Abstract
Nuclear factor-erythroid 2-related factor 3 (NRF3), a nuclear transcription factor, has been implicated in various cellular processes including carcinogenesis. However, mechanisms underlying its regulation in carcinogenesis are unclear. Herein, we found that NRF3 is lowly expressed in colorectal cancer (CRC) tissues and cells, and NRF3 low-expressions in CRC tissue samples are associated with CRC carcinogenesis and poor patient outcomes. Nrf3-knockdown increased CRC cell growth, colony formation, and cell motility and invasion, and Nrf3-knockin dramatically decreased CRC cell growth and colony formation. Mechanistically, NRF3 increased CRC cell apoptosis and arrested cell G2/M stage. NRF3 was found to be reversely with epidermal growth factor receptor (EGFR) and p38. Strikingly, Nrf3-knockin dramatically decreased phosphorylated-EGFR at Tyrosine845 (pEGFR Tyr845) and phosphorylated-p38 at Threonine180/Tyrosine182 (p-p38 Thr180/Tyr182) expressions, and Nrf3-knockdown increased pEGFR Tyr845 and p-p38 Thr180/Tyr182. Moreover, NRF3 regulated EGFR and p38 down-stream molecules, protein kinase B (AKT), activating transcription factor (ATF) 2, and C/EBP homologous protein (CHOP) expressions. NRF3 loss-increased CRC growth through EGFR and p38 was confirmed in nude mice. Collectively, NRF3-loss in CRC cell increases EGFR and p38 phosphorylation activation, enhances cell proliferation and decreases cell apoptosis, and finally promotes CRC malignance.
Keywords: NRF3, colorectal cancer, EGFR, p38, tumorigenesis
Introduction
Colorectal cancer (CRC) is one of the most common malignant tumors in worldwide, with approximately 1.2 million new cases and 608,700 deaths per year [1]. CRC is the third most frequent cancer in males and the second in females. The majority of CRC patients have sporadic disease with no apparent evidence of inheritance, while the remaining 25% have a family history of CRC. Genetic mutations have been identified as the cause of inherited cancer risk in some tumor-prone families; these mutations are estimated to account for only 5% to 6% of CRC cases overall [2]. Various factors including nuclear factor-erythroid 2-related factor (NRF) 3 are involved in CRC incidence [3]. Sporadic CRCs are caused by somatic mutations, and account for approximately 75% of all CRCs. Hereditary CRCs are caused by germline-inactivating mutations in oncogenes or tumor suppressor genes, and familial CRCs are caused by minor variant or single nucleotide polymorphisms genes [4-7].
NRF3 belongs to the cap ‘n’ collar (CNC) family comprising NRF1 and NRF2 [8-11]. Recently, a physiological relationship between NRF3 and cancers has been reported. The human cancer genome project has identified Nrf3 as one of 127 significantly mutated genes, and reported its significant gene induction in human cancers including colorectal adenocarcinoma [12]. Extensive biochemical studies have elucidated a part of the regulatory mechanisms of NRF3. Under physiological conditions, the transcriptional activity of NRF3 is repressed by its sequestration in the endoplasmic reticulum (ER), thereby preventing its unnecessary gene expression [13]. Upon exposure to a stress and/or a signal, NRF3 translocates into the nucleus, and exerts its transcriptional activity through the antioxidant response element (ARE) or Maf recognition elements (MARE) by heterodimerizing with small Maf proteins. These observations imply that NRF3 functions as an inducible transcription factor in response to certain activation signal(s). To understand the comprehensive biological function of NRF3 in cancer cells, further elucidation of its regulatory mechanisms, including its nuclear entry from the ER, and identification of its target gene(s) are indispensable.
The role of epidermal growth factor receptor (EGFR) in cancer development and treatment is well known [14-16]. EGFR belongs to ErbB family of receptor tyrosine kinases. Upon ligand stimulation, EGFR dimerizes, and dimerization is then followed by receptor internalization and autophosphorylation, which serves as binding sites for recruiting signal transducers and activators of intracellular signal transduction cascade. Ligation of EGFR activates mitogen-activated protein kinase (MAPK) cascades, and regulates downstream molecular, extracellular signal-regulated kinases (ERKs) and protein kinase B (AKT) [17,18]. p38/MAPK has been implicated in the regulation of different cancerous and noncancerous cell [19,20]. p38/MAPK is relatively inactive in its non-phosphorylated form, and becomes rapidly activated by phosphorylation of two Thr-GlyTyr motifs [21,22]. Phosphorylated p38 proteins can activate several transcription factors, such as activating transcription factor (ATF) 2, and C/EBP homologous protein (CHOP). p38/MAPK activation and overexpression were reported in human cancers including colorectal cancer, and correlated with a poor prognosis [23-25]. Herein, we showed that NRF3 is lowly expressed in CRC tissues, and its lowexpression is associated with CRC carcinogenesis and poor patient outcomes. Nrf3-knockin decreases EGFR and p38 activation. NRF3 was found to decrease CRC cell growth and increase cell apoptosis, and finally inhibit CRC malignance. NRF3-reduced EGFR expression represents a novel molecular target in CRC therapy.
Materials and methods
Reagents and antibodies
Chemical reagents for molecular biology experiments, AG1478 and SB203580 were purchased from Sigma-Aldrich (St. Louis, MO). Dulbecco’s modified Eagle medium (DMEM), 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide (MTT), and other supplements were obtained from Life Technologies (Rockville, MD). Antibodies against NRF3, p38, RAD51, BAX, cyclin D1, cyclin-dependent kinase (CDK) 4, or CDK6 were purchased from Abnova Company (Shanghai, China). Antibodies against AKT, ATF2, CHOP or GAPDH were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA) and Cell Signal Technology, Inc. (Beverly, MA). Antibody against phosphorylated-p38 at Threonine180/Tyrosine182 (p-p38 Thr180/Tyr182) was purchased from Cell Signaling Technology (MA, USA). The antibody against phosphorylated-EGFR at Tyrosine845 (pEGFR Tyr845) was purchased from UBI (Lake Placid, NY).
Tissue microarray and immunohistochemical staining
Human tissue microarrays (HColA180Su12) containing 80 pairs of CRC tissues, corresponding adjacent non-tumor tissues, and 20 cases of CRC cancers at various stages were purchased from Outdo Biotech Company (Shanghai, China). 100 patients enrolled into this study contained 57 males and 43 females. The median age of the patients was 46.5 age years (range 35-76), < 50 age year patients were 34 cases, > 50 age year patients were 66 cases. The tumor histology and stages were classified according to WHO classification and TNM staging system of UICC, respectively. The patients in T1-T2 stages were 37 cases, and T3-T4 patients were 63 cases. The patients in N0 stage were 39 cases, and N1-N3 patients were 61 cases. The patients in M0 stage were 34 cases, and M1 patients were 66 cases. These tissue microarrays were stained with NRF3 antibody (dilution 1:2000) as described previously [26]. The stained tissue microarrays were evaluated independently by two pathologists who were blinded to the clinical features and clinical outcome. Each case was scored based on intensity and percentage of the positive cells. At least 10 high-power fields were chosen randomly, and > 1000 cells were counted for each section. The intensity of NRF3 staining was scored as 0 (no signal), 1+ (weak), 2 (moderate), and 3 (marked). Percentage scores were assigned as 0, negative; 1, 1-25%; 2, 26-50%; 3, 51-75%; and 4, 76-100%. The summed (extension + intensity) was used as the total score. We grouped all samples into the high expression group (total score ≥ 2) and the low one (total score < 2) according to the protein expression [27]. Immunohistochemical (IHC) staining for NRF3 was quantified using German semiquantitative scoring system as described previously [28]. Immunoreactive score (IRS) was determined using the product of extent score and the staining intensity score.
Cell culture and treatment
CRC cell lines, SW480, HT-29, SW620, DLD1, HCT116, loVo, RK0, and HEK293T (an embryonic kidney cell line 293T) were obtained from American Type Culture Collection (Maryland). All the cell lines were grown in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) at 37°C and in 5% CO2. The cells in logarithmic growth were inoculated in a 12-well culture plate (3 × 105 cells/well) for 24 h, and then treated with AG1478 or SB20358 at the indicated concentrations. AG1478 and SB20358 were dissolved in dimethyl sulfoxide (DMSO) as stock solutions, and appropriate amounts of the stock solution were added to the culture cells to achieve the indicated concentrations.
Plasmids and vectors constructing
Nrf3 DNA fragment was generated by polymerase chain reaction (PCR) and cloned into pSIN-vector. Short hairpin RNAs (sh) NRF3#1 and shNRF3#2 were designed to target Nrf3, and shNRF3#1 and shNRF3#2 sequences are shown in Table S1. They were synthetized by GenePharma (Shanghai, China) and cloned into pLVX, and then pLVX-shNRF3#1 and pLVX-shNRF3#1 were obtained. PCR primers used are listed in Table S2. All the plasmids were verified by performing sequencing.
