Abstract
HECT domain and ankyrin repeat containing E3 ubiquitin protein ligase 1 (HACE1) is frequently downregulated or lost in numerous types of cancer, including liver cancer. The aim of the present study was to examine whether demethylation of the HACE1 gene could inhibit tumour progression. The expression of HACE1 was detected in liver cancer cell lines. Clustered regularly interspaced short palindromic repeats (CRISPR)-CRISPR-associated (Cas)-based demethylation single guide RNAs for the HACE1 gene promoter were designed and transfected into liver cancer cells. Subsequently, proliferation was detected by MTT and colony formation assays, and optineurin (OPTN) ubiquitination and microtubule-associated proteins 1A/1B light chain 3B protein levels were detected by immunoblotting. The levels of HACE1 were significantly reduced in liver cancer cell lines compared with in a normal liver cell line. Demethylation of the HACE1 gene promoter increased HACE1 expression, inhibited the proliferation of liver cancer cells, and promoted OPTN ubiquitination and autophagy activity in liver cancer cells. In conclusion, activation of HACE1 expression by promoter demethylation may provide a suitable approach for anticancer therapy.
Keywords: liver cancer cells, HECT domain and ankyrin repeat containing E3 ubiquitin protein ligase 1, demethylation, proliferation
Introduction
Liver cancer has the fourth highest cancer mortality rate worldwide, and China is a country with a high incidence rate of liver cancer, where it accounts for ~40% of the total number of cancer cases and cases of cancer-associated mortality (1,2). Surgery remains the most effective treatment option for patients with liver cancer (3,4). The majority of liver cancer cases are diagnosed at an advanced or unresectable stage; therefore, novel therapeutic strategies and therapeutic targets are required. Current studies mainly focus on the pathogenic genes and molecular mechanisms involved in liver cancer (5,6).
HECT domain and ankyrin repeat containing E3 ubiquitin protein ligase 1 (HACE1) is a member of the HECT domain-containing E3 ligase family and was originally identified to be associated with the occurrence of Wilms' tumour (7). Furthermore, additional studies revealed that lower expression or mutations of HACE1 are associated with numerous types of human malignant tumours, including breast cancer, colorectal cancer, lung cancer, liver cancer, gastric cancer and lymphoma (5,8–12), which suggests that HACE1 functions as a tumour suppressor. Additional research concerning HACE1 has focused on a variety of downstream pathways; for example, HACE1 has been reported to act as a tumour suppressor by ubiquitinating optineurin (OPTN) and activating selective autophagy (10).
In mammalian cells, DNA methylation is critical for the regulation of gene expression and, therefore, serves a pivotal role in numerous physiological and pathological processes (13). DNA hypermethylation of tumour suppressor genes silences their expression and contributes to several types of human cancer (9,14). Targeted DNA demethylation via the widely used clustered regularly interspaced short palindromic repeat (CRISPR)-CRISPR-associated (Cas) system has been widely reported on in recent years (15–17). A strategy for targeted demethylation of specific genomic loci by tethering Tet1-CD to the MS2 RNA element-containing single guide RNA (sgRNA) 2.0 system-guided dCas9 and MS2 bacteriophage coat proteins was the first sgRNA and Cas9-mediated demethylation system to be reported (15,17).
In the present study, low HACE1 expression was identified in human liver cancer cell lines compared with in a normal liver cell line. Subsequently, HACE1 expression was activated via targeted DNA demethylation using a two-plasmid system. Finally, in the present study, increased HACE1 expression was revealed to inhibit proliferation and activate selective autophagy in liver cancer cells.
Materials and methods
Plasmid construction
The pdCas9-Tet1-CD and pcDNA3.1-MS2-Tet1-CD plasmids were provided by Professor Ronggui Hu (Chinese Academy of Sciences, Shanghai, China). sgRNAs targeting HACE1 were designed using an online tool (version 1.2; http://crispr.mit.edu/) as previously described (18). The designed sgRNAs (Table I) were synthesized as oligonucleotides (Sangon Biotech Co., Ltd., Shanghai, China), annealed and inserted into the pdCas9-Tet1-CD vector, which was digested with BbsI.
Table I.
Sequences of the sgRNA target sites used in construction of the pdCas9-Tet1-CD expression plasmids.
| sgRNA for HACE1 | Target site sequence (protospacer adjacent motif region) |
|---|---|
| sgRNA1 | 5′-GCCCTGGGCGGAGTCACGTTGGG-3′ |
| sgRNA2 | 5′-GCGCCCAGGCCACGCCAACGCGG-3′ |
| sgRNA3 | 5′-TGGGCGTACTCCTAAGCTTCTGG-3′ |
| sgRNA4 | 5′-GAGTACGCCCAGTCGCTGCGTGG-3′ |
| sgRNA5 | 5′-CCTGCCGGGCGGCTTTATGAGGG-3′ |
| sgRNA6 | 5′-CCCTCATAAAGCCGCCCGGCAGG-3′ |
| sgRNA7 | 5′-CGTTGATGATGTATGTTGGCTGG-3′ |
HACE1, HECT domain and ankyrin repeat containing E3 ubiquitin protein ligase 1; sgRNA, single guide RNA.
