Abstract
Long non-coding RNAs (lncRNAs) have been shown to play important roles in many human diseases. However, their functions and mechanisms in tumorigenesis and development remain largely unknown. Here, we demonstrated that focally amplified lncRNA in epithelial cancer (FALEC) was upregulated and significantly correlated with lymph node metastasis, TNM stage in gastric cancer (GC). Further experiments revealed that FALEC knockdown significantly inhibited GC cells migration and invasion in vitro. Mechanistic investigations demonstrated that small interfering RNA-induced silencing of FALEC decreased expression of the nearby gene extracellular matrix protein 1 (ECM1) in cis. Additionally, ECM1 and FALEC expression were positively correlated, and high levels of ECM1 predicted shorter survival time in GC patients. Our results suggest that the downregulation of FALEC significantly inhibited the migration and invasion of GC cells through impairing ECM1 expression by exerting an enhancer-like function. Our work provides valuable information and a novel promising target for developing new therapeutic strategies in GC.
Keywords: lncRNA, FALEC, ECM1, migration and invasion, gastric cancer
Introduction
Gastric cancer (GC) is the third leading cause of cancer-related death worldwide, and imposes a significant health burden worldwide, especially in East Asia (Rahman et al., 2014; Ferlay et al., 2015; Torre et al., 2015). In spite of the rapid advances in medical technology such as surgery, radiation and chemotherapy during recent decades, GC patients are usually diagnosed at an advanced stage with distant metastases which results in a low 5-year survival rate (Macdonald et al., 2001; Gupta and Massague, 2006; Ferlay et al., 2015). Metastasis is responsible for as much as 90% of cancer-associated mortality, yet it remains one of the most enigmatic aspects and is a poorly understood component of cancer pathogenesis (Chaffer and Weinberg, 2011). Therefore, identifying metastasis-related genes and unraveling their underlying molecular mechanism involved in metastasis will provide potential molecular targets and may help deliver new effective therapies for GC patients with pre-established metastases.
Carcinogenesis and development of GC are complex biological process characterized by various molecular abnormalities, including both genetic and epigenetic alterations. With the advancement of high-throughput sequencing technology, common long non-coding RNA species exceeding 200 nucleotides in length and without protein-coding potential have recently aroused great interest (Ponting et al., 2009). Mounting evidences have indicated that lncRNAs involved in the occurrence and development of malignant tumors (Guttman et al., 2011; Wang and Chang, 2011; Schmitt and Chang, 2016) through a variety of mechanisms, including chromosome remodeling, RNA processing and stability, transcription and post-transcriptional regulation (Kapranov et al., 2007; Khalil et al., 2009; Cesana et al., 2011; Heo and Sung, 2011; Lee, 2012; Xing et al., 2014). Indeed, several lncRNAs associated with tumor development and progression such as HOTAIR, ANRIL, H19, PVT1 and GAPLINC, have been well characterized in GC and shown to have aberrant expression (Hu Y. et al., 2014; Li et al., 2014; Fang et al., 2015; Nasrollahzadeh-Khakiani et al., 2017). However, although the functions of lncRNAs are gradually being understood, the underlying mechanisms of lncRNA-related regulation are still remains largely uncharacterized.
Recently, non-coding RNAs with enhancer-like (eRNA) activity have become one of the most interesting fields in the understanding of gene transcription because of their wide transcription from enhancer DNA elements (Kim et al., 2010; Lam et al., 2014; Mao et al., 2018), and the knockdown of eRNAs would decrease nearby or target gene expression by cis-regulation (Orom et al., 2010). The enhancer-like lncRNA FALEC, located in close proximity to the tumor metastasis-associated ECM1 gene, has attracted our attention (Orom et al., 2010; Lee et al., 2015). Retrospective analysis showed that FALEC contributed to carcinogenesis by regulating the expression of AKT1, p21, E2F, and acting as a ceRNA of miR-1236, affecting the PTEN/AKT signaling pathway and targeting epithelial-mesenchymal transition-related genes by trans-regulation (Hu X. et al., 2014; Jeong et al., 2016; Pan et al., 2017; Li B. et al., 2018; Li Y. et al., 2018; Wang et al., 2018). In the present study, we found that the expression of FALEC was significantly upregulated in GC tissues compared with paired adjacent non-tumor tissues. RNA interference-based loss-of-function assays found that FALEC knockdown inhibited the migration and invasion ability of GC cells through impairing ECM1 expression by exerting its enhancer-like function in cis. Our results not only reveal a novel molecular regulatory mechanism of FALEC in GC, but also indicate a potential biomarker and therapeutic target for individualized treatment of GC patients.
Materials and Methods
Tissue Collection and Ethics Statement
A total of 60 matched pairs of GC tissues and adjacent normal tissues were obtained from patients who underwent surgical resection without any neoadjuvant treatment between May 2016 and July 2018 at the First Affiliated Hospital of Wannan Medical College, China. Tumor–node–metastasis (TNM) staging of GC samples was performed by two senior pathologists. All tissue specimens after surgery were immediately frozen in liquid nitrogen for subsequent RNA isolation. Written informed consent was obtained from all GC patients, and the study was performed with the approval of the Medical Ethical Committee of Wannan Medical College.
Cells Culture and Transfection
The human GC cell lines SGC-7901, BGC-823, AGS, MKN-45, MGC-803, HGC-27 and the immortalized human gastric epithelial mucosa cell line GES-1 were purchased from the Institute of Biochemistry and Cell Biology of the Chinese Academy of Sciences (Shanghai, China) and the Cell Resource Center of Institute of Basic Medical Sciences, Chinese Academy of Medical Sciences and Peking Union Medical College (Beijing, China). Cells were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS, Wisent, St-Bruno, PQ, Canada), 100 U/ml penicillin and 100 mg/ml streptomycin (Invitrogen, Carlsbad, CA, United States) in a 37°C incubator supplied with humidified air containing 5% CO2. SiRNA and antisense oligodeoxy nucleotides (ASO) sequences were designed and synthesized by GenePharma (GenePharma, Shanghai, China) and RiboBio (RiboBio, Ltd., Guangzhou, China), all the sequences for siRNAs and ASO were listed in Supplementary Table S1. GC cells were transfected with the siRNA at a final concentration of 25 nmol/L using Lipofectamine 2000 (Invitrogen, CA, United States) and ASOs at the final concentration of 100 nM using Oligofectamine transfection reagent (Invitrogen) according to the manufacturer’s instructions.
