Abstract
Nano-sized hydroxyapatite (nHA) particles have been demonstrated to exert anti-cancer effects on multiple cancer cell lines and animal models of cancer biology. However, the molecular mechanism underlying the effects of nHA particles on glioma cells remains unclear. The present study aimed to examine the effects of nHA on the behavior of glioma cells and investigate its underlying molecular mechanism. Rat glioma C6 cells and human glioma U87MG ATCC cells were exposed to nHA (20–100 µg/ml), and its effects on cell morphology, viability, apoptosis, cell cycle, invasion and nuclear factor (NF)-κB signaling were analyzed. Exposure of C6 and U87MG ATCC cells to 20 µg/ml nHA for 24 h caused cell detachment. Viability of C6 and U87MG ATCC cells were significantly reduced by nHA in a dose-dependent manner (P<0.05). Nuclear staining with Hoechst 33258 exhibited clear chromatin condensation in C6 cells following 24 h exposure to ≥25 µg/ml nHA. Flow cytometry revealed that nHA (20–100 µg/ml) significantly induced apoptosis and cell cycle G2/M arrest in C6 and U87MG ATCC cells (P<0.05). Transwell invasion assay demonstrated that nHA (20–60 µg/ml) significantly inhibited invasion of U87MG ATCC cells (P<0.05). Furthermore, western blotting and confocal immunofluorescence microscopy revealed that nHA (20–100 µg/ml) decreased NF-κB p65 protein expression and blocked NF-κB p65 nuclear translocation in C6 cells. The protein expression of NF-κB target molecules, such as B cell lymphoma 2, cyclooxygenase-2 and survivin, were also significantly reduced by nHA in a dose-dependent manner in both C6 and U87MG ATCC cells (P<0.05). In conclusion, it was demonstrated that the inhibitory effect of nHA on glioma cells is likely associated with the downregulation of NF-κB signaling.
Keywords: glioma cancer, nano-hydroxyapatite, apoptosis, nuclear factor-κB
Introduction
Hydroxyapatite [HA; Ca10(PO4)6(OH)2] has a superior biocompatibility to human tissue, slow biodegradability in situ, and excellent osteoconductivity and osteoinductivity. Therefore, it has become one of the most favorable biomaterials in many tissue-engineering applications, such as scaffolds for bone repair and bioactive coating on metallic prosthetic implants (1–4). Recently, nano-sized HA (nHA) particles have also been tested as a potential biomaterial for drug delivery. An arginine-modified nHA delivery system has previously been developed and demonstrated to successfully deliver intracellular DNAzymes in both in vitro cell lines and in vivo mouse xenograft (5–7).
In addition to being used for drug delivery, nHA particles have been demonstrated to have an anti-cancer effect on multiple types of cancer cell lines and animal models of cancer. In a rabbit model of hepatic VX2 tumor implant, Hu et al (8) demonstrated that intravenous injection of 20 mg/kg nHA collosol significantly reduced tumor growth. In vitro studies have demonstrated that nHA particles significantly inhibited the proliferation of human colon carcinoma HCT116 cells, lymphatic leukemia P388 cells, gastric cancer SGC-7901 cells, osteoblast-like MG-63 cells, hepatoma HepG2 cells and breast cancer MCF-7 cells (9–14). The molecular and cellular mechanism underlying the anti-cancer effect appears to be associated with induction of apoptosis and cell cycle arrest, downregulation of oncogenes and upregulation of tumor suppressor genes (9–15). Chen et al (9) demonstrated that nHA induced mitochondria-dependent apoptosis in human gastric cancer cells by upregulating the expression of pro-apoptotic protein B cell lymphoma (Bcl)-2-associated X protein and reducing mitochondrial membrane potential and the release of cytochrome C. The expression of oncogene c-Myc in tumor tissue of a rabbit model of implanted hepatic VX2 tumors was significantly reduced by nHA (8), and the expression of the tumor suppressor p53 in human breast cancer MCF-7 cells was increased by nHA (15).
The effect of nHA on glioma cells has also been investigated. Chu et al (16) demonstrated that nHA significantly inhibited the proliferation of human glioma U251 and SHG44 cells in a dose- and time-dependent manner, and tail vein injection of nHA collosol significantly reduced the tumor volume in nude mice transplanted with the human glioma cells. Consistent with the findings from other human cell lines, the inhibitory effect of nHA on glioma cell growth was associated with the stimulation of apoptosis. It has been demonstrated that genetic and morphological characteristics of different glioma cell lines are varied (17). For instance, U87MG ATCC cells express wild type p53, whereas p53 in U251 cells is mutated (18). In addition, the behaviors of U251 and U87MG ATCC cells in mouse xenograft model are also different (17). Therefore, the intracellular signaling pathways regulating key cellular processes, such as proliferation and cell cycle, may vary in these cell lines, resulting in different response to anti-cancer reagents. Therefore, to further investigate the role of nHA in the regulation of glioma development and the underlying molecular mechanism, the effect of nHA was examined on the behavior human glioma U87MG ATCC cells and rat glioma C6 cells.
Materials and methods
Cell culture
Rat glioma cell line C6 and human glioma cell line U87MG ATCC were purchased from American Type Culture Collection (ATCC; Manassas, VA, USA). The cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS; Thermo Fisher Scientific, Inc.), 100 µg/ml penicillin and 100 µg/ml streptomycin and incubated at 37°C with 5% CO2. The U87MG ATCC cell line is known to be misidentified, but a previous study has indicated that this cell line is likely to be derived from a glioblastoma (19).
