Abstract
Most patients with multiple myeloma (MM) would finally relapse despite the fact that initial MM may turn to remission by conventional chemotherapy. Current chemotherapy regimens have limited effect on relapse MM patients. As a new histone deacetylase inhibitor, chidamide has been used in malignancy treatment. However, it is still unknown if chidamide can be used in MM. Here, by RNA sequencing, succinate dehydrogenase subunit A (SDHA) was screened to be the key molecule modulated by chidamide. SDHA was downregulated in MM patients and the expression of SDHA had negative correlation with the severity of MM. In vitro, chidamide inhibited proliferation and invasion of MM cells, and this effect vanished after knocking down SDHA. Lenalidomide and low dose of bortezomib had synergistic effect with chidamide, and similarly this effect was attenuated by SDHA siRNA. Moreover, chidamide decreased the production of reaction oxygen species (ROS) via SDHA. In a word, by targeting the key molecule SDHA, chidamide may represent a promising strategy in MM treatment.
Keywords: Multiple myeloma (MM), chidamide, succinate dehydrogenase subunit A (SDHA)
Introduction
Multiple myeloma (MM) is an incurable plasma cell malignancy characterized by proliferation of monoclonal plasma cells in bone marrow [1]a. These abnormal malignant plasma cells frequently lead to widely osteolytic damage. MM is the second most frequent hematological cancer following lymphoma, accounting for approximately 10% of all hematological malignancies [2]. In addition, MM is usually developed from a pre-malignant state, which is so called as monoclonal gammopathy of undetermined significance (MGUS) [3]. Although hematopoietic stem cell transplantation [4] and various chemotherapy agents, such as cytotoxic drug doxorubicin, proteasome inhibitor bortezomib and immunomodulatory drug lenalidomide [5], have significantly achieved longer period of remission and increased survival time in patients with MM, almost every patient would eventually relapse, and traditional agents would be probably useless [6]. Therefore, a new therapy approach should be soon developed and applied in clinical treatment.
Chidamide is a novel benzamide chemical class of histone deacetylase inhibitor (HDACi) [7]. The main target of chidamide is HDAC I which is closely related to tumorgenesis and progression [8]. Chidamide not only inhibits proliferation of tumor cells directly, but also enhances the cellular immunity ability in various kinds of tumors [7]. To date, chidamide has been approved in China for the oral treatment of recurrent or refractory peripheral T-cell lymphoma (PTCL) [9]. But so far, the indications of chidamide still remained limited. Therefore, it is urgent to explore the effect of chidamide in MM and clarify the mechanism, and this would have positive effect on clinical prognosis of MM in the future.
In the present study, we performed a RNA sequencing program and chose SDHA as a gene closely related to chidamide in MM. Chidamide inhibited proliferation and invasion of MM cells via SDHA. In addition, we determined the synergistic effect of chidamide and traditional agents in MM, which was also attenuated by knocking down SDHA. Inhibition of SDH-HIFα axis might be the underlying mechanism of chidamide in treating MM. This study described for the first time the application and mechanism of chidamide in MM.
Materials and methods
Patients
Twenty-three patients diagnosed as MM and thirteen as MGUS were included in this study. The histological diagnosis was established according to IMWG 2014 criteria. The study was approved by Shanghai Zhongshan Hospital Review Board and informed consent was obtained from patients in accordance with the Declaration of Helsinki.
Bone marrow samples were obtained from MM and MGUS patients and healthy individuals who underwent bone marrow aspiration for suspected hematological disease. Bone marrow mononuclear cells (BMMCs) were isolated from the bone marrow samples by Ficoll-isopaque centrifugation.
Cells and reagents
OPM-2 and H929 cell lines were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). Cells were cultured in RPMI-1640 medium with 10% heat-inactivated fetal bovine serum in a humidified atmosphere of 95% air and 5% CO2 at 37°C. Lenalidomide was from Celgene (NJ, USA). Bortezomib was from Jassen Pharmaceuticals (Xian, China). Chidamide was kindly provided by Chipscreen Biosciences (Shenzhen, China).
RNA sequencing
Total RNA was extracted from frozen sections using Trizol agent (Invitrogen, Carlsbad, CA, USA) and cDNA was reverse transcribed by the reverse transcription kit (TAKARA, Japan). The products were purified and enriched with PCR to create the final cDNA library. The clusters of the cDNA library were generated on the flow cell using TruSeq PE Cluster kit and HiSeq PE flow cell. The clusters were finally sequenced on HiSeq. 2000 system using TruSeq SBS kit.
Genes detection
SDHA expression was analyzed by real-time quantitative RT-PCR using 7500HT Fast Real-time PCR system (Applied Biosystem, Foster City, CA, USA). SDHA: Forward, 5’-AACCCGTGTTAATGCCTCTG-3’ and Reverse, 5’-CAAACGAGGGAACATGTGTG-3’. LY75: Forward, 5’-TGCCTTGGCCTCGATATTAC-3’ and Reverse, 5’-TCTGAGCCTCCTTTCTTCCA-3’. ITGA7: Forward, 5’-ATCAAGATTTGGCAGGATCG-3’ and Reverse, 5’-ACACAGGGTGAATGGGAGAG-3’. MEIS3: Forward, 5’-CTCACACCTGCCTCTGGTTC-3’ and Reverse, 5’-GCAGAGGTGAAGGCAGAAGT-3’. FCER2: Forward, 5’-ACACATCTCCCGCTCCTCTA-3’ and Reverse, 5’-CACCTGAGCTGGGGATACTC-3’. MRPL30: Forward, 5’-ACGGTGGCTCACCTGTAATC-3’ and Reverse, 5’-TACAGTGGCACCATCTCAGC-3’. GAPDH: Forward, 5’-GAAGGTGAAGGTCGGAGTC-3’ and Reverse, 5’-GAAGATGGTGATGGGATTTC-3. Relative expressions were calculated by the method of ΔΔCT.
