Abstract
Although microRNA-425-5p (miR-425-5p) has been previously revealed to be upregulated in cervical cancer, the cellular function of miR-425-5p in cervical cancer remains unknown. The aim of the current study was to investigate the cellular function of miR-425-5p and its underlying mechanism in cervical cancer. Reverse transcription-quantitative polymerase chain reaction was used to measure miR-425-5p expression in several cervical cancer cell lines. TargetScan bioinformatics analysis was used to predict apoptosis-inducing factor mitochondria-associated 1 (AIFM1) as a novel target of miR-425-5p, and this was verified by dual-luciferase reporter assay. Furthermore, cell transfections were used to investigate the role of miR-425-5p in cervical cancer. The effect of miR-425-5p on cell viability and apoptosis in HeLa cells was detected by MTT assay and flow cytometry, respectively. The present study demonstrated that miR-425-5p was significantly upregulated in cervical cancer cell lines. In addition, AIFM1 was identified as a direct target of miR-425-5p and negatively regulated by miR-425-5p. Downregulation of miR-425-5p inhibited HeLa cell viability and induced cell apoptosis. Furthermore, downregulation of miR-425-5p significantly increased the protein and mRNA expression levels of cytochrome c, caspase-3, caspase-9 and DNA damage regulated autophagy modulator 1. The effects of miR-425-5p inhibition on HeLa cell viability and apoptosis were significantly reversed by AIFM1 knockdown. In conclusion, the present study demonstrated that miR-425-5p was upregulated in cervical cancer, and downregulation of miR-425-5p inhibited cervical cancer cell growth by targeting AIFM1.
Keywords: cervical cancer, microRNA-425-5p, apoptosis-inducing factor mitochondria-associated 1, cell proliferation, apoptosis
Introduction
Cervical cancer is one of the most important reproductive health problems for adult women worldwide (1) and the second most common cancer among women worldwide (2). Approximately 500,000 newly diagnosed cases of cervical cancer are identified each year and at least 200,000 cases succumb to the disease (3). Cervical cancer incidence rates are high in developing countries, where more than 80% of the cervical cancer cases occur in the world (4). It is widely recognized that the leading cause of cervical cancer is persistent infection with specific types of the human papillomavirus (5). The complex process from normal tissue to cervical cancer involves going through mild dysplasia, moderate dysplasia, severe dysplasia, carcinoma in situ, and infiltrating carcinoma (6). This complex progression involves the abnormal expression of numerous oncogenes and tumor suppressor genes (7). The current primary treatment for cervical cancer is surgery, radiotherapy and chemotherapy (8). Although improvements have been made, there remain limitations associated with the current treatment options (9). Recurrent cervical cancer and metastasis occur frequently in patients with advanced cervical cancer (10). It is therefore important to investigate new and effective therapeutic targets for the treatment of cervical cancer.
MicroRNAs (miRNAs) are a class of small non-coding, single-stranded RNAs 19–25 nucleotides in length, found in eukaryotic cells. miRNAs can regulate post-transcriptional gene expression through mRNA degradation or translation inhibition of target mRNAs through the complete or partial binding in the 3′ untranslated regions (3′UTR) of target mRNAs (11,12). Previous studies have demonstrated that miRNAs are involved in the development of various types of cancer (13–16). Abnormal expression of miRNAs is associated with the occurrence and development of cervical cancer through the regulation of target gene expression, which include oncogenes and tumor suppressor genes (17–21). MicroRNA-425-5p (miR-425-5p) was previously revealed to be upregulated in cervical cancer (22), however the cellular function of miR-425-5p in cervical cancer remains unknown. The aim of the current study was to investigate the cellular function of miR-425-5p and its underlying mechanism in cervical cancer.
Materials and methods
Cell culture
Human cervical cancer cell lines HeLa, SiHa, C-33A and ME-180, as well as the human cervical epithelium cell line End1/E6E7 were purchased from American Type Culture Collection (Manassas, VA, USA). HeLa, SiHa and C-33A cells were grown in high glucose Dulbeco's modified Eagle medium and ME-180 cells were grown in RPMI 1640 medium (both Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (Invitrogen; Thermo Fisher Scientific, Inc.), 1% streptomycin-penicillin solution and maintained at 37°C in a 5% CO2-humidified incubator. Cells were passaged every 2–3 days. End1/E6E7 cells were grown in Keratinocyte serum-free medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 0.1 ng/ml human recombinant epithelial growth factor, 0.05 mg/ml bovine pituitary extract (both Santa Cruz Biotechnology, Inc., Dallas, TX, USA), 1% streptomycin-penicillin solution and maintained at 37°C in a 5% CO2-humidified incubator.