Cell culture and transfection
The human liver cancer cell lines HepG2 and Huh7, and the normal liver cell line L02 were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). The human liver cancer cell line Hep3B was purchased from the American Type Culture Collection (Manassas, VA, USA). All four cell lines have been authenticated by short tandem repeat profiling within the last 2 years. All cell lines were cultured in Dulbecco's modified Eagle's medium supplemented with 10% foetal bovine serum, 100 U/ml penicillin and 100 mg/ml streptomycin (all Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) in a 37°C humidified atmosphere with 5% CO2. For the demethylation experiments, Hep3B or HepG2 cells (104 cells/well) were seeded into plates and the pdCas9-Tet1-CD (10 µg) and pcDNA3.1-MS2-Tet1-CD (8 µg) plasmids were transfected into cells using Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.). The subsequent experiments were performed 48 h post-transfection. For colony formation assays, cell lines with stable expression of the demethylation system were established as previously described (15).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from cells using a total RNA kit (Tiangen Biotech Co., Ltd., Beijing, China). cDNA was synthesized using the ReverTra Ace qPCR RT Master Mix (Toyobo Life Science, Osaka, Japan). The temperature protocol was as follows: Incubation at 37°C for 15 min, 50°C for 5 min, and then 98°C for 5 min. qPCR was performed to assess the relative abundance of HACE1 mRNA using specific primers (Table II) and SYBR Green dye (Toyobo Life Science) on an ABI 7500 fast real-time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.). Thermocycling conditions were as follows: Initial denaturing at 94°C for 2 min followed by 40 cycles of 95°C for 15 sec, 58°C for 15 sec and 72°C for 30 sec. The relative abundance of HACE1 was normalized to that of GAPDH using the 2−ΔΔCq method (19). All data were obtained from at least three independent experiments.
Table II.
Sequences of the primers used in RT-qPCR and bisulphite DNA sequencing.
| Target gene | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|
| Primers for RT-qPCR | ||
| GAPDH | GAGTCAACGGATTTGGTCGTATTG | ATTTGCCATGGGTGGAATCATATTG |
| HACE1 | GCAAGAAATGGGCAGAAGAAATGTA | CATCCTCAACATCAACATCACTGAC |
| Primers for bisulphite | ||
| DNA sequencing | ||
| HACE1 promoter | ATAGGGATATAATATAGTTTAA | AAAAACTATAATTTCCAACTA |
HACE1, HECT domain and ankyrin repeat containing E3 ubiquitin protein ligase 1; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
Bisulphite DNA sequencing
Genomic DNA (gDNA) was extracted from cells of the indicated groups (control cells and cells transfected with sgRNA 1, 4 and 5) using the standard phenol-chloroform extraction method (20). gDNA was treated with bisulphite using the CpGenome Turbo Bisulphite Modification kit (EMD Millipore, Billerica, MA, USA) according to the manufacturer's protocol. The modified DNA was amplified using Platinum Taq DNA Polymerase (Thermo Fisher Scientific, Inc.) with the respective primer sets that recognize bisulphite-modified DNA only (Table II). The cycling parameters were as follows: Initial denaturing at 94°C for 2 min, followed by 33 cycles of 95°C for 15 sec, 58°C for 15 sec and 72°C for 30 sec. Subsequently, the PCR products were cloned into the pMD18-T vector (Takara Bio, Inc., Otsu, Japan) and were sent for Sanger sequencing by Biosune Biotechnology, Co. (Shanghai, China).
Cell proliferation assay
Cells stably transfected with the demethylation system were seeded into a 96-well plate (3,000 cells/well). For this experiment, 6 h post-cell seeding was defined as the 0 h time point. After 0, 24 or 48 h, the cells were incubated with MTT solution (cat. no. C0009; Beyotime Institute of Biotechnology, Haimen, China) for 4 h at 37°C. The formazan product was then dissolved in dimethyl sulfoxide and quantified spectrophotometrically at a wavelength of 570 nm using a microplate reader (Bio-Rad Laboratories, Inc., Hercules, CA, USA). Experiments were performed in triplicate and repeated three times.
Colony formation assay
Cells stably transfected with the demethylation system were seeded into 6-well plates (1,000 cells/well). After 7 days, the cells were fixed with 4% paraformaldehyde (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) on ice for 30 min and stained with 0.1% crystal violet (cat. no. C0121; Beyotime Institute of Biotechnology) for 20 min at room temperature.