Cell transfection
Small interfering RNA (siRNA) with SDHA targeted or control siRNA for cell transfection were synthesized by Biotend (Shanghai, China). OPM-2 and H929 cells were transfected by siRNA with an ultimate concentration of 100 nM according to the manufacturer’s protocol. The transfected clones were detected by 48 hours after transfection.
Western blot
Western blot was performed as described previously [10]. Antibodies against HIFα was from Abcam (ab16066, Cambridge, MA, USA). Antibody against SDHA was from Abcam (ab14715). Actin (Cell Signaling technology, Beverly, MA, USA) was used to ensure equivalent protein loading.
Cell invasion assay
Cell invasion was tested in the Matrigel Invasion Chamber (BD Pharmingen, Franklin Lakes, NJ, USA), which is composed of the upper and lower compartment separated by the polycarbonate membranes (8 μm pore size). 6 × 104 cells were incubated with RPMI-1640 medium (FBS-free, 200 μl) for 24 hours and added to the upper compartment, while RPMI-1640 with 10% FBS (500 μl) was added to the lower compartment. After incubation with 5% CO2 at 37°C for 24 hours, cell invasion was tested as described previously [11]. The membrane was stained by Wright-Giemsa staining and the invading cells were observed under the microscope at 40 × magnifications and counted in different fields of membranes in triplicate.
Cell proliferation assay
Cells were seeded at a density of 2 × 105 cells per well in 6-well plates and incubated at 37°C with chidamide or SDHA siRNA alone or in combination with chidamide and siRNA. Cell counts were calculated after 24 hours and 48 hours.
CCK8 assay
Cells were seeded at a density of 5 × 105 cells per well in 96-well plates and incubated at 37°C with doxorubicin, bortezomib, lenalidomide alone or in combination with chidamide. After 24-h incubation, 0.1 mg CCK8 was added to each well and the absorbance was measured at 490 nm by spectrophotometry.
Synergistic analysis
To determine the synergistic effect of chidamide combined with other chemotherapeutic agents, the combination index (CI) method was described by Chou and Talalay (CI = DA/ICXA+DB/ICXB+DA*DB/ICXA*ICXB) [12]. This method allows quantitative determination of drug interactions, where CI < 1, = 1, and > 1 indicate synergism, additive effect, and antagonism, respectively.
Detection of ROS accumulation
Mitochondrial ROS production was measured as described [10]. Cell lines or BMMCs were treated with chidamide for 24 hours or not for the indicated time periods. CMH2DCFDA (5 mM) was added 30 min before collecting cells. Flow cytometry was used to analyze ROS production.
Statistical analysis
Differences of gene expression among groups were assessed by the Mann-Whitney U test. In vitro experimental results were expressed as mean ± SEM. of data obtained from three separate experiments and determined using a t-test to compare variance. P < 0.05 was considered statistically significant.
Results
SDHA was downregulated in MM patients
Firstly, RNA sequencing was performed to screen target genes of chidamide in MM patients (Figure 1A). Three MM patients were included in this test. Their BMMCs were cultured with 6 μM chidamide or not, and six of the most significantly changed coding genes were selected (Figure 1B). Five of them were upregulated genes after chidamide treated, while the other one was downregulated. The expression status of these genes was validated by realtime RT-PCR in patients BMMCs. Data showed that compared with DMSO-treated cells, after adding 6 μM chidamide, the expression of SDHA and FCER2 was increased (P = 0.0412 and P = 0.0207, respectively), and MRPL30 decreased (P = 0.0405) in patients’ BMMCs, which corresponded with the results of RNA sequencing (Figure 2A). Then we used realtime RT-PCR to compare the expression of the six genes between patients and normal individuals. The expression of SDHA was downregulated and ITGA7 was upregulated in MM patients (P = 0.0292 and P = 0.0002, respectively, Figure 2B). Based on the data above, SDHA was considered as the most valuable target gene of chidamide in MM. In order to determine the relationship between expression of SDHA and severity of the disease, we chose 10 relapsed MM patients, 13 newly diagnosed (initial) MM patients, 13 MGUS patients and 9 normal individuals to examine the expression of SDHA. Realtime RT PCR showed that SDHA expression in healthy individuals was the highest and followed by patients with MGUS and initial MM. Patients with relapsed MM had the lowest SDHA expression (P = 0.04 and 0.05 compared with normal individuals and MGUS patients, respectively, Figure 2C).
Figure 1.
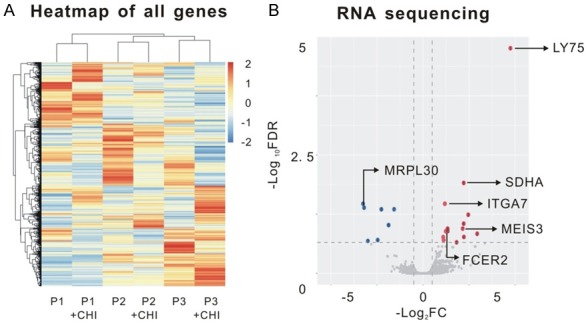
Genes regulated by chidamide in MM which were screened by RNA sequencing. A. Schematic of RNA sequencing analysis on bone marrow samples of 3 MM patients. BMMCs were isolated by Ficoll centrifugation from patients and then cultured with 6 μM chidamide or DMSO for 24 hours. B. Volcano plots of the genes of differential expression. The differentially expressed genes were plotted as blue (down-regulated genes after chidamide treatment) and red (up-regulated genes after treatment) points and other genes were indicated as gray points. Non-coding genes were excluded and top six genes which were relevant to malignancies were chosen by previous references.