Dual-luciferase reporter assay
TargetScan bioinformatics software (www.targetscan.org/vert_71) was used to predict the putative target genes of miR-425–5. Apoptosis-inducing factor mitochondria-associated 1 (AIFM1) was identified as a potential target of miR-425-5p. The QuikChange Site-Directed Mutagenesis kit (Agilent Technologies, Inc., Santa Clara, CA, USA) was used, according to the manufacturer's protocol, to make a point mutation in the miR-425-5p binding domain on the 3′UTR of AIFM1. To confirm direct target binding, the wild-type (WT) 3′UTR AIFM1 or the mutant (MUT) 3′UTR AIFM1 were cloned into the dual-luciferase reporter vector pmiR-RB-REPORT™ (Guangzhou RiboBio Co., Ltd., Guangzhou, China). HeLa cells were co-transfected with 100 ng WT-AIFM1 or 100 ng MUT-AIFM1 and 50 nM miR-425-5p mimic (forward, 5′-AAUGACACGAUCACUCCCGUUGA-3′ and reverse, 5′-AACGGGAGUGAUCGUGUCAUUUU-3′) or 50 nM mimic control (forward, 5′-UUCUCCGAACGUGUCACGUTT-3′ and reverse, 5′-ACGUGACACGUUCGGAGAATT-3′) using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. MiR-425-5p mimic and mimic control were purchased from GenePharma Co., Ltd. (Shanghai, China). Following incubation for 48 h, luciferase activity was detected using a Dual-Luciferase® Reporter assay system (Promega Corporation, Madison, WI, USA), according to the manufacturer's protocol. Firefly luciferase activity was normalized to Renilla luciferase activity.
Cell transfection
HeLa cells were seeded into a 6-well plate at a density of 1×106 cells/well and cultured at 37°C for 24 h. MiR-425-5p inhibitor and inhibitor control were obtained from GenePharma Co., Ltd. Cells were transfected with 100 nM miR-425-5p inhibitor (5′-AGGCGAAGGAUGACAAAGGGAA-3′), 100 nM inhibitor control (5′-CAGUACUUUUGUGUAGUACAA-3′), 10 µM control-siRNA (cat. no. 36869; Santa Cruz Biotechnology, Inc.), 10 µM AIFM1-siRNA (cat. no. 26926; OriGene Technologies, Inc., Rockville, MD, USA), miR-425-5p inhibitor + control-siRNA, or miR-425-5p inhibitor + AIFM1-siRNA using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. Following incubation for 48 h, transfection efficiency was measured.
Cell proliferation assay
Cell viability was measured by MTT assay. Following a 48-h cell transfection, HeLa cells were seeded into 96-well plates at a density of 1×104 cells/per well and cultured for 24 h. Following incubation, 20 ml MTT solution (0.5 mg/ml; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) was added to each well and further incubated at 37°C for 4 h. Cell viability was determined by measuring the absorbance at a wavelength of 570 nm using a FLUOstar® Omega Microplate Reader (BMG Labtech GmbH, Ortenberg, Germany).
Flow cytometric analysis of apoptosis
Cell apoptosis was analyzed using the Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) apoptosis detection kit (cat. no. 70-AP101-100; MultiSciences, Hangzhou, Zhejiang, China). Following a 48-h cell transfection, HeLa cells were harvested with 0.25% trypsin, washed with PBS and subsequently stained with 5 µl Annexin V-FITC and 5 µl PI for 30 min at room temperature without light. Early and late apoptotic cells were subsequently analyzed using a flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA), and data were analyzed using WinMDI software (version 2.5; Purdue University Cytometry Laboratories; www.cyto.purdue.edu/flowcyt/software/Catalog.htm).
Western blot analysis
Total protein was extracted from cells using radioimmunoprecipitation assay buffer (Beyotime Institute of Biotechnology, Haimen, China), according to the manufacturer's protocol. Total protein was quantified using a bicinchoninic acid assay kit (Pierce; Thermo Fisher Scientific, Inc.) and 30 mg protein/lane was separated via SDS-PAGE on a 12% gel. The separated proteins were transferred onto polyvinylidene fluoride membranes (EMD Millipore, Billerica, MA, USA) and blocked for 1 h at room temperature with 5% skimmed milk. The membranes were incubated with primary antibodies against AIFM1 (1:1,000; cat. no. BA3715-1; Boster Biological Technology, Pleasanton, CA, USA), DNA damage regulated autophagy modulator 1 (DRAM; 1:1,000; cat. no. 208160; Abcam, Cambridge, MA, USA), cytochrome c (1:1,000; cat. no. 11940), caspase-3 (1:1,000; cat. no. 9665), caspase-9 (1:1,000; cat. no. 9502) and β-actin (1:1,000; cat. no. 4970; all Cell Signaling Technology Inc., Danvers, MA, USA) overnight at 4°C. Following primary incubation, membranes were incubated with horseradish peroxidase-conjugated secondary antibody, anti-rabbit IgG (1:2,000; cat. no. 7074; Cell Signaling Technology, Inc.) for 2 h at room temperature. Protein bands were visualized using the enhanced chemiluminescence Western Blotting Detection kit (EMD Millipore).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from cells using TRIzol® reagent (Invitrogen, Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. Total RNA was reverse transcribed into cDNA using the TaqMan™ MicroRNA Reverse Transcription kit (Applied Biosystems; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. qPCR was subsequently performed using the SYBR® Premix Ex Taq™ II kit (Takara Bio, Inc., Otsu, Japan). The following primer pairs were used for the qPCR: GAPDH forward, 5′CTTTGGTATCGTGGAAGGACTC3′ and reverse, 5′GTAGAGGCAGGGATGATGTTCT3′; and U6 forward, 5′GCTTCGGCAGCACATATACTAAAAT3′ and reverse, 5′CGCTTCACGAATTTGCGTGTCAT3′; miR-425-5p forward, 5′TGCGGAATGACACGATCACTCCCG3′ and reverse, 5′CCAGTGCAGGGTCCGAGGT3′; AIFM1 forward, 5′TTGAGAATGGTGGTGTGGCT3′ and reverse, 5′AGACTTCTTGGAGTACCTCCTGT3′; caspase-3 forward, 5′AGAACTGGACTGTGGCATTG3′ and reverse, 5′CACAAAGCGACTGGATGAAC3′; caspase-9 forward, 5′TGTTTCCGAGCGAGGGATTT3′ and reverse, 5′CGCAGGAAGGTTTTGGGGTA3′; DRAM forward, 5′AGACTCCATCTTTTCACCCAAA3′ and reverse, 5′GCTCTTCACCTTTCAAGCCTAA3′; cytochrome c forward, 5′TGCCACACTGTTGAAGCCGGT3′ and reverse, 5′GATCTGCACGCTCGTTTGCCT3′. The following thermocycling conditions were used for the qPCR: Initial denaturation at 95°C for 10 min; 35 cycles of 95°C for 15 sec and 55°C for 40 sec. The relative mRNA expression levels were quantified using the 2−ΔΔCq method and normalized to the internal reference gene, U6 or GAPDH (23).