Gene transfection and stable transfect of cells
Gene transfection and stable cell line establishment were performed as described previously [29]. Briefly, 1 × 104 of SW480 and LoVo cells were respectively transfected with 2 μg DNA of pLVX-shscramble, pLVX-shNRF3#1 or pLVX-shNRF3#2, and HCT116 cells were respectively transfected with pSIN or pSIN-NRF3 following the manufacture’s suggested protocol. The stably transfected cell lines, pSIN-HCT116, NRF3-HCT116, shNRF3#1-SW480, shNFR3#2-SW480, shscramble-SW480, shNRF3#1-LoVo, shNRF3#2-loVo were obtained by selection and further confirmed by assessing NRF3 expression.
Western-blotting
Western-blotting was performed as described previously [29]. Briefly, 1 × 106 cells were lysed with lysis buffer (1 × PBS, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate, and freshly added 100 μg/ml phenylmethanesulfonyl fluoride, 10 μg/ml aprotinin, and 1 mM sodium orthovanadate). Cell lysates obtained were centrifuged, and protein concentration of the clarified lysates was measured using Easy II Protein Quantitative Kit (BCA). 40 μg of the supernatant protein was separated by 10% SDS-PAGE and transferred onto a nitrocellulose membrane. The protein membrane was blocked with 5% non fat milk, incubated with the indicated antibody, and then incubated with an appropriate peroxidase conjugated secondary antibody. The signal was developed using 4-chloro-1-napthol/3,3-o-diaminobenzidine, and relative photographic density was quantified by a gel documentation and analysis system. GAPDH was used as an internal control to verify basal expression levels and equal protein loading. The ratio of the specific proteins to GAPDH was calculated.
Apoptosis analysis
Apoptosis analysis was performed as described previously [30]. Briefly, 1 × 104 cells of pSIN-HCT116, NRF3-HCT116, shNRF3#1-SW480, shNFR3#2-SW480, shscramble-SW480, shNRF3#1-LoVo or shNRF3#2-loVo were seeded on six-well plates and cultured to reach 70% confluence, and were treated with 10 or 80 μg/ml 5-fluorouracil (5-FU). After 24 h treatment, the cells were collected by 0.02% trypsin without eathylene diamine tetra acetic acid (EDTA), and stained with annexin V-EGFP (Enhanced Green Fluorescent Protein) and propidium iodide (KeyGen Biotec) according to the manufacturer’s recommendations, and analyzed by flow cytometry.
MTT and colony formation assays
Cell growth was determined by performing MTT assays as described previously [31]. Briefly, pSIN-HCT116, pSIN-NRF3-HCT116, shNRF3#1-SW480, shNRF3#2-W480, shNRF3#1-LoVo shNRF3#2-LoVo (1 × 103) were seeded in 96-well microplates. The cells were cultured for the indicated time, followed by incubation with MTT for 4 h. Optical density (OD) was determined at 450 nm using a microplate reader. Measurements were acquired once per day for 5 d. For the colony-formation assay, pSIN-HCT116 and pSIN-NRF3-HCT116 cells were plated at a density of 500 cells/well in six-well plates and were cultured for 12 d. Colonies were fixed in methanol, stained with 0.5% gentian violet, and counted [32]. Results are presented as mean ± SD of three independent experiments.
Real-time PCR
Real-time PCR was performed as described previously [33]. Briefly, 1 μg DNase-treated RNA was reverse transcribed using Revert AidTM First-Strand cDNA Synthesis Kit (MBI Fermentas, USA) according to the manufacturer’s instructions. Threshold cycle (Ct) value of each sample was determined using Platinum SYBR Green qPCR SuperMix-UDG with ROX (Invitrogen) in ABI 7900HT Real-Time PCR System (Applied Biosystems, Foster City, CA). Sequences of primers used are shown in Table S3. Relative mRNA expression of each target gene was normalized to expression of the housekeeping gene GAPDH. Relative mRNA level was calculated as two power values of ΔCt (Ct value of GAPDH Ct of target gene).
Tumor growth assays in vivo
In vivo tumor growth assays were performed as described previously [34]. Briefly, female BABL/c athymic nude mice (age 4 w) were obtained from an animal center of Guangdong Province (Guangzhou, China). All animal experiments were performed according to the National Institutes of Health Animal Use Guidelines on the Use of Experimental Animals. The nude mice were subcutaneously injected with 2 × 106 cells of shscramble-sw480 and shNRF3#1-SW480, 6 mice per group. The tumors of mice were measured per 2 d. After 17 days, the mice were euthanized, and tumor weights were measured. shNRF3#1-SW480 cells were treated with DMSO, AG1478 (EGFR specific inhibitor) at 10 μM [35] or SB203580 (p38 inhibitor) at 10 μM [36]. These treated cells were subcutaneously injected into nude mice, 6 mice per group. After 17 days, the mice were euthanized. Tumors in the mice were removed and weighed.
Cell invasion and motility assay
Cell invasion and motility were assayed according to the methods described previously with minor modifications [37]. Cell invasion and motility of shscramble-SW480, shNRF3#1-SW480, shNRF3#2-W480 cells were detected using Boyden chamber invasion assay in vitro. Briefly, for invasion assay, matrigel (25 mg/50 ml, Collaborattiv Biomedical Products, Bedford, MA) was added into the chamber to be 8 mm pore size polycarbonate membrane filters. The cells were trypsinized to be suspension cells, and were seeded into the Boyden chamber (Neuro Probe, cabin John, MD) at the upper part at a density of 1.5 × 104 cells/well in 50 μl of serum-free medium, and then incubated for 12 h at 37°C. The bottom chamber also contained standard medium with 20% FBS. The cells invaded to the low surface of membrane were fixed with methanol, and stained with hematoxylin and eosin. The invaded cell numbers were counted under a light microscope. The motility assay was carried out as described in the invasion assay with no coating of matrigel.
Results
NRF3 is lowly expressed in CRC cells and tissues
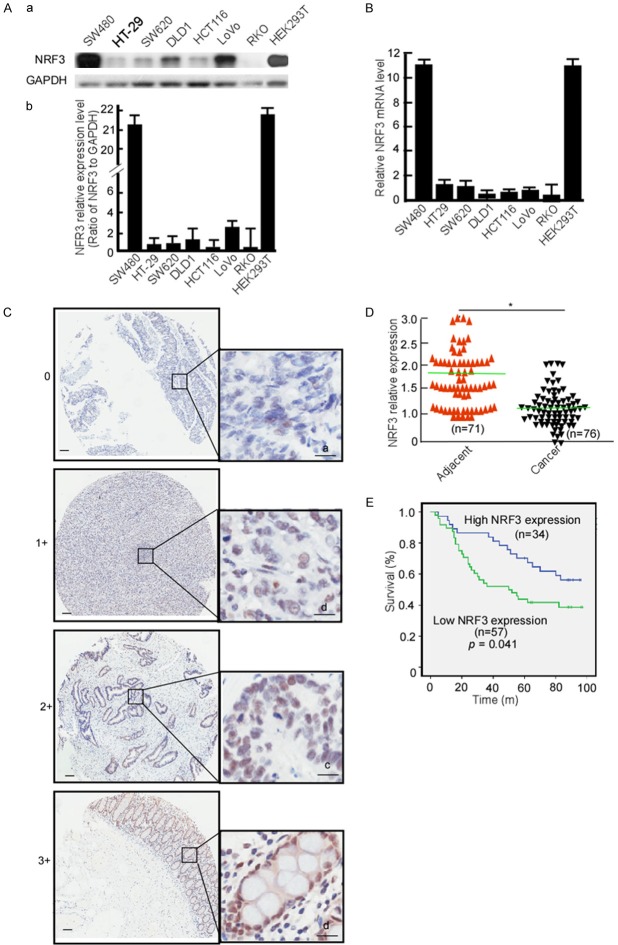
To clarify NRF3 expression in CRC cell, CRC cell lines, SW480, HT-29, SW620, DLD1, HCT116, LoVo, and RK0 cells were detected NRF3 expression using Western-blotting. HEK293T cell served as a control. Compared with HEK293T, NRF3 protein levels were mostly low-expression in HT-29, SW620, DLD1, HCT116, and RK0 except for SW480 and LoVo (Figure 1Aa and 1Ab). HCT116, SW620 and DLD1 had a relatively low NRF3 expression, and SW480 and LoVo had a high expression, they were used to perform the next experiments. Simultaneously, Nrf3 mRNA was detected in these cell lines using real-time PCR, the same results with NRF3 protein expression were obtained (Figure 1B). To clarify NRF3 expression in CRC tissue, a tissue microarray containing 80 pairs of CRC, adjacent non-tumor tissues, and other 20 CRC tissue samples was used to detect NRF3 expression. The IHC results showed that NRF3 was significantly low in CRC tissues when compared with the matched adjacent normal tissues (Figure 1C, 1D, P < 0.05). The positive rate of NRF3 was 78.8% in normal tissues, 47.1% in primary CRC and 29.3% in metastatic CRC tissues, respectively (Table 1). The positive rate of NRF3 was low in primary CRC tissues (Table 1, P < 0.05) and in metastatic CRC (Table 1, P < 0.05) when compared with the normal tissues, but no difference between primary CRC and metastatic CRC tissues. The association of NRF3 expression with CRC stages was analyzed. NRF3 expression was not correlated with T stage (original tumor size and nearby tissue invasion) (Table 2, P < 0.05), N stage (lymph node metastasis) (Table 2, P = 0.191), nor M stage (distant metastasis) (Table 2, P < 0.05). The patients with high NRF3 displayed longer overall survival than low NRF3 expression (Figure 1E, < 0.05). These data strongly suggest that low NRF3 is associated with CRC development.