RNA Extraction, cDNA Synthesis and Quantitative Real-Time (qRT) PCR Analysis
DNA extraction of GC cells was performed with the Genomic DNA Mini Preparation Kit (Beyotime Biotechnology, China) following the manufacturer’s instructions. Total RNA from GC cell lines and tissues was extracted using the TRIzol reagent (Invitrogen, Carlsbad, CA, United States), and reverse transcription was performed with the PrimeScriptTM RT reagent Kit with gDNA Eraser (Takara, Dalian, China) with random hexamers on 1 μg of RNA. Relative expression levels of FALEC and ECM1 were determined by qRT-PCR using the YBR® Premix Ex TaqTM II kit (Takara, Dalian, China) according to the manufacturer’s instructions. Primers used for qRT-PCR analyses were synthesized by GENEWIZ (Suzhou, China) and are listed in Supplementary Tables S2, S3. The relative expressions of interested genes were normalized to the expression of β-actin. Gene expression was analyzed using the 2-ΔΔCt method, and all experiments were performed in triplicate.
In vitro Cell Migration and Invasion Assays
Wound healing and transwell invasion and migration assays were performed as described previously (Cui et al., 2015). Briefly, a scratch was generated across confluent cell monolayers using a 10 μl sterile pipette tip followed by supplemented with serum-free medium after washed with phosphate buffered solution (PBS) to remove floating cells. In vitro cell migration and matrigel (BD Biosciences, San Jose, CA, United States) invasion assays were performed using the transwell system (8-μm pore size with polycarbonate membrane; Corning Costar, MA, United States). Cells were seeded into the top chamber in serum-free medium, and medium containing 10% fetal bovine serum was placed in the bottom chamber as an attractant. Cells migrating through the pores or invading through the matrigel were fixed with methanol and stained with 0.5% crystal violet after 36 h of incubation. Images were obtained at 20x magnification by using a microscope (Olympus, Tokyo, Japan).
Cloning of pGL3-Promoter-FALEC-Reverse Reporters and Luciferase Assay
pGL3-Promoter was digested with BamHI and SalI, and FALEC coding sequence was synthesized by General Biosystems (General Biosystems, Inc., Chuzhou, China) was cloned into the restriction enzyme sites sites 5′ upstream to the luciferase gene. Luciferase assays were performed in 96-well plates using the Dual-Luciferase® Reporter (DLRTM) Assay (Promega, Madison, WI, United States) according to the manufacturer’s protocol, and the relative luciferase activity was normalized to the Renilla luciferase activity.
Western Blot Assay
Whole-cell lysates were separated by 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred to 0.22 μm polyvinylidene fluoride membranes (Millipore). After blocking the membrane with 5% skimmed milk for 2 h at room temperature, it was incubated with specific anti-ECM1 (Bioworld Technology, Inc.) and anti-β-actin (Sigma-Aldrich) antibodies overnight at 4°C. Protein detection was performed with the enhanced chemiluminescence (ECL) system (SuperSignal; Pierce, United States).
Bioinformatic Analysis
Comparative analysis of FALEC and ECM1 expression levels and correlation analysis were obtained from Gene Expression Profiling Interactive Analysis (GEPIA1, a web server for cancer and normal gene expression profiling and interactive analyses) (Tang et al., 2017). The expression of ECM1 in GC and its correlation with clinical information such as GC grade and subtype was evaluated using online analysis tool UALCAN (Chandrashekar et al., 2017). Kaplan-Meier survival analysis2 was used to determine the influence of ECM1 expression on GC prognosis, and the log-rank test was utilized to compare survival curves between high and low ECM1 expression groups (Szasz et al., 2016).
Statistical Analysis
All data are presented as means ± SD and all statistical analyses were performed using SPSS 20.0 software (IBM Corp., Armonk, NY, United States) or GraphPad Prism 5 software (GraphPad Software, Inc., La Jolla, CA, United States). Differences in the expression levels of FALEC and ECM1 between tumor and non-tumor tissues were analyzed using the chi-squared test (χ2 test). The difference in ECM1 expression regulated by FALEC was estimated using the Student’s t-test. The association between FALEC expression and pathological features of GC was analyzed with the chi-squared (χ2) test. The correlation between FALEC and ECM1 expression levels was analyzed using Pearson’s correlation coefficient test. Survival curves were conducted using the Kaplan-Meier method and the difference was analyzed by log-rank test. All p-values presented were two-sided and the difference was considered to be statistically significant at ∗p < 0.05 and ∗∗p < 0.01.
Results
FALEC Was Upregulated in Human GC Cell Lines and Gastric Cancer Tissue Specimens
To investigate the possible role of FALEC in the progression of GC, we first detected the expression of FALEC in six human GC cell lines and 60 pairs of GC tissues and paired non-tumorous tissues. As shown by qRT-PCR analysis, FALEC expression was higher in GC cell lines and tumor tissues than controls and non-tumorous tissues (Figure 1A,B). The highest and lowest levels of FALEC were detected in HGC-27 (cell line established from lymph node with metastasis from GC) and AGS (established from human primary GC) cells indicated that FALEC may be a putative driver of metastasis (Figure 1A). Further statistical analysis found that FALEC expression was remarkably higher in tumor tissues compared with adjacent non-tumor tissues (Figure 1C). Next, we divided the samples into high (≥2-fold increase in GC tissues compared with adjacent non-tumor tissues) and low (less than 2-fold) FALEC expression groups to explore correlations between FALEC expression and clinicopathological characteristics in GC patients. As shown in Table 1, although no significant associations between FALEC expression and gender, age, Lauren’s classification, or histological type were detected, the expression of FALEC was significantly correlated with TNM and lymph node metastasis stage. An in-depth analysis found that FALEC expression significantly higher in GC tissues with lymph node metastasis than in non-lymph node metastasis (Figure 1D).
FIGURE 1.
