Abstract
As a core kinase in the Hippo pathway, large tumor suppressor kinase 2 (LATS2) regulates cell proliferation, migration and invasion through numerous signaling pathways. However, its functions on cell proliferation, migration and invasion in glioma have yet to be elucidated. The present study revealed that LATS2 was downregulated in glioma tissues and cells, as determined by reverse transcription-quantitative polymerase chain reaction and immunohistochemistry. In addition, Cell Counting Kit-8, scratch wound healing and Transwell assays revealed that overexpression of LATS2 in U-372 MG cells inhibited cell proliferation, migration and invasion. Furthermore, western blot analysis indicated that the expression levels of phosphorylated (p)-yes-associated protein and p-tafazzin were increased in cells with LATS2 overexpression. These results indicated that LATS2 is a potential tumor suppressor, and downregulation of LATS2 in glioma may contribute to cancer progression.
Keywords: LATS2, glioma, cell proliferation, cell migration and invasion, YAP/TAZ
Introduction
Gliomas are the most common tumors in the central nervous system (CNS), which are associated with a poor prognosis. Due to the ability of gliomas to locally invade the surrounding brain parenchyma, the median survival time is only 12–21 months, even in patients that have received appropriate diagnosis and treatment, such as surgery, radiation and chemotherapy (1,2). Although considerable effort and progress has been made to coordinate effective therapies, the heterogeneity of gliomas, with regards to cytology and gene expression, makes it a complex task (3,4). Therefore, understanding the molecular mechanism and identifying specific glioma-associated genes as pharmacological targets for treatment is critically important.
It has previously been reported that the evolutionarily conserved Hippo pathway serves an important role in cellular proliferation, transformation and tumorigenesis. The core of the Hippo pathway is a kinase cascade involving the macrophage-stimulating (MST)1/2 kinases (also known as serine threonine kinase 4/3) of the STE-20 family, and their downstream kinases, large tumor suppressor kinase (LATS)1/2 of the AGC family (5). Inhibition of the Hippo pathway results in hyperactivation of the main downstream effector yes-associated protein (YAP)/tafazzin (TAZ)/TEA domain transcription factor (TEAD), which in turn promotes the expression of cell proliferation-, epithelial-mesenchymal transition (EMT)- and anti-apoptosis-associated genes, leading to accelerated tumor development and malignancy (6). Therefore, in cancer, various signaling cascades that suppress the Hippo pathway and activate YAP/TAZ/TEAD have been described. Notably, dysregulation of the Hippo pathway has been revealed to participate in various types of cancers, including liver, lung, colorectal, ovarian and prostate cancers (7). In glioma, previous studies have reported that YAP and TAZ are highly expressed, thus increasing tumor initiation and invasion (8,9). Gene Expression Omnibus (GSE15824) data revealed the downregulation of LATS2 in glioma (10); however, whether downregulation of LATS2 mediates tumorigenesis through activation of YAP/TAZ in glioma remains unknown.
The present study confirmed that LATS2 expression was downregulated in glioma clinical specimens. Notably, in vitro overexpression of LATS2 suppressed proliferation, migration and invasion of U-372 MG cells, via increasing the levels of phosphorylated (p)-YAP/TAZ, resulting in their cytoplasmic retention. Taken together, these findings indicated the role of LATS2-mediated phosphorylation of YAP/TAZ in glioma tumorigenesis.
Materials and methods
Tissue specimens and clinical data
The present study was approved by the Ethics Committee of The Second Hospital of Hebei Medical University (Shijiazhuang, China), and all patients provided written informed consent. The patients included 40 men and 28 women, with a mean age of 39.5 years (age range, 8–73 years), who were recruited to the study between January 2016 and December 2017. A total of 80 tissue samples (19 astrocytoma, 24 oligodendroglioma, 25 glioblastoma samples and 12 normal samples) were obtained from the Department of Pathology of The Second Hospital of Hebei Medical University, and were used in accordance with the guidelines approved by The Second Hospital of Hebei Medical University Ethics Committee. No patients underwent radiation or chemotherapy prior to surgery. The 12 normal cortex tissue specimens were obtained from individuals who died in traffic accidents; their families provided informed consent for the use of these tissues. The use of these 12 normal cortex tissue specimens was also approved by the Ethics Committee of The Second Hospital of Hebei Medical University.