Statistical analysis
Data are presented as the mean ± standard deviation of at least three independent experiments. All statistical analyses were performed using SPSS software (version 17.0; SPSS, Inc., Chicago. IL, USA). Student's t-test was performed for comparison analysis between two groups. One-way analysis of variance followed by Tukey's post hoc test was performed for analyze differences among multiple groups. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-425-5p expression is upregulated in cervical cancer
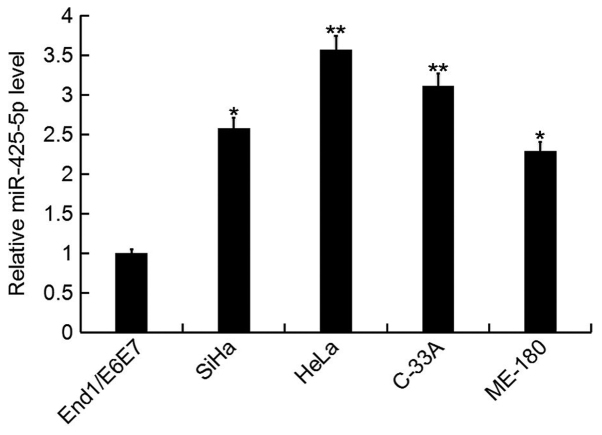
In the current study, the expression level of miR-425-5p was determined by RT-qPCR in several human cervical cancer cell lines (HeLa, SiHa, C-33A and ME-180), as well as the human normal cervical epithelium cell line End1/E6E7. The miR-425-5p expression level was significantly increased in human cervical cancer cell lines compared with the normal cervical epithelium cell line (Fig. 1). As the highest level of miR-425-5p expression was detected in HeLa cells, these were selected for all subsequent experiments.
Figure 1.
miR-425-5p expression in human cervical cancer cell lines. The relative expression level of miR-425-5p was determined by reverse transcription-quantitative polymerase chain reaction in human cervical cancer cell lines HeLa, SiHa, C-33A and ME-180, and the normal cervical epithelium cell line End1/E6E7. Data are presented as the mean ± standard deviation. *P<0.05 and **P<0.01 vs. End1/E6E7. miR, microRNA.
AIFM1 is a target gene of miR-425-5p
To further investigate the role of miR-425-5p in human cervical cancer, potential targets of miR-425-5p were examined. TargetScan bioinformatics software was used to identify AIFM1 as a putative target gene of miR-425-5p (Fig. 2A). To confirm whether miR-425-5p directly regulates AIFM1 expression via interaction with the predicted binding sites, luciferase reporter assays were performed. Following co-transfection with miR-425-5p mimic, the luciferase reporter activity of WT-AIFM1 was significantly decreased compared with the luciferase reporter activity of MUT-AIFM1 (Fig. 2B). The results suggest that AIFM1 is a direct target of miR-425-5p.
Figure 2.
