tissue; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; BC, bladder cancer; NOTCH2, Notch receptor 2.
miR-758-3p suppresses BC cell proliferation, migration and invasion through targeting NOTCH2
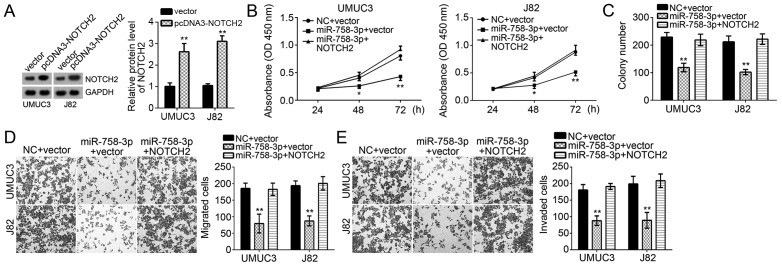
To determine whether suppression of cell proliferation, migration and invasion by miR-758-3p relies on NOTCH2, UMUC3 and J82 cells transduced with miR-758-3p mimics were subjected to ectopic overexpression of NOTCH2. Western blot analysis confirmed that the levels of NOTCH2, which were decreased by miR-758-3p mimics, were restored by co-transfection with NOTCH2 overexpression vector (Fig. 4A). Functional experiments indicated that restoration of NOTCH2 promoted the proliferation and colony formation ability of UMUC3 and J82 cells transfected with miR-758-3p mimics (Fig. 4B and C). Furthermore, overexpression of NOTCH2 also rescued the migration and invasion of UMUC3 and J82 cells transfected with miR-758-3p mimics (Fig. 4D and E). In conclusion, miR-758-3p inhibited BC cell proliferation, migration and invasion at least in part through targeting the mRNA of NOTCH2 and promoting its degradation.
Figure 4.
miR-758-3p suppresses bladder cancer cell proliferation, migration and invasion through targeting NOTCH2. (A) Western blot analysis was used to measure the protein levels of NOTCH2 in UMUC3 and J82 cells. (B) A Cell Counting Kit-8 assay was performed to assess the proliferation and (C) a clonogenic assay was used to determine the colony formation ability of UMUC3 and J82 cells transfected with miR-758-3p mimics as well as NOTCH2-overexpressing plasmid or control. (D and E) Transwell assays were utilized to evaluate the migration and invasion of UMUC3 and J82 cells transfected with miR-758-3p mimics as well as NOTCH2-overexpressing plasmid or control (magnification, ×100). *P<0.05; **P<0.01 vs. control group. NOTCH2, Notch receptor 2; miR, microRNA; NC, negative control; OD, optical density.
Discussion
BC has become the most common malignancy of the urinary tract, originating from bladder mucosa, worldwide (17). Each year, there are large numbers of BC cases and increasing BC-related mortality rates (17). Thus, it is vital to reveal the underlying mechanisms of the genesis and progression of BC and develop effective therapeutic methods. Accumulating evidence indicates that miRNAs are potential biomarkers for diagnosis and prognosis in numerous cancer types (18,19). For instance, miR-122 and miR-224 have been reported to serve as biomarkers for early diagnosis of hepatocellular carcinoma (20). The present study demonstrated that miR-758-3p has a tumor suppressor function in BC and therefore miR-758-3p may be a promising therapeutic target.
In the past decades, miRNAs have attracted wide attention, and a vast number of studies have demonstrated their essential and general functions in a diversity of biological processes, including cell migration, proliferation and invasion (21,22). For instance, Ding et al (23) reported that miR-367 suppresses clear-cell renal cell cancer progression. Wu et al (24) reported that miR-21 contributes to colorectal cancer progression via targeting phosphatase and tensin homolog. Another previous study indicated that miR-758-3p suppresses liver cancer development by suppressing MDM2 and mammalian target of rapamycin (13). In cervical cancer patients, the levels of miR-758 were reported to be decreased in the tumor tissues, blood and cervical exfoliated cells, and it was indicated that miR-758 may regulate the infiltration and invasion of cervical cancer by targeting matrix extracellular phosphoglycoprotein (14). These studies suggest a tumor suppressor role for miR-758-3p. However, the effect of miR-758-3p in BC has remained elusive. The present results indicated that miR-758-3p was downregulated in BC tissues compared with that in matched normal tissues. Furthermore, CCK-8, colony formation and Transwell assays suggested that transfection with miR-758-3p mimics markedly inhibited the malignant behavior of BC cells. In addition, NOTCH2 was identified as a direct target of miR-758-3p in BC cells.