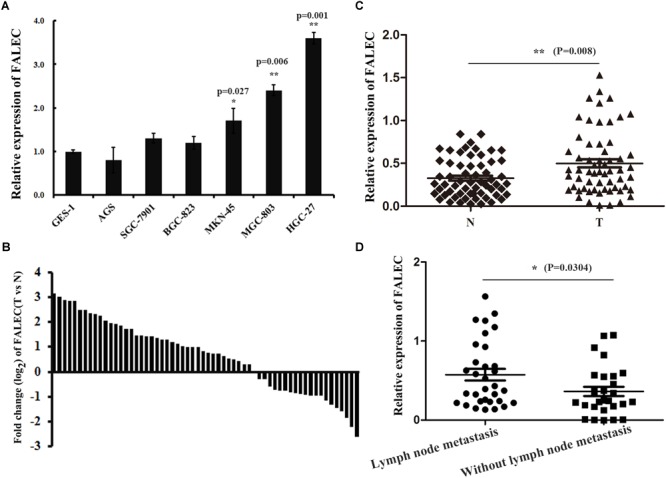
The expression of FALEC was upregulated in GC cell lines and tissues. (A) Relative expression of FALEC in GC cell lines and immortalized normal gastric epithelial cell line (GES-1) was examined by qRT-PCR. (B) The fold change (log2) of FALEC expression between 60 pairs of GC and adjacent normal tissues was measured by qRT-PCR. (C) FALEC expression in GC tissues and paired adjacent normal tissues were examined by qRT-PCR (n = 60), T (Tumor) and N (Non-tumor). (D) Patients with lymph node metastasis showed significantly higher levels of FALEC compared with no lymph node metastasis. ∗P < 0.05, ∗∗P < 0.01.
Table 1.
The relationship of FALEC with the clinicopathological characteristics in patients with GC (n = 60).
| Feature | Expression of FALEC |
F | p-value | |
|---|---|---|---|---|
| T > N | T ≤ N | |||
| Age | 0.019 | 0.891 | ||
| <60 | 12 | 9 | ||
| ≥60 | 23 | 16 | ||
| Gender | 0.099 | 0.753 | ||
| Female | 14 | 9 | ||
| Male | 21 | 16 | ||
| Lauren’s classification | 0.096 | 0.757 | ||
| Intestinal type | 14 | 11 | ||
| Diffuse type | 21 | 14 | ||
| Lymph node metastasis | 5.173 | 0.023* | ||
| No | 12 | 16 | ||
| Yes | 23 | 9 | ||
| Histological grade | 5.813 | 0.055 | ||
| High | 6 | 10 | ||
| Moderate | 9 | 8 | ||
| Poor | 20 | 7 | ||
| TNM staging | 6.251 | 0.012* | ||
| Stage I/II | 11 | 16 | ||
| Stage III/IV | 24 | 9 | ||
Chi-square test, ∗indicates P-value < 0.05.
Reduction of FALEC Expression Suppressed GC Cell Migration and Invasion in vitro
The amplification of a gene can lead to its overexpression such as EGFR and MYC are contained in frequently-amplified regions (Beroukhim et al., 2010). Hu et al. found that FALEC exhibited copy-number gains and losses in cancer (Athie and Huarte, 2014). But, we did not detect significant amplification of FALEC DNA (Supplementary Table S3), therefore, the overexpression of FALEC may not due to the DNA amplification in GC cells.
To explore the potential role of FALEC in GC progression, three siRNA pairs and two ASO targeting FALEC were synthesized to screen for the most effective sequence in HGC-27 and MGC-803 cells. The most efficient siRNA 2 and ASO 1 were chosen for subsequent functional analysis (Figure 2A,B). Because the over-expression of FALEC is significantly associated with lymph node metastasis in GC patients, we first explored its effect on the migration and invasion of GC cells. After transiently transfecting with siRNA or negative control into the GC cell lines, qRT-PCR was used to detect the interference efficiency before cell scratch and transwell assays were performed (Figure 2C). As shown in Figure 2D, the FALEC knockdown resulted in a slower recovery after wounding in HGC-27 (left) and MGC-803 (right) cells compared with the control. Also as expected, FALEC knockdown produced a significant reduction in migration and invasive ability in HGC-27 (Figure 2E) and MGC-803 (Figure 2F) cell lines compared with control by transwell assays. Taken together, these results indicate that knockdown of FALEC expression attenuates migration and invasion in GC cell lines, thereby suppressing the malignant progression of GC.
FIGURE 2.
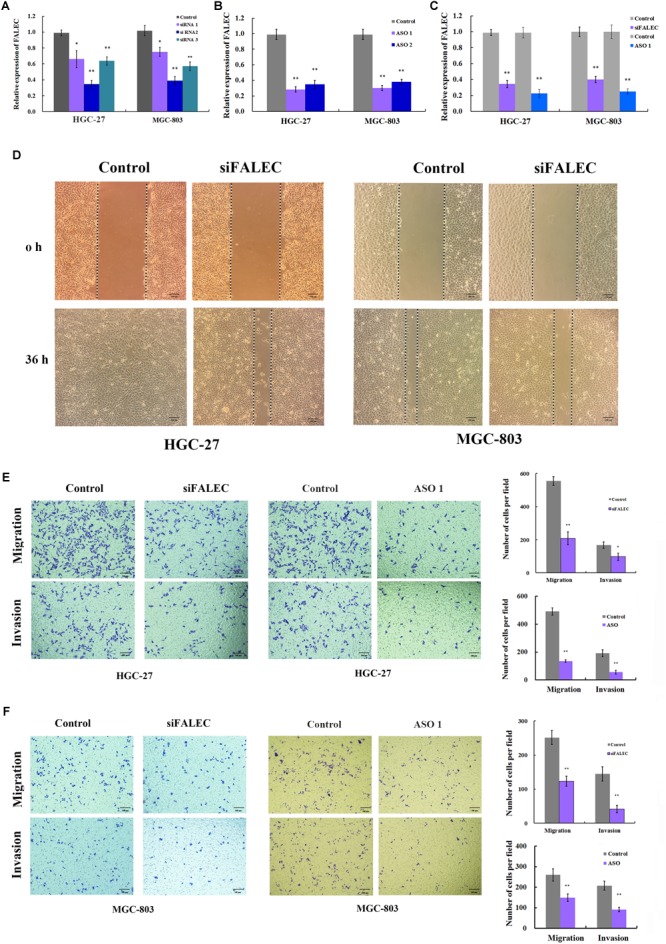
Knockdown of FALEC inhibited the cell migration and invasion in GC cells. (A,B) The interference efficiency of three siRNAs and two ASOs targeting to FALEC was analyzed by qRT-PCR in HGC-27 and MGC-803 cells. (C) The knockdown efficiency was measured by qRT-PCR when cell scratch and transwell assays were performed. (D) Knockdown of FALEC attenuated cell migration in HGC-27(left) and MGC-803 (right) cell lines. Images were acquired at 0 and 36 h after scratches were generated. Depletion of FALEC significantly inhibited HGC-27 (E) and MGC-803 (F) cells migration and invasion. Representative images of transwell results were taken under 100× original magnifications. The numbers of migrated or invaded cells were counted in five fields under the microscope from three independent experiments. ∗P < 0.05, ∗∗P < 0.01.