Figure 2.
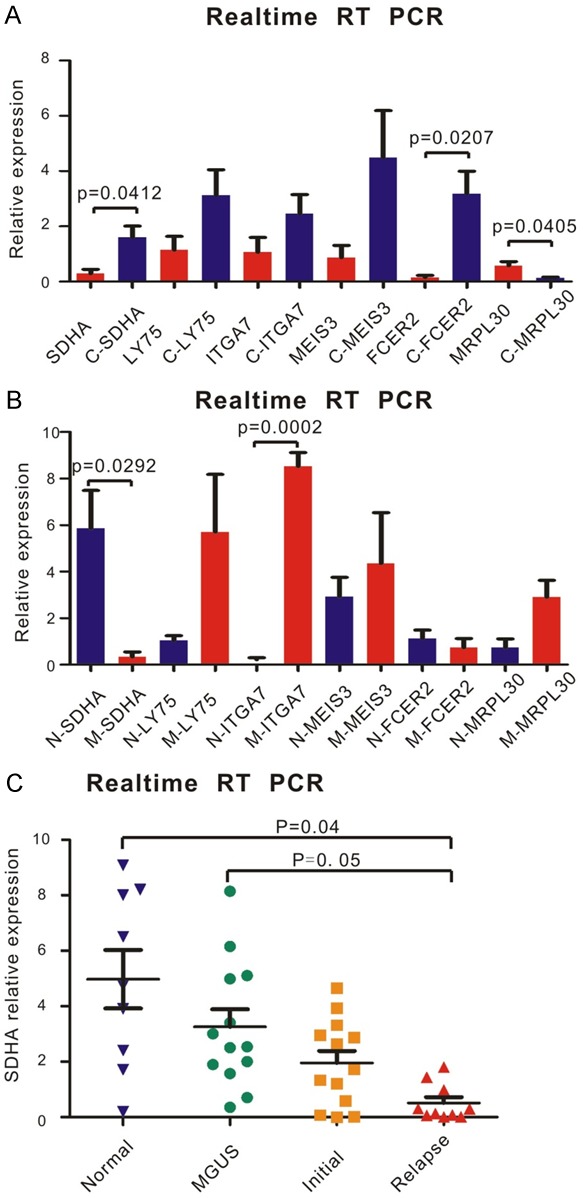
SDHA was sensitive to chidamide and its ectopic expression was related to disease development in MM. A. RNA was isolated from normal volunteers and MM patients. cDNA used in Realtime RT PCR was reverse transcribed from the RNA above. The six most differentially expressed genes were validated by Real-time RT PCR. After adding 6 μM chidamide, the expression of SDHA and FCER2 was increased, and MRPL30 decreased in patients’ BMMCs (n = 3) compared with cells adding DMSO, (C stood for cells with chidamide treatment). B. mRNA levels of the six most differentially expressed genes between normal volunteers (n = 3) and MM patients (n = 3) was detected by Realtime RT PCR. The expression of SDHA was upregulated and ITGA7 was downregulated in MM patients (N stood for normal volunteers; M stood for MM patients). C. RNA was isolated from normal volunteers (n = 9), MGUS (n = 13) and MM patients (n = 23). By Realtime RT PCR, SDHA expression in normal volunteers was the highest and followed by patients with MGUS and initial MM. Patients with relapse MM (n = 10) had the lowest SDHA expression.
Chidamide inhibited proliferation and invasion of MM cells through SDHA
Ability of proliferation and invasion was the most important characteristics of tumor cells. Myeloma cell lines (OPM-2 and H929) were cultured with an original concentration of 2*105/L in order to find out the effect of chidamide on inhibiting proliferation of MM cells. Chidamide dramatically inhibited proliferation of both H929 and OPM-2 cell lines (P = 0.0001 and P = 0.0052, respectively, Figure 3A). However, when SDHA was knocked down by siRNA, this inhibition effect was not as powerful as before in H929 cells (Figure 3C). Similarly, as revealed by cell invasion assay, chidamide-treated H929 cells achieved a notably lower percentage of cell invasion than those treated with DMSO (P = 0.0064, Figure 3B). Interestingly, when SDHA was knocked down, this invasive ability was not significantly changed no matter chidamide was added or not (Figure 3D). Above results revealed that SDHA acted as the key molecule in chidamide inhibiting proliferation and invasion in MM cell lines.
Figure 3.