NOTCH2, a member of the NOTCH family, has a role in developmental processes. NOTHC2 signaling is evolutionarily conserved and is involved in cell fate decisions (25). Increasing evidence has indicated that NOTCH signaling is involved in the development and progression of numerous human cancer types, including BC (16,26). Furthermore, a recent review also indicated that NOTCH2 acts as an oncogene that promotes cell proliferation and metastasis through epithelial-to-mesenchymal transition, cell cycle progression and maintenance of stem cells in BC (27). In the present study, NOTCH2 was identified to be downregulated by miR-758-3p in BC cells. Furthermore, the expression of NOTCH2 was negatively correlated with that of miR-758-3p in BC tissues. Notably, restoration of NOTCH2 reversed the effects of miR-758-3p mimics on BC cell proliferation, migration and invasion.
In conclusion, the present study indicated a tumor suppressive role of miR-758-3p in BC, as indicated by its inhibitory effect on cell proliferation, migration and invasion through repression of NOTCH2 expression.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
XW, BC and XS designed the study, analyzed and interpreted the results and prepared the manuscript. HS, JZ, FZ and JC performed the experiments. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The protocol used in the present study was approved by the Institutional Ethics Committee of Xiangyang Central Hospital (Xiangyang, China). All patients provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Tan M, Mu X, Liu Z, Tao L, Wang J, Ge J, Qiu J. microRNA-495 promotes bladder cancer cell growth and invasion by targeting phosphatase and tensin homolog. Biochem Biophys Res Commun. 2017;483:867–873. doi: 10.1016/j.bbrc.2017.01.019. [DOI] [PubMed] [Google Scholar]
- 2.Jiang Z, Zhang Y, Cao R, Li L, Zhong K, Chen Q, Xiao J. miR-5195-3p inhibits proliferation and invasion of human bladder cancer cells by directly targeting Oncogene KLF5. Oncol Res. 2017;25:1081–1087. doi: 10.3727/096504016X14831120463349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang H, Li Q, Niu X, Wang G, Zheng S, Fu G, Wang Z. miR-143 inhibits bladder cancer cell proliferation and enhances their sensitivity to gemcitabine by repressing IGF-1R signaling. Oncol Lett. 2017;13:435–440. doi: 10.3892/ol.2016.5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li P, Yang X, Cheng Y, Zhang X, Yang C, Deng X, Li P, Tao J, Yang H, Wei J, et al. MicroRNA-218 increases the sensitivity of bladder cancer to cisplatin by targeting Glut1. Cell Physiol Biochem. 2017;41:921–932. doi: 10.1159/000460505. [DOI] [PubMed] [Google Scholar]
- 5.Guo YW, Ying L, Tian Y, Yang PQ, Zhu YC, Wang ZP, Qiu F, Lin J. miR-144 downregulation increases bladder cancer cell proliferation by targeting EZH2 and regulating Wnt signaling. FEBS J. 2013;280:4531–4538. doi: 10.1111/febs.12417. [DOI] [PubMed] [Google Scholar]
- 6.Xu X, Chen H, Lin YW, Hu ZH, Mao YQ, Wu J, Xu XL, Zhu Y, Li SQ, Zheng XY, Xie LP. MicroRNA-409-3p inhibits migration and invasion of bladder cancer cells via targeting c-Met. Mol Cells. 2013;36:62–68. doi: 10.1007/s10059-013-0044-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tang SL, Gao YL, Chen XB. MicroRNA-214 targets PCBP2 to suppress the proliferation and growth of glioma cells. Int J Clin Exp Pathol. 2015;8:12571–12576. [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao J, Xu J, Zhang R. MicroRNA-411 inhibits malignant biological behaviours of colorectal cancer cells by directly targeting PIK3R3. Oncol Rep. 2018;39:633–642. doi: 10.3892/or.2017.6135. [DOI] [PubMed] [Google Scholar]
- 9.Yu Z, Lin X, Tian M, Chang W. microRNA196b promotes cell migration and invasion by targeting FOXP2 in hepatocellular carcinoma. Oncol Rep. 2018;39:731–738. doi: 10.3892/or.2017.6130. [DOI] [PubMed] [Google Scholar]
- 10.Shang A, Yang M, Shen F, Wang J, Wei J, Wang W, Lu W, Wang C, Wang C. miR-1-3p suppresses the proliferation, invasion and migration of bladder cancer cells by up-regulating SFRP1 expression. Cell Physiol Biochem. 2017;41:1179–1188. doi: 10.1159/000464379. [DOI] [PubMed] [Google Scholar]
- 11.Yuan Q, Sun T, Ye F, Kong W, Jin H. MicroRNA-124-3p affects proliferation, migration and apoptosis of bladder cancer cells through targeting AURKA. Cancer Biomark. 2017;19:93–101. doi: 10.3233/CBM-160427. [DOI] [PubMed] [Google Scholar]