Depletion of FALEC Attenuated Nearby ECM1 Expression in GC Cell Lines
Previous studies demonstrated that a subset of enhancers was transcribed into a class of eRNAs, and that eRNA knockdown would decrease expression of nearby or target genes by cis-regulation (Kim et al., 2010; Li et al., 2013; Hsieh et al., 2014). To test this hypothesis, we interrogated the FALEC locus and found that protein-coding genes TARS2, RPRD2, ECM1, ADAMTSL4, MCL1 and ENSA were within this genomic region (Figure 3A). To determine whether FALEC has an enhancer-like function, we explored the effect of FALEC on the transcription of nearby genes in HGC-27 and MGC-803 cell lines. As shown in Figure 3B, the depletion of FALEC led to a significantly reduced the expression of its adjacent protein-coding gene ECM1, but did not affect the other genes surrounding FALEC in mRNA level in HGC-27 and MGC-803 cells. These results implied that FALEC may functions as an enhancer element in specifically regulating the expression of ECM1 by cis, and this effect was specific to the ECM1, as we did not detect any changes in the other genes surrounding FALEC. Moreover, ECM1 knockdown did not affect the expression level of FALEC or any of the other adjacent genes further supporting the fact that FALEC is an independent regulator of ECM1 (Figure 3C). Together, these results implied that FALEC might functions as an eRNA to activate ECM1 expression in GC cell lines.
FIGURE 3.
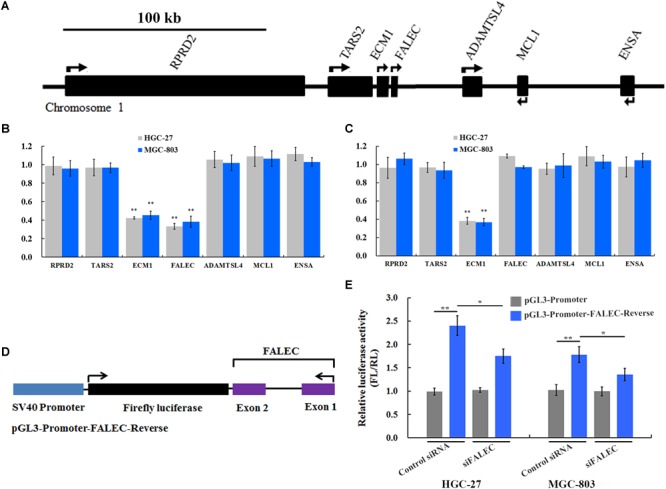
FALEC modulated ECM1 expression by exerting its enhancer-like function in cis. (A) The relative loci of genes adjacent to FALEC. The scale bar represents 100 kb. (B) Depletion of FALEC specifically decreased expression of protein-coding gene ECM1 in HGC-27 and MGC-803 cells. (C) siRNA-induced knockdown of the ECM1 did not affect the transcription of its neighboring genes. ∗∗P < 0.01. (D) Diagram of the FALEC DNA sequence cloned down-stream of Firefly luciferase in a pGL3-Promoter vector (pGL3-Promoter-FALEC-Reverse). (E) FALEC inserts result in a significant enhancement of transcription in GC cells, and depletion of FALEC resulted in a significant decrease in transcriptional enhancement compared with control in HGC-27 and MGC-803 cells. ∗P < 0.05, ∗∗P < 0.01.
FALEC Modulated the Expression of ECM1 by Exerting Its Enhancer-Like Function
Previous studies showed that an important property of enhancers that stimulates transcriptional activity is independent of the orientation of the DNA sequence (Khoury and Gruss, 1983). To dissect the regulatory mechanisms of the FALEC-activation on the expression of ECM1, we constructed a vector in which the FALEC sequence is reversed (pGL3-Promoter-FALEC-Reverse) (Figure 3D) and cloned down-stream of Firefly luciferase in the pGL3-Promoter vector to assess its orientation independence and enhancement of transcription. As shown in Figure 3E, FALEC inserts result in a significant enhancement of transcription in HGC-27 and MGC-803 cells compared with control. To further demonstrate that the observed potentiation of gene expression is mediated through the action of FALEC, we knocked down the FALEC using siRNAs, depletion of FALEC resulted in a significant decrease of in transcriptional enhancement compared with control in HGC-27 and MGC-803 cells (Figure 3E). Taken together, these experiments demonstrated that FALEC modulated ECM1 expression by exerting its enhancer-like function although the underlying nature of FALEC in the regulation of ECM1 needs further investigation.
Knockdown of FALEC Inhibited GC Cell Migration and Invasion Partly by Downregulating ECM1
Previous research suggested that ECM1 plays pivotal roles in cancer cell migration and invasion (Xiong et al., 2012; Gomez-Contreras et al., 2017), so we examined whether ECM1 affected the migration and invasion ability of the GC cells. SiRNA was used to knockdown the expression of ECM1, and the silencing efficiency was assessed by qRT-PCR (Figure 4A) and western blotting (Figure 4B) in HGC-27 and MGC-803 cells. As shown in Figure 4C,D, ECM1 knockdown significantly impaired the migration and invasion ability of GC cells compared with control. To further investigate whether FALEC affected cell migration and invasion by down-regulating ECM1, we used siRNA and transwell assays to evaluate the cell migration and invasion abilities of the two genes. Compared with the control group, simultaneous silencing of ECM1 and FALEC significantly inhibited the migration and invasion ability of GC cells more than ECM1 alone or the control group in GC cells (Figure 4C,D). These results indicated that FALEC knockdown inhibits cell migration and invasion partly by down-regulating ECM1 in GC cells.