Preparation of nHA suspension
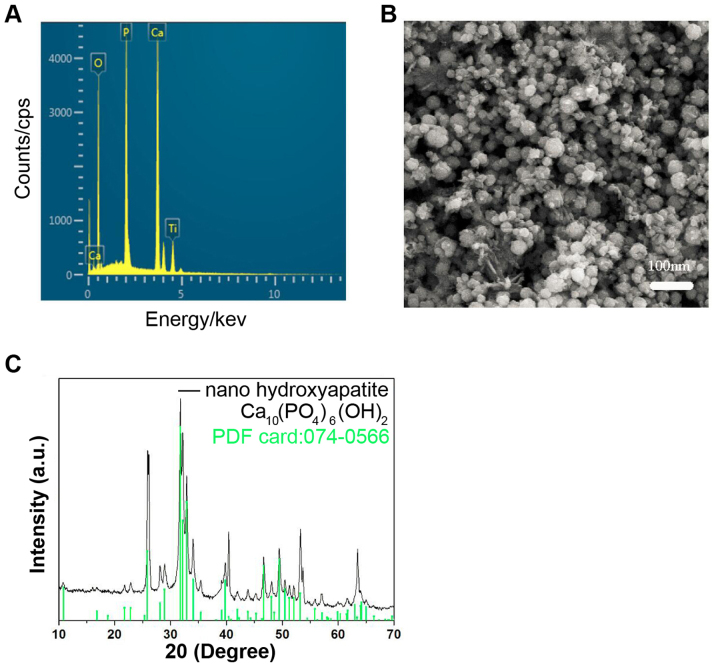
nHA was prepared using the electrode position method. A titanium sheet was used as the cathode, and Pt was used as the anode. The electrolyte solution contained 0.04 M Ca(NO3)2 and 0.01 M NH4H2PO4 with a pH at 5.0–6.0. The preparation temperature was 80°C and the density was 10 mA/cm2. The deposition time was 10 min. The characteristics of the nHA are presented in Fig. 1. Autoclaved nHA (1 mg/ml) was suspended in DMEM media containing 1.5 mg/ml dispersant agent polyethylene glycol-200 (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) and mixed by ultrasound sonication. The nHA suspension was stored at 4°C for future use and sonicated at 24 kHz and 400 W for 5 min prior to use. Following sputter-coating with gold, the nHA was observed using scanning electron microscopy (KYKY-1000B; Chinese Academy of Sciences, Beijing Science Instrument Development Center, Beijing, China) and photographed. The molecular weight of nHA is 1004 and the standard hydroxyapatite XRD profile (file no. 074-0566) was used.
Figure 1.
Characteristics of nHA. (A) The energy dispersive X-ray spectrometry of nHA. (B) Scanning electron microscopy of nHA. (C) X-ray diffraction of nHA. nHA, nano-hydroxyapatite.
Determination of cell viability
Cell viability was determined via MTT assay (Sigma-Aldrich; Merck KGaA). Briefly, C6 and U87MG ATCC cells in the exponential growth phase were collected, re-suspended in DMEM, seeded in 96-well plate at 4×103 cells/100 µl DMEM/well, and incubated at 5% CO2 and 37°C overnight. Subsequently, the cells were exposed to nHA at 0, 20, 40, 60, 80 or 100 µg/ml (0, 19.9, 39.8, 59.7, 79.6 and 99.6 µM, respectively) and incubated for a further 48 h. At 4 h prior to the end of the incubation, 20 µl MTT solution (0.5 mg/ml) was added to the cells. At the end of the incubation, 150 µl dimethylsulfoxide (Sigma-Aldrich; Merck KGaA) was added to terminate the MTT reaction, and the plate was then kept at room temperature (22-25°C) in darkness on a shaker at 80 rpm for 10 min. Absorbance at 490 nm was determined in a plate reader (Thermo Fisher Scientific, Inc.). Six wells were used for each treatment and the experiment was repeated at least 3 times. Wells containing no cells were used as blank controls.
Hoechst staining
Cover glasses were put into a 12-well plate. C6 cells in exponential growth phase were seeded at 5×104 cells/1 ml DMEM/well and incubated at 37°C for 24 h. The cells were then exposed to nHA at 0, 20, 60 or 100 µg/ml in fresh DMEM media and incubated for a further 24 h. Culture media were removed and the cells were then fixed with 2.5% glutaraldehyde (0.5 ml/well) at room temperature for 10 min or at 4°C overnight. The cells were subsequently washed twice with PBS, and 0.5 ml Hoechst 33258 (0.5 µg/ml; Sigma-Aldrich; Merck KGaA) was added in each well. The plate was incubated at room temperature (22-25°C) for 20–30 min and washed 2–3 times at room temperature (22-25°C) with PBS. The cells were observed and images were captured under fluorescent microscopy (magnification, ×100).
Flow cytometry
C6 and U87MG ATCC cells were seeded in 6-well plates at a density of 2×105 cells/well overnight at 37°C and treated with nHA at 0, 20, 40, 60, 80 or 100 µg/ml for 48 h at 37°C. Floating cells in the culture media were collected by centrifugation (1,500 × g at 4°C for 3 min). The attached cells were harvested by trypsinization. The collected cells were washed twice with PBS following centrifugation at 4°C (1,000 × g; 5 min), re-suspended in 250 µl binding buffer (Becton, Dickinson and Company, Franklin, Lakes, NJ, USA) containing 5 µl Annexin V-fluorescein isothiocyanate and 10 µl propidium iodide (Becton, Dickinson and Company), and incubated at room temperature (22-25°C) for 15 min in darkness. The cell suspension was then diluted with 200 µl PBS, filtered through 300-mesh filters, and analyzed using a BD FACSCalibur™ flow cytometer. Apoptosis and cell cycle were analyzed using FlowJo software (Version 10, Tree Star, Inc., Ashland, OR, USA). The experiment was repeated at least 3 times.