Figure 1.
NRF3 expressions in CRC cells and tissues. (A) NRF3 protein was detected in SW480, HT-29, SW620, DLD1, HCT116, LoVo RK0 and HEK329T using Western-blotting (a), abundances of NRF3 in CRC cells were analyzed (b). (B) Nrf3 mRNA was analyzed in CRC cell lines using Real-time PCR. (C) NRF3 expression in CRC and non-tumor tissues microarray was detected using IHC. a, negative, scored as 0; b, weakly positive, scored as 1; c, moderately positive, scored as 2; d, positive, scored as 3; Scale Bar, 100 μm. (D) NRF3 expression levels in CRC and adjacent tissue samples were quantified using a German semiquantitative scoring system. Relative expressions in CRC tissues and adjacent non-tumor tissues were statistically analyzed using Mann-Whitney U test. The dots represent scores. (E) The overall survival of CRC patients with high (34) or low (57) NRF3 expression. *, P < 0.05.
Table 1.
NRF3 expressions in normal colorectal, primary CRC, and metastatic CRC tissues
Note: NC, normal colorectal tissue; CRC, primary CRC; MCRC, metastatic CRC tissues.
CRC versus NC;
MCRC versus NC.
Table 2.
NRF3 expressions in CRC samples at various clinical stages
| Characteristic | Cases | NRF3 expression | p | |
|---|---|---|---|---|
|
| ||||
| Low (%) | High (%) | |||
| All patients | ||||
| Gender | ||||
| Male | 40 | 24 (60.0) | 16 (40.0) | 0.677 |
| Female | 51 | 33 (64.7) | 18 (35.3) | |
| Age (yrs) | ||||
| < 50 | 32 | 18 (56.3) | 14 (43.7) | 0.781 |
| ≥ 50 | 59 | 39 (66.1) | 20 (66.9) | |
| T stage | ||||
| T1-T2 | 34 | 21 (61.8) | 13 (38.2) | 0.892 |
| T3-T4 | 57 | 36 (63.2) | 21(36.8) | |
| N stage | ||||
| N0 | 33 | 17 (51.5) | 16 (48.5) | 0.191 |
| N1-N3 | 58 | 40 (69.0) | 18 (31.0) | |
| M stage | ||||
| M0 | 34 | 18 (52.9) | 16 (47.1) | 0.897 |
| M1 | 57 | 39 (68.4) | 18 (31.6) | |
NRF3 inhibits CRC cell growth
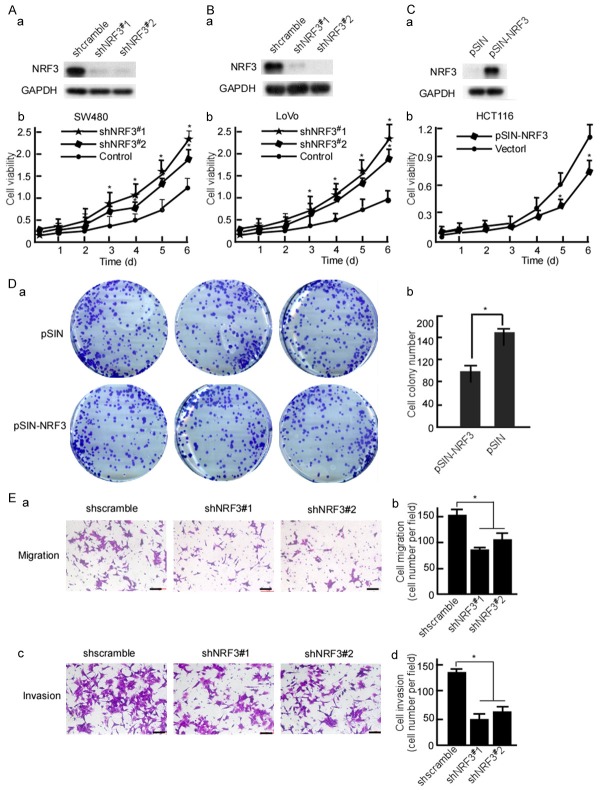
To investigate NRF3 functions in CRC cell growth, shNRF3#1 and shNRF3#2 were designed to block NRF3 expression, and pLVX-shNRF3#1 and pLVX-shNRF3#2 were constructed. SW480 and LoVo cells were stable transfected with pLVX-shNRF3#1 or pLVX-shNRF3#2, respectively. Western-blotting was performed to evaluate the efficiency of shNRF3#1, 2. The results showed that the shNRF3#1, 2 effectively blocked NRF3 protein expression (Figure 2Aa and 2Ba). Viability of the transfected cells was determined by performing MTT assay. After NRF3 was knockdown, SW480 (Figure 2Ab, P < 0.05) and LoVo (Figure 2Bb, P < 0.05) growths were increased when compared with the scramble control. To further determine NRF3’s function, NRF3 expression vector, pSIN-NRF3 was constructed, and was transfected into HCT116 cell. NRF3 was effectively expressed in the transfected cells (Figure 2Ca). The growth kinetics of HCT116 was decreased when being transfected with NRF3 (Figure 2Cb, P < 0.05). Further, cell colony formation of the transfected cells was detected. NRF3 dramatically decreased the colony formation of HCT116 (Figure 2Da and 2Db, P < 0.05). Next, motility and invasion of the transfected cells were also observed, 10 fields were randomly selected and counted the invaded cells per cell well. The results showed that motility (Figure 2Ea and 2Eb, P < 0.05) and invasion (Figure 2Ec and 2Ed, P < 0.05) of the NRF3-transfected cells were dramatically decreased when compared with the control group.
Figure 2.
NRF3 decreases CRC cell growth and Nrf3-knockdown increases cell growth. SW480 (A) and LoVo (B) cells were transfected with pLVX-shscramble, pLVX-shNRF3#1 or pLVX-shNRF3#2 respectively. NRF3 expression in the transfected cells was detected using Western-blotting (a). Viability of the transfected cells was measured using MTT (b). (C) HCT116 cells were stable transfected with pSIN or pSIN-NRF3. NRF3 expression in the transfected cells was detected (a). Viability of the transfected cells was measured (b). (D) Colony formation of NRF3-transfected HCT116 cells was detected using colony formation assay. (E) SW480 cells were transfected with pLVX-shscramble, pLVX-shNRF3#1 or pLVX-shNRF3#2 respectively. Migration (a) and invasion (c) of the transfected cells were detected with Boyden chamber invasion assay. The invaded cells were counted (b, d). All experiments were repeated three times. Data are presented as means ± S.D. of three independent experiments and were statistically analyzed using Student’s t test. Scale bar, 50 μm. *, P < 0.05.
NRF3 expression is negatively related with EGFR and p38 in CRC tissues
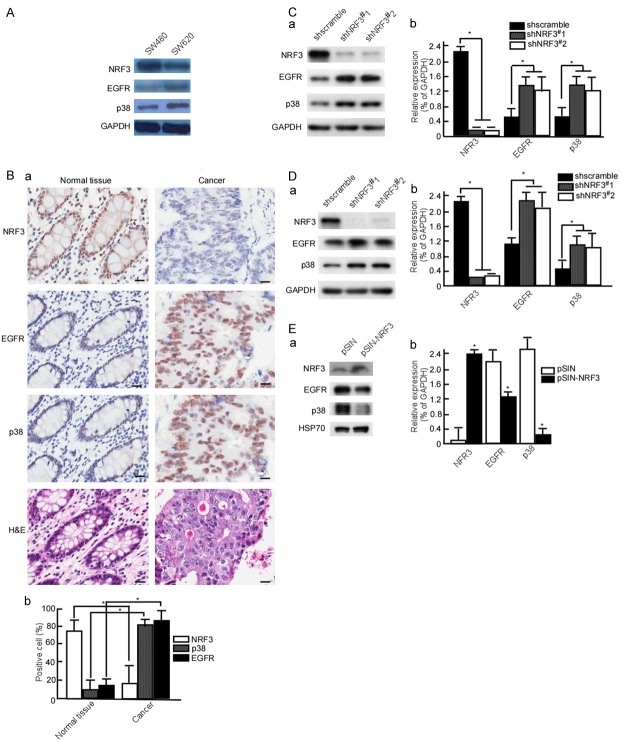
The above results showed that NRF3 is lowly expressed in CRC tissue and cell. EGFR and p38 are highly expressed in CRC, and served as therapeutic targets for CRC therapy [38-41]. To clarify the association of NRF3 with EGFR and p38, SW480 with high NRF3 and SW620 with low NRF3 were used to detect p38 and EGFR expression. The results showed that SW480 cells had low EGFR and p38 expression, while SW620 cells had high EGFR (Figure 3A). This implies that NFR3 expression is negatively associated with EGFR and p38. To confirm this finding, CRC samples were used to determine the relationship of NRF3 with EGFR and p38 expression. The results showed that normal tissues had high NRF3, low EGFR and p38 expression, while CRC tissues had low NRF3, high EGFR and p38 expression (Figure 3Ba and 3Bb, P < 0.05). To further investigate whether NRF3 negatively regulates EGFR and p38, SW480 and LoVo cells were knockdown NFR3 expression using shNRF3#1, 2, and then EGFR and p38 expression were observed. When Nrf3-knockdown, EGFR and p38 were increased in SW480 cells (Figure 3Ca and 3Cb, P < 0.05), and LoVo cells (Figure 3Da and 3Db, P < 0.05). Simultaneously, we up-regulated NRF3 expression, and observed EGFR and p38 expression. HCT116 cells were stable transfected with pSIN-NRF3 to observe EGFR and p38 expression. The results displayed that EGFR and expression were down-regulated when Nrf3-transfect (Figure 3Ea and 3Eb, P < 0.05).
