Abstract
Pancreatic cancer (PC) is one of the most malignant types of human cancer and has an extremely poor prognosis. MicroRNAs (miRs) reportedly serve a critical role in pancreatic ductal adenocarcinoma (PDAC) progression. Understanding the expression patterns and functions of miRs may provide strategies for the diagnosis and treatment of patients with PC. In particular, miR-634 is attracting interest due to its critical role in regulating the biology of some types of cancer. However, the expression patterns, biological function and molecular mechanism of miR-634 in PC remain unknown. In the present study, miR-634 expression levels in PC tissues and cell lines were significantly downregulated. Notably, the ectopic overexpression of miR-634 in PC cells inhibited tumor progression, whereas miR-634 silencing reversed these effects. Furthermore, reverse transcription-quantitative polymerase chain reaction, western blot analysis and the dual-luciferase assay revealed that miR-634 regulated heat shock-related 70 kDa protein 2 (HSPA2) by directly binding to its 3-untranslated region. In clinical samples of PC, miR-634 was inversely correlated with HSPA2, which was upregulated in PC. In the rescue experiment, HSPA2 overexpression partially abrogated the effects of miR-634 mimicry on biological function. In conclusion, miR-634 functioned as a tumor suppressor in regulating PC progression by targeting HSPA2 and may therefore be a novel potential therapeutic target for PC.
Keywords: microRNA-634, pancreatic cancer, proliferation, migration and invasion, heat shock-related 70 kDa protein 2
Introduction
Pancreatic cancer (PC) is one of the most malignant solid tumors in humans that is characterized by its late diagnosis, rapid progression, early metastasis and chemoresistance (1,2). Although an increasing number of therapies, including surgical resection, chemotherapy and radiotherapy, have been used in recent years, the overall 5-year survival rate is still <5% (2,3). Recent studies have indicated that microRNAs (miRs) have a critical role in the progression of PC (4,5). Therefore, a detailed understanding of the miR-based molecular mechanisms of PC malignancy may provide useful insights into the identification of biomarkers and development of novel therapeutic strategies for PC.
miRs are small, non-coding RNA molecules, which post-transcriptionally regulate gene expression by directly binding to the 3′-untranslated region (UTR) of their target genes and induce target mRNA degradation or suppress target mRNA translation (6). An individual miR typically has multiple target genes with partially complementary mRNA sequences, whereas a single gene can be targeted by several miRs. Previous studies have demonstrated that miR-634 acts as tumor suppressor in glioma, gastric carcinoma, hepatocellular carcinoma, nasopharyngeal carcinoma, ovarian cancer and cervical cancer (7–12). However, to the best of our knowledge, the function and mechanism of miR-634 in PC progression has not been fully elucidated. Therefore, elucidating the function and mechanism of miR-634 in PC is important.
The aim of the present study was to illustrate the expression level of miR-634 in PC tissues and cell lines, its association with PC progression and the underlying molecular mechanisms in order to identify whether miR-634 serves as a tumor suppressor in PC.
Materials and methods
Cell culture
Human pancreatic cancer lines (Capan-2, PANC-1, SW1990 and COLO357) were purchased from the Cell Resource Center, Chinese Academy of Science Committee (Shanghai, China). The human immortal ductal cell line HPDE was obtained from American Type Culture Collection (Manassas, VA, USA). Cancer cells were cultured in Dulbecco's modified Eagle's medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.). HPDE cells were maintained in keratinocyte serum-free medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented with bovine pituitary extract and epidermal growth factor (Gibco; Thermo Fisher Scientific, Inc.). All cells were maintained in a humidified incubator at 37°C with 5% CO2.
Human tissue samples
A total of 30 paired PC and normal adjacent tissues were collected from patients with PC (19 males and 11 females; age range, 41–69 years old) who had undergone pancreaticoduodenectomy at LanLing County Hospital (Linyi, China) between June 2015 and December 2016. A total of 28 patients had PDAC and two patients had adenosquamous carcinoma of the pancreas. According to the criteria of the American Joint Commission on Cancer (13), these patients were divided into three stages (stage I, n=6; stage II, n=16; stage III, n=8). Furthermore, 20 patients had lymph node metastasis and 10 patients had no lymph node metastasis (no other organ metastasis was indicated). The patients had not received chemotherapy or radiation therapy prior to surgery. The samples were frozen and stored at −80°C until total RNA extraction was performed. The Research Ethics Committee of LanLing County Hospital (Linyi, China) approved the present study. All patients provided their signed consent to the research.