Transwell invasion assay
U87MG ATCC cell invasion assay was performed using 24-well Transwell plates (8-µm pore size; Corning Inc., Corning, NY, USA) coated with Matrigel (1 mg/ml; Becton, Dickinson and Company). Cells (5×104/well) were seeded in the upper chambers of the wells in 200 µl serum-free media containing HA at 0, 20, 40 or 60 µg/ml. The lower chambers contained 500 µl DMEM supplemented with 10% FBS. The cells were incubated at 4°C for overnight, and the cells remaining on the upper surface of the chamber were removed using cotton swabs. The cells on the other side of the membrane were fixed at room temperature (22-25°C) with methanol for 10 min and stained with Giemsa solution (Beyotime Institute of Biotechnology, Beijing, China) at room temperature (22-25°C) for 15 min. The penetrated cells at the lower surface of the membrane were counted under a microscope (magnification, ×100; BX51; Olympus Corporation, Tokyo, Japan). A total of 5 fields of view for each membrane were randomly selected and the average cell number of the 5 fields was calculated. The experiment was repeated at least 3 times.
Subcellular localization of nuclear factor (NF)-κB p65
C6 cells in exponential growth phase were cultured in 6-well plates containing cover glasses at a density of 5×104 cells/1 ml DMEM/well and incubated at 37°C for 24 h. The cells were then exposed to nHA at 0, 20, 60, or 100 µg/ml and incubated for a further 24 h. The culture media were removed and the plate was washed with PBS twice for 5 min each. The cells were fixed with 2.5% glutaraldehyde (0.5 ml/well) at room temperature for 15 min, washed with PBS three times for 5 min each, and permeabilized with 0.5 ml 0.2% Triton X-100 at room temperature for 15 min. After the cells were blocked with blocking buffer [10% bovine serum albumin (Beyotime Institute of Biotechology) in PBS] for 1 h at room temperature, the cells were incubated with rabbit anti-rat NF-κB p65 antibody (cat. no. PA5 16545; 1:800 dilution; Thermo Fisher Scientific, Inc.) at room temperature for 1 h. The cells were washed with PBS three times for 5 min each and incubated with anti-rabbit IgG conjugated with Cy3 (cat. no. D111107; 1:1,000 dilution; Boster Biological Technology, Wuhan, China) at room temperature for 1 h in the dark. Following washing with PBS twice for 10 min each, the cells were exposed to Hoechst 33258 (0.5 µg/ml; Sigma-Aldrich; Merck KGaA) at 37°C for 5 min in dark. The cover glass was washed 3 times with PBS, mounted on a slide, and observed under a confocal fluorescent microscope (magnification, ×200, Nikon Corporation, Tokyo, Japan) and images of fluorescence staining were captured.
Western blotting
C6 and U87MG ATCC cells in exponential growth phase were exposed to nHA at 0, 20, 60 or 100 µg/ml for 24 h at 37°C and collected using a cell scraper and centrifugation at 4°C at 300 × g for 5 min. The collected cells were lysed in ice-cold radioimmunoprecipitation assay buffer containing protease inhibitor phenylmethane sulfonyl fluoride and phosphatase inhibitor cocktail (Sigma-Aldrich; Merck KGaA) and centrifuged at 10,000 × g at 4°C for 10 min. The supernatant was collected and protein concentration of the supernatant was determined by bicinchoninic acid assay. A total of 50 µg protein from each sample was loaded in 12% SDS-PAGE gel for electrophoresis. Proteins were then transferred onto a polyvinylidene difluoride membrane. The membrane was blocked with 5% dry skimmed milk in PBS at room temperature for 2 h, washed, and incubated with the following primary antibodies: Rabbit anti-rat or human Bcl-2 (cat. no. 12789-1-AP; 1:1,000 dilution; ProteinTech Group, Inc., Chicago, IL, USA), rabbit anti-rat or human cyclooxygenase (Cox)-2 (cat. no. 12375-1-AP; 1:800 dilution; ProteinTech Group, Inc.), mouse anti-rat or human survivin (cat. no. sc-101433; 1:800 dilution; Santa Cruz Biotechnology, Inc., Dallas, TX, USA), rabbit anti-rat or human vascular endothelial growth factor (VEGF; cat. no. 19003-1-AP; 1:1,000 dilution; ProteinTech Group, Inc.), mouse anti-rat or human GAPDH (cat. no. 60004-1-Ig; 1:2,000 dilution; ProteinTech Group, Inc.), rabbit anti-rat or human matrix metalloproteinase (MMP)-2 (cat. no. 87809; 1:800 dilution; Cell Signaling Technology, Inc., Danvers, MA, USA), rabbit anti-rat or human MMP-9 (cat. no. 13667; 1:1,000 dilution; Cell Signaling Technology, Inc.), rabbit anti-rat or human NF-κB p65 (cat. no. PA5 16545; 1:1,000 dilution; Thermo Fisher Scientific, Inc.), phospho NF-κB p65 (cat. no. 3022; 1:1,000 dilution; Cell Signaling Technology, Inc.) and mouse anti-rat or human β-actin (cat. no. 66009-1-Ig; 1:2,000 dilution; ProteinTech Group, Inc.) at 4°C overnight. Following thorough washing, the membrane was incubated with horseradish peroxidase-conjugated secondary antibodies (cat. nos. ZDR5306 and ZDR5307; 1:2,000 dilution; Beijing Golden Bridge Biotechnology Co., Ltd, Beijing, China) at room temperature for 1 h. The membrane was then washed thoroughly 4 times for 15 min each. The protein signal was developed using an enhanced chemiluminescence detection kit (Thermo Fisher Scientific, Inc.) and exposed on films. Quantity One software (v4.6.6, Bio-Rad Laboratories, Inc., Hercules, USA) was used to quantify the intensity of protein signals. The experiment was repeated at least 3 times.