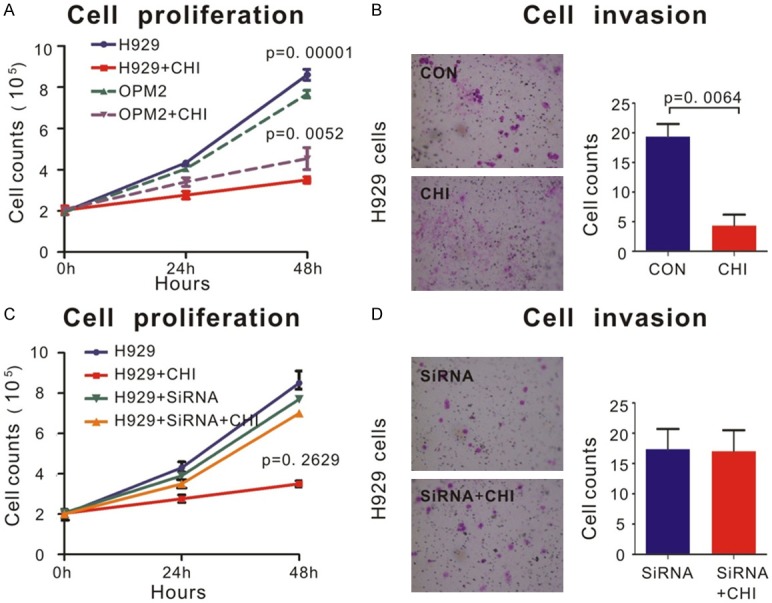
Chidamide inhibited proliferation and invasion of MM cells via SDHA. A. OPM2 and H929 cells lines treated by 6 μM chidamide or DMSO for 48 h. Cell proliferation was measured as described in Materials and Methods. Cells treated by chidamide proliferated much slower than cells treated by DMSO, especially in H929 cells. B. By transwell invasion assay, invasion ability of H929 cells was significantly inhibited by 6 μM chidamide treatment than that by DMSO. Quantifications of cell invasion were shown in the right panel. C. SDHA siRNA and control siRNA were used as described in Materials and Methods. The ability of proliferation of H929 cells transfected with SDHA siRNA remained a same level no matter treated by chidamide or DMSO. D. By transwell invasion assay, SDHA siRNA transfected cells acquired no decrease in cell invasion after chidamide treatment. Quantifications of cell invasion were shown in the right panel.
Bortezomib and lenalidomide had a synergistic effect with chidamide via SDHA
CCK8 assay was used to determine dose-response curves of chemotherapeutic agents and synergistic effect of chidamide combined with other agents. Dose-response curves of doxorubicin, bortezomib, lenalidomide and chidamide were shown in Figure 4. Compared with each agent alone, a significant increase in cell death was observed in several circumstances of combined treatment (Figure 5A and 5B). As shown in the right panel of Figure 3A, 2.5 mg/L bortezomib and 0.6 μM chidamide induced less than 10% reduction in cell viability; however, in combination they achieved over 20% cell reduction, which indicated a synergistic effect. The similar synergistic effect happened on 0.125 g/L or 12.5 mg/L lenalidomide and 0.6 μM chidamide (Figure 5B). However, when concentration of bortezomib was 25 mg/L, there was no synergistic effect any more (Figure 5A, left panel). On the whole, the CI of lenalidomide and low concentration of bortezomib yielded many of the data points to the area < 1 when combined with chidamide treatment, denoting synergistic interactions in MM cell line (Figure 5C and 5D). However, we did not found any synergistic effect between doxorubicin and chidamide.
Figure 4.
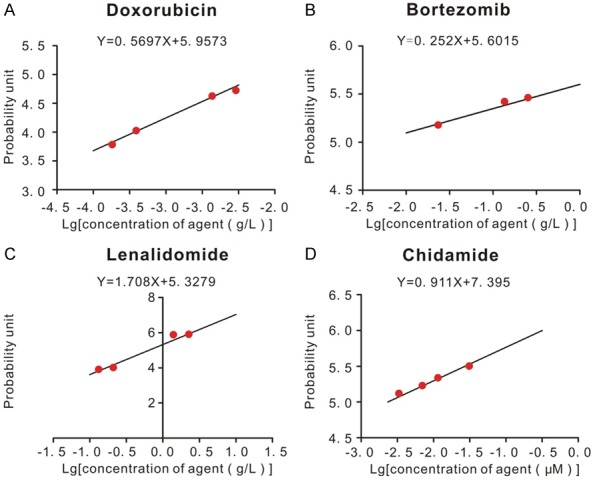
Dose-response curves of doxorubicin, bortezomib, lenalidomide and chidamide in H929 cells. CCK8 assay was used to determine the dose-response curves of agents. 100 μL 5*105/L H929 cells were put into 96-wells plate. Different concentrations of agents were added to wells respectively for 24 h. CCK8 assay was used as described in Materials and Methods. Apoptosis rates of cells induced by agents were converted to probability unit based on percentage-probability unit chart. The IC50 concentration of each agent was the corresponding value at the probability unit of 5. A. Dose-response curve of doxorubicin. B. Dose-response curve of bortezomib. C. Dose-response curve of lenalidomide. D. Dose-response curve of chidamide.
Figure 5.
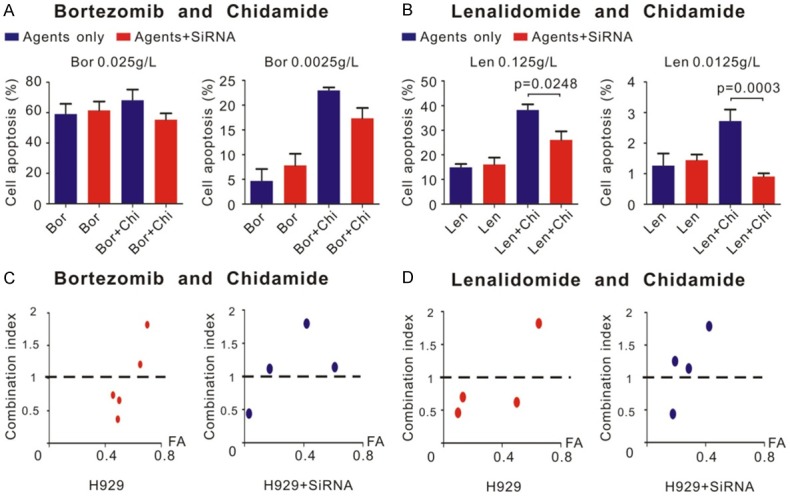
Bortezomib and lenalidomide had synergistic effect with chidamide via SDHA. CCK8 assay was used to detect the synergistic effect between agents. Different concentrations of agents were added to wells respectively for 24 h. CCK8 was used as described in Materials and Methods. A. Left panel showed 0.025 g/L bortezomib and 0.6 μM chidamide have no synergistic effect and not regulated by SDHA siRNA. Right panel showed 0.0025 g/L bortezomib and 0.6 μM chidamide have synergistic effect (calculated as described in materials and methods). But the regulation effect of SDHA siRNA was not significant. B. 0.125 g/L and 0.0125 g/L lenalidomide both have synergistic effect with chidamide. And this synergistic effect was reversed by SDHA siRNA. C. CI distribution between bortezomib and chidamide in different inhibition rate (FA). D. CI distribution between lenalidomide and chidamide in different inhibition rate (FA).