Cell transfection
miR-634 mimic (5-AACCAGCACCCCAACUUUGGACGGTATTCGCACTGGATACGACGAACTTT-3), miR-negative control (NC; 5-ACUACUGAGUGACAGUAGA-3), miR-634 inhibitor (5-CACUACUUUUGUGUCCCACUU-3) and antiNC (5-CAGUACUUUUGUGUAGUACAA-3) were purchased from Guangzhou RiboBio Co., Ltd. (Guangzhou, China). The open reading frame of heat shock-related 70 kDa protein 2 (HSPA2) was inserted into pcDNA3.1 vector (Invitrogen; Thermo Fisher Scientific, Inc.) to generate the pcDNA3.1/HSPA2 overexpression vector. A total of 5×105 cells were transfected with 2.5 µg NC, miR-634 mimic, antiNC, miR-634 inhibitor, empty vector or HSPA2 overexpression vector using Lipofectamine 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Overexpression efficiency was analyzed via western blot analysis. After 48 h of transfection, the cells were harvested and used for further experiments.
Colony formation assay
PANC-1 cells transfected with miR-634 mimic, miR-634 inhibitor or miR-634 mimic and HSPA2 plasmid were seeded at 400 cells/well in 6-well plates at 37°C. After 10 days, the colonies were fixed with 4% paraformaldehyde at room temperature for 10 min and stained with 0.5% crystal violet at room temperature for 10 min. The number of colonies (>50 cells) was counted under a light microscope (magnification, ×100).
Cell Counting Kit-8 (CCK) assay
Cell viability was analyzed using CCK-8 (Beyotime Institute of Biotechnology, Haimen, China). Briefly, PANC-1 cells (3,000 cells/well) transfected with miR-634 mimic, miR-634 inhibitor or miR-634 mimic and HSPA2 plasmid in 200 µl DMEM were plated into 96-well plates. After 0, 24, 48 and 72 h of seeding, 10 µl of CCK-8 solution was added into each well. Following 2 h of incubation, the optical density values at 450 nm of each well were measured using a microplate reader.
Transwell assay
Cell metastasis was determined using transwell chambers (Corning, Inc., Corning, NY, USA). For the invasion assay, the upper sides of the filters were coated with 50 µl Matrigel (BD Biosciences, San Jose, CA, USA). For the invasion and migration assays, 5×104 cells in 200 µl of serum-free DMEM were seeded in the upper chamber. The lower chamber was filled with DMEM supplemented with 5% FBS. Following incubation at 37°C in an atmosphere containing 5% CO2 for 24 h, cells on the lower filter were fixed with 10% methanol at room temperature for 20 min, stained with 0.5% crystal violet at room temperature for 10 min and counted under a light microscope (magnification, ×100).
Flow cytometry
Cell apoptosis was assessed using flow cytometry with staining of the cells using the Annexin V/propidium iodide (PI) kit (KeyGEN BioTHCH, Nanjing, Jiangsu, China) according to the manufacturer's protocols. The samples were analyzed using a fluorescence-activated cell sorter system (Cytomics FC 500 MPL; Beckman Coulter, Inc., Brea, CA, USA) to evaluate the apoptotic levels according to the manufacturer's protocol. Data were analyzed using ModFit LT 3.0 (Verity Software House, Inc., Topsham, ME, USA).
RNA extraction and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA from cells was extracted using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and then reverse-transcribed using a reverse transcription kit (Takara Biotechnology Co., Ltd., Dalian, China) following the manufacturer's protocol. qPCR was conducted with All-in-One miRNA qPCR Detection kit (AOMD-Q020, GeneCopoeia, Inc., Rockville, MD, USA) on a CFX96 Real-Time PCR Detection System supplied with analytical software (Bio-Rad Laboratories, Inc., Hercules, CA, USA). PCR conditions were as follows: 40 cycles of 95°C for 15 sec, 60°C for 15 sec and 72°C for 45 sec. To determine HSPA2 mRNA expression levels, GAPDH served as an internal control. To examine the expression levels of miR-634, U6 acted as the internal control. The relative expression of miR-634 and HSPA2 was calculated utilizing the comparative 2ΔΔCq method (14). The primers used for amplification were as follows: U6, forward 5′-CTCGCTTCGGCAGCACA-3′ and reverse 5′-AACGCTTCACGAATTTGCGT-3′; GAPDH, forward 5′-AGAAGGCTGGGGCTCATTTG-3′ and reverse 5′-AGGGGCCATCCACAGTCTTC-3′; miR-634, forward 5′-CAGTCTCAAACCAGCACC-3′ and reverse 5′-TATGGTTGTTCACGACTCCTTCAC-3′; and HSPA2, forward 5′-AAACTTTACCAAGGTGGTCCTG-3′ and reverse 5′-GCTTAGTCCACTTCTTCGATGG-3′.