Statistical analysis
Data are presented as mean ± standard deviation. Statistical analysis was performed using SPSS software (version 17.0; SPSS, Inc., Chicago, IL, USA). The statistical significance of differences was determined using Student's two-tailed t-test in two groups and one-way analysis of variance followed by Dunnett's test in multiple groups. P-values were 2-sided and P<0.05 was considered to indicate a statistically significant difference.
Results
Characteristics of nHA
The energy dispersive X-ray spectrometry demonstrated that the predominant components in the nHA were Ca, P and O, and Ti in the spectrum was from the cathode substrate (Fig. 1A). The particle size of nHA was ~25 nm (Fig. 1B). The X-ray diffraction (XRD) (Fig. 1C) of the nHA revealed that the characteristic peaks of the prepared nHA matched the standard hydroxyapatite XRD profile (standard nHA powder XRD file: 074-0566) well. These results suggested that the nHA was prepared successfully.
nHA significantly reduces glioma cell growth
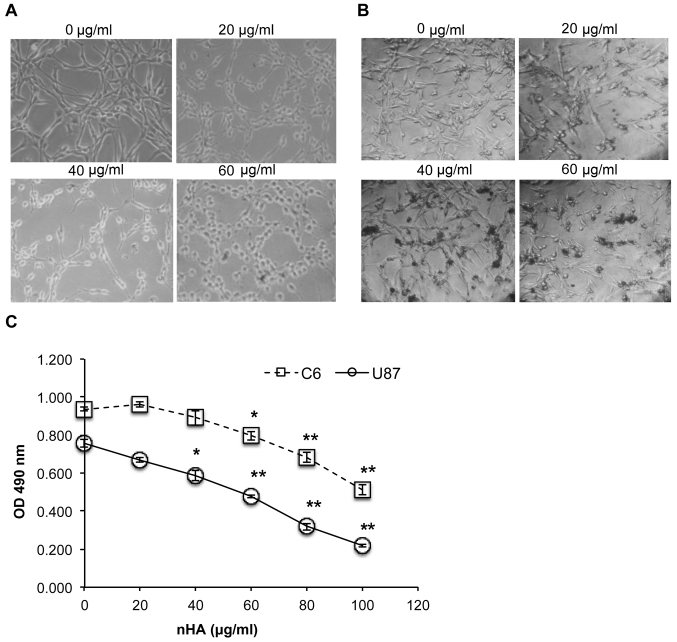
As presented in Fig. 2A and B, the cell morphology of both C6 cells and U87MG ATCC cells was markedly damaged by nHA at ≥20 µg/ml. Following exposure to nHA for 24 h, cells became rounded and began to detach from the tissue culture surface (Fig. 2A and B). MTT assay revealed that cell viability of C6 and U87MG ATCC cells was significantly reduced by nHA in a dose-dependent manner (Fig. 2C). Collectively, the anti-tumor activity of nHA was demonstrated in glioma cells.
Figure 2.
Treatment with nHA damaged cell morphology and reduced cell viability. (A) nHA damaged C6 cell morphology (magnification, ×200). C6 cells were treated with 0, 20, 40 or 60 µg/ml nHA and incubated for 24 h. (B) nHA damaged U87MG ATCC cell morphology. U87MG ATCC cells were treated with 0, 20, 40 or 60 µg/ml nHA and incubated for 24 h. (C) nHA significantly reduced the viability of C6 cells and U87MG ATCC cells. Cells were treated with nHA at the indicated concentration for 48 h and analyzed via MTT assay. The experiment was repeated at least 3 times. *P<0.05; **P<0.01 vs. 0 µg/ml nHA. nHA, nano-hydroxyapatite; OD, optical density.
nHA significantly regulates glioma cell apoptosis, cell cycle and invasion