Interestingly, as shown in Figure 5A and 5B, when SDHA was knocked down, cells apoptosis induced by bortezomib and lenalidomide combined with chidamide greatly decreased, especially by lenalidomide (P = 0.0248 and P = 0.0003, respectively at the concentration of 0.125 g/L and 12.5 mg/L, Figure 5B), which indicated the synergistic effect between chidamide and other agents was induced by SDHA. When SDHA was knocked down by siRNA, not only the percentage of cell apoptosis, but also most of CI between chidamide and bortezomib or lenalidomide raised above 1, which indicated the synergistic effect tended to disappear (Figure 5C and 5D).
Chidamide decreased ROS production through increasing the expression of SDHA
In order to gain further mechanism of chidamide-SDHA-MM axis as SDHA was reported to be closely related to HIFA and its downstream effect such as the production of ROS, we used Western Blot to determine how they interacted. Our result showed that when SDHA was knocked down in H929 cells by siRNA, expression of HIFα protein was increased (P = 0.0068, Figure 6B). HIFα decreased after adding 6 μM chidamide (P = 0.0488, Figure 6B, right panel). However, when SDHA was knocked down, chidamide did not affect the expression of HIFα any longer (Figure 6B, P = 0.142). ROS in MM patients were much higher than that in normal people, which caused by higher HIFα expression in large extent (Figure 6D, right panel). Chidamide inhibited ROS production. Similar to HIFα, the ROS production was not sensitive to chidamide any more when SDHA was knocked down (Figure 6C, left panel, P = 0.142).
Figure 6.
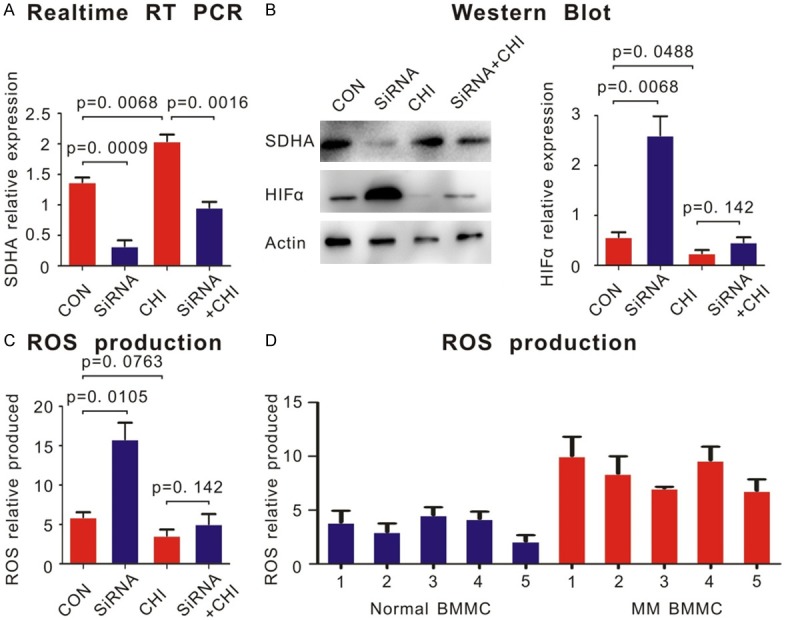
Chidamide decreased ROS production through increasing the expression of SDHA. A. Chidamide could increase the level of SDH in H929 cells showed by realtime RT PCR. B. Western blot showed that after knocking down SDHA by siRNA in H929 cells, the level of HIFα protein increased. Chidamide could increase the expression of SDHA and decrease the level of HIFα. But when SDHA knocked down, chidamide had little effect on HIFα expression. The quantifications of HIFα expression were shown in the right panel. C. Mitochondrial ROS production was determined by CM-H2DCFDA staining and flow cytometry. The production of ROS was decreased after knocking down SDHA in H929 cells. Chidamide decreased the production of ROS. But when SDHA knocked down, chidamide had little effect on ROS production. D. BMMCs of patients with MM produced more ROS than normal volunteers.