- 12.Feng C, Sun P, Hu J, Feng H, Li M, Liu G, Pan Y, Feng Y, Xu Y, Feng K, Feng Y. miRNA-556-3p promotes human bladder cancer proliferation, migration and invasion by negatively regulating DAB2IP expression. Int J Oncol. 2017;50:2101–2112. doi: 10.3892/ijo.2017.3969. [DOI] [PubMed] [Google Scholar]
- 13.Jiang D, Cho W, Li Z, Xu X, Qu Y, Jiang Z, Guo L, Xu G. miR-758-3p suppresses proliferation, migration and invasion of hepatocellular carcinoma cells via targeting MDM2 and mTOR. Biomed Pharmacother. 2017;96:535–544. doi: 10.1016/j.biopha.2017.10.004. [DOI] [PubMed] [Google Scholar]
- 14.Meng X, Zhao Y, Wang J, Gao Z, Geng Q, Liu X. Regulatory roles of miRNA-758 and matrix extracellular phosphoglycoprotein in cervical cancer. Exp Ther Med. 2017;14:2789–2794. doi: 10.3892/etm.2017.4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 16.Hayashi T, Gust KM, Wyatt AW, Goriki A, Jäger W, Awrey S, Li N, Oo HZ, Altamirano-Dimas M, Buttyan R, et al. Not all NOTCH Is created equal: The oncogenic role of NOTCH2 in bladder cancer and its implications for targeted therapy. Clin Cancer Res. 2016;22:2981–2992. doi: 10.1158/1078-0432.CCR-15-2360. [DOI] [PubMed] [Google Scholar]
- 17.He X, Ping J, Wen D. MicroRNA-186 regulates the invasion and metastasis of bladder cancer via vascular endothelial growth factor C. Exp Ther Med. 2017;14:3253–3258. doi: 10.3892/etm.2017.4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Souza MF, Kuasne H, Barros-Filho MC, Cilião HL, Marchi FA, Fuganti PE, Paschoal AR, Rogatto SR, Cólus IMS. Circulating mRNAs and miRNAs as candidate markers for the diagnosis and prognosis of prostate cancer. PLoS One. 2017;12:e0184094. doi: 10.1371/journal.pone.0184094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Masuda T, Hayashi N, Kuroda Y, Ito S, Eguchi H, Mimori K. MicroRNAs as Biomarkers in Colorectal Cancer. Cancers (Basel) 2017;9:124. doi: 10.3390/cancers9090124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Amr KS, Elmawgoud Atia HA, Elazeem Elbnhawy RA, Ezzat WM. Early diagnostic evaluation of miR-122 and miR-224 as biomarkers for hepatocellular carcinoma. Genes Dis. 2017;4:215–221. doi: 10.1016/j.gendis.2017.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shao LP, Shen ZJ, Qian H, Zhou SF, Chen YG. Knockdown of miR-629 inhibits ovarian cancer malignant behaviors by targeting testis-specific Y-like protein 5. DNA Cell Biol. 2017;36:1108–1116. doi: 10.1089/dna.2017.3904. [DOI] [PubMed] [Google Scholar]
- 22.Zheng YG, Lu XW, Xu LP, Chen Z, Li QX, Yuan J. MicroRNA-675 promotes glioma cell proliferation and motility by negatively regulating retinoblastoma 1. Hum Pathol. 2017;69:63–71. doi: 10.1016/j.humpath.2017.09.006. [DOI] [PubMed] [Google Scholar]
- 23.Ding D, Zhang Y, Wen L, Fu J, Bai X, Fan Y, Lin Y, Dai H, Li Q, Zhang Y, An R. miR-367 regulates cell proliferation and metastasis by targeting metastasis-associated protein 3 (MTA3) in clear-cell renal cell carcinoma. Oncotarget. 2017;8:63084–63095. doi: 10.18632/oncotarget.18647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu YY, Song Y, Xiong Y, Wang X, Xu K, Han B, Bai Y, Li L, Zhang Y, Zhou L. MicroRNA-21 (Mir-21) promotes cell growth and invasion by repressing tumor suppressor PTEN in colorectal cancer. Cell Physiol Biochem. 2017;43:945–958. doi: 10.1159/000481648. [DOI] [PubMed] [Google Scholar]
- 25.Jiang L, Lin TS, Xu CC, Hu SK, Pan YY, Jin R. miR-124 interacts with the Notch1 signalling pathway and has therapeutic potential against gastric cancer. J Cell Mol Med. 2016;20:313–322. doi: 10.1111/jcmm.12724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen X, Xiao W, Chen W, Liu X, Wu M, Bo Q, Luo Y, Ye S, Cao Y, Liu Y. MicroRNA-26a and −26b inhibit lens fibrosis and cataract by negatively regulating Jagged-1/Notch signaling pathway. Cell Death Differ. 2017;24:1990. doi: 10.1038/cdd.2017.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goriki A, Seiler R, Wyatt AW, Contreras-Sanz A, Bhat A, Matsubara A, Hayashi T, Black PC. Unravelling disparate roles of NOTCH in bladder cancer. Nat Rev Urol. 2018;15:345–357. doi: 10.1038/s41585-018-0005-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.