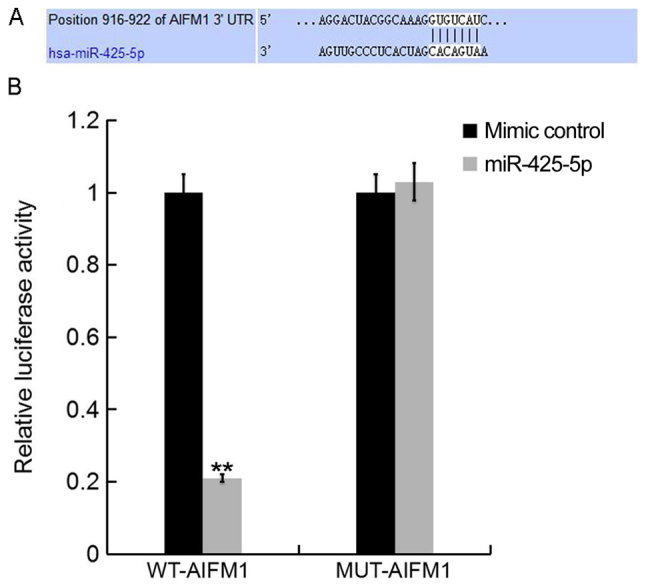
AIFM1 is a direct target gene of miR-425-5p. (A) TargetScan software was used to predict a binding site for miR-425-5p in the 3′UTR of AIFM1. (B) Luciferase activity of a dual-luciferase reporter vector containing wild-type 3′UTR-AIFM1 or a mutant 3′UTR-AIFM1. Data are presented as the mean ± standard deviation of three independent experiments. **P<0.01 vs. mimic control. AIFM1, apoptosis-inducing factor mitochondria-associated 1; UTR, untranslated region; miR, microRNA; WT, wild-type; MUT, mutant-type; WT-AIFM1, HeLa cells co-transfected with WT 3′UTR-AIFM1 and either mimic control or miR-425-5p; MUT-AIFM1, HeLa cells co-transfected with MUT 3′UTR-AIFM1 and either mimic control or miR-425-5p.
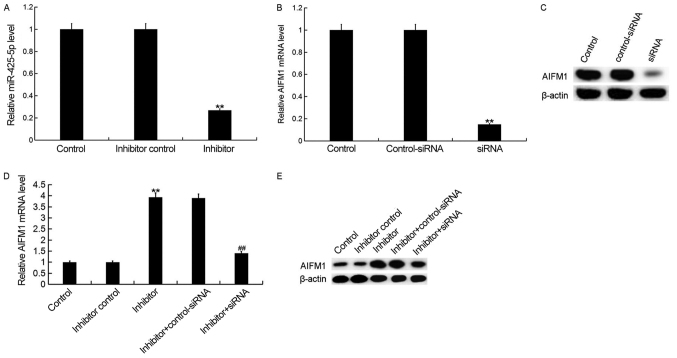
Downregulation of miR-425-5p enhances AIFM1 expression in HeLa cells
To investigate the effect of miR-425-5p on cervical cancer, HeLa cells were transfected with miR-425-5p inhibitor, inhibitor control, AIFM1-siRNA, control-siRNA, miR-425-5p inhibitor + AIFM1-siRNA or miR-425-5p inhibitor + control-siRNA, respectively. Following incubation for 48 h, transfection efficiency was measured. The current study demonstrated that the miR-425-5p inhibitor significantly decreased miR-425-5p expression in HeLa cells (Fig. 3A). In addition, the mRNA and protein expression levels of AIFM1 were decreased in HeLa cells following transfection with AIFM1-siRNA (Fig. 3B and C). Furthermore, the mRNA and protein expression levels of AIFM1 were increased following transfection with miR-425-5p inhibitor compared with the control group. However, AIFM1-siRNA reversed the effects observed with miR-425-5p inhibitor (Fig. 3D and E).
Figure 3.
miR-425-5p inhibitor enhances AIFM1 expression in HeLa cells. (A) The relative expression level of miR-425-5p was determined by reverse transcription-quantitative polymerase chain reaction in HeLa cells following transfection with miR-425-5p inhibitor and inhibitor control. The (B) mRNA and (C) protein expression level of AIFM1 was analyzed following transfection with AIFM1-siRNA or control-siRNA. The (D) mRNA and (E) protein expression level of AIFM1 was analyzed following transfection with miR-425-5p inhibitor, inhibitor control, miR-425-5p inhibitor + AIFM1-siRNA or miR-425-5p inhibitor + control-siRNA. Data are presented as the mean ± standard deviation. **P<0.01 vs. control group; ##P<0.01 vs. inhibitor group. AIFM1, apoptosis-inducing factor mitochondria-associated 1; miR, microRNA; siRNA, small interfering RNA; control, untransfected HeLa cells; inhibitor control, HeLa cells transfected with inhibitor control; inhibitor, HeLa cells transfected with miR-425-5p inhibitor; control-siRNA, HeLa cells transfected with control siRNA; siRNA, HeLa cells transfected with AIFM1-siRNA; inhibitor + control-siRNA, HeLa cells co-transfected with miR-425-5p inhibitor and control siRNA; inhibitor + siRNA, HeLa cells co-transfected with miR-425-5p inhibitor and AIFM1-siRNA.
Downregulation of miR-425-5p inhibits HeLa cell viability
To investigate the cellular function of miR-425-5p in cervical cancer, the MTT assay was used to examine the viability of HeLa cells following transfection with miR-425-5p inhibitor, inhibitor control, miR-425-5p inhibitor + AIFM1-siRNA or miR-425-5p inhibitor + control-siRNA. The current study demonstrated that the miR-425-5p inhibitor significantly decreased HeLa cell viability compared with the control group. However, AIFM1-siRNA significantly reversed the effect of miR-425-5p inhibitor on HeLa cell viability (Fig. 4).
Figure 4.