Cell culture
The U-372 MG (TPBT001265C; Tongpai Biological Technology, Shanghai, China), LN-229 (BNCC341218; BeNa Culture Collection, Kunshan, China), U-251 MG (CBP60300; BeNa Culture Collection) and A172 human glioma cell lines (CRL-1620; American Type Culture Collection, Manassas, VA, USA), and the HEB human normal glial cell line (BeNa Culture Collection) were cultured in Dulbecco's modified Eagle's medium with high glucose (cat. no. 11965–092; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with glutamine, penicillin/streptomycin (cat. no. 15070063; Gibco; Thermo Fisher Scientific, Inc.) and 10% fetal bovine serum (cat. no. 12483020; Gibco; Thermo Fisher Scientific, Inc.) at 37°C in an incubator containing 5% CO2. Plasmids (3 µg/106 cells) or small interfering (si)RNA (30 pmol/106 cells) were transfected into cells using Lipofectamine® 3000 (cat. no. L3000015; Invitrogen; Thermo Fisher Scientific, Inc.) or Lipofectamine® RNAiMAX (cat. no. 13778150; Invitrogen; Thermo Fisher Scientific, Inc.) respectively, according to the manufacturer's protocols. Cells were cultured for another 48 h, or for the indicated durations, prior to further analysis.
Plasmids and siRNA
The entire human LATS2 (Gene ID, 26524; size, 3,267 bp) and YAP (Gene ID, 10413; size, 1,515 bp) genes were amplified by polymerase chain reaction (PCR) using KOD DNA polymerase (Toyobo Life Science, Osaka, Japan), according to the manufacturer's protocol. The fragments were then cloned into pcDNA3.1 plasmids (cat. no. V79020; Invitrogen; Thermo Fisher Scientific, Inc.) by restriction enzyme cutting and ligation, and were sequenced by GENEWIZ, Inc. (Suzhou, China) through Sanger sequencing for validation; the empty pcDNA3.1 vector was used as a control plasmid. The primers used for PCR are listed in Table I, and restriction enzyme recognition sites are underlined. Site-directed mutagenesis for YAP S127A was introduced using the Site-Directed Mutagenesis kit (Beijing SBS Genetech Co., Ltd., Beijing, China), according to the manufacturer's protocol. The sequences of LATS2 siRNA and scramble siRNA, which was used as a negative control, are described in a previous study (11) and were purchased from Invitrogen; Thermo Fisher Scientific, Inc.
Table I.
Polymerase chain reaction primers and siRNA sequences.
| Primers/siRNAs | Sequences (5′-3′) |
|---|---|
| LATS2-forward | CGGGATCCATGAGGCCAAAGACTTTTCCTGCC |
| LATS2-reverse | CCGCTCGAGCTACACGTACACAGGCTGGCAGCC |
| YAP-forward | CGGAATTCATGGATCCCGGGCAGCAG |
| YAP-reverse | CGTCTAGACTATAACCATGTAAGAAAGCTTTCTTTATCTAGC |
| siLAST2 | UACCAUAAAUACAAUCUUCTT |
| Scramble | UUCUCCGAACGUGUCACGUTT |
Restriction enzyme recognition sites (BamHI in LATS2-forward primer and XhoI in LATS2-reverse primer; EcoRI in YAP-forward primer and XbaI in YAP-reverse primer) are underlined. LATS2, large tumor suppressor kinase 2; si/siRNA, small interfering RNA; YAP, yes-associated protein.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.), and the concentration of total RNA was measured using NanoDrop 2000 (NanoDrop; Thermo Fisher Scientific, Inc., Wilmington, DE, USA). Subsequently, RNA (1 µg) was reverse transcribed using the PrimeScript™ RT reagent kit with gDNA Eraser (Takara Biotechnology Co., Ltd., Dalian, China), according to the manufacturer's protocol. RT-qPCR was conducted using SYBR Premix Ex Taq (Takara Biotechnology Co., Ltd.) on an Applied Biosystems 7500 Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA). The thermal cycling conditions were as follows: Pre-denaturation at 95°C for 30 sec, followed by 40 cycles at 95°C for 5 sec and 60°C for 30 sec. Nucleotide sequences of the primers used for RT-qPCR were as follows (5′-3′): LATS2, forward TGGCACCTACTCCCACAG, and reverse CCAAGGGCTTTCTTCATCT; GAPDH, forward TCATGGGTGTGAACCATGAGAA; and reverse GGCATGGACTGTGGTCATGAG. The relative mRNA expression levels of LATS2 were normalized to GAPDH and were calculated as 2ΔCq as described in previous study, where ΔCq = CqLATS2 - CqGAPDH (12).