Discussion
Chidamide is an independently developed agent by Chinese and proved to be effective in various malignancies. In lung cancer cell lines, Lin SH et al. discovered chidamide alleviated TGF-β induced epithelial-mesenchymal transition [13]. Zhao B et al. found that chidamide inhibited pancreatic cancer development by modulating the ratio of Bax/Bcl-2 and p21 [14]. In hepatic cancer, chidamide also had its treatment efficacy [15]. Besides solid tumors, the main application of chidamide was treatment of refractory hematologic malignancies. Chidamide has been already used as a conventional therapy approach for the treatment of recurrent or refractory PTCL in China. A multicenter real-world study of chidamide application in relapsed and refractory PTCL demonstrated that chidamide had a favorable efficacy and an acceptable safty [16]. Li Y et al. proved chidamide could target stem and progenitor cells of acute myeloid leukemia [17]. In NK/T cell lymphoma, chidamide induced apoptosis through ATM-Chk2-p53-p21 pathway [18]. However, only limited case reports of chidamide were published to show chidamide treatment effect to MM [19]. The mechanisms and synergistic effect agents of chidamide in MM still remained vacuum. In this study, we found a probable target, SDHA, as a marker negatively correlated with the severity of MM and was targeted by chidamide.
SDHA encodes a subunit of succinate dehydrogenase (SDH), a mitochondrial enzyme involved in two essential energy-producing metabolic processes of the cell, the Krebs cycle and the electron transport chain [20]. Owing to the central function of SDH in cellular energy metabolism, it plays an important role in tumor suppression. SDH was mainly considered as a susceptibility gene for paraganglioma/phaeochromocytoma syndrome [21]. In addition, SDH is also involved in gastrointestinal stromal tumors (GISTs), renal tumors, thyroid tumors, testicular seminoma and neuroblastomas [22].
SDHA was chosen by RNA sequencing from more than 16,000 genes. Non-coding RNAs (ncRNAs) such as long non-coding RNAs (lncRNAs) were not considered in this study. Nevertheless, it does not mean that ncRNAs was unimportant. On the contrary, many lncRNAs may be vitally essential in biological process regulation. As a hot spot nowadays, what role ncRNAs play in chidamide regulating MM cells needs to be furtherly discovered. At first, the top six chidamide-regulating mRNAs were screened including SDHA. SDHA emerged as the best candidate mainly because of the two step validation which confirmed sensitive to chidamide and differentially expressed between normal volunteers and MM patients. SDHA was upregulated in BMMCs of patients treated by chidamide; and its expression decreased in MM patients. These suggested that SDH was a potential tumor suppressor which was targeted by chidamide.
There is a leading biochemical mechanism of how ectopic expression of SDH lead to tumor formation and development. Loss of abundant expression of SDH leads to accumulation of succinate in cells [23]. Cells with excess amount of succinate which acts as an intracellular messenger between mitochondria to cytosol cannot hydrate and degrade HIFα. This phenomenon eventually causes accumulation of ROS, which macroscopically leads to angiogenesis and proliferation of tumors [24]. In our study, chidamide raised the expression of SDHA to inhibit accumulation of HIFα and ROS. As a HDAC inhibitor, the underlying molecular mechanism of chidamide regulating expression of SDH could be acetylation of histone of SDHA or its upstream regulators. In normal conditions, von Hippel-Lindau (VHL) protein recognized hydroxylated HIFα by the PHD enzymes and then HIFα will be polyubiquitylated and degraded. However, in SDHA deficient conditions, the activity of PHD enzymes is inhibited by accumulated succinate. HIFα is not hydroxylated and eventually escape degradation [25]. The excess ROS produced due to these abnormal HIFα induces the expression of genes involved in cell survival, proliferation, angiogenesis and so on [26]. In vitro, chidamide decrease the level of MM cells proliferation and invasion, but these effects almost vanished after adding SDHA siRNA. We inferred chidamide-SDHA-HIFα axis as the main pathway of chidamide in MM cells from the above phenomenon. There is also a study showed chidamide suppressed the pathway of HIFα and decreased the production of ROS [27].
Chidamide was reported to have synergistic effect with decitabine in Hodgkin lymphoma by Jiang T et.al. As conventional chemotherapeutic drugs, doxorubicin, bortezomib and lenalidomide were widely used in MM patients. Zhang H et al. found synergistic antitumor effect of chidamide and doxorubicin in peripheral T-cell lymphoma [28]. However, we have no evidence that chidamide have synergistic effect with doxorubicin in MM. Lenalidomide and bortezomib showed synergistic effect under most of the concentrations in vitro which could be attenuated by knocking down SDHA. To our knowledge, we provided first evidence that the synergistic effect of chidamide in MM. Further research should be performed to determine the effect of chidamide in MM patients in order to broaden the indication of chidamide and offer patients better quality of life.
Conclusions
In summary, our findings identified SDHA as a novel target of chidamide in MM cells. SDHA was low expressed in MM patients and low level of SDHA led to high proliferation and invasion abilities in MM cells. By targeting the key molecule SDHA, chidamide inhibited proliferation and invasion of MM cells and showed synergistic effect with lenalidomide and bortezomib. Chidamide could also decrease ROS production through increasing the expression of SDHA. Through chidamide-SDHA pathway, chidamide combines with traditional chemical agents such as lenalidomide and bortezomib might represent a promising strategy in MM treatment.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (81570123), the National Key New Drug Creation Special Programs (2017ZX09304-021) and Academic Pacesetters Program of Shanghai Healthcare System (2017BR033). YS and PL designed the experiments; YS, ZX, JX and MS performed the experiments; YS analyzed the data; YS, JL and PL provided professional writing services and materials and wrote the manuscript. All authors meet the criteria for authorship and approved the final manuscript for publication. We would like to thank everyone in the department of hematology of Zhongshan Hospital and Hongyan Qiu for supporting this work.
Disclosure of conflict of interest
None.