IP and immunoblotting
For IP, ~6 million cells transfected with the demethylation system were lysed in IP buffer (50 mM Tris-HCl, 5 mM EDTA, 0.1% SDS and 1% Nonidet P-40) supplemented with a protease inhibitor cocktail (Roche Diagnostics, Basel, Switzerland). Protein concentration of cell lysates was determined using the Pierce™ Bicinchoninic Acid Protein Assay Kit (Thermo Fisher Scientific, Inc.) and 2 mg protein of the whole-cell-lysate was incubated with OPTN antibody (1:100 dilution; cat. no. 10837-1-AP; Proteintech Group, Inc., Chicago, IL, USA) and protein G agarose beads (cat. no. 16-266, Merck KGaA) overnight at 4°C. The immunoprecipitants were enriched and denatured at 100°C for 10 min in 2X SDS-PAGE loading buffer. The whole-cell-lysate input (20 µg), and immunoprecipitants were then separated by SDS-PAGE (4% concentrating gel and 10% separating gel), and transferred onto polyvinylidene difluoride membranes (Bio-Rad Laboratories, Inc.). Membranes were blocked in 5% milk in TBS with 0.1% Tween-20 for 1 h at room temperature. The membranes were incubated at room temperature for 2 h with primary antibodies against ubiquitin (1:1,000 dilution; cat. no. sc-47721; Santa Cruz Biotechnology, Inc., Dallas, TX, USA), OPTN (1:2,000 dilution), HACE1 (1:1,000 dilution; cat. no. ab133637; Abcam, Cambridge, UK), GAPDH (1:5,000 dilution; cat. no. 60004-1-Ig; Proteintech Group, Inc.) and microtubule-associated proteins 1A/1B light chain 3B (LC3; 1:500 dilution; cat. no. L7543; Sigma-Aldrich; Merck KGaA). Membranes were incubated with horseradish peroxidase(HRP)-conjugated goat anti-mouse IgG (H+L) or HRP-conjugated goat anti-rabbit IgG (H+L) secondary antibodies (1:5,000 dilution; cat. nos. G-21040 and 31460; Thermo Fisher Scientific, Inc.) for 1 h at room temperature. Protein was then labelled by adding 1 ml SuperSignal™ West Pico PLUS Chemiluminescent Substrate (cat. no. 34577; Thermo Fisher Scientific, Inc.). The signal was visualized using the Tanon 5200 Imaging System (Tanon Science and Technology Co., Ltd., Shanghai, China).
Statistical analysis
All experiments were performed in triplicate. All values are presented as the means ± standard deviation. One-way analysis of variance was performed with Tukey's post hoc multiple comparisons test using GraphPad Prism software (version 5; GraphPad Software, Inc., La Jolla, CA, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
Lower expression of HACE1 in human liver cancer cell lines
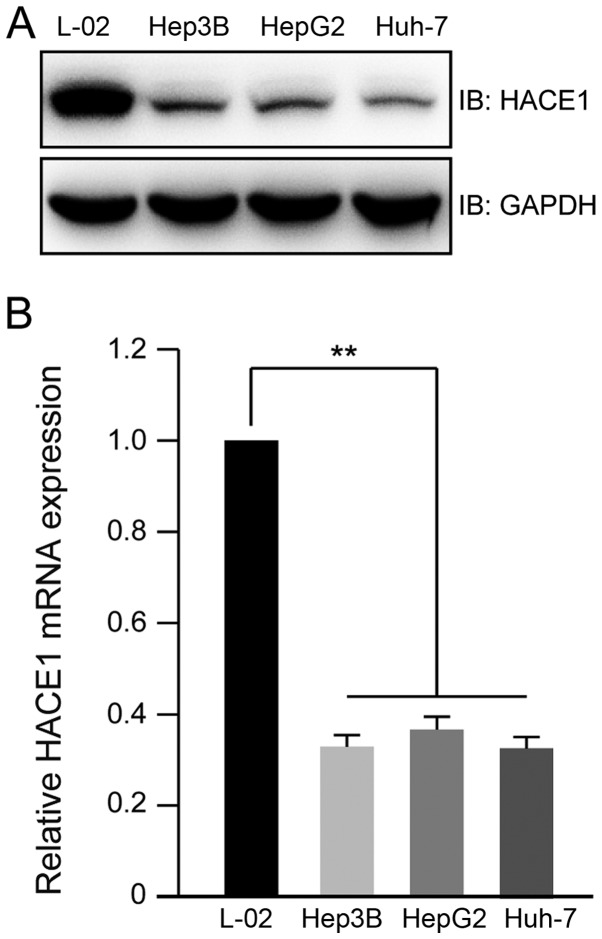
To explore the expression profile of HACE1 in human liver cancer cell lines, lysates from three liver cancer cell lines (Hep3B, HepG2 and Huh-7) and one normal liver cell line (L-02) were prepared. Immunoblot analysis indicated that the protein levels of HACE1 decreased in the three liver cancer cell lines compared with in the normal liver cell line (Fig. 1A). The mRNA expression levels of HACE1 in the normal liver cell line were ~4 times higher than those in the liver cancer cell lines (Fig. 1B).
Figure 1.