Figure 3.
NRF3 decreases EGFR and p38 expressions. (A) NRF3, EGFR and p38 expressions were detected in SW480 and SW620 cells using Western-blotting. (B) NRF3, EGFR and p38 expressions were analyzed in CRC samples using IHC (a), the positive cells were counted in IHC stained slides (b). SW480 (C) and LoVo (D) cells were stable transfected with pLVX-shscramble, pLVX-shNRF3#1 or pLVX-shNRF3#2 respectively. NRF3, EGFR and p38 expressions were detected in the transfected cells (a), abundances of NRF3, EGFR and p38 in CRC cells were analyzed (b). (E) HCT116 cells were transfected with pSIN or pSIN-NRF3, NRF3, EGFR and p38 expressions were analyzed in the transfected cells (a), abundances of NRF3, EGFR and p38 in CRC cells were analyzed (b). All experiments were repeated three times. Data are presented as means ± S.D. of three independent experiments and were statistically analyzed using Student’s t test. Scale bar, 50 μm. *, P < 0.05.
NRF3 decreases EGFR and p38 activation and downregulates its downstream genes
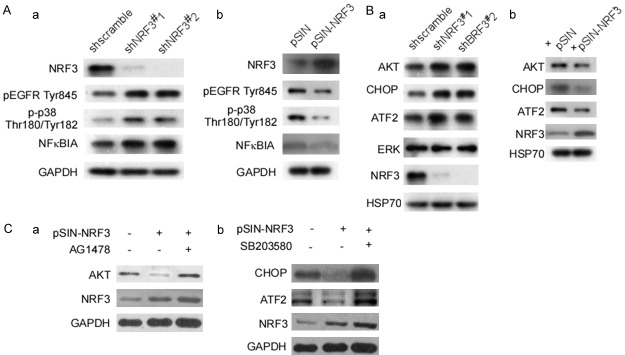
pEGFR Tyr845 is an important manner of EGFR activation, The phosphorylation of EGFR at Tyr845 regulates propensity of the downstream signaling network to participate in cell proliferation, survival, migration, and angiogenesis [42]. The next step, we investigated whether NRF3 affects EGFR phosphorylation. At first, shNRF3#1,2-LoVo cells were detected pEGFR Tyr845 expression. The results showed that pEGFR Tyr845 expression significantly increased when Nrf3-knockdown (Figure 4Aa). p38 is activated by phosphorylation of two Thr-GlyTyr motifs, p38/MAPK phosphorylated-activation plays a important role in human cancer [43]. p-p38 Thr180/Tyr182 and NFκBIA was analyzed. Nrf3-knockdown significantly increased p-p38 Thr180/Tyr182 and NFκBIA expressions (Figure 4Aa). These data imply that Nrf3-knockdown activates p38/MAPK/NFκB and EGFR signal pathways. To further confirm NRF3 regulating EGFR and p38/MAPK, HCT116 cells were transfected with pSIN-NRF3, and then EGFR and p38 were detected. pEGFR Try845 and p-p38 Thr180/Tyr182 significantly decreased in Nrf3-transfected cells when compared with the control (Figure 4Ab).
Figure 4.
NRF3 activates EGFR and p38/MAPK signal pathway. (A) SW480 cells were stable transfected with pLVX-shscramble, pLVX-shNRF3#1 or pLVX-shNRF3#2 respectively. pEGFR Try845, p-p38 Thr180/Tyr182, and NFκBIA expressions were detected in SW480-shNRF3#1, SW480-shNRF3#2 using Western-blotting (a). HCT116 cells were stable transfected with pSIN or pSIN-NRF3. NRF3, pEGFR Tyr845, p-p38 Thr180/Tyr182, and NFκBIA expressions were detected in pSIN-HCT116 and NRF3-HCT116 (b). (B) AKT, CHOP, ATF2, ERK, and NRF3 in shNRF3#1-SW480, shNRF3#2-SW480 cells were detected (a). AKT, CHOP, ATF2, ERK, and NRF3 in pSIN-HCT116 and NRF3-HCT116 cells were detected (b). (C) shNRF3#1-SW480, shNRF3#2-SW480 cells were treated with AG1478, and AKT and NRF3 were tested (a). shNRF3#1-SW480, shNRF3#2-SW480 cells were treated with SB203580, and CHOP, ATF2, ERK and NRF3 were tested (b). pEGFR Tyr845, phosphorylated EGFR Tyrosine 845; p-p38 Thr180/Tyr182, phosphorylation p38 Threonine180/Tyrosine182.
ERK1/2 and AKT are EGFR’s downstream molecules, and play an important role in EGFR-signal pathway [44,45]. CHOP and ATF2 are p38’s downstream molecules, and participate in p38 functions [46,47]. Next, we investigated whether NRF3 regulates ERK1/2, AKT, CHOP and ATF2 expressions. shNRF3#1-SW480 and shNRF3#2-SW480 cells were used to analyze ERK1/2, AKT, CHOP and ATF2 expressions. The results showed that Nrf3-knockdown increased the amount of AKT, CHOP and ATF2, did not ERK (Figure 4Ba). Over-expression of NRF3 in HCT116 cells decreased AKT, CHOP, and ATF2 expression (Figure 4Bb). To further confirm whether NRF3 regulates AKT via EGFR, and regulates CHOP and ATF2 via p38, AG1478 was used to block EGFR expression and SB203580 was used to block p38, and then AKT, CHOP and ATF2 were analyzed. NRF3-mediated AKT decrease was reversed by AG1478 (Figure 4Ca), similarly, CHOP and ATF2 were also rescued by SB203580 (Figure 4Cb). These results showed that EGFR and p38 signal pathways activations are important for NRF3 functions.
NRF3 induces apoptosis and rest cell cycle at G2/M
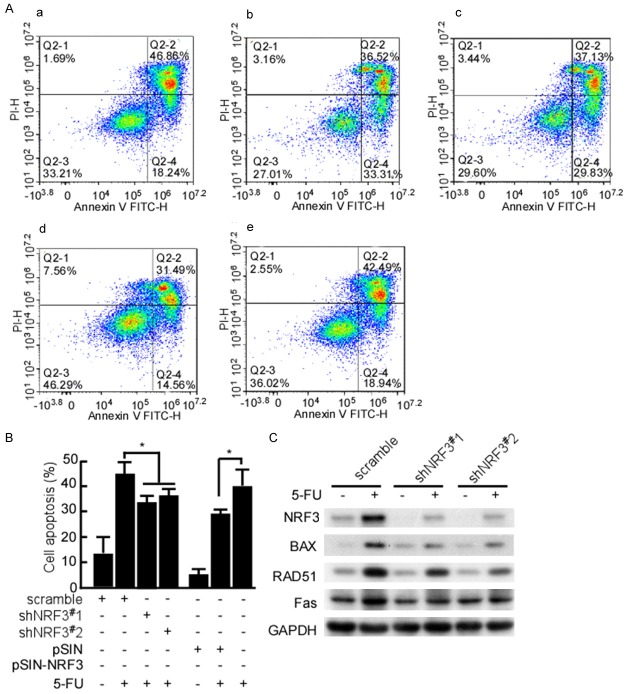
The above results showed that NRF3 inhibited cell growth and colony formation. The next step is to probe whether NRF3 induces cell apoptosis. The stable transfect cells with shNRF3#1, 2, shNRF3#1-SW480 and shNRF3#2-SW480 cells were stained using Annexin V-FITC/PI, and the cell apoptosis was analyzed by flow cytometry. There was no difference in apoptosis of shNRF3#1,2-SW480 and shcramble-SW480 (data not shown). 5-FU was used to treat the above transfect cells, and then the cell apoptosis was detected. The cytometry data showed that shNRF3#1,2 significantly decreased 5-FU-induced apoptosis (Figure 5Ab, 5Ac and 5B, P < 0.05). HCT116 cells were stable transfected with pSIN-NRF3, and NRF3-HCT116 cells were treated with 5-FU, and then the cell apoptosis was analyzed. The cytometry analysis data showed Nrf3-transfect significantly increased 5-FU-induced apoptosis (Figure 5Ae and 5B, P < 0.05). Simultaneously, apoptotic molecules, BAX, RAD51, and Fas were detected in these treated cells. BAX, RAD51, and Fas expressions were decreased in shNRF3#1, 2 groups (Figure 5C). The results showed that NRF3 is involved in CRC cell apoptosis.