Western blot analysis
Proteins were extracted using radioimmunoprecipitation lysis buffer (Thermo Fisher Scientific, Inc.) and the protein content was determined using the Bicinchoninic Acid Protein assay kit (Beyotime Institute of Biotechnology). Equal amounts of the protein (50 µg/lane) from lysates of PANC-1 cells were subjected to SDS-PAGE (10% gels) and then transferred to polyvinylidene difluoride membranes. The membranes were blocked with 5% non-fat milk for 60 min at room temperature, followed by incubation with primary antibodies against HSPA2 (cat. no. ab108416; 1:500; Abcam, Cambridge, MA, USA) and GAPDH (cat. no. AF0006; 1:1,000; Beyotime Institute of Biotechnology) overnight at 4°C. Membranes were then washed with 0.1% Tween-20 in PBS (PBST) three times at room temperature. Then, they were incubated with horseradish peroxidase-conjugated goat anti-rabbit secondary antibodies (cat. no. sc-2004; 1:3,000; Santa Cruz Biotechnology, Inc. Santa Cruz, CA, USA) at room temperature for 1 h. The proteins of interest were detected by the enhanced chemiluminescence detection system (Sea Biotech, Shanghai, China). Finally, the intensity of protein bands was detected using Image-Pro Plus 6.0 software (Media Cybernetics, Inc., Rockville, MD, USA). GAPDH served as the loading control.
Target prediction
The TargetScanHuman database and TargetScanHuman Release 7.1 software (http://www.targetScan.org) were used to predict the potential target gene of miR-634.
RNA immunoprecipitation (RIP)
RIP assays were performed using the Imprint RNA Immunoprecipitation kit (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) with the protein argonaute-2 (AGO2) antibodies (cat. no. 2897; 1:500; Cell Signaling Technology, Inc., Danvers, MA, USA) or IgG antibodies (cat. no. A6066; 1:500; Sigma-Aldrich; Merck KGaA) for 2 h at room temperature. The mRNA expression levels of miR-634 and HSPA2 in the immunoprecipitates were detected using qPCR analysis.
Dual-luciferase reporter assay
The wild-type (WT) or mutant (MUT) HSPA2-3′UTR which contained the miR-634 binding sites was inserted into the luciferase genes in the pGL3 vectors (Promega Corporation, Madison, WI, USA). PANC-1 cells were co-transfected with 0.1 mg pGL3-WT HSPA2-3′-UTR or 0.1 mg pGL3-MUT HSPA2-3′-UTR and 10 nM miR-634 mimic or 10 nM miR-634 inhibitor using Lipofectamine 2000 reagent. At 48 h after transfection, the cells were gathered and analyzed via Dual-Luciferase Reporter Assay system (GeneCopoeia, Inc.) according to the manufacturer's protocol. The activity of firefly luciferase was normalized to the corresponding Renilla luciferase activity.
Statistical analysis
All the experiments were performed at least three times. Quantitative values were expressed as the mean ± standard error of the mean. Statistical analyses were performed using SPSS 13.0 software (SPSS, Inc., Chicago, IL, USA). Data of more than two groups were analyzed using one-way analysis of variance with Tukey's post hoc test. Statistical analysis of miR-634 and HSPA2 expression between PC tissues and control tissues was evaluated using the paired Student's t-test. The statistical analysis of unpaired two groups was evaluated using an unpaired Student's t-test. The correlations between miR-634 expression levels and the HSPA2 mRNA expression levels in PC tissues were analyzed using Spearman's rank test. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression levels of miR-634 in human PC tissues and cell lines
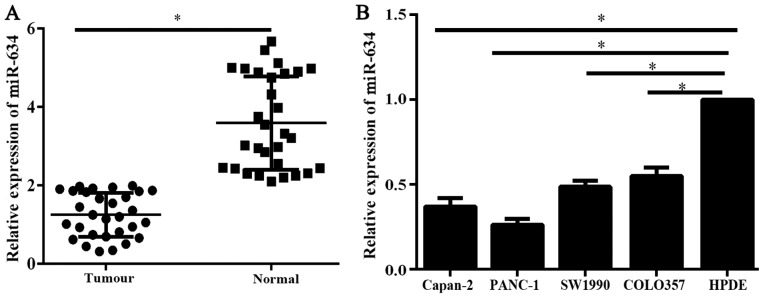
Expression levels of miR-634 in 30 PC tissues and corresponding adjacent normal tissues were analyzed using qPCR. The expression levels of miR-634 in PC tissues was significantly reduced compared with that in adjacent normal tissues (Fig. 1A). Furthermore, the miR-634 expression levels were assessed by RT-qPCR in PC cell lines and the immortal human pancreatic ductal cell line HPDE. Results of RT-qPCR indicated that miR-634 expression levels were significantly reduced in the PC cells compared with HPDE cells (Fig. 1B). These data suggested that the low expression of miR-634 may be associated with the malignant process of PC.