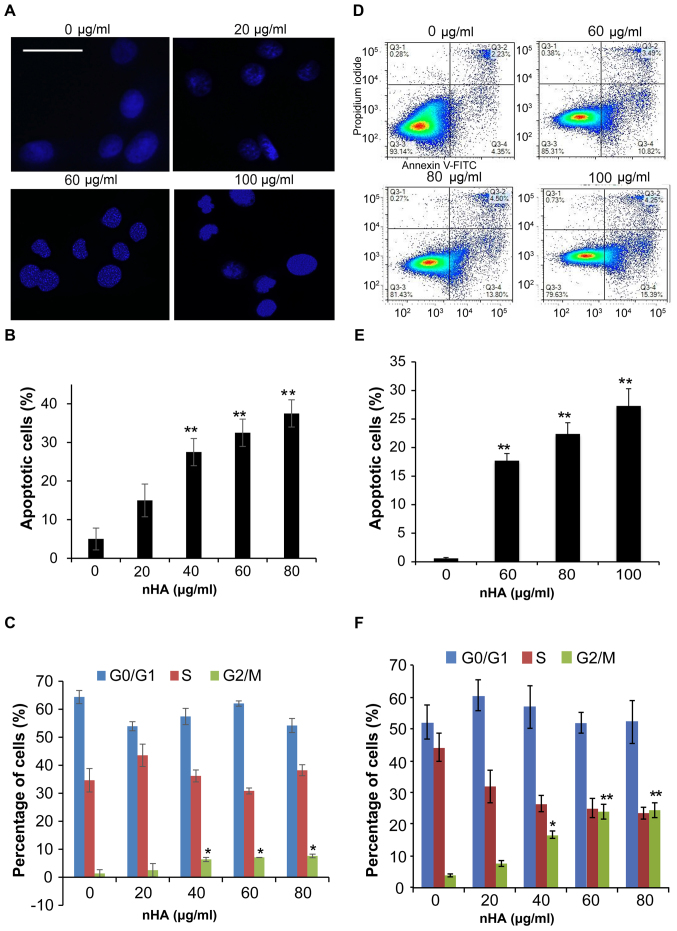
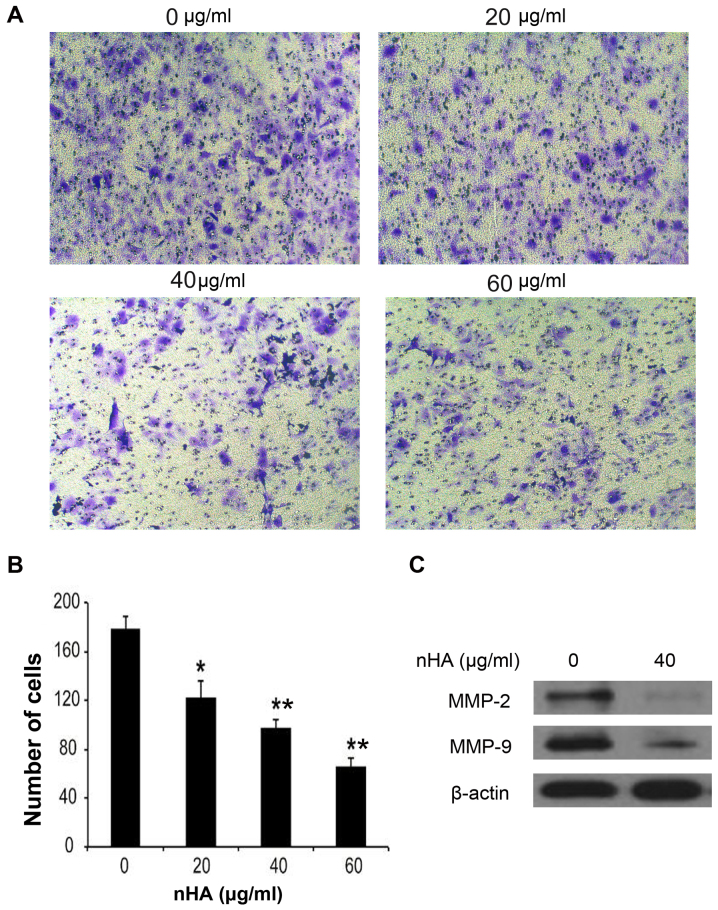
Staining of nuclei of C6 cells with Hoechst 33258 demonstrated that nuclear condensation appeared in C6 cells following 24 h treatment with nHA ≥20 µg/ml (Fig. 3A), and flow cytometry analysis revealed that treatment with nHA for 48 h increased the proportion of apoptotic C6 cells in a dose-dependent manner (Fig. 3B). However, treatment with ≥40 µg/ml nHA for 48 h significantly increased the proportion of cells in G2/M phase, supporting that nHA induces cell cycle G2/M arrest in C6 cells (Fig. 3C). Consistent with the results of C6 cells, exposure of U87MG ATCC cells to nHA induced apoptosis in a dose-dependent manner (Fig. 3D and E). In addition, nHA also induces cell cycle G2/M arrest in U87MG cells (Fig. 3F). In addition to inhibiting cell proliferation and stimulating apoptosis, nHA significantly inhibited the invasion of U87MG ATCC cells in a dose-dependent manner (Fig. 4A and B). The MMP-2 and MMP-9 expression in U87MG ATCC cells were also decreased by nHA (Fig. 4C). These results suggested that nHA functioned as a multiple regulatory factors in glioma cells.
Figure 3.
nHA induced apoptosis. (A) Images of Hoechst 33258 staining of C6 cells. C6 cells were treated with 0, 20, 60 or 100 µg/ml nHA for 24 h and stained with Hoechst 33258. The scale bar represents 30 µm. (B) nHA induced apoptosis of C6 Cells. (C) nHA caused G2/M arrest of C6 cells. (D) Flow cytometry analysis of U87MG ATCC cells treated with nHA. (E) nHA induced apoptosis of U87MG ATCC Cells. (F) nHA caused G2/M arrest of U87MG ATCC cells. *P<0.05; **P<0.01 vs. 0 µg/ml nHA. nHA, nano-hydroxyapatite.
Figure 4.
nHA reduced the invasion of U87MG ATCC cells. (A) Images of invaded U87MG ATCC cells. (B) Number of invaded U87MG ATCC cells following treatment with nHA. U87MG ATCC cells were treated with 0, 20, 40 or 60 µg/ml nHA overnight. (B) The cells in the upper chambers of a Transwell plate were removed and the cells passing through the pore were stained and counted. (C) Images of western blot analysis for MMP-2 and MMP-9. β-actin was used as a loading control. *P<0.05; **P<0.01 vs. 0 µg/ml nHA. nHA, nano-hydroxyapatite; MMP, matrix metalloproteinase.