FIGURE 4.
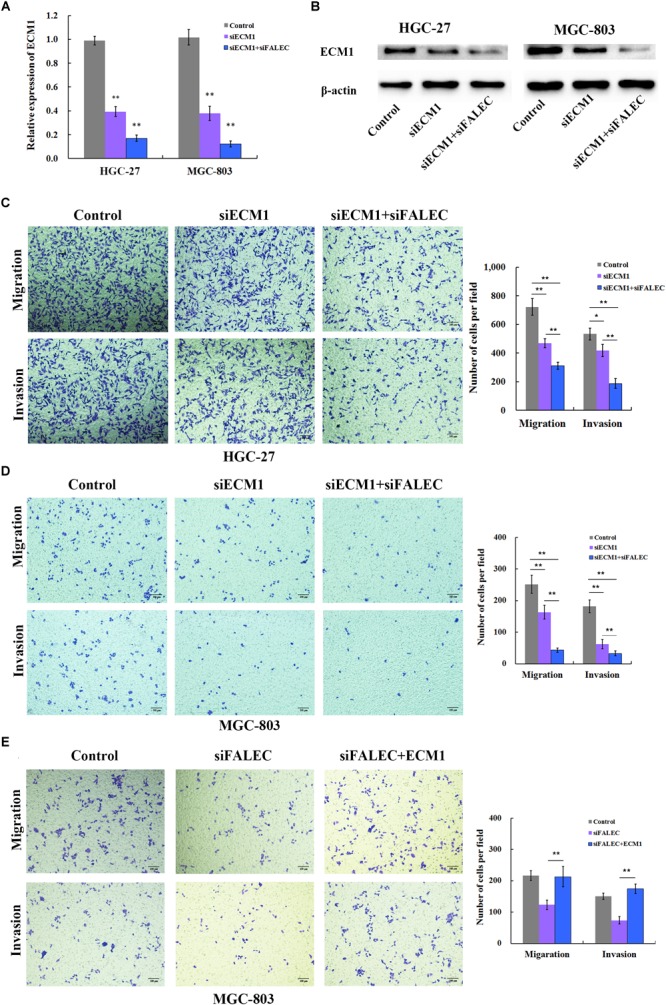
FALEC knockdown decreased migration and invasion partly by down regulating ECM1 in GC cells. (A,B) Relative expression of ECM1 was examined after ECM1 silencing alone or simultaneously silencing FALEC and ECM1 in HGC-27 and MGC-803 cells by qRT-PCR and western blotting assay. Compared with control, ECM1 knockdown significantly impaired cell migration and invasion ability of HGC-27 and MGC-803 cells (C,D). Similarly, the knockdown of FALEC and ECM1 simultaneously resulted in significant decreased the migration and invasion in GC cells than sciencing ECM1 alone or control group (C,D). (E) ECM1 overexpression markedly restored the abrogated migration and invasion of GC cells inhabited by FALEC.
To clarify that ECM1 was the functional target of FALEC, a rescue experiment was conducted in GC cells. We ectopically expressed ECM1 by transfecting of pcDNA3.1(+) containing the ECM1-coding sequence together with siRNA for FALEC into MGC-803 cells. The transwell assay clearly demonstrated that ECM1 overexpression markedly restored the abrogated migration and invasion of GC cells inhabited by FALEC (Figure 4E). This result further confirmed that FALEC-targeted ECM1 is involved in GC cell migration and invasion.
Elevated ECM1 Expression Is Significantly Positively Associated With Increased FALEC Expression and Predicted Poor Prognosis in GC
To explore the relationship between ECM1 and FALEC mRNA expression and their clinical significance, we investigated ECM1 expression levels in GC specimens using the GEPIA online database. As shown in Figure 5A, ECM1 was significantly upregulated in tumor tissues compared with non-tumorous tissues. A remarkable positive correlation was found between ECM1 and FALEC mRNA expression in GC (Figure 5B), further indicating that FALEC exerts enhancer effects to promote ECM1 expression, thereby promoting migration and invasion in GC cells. Next, we employed the UALCAN online database to analyze the expression of ECM1 in different stages and histological subtypes of GC patients. Box plot showing that the expression of ECM1 in subtypes of GC samples were significantly higher than normal except for intestinal adenocarcinoma (Mucinous) (Figure 5C), and the expression level of ECM1 was relatively high compared to the normal gastric tissues for GC patients (Figure 5D). Finally, we performed a correlation analysis of ECM1 expression and clinical outcome of GC patients using a Kaplan–Meier plotter. Kaplan-Meier survival plots showed that high ECM1 expression in GC patients was associated with shorter overall survival (OS) (Figure 5E) and progression-free survival rates (PFS) (Figure 5F) compared with patients with low ECM1 expression. These results above suggested that FALEC exerts enhancer activity to promote the ECM1 expression and participate in the development of GC.
FIGURE 5.