Figure 1.
Expression levels of miR-634 in human PC tissues and cells (A) Relative expression of miR-634 in PC tissues and paired adjacent noncancerous tissues (n=30). (B) Relative expression of miR-634 in PC cell lines and immortal human pancreatic ductal cell line HPDE. *P<0.05 as indicated. miR, microRNA; PC, pancreatic cancer.
miR-634 inhibits the viability and proliferation of PANC-1 cells
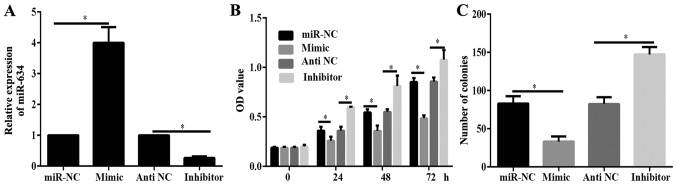
PANC-1 cells were identified to have the lowest expression level of miR-634 (Fig. 1B). Therefore, PANC-1 cells were used for functional testing. PANC-1 cells were transfected with miR-634 mimics, miR-NC, miR-634 inhibitor or antiNC. Results of RT-qPCR indicated that miR-634 mimic could significantly increase miR-634 expression, while miR-634 inhibitor significantly decreased miR-634 expression in PANC-1 cells (Fig. 2A). Results of CCK-8 and colony formation assays demonstrated that the viability and proliferation of cells in the miR-634 mimics group was significantly decreased compared with that in the miR-NC group (Fig. 2B and C). By contrast, the viability and proliferation of cells in the miR-634 inhibitor group was significantly increased than that of cells in the antiNC group (Fig. 2B and C). These data demonstrated that miR-634 inhibited PC growth.
Figure 2.
miR-634 inhibits the viability and proliferation of PANC-1 cells. (A) Relative expression of miR-634 in PANC-1 cells transfected with miR-634 mimics, miR-NC, miR-634 inhibitor or antiNC, respectively. (B) Viability of PANC-1 cells was detected using the Cell Counting Kit-8 assay. (C) Proliferation of PANC-1 cells was detected by colony formation assay. *P<0.05 as indicated. miR, microRNA; NC, negative control; OD, optical density.
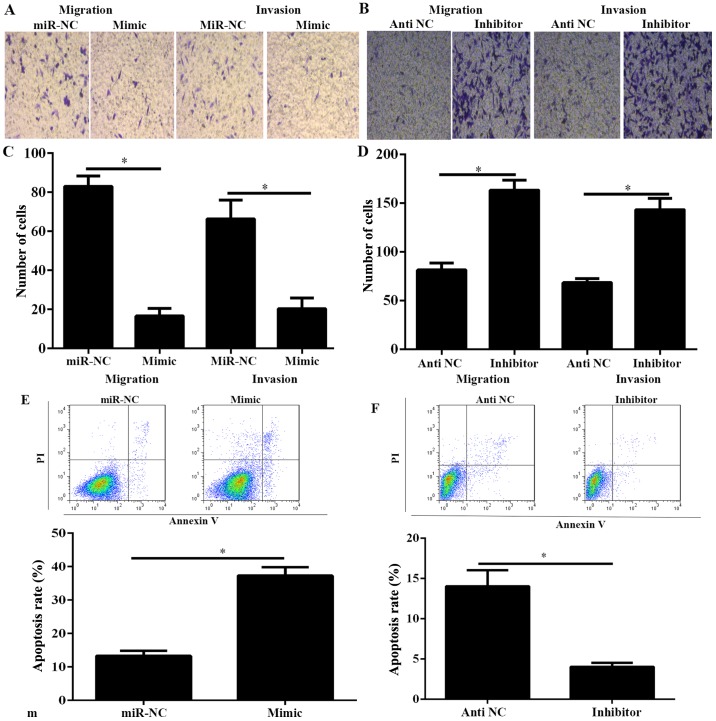
miR-634 inhibits the migration and invasion of PANC-1 cells and enhances the apoptosis rate
PC is characterized by early metastasis (2). Therefore, to demonstrate the function of miR-634 on the migration and invasion of PANC-1 cells, the migration and invasion of PANC-1 cells was investigated using Transwell migration and invasion assays. Results indicated that miR-634 mimic significantly decreased the capability of migration and invasion, whereas miR-634 inhibitor significantly increased the capability of migration and invasion (Fig. 3A-D). Furthermore, cell apoptosis was investigated in PANC-1 cells transfected with miR-634 mimic or inhibitor. As demonstrated in Fig. 3E and F, overexpression of miR-634 could significantly increase the cell apoptosis rate in PANC-1 cells, whereas miR-634 silencing promoted the opposite results.