Figure 5.
NRF3 increases CRC cell apoptosis. (A) SW480 cells were stable transfected with pLVX-shscramble (a), pLVX-shNRF3#1 (b) or pLVX-shNRF3#2 (c), respectively. HCT116 cells were respectively transfected pSIN (d) or pSIN-NRF3 (e). The stable cell lines, shNRF3#1-SW480, shNRF3#2-SW480, pSIN-HCT116, and NRF3-HCT116 were were treated with 5-FU. The treated cells were stained with Annexin V/PI staining, and then analyzed with flow cytometry. (B) The apoptosis cells were counted in the cell lines treated with Dox. The experiments were repeated three times. Data are presented as means ± S.D. of three independent experiments and were statistically analyzed using Student’s t test. Scale bar, 50 μm. *, P < 0.05. (C) FRF3, BAX, RAD51, and Fas in the treated cells treated with Dox were analyzed using Western-blotting.
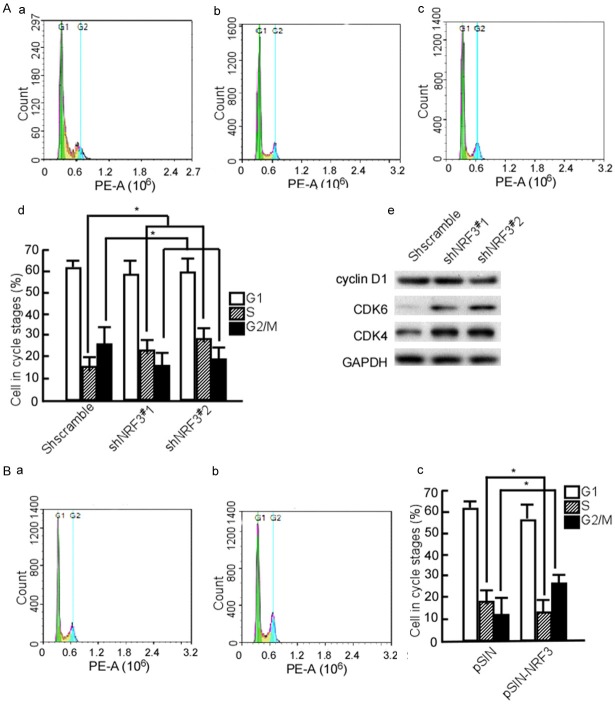
Next, we synchronized shNRF3#1-SW480 and shNRF3#2-SW480 cells with thymidine to determine cell cycle progression, and found that most of shNRF3#1-SW480 and shNRF3#2-SW480 cells stayed in S phase (Figure 6Ab, 6Ac and 6Ad, P < 0.05), while shscramble-SW480 cells stayed at G2/M phase (Figure 6Ba and 6Bd, P < 0.05). Simultaneously, cyclin D1, CDK4, and CDK6 were detected. The results showed that CDK4 and CDK6 expressions were increased in shNRF3#1-SW480 and shNRF3#2-SW480 cells (Figure 6Ae). In additionally, NRF3-HCT116 cells were used to further analyze the effect of NFR3 on the cell cycle, and the data showed that G2/M phase cells significantly increased in the NFR3 transfect group (Figure 6Bb and 6Bc, P < 0.05). These results suggest that NFR3 may suppress G1/S transition.
Figure 6.
NRF3 arrests CRC cell at G2/M. (A) SW480 cells were stable transfected with pLVX-shscramble, pLVX-shNRF3#1 or pLVX-shNRF3#2. The stable cell lines, shscramble-SW480 (a), shNRF3#1-SW480 (b), and shNRF3#2-SW480 (c) were synchronized at G1/S boundary by double thymidine, and then were released in a fresh medium. Cell cycle profiles were analyzed by flow cytometry. The cells in various cycle stages were counted (d). Cyclin D1, CDK4, and CDK6 were detected in the transfected cells using Western-blotting (e). (B) HCT116 cells were stable transfected with pSIN (a) or pSIN-NRF3 (b). The stable transfected cells were synchronized by thymidine, and then were released. Cell cycle profiles were analyzed. The cells in various cycle stages were counted (c). The experiments were repeated three times. Data are presented as means ± S.D. of three independent experiments and were statistically analyzed using Student’s t test. *, P < 0.05.
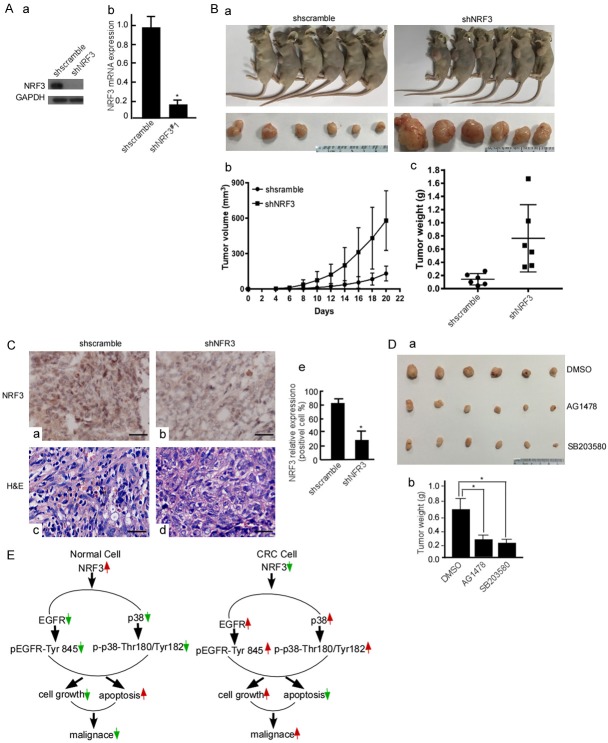
Nrf3-knockdown promotes CRC growth in vivo
The above-mentioned results showed that Nrf3-knockdown increased CRC cell growth and colony formation, and also found that NRF3-mediated EGFR decrease regulated its downstream molecules. In the next step, we further confirmed whether NRF3-loss increases CRC growth using nude mice. Data showed that the tumors of mice injected with shNRF3#1-SW480 were bigger than that of the shscramble mice (Figure 7Ba, 7Bb and 7Bc, P < 0.05). NRF3 expression was detected in the mice tumor tissues using IHC, and positive cells were counted. The IHC results showed that NRF3 expression was lower in the tumor tissues of shNRF3#1-SW480 (Figure 7Cb) than that in the shscramble group (Figure 7Ca and 7Ce, P < 0.05). To further confirm whether EGFR and p38 play an important role in shNRF3-increased CRC growth, shNRF3#1-SW480 cells were treated with AG1478 to block EGFR expression, and SB203580 was used to inhibit p38 expression, and CRC cell growth was observed. The solvent for AG1478 and SB20358, DMSO served as a blank control. The results showed that when EGFR and p38 expressions were blocked, shNRF3-increased CRC growth was blocked (Figure 7Da and 7Db, P < 0.05). Collectively, EGRF and p38 exert a major role on NRF3 loss-mediated CRC growth.
Figure 7.
Nrf3-knockdown promotes CRC growth in vivo. (A) NRF3 protein (a) and Nrf3 mRNA (b) in shscramble-SW480 and shNRF3#1-SW480 cells were analyzed using Western-blotting and Real-time PCR, respectively. (B) Shscramble-SW480 and shNRF3#1-SW480 cells were subcutaneously injected into the dorsal flanks of mice. The tumors of mice were measured per 2 d. After 17 days, the mice were euthanized. Representative images are shown (a). The tumor growth of shscramble-SW480 and shNRF3#1-SW480 cells in vivo was calculated by tumor volume (b). The removed tumors were weighed (c). (C) NRF3 expressions in the implanted tumors was detected using IHC (a, b, c, d), positive cells were counted in 10 fields of the IHC stained section under microscope (e). (D) shNRF3#1-SW480 cells were treated with DMSO, AG1478 or SB203580. These treated cells were subcutaneously injected into nude mice, 6 mice per group. After 17 days, the mice were euthanized, the tumors of mice were removed (a). The removed tumors were weighed (b). *, P < 0.05. (E) Schematic illustration of NRF3 mechanism involved in CRC tumorigenesis. In normal cell, NRF3 inhibits EGFR and p38 activation through decreasing phosphorylation, and regulates the balance of cell growth and apoptosis, which is involved colorectal cell maintenance. In CRC cell, NRF3-loss increases EGFR and p38 phosphorylation activation, enhances cell proliferation and decreases cell apoptosis, and finally promotes CRC malignance.