References
- 1.Kariyawasan CC, Hughes DA, Jayatillake MM, Mehta AB. Multiple myeloma: causes and consequences of delay in diagnosis. QJM. 2007;100:635–40. doi: 10.1093/qjmed/hcm077. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 3.Stelmach-Gołdyś A, Czarkowska-Paczek B. Monoclonal gammopathy of undetermined significance--potential risk factor of multiple myeloma. Przegl Lek. 2012;69:194–6. [PubMed] [Google Scholar]
- 4.Giralt S, Garderet L, Durie B, Cook G, Gahrton G, Bruno B, Hari P, Lokhorst H, McCarthy P, Krishnan A, Sonneveld P, Goldschmidt H, Jagannath S, Barlogie B, Mateos M, Gimsing P, Sezer O, Mikhael J, Lu J, Dimopoulos M, Mazumder A, Palumbo A, Abonour R, Anderson K, Attal M, Blade J, Bird J, Cavo M, Comenzo R, de la Rubia J, Einsele H, Garcia-Sanz R, Hillengass J, Holstein S, Johnsen HE, Joshua D, Koehne G, Kumar S, Kyle R, Leleu X, Lonial S, Ludwig H, Nahi H, Nooka A, Orlowski R, Rajkumar V, Reiman A, Richardson P, Riva E, San Miguel J, Turreson I, Usmani S, Vesole D, Bensinger W, Qazilbash M, Efebera Y, Mohty M, Gasparreto C, Gajewski J, LeMaistre CF, Bredeson C, Moreau P, Pasquini M, Kroeger N, Stadtmauer E. American society of blood and marrow transplantation, european society of blood and marrow transplantation, blood and marrow transplant clinical trials network, and international myeloma working group consensus conference on salvage hematopoietic cell transplantation in patients with relapsed multiple myeloma. Biol Blood Marrow Transplant. 2015;21:2039–2051. doi: 10.1016/j.bbmt.2015.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anderson KC, Kyle RA, Rajkumar SV, Stewart AK, Weber D, Richardson P ASH/FDA Panel on Clinical Endpoints in Multiple Myeloma. Clinically relevant end points and new drug approvals for myeloma. Leukemia. 2008;22:231–9. doi: 10.1038/sj.leu.2405016. [DOI] [PubMed] [Google Scholar]
- 6.Kumar SK, Lee JH, Lahuerta JJ, Morgan G, Richardson PG, Crowley J, Haessler J, Feather J, Hoering A, Moreau P, LeLeu X, Hulin C, Klein SK, Sonneveld P, Siegel D, Bladé J, Goldschmidt H, Jagannath S, Miguel JS, Orlowski R, Palumbo A, Sezer O, Rajkumar SV, Durie BG International Myeloma Working Group. Risk of progression and survival in multiple myeloma relapsing after therapy with IMiDs and bortezomib: a multicenter international myeloma working group study. Leukemia. 2012;26:149–57. doi: 10.1038/leu.2011.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ning ZQ, Li ZB, Newman MJ, Shan S, Wang XH, Pan DS, Zhang J, Dong M, Du X, Lu XP. Chidamide (CS055/HBI-8000): a new histone deacetylase inhibitor of the benzamide class with antitumor activity and the ability to enhance immune cell-mediated tumor cell cytotoxicity. Cancer Chemother Pharmacol. 2012;69:901–9. doi: 10.1007/s00280-011-1766-x. [DOI] [PubMed] [Google Scholar]
- 8.Hesham HM, Lasheen DS, Abouzid KAM. Chimeric HDAC inhibitors: comprehensive review on the HDAC-based strategies developed to combat cancer. Med Res Rev. 2018;38:2058–2109. doi: 10.1002/med.21505. [DOI] [PubMed] [Google Scholar]
- 9.Lu X, Ning Z, Li Z, Cao H, Wang X. Development of chidamide for peripheral T-cell lymphoma, the first orphan drug approved in China. Intractable Rare Dis Res. 2016;5:185–91. doi: 10.5582/irdr.2016.01024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu P, Xu B, Shen W, Zhu H, Wu W, Fu Y, Chen H, Dong H, Zhu Y, Miao K, Xu W, Li J. Dysregulation of TNFalpha-induced necroptotic signaling in chronic lymphocytic leukemia: suppression of CYLD gene by LEF1. Leukemia. 2012;26:1293–300. doi: 10.1038/leu.2011.357. [DOI] [PubMed] [Google Scholar]
- 11.Xu PP, Sun YF, Fang Y, Song Q, Yan ZX, Chen Y, Jiang XF, Fei XC, Zhao Y, Leboeuf C, Li B, Wang CF, Janin A, Wang L, Zhao WL. JAM-A overexpression is related to disease progression in diffuse large B-cell lymphoma and downregulated by lenalidomide. Sci Rep. 2017;7:7433. doi: 10.1038/s41598-017-07964-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chou TC. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol Rev. 2006;58:621–81. doi: 10.1124/pr.58.3.10. [DOI] [PubMed] [Google Scholar]
- 13.Lin SH, Wang BY, Lin CH, Chien PJ, Wu YF, Ko JL, Chen JJ. Chidamide alleviates TGF-beta-induced epithelial-mesenchymal transition in lung cancer cell lines. Mol Biol Rep. 2016;43:687–95. doi: 10.1007/s11033-016-4005-z. [DOI] [PubMed] [Google Scholar]
- 14.Zhao B, He T. Chidamide, a histone deacetylase inhibitor, functions as a tumor inhibitor by modulating the ratio of Bax/Bcl-2 and P21 in pancreatic cancer. Oncol Rep. 2015;33:304–10. doi: 10.3892/or.2014.3595. [DOI] [PubMed] [Google Scholar]