nHA inhibits NF-κB p65 nuclear translocation
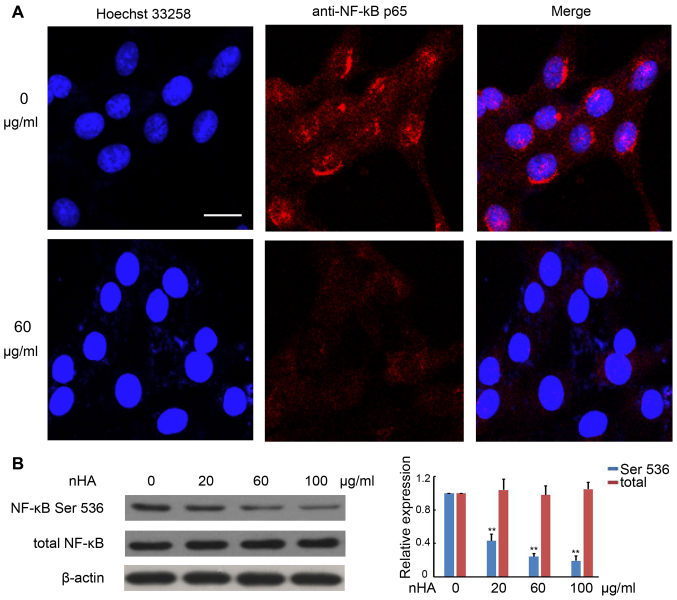
It has been demonstrated that the NF-κB signaling pathway promotes cancer development by suppressing apoptosis (20). Immunofluorescent staining with anti-NF-κB p65 antibody revealed strong punctate staining signals in the nuclei of C6 cells without exposure to nHA, whereas cells treated with nHA exhibited weak diffused staining signals (Fig. 5A). Western blotting further confirmed that protein expression of phosphorylated NF-κB p65 in C6 cells was substantially reduced by nHA treatment in a dose-dependent manner, whereas nHA had no significant effect on total NF-κB expression in C6 cells (Fig. 5B). These results suggest that nHA may reduce NF-κB expression and block its nuclear translocation.
Figure 5.
nHA inhibited NF-κB p65 nuclear translocation and protein expression. (A) Images of fluorescent staining with Hoechst 33258 and anti-NF-κB p65 antibody. C6 cells were treated with 60 µg/ml nHA for 24 h and stained with Hoechst 33258 and anti-NF-κB p65 antibody. Images (original magnification, ×20) were captured using a confocal fluorescent microscope. Experiments were repeated at least 3 times and representative images are presented. The scale bar represents 15 µm. (B) Images of western blotting analysis for NF-κB p65 (Ser 536) and total NF-κB p65 in C6 cells that were treated with 0, 20, 60 or 100 µg/ml for 24 h. β-actin was used as a loading control. The experiment was repeated 3 times. The relative expression of NF-κB was analyzed. **P<0.01 vs. 0 µg/ml nHA. nHA, nano-hydroxyapatite; NF, nuclear factor.
nHA inhibits the protein expression of NF-κB targeting genes
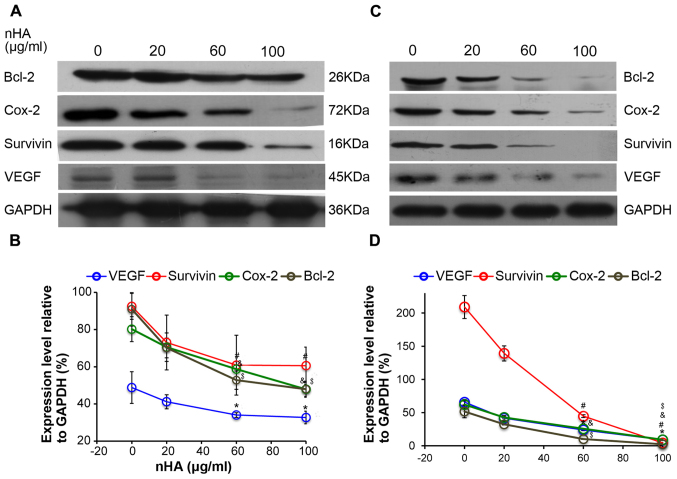
Consistently, the expressions of the downstream NF-κB targeting molecules including Bcl-2, Cox-2, survivin, and VEGF in C6 cells were also reduced by nHA in a dose-dependent manner (Fig. 6A and B). Similarly, nHA markedly reduced the protein expression of the NF-κB targeting molecules Bcl-2, Cox-2, VEGF and survivin in C6 and U87MG ATCC cells (Fig. 6C and D). These findings indicate that nHA may stimulate apoptosis in the glioma cells by inhibiting the NF-κB signaling pathway.
Figure 6.
nHA reduced protein expression of nuclear factor-κB p65 target molecules. (A) Representative images of western blotting of C6 cells. (B) Densitometry analysis of the western blotting of C6 cells. (C) Representative images of western blotting of U87MG ATCC cells. (D) Densitometry analysis of the western blotting of U87MG ATCC cells. C6 and U87MG ATCC cells were treated with 0, 20, 60 or 100 µg/ml nHA for 24 h. The experiment was repeated 3 times. Representative images are presented. *P<0.05, VEGF expression vs. 0 group; #P<0.05, Survivin expression vs. 0 group; &P<0.05, Cox-2 expression vs. 0 group; $P<0.05, Bcl-2 expression vs. with 0 group. nHA, nano-hydroxyapatite; Bcl-2, B cell lymphoma-2; Cox-2, cyclooxygenase-2; VEGF, vascular endothelial growth factor.
Discussion
In the present study, it was demonstrated that nHA significantly inhibited the proliferation and invasion of glioma cells and induced apoptosis and cell cycle G2/M arrest. These results are consistent with the findings of previous studies using the same or different glioma cell lines. Chu et al (16) demonstrated that 48 h exposure to nHA at 120 and 240 µg/ml significantly induced apoptosis of glioma U251 and SHG44 cells, respectively. Glioma U87MG ATCC and C6 cells appeared to be more sensitive to nHA as the present study revealed that nHA induced marked apoptotic morphologic changes in C6 cells and U87MG ATCC cells at 20 µg/ml, and flow cytometry revealed substantial apoptosis of C6 and U87MG ATCC cells at 20 and 60 µg/ml nHA, respectively. Similar to the results of the present study, Xu et al (21) also demonstrated that exposure of C6 cells to nHA at 10 µg/ml for 48 h significantly stimulated apoptosis and reduced cell viability. In addition to inhibiting cell proliferation, it was demonstrated that nHA 20–60 µg/ml reduced U87MG ATCC cell invasion significantly. In an in vitro study to investigate the effects of poly(lactide-co-glycolide) (PLGA)/nHA microspheres as a drug delivery system for temozolomide (TMZ) on U87MG ATCC cellular behavior, Zhang et al (22) demonstrated that addition of nHA in TMZ/PLGA microspheres further reduced U87MG ATCC cell invasion, suggesting that nHA may inhibit U87MG ATCC cell invasion either directly or indirectly by enhancing the anti-cancer effects of TMZ in the microspheres.