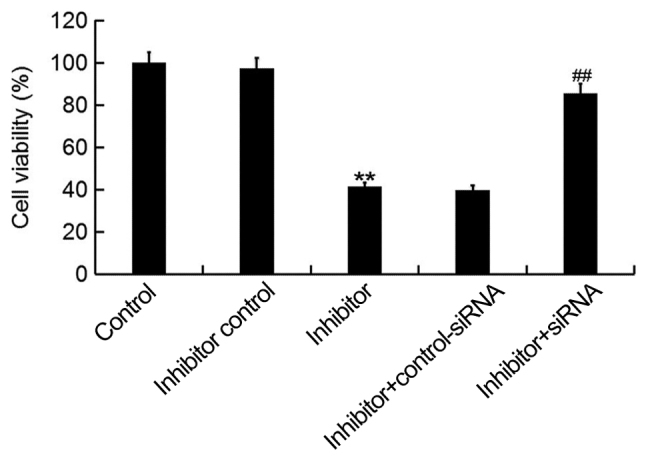
miR-425-5p inhibitor suppresses HeLa cell viability. MTT assay was used to examine cell viability of HeLa cells following transfection with miR-425-5p inhibitor, inhibitor control, miR-425-5p inhibitor + AIFM1-siRNA or miR-425-5p inhibitor + control-siRNA. Data are presented as the mean ± standard deviation. **P<0.01 vs. control group; ##P<0.01 vs. inhibitor group. siRNA, small interfering RNA; control, untransfected HeLa cells; inhibitor control, HeLa cells transfected with inhibitor control; inhibitor, HeLa cells transfected with miR-425-5p inhibitor; inhibitor + control-siRNA, HeLa cells co-transfected with miR-425-5p inhibitor and control siRNA; inhibitor + siRNA, HeLa cells co-transfected with miR-425-5p inhibitor and AIFM1-siRNA.
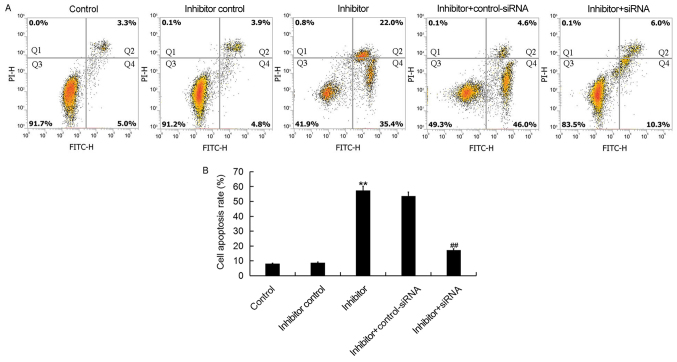
Downregulation of miR-425-5p induces HeLa cell apoptosis
To further investigate the cellular function of miR-425-5p in cervical cancer, flow cytometry was used to examine cell apoptosis in HeLa cells following transfection with miR-425-5p inhibitor, inhibitor control, miR-425-5p inhibitor + AIFM1-siRNA or miR-425-5p inhibitor + control-siRNA. The current study demonstrated that the miR-425-5p inhibitor significantly increased HeLa cell apoptosis compared with the control group. However, AIFM1-siRNA significantly reversed the effect of miR-425-5p inhibitor on HeLa cell apoptosis (Fig. 5A and B).
Figure 5.
miR-425-5p inhibitor induces HeLa cell apoptosis. (A) Early apoptosis (Q2) and late apoptosis (Q4) was detected by flow cytometry in HeLa cells following transfection with miR-425-5p inhibitor, inhibitor control, miR-425-5p inhibitor + AIFM1-siRNA or miR-425-5p inhibitor + control-siRNA. (B) The effect of miR-425-5p knockdown on HeLa cell apoptosis was examined. Data are presented as the mean ± standard deviation. **P<0.01 vs. control group; ##P<0.01 vs. inhibitor group. siRNA, small interfering RNA; FITC, fluorescein isothiocyanate; control, untransfected HeLa cells; inhibitor control, HeLa cells transfected with inhibitor control; inhibitor, HeLa cells transfected with miR-425-5p inhibitor; inhibitor + control-siRNA, HeLa cells co-transfected with miR-425-5p inhibitor and control siRNA; inhibitor + siRNA, HeLa cells co-transfected with miR-425-5p inhibitor and AIFM1-siRNA; PI, propidium iodide.
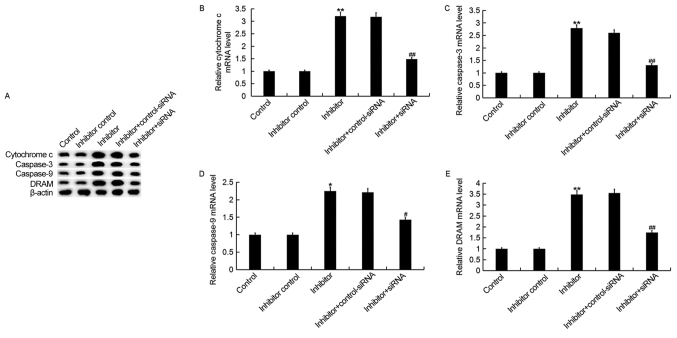
Downregulation of miR-425-5p upregulates the expression of cytochrome c, caspase-3, caspase-9 and DRAM in HeLa cells
To investigate the regulatory effect of miR-425-5p in cervical cancer, the expression of four pro-apoptotic genes, cytochrome c, caspase-3, caspase-9 and DRAM were examined in HeLa cells following transfection with miR-425-5p inhibitor, inhibitor control, miR-425-5p inhibitor + AIFM1-siRNA or miR-425-5p inhibitor + control-siRNA (Fig. 6). The current study demonstrated that the miR-425-5p inhibitor significantly upregulated both the protein and mRNA expression levels of all four pro-apoptotic genes analyzed, compared with the control group. However, the enhanced expression of these pro-apoptotic genes was significantly reversed by AIFM1 knockdown.