- 15.Wang H, Guo Y, Fu M, Liang X, Zhang X, Wang R, Lin C, Qian H. Antitumor activity of Chidamide in hepatocellular carcinoma cell lines. Mol Med Rep. 2012;5:1503–8. doi: 10.3892/mmr.2012.858. [DOI] [PubMed] [Google Scholar]
- 16.Shi Y, Jia B, Xu W, Li W, Liu T, Liu P, Zhao W, Zhang H, Sun X, Yang H, Zhang X, Jin J, Jin Z, Li Z, Qiu L, Dong M, Huang X, Luo Y, Wang X, Wang X, Wu J, Xu J, Yi P, Zhou J, He H, Liu L, Shen J, Tang X, Wang J, Yang J, Zeng Q, Zhang Z, Cai Z, Chen X, Ding K, Hou M, Huang H, Li X, Liang R, Liu Q, Song Y, Su H, Gao Y, Liu L, Luo J, Su L, Sun Z, Tan H, Wang H, Wang J, Wang S, Zhang H, Zhang X, Zhou D, Bai O, Wu G, Zhang L, Zhang Y. Chidamide in relapsed or refractory peripheral T cell lymphoma: a multicenter real-world study in China. J Hematol Oncol. 2017;10:69. doi: 10.1186/s13045-017-0439-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Y, Chen K, Zhou Y, Xiao Y, Deng M, Jiang Z, Ye W, Wang X, Wei X, Li J, Liang J, Zheng Z, Yao Y, Wang W, Li P, Xu B. A new strategy to target acute myeloid leukemia stem and progenitor cells using chidamide, a histone deacetylase inhibitor. Curr Cancer Drug Targets. 2015;15:493–503. doi: 10.2174/156800961506150805153230. [DOI] [PubMed] [Google Scholar]
- 18.Zhou J, Zhang C, Sui X, Cao S, Tang F, Sun S, Wang S, Chen B. Histone deacetylase inhibitor chidamide induces growth inhibition and apoptosis in NK/T lymphoma cells through ATM-Chk2-p53-p21 signalling pathway. Invest New Drugs. 2018;36:571–580. doi: 10.1007/s10637-017-0552-y. [DOI] [PubMed] [Google Scholar]
- 19.Wang DY, Cui YS, Liu YZ, Liu LN, Song YP, Fang BJ. Successful treatment of one case with relapsed refractory multiple myeloma by chidamide in combination with bortezomib and dexamethasone. Zhonghua Xue Ye Xue Za Zhi. 2016;37:463. doi: 10.3760/cma.j.issn.0253-2727.2016.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Warburg O, Wind F, Negelein E. The metabolism of tumors in the body. J Gen Physiol. 1927;8:519–30. doi: 10.1085/jgp.8.6.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burnichon N, Brière JJ, Libé R, Vescovo L, Rivière J, Tissier F, Jouanno E, Jeunemaitre X, Bénit P, Tzagoloff A, Rustin P, Bertherat J, Favier J, Gimenez-Roqueplo AP. SDHA is a tumor suppressor gene causing paraganglioma. Hum Mol Genet. 2010;19:3011–20. doi: 10.1093/hmg/ddq206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bardella C, Pollard PJ, Tomlinson I. SDH mutations in cancer. Biochim Biophys Acta. 2011;1807:1432–43. doi: 10.1016/j.bbabio.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 23.Selak MA, Armour SM, MacKenzie ED, Boulahbel H, Watson DG, Mansfield KD, Pan Y, Simon MC, Thompson CB, Gottlieb E. Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-alpha prolyl hydroxylase. Cancer Cell. 2005;7:77–85. doi: 10.1016/j.ccr.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 24.Kaelin WG Jr, Ratcliffe PJ. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell. 2008;30:393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 25.Pollard PJ, Brière JJ, Alam NA, Barwell J, Barclay E, Wortham NC, Hunt T, Mitchell M, Olpin S, Moat SJ, Hargreaves IP, Heales SJ, Chung YL, Griffiths JR, Dalgleish A, McGrath JA, Gleeson MJ, Hodgson SV, Poulsom R, Rustin P, Tomlinson IP. Accumulation of Krebs cycle intermediates and over-expression of HIF1alpha in tumours which result from germline FH and SDH mutations. Hum Mol Genet. 2005;14:2231–9. doi: 10.1093/hmg/ddi227. [DOI] [PubMed] [Google Scholar]
- 26.Pollard P, Wortham N, Barclay E, Alam A, Elia G, Manek S, Poulsom R, Tomlinson I. Evidence of increased microvessel density and activation of the hypoxia pathway in tumours from the hereditary leiomyomatosis and renal cell cancer syndrome. J Pathol. 2005;205:41–9. doi: 10.1002/path.1686. [DOI] [PubMed] [Google Scholar]
- 27.Zhang H, Li L, Li M, Huang X, Xie W, Xiang W, Yao P. Combination of betulinic acid and chidamide inhibits acute myeloid leukemia by suppression of the HIF1alpha pathway and generation of reactive oxygen species. Oncotarget. 2017;8:94743–94758. doi: 10.18632/oncotarget.21889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang H, Dong L, Chen Q, Kong L, Meng B, Wang H, Fu K, Wang X, Pan-Hammarström Q, Wang P, Wang X. Synergistic antitumor effect of histone deacetylase inhibitor and doxorubicin in peripheral T-cell lymphoma. Leuk Res. 2017;56:29–35. doi: 10.1016/j.leukres.2017.01.025. [DOI] [PubMed] [Google Scholar]