Expression profile of HACE1 in normal liver and liver cancer cell lines. (A) Protein expression profile of HACE1 in normal liver and liver cancer cell lines detected by immunoblotting. L-02 is a normal liver cell line; Hep3B, HepG2 and Huh-7 are liver cancer cell lines. (B) mRNA expression levels of HACE1 in normal liver and liver cancer cell lines detected by reverse transcription-quantitative polymerase chain reaction. Data are presented as the means ± standard deviation of three independent experiments and were analysed using one-way analysis of variance with Tukey's post hoc test. **P<0.01. HACE1, HECT domain and ankyrin repeat containing E3 ubiquitin protein ligase 1; IB, immunoblotting.
CRISPR-Cas-based HACE1 promoter demethylation sgRNA design and activity detection
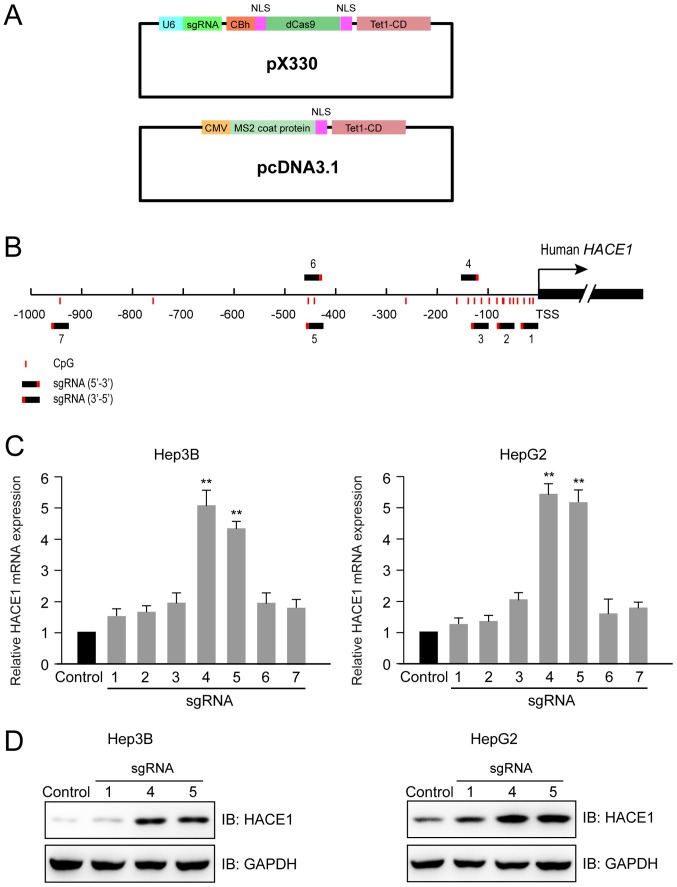
Using the previously described (14) two-plasmid demethylation system (Fig. 2A), seven sgRNAs were designed that targeted regions between −1,000 bp and the transcription start site (TSS) of the human HACE1 gene (Fig. 2B). At 48 h after the transfection of Hep3B and HepG2 cells with dCas9-Tet1-CD (sgRNAs 1–7) and MS2-Tet1-CD, two sgRNAs (4 and 5) were identified to increase the transcription of HACE1 mRNA by 4–5 times compared with the control group with no sgRNA (Fig. 2C). Furthermore, immunoblot analysis indicated that the protein levels of HACE1 were evidently increased in cells transfected with the aforementioned two sgRNAs (4 and 5) compared with the control group or sgRNA 1, which had little effect on HACE1 gene expression and was used as a negative control (Fig. 2D).
Figure 2.
CRISPR-Cas-based HACE1 promoter demethylation sgRNA design and activity detection. (A) Schematic representation of the sgRNA-guided demethylation system expression cassettes. (B) Seven sgRNAs were designed to target regions between −1,000 bp and the TSS of the human HACE1 gene. sgRNAs recognizing their respective target sites were shown in a black-red colour (red colour represents the protospacer adjacent motif region), and the CpG sites were indicated with red lines. (C) HACE1 mRNA expression was detected by reverse transcription-quantitative polymerase chain reaction 48 h after co-transfection with sgHACE1(1–7)-guided dCas9-Tet1-CD and MS2-Tet1-CD in Hep3B and HepG2 cells. The results subsequent to normalization for the control group (without sgRNA) are presented. (D) HACE1 protein expression was detected by immunoblotting 48 h after co-transfection with HACE1 sgRNA (1, 4 and 5)-guided dCas9-Tet1-CD and MS2-Tet1-CD in Hep3B and HepG2 cells. Data are presented as the means ± standard deviation of three independent experiments and were analysed using one-way analysis of variance with Tukey's post hoc test. **P<0.01 compared with control group. Cas, CRISPR-associated; CRISPR, clustered regularly interspaced short palindromic repeats; HACE1, HECT domain and ankyrin repeat containing E3 ubiquitin protein ligase 1; IB, immunoblotting; sgRNA, single guide RNA; TSS, transcription start site.