Figure 3.
miR-634 inhibits migration and invasion, and enhances the apoptosis rate of PANC-1 cells. Transwell assays were used to detect the migration and invasion capability of PANC-1 cells. (A and B) Migrant and invasive PANC-1 cells were fixed, stained and counted, and images were captured (magnification, ×100). (C and D) Quantification of migrant and invasive PANC-1 cells in the lower chamber. (E and F) The apoptosis rate of PANC-1 cells were detected by flow cytometry. *P<0.05 as indicated. miR, microRNA; NC, negative control; PI, propidium iodide.
HSPA2 is a direct target of miR-634 in PANC-1 cells
To identify the molecular mechanism underlying the suppressive role of miR-634 in PC progression, Targetscan was used to predict the potential target genes of miR-634. Analysis indicated that the 3′-UTR of HSPA2 contained predicted binding sites for miR-634 (Fig. 4A). Notably, previous studies have revealed that HSPA2 correlates with PC development and progression (15,16). The protein expression levels of HSPA2 in PANC-1 cells under the regulation of miR-634 were investigated. Results demonstrated that miR-634 mimic markedly decreased the protein expression of HSPA2, whereas miR-634 inhibitor markedly enhanced the protein expression of HSPA2 in PANC-1 cells (Fig. 4B). Furthermore, the dual-luciferase reporter assay was performed to confirm that miR-634 could direct target HSPA2. Results indicated that the luciferase activity in cells transfected with miR-634 mimic was significantly decreased compared with that of cells transfected miR-NC in the HSPA2-3′-UTR-wild type (WT) group; however, there was no significant difference in the HSPA2-3′-UTR-mutant (MUT) group (Fig. 4C). By contrast, luciferase activity was significantly increased in cells treated with miR-634 inhibitor compared with those treated with antiNC in the HSPA2-3′-UTR-WT group (Fig. 4D). However, luciferase activity was not significantly different in the HSPA2-the 3′-UTR-MUT group (Fig. 4D).
Figure 4.
HSPA2 is a direct target of miR-634. (A) Predicted binding sites for miR-634 in the 3′-UTR of HSPA2. (B) The protein expression levels of HSPA2 in PANC-1 cells were detected by western blot anlaysis. (C and D) Luciferase activity was detected using the Dual-Luciferase reporter assay. (E) RNA immunoprecipitation assay indicated the association between miR-634 and HSPA2 transcripts in PANC-1 cells. (F) Quantitative polymerase chain reaction was used to detect the expression levels of HSPA2 in the PC tissues and adjacent normal tissues. (G) Expression levels of miR-634 and HSPA2 mRNA were negatively correlated in PDAC tissues according to Pearson correlation analysis. *P<0.05 as indicated. miR, microRNA; NC, negative control; WT, wild-type; MUT, mutant; 3′-UTR, 3′-untranslated region; HSPA2, heat shock-related 70 kDa protein 2; PDAC, pancreatic ductal adenocarcinoma.