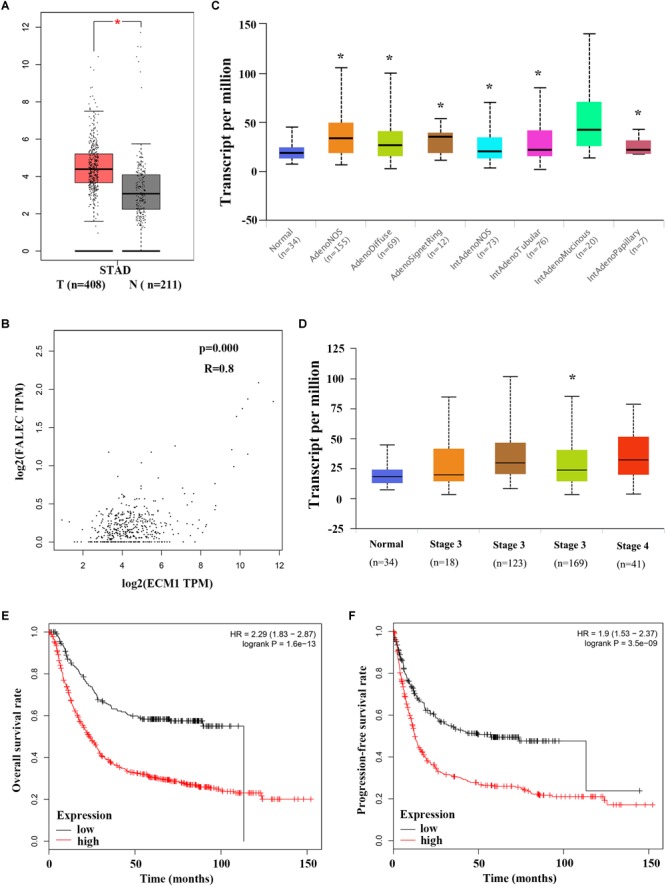
Elevated ECM1 expressions is significantly positively associated with increased FALEC expression in patients with GC. (A) ECM1 expression in GC tumor tissues was significantly higher than paired adjacent non-tumor samples, and a remarkable positive correlation was found between ECM1 and FALEC mRNA expression in GC by using GEPIA database (B). (C,D) shows a box plot that ECM1 expression in different histological subtypes and stages in GC patients using UALCAN database. Kaplan-Meier survival plots shown that GC patients with high ECM1 expression have reduced lower overall (E) and progression-free survival (F) rates compared with patients with low expression level of FALEC. ∗P < 0.05, ∗∗P < 0.01.
Discussion
Aberrant expression of lncRNAs that involved in the regulation of critical malignant biological behaviors such as cell proliferation and metastasis has been comprehensively reported in human cancers (Yan et al., 2015; Li et al., 2016; Hao et al., 2017; Chiu et al., 2018). However, although previous studies have demonstrated that abnormal lncRNA expression is involved in GC pathogenesis, but the regulators involved in lncRNAs dysregulation and underlying mechanisms in GC remained to be elucidated (Sun et al., 2016).
Long non-coding RNAs exert their regulatory functions in cis or in trans and are involved in various biological processes (Ulitsky and Bartel, 2013; Cech and Steitz, 2014). Previous studies revealed that FALEC contributed to carcinogenesis by trans-regulating the expression of genes such as P21 and AKT1 involved in the malignant proliferation, invasion, and metastasis of tumor cells (Hu X. et al., 2014; Zhong et al., 2015; Jeong et al., 2016; Ni et al., 2017; Pan et al., 2017; Zhao et al., 2017; Li B. et al., 2018; Li Y. et al., 2018; Wang et al., 2018). FALEC is a validated 566 nt lncRNA located at 1q21.2, which has been confirmed to be upregulated in carcinogenesis. However, little is known about the detailed function and mechanism of FALEC in cis in tumorigenesis and tumor development.
In our present study, we found the expression level of FALEC in GC tissues was significantly higher than in paired non-tumor tissues (Figure 1B). The high expression was positively correlated with lymph node metastasis and TNM stage (Table 1). Our results further proved that FALEC regulated the expression of ECM1 by exerting its eRNA function in cis, thus affecting the migration and invasion ability of GC cells (Figure 2D, 3B,C,E, 4B–D).
Generally, eRNAs are transcribed from putative enhancer regions and play a critical role in the transcription of targeted genes (Heintzman et al., 2009), and the expression level of eRNAs is correlated with that of nearby genes (Mao et al., 2018). Our current research on FALEC and ECM1 in GC supports these conclusions (Figure 5B). However, it remains to be determined how eRNAs regulate gene expression. Some evidence suggests that eRNAs affect chromatin states and enhancer–promoter looping, increase pol II binding, and act as a decoy for the negative elongation factor complex (Maruyama et al., 2014; Schaukowitch et al., 2014; Liang et al., 2016). These models remain controversial, and the underlying nature of eRNA in the regulation of gene expression requires further investigation. As for FALEC, our studies found that FALEC exerts its regulatory function in cis in GC, but the detailed mechanism of how FALEC regulates the expression of ECM1, and other targets may be influenced by FALEC in cis or in trans requires further investigation.
In summary, we showed that FALEC functions as an eRNA to activate ECM1 expression in the progression of GC. FALEC could serve as a new potential biomarker and therapeutic target for individualized treatment of GC patients.
Author Contributions
HW and ML conceived, designed the study, and wrote the manuscript. FQ, HW, and YZ performed the experiments and helped to draft the manuscript. JZ provided assistance for clinical sample collection, preservation, data and statistical analysis. SW, MH, TW, FH, WC, and DS provided statistical analysis of the data, contributed to some experiments, and helped to draft the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This work was supported by National Natural Science Foundation of China (21707002 and 81602300) and Foundation for Young Talents and Natural Science in Higher Education of Anhui Province (KJ2017A213, KJ2017A219, gxyq2018035, and BYKY1615ZD).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2019.00255/full#supplementary-material
References
- Athie A., Huarte M. (2014). FAL1ing inside an amplicon. Cancer Cell 26 303–304. 10.1016/j.ccr.2014.08.009 [DOI] [PubMed] [Google Scholar]
- Beroukhim R., Mermel C. H., Porter D., Wei G., Raychaudhuri S., Donovan J., et al. (2010). The landscape of somatic copy-number alteration across human cancers. Nature 463 899–905. 10.1038/nature08822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech T. R., Steitz J. A. (2014). The noncoding RNA revolution-trashing old rules to forge new ones. Cell 157 77–94. 10.1016/j.cell.2014.03.008 [DOI] [PubMed] [Google Scholar]
- Cesana M., Cacchiarelli D., Legnini I., Santini T., Sthandier O., Chinappi M., et al. (2011). A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 147 358–369. 10.1016/j.cell.2011.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaffer C. L., Weinberg R. A. (2011). A perspective on cancer cell metastasis. Science 331 1559–1564. 10.1126/science.1203543 [DOI] [PubMed] [Google Scholar]