Upregulation of the target HACE1 gene by specific DNA demethylation
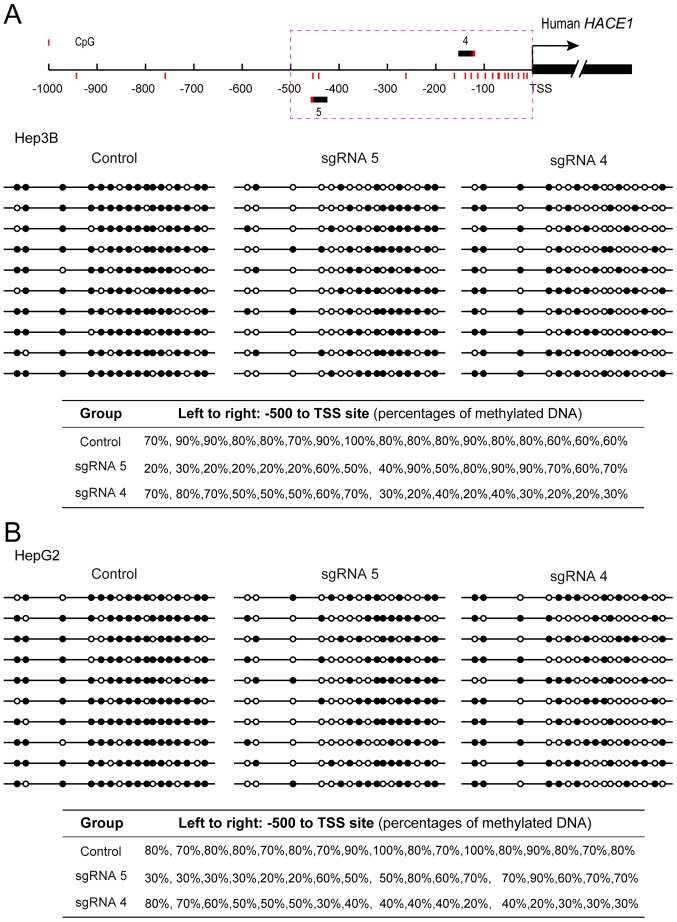
To determine whether the upregulated HACE1 gene transcription was a direct result of targeted demethylation that occurred at a specific HACE1 promoter sequence, the methylation status of sgRNA-targeted loci (between −500 bp and TSS) was examined using a bisulphite sequencing approach. As shown in Fig. 3A and B, expression of the two-plasmid demethylation system resulted in the removal of methyl groups from the neighbouring CpG islands of the HACE1 gene promoter in the Hep3B and HepG2 cell lines. Additionally, the percentages of methylated CpG for each CpG island are shown.
Figure 3.
CRISPR-Cas-based sgRNA system promotes HACE1 promoter demethylation. Percentages of methylated DNA at each site in the HACE1 promoter (between −500 bp and TSS) were determined by bisulphite sequencing 48 h after co-transfection with sgHACE1 (1, 4 and 5)-guided dCas9-Tet1-CD and MS2-Tet1-CD in (A) Hep3B and (B) HepG2 cells. Each line represents an individual clone, and each circle represents an individual CpG island. A black circle represents a methylated CpG and a white circle represents an unmethylated CpG. The percentages of methylated CpG in each CpG island are shown. Cas, CRISPR-associated; CRISPR, clustered regularly interspaced short palindromic repeats; HACE1, HECT domain and ankyrin repeat containing E3 ubiquitin protein ligase 1; sgRNA, single guide RNA; TSS, transcription start site.
Demethylation of HACE1 inhibits human liver cancer cell proliferation and colony formation
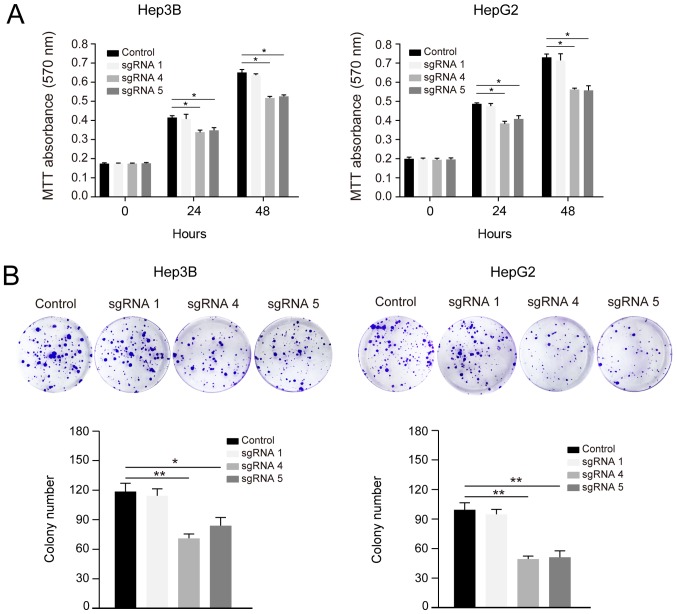
A previous study reported that HACE1 has a tumour-suppressive role in hepatocellular carcinoma (5). To evaluate whether CRISPR-Cas-induced upregulation of HACE1 resulted in physiologically relevant effects, the proliferation of Hep3B and HepG2 cells was detected by MTT assays at 24 and 48 h post transfection. sgRNAs 4 and 5-induced HACE1 upregulation significantly inhibited liver cancer cell proliferation compared with the control and sgRNA 1 groups (Fig. 4A).