Discussion
The knowledge derived from the study of syndromes with Mendelian dominant inheritance, which are characterized by a primary predisposition to benign or malignant tumors of the large bowel, has provided important clues regarding the molecular events driving CRC progression from normal epithelium to adenoma to carcinoma [48]. Overexpression of specific oncogenes or/and low expression of tumor suppressor genes in the epithelium result in the formation of hyperproliferative mucosa, produce a benign adenoma, and eventually form a carcinoma [48-50]. The present study suggests that NRF3 loss is involved in CRC carcinogenesis, it is based upon the following three results: (1) NRF3 was lowly expressed in CRC tissues and cells, and its low-expression was associated with advanced CRC development and low 5-year survival rate. (2) Nrf3-knockdown increased CRC cell malignant activity, and Nrf3-knockin decreased cell malignance. (3) In vivo, Nrf3-knockdown enhanced CRC tumor growth. These results suggest that NRF3-loss promotes CRC development. Determination of the underlying mechanism indicated that NRF3 loss is involved CRC development through activating EGFR and p38.
Over-expression and activation of EGFR play a positive role on cancer cell growth and metastasis in variety of solid tumors including CRC [51-54]. EGFR tyrosine phosphorylation leads to activation of numerous of intracellular signals, which are critical to tumor progression including cell growth, epithelial-mesenchymal transition (EMT), metastasis, and angiogenesis. These biological functions are mediated by numerous of EGFR downstream genes including extracellular signal-regulated kinase 1/2 (ERK1/2) and AKT protein kinase [44,45]. Our data showed Nrf3-knockin down-regulated pEGFR Tyr845, and Nrf3-knockdown increased it, while pEGFR Tyr845 is an important manner of EGFR activation. In additional, NRF3 regulated EGFR downstream molecules, ERK and AKT. So, we think that EGFR phosphorylated-activation may be an important mechanism of NRF3-mediated CRC.
p38/MAPKs are a family of serine/threonine-directed kinases classified as “stress-activated” kinases. p38 phosphorylation at Thr180/Tyr182 is an important manner of p38 pathway activation. p38 pathway is involved in the response to extracellular stress stimuli and cell cycle-related events [55,56]. p38/MAPKs activation allows cells to interpret a wide range of external signals and respond appropriately by generating a plethora of different biological effects, such as inflammation, differentiation, cell proliferation and survival [57,58]. p38/MAPKs pathway, together with various signaling cascades such as JNK, ERK, AMPK and PI3K [59-61], regulates the balance between cell survival and cell death through direct effects on cancer development. The tight regulation of survival/death signals by p38/MAPKs can result in opposite molecular functions in tumor development. p38/MAPKs is required for CRC cell proliferation and survival, and its genetic depletion or the pharmacological blockade on its activity induces cell growth arrest, autophagy and death in a cell type-specific manner [62-64]. p38/MAPKs are involved in sustaining tumor growth in some types cancers including follicular lymphoma, lung, thyroid, colorectal, ovarian [65,66] and breast carcinomas, as well as colorectal cancer [67]. In various cancer cell lines, p38 has been shown to enhance tumor cell growth after acquisition of the malignant phenotype. Enhanced p38/MAPK phosphorylation has been correlated with poor overall survival [68,69]. Aberrant activation of p38 is in high grade CRC biopsies [70]. In this study, we found that p38 is highly expressed in CRC samples, and is negatively associated with NRF3. Mechanistically, NFR3 up-regulates p-p38 Thr180/Tyr182 expression, and regulated p38 downstream molecule. Activated p38 activation by NRF3 is involved in CRC development.
In conclusion, our results showed that NRF3-loss in CRC cell enhances EGFR and p38 phosphorylated-expression, which activates EGFR and p38 pathways. The activated EGFR and p38 pathway by NRF3-loss reduced CRC cell apoptosis and increases CRC cell growth, thus promoting CRC malignance (Figure 7E). NRF3 may be a novel molecular target for CRC therapy.
Acknowledgements
This work was supported in part by the National Natural Science Foundation of China (81872226, 81502346), Hunan Provincial Natural Science Foundation of China (2018JJ6131, 2019JJ40175), Changsha Science and Technology Project (kg1801107), Research Projects of Hunan Health Commission (B2019084).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Kemp Z, Thirlwell C, Sieber O, Silver A, Tomlinson I. An update on the genetics of colorectal cancer. Hum Mol Genet. 2004;13:R177–185. doi: 10.1093/hmg/ddh247. [DOI] [PubMed] [Google Scholar]
- 3.Chowdhury A, Katoh H, Hatanaka A, Iwanari H, Nakamura N, Hamakubo T, Natsume T, Waku T, Kobayashi A. Multiple regulatory mechanisms of the biological function of NRF3 (NFE2L3) control cancer cell proliferation. Sci Rep. 2017;7:12494. doi: 10.1038/s41598-017-12675-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jasperson KW, Tuohy TM, Neklason DW, Burt RW. Hereditary and familial colon cancer. Gastroenterology. 2010;138:2044–2058. doi: 10.1053/j.gastro.2010.01.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duraturo F, Liccardo R, Cavallo A, De Rosa M, Grosso M, Izzo P. Association of low-risk MSH3 and MSH2 variant alleles with Lynch syndrome: probability of synergistic effects. Int J Cancer. 2011;129:1643–1650. doi: 10.1002/ijc.25824. [DOI] [PubMed] [Google Scholar]
- 6.Rustgi AK. The genetics of hereditary colon cancer. Genes Dev. 2007;21:2525–2538. doi: 10.1101/gad.1593107. [DOI] [PubMed] [Google Scholar]
- 7.De Rosa M, Pace U, Rega D, Costabile V, Duraturo F, Izzo P, Delrio P. Genetics, diagnosis and management of colorectal cancer (Review) Oncol Rep. 2015;34:1087–1096. doi: 10.3892/or.2015.4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sykiotis GP, Bohmann D. Stress-activated cap’n’collar transcription factors in aging and human disease. Sci Signal. 2010;3:re3. doi: 10.1126/scisignal.3112re3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chevillard G, Blank V. NFE2L3 (NRF3): the Cinderella of the Cap’n’Collar transcription factors. Cell Mol Life Sci. 2011;68:3337–3348. doi: 10.1007/s00018-011-0747-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bugno M, Daniel M, Chepelev NL, Willmore WG. Changing gears in Nrf1 research, from mechanisms of regulation to its role in disease and prevention. Biochim Biophys Acta. 2015;1849:1260–1276. doi: 10.1016/j.bbagrm.2015.08.001. [DOI] [PubMed] [Google Scholar]
- 11.Kobayashi A, Ito E, Toki T, Kogame K, Takahashi S, Igarashi K, Hayashi N, Yamamoto M. Molecular cloning and functional characterization of a new Cap’n’ collar family transcription factor Nrf3. J Biol Chem. 1999;274:6443–6452. doi: 10.1074/jbc.274.10.6443. [DOI] [PubMed] [Google Scholar]
- 12.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y, Kobayashi A, Yamamoto M, Hayes JD. The Nrf3 transcription factor is a membrane-bound glycoprotein targeted to the endoplasmic reticulum through its N-terminal homology box 1 sequence. J Biol Chem. 2009;284:3195–3210. doi: 10.1074/jbc.M805337200. [DOI] [PubMed] [Google Scholar]
- 14.Shepard HM, Brdlik CM, Schreiber H. Signal integration: a framework for understanding the efficacy of therapeutics targeting the human EGFR family. J Clin Invest. 2008;118:3574–3581. doi: 10.1172/JCI36049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mitsudomi T, Yatabe Y. Epidermal growth factor receptor in relation to tumor development: EGFR gene and cancer. FEBS J. 2010;277:301–308. doi: 10.1111/j.1742-4658.2009.07448.x. [DOI] [PubMed] [Google Scholar]