To further confirm the interaction between miR-634 and the HSPA2 3′-UTR, RIP assay was performed. The RIP assay was performed with the AGO2 antibody. In the RNA extracted from the precipitated AGO2 protein, it was possible to detect significantly increased enrichment of the miR-634 and HSPA2 3′-UTR in the Ago2 group compared with IgG in PANC-1 cells (Fig. 4E), indicating that miR-634 and the HSPA2 3′-UTR existed in an RNA-induced silencing complex. These data suggested that miR-634 directly targets HSPA2 by binding to its 3′-UTR region in PANC-1 cells. In addition, RT-qPCR results indicated that the expression level of HSPA2 in PC tissues was significantly increased compared with adjacent normal tissues (Fig. 4F). Furthermore, Pearson correlation analysis demonstrated that miR-634 was negatively correlated with HSPA2 mRNA expression (Fig. 4G). These data demonstrated that HSPA2 is a direct target of miR-634 in PANC-1 cells.
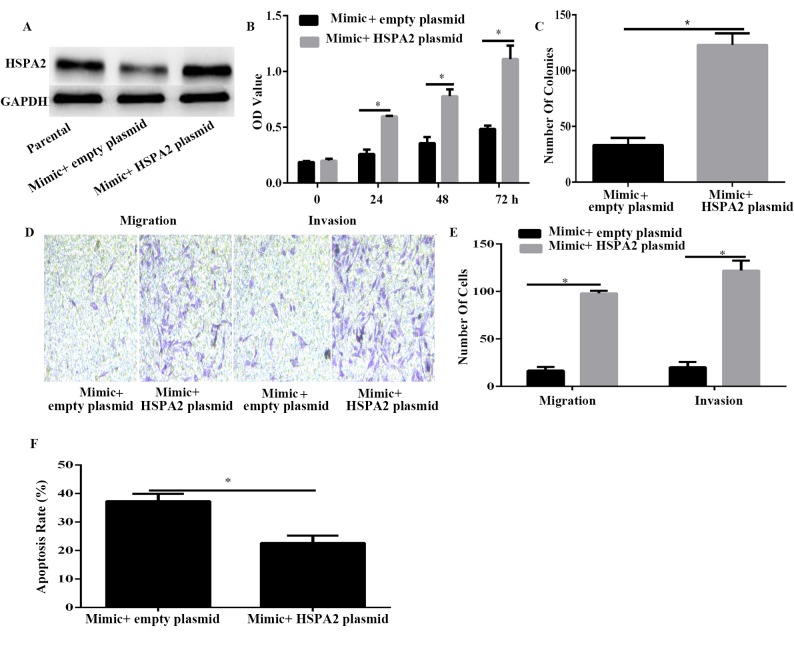
HSPA2 is associated with miR-634-induced suppression in PANC-1 cells
To confirm that HSPA2 was a functional target of miR-634, HSPA2 was overexpressed using HSPA2 plasmid in miR-634-overexpressed PANC-1 cells (Fig. 5A). Results of CCK-8 and colony formation assays demonstrated that HSPA2 overexpression significantly reversed the inhibitory effect induced by miR-634 mimic on cell viability and proliferation (Fig. 5B and C). Furthermore, the Transwell assay results demonstrated that HSPA2 overexpression significantly reversed the inhibitory effect induced by miR-634 mimic on cell migration and invasion (Fig. 5D and E). Furthermore, the results of flow cytometry indicated that HSPA2 overexpression could decrease PANC-1 cell apoptosis induced by miR-634 overexpression. These data indicated that HSPA2 may be a functional mediator of miR-634 in PANC-1 cells.
Figure 5.
HSPA2 is a functional target of miR-634 in PANC-1 cells. PANC-1 cells were transfected with miR-634 and empty plasmid or HSPA2 plasmid. (A) Western blot analysis was used to detect the expression level of HSPA2 in PANC-1 cells. (B) Cell Counting Kit-8 was used to detect the viability of PANC-1 cells. (C) The colony formation assay was used to detect the proliferation of PANC-1 cells. (D) Migrant and invasive PANC-1 cells were fixed, stained and counted, and images were captured (magnification, ×100). (E) Quantification of migrant and invasive PANC-1 cells in the lower chamber. (F) The apoptosis rate of PANC-1 cells was detected by flow cytometry. *P<0.05 as indicated. miR, microRNA; HSPA2, heat shock-related 70 kDa protein 2.
Discussion
miRs act as post-transcriptional gene regulators in the etiology of various pathological events, including apoptosis, proliferation, migration and invasion (6). Abnormal expression of miRs has been considered to impact cancer progression. Several studies have revealed that miR-634 functions as a biomarker and has significant roles in various types of cancer, including glioma, gastric carcinoma, hepatocellular carcinoma, nasopharyngeal carcinoma, ovarian cancer and cervical cancer (7–12). However, to the best of our knowledge, no study has investigated the role of miR-634 in PC thus far. In the present study, it was revealed that miR-634 was significantly downregulated in PC. Furthermore, miR-634 overexpression significantly inhibited the viability, proliferation, migration and invasion of PC cells, whereas its inhibition led to opposite effects. The findings demonstrated that miR-634 acts as a tumor suppressor by suppressing its target gene HSPA2.