Figure 4.
Demethylation of HACE1 inhibits liver cancer cell proliferation and colony formation. (A) sgRNA-mediated HACE1 demethylation inhibited liver cancer cell proliferation. Hep3B and HepG2 cells stably co-transfected with sgHACE1 (1, 4 and 5)-guided dCas9-Tet1-CD and MS2-Tet1-CD were seeded into 96-well plates, and cell proliferation was detected by an MTT assay at different time points. For this assay, 6 h after the cells were seeded was considered as the 0 h time point. Three independent experiments were performed. (B) sgRNA-mediated HACE1 demethylation inhibited liver cancer cell colony formation. Hep3B or HepG2 cells (1,000) stably co-transfected with sgHACE1 (1, 4 and 5)-guided dCas9-Tet1-CD and MS2-Tet1-CD were seeded into 6-well plates. The colonies were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet 7 days after seeding. The colony numbers were counted and calculated using three samples from each group. Data are expressed as the means ± standard deviation and were analysed using one-way analysis of variance with Tukey's post hoc test. *P<0.05, **P<0.01. Cas, clustered regularly interspaced short palindromic repeats-associated; HACE1, HECT domain and ankyrin repeat containing E3 ubiquitin protein ligase 1; sgRNA, single guide RNA.
The demethylation systems containing sgRNAs 4 and 5 were stably expressed in Hep3B and HepG2 cells; subsequently, these cells underwent colony formation assays. Decreased colony numbers were observed in the sgRNA 4 and 5 groups compared with in the control and sgRNA 1 groups (Fig. 4B). These data suggested that CRISPR-Cas-based upregulation of HACE1 resulted in physiologically relevant effects and inhibited liver cancer cell proliferation.
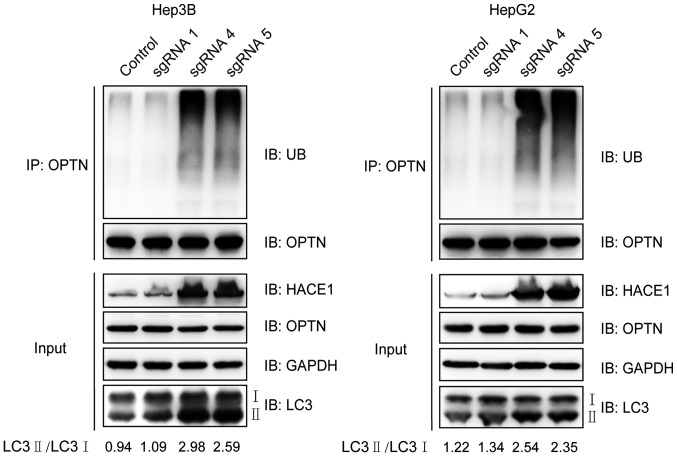
Demethylation of HACE1 promotes OPTN ubiquitination and autophagy in liver cancer cells
Ubiquitination of the autophagy receptor OPTN by HACE1 has been demonstrated to activate selective autophagy, resulting in tumour suppression in lung cancer (10). In the present study, it was investigated if this phenomenon could also occur in liver cancer cells. The ubiquitination of OPTN markedly increased when cells were transfected with demethylation system vectors containing sgRNAs 4 or 5 (Fig. 5). LC3 is a central protein in the autophagy pathway, where it functions in autophagy substrate selection and autophagosome biogenesis. As a result, LC3 is the most widely used marker of autophagosomes. The ratio of LC3 II to LC3 I clearly increased when Hep3B and HepG2 cells were transfected with demethylation system vectors containing sgRNAs 4 or 5 compared with the control and sgRNA 1 groups (Fig. 5). Collectively, these results suggested that increased expression of HACE1 by targeted demethylation may promote OPTN ubiquitination and autophagy activity in liver cancer cells.
Figure 5.
Demethylation of HACE1 promotes OPTN ubiquitination and autophagy. sgRNA-mediated HACE1 demethylation promoted OPTN ubiquitination. Hep3B or HepG2 cells were co-transfected with sgHACE1 (1, 4 and 5)-guided dCas9-Tet1-CD and MS2-Tet1-CD for 48 h. Subsequently, cell lysates were immunoprecipitated with an anti-OPTN antibody and subjected to immunoblotting. LC3 II/I expression was also detected. Inputs refers to whole cell lysates, and GAPDH was used as a loading control. Cas, clustered regularly interspaced short palindromic repeats-associated; HACE1, HECT domain and ankyrin repeat containing E3 ubiquitin protein ligase 1; IB, immunoblotting; LC3, microtubule-associated proteins 1A/1B light chain 3B; OPTN, optineurin; sgRNA, single guide RNA; UB, ubiquitin.