Immunohistochemistry
Tissues were fixed in 10% neutral formalin for ≤24 h at 25°C, and were then embedded in paraffin. Paraffin-embedded tissue sections (4 µm) were cut, and were then dewaxed in xylene and rehydrated in graded ethanol. After rinsing with water, the sections were heated in antigen retrieval solution (cat. no. P0081; Beyotime Institute of Biotechnology, Shanghai, China) for 10 min, incubated with 3% hydrogen peroxide for 5 min at 25°C to inactivate endogenous peroxidase and blocked with 3% bovine serum albumin (BSA; cat. no. ST023-200g; Beyotime Institute of Biotechnology) at 25°C for 30 min. Subsequently, samples were incubated with LATS2 antibody (1:100; cat. no. ab110780; Abcam, Cambridge, MA, USA) at 4°C overnight. After three 5-min washes in Tris-buffered saline-0.1% Tween-20 (TBST), the sections were incubated with biotin-conjugated goat anti-rabbit immunoglobulin G (IgG) (1:500; cat. no. SA00004-2; Wuhan Sanying Biotechnology, Wuhan, Hubei, China) at 25°C for 45 min, and were subsequently incubated with streptavidin-biotin complex containing horseradish peroxidase (HRP) (cat. no. P0603; Beyotime Institute of Biotechnology) at 25°C for 30 min. Finally, sections were incubated with DAB until color developed and images were captured under a bright field microscope. Finally, a semi-quantitative assessment of LATS2 relative expression was conducted using ImageJ software (ImageJ bundled with 64-bit Java 1.8.0_112; National Institutes of Health, Bethesda, MD, USA). Briefly, 10 visual fields were randomly observed and 200 cells were counted in each field. The positively stained cells were calculated for each section.
Western blot analysis
Cells were collected and lysed in radioimmunoprecipitation assay buffer (cat. no. 9806; Cell Signaling Technology, Inc., Danvers, MA, USA), and total protein concentration was determined using the bicinchoninic acid assay. Protein samples (20 µg) were loaded, separated by 10% SDS-PAGE and transferred to polyvinylidene difluoride membranes. Blots were blocked in 5% (w/v) dry skimmed milk at 25°C for 60 min and were then incubated with the following primary antibodies: Anti-LATS2 (1:1,000; cat. no. ab110780; Abcam), anti-YAP (1:5,000; cat. no. ab52771; Abcam), anti-p-YAP S127 (1:5,000; cat. no. ab76252; Abcam), anti-TAZ (1:1,000; cat. no. ab84927; Abcam), anti-p-TAZ Ser89 (1:1,000; cat. no. 75275; Cell Signaling Technology, Inc.), anti-Cyclin D1 (1:200; cat. no. ab16663; Abcam), anti-cyclin-dependent kinase (CDK)4 (1:1,000; cat. no. ab108357; Abcam), anti-CDK6 (1:5,000; cat. no. ab124821; Abcam) anti-β-actin (1:5,000; cat. no. ab8226; Abcam) at 4°C overnight. After washing three times in TBST (10 min/wash), blots were incubated with HRP-conjugated goat anti-mouse IgG (1:5,000; cat. no. SA00001-1; Wuhan Sanying Biotechnology) or goat anti-rabbit IgG (1:5,000; cat. no. SA00001-2; Wuhan Sanying Biotechnology) for 45 min at room temperature. The blots were then developed using enhanced chemiluminescence substrate (cat. no. 32209; Pierce; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol.
Cell proliferation analysis
Cell growth curves were plotted using the Cell Counting Kit-8 (CCK-8) assay (Dojindo Molecular Technologies, Inc., Kumamoto, Japan), according to the manufacturer's protocol. A total of 104 cells were seeded in 96-well plates with six replicates for each condition. Following incubation with the CCK-8 reagent, absorbance was measured at 450 nm. Each experiment was performed in triplicate.