To date, few miR-634 targets have been experimentally validated. In the present study, luciferase reporter assays and western blot analysis demonstrated that HSPA2 is a direct miR-634 target. HSPA2, as a testis-specific protein, serves a critical role in spermatogenesis (17). A previous study demonstrated that human tumor tissues can express HSPA2 at high levels (18). The polymorphism of HSPA2 at position 1,267 has been suggested to be associated with carcinogenesis in some types of malignant cancer tissues, including lung cancer, cervical cancer, oesophageal squamous cell carcinoma and hepatocellular carcinoma (19–22). A recent study has indicated that overexpressed HSPA2 is correlated with tumor angiogenesis and poor prognosis in PC, and may have an important role in PC progression and serve as a potential biomarker for the prediction of adverse prognosis in pancreatic carcinoma (15). However, the specific molecular mechanisms underlying the observed increase of HSPA2 in PC remain unclear. In the present study, it was demonstrated that miR-634 specifically targets HSPA2 in human PC cells. HSPA2 expression was upregulated in PC tissues compared with adjacent normal tissues, and the levels were inversely correlated with miR-634 expression in PC tissues. Furthermore, HSPA2 expression was significantly decreased in miR-634-overexpressing cells, and forced HSPA2 expression reversed the phenotypes associated with miR-634 overexpression. Thus, to the best of our knowledge, the present study provided evidence for the first time that miR-634 can suppress PC cell growth by inhibiting HSPA2. Notably, miR-634 can regulate other genes, including CYR61, JAG1, Rab1A and DHX33 (7–9); therefore, further assessment of the molecular mechanism of miR-634 in PC is required. A recent study has revealed that miR-634 could directly regulate Jagged 1 expression in gastric cancer (8); however, the present study failed to identify any significant association between miR-634 and JAG1 in PC (data not shown). This may be due to tissue specificity. Future studies should be performed to further identify the mechanism of miR-634 inhibition on the progression of PC and the biological function of miR-634 in other types of cancer.
In conclusion, the present findings suggested that miR-634 is downregulated in PC and serves as a tumor suppressor by directly targeting HSPA2. Additionally, the results further clarified the importance of the miR-634/HSPA2 molecular network in PC development and may provide a novel therapeutic approach for treating PC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
DRC and XWZ conceived and designed the experiments. DRC, XLW, JWZ and XWZ conducted all of the experiments. JWZ and XWZ wrote and revised the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Medical Ethics Committee of LanLing County Hospital (Shandong, China). All patients included in this research were required to provide written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Li D, Xie K, Wolff R, Abbruzzese JL. Pancreatic cancer. Lancet. 2004;363:1049–1057. doi: 10.1016/S0140-6736(04)15841-8. [DOI] [PubMed] [Google Scholar]
- 3.Oettle H, Post S, Neuhaus P, Gellert K, Langrehr J, Ridwelski K, Schramm H, Fahlke J, Zuelke C, Burkart C, et al. Adjuvant chemotherapy with gemcitabine vs. observation in patients undergoing curative-intent resection of pancreatic cancer: A randomized controlled trial. JAMA. 2007;297:267–277. doi: 10.1001/jama.297.23.2582-a. [DOI] [PubMed] [Google Scholar]
- 4.Diab M, Muqbil I, Mohammad RM, Azmi AS, Philip PA. The role of microRNAs in the diagnosis and treatment of pancreatic adenocarcinoma. J Clin Med. 2016;5:E59. doi: 10.3390/jcm5060059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yonemori K, Kurahara H, Maemura K, Natsugoe S. MicroRNA in pancreatic cancer. J Hum Genet. 2017;62:33–40. doi: 10.1038/jhg.2016.59. [DOI] [PubMed] [Google Scholar]