- 16.Llovet JM, Bruix J. Molecular targeted therapies in hepatocellular carcinoma. Hepatology. 2008;48:1312–1327. doi: 10.1002/hep.22506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takeuchi K, Ito F. EGF receptor in relation to tumor development: molecular basis of responsiveness of cancer cells to EGFR-targeting tyrosine kinase inhibitors. FEBS J. 2010;277:316–326. doi: 10.1111/j.1742-4658.2009.07450.x. [DOI] [PubMed] [Google Scholar]
- 18.Eguchi S, Dempsey PJ, Frank GD, Motley ED, Inagami T. Activation of MAPKs by angiotensin II in vascular smooth muscle cells. Metalloprotease-dependent EGF receptor activation is required for activation of ERK and p38 MAPK but not for JNK. J Biol Chem. 2001;276:7957–7962. doi: 10.1074/jbc.M008570200. [DOI] [PubMed] [Google Scholar]
- 19.Alspach E, Flanagan KC, Luo X, Ruhland MK, Huang H, Pazolli E, Donlin MJ, Marsh T, Piwnica-Worms D, Monahan J, Novack DV, McAllister SS, Stewart SA. p38MAPK plays a crucial role in stromal-mediated tumorigenesis. Cancer Discov. 2014;4:716–729. doi: 10.1158/2159-8290.CD-13-0743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Igea A, Nebreda AR. The stress kinase p38alpha as a target for cancer therapy. Cancer Res. 2015;75:3997–4002. doi: 10.1158/0008-5472.CAN-15-0173. [DOI] [PubMed] [Google Scholar]
- 21.Zhang F, Strand A, Robbins D, Cobb MH, Goldsmith EJ. Atomic structure of the MAP kinase ERK2 at 2.3 a resolution. Nature. 1994;367:704–711. doi: 10.1038/367704a0. [DOI] [PubMed] [Google Scholar]
- 22.Wilson KP, Fitzgibbon MJ, Caron PR, Griffith JP, Chen W, McCaffrey PG, Chambers SP, Su MS. Crystal structure of p38 mitogen-activated protein kinase. J Biol Chem. 1996;271:27696–27700. doi: 10.1074/jbc.271.44.27696. [DOI] [PubMed] [Google Scholar]
- 23.Greenberg AK, Basu S, Hu J, Yie TA, Tchou-Wong KM, Rom WN, Lee TC. Selective p38 activation in human non-small cell lung cancer. Am J Respir Cell Mol Biol. 2002;26:558–564. doi: 10.1165/ajrcmb.26.5.4689. [DOI] [PubMed] [Google Scholar]
- 24.Elenitoba-Johnson KS, Jenson SD, Abbott RT, Palais RA, Bohling SD, Lin Z, Tripp S, Shami PJ, Wang LY, Coupland RW, Buckstein R, Perez-Ordonez B, Perkins SL, Dube ID, Lim MS. Involvement of multiple signaling pathways in follicular lymphoma transformation: p38-mitogen-activated protein kinase as a target for therapy. Proc Natl Acad Sci U S A. 2003;100:7259–7264. doi: 10.1073/pnas.1137463100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pomerance M, Quillard J, Chantoux F, Young J, Blondeau JP. High-level expression, activation, and subcellular localization of p38-MAP kinase in thyroid neoplasms. J Pathol. 2006;209:298–306. doi: 10.1002/path.1975. [DOI] [PubMed] [Google Scholar]
- 26.Hong J, Hu K, Yuan Y, Sang Y, Bu Q, Chen G, Yang L, Li B, Huang P, Chen D, Liang Y, Zhang R, Pan J, Zeng YX, Kang T. CHK1 targets spleen tyrosine kinase (L) for proteolysis in hepatocellular carcinoma. J Clin Invest. 2012;122:2165–2175. doi: 10.1172/JCI61380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao SH, Wang Y, Wen L, Zhai ZB, Ai ZH, Yao NL, Wang L, Liu WC, Chen BL, Li Y, Yang H. Basigin-2 is the predominant basigin isoform that promotes tumor cell migration and invasion and correlates with poor prognosis in epithelial ovarian cancer. J Transl Med. 2013;11:92. doi: 10.1186/1479-5876-11-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klopfleisch R. Multiparametric and semiquantitative scoring systems for the evaluation of mouse model histopathology--a systematic review. BMC Vet Res. 2013;9:123. doi: 10.1186/1746-6148-9-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kang T, Wei Y, Honaker Y, Yamaguchi H, Appella E, Hung MC, Piwnica-Worms H. GSK-3 beta targets Cdc25A for ubiquitin-mediated proteolysis, and GSK-3 beta inactivation correlates with Cdc25A overproduction in human cancers. Cancer Cell. 2008;13:36–47. doi: 10.1016/j.ccr.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu M, Tang X, Guo J, Sun W, Tang F. Reversal effect of adenovirus-mediated human interleukin 24 transfection on the cisplatin resistance of A549/DDP lung cancer cells. Oncol Rep. 2017;38:2843–2851. doi: 10.3892/or.2017.6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang J, Wang G, Zhang M, Li FY, Sang Y, Wang B, Hu K, Wu Y, Luo R, Liao D, Cao J, Wang X, Wang L, Zhang R, Zhang X, Deng WG, Xie D, Xu RH, Kang T. Paradoxical role of CBX8 in proliferation and metastasis of colorectal cancer. Oncotarget. 2014;5:10778–10790. doi: 10.18632/oncotarget.2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He Z, Tang F, Ermakova S, Li M, Zhao Q, Cho YY, Ma WY, Choi HS, Bode AM, Yang CS, Dong Z. Fyn is a novel target of (-)-epigallocatechin gallate in the inhibition of JB6 Cl41 cell transformation. Mol Carcinog. 2008;47:172–183. doi: 10.1002/mc.20299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu S, Wu Y, Chen Q, Cao J, Hu K, Tang J, Sang Y, Lai F, Wang L, Zhang R, Li SP, Zeng YX, Yin Y, Kang T. hSSB1 regulates both the stability and the transcriptional activity of p53. Cell Res. 2013;23:423–435. doi: 10.1038/cr.2012.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang FQ, Duan CJ, Huang DM, Wang WW, Xie CL, Meng JJ, Wang L, Jiang HY, Feng DY, Wu SH, Gu HH, Li MY, Deng FL, Gong ZJ, Zhou H, Xu YH, Tan C, Zhang X, Cao Y. HSP70 and mucin 5B: novel protein targets of N,N’-dinitrosopiperazine-induced nasopharyngeal tumorigenesis. Cancer Sci. 2009;100:216–224. doi: 10.1111/j.1349-7006.2008.01028.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dorobantu CM, Harak C, Klein R, van der Linden L, Strating JR, van der Schaar HM, Lohmann V, van Kuppeveld FJ. Tyrphostin AG1478 inhibits encephalomyocarditis virus and hepatitis c virus by targeting phosphatidylinositol 4-kinase IIIalpha. Antimicrob Agents Chemother. 2016;60:6402–6406. doi: 10.1128/AAC.01331-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang SF, Li HC, Huang YP, Tasi WJ, Chou YY, Lu SC. SB203580 increases G-CSF production via a stem-loop destabilizing element in the 3’ untranslated region in macrophages independently of its effect on p38 MAPK activity. J Biomed Sci. 2016;23:3. doi: 10.1186/s12929-016-0221-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang F, Zou F, Peng Z, Huang D, Wu Y, Chen Y, Duan C, Cao Y, Mei W, Tang X, Dong Z. N,N’-dinitrosopiperazine-mediated ezrin protein phosphorylation via activation of Rho kinase and protein kinase C is involved in metastasis of nasopharyngeal carcinoma 6-10B cells. J Biol Chem. 2011;286:36956–36967. doi: 10.1074/jbc.M111.259234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Misale S, Di Nicolantonio F, Sartore-Bianchi A, Siena S, Bardelli A. Resistance to anti-EGFR therapy in colorectal cancer: from heterogeneity to convergent evolution. Cancer Discov. 2014;4:1269–1280. doi: 10.1158/2159-8290.CD-14-0462. [DOI] [PubMed] [Google Scholar]
- 39.Lien K, Berry S, Ko YJ, Chan KK. The use of EGFR inhibitors in colorectal cancer: is it clinically efficacious and cost-effective? Expert Rev Pharmacoecon Outcomes Res. 2015;15:81–100. doi: 10.1586/14737167.2015.982100. [DOI] [PubMed] [Google Scholar]
- 40.Chocry M, Leloup L, Kovacic H. Reversion of resistance to oxaliplatin by inhibition of p38 MAPK in colorectal cancer cell lines: involvement of the calpain/Nox1 pathway. Oncotarget. 2017;8:103710–103730. doi: 10.18632/oncotarget.21780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zou X, Blank M. Targeting p38 MAP kinase signaling in cancer through post-translational modifications. Cancer Lett. 2017;384:19–26. doi: 10.1016/j.canlet.2016.10.008. [DOI] [PubMed] [Google Scholar]
- 42.Sato K. Cellular functions regulated by phosphorylation of EGFR on Tyr845. Int J Mol Sci. 2013;14:10761–10790. doi: 10.3390/ijms140610761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cuenda A, Rousseau S. p38 MAP-kinases pathway regulation, function and role in human diseases. Biochim Biophys Acta. 2007;1773:1358–1375. doi: 10.1016/j.bbamcr.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 44.Chang ZG, Wei JM, Qin CF, Hao K, Tian XD, Xie K, Xie XH, Yang YM. Suppression of the epidermal growth factor receptor inhibits epithelial-mesenchymal transition in human pancreatic cancer PANC-1 cells. Dig Dis Sci. 2012;57:1181–1189. doi: 10.1007/s10620-012-2036-4. [DOI] [PubMed] [Google Scholar]