Cell cycle analysis
Cells were collected, washed twice in cold PBS and fixed with 75% ethanol at −20°C. A total of 5×106 fixed cells were then incubated with propidium iodide (PI) staining solution containing 0.5% Triton X-100, 20 µg/ml PI (cat. no. p4170; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) and 0.2 mg/ml RNase A (Sigma-Aldrich; Merck KGaA) in PBS at 4°C for 30 min. Cells were analyzed by FACSCalibur flow cytometry (BD Biosciences, Franklin Lakes, NJ, USA), and the DNA content of labeled cells was calculated. Each experiment was performed in triplicate.
Scratch wound healing assay
Cells were cultured on 35-mm glass base dishes as ~90% confluent monolayers. Subsequently, the cells were scraped with a 200-µl pipette tip and the cells were washed twice with PBS to remove cell debris. To analyze scratch wound closure, optical images were captured at 0 and 24 h time points using a microscope (Olympus IX71 microscope; Olympus Corporation, Tokyo, Japan) and were analyzed with ImageJ (ImageJ bundled with 64-bit Java 1.8.0_112; National Institutes of Health).
Invasion assay
In vitro invasion assay was performed in transwell inserts (pore size, 8-µm pore; Costar; Corning Incorporated, Corning, NY, USA). Cell suspensions (1×106 cells/ml; 200 µl) were seeded on fibronectin-coated polycarbonate membranes precoated with 50 µl Matrigel (1 mg/ml; BD Biosciences). After 18 h, the cells on the lower side of the membrane were fixed with pre-chilled 100% methanol at 4°C for 30 min and stained with Giemsa at 25°C for 10 min. The stained cells were captured under a microscope (Olympus IX71 microscope; Olympus Corporation) at ×200 magnification and were counted. Experiments were performed in triplicate.
Immunofluorescence
Cells transfected with LATS2 plasmids were grown on glass coverslips. After gentle washing with PBS, cells were fixed in pre-chilled 4% paraformaldehyde for 30 min at 4°C. Subsequently, fixed cells were permeabilized with 0.05% NP-40 in PBS containing 1% BSA for 30 min at 4°C, and blocked in 3% BSA and 10% normal goat serum (cat. no. C0265; Beyotime Institute of Biotechnology) for 30 min at 25°C. Cells were incubated with anti-YAP (1:200; cat. no. ab52771; Abcam) and anti-TAZ (1:200; cat. no. ab84927; Abcam) at 4°C overnight. After washing three times in 0.1% PBS-Tween-20 (3 min/wash), the coverslips were incubated with Alexa Fluor® 488-conjugated goat anti-rabbit IgG (1:500; cat. no. ab150077; Abcam) for 30 min at room temperature. Subsequently, slides were incubated with DAPI (cat. no. C1005; Beyotime Institute of Biotechnology) for 5 min at 25°C and were washed three times in PBS (5 min/wash). Fluorescence was examined under an Olympus fluorescence microscope (Olympus Corporation).
Statistical analyses
All statistical analyses were performed using SPSS 17.0 (SPSS, Inc., Chicago, IL, USA) software. Data are presented as the means ± standard error of the mean, and differences between groups were evaluated using Student's t-test for two groups, or one-way analysis of variance followed by Tukey's multiple comparisons test for three or more groups. P<0.05 was considered to indicate a statistically significant difference.