Figure 6.
miR-425-5p inhibitor upregulates pro-apoptotic gene expression in HeLa cells. (A) The protein expression levels of cytochrome c, caspase-3, caspase-9 and DRAM were determined by western blot analysis in HeLa cells following transfection with miR-425-5p inhibitor, inhibitor control, miR-425-5p inhibitor + AIFM1-siRNA or miR-425-5p inhibitor + control-siRNA. The mRNA expression levels of (B) cytochrome c, (C) caspase-3, (D) caspase-9 and (E) DRAM were determined by reverse transcription-quantitative polymerase chain reaction in HeLa cells following transfection with miR-425-5p inhibitor, inhibitor control, miR-425-5p inhibitor + AIFM1-siRNA or miR-425-5p inhibitor + control-siRNA. Data are presented as the mean ± standard deviation. *P<0.05 and **P<0.01 vs. control group; #P<0.05 and ##P<0.01 vs. inhibitor group. DRAM, DNA damage regulated autophagy modulator 1; siRNA, small interfering RNA; control, untransfected HeLa cells; inhibitor control, HeLa cells transfected with inhibitor control; inhibitor, HeLa cells transfected with miR-425-5p inhibitor; inhibitor + control-siRNA, HeLa cells co-transfected with miR-425-5p inhibitor and control siRNA; inhibitor + siRNA, HeLa cells co-transfected with miR-425-5p inhibitor and AIFM1-siRNA.
Discussion
Increasing evidence suggests that miRNAs are involved in the development of various types of cancer (13–16). Previous reports have indicated that miRNAs serve a regulatory role in cell proliferation, differentiation, apoptosis, migration and metabolism (12,24,25). Studies have revealed that enhanced expression levels of miR-425-5p exist in different types of cancer, which suggest that miR-425-5p may have multiple functions in the development of cancer (22,26). Studies previously demonstrated that miR-425-5p can promote invasion and metastasis in hepatocellular carcinoma, colorectal cancer and gastric cancer (27–29).
The aim of the current study was to investigate the cellular function of miR-425-5p and its underlying mechanism in cervical cancer. In the current study, the expression level of miR-425-5p was determined by RT-qPCR in several human cervical cancer cell lines including HeLa, SiHa, C-33A and ME-180, as well as the human normal cervical epithelium cell line End1/E6E7. miR-425-5p expression was significantly increased in human cervical cancer cell lines compared with the normal cervical epithelium cell line. In addition, HeLa cells expressed the highest level of miR-425-5p and were selected for all subsequent experiments.
To further investigate the role of miR-425-5p in human cervical cancer, potential targets of miR-425-5p were examined in HeLa cells using the TargetScan software. Bioinformatic analysis identified AIFM1 as a putative target gene of miR-425-5p and this was verified by dual-luciferase reporter assay. AIFM1, which is located in the mitochondrion intermembrane space, serves a role in the regulation of cell apoptosis (30–33).
The effect of miR-425-5p on cervical cancer cell viability and apoptosis was examined in HeLa cells. Downregulation of miR-425-5p significantly decreased cell viability and induced cell apoptosis, however, the inhibitory effect exerted by the miR-425-5p inhibitor was significantly reversed by AIFM1 knockdown. These results suggest that miR-425-5p inhibitor may function as a tumor suppressor in cervical cancer by enhancing the expression of AIFM1. AIFM1 is a phylogenetically conserved mitochondrial flavoprotein with NADH oxidation and potent apoptosis-inducing activity (30–32). AIFM1 induces mitochondria to release the apoptogenic proteins cytochrome c and caspase-9, thereby initiating apoptosis (31,33). A previous study demonstrated that overexpression of AIFM1 induced apoptosis by promoting the transcription of caspase-3 and DRAM in hepatoma cells (33).
To investigate the underlying molecular mechanism of miR-425-5p in cervical cancer, the expression of pro-apoptotic genes including cytochrome c, caspase-3, caspase-9 and DRAM was examined in HeLa cells. The current study demonstrated that both the protein and mRNA expression levels of cytochrome c, caspase-3, caspase-9 and DRAM were significantly upregulated following transfection with miR-425-5p inhibitor. However, the enhanced expression of all four pro-apoptotic genes was significantly reversed by AIFM1 knockdown.
The present study investigated the cellular function of miR-425-5p and its underlying mechanism in cervical cancer, however in vivo studies and clinical trial data are required to validate the preliminary in vitro results obtained in the current study. Furthermore, the cellular function of AIFM1 alone in cervical cancer needs to be further investigated.