- Chandrashekar D. S., Bashel B., Balasubramanya S. A. H., Creighton C. J., Ponce-Rodriguez I., Chakravarthi B., et al. (2017). UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 19 649–658. 10.1016/j.neo.2017.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu H. S., Somvanshi S., Patel E., Chen T. W., Singh V. P., Zorman B., et al. (2018). Pan-cancer analysis of lncRNA regulation supports their targeting of cancer genes in each tumor context. Cell Rep. 23:297–312.e12. 10.1016/j.celrep.2018.03.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H., Wang L., Gong P., Zhao C., Zhang S., Zhang K., et al. (2015). Deregulation between miR-29b/c and DNMT3A is associated with epigenetic silencing of the CDH1 gene, affecting cell migration and invasion in gastric cancer. PLoS One 10:e0123926. 10.1371/journal.pone.0123926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X. Y., Pan H. F., Leng R. X., Ye D. Q. (2015). Long noncoding RNAs: novel insights into gastric cancer. Cancer Lett. 356(2 Pt B) 357–366. 10.1016/j.canlet.2014.11.005 [DOI] [PubMed] [Google Scholar]
- Ferlay J., Soerjomataram I., Dikshit R., Eser S., Mathers C., Rebelo M., et al. (2015). Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 136 E359–E386. 10.1002/ijc.29210 [DOI] [PubMed] [Google Scholar]
- Gomez-Contreras P., Ramiro-Diaz J. M., Sierra A., Stipp C., Domann F. E., Weigel R. J., et al. (2017). Extracellular matrix 1 (ECM1) regulates the actin cytoskeletal architecture of aggressive breast cancer cells in part via S100A4 and Rho-family GTPases. Clin. Exp. Metastasis 34 37–49. 10.1007/s10585-016-9827-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta G. P., Massague J. (2006). Cancer metastasis: building a framework. Cell 127 679–695. 10.1016/j.cell.2006.11.001 [DOI] [PubMed] [Google Scholar]
- Guttman M., Donaghey J., Carey B. W., Garber M., Grenier J. K., Munson G., et al. (2011). lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 477 295–300. 10.1038/nature10398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao N. B., He Y. F., Li X. Q., Wang K., Wang R. L. (2017). The role of miRNA and lncRNA in gastric cancer. Oncotarget 8 81572–81582. 10.18632/oncotarget.19197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman N. D., Hon G. C., Hawkins R. D., Kheradpour P., Stark A., Harp L. F., et al. (2009). Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459 108–112. 10.1038/nature07829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo J. B., Sung S. (2011). Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 331 76–79. 10.1126/science.1197349 [DOI] [PubMed] [Google Scholar]
- Hsieh C. L., Fei T., Chen Y., Li T., Gao Y., Wang X., et al. (2014). Enhancer RNAs participate in androgen receptor-driven looping that selectively enhances gene activation. Proc. Natl. Acad. Sci. U.S.A. 111 7319–7324. 10.1073/pnas.1324151111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X., Feng Y., Zhang D., Zhao S. D., Hu Z., Greshock J., et al. (2014). A functional genomic approach identifies FAL1 as an oncogenic long noncoding RNA that associates with BMI1 and represses p21 expression in cancer. Cancer Cell 26 344–357. 10.1016/j.ccr.2014.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Wang J., Qian J., Kong X., Tang J., Wang Y., et al. (2014). Long noncoding RNA GAPLINC regulates CD44-dependent cell invasiveness and associates with poor prognosis of gastric cancer. Cancer Res. 74 6890–6902. 10.1158/0008-5472.CAN-14-0686 [DOI] [PubMed] [Google Scholar]
- Jeong S., Lee J., Kim D., Seol M. Y., Lee W. K., Jeong J. J., et al. (2016). Relationship of focally amplified long noncoding on chromosome 1 (FAL1) lncRNA with E2F transcription factors in thyroid cancer. Medicine 95:e2592. 10.1097/MD.0000000000002592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P., Cheng J., Dike S., Nix D. A., Duttagupta R., Willingham A. T., et al. (2007). “RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 316 1484–1488. 10.1126/science.1138341 [DOI] [PubMed] [Google Scholar]
- Khalil A. M., Guttman M., Huarte M., Garber M., Raj A., Rivea Morales D., et al. (2009). Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. U.S.A. 106 11667–11672. 10.1073/pnas.0904715106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury G., Gruss P. (1983). Enhancer elements. Cell 33 313–314. 10.1016/0092-8674(83)90410-5 [DOI] [PubMed] [Google Scholar]
- Kim T. K., Hemberg M., Gray J. M., Costa A. M., Bear D. M., Wu J., et al. (2010). Widespread transcription at neuronal activity-regulated enhancers. Nature 465 182–187. 10.1038/nature09033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam M. T., Li W., Rosenfeld M. G., Glass C. K. (2014). Enhancer RNAs and regulated transcriptional programs. Trends Biochem. Sci. 39 170–182. 10.1016/j.tibs.2014.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. T. (2012). Epigenetic regulation by long noncoding RNAs. Science 338 1435–1439. 10.1126/science.1231776 [DOI] [PubMed] [Google Scholar]
- Lee K. M., Nam K., Oh S., Lim J., Kim R. K., Shim D., et al. (2015). ECM1 regulates tumor metastasis and CSC-like property through stabilization of beta-catenin. Oncogene 34 6055–6065. 10.1038/onc.2015.54 [DOI] [PubMed] [Google Scholar]
- Li B., Mao R., Liu C., Zhang W., Tang Y., Guo Z. (2018). LncRNA FAL1 promotes cell proliferation and migration by acting as a CeRNA of miR-1236 in hepatocellular carcinoma cells. Life Sci. 197 122–129. 10.1016/j.lfs.2018.02.006 [DOI] [PubMed] [Google Scholar]