Discussion
HACE1 downregulation has been identified in numerous types of cancer, including hepatocellular carcinoma, breast cancer, colorectal cancer, gastric cancer and leukaemia (5,11). Previous studies have demonstrated that decreased expression or deletion of HACE1 caused by HACE1 methylation or ubiquitination is associated with the occurrence and invasion of various types of carcinoma (18,21–23). HACE1 is a candidate tumour suppressor gene and a potential therapeutic target for several types of human cancer, including liver cancer. The results of the present study indicated that demethylation of the HACE1 promoter enhanced its expression, and inhibited proliferation and colony formation of liver cancer cells. However, a previous study demonstrated that HACE1 promotes melanoma cell migration and adhesion in vitro and that it is required for mouse lung colonization by melanoma cells in vivo (24). These findings indicated that whether HACE1 acts as a tumour suppressor gene or oncogene may depend on the cancer type.
HACE1 was first studied in Wilms' tumour and was later identified to be frequently lost or downregulated in a variety of tumours. Its role in tumour suppression has been extensively investigated, and a number of studies have indicated that HACE1 can restrain reactive oxygen species generation, control cell fate by regulating TNF receptor superfamily member 1A, and impede tumour growth by accelerating the ubiquitination of Rac family small GTPase 1 (11,25,26). Notably, it has been reported that HACE1 acts as a tumour suppressor by ubiquitinating OPTN, and that it activates selective autophagy (10). The present study indicated that demethylation of the HACE1 promoter may lead to OPTN ubiquitination and elevated protein levels of LC3 II.
A previous study indicated that the dCas9-based demethylation system has non-additive effects (14). In the present study, the two-plasmid demethylation system efficiently removed the methyl groups from neighbouring CpG islands on the HACE1 gene promoter in liver cancer cells.
In conclusion, further efforts are required to apply the dCas9-based demethylation system in animal models to investigate liver cancer that is aetiologically caused by HACE1 gene hypermethylation. Activating the expression of HACE1 may be a promising approach for anticancer therapy.
Acknowledgements
The authors would like to thank Professor Ronggui Hu (Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences) for providing the demethylation system plasmids (pdCas9-Tet1-CD and pcDNA3.1-MS2-Tet1-CD) and technical support.
Funding
This study was supported by research grants from the National Natural Science Foundation Youth Fund of China (grant no. 81702622) and Liaoning Province Doctoral Startup Fund (grant no. 201501022).
Availability of data and materials
All data generated or analysed during this study are included in the published article.
Authors' contributions
ZY and ZL conceived and designed the experiments. ZY, YL and TH performed the experiments, collected the data and analysed the results. ZY and ZL wrote the paper.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clinicians. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 3.Waller LP, Deshpande V, Pyrsopoulos N. Hepatocellular carcinoma: A comprehensive review. World J Hepatol. 2015;7:2648–2663. doi: 10.4254/wjh.v7.i26.2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Waghray A, Murali AR, Menon KN. Hepatocellular carcinoma: From diagnosis to treatment. World J Hepatol. 2015;7:1020–1029. doi: 10.4254/wjh.v7.i8.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gao ZF, Wu YN, Bai ZT, Zhang L, Zhou Q, Li X. Tumor-suppressive role of HACE1 in hepatocellular carcinoma and its clinical significance. Oncol Rep. 2016;36:3427–3435. doi: 10.3892/or.2016.5205. [DOI] [PubMed] [Google Scholar]
- 6.Villanueva A, Minguez B, Forner A, Reig M, Llovet JM. Hepatocellular carcinoma: Novel molecular approaches for diagnosis, prognosis, and therapy. Annu Rev Med. 2010;61:317–328. doi: 10.1146/annurev.med.080608.100623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anglesio MS, Evdokimova V, Melnyk N, Zhang L, Fernandez CV, Grundy PE, Leach S, Marra MA, Brooks-Wilson AR, Penninger J, Sorensen PH. Differential expression of a novel ankyrin containing E3 ubiquitin-protein ligase, Hace1, in sporadic Wilms' tumor versus normal kidney. Hum Mol Genet. 2004;13:2061–2074. doi: 10.1093/hmg/ddh215. [DOI] [PubMed] [Google Scholar]