A number of previous studies on cancer cell lines have demonstrated that nHA affected the protein expression of molecules involved in apoptosis (9,12–14). The anti-apoptotic protein, Bcl-2, has been consistently demonstrated to be downregulated, whereas caspase-3 and −9 were activated by nHA in human gastric cancer cells, osteoblast-like cells and hepatoma cells (9,12–14). In the present study, the protein level of Bcl-2 in C6 and U87MG ATCC cells was also significantly reduced by nHA in a dose-dependent manner. To further investigate the molecular mechanism underlying nHA-mediated stimulation of apoptosis of glioma cells, the expression and activity of the nuclear factor NF-κB was examined. It has been demonstrated that activation of NF-κB signaling can suppress apoptosis, leading to cancer development (20). In the present study, C6 cells without exposure to nHA presented clear NF-κB p65 activity and nHA treatment reduced NF-κB P65 level and blocked its nuclear translocation. One of the mechanisms by which NF-κB signaling pathway inhibits apoptosis is to neutralize reactive oxygen species (ROS) (23). Xu et al (21) demonstrated that exposure of C6 cells to nHA resulted in intracellular accumulation of ROS. These findings suggest that nHA may induce apoptosis by downregulating the NF-κB signaling pathway. Bcl-2, Cox-2, survivin and VEGF are target genes of NF-κB and contribute to cancer development (24). The present results demonstrated that nHA significantly reduced the protein expressions of these genes in C6 and U87MG ATCC cells. Furthermore, as NF-κB has essential roles in inflammatory responses (20), nHA-mediated inhibition of NF-κB suggests that nHA could have anti-inflammation effects. Future studies are required to further evaluate this hypothesis.
The findings of the present study and previous studies clearly demonstrate that nHA can directly induce apoptosis and inhibit glioma cell proliferation in vitro and in animal models (16,21). In addition, nHA has also been demonstrated to exert therapeutic beneficial effects indirectly. Chu et al (16) demonstrated that combination of nHA and 1,3-bis(2-chloroethyl)-1-nitrosourea (BCNU) significantly reduced BCNU-associated adverse reactions in a mouse xenograft model transplanted with human glioma U251 cells. The same research group also demonstrated that pre-exposure of U251 cells to nHA or pre-treatment of nude mice bearing U251 tumors with nHA prior to irradiation enhanced radiosensitivity of the glioma cells and improved irradiation efficacy (25).
The present study demonstrated that nHA significantly inhibited glioma cell proliferation, induced apoptosis and cell cycle G2/M arrest, and reduced invasion. The molecular mechanism underlying the nHA-mediated anti-cancer effect appeared to be associated with downregulation of the NF-κB signaling pathway.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Natural Science Foundation of China (grant no. 81602186), China Postdoctoral Science Foundation (grant no. 2017M612216) and the Research Award Fund for Outstanding Middle-aged and Young scientists of Shandong Province, China (grant no. BS2011SW004).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
GG and AT conducted all the experiments. ZY and CW participated in the design of the study and assisted in drafting the manuscript. XL conducted the statistical analysis. CF participated in the cell staining. CW designed the project and finalized the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Billström GH, Blom AW, Larsson S, Beswick AD. Application of scaffolds for bone regeneration strategies: Current trends and future directions. Injury. 2013;44(Suppl 1):S28–S33. doi: 10.1016/S0020-1383(13)70007-X. [DOI] [PubMed] [Google Scholar]
- 2.Diez M, Kang MH, Kim SM, Kim HE, Song J. Hydroxyapatite (HA)/poly-L-lactic acid (PLLA) dual coating on magnesium alloy under deformation for biomedical applications. J Mater Sci Mater Med. 2016;27:34. doi: 10.1007/s10856-015-5643-8. [DOI] [PubMed] [Google Scholar]
- 3.Habibovic P, de Groot K. Osteoinductive biomaterials-properties and relevance in bone repair. J Tissue Eng Regen Med. 2007;1:25–32. doi: 10.1002/term.5. [DOI] [PubMed] [Google Scholar]
- 4.Hutmacher DW, Schantz JT, Lam CX, Tan KC, Lim TC. State of the art and future directions of scaffold-based bone engineering from a biomaterials perspective. J Tissue Eng Regen Med. 2007;1:245–260. doi: 10.1002/term.24. [DOI] [PubMed] [Google Scholar]
- 5.Yin B, Ma P, Chen J, Wang H, Wu G, Li B, Li Q, Huang Z, Qiu G, Wu Z. Hybrid macro-porous titanium ornamented by degradable 3D Gel/nHA micro-scaffolds for bone tissue regeneration. Int J Mol Sci. 2016;17:575. doi: 10.3390/ijms17040575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mohamadyar-Toupkanlou F, Vasheghani-Farahani E, Hanaee-Ahvaz H, Soleimani M, Dodel M, Havasi P, Ardeshirylajimi A, Taherzadeh ES. Osteogenic differentiation of MSCs on fibronectin-coated and nHA-modified scaffolds. ASAIO J. 2017;63:684–691. doi: 10.1097/MAT.0000000000000551. [DOI] [PubMed] [Google Scholar]