- Li Y., Zhou L., Lu C., Shen Q., Su Y., Zhi Z., et al. (2018). Long non-coding RNA FAL1 functions as a ceRNA to antagonize the effect of miR-637 on the down-regulation of AKT1 in Hirschsprung’s disease. Cell Prolif. 51:e12489. 10.1111/cpr.12489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Yu B., Li J., Su L., Yan M., Zhu Z., et al. (2014). Overexpression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancer. Oncotarget 5 2318–2329. 10.18632/oncotarget.1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Meng H., Bai Y., Wang K. (2016). Regulation of lncRNA and its role in cancer metastasis. Oncol. Res. 23 205–217. 10.3727/096504016X14549667334007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Notani D., Ma Q., Tanasa B., Nunez E., Chen A. Y., et al. (2013). Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature 498 516–520. 10.1038/nature12210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J., Zhou H., Gerdt C., Tan M., Colson T., Kaye K. M., et al. (2016). Epstein-barr virus super-enhancer eRNAs are essential for MYC oncogene expression and lymphoblast proliferation. Proc. Natl. Acad. Sci. U.S.A. 113 14121–14126. 10.1073/pnas.1616697113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald J. S., Smalley S. R., Benedetti J., Hundahl S. A., Estes N. C., Stemmermann G. N., et al. (2001). Chemoradiotherapy after surgery compared with surgery alone for adenocarcinoma of the stomach or gastroesophageal junction. N. Engl. J. Med. 345 725–730. 10.1056/NEJMoa010187 [DOI] [PubMed] [Google Scholar]
- Mao R., Wu Y., Ming Y., Xu Y., Wang S., Chen X., et al. (2018). Enhancer RNAs: a missing regulatory layer in gene transcription. Sci. China Life Sci. 10.1007/s11427-017-9370-9 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Maruyama A., Mimura J., Itoh K. (2014). Non-coding RNA derived from the region adjacent to the human HO-1 E2 enhancer selectively regulates HO-1 gene induction by modulating Pol II binding. Nucleic Acids Res. 42 13599–13614. 10.1093/nar/gku1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasrollahzadeh-Khakiani M., Emadi-Baygi M., Schulz W. A., Nikpour P. (2017). Long noncoding RNAs in gastric cancer carcinogenesis and metastasis. Brief. Funct. Genom. 16 129–145. 10.1093/bfgp/elw011 [DOI] [PubMed] [Google Scholar]
- Ni N., Song H., Wang X., Xu X., Jiang Y., Sun J. (2017). Up-regulation of long noncoding RNA FALEC predicts poor prognosis and promotes melanoma cell proliferation through epigenetically silencing p21. Biomed. Pharmacother. 96 1371–1379. 10.1016/j.biopha.2017.11.060 [DOI] [PubMed] [Google Scholar]
- Orom U. A., Derrien T., Beringer M., Gumireddy K., Gardini A., Bussotti G., et al. (2010). Long noncoding RNAs with enhancer-like function in human cells. Cell 143 46–58. 10.1016/j.cell.2010.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan C., Yao G., Liu B., Ma T., Xia Y., Wei K., et al. (2017). Long noncoding RNA FAL1 promotes cell proliferation, invasion and epithelial-mesenchymal transition through the PTEN/AKT signaling axis in non-small cell lung cancer. Cell Physiol. Biochem. 43 339–352. 10.1159/000480414 [DOI] [PubMed] [Google Scholar]
- Ponting C. P., Oliver P. L., Reik W. (2009). Evolution and functions of long noncoding RNAs. Cell 136 629–641. 10.1016/j.cell.2009.02.006 [DOI] [PubMed] [Google Scholar]
- Rahman R., Asombang A. W., Ibdah J. A. (2014). Characteristics of gastric cancer in Asia. World J. Gastroenterol. 20 4483–4490. 10.3748/wjg.v20.i16.4483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaukowitch K., Joo J. Y., Liu X., Watts J. K., Martinez C., Kim T. K. (2014). “Enhancer RNA facilitates NELF release from immediate early genes.” Mol. Cell. 56 29–42. 10.1016/j.molcel.2014.08.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt A. M., Chang H. Y. (2016). Long noncoding RNAs in cancer pathways. Cancer Cell 29 452–463. 10.1016/j.ccell.2016.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M., Nie F. Q., Wang Z. X., De W. (2016). Involvement of lncRNA dysregulation in gastric cancer. Histol. Histopathol. 31 33–39. 10.14670/HH-11-655 [DOI] [PubMed] [Google Scholar]
- Szasz A. M., Lanczky A., Nagy A., Forster S., Hark K., Green J. E., et al. (2016). Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget 7 49322–49333. 10.18632/oncotarget.10337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z., Li C., Kang B., Gao G., Li C., Zhang Z. (2017). GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 45 W98–W102. 10.1093/nar/gkx247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torre L. A., Bray F., Siegel R. L., Ferlay J., Lortet-Tieulent J., Jemal A. (2015). Global cancer statistics, 2012. CA Cancer J. Clin. 65 87–108. 10.3322/caac.21262 [DOI] [PubMed] [Google Scholar]
- Ulitsky I., Bartel D. P. (2013). lincRNAs: genomics, evolution, and mechanisms. Cell 154 26–46. 10.1016/j.cell.2013.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K. C., Chang H. Y. (2011). Molecular mechanisms of long noncoding RNAs. Mol. Cell. 43 904–914. 10.1016/j.molcel.2011.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhao Z., Zhang S., Li Z., Li D., Yang S., et al. (2018). LncRNA FAL1 is a negative prognostic biomarker and exhibits pro-oncogenic function in osteosarcoma. J Cell Biochem. 106 46–56. 10.1002/jcb.27074 [DOI] [PubMed] [Google Scholar]
- Xing Z., Lin A., Li C., Liang K., Wang S., Liu Y., et al. (2014). lncRNA directs cooperative epigenetic regulation downstream of chemokine signals. Cell 159 1110–1125. 10.1016/j.cell.2014.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong G. P., Zhang J. X., Gu S. P., Wu Y. B., Liu J. F. (2012). Overexpression of ECM1 contributes to migration and invasion in cholangiocarcinoma cell. Neoplasma 59 409–415. 10.4149/neo_2012_053 [DOI] [PubMed] [Google Scholar]
- Yan X., Hu Z., Feng Y., Hu X., Yuan J., Zhao S. D., et al. (2015). Comprehensive genomic characterization of long non-coding RNAs across human cancers. Cancer Cell 28 529–540. 10.1016/j.ccell.2015.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R., Sun F., Bei X., Wang X., Zhu Y., Jiang C., et al. (2017). Upregulation of the long non-coding RNA FALEC promotes proliferation and migration of prostate cancer cell lines and predicts prognosis of PCa patients. Prostate 77 1107–1117. 10.1002/pros.23367 [DOI] [PubMed] [Google Scholar]
- Zhong X., Hu X., Zhang L. (2015). Oncogenic long noncoding RNA FAL1 in human cancer. Mol. Cell. Oncol. 2:e977154. 10.4161/23723556.2014.977154 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