- 45.Gan Y, Shi C, Inge L, Hibner M, Balducci J, Huang Y. Differential roles of ERK and Akt pathways in regulation of EGFR-mediated signaling and motility in prostate cancer cells. Oncogene. 2010;29:4947–4958. doi: 10.1038/onc.2010.240. [DOI] [PubMed] [Google Scholar]
- 46.Shatanawi A, Lemtalsi T, Yao L, Patel C, Caldwell RB, Caldwell RW. Angiotensin II limits NO production by upregulating arginase through a p38 MAPK-ATF-2 pathway. Eur J Pharmacol. 2015;746:106–114. doi: 10.1016/j.ejphar.2014.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Do MT, Na M, Kim HG, Khanal T, Choi JH, Jin SW, Oh SH, Hwang IH, Chung YC, Kim HS, Jeong TC, Jeong HG. Ilimaquinone induces death receptor expression and sensitizes human colon cancer cells to TRAIL-induced apoptosis through activation of ROS-ERK/p38 MAPK-CHOP signaling pathways. Food Chem Toxicol. 2014;71:51–59. doi: 10.1016/j.fct.2014.06.001. [DOI] [PubMed] [Google Scholar]
- 48.Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, Leppert M, Nakamura Y, White R, Smits AM, Bos JL. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319:525–532. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- 49.Pancione M, Remo A, Colantuoni V. Genetic and epigenetic events generate multiple pathways in colorectal cancer progression. Patholog Res Int. 2012;2012:509348. doi: 10.1155/2012/509348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ewing I, Hurley JJ, Josephides E, Millar A. The molecular genetics of colorectal cancer. Frontline Gastroenterol. 2014;5:26–30. doi: 10.1136/flgastro-2013-100329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Suriyo T, Tachachartvanich P, Visitnonthachai D, Watcharasit P, Satayavivad J. Chlorpyrifos promotes colorectal adenocarcinoma H508 cell growth through the activation of EGFR/ERK1/2 signaling pathway but not cholinergic pathway. Toxicology. 2015;338:117–129. doi: 10.1016/j.tox.2015.10.009. [DOI] [PubMed] [Google Scholar]
- 52.Chen D, Huang X, Cai J, Guo S, Qian W, Wery JP, Li QX. A set of defined oncogenic mutation alleles seems to better predict the response to cetuximab in CRC patient-derived xenograft than KRAS 12/13 mutations. Oncotarget. 2015;6:40815–40821. doi: 10.18632/oncotarget.5886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Linardou H, Dahabreh IJ, Kanaloupiti D, Siannis F, Bafaloukos D, Kosmidis P, Papadimitriou CA, Murray S. Assessment of somatic k-RAS mutations as a mechanism associated with resistance to EGFR-targeted agents: a systematic review and meta-analysis of studies in advanced non-small-cell lung cancer and metastatic colorectal cancer. Lancet Oncol. 2008;9:962–972. doi: 10.1016/S1470-2045(08)70206-7. [DOI] [PubMed] [Google Scholar]
- 54.Wang RY, Chen L, Chen HY, Hu L, Li L, Sun HY, Jiang F, Zhao J, Liu GM, Tang J, Chen CY, Yang YC, Chang YX, Liu H, Zhang J, Yang Y, Huang G, Shen F, Wu MC, Zhou WP, Wang HY. MUC15 inhibits dimerization of EGFR and PI3K-AKT signaling and is associated with aggressive hepatocellular carcinomas in patients. Gastroenterology. 2013;145:1436–1448. e1431–1412. doi: 10.1053/j.gastro.2013.08.009. [DOI] [PubMed] [Google Scholar]
- 55.Brewster JL, de Valoir T, Dwyer ND, Winter E, Gustin MC. An osmosensing signal transduction pathway in yeast. Science. 1993;259:1760–1763. doi: 10.1126/science.7681220. [DOI] [PubMed] [Google Scholar]
- 56.Shiozaki K, Russell P. Conjugation, meiosis, and the osmotic stress response are regulated by Spc1 kinase through Atf1 transcription factor in fission yeast. Genes Dev. 1996;10:2276–2288. doi: 10.1101/gad.10.18.2276. [DOI] [PubMed] [Google Scholar]
- 57.Wagner EF, Nebreda AR. Signal integration by JNK and p38 MAPK pathways in cancer development. Nat Rev Cancer. 2009;9:537–549. doi: 10.1038/nrc2694. [DOI] [PubMed] [Google Scholar]
- 58.Chiacchiera F, Simone C. Signal-dependent regulation of gene expression as a target for cancer treatment: inhibiting p38alpha in colorectal tumors. Cancer Lett. 2008;265:16–26. doi: 10.1016/j.canlet.2008.02.061. [DOI] [PubMed] [Google Scholar]
- 59.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 60.Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C, Edkins S, O’Meara S, Vastrik I, Schmidt EE, Avis T, Barthorpe S, Bhamra G, Buck G, Choudhury B, Clements J, Cole J, Dicks E, Forbes S, Gray K, Halliday K, Harrison R, Hills K, Hinton J, Jenkinson A, Jones D, Menzies A, Mironenko T, Perry J, Raine K, Richardson D, Shepherd R, Small A, Tofts C, Varian J, Webb T, West S, Widaa S, Yates A, Cahill DP, Louis DN, Goldstraw P, Nicholson AG, Brasseur F, Looijenga L, Weber BL, Chiew YE, DeFazio A, Greaves MF, Green AR, Campbell P, Birney E, Easton DF, Chenevix-Trench G, Tan MH, Khoo SK, Teh BT, Yuen ST, Leung SY, Wooster R, Futreal PA, Stratton MR. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, Shen D, Boca SM, Barber T, Ptak J, Silliman N, Szabo S, Dezso Z, Ustyanksky V, Nikolskaya T, Nikolsky Y, Karchin R, Wilson PA, Kaminker JS, Zhang Z, Croshaw R, Willis J, Dawson D, Shipitsin M, Willson JK, Sukumar S, Polyak K, Park BH, Pethiyagoda CL, Pant PV, Ballinger DG, Sparks AB, Hartigan J, Smith DR, Suh E, Papadopoulos N, Buckhaults P, Markowitz SD, Parmigiani G, Kinzler KW, Velculescu VE, Vogelstein B. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 62.Comes F, Matrone A, Lastella P, Nico B, Susca FC, Bagnulo R, Ingravallo G, Modica S, Lo Sasso G, Moschetta A, Guanti G, Simone C. A novel cell type-specific role of p38alpha in the control of autophagy and cell death in colorectal cancer cells. Cell Death Differ. 2007;14:693–702. doi: 10.1038/sj.cdd.4402076. [DOI] [PubMed] [Google Scholar]
- 63.Simone C. Signal-dependent control of autophagy and cell death in colorectal cancer cell: the role of the p38 pathway. Autophagy. 2007;3:468–471. doi: 10.4161/auto.4319. [DOI] [PubMed] [Google Scholar]
- 64.Madia F, Grossi V, Peserico A, Simone C. Updates from the intestinal front line: autophagic weapons against inflammation and cancer. Cells. 2012;1:535–557. doi: 10.3390/cells1030535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Matrone A, Grossi V, Chiacchiera F, Fina E, Cappellari M, Caringella AM, Di Naro E, Loverro G, Simone C. p38alpha is required for ovarian cancer cell metabolism and survival. Int J Gynecol Cancer. 2010;20:203–211. doi: 10.1111/igc.0b013e3181c8ca12. [DOI] [PubMed] [Google Scholar]
- 66.Grossi V, Simone C. Special agents hunting down women silent killer: the emerging role of the p38alpha kinase. J Oncol. 2012;2012:382159. doi: 10.1155/2012/382159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Grossi V, Peserico A, Tezil T, Simone C. p38alpha MAPK pathway: a key factor in colorectal cancer therapy and chemoresistance. World J Gastroenterol. 2014;20:9744–9758. doi: 10.3748/wjg.v20.i29.9744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Esteva FJ, Sahin AA, Smith TL, Yang Y, Pusztai L, Nahta R, Buchholz TA, Buzdar AU, Hortobagyi GN, Bacus SS. Prognostic significance of phosphorylated P38 mitogen-activated protein kinase and HER-2 expression in lymph node-positive breast carcinoma. Cancer. 2004;100:499–506. doi: 10.1002/cncr.11940. [DOI] [PubMed] [Google Scholar]
- 69.Wang SN, Lee KT, Tsai CJ, Chen YJ, Yeh YT. Phosphorylated p38 and JNK MAPK proteins in hepatocellular carcinoma. Eur J Clin Invest. 2012;42:1295–1301. doi: 10.1111/eci.12003. [DOI] [PubMed] [Google Scholar]
- 70.Chiacchiera F, Grossi V, Cappellari M, Peserico A, Simonatto M, Germani A, Russo S, Moyer MP, Resta N, Murzilli S, Simone C. Blocking p38/ERK crosstalk affects colorectal cancer growth by inducing apoptosis in vitro and in preclinical mouse models. Cancer Lett. 2012;324:98–108. doi: 10.1016/j.canlet.2012.05.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.