- 7.Chen Y, Yang L, Huang S, Li Z, Zhang L, He J, Xu Z, Liu L, Cao Y, Sun L. Delivery system for DNAzymes using arginine-modified hydroxyapatite nanoparticles for therapeutic application in a nasopharyngeal carcinoma model. Int J Nanomedicine. 2013;8:3107–3118. doi: 10.2147/IJN.S48321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu J, Liu ZS, Tang SL, He YM. Effect of hydroxyapatite nanoparticles on the growth and p53/c-Myc protein expression of implanted hepatic VX2 tumor in rabbits by intravenous injection. World J Gastroenterol. 2007;13:2798–2802. doi: 10.3748/wjg.v13.i20.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen X, Deng C, Tang S, Zhang M. Mitochondria-dependent apoptosis induced by nanoscale hydroxyapatite in human gastric cancer SGC-7901 cells. Biol Pharm Bull. 2007;30:128–132. doi: 10.1248/bpb.30.128. [DOI] [PubMed] [Google Scholar]
- 10.Dey S, Das M, Balla VK. Effect of hydroxyapatite particle size, morphology and crystallinity on proliferation of colon cancer HCT116 cells. Mater Sci Eng C Mater Biol Appl. 2014;39:336–339. doi: 10.1016/j.msec.2014.03.022. [DOI] [PubMed] [Google Scholar]
- 11.Li G, Huang J, Li Y, Zhang R, Deng B, Zhang J, Aoki H. In vitro study on influence of a discrete nano-hydroxyapatite on leukemia P388 cell behavior. Biomed Mater Eng. 2007;17:321–327. [PubMed] [Google Scholar]
- 12.Shi Z, Huang X, Cai Y, Tang R, Yang D. Size effect of hydroxyapatite nanoparticles on proliferation and apoptosis of osteoblast-like cells. Acta Biomater. 2009;5:338–345. doi: 10.1016/j.actbio.2008.07.023. [DOI] [PubMed] [Google Scholar]
- 13.Shi Z, Huang X, Liu B, Tao H, Cai Y, Tang R. Biological response of osteosarcoma cells to size-controlled nanostructured hydroxyapatite. J Biomater Appl. 2010;25:19–37. doi: 10.1177/0885328209339396. [DOI] [PubMed] [Google Scholar]
- 14.Yuan Y, Liu C, Qian J, Wang J, Zhang Y. Size-mediated cytotoxicity and apoptosis of hydroxyapatite nanoparticles in human hepatoma HepG2 cells. Biomaterials. 2010;31:730–740. doi: 10.1016/j.biomaterials.2009.09.088. [DOI] [PubMed] [Google Scholar]
- 15.Meena R, Kesari KK, Rani M, Paulraj R. Effects of hydroxyapatite nanoparticles on proliferation and apoptosis of human breast cancer cells (MCF-7) J Nanopart Res. 2012;14:712. doi: 10.1007/s11051-011-0712-5. [DOI] [Google Scholar]
- 16.Chu SH, Feng DF, Ma YB, Li ZQ. Hydroxyapatite nanoparticles inhibit the growth of human glioma cells in vitro and in vivo. Int J Nanomedicine. 2012;7:3659–3666. doi: 10.2147/IJN.S33584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacobs VL, Valdes PA, Hickey WF, De Leo JA. Current review of in vivo GBM rodent models: Emphasis on the CNS-1 tumour model. ASN Neuro. 2011;3:e00063. doi: 10.1042/AN20110014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Radaelli E, Ceruti R, Patton V, Russo M, Degrassi A, Croci V, Caprera F, Stortini G, Scanziani E, Pesenti E, Alzani R. Immunohistopathological and neuroimaging characterization of murine orthotopic xenograft models of glioblastoma multiforme recapitulating the most salient features of human disease. Histol Histopathol. 2009;24:879–891. doi: 10.14670/HH-24.879. [DOI] [PubMed] [Google Scholar]
- 19.Allen M, Bjerke M, Edlund H, Nelander S, Westermark B. Origin of the U87MG glioma cell line: Good news and bad news. Sci Transl Med. 2016;8:354re3. doi: 10.1126/scitranslmed.aaf6853. [DOI] [PubMed] [Google Scholar]
- 20.Van Antwerp DJ, Martin SJ, Kafri T, Green DR, Verma IM. Suppression of TNF-alpha-induced apoptosis by NF-kappaB. Science. 1996;274:787–789. doi: 10.1126/science.274.5288.787. [DOI] [PubMed] [Google Scholar]
- 21.Xu J, Xu P, Li Z, Huang J, Yang Z. Oxidative stress and apoptosis induced by hydroxyapatite nanoparticles in C6 cells. J Biomed Mater Res A. 2012;100:738–745. doi: 10.1002/jbm.a.33270. [DOI] [PubMed] [Google Scholar]
- 22.Zhang D, Tian A, Xue X, Wang M, Qiu B, Wu A. The effect of temozolomide/poly(lactide-co-glycolide) (PLGA)/nano-hydroxyapatite microspheres on glioma U87 cells behavior. Int J Mol Sci. 2012;13:1109–1125. doi: 10.3390/ijms13011109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karin M. Nuclear factor-kappaB in cancer development and progression. Nature. 2006;441:431–436. doi: 10.1038/nature04870. [DOI] [PubMed] [Google Scholar]
- 24.Naugler WE, Karin M. NF-kappaB and cancer-identifying targets and mechanisms. Curr Opin Genet Dev. 2008;18:19–26. doi: 10.1016/j.gde.2008.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chu SH, Karri S, Ma YB, Feng DF, Li ZQ. In vitro and in vivo radiosensitization induced by hydroxyapatite nanoparticles. Neuro Oncol. 2013;15:880–890. doi: 10.1093/neuonc/not030. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.