- 6.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 7.Tan Z, Zhao J, Jiang Y. MiR-634 sensitizes glioma cells to temozolomide by targeting CYR61 through Raf-ERK signaling pathway. Cancer Med. 2018;7:913–921. doi: 10.1002/cam4.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo J, Zhang CD, An JX, Xiao YY, Shao S, Zhou NM, Dai DQ. Expression of miR-634 in gastric carcinoma and its effects on proliferation, migration, and invasion of gastric cancer cells. Cancer Med. 2018;7:776–787. doi: 10.1002/cam4.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang CZ, Cao Y, Fu J, Yun JP, Zhang MF. miR-634 exhibits anti-tumor activities toward hepatocellular carcinoma via Rab1A and DHX33. Mol Oncol. 2016;10:1532–1541. doi: 10.1016/j.molonc.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Jaarsveld MT, van Kuijk PF, Boersma AW, Helleman J, van IJcken WF, Mathijssen RH, Pothof J, Berns EM, Verweij J, Wiemer EA. miR-634 restores drug sensitivity in resistant ovarian cancer cells by targeting the Ras-MAPK pathway. Mol Cancer. 2015;14:196. doi: 10.1186/s12943-015-0464-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cong J, Liu R, Wang X, Jiang H, Zhang Y. MiR-634 decreases cell proliferation and induces apoptosis by targeting mTOR signaling pathway in cervical cancer cells. Artif Cells Nanomed Biotechnol. 2016;44:1694–1701. doi: 10.3109/21691401.2015.1080171. [DOI] [PubMed] [Google Scholar]
- 12.Peng X, Cao P, He D, Han S, Zhou J, Tan G, Li W, Yu F, Yu J, Li Z, Cao K. miR-634 sensitizes nasopharyngeal carcinoma cells to paclitaxel and inhibits cell growth both in vitro and in vivo. Int J Clin Exp Pathol. 2014;7:6784–6791. [PMC free article] [PubMed] [Google Scholar]
- 13.Chun YS, Pawlik TM, Vauthey JN. 8th edition of the AJCC cancer staging manual: Pancreas and hepatobiliary cancers. Ann Surg Oncol. 2018;25:845–847. doi: 10.1245/s10434-017-6025-x. [DOI] [PubMed] [Google Scholar]
- 14.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 15.Zhai LL, Xie Q, Zhou CH, Huang DW, Tang ZG, Ju TF. Overexpressed HSPA2 correlates with tumor angiogenesis and unfavorable prognosis in pancreatic carcinoma. Pancreatology. 2017;17:457–463. doi: 10.1016/j.pan.2017.04.007. [DOI] [PubMed] [Google Scholar]
- 16.Zhang H, Gao H, Liu C, Kong Y, Wang C, Zhang H. Expression and clinical significance of HSPA2 in pancreatic ductal adenocarcinoma. Diagn Pathol. 2015;10:13. doi: 10.1186/s13000-015-0253-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dix DJ, Allen JW, Collins BW, Poorman-Allen P, Mori C, Blizard DR, Brown PR, Goulding EH, Strong BD, Eddy EM. HSP70-2 is required for desynapsis of synaptonemal complexes during meiotic prophase in juvenile and adult mouse spermatocytes. Development. 1997;124:4595–4603. doi: 10.1242/dev.124.22.4595. [DOI] [PubMed] [Google Scholar]
- 18.Scieglinska D, Krawczyk Z. Expression, function, and regulation of the testis-enriched heat shock HSPA2 gene in rodents and humans. Cell Stress Chaperones. 2015;20:221–235. doi: 10.1007/s12192-014-0548-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fu Y, Zhao H, Li XS, Kang HR, Ma JX, Yao FF, Du N. Expression of HSPA2 in human hepatocellular carcinoma and its clinical significance. Tumour Biol. 2014;35:11283–11287. doi: 10.1007/s13277-014-2430-y. [DOI] [PubMed] [Google Scholar]
- 20.Zhang H, Chen W, Duan CJ, Zhang CF. Overexpression of HSPA2 is correlated with poor prognosis in esophageal squamous cell carcinoma. World J Surg Oncol. 2013;11:141. doi: 10.1186/1477-7819-11-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scieglinska D, Gogler-Piglowska A, Butkiewicz D, Chekan M, Malusecka E, Harasim J, Habryka A, Krawczyk Z. HSPA2 is expressed in human tumors and correlates with clinical features in non-small cell lung carcinoma patients. Anticancer Res. 2014;34:2833–2840. [PubMed] [Google Scholar]
- 22.Garg M, Kanojia D, Saini S, Suri S, Gupta A, Surolia A, Suri A. Germ cell-specific heat shock protein 70-2 is expressed in cervical carcinoma and is involved in the growth, migration, and invasion of cervical cells. Cancer. 2010;116:3785–3796. doi: 10.1002/cncr.25218. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.