In conclusion, the current study demonstrated that miR-425-5p was upregulated in cervical cancer, and this may contribute to cervical cancer development by inhibiting AIFM1 expression. Furthermore, inhibition of miR-425-5p decreased cervical cancer cell viability and induced cell apoptosis. Therefore, the miR-425-5p/AIFM1 axis may serve a role in cervical cancer progression and this may be a promising therapeutic target for the treatment of patients.
Acknowledgements
Not applicable.
Funding
The present study was supported by a grant from the Shijiazhuang Science and Technology Bureau of China 2013 Science and Technology Project (grant no. 131462543).
Availability of data and materials
All datasets used and/or generated during the current study are available from the corresponding author on reasonable request.
Authors' contributions
YZ designed the study. YZ, YY, and RL analyzed the data. YM, GT and QC analyzed the data and prepared the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Forouzanfar MH, Foreman KJ, Delossantos AM, Lozano R, Lopez AD, Murray CJ, Naghavi M. Breast and cervical cancer in 187 countries between 1980 and 2010: A systematic analysis. Lancet. 2011;378:1461–1484. doi: 10.1016/S0140-6736(11)61351-2. [DOI] [PubMed] [Google Scholar]
- 2.De Sanjose S, Quint WG, Alemany L, Geraets DT, Klaustermeier JE, Lloveras B, Tous S, Felix A, Bravo LE, Shin HR, et al. Human papillomavirus genotype attribution in invasive cervical cancer: A retrospective cross-sectional worldwide study. Lancet Oncol. 2010;11:1048–1056. doi: 10.1016/S1470-2045(10)70230-8. [DOI] [PubMed] [Google Scholar]
- 3.Li Z, Wang H, Wang Z, Cai H. MiR-195 inhibits the proliferation of human cervical cancer cells by directly targeting cyclin D1. Tumor Biol. 2016;37:6457–6463. doi: 10.1007/s13277-015-4540-6. [DOI] [PubMed] [Google Scholar]
- 4.zur Hausen H. Papillomaviruses and cancer: From basic studies to clinical application. Nat Rev Cancer. 2002;2:342–350. doi: 10.1038/nrc798. [DOI] [PubMed] [Google Scholar]
- 5.Khan MJ, Castle PE, Lorincz AT, Wacholder S, Sherman M, Scott DR, Rush BB, Glass AG, Schiffman M. The elevated 10-year risk of cervical precancer and cancer in women with human papillomavirus (HPV) type 16 or 18 and the possible utility of type-specific HPV testing in clinical practice. J Natl Cancer Inst. 2005;97:1072–1079. doi: 10.1093/jnci/dji187. [DOI] [PubMed] [Google Scholar]
- 6.Mariuzzi G, Santinelli A, Valli M, Sisti S, Montironi R, Mariuzzi L, Alberti R, Pisani E. Cytometric evidence that cervical intraepithelial neoplasia I and II are dysplasias rather than true neoplasias. An image analysis study of factors involved in the progression of cervical lesions. Anal Quant Cytol Histol. 1992;14:137–147. [PubMed] [Google Scholar]
- 7.Zou DL, Zhou Q, Wang D, Guan L, Yuan L, Li S. The downregulation of microRNA-10b and its role in cervical cancer. Oncol Res. 2016;24:99–108. doi: 10.3727/096504016X14611963142173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dueñas-Gonzalez A, Cetina L, Mariscal I, de la Garza J. Modern management of locally advanced cervical carcinoma. Cancer Treat Rev. 2003;29:389–399. doi: 10.1016/S0305-7372(03)00068-9. [DOI] [PubMed] [Google Scholar]
- 9.Ebina Y, Mikami M, Nagase S, Tabata T, Kaneuchi M, Tashiro H, Mandai M, Enomoto T, Kobayashi Y, Katabuchi H, et al. Japan Society of Gynecologic Oncology guidelines 2017 for the treatment of uterinecervical cancer. Int J Clin Oncol. 2018 Oct 5; doi: 10.1007/s10147-018-1351-y. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 10.Glick SB, Clarke AR, Blanchard A, Whitaker AK. Cervical cancer screening, diagnosis and treatment interventions for racial and ethnic minorities: A systematic review. J Gen Intern Med. 2012;27:1016–1032. doi: 10.1007/s11606-012-2052-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- 12.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 13.Hu L, Ai J, Long H, Liu W, Wang X, Zuo Y, Li Y, Wu Q, Deng Y. Intergrative microRNA and gene profiling data analysis reveals novel biomarkers and mechanisms for lung cancer. Oncotarget. 2016;7:8441–8454. doi: 10.18632/oncotarget.7264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsai MM, Wang CS, Tsai CY, Huang HW, Chi HC, Lin YH, Lu PH, Lin KH. Potential diagnostic, prognostic and therapeutic targets of microRNAs in human gastric cancer. Int J Mol Sci. 2016;17(pii):E945. doi: 10.3390/ijms17060945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao H, Li M, Li L, Yang X, Lan G, Zhang Y. MiR-133b is down-regulated in human osteosarcoma and inhibits osteosarcoma cells proliferation, migration and invasion, and promotes apoptosis. PLoS One. 2013;8:e83571. doi: 10.1371/journal.pone.0083571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaukoniemi KM, Rauhala HE, Scaravilli M, Latonen L, Annala M, Vessella RL, Nykter M, Tammela TL, Visakorpi T. Epigenetically altered miR-193b targets cyclin D1 in prostate cancer. Cancer Med. 2015;4:1417–1425. doi: 10.1002/cam4.