- 8.Goka ET, Lippman ME. Loss of the E3 ubiquitin ligase HACE1 results in enhanced Rac1 signaling contributing to breast cancer progression. Oncogene. 2015;34:5395–5405. doi: 10.1038/onc.2014.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hibi K, Sakata M, Sakuraba K, Shirahata A, Goto T, Mizukami H, Saito M, Ishibashi K, Kigawa G, Nemoto H, Sanada Y. Aberrant methylation of the HACE1 gene is frequently detected in advanced colorectal cancer. Anticancer Res. 2008;28:1581–1584. [PubMed] [Google Scholar]
- 10.Liu Z, Chen P, Gao H, Gu Y, Yang J, Peng H, Xu X, Wang H, Yang M, Liu X, et al. Ubiquitylation of autophagy receptor optineurin by HACE1 activates selective autophagy for tumor suppression. Cancer Cell. 2014;26:106–120. doi: 10.1016/j.ccr.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen YL, Li DP, Jiang HY, Yang Y, Xu LL, Zhang SC, Gao H. Overexpression of HACE1 in gastric cancer inhibits tumor aggressiveness by impeding cell proliferation and migration. Cancer Med. 2018;7:2472–2484. doi: 10.1002/cam4.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang Y, de Reynies A, de Leval L, Ghazi B, Martin-Garcia N, Travert M, Bosq J, Brière J, Petit B, Thomas E, et al. Gene expression profiling identifies emerging oncogenic pathways operating in extranodal NK/T-cell lymphoma, nasal type. Blood. 2010;115:1226–1237. doi: 10.1182/blood-2009-05-221275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robertson KD. DNA methylation and human disease. Nature reviews. Genetics. 2005;6:597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- 14.Sakata M, Kitamura YH, Sakuraba K, Goto T, Mizukami H, Saito M, Ishibashi K, Kigawa G, Nemoto H, Sanada Y, Hibi K. Methylation of HACE1 in gastric carcinoma. Anticancer Res. 2009;29:2231–2233. [PubMed] [Google Scholar]
- 15.Xu X, Tao Y, Gao X, Zhang L, Li X, Zou W, Ruan K, Wang F, Xu GL, Hu R. A CRISPR-based approach for targeted DNA demethylation. Cell Discov. 2016;2:16009. doi: 10.1038/celldisc.2016.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morita S, Noguchi H, Horii T, Nakabayashi K, Kimura M, Okamura K, Sakai A, Nakashima H, Hata K, Nakashima K, Hatada I. Targeted DNA demethylation in vivo using dCas9-peptide repeat and scFv-TET1 catalytic domain fusions. Nat Biotechnol. 2016;34:1060–1065. doi: 10.1038/nbt.3658. [DOI] [PubMed] [Google Scholar]
- 17.Liu XS, Wu H, Krzisch M, Wu X, Graef J, Muffat J, Hnisz D, Li CH, Yuan B, Xu C, et al. Rescue of fragile X syndrome neurons by DNA methylation editing of the FMR1 gene. Cell. 2018;172:979–992 e976. doi: 10.1016/j.cell.2018.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu X, Li C, Gao X, Xia K, Guo H, Li Y, Hao Z, Zhang L, Gao D, Xu C, et al. Excessive UBE3A dosage impairs retinoic acid signaling and synaptic plasticity in autism spectrum disorders. Cell Res. 2018;28:48–68. doi: 10.1038/cr.2017.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Smith-Ravin J, England J, Talbot IC, Bodmer W. Detection of c-Ki-ras mutations in faecal samples from sporadic colorectal cancer patients. Gut. 1995;36:81–86. doi: 10.1136/gut.36.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mettouchi A, Lemichez E. Ubiquitylation of active Rac1 by the E3 ubiquitin-ligase HACE1. Small GTPases. 2012;3:102–106. doi: 10.4161/sgtp.19221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gacon G, Mettouchi A, Lemichez E. The tumor suppressor HACE1 targets Rac1 to ubiquitin-mediated proteasomal degradation. Med Sci (Paris) 2012;28:39–41. doi: 10.1051/medsci/2012281014. [DOI] [PubMed] [Google Scholar]
- 23.Lachance V, Degrandmaison J, Marois S, Robitaille M, Génier S, Nadeau S, Angers S, Parent JL. Ubiquitylation and activation of a Rab GTPase is promoted by a beta(2)AR-HACE1 complex. J Cell Sci. 2014;127:111–123. doi: 10.1242/jcs.132944. [DOI] [PubMed] [Google Scholar]
- 24.El-Hachem N, Habel N, Naiken T, Bzioueche H, Cheli Y, Beranger GE, Jaune E, Rouaud F, Nottet N, Reinier F, et al. Uncovering and deciphering the pro-invasive role of HACE1 in melanoma cells. Cell Death Differ. 2018;25:2010–2022. doi: 10.1038/s41418-018-0090-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cetinbas N, Daugaard M, Mullen AR, Hajee S, Rotblat B, Lopez A, Li A, De Berardinis RJ, Sorensen PH. Loss of the tumor suppressor Hace1 leads to ROS-dependent glutamine addiction. Oncogene. 2015;34:4005–4010. doi: 10.1038/onc.2014.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tortola L, Nitsch R, Bertrand MJM, Kogler M, Redouane Y, Kozieradzki I, Uribesalgo I, Fennell LM, Daugaard M, Klug H, et al. The tumor suppressor hace1 is a critical regulator of TNFR1-mediated cell fate. Cell Rep. 2016;16:3414. doi: 10.1016/j.celrep.2016.08.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analysed during this study are included in the published article.