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang X, Tang S, Le SY, Lu R, Rader JS, Meyers C, Zheng ZM. Aberrant expression of oncogenic and tumor-suppressive microRNAs in cervical cancer is required for cancer cell growth. PLoS One. 2008;3:e2557. doi: 10.1371/journal.pone.0002557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilting SM, van Boerdonk RA, Henken FE, Meijer CJ, Diosdado B, Meijer GA, le Sage C, Agami R, Snijders PJ, Steenbergen RD. Methylation-mediated silencing and tumor suppressive function of hsa-miR-24 in cervical cancer. Mol Cancer. 2010;9:167. doi: 10.1186/1476-4598-9-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Long MJ, Wu FX, Li P, Liu M, Li X, Tang H. MicroRNA-10a targets CHL1 and promotes cell growth, migration and invasion in human cervical cancer cells. Cancer Lett. 2012;324:186–196. doi: 10.1016/j.canlet.2012.05.022. [DOI] [PubMed] [Google Scholar]
- 20.Kogo R, How C, Chaudary N, Bruce J, Shi W, Hill RP, Zahedi P, Yip KW, Liu FF. The microRNA-218~Survivin axis regulates migration, invation, and lymph node metastasis in cervical cancer. Oncotarget. 2015;6:1090–1100. doi: 10.18632/oncotarget.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deng B, Zhang Y, Zhang S, Wen F, Miao Y, Guo K. microRNA-142-3p inhibits cell proliferation and invasion of cervical cancer cells by targeting FZD7. Tumor Biol. 2015;36:8065–8073. doi: 10.1007/s13277-015-3483-2. [DOI] [PubMed] [Google Scholar]
- 22.Sun L, Jiang R, Li J, Wang B, Ma C, Lv Y, Mu N. MicoRNA-425-5p is a potential prognostic biomarker for cervical cancer. Ann Clin Biochem. 2017;54:127–133. doi: 10.1177/0004563216649377. [DOI] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Ebert MS, Sharp PA. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012;149:515–524. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rogers K, Chen X. Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell. 2013;25:2383–2399. doi: 10.1105/tpc.113.113159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quan J, Li Y, Pan X, Lai Y, He T, Lin C, Zhou L, Zhao L, Sun S, Ding Y, et al. Oncogenic miR-425-5p is associated with cellular migration, proliferation and apoptosis in renal cell carcinoma. Oncol Lett. 2018;16:2175–2184. doi: 10.3892/ol.2018.8948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fang F, Song T, Zhang T, Cui Y, Zhang G, Xiong Q. MiR-425-5p promotes invasion and metastasis of hepatocellular carcinoma cells through SCAI-mediated dysregulation of multiple signaling pathways. Oncotarget. 2017;8:31745–31757. doi: 10.18632/oncotarget.15958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cristóbal I, Madoz-Gúrpide J, Rojo F, García-Foncillas J. Potential therapeutic value of miR-425-5p in metastatic colorectal cancer. J Cell Mol Med. 2016;20:2213–2214. doi: 10.1111/jcmm.12902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Z, Wen M, Guo J, Shi J, Wang Z, Tan B, Zhang G, Zheng X, Zhang A. Clinical value of miR-425-5p detection and its association with cell proliferation and apoptosis of gastric cancer. Pathol Res Pract. 2017;213:929–937. doi: 10.1016/j.prp.2017.05.009. [DOI] [PubMed] [Google Scholar]
- 30.Susin SA, Lorenzo HK, Zamzami N, Marzo I, Snow BE, Brothers GM, Mangion J, Jacotot E, Costantini P, Loeffler M, et al. Molecular characterization of mitochondrial apoptosis-inducing factor. Nature. 1999;397:441–446. doi: 10.1038/17135. [DOI] [PubMed] [Google Scholar]
- 31.Joza N, Susin SA, Daugas E, Stanford WL, Cho SK, Li CY, Sasaki T, Elia AJ, Cheng HY, Ravagnan L, et al. Essential role of the mitochondrial apoptosis-inducing factor in programmed cell death. Nature. 2001;410:549–554. doi: 10.1038/35069004. [DOI] [PubMed] [Google Scholar]
- 32.Cregan SP, Dawson VL, Slack RS. Role of AIF in caspase-dependent and caspase-independent cell death. Oncogene. 2004;23:2785–2796. doi: 10.1038/sj.onc.1207517. [DOI] [PubMed] [Google Scholar]
- 33.Liu D, Liu M, Wang W, Pang L, Wang Z, Yuan C, Liu K. Overexpression of apoptosis-inducing factor mitochondrion-associated 1 (AIFM1) induces apoptosis by promoting the transcription of caspase-3 and DRAM in hepatoma cells. Biochem Biophys Res Commun. 2018;498:453–457. doi: 10.1016/j.bbrc.2018.02.203. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All datasets used and/or generated during the current study are available from the corresponding author on reasonable request.