Results
LATS2 is downregulated in glioma
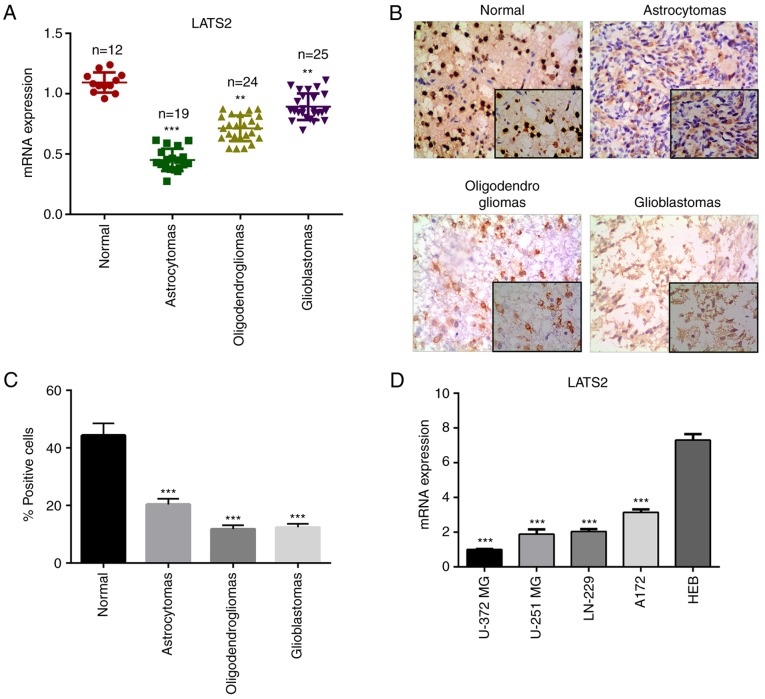
To investigate the function of LATS2 in glioma, the present study determined whether the relative expression levels of LATS2 were altered in glioma tissues (astrocytomas, oligodendrogliomas and glioblastomas) compared with in normal tissue samples using RT-qPCR. The results revealed that the mRNA expression levels of LATS2 were significantly reduced in astrocytomas, oligodendrogliomas and glioblastomas compared with in normal tissue samples (Fig. 1A). For further validation, immunohistochemistry was performed to examine the expression levels of LATS2 in clinical specimens. Consistent with the RT-qPCR results, a lower signal was detected in glioma samples compared with in normal tissues (Fig. 1B and C). These findings suggested that LATS2 is downregulated in glioma, and prompted further investigation into the biological functions of LATS2 in glioma. In order to assess the function of LATS2 in vitro, the expression levels of LATS2 were analyzed in a panel of glioma cell lines, including U-372 MG, LN-229, U-251 MG and A172, and a normal glial cell line (HEB). Of all the glioma cell lines, U-372 MG had the lowest LATS2 expression, as revealed using RT-qPCR analysis; this cell line was chosen for subsequent functional analysis (Fig. 1D).
Figure 1.
LATS2 was downregulated in glioma. (A) Expression levels of LATS2 were significantly lower in astrocytomas, oligodendrogliomas and glioblastomas compared with in normal tissues, as detected by reverse transcription-quantitative polymerase chain reaction. (B) Representative immunohistochemistry images (magnification, ×200) of LATS2 in astrocytomas, oligodendrogliomas and primary glioblastomas. (C) Number of positive cells in immunohistochemistry was calculated. (D) mRNA expression levels of LATS2 in glioma cell lines (U-372 MG, LN-229, U-251 MG and A172) were lower than in the normal glial cell line (HEB); U-372 MG cells had the lowest expression of LATS2. Data are presented as the means ± standard deviation. **P<0.01; ***P<0.001 compared with the normal group or HEB cells. LATS2, large tumor suppressor kinase 2.
LATS2 suppresses cell proliferation, migration and invasion in vitro
Since LATS2 functions as a core kinase in the Hippo pathway regulating cell proliferation and EMT, the present study assessed whether manipulating the expression levels of LATS2, by plasmid or siRNA transfection, affected cell proliferation, migration and invasion of U-372 MG cells. Initially, the transfection efficiency of LATS2 plasmids and LATS2 siRNA was confirmed. The mRNA and protein expression levels of LATS2 were significantly upregulated in cells transfected with LATS2 plasmids and were downregulated in cells transfected with siLATS2, compared with in cells transfected with an empty vector or scramble siRNA (Fig. 2A). To determine the effects of LATS2 on cell proliferation, cell growth was measured using a CCK-8 assay. The growth curves revealed that LATS2 overexpression significantly inhibited cell growth, whereas LATS2 knockdown resulted in opposite effects (Fig. 2B). Subsequently, the effects of LATS2 on cell cycle progression were determined through flow cytometry using PI staining. As shown in Fig. 2C, overexpression of LATS2 increased the number of cells in the G1 phase and decreased the number of cells in the S phase, whereas LATS2 knockdown exhibited the opposite effect; neither of them had any effect on the number of cells in the G2/M phase (Fig. 2C). These results indicated that expression of LATS2 delayed cell cycle progression in the G1 phase. Given the known functions of cyclin D1, CDK4 and CDK6 in G1/S transition, the present study measured the expression levels of these proteins by western blotting. Notably, overexpression of LATS2 decreased the expression levels of cyclin D1, CDK4 and CDK6, where LATS2 knockdown had the opposite effect; these proteins are required for progression through the G1 phase of the cell cycle (Fig. 2D).
Figure 2.
Overexpression of LATS2 suppresses cell proliferation by delaying G1/S transition in U-372 MG cells. (A) Efficiency of LATS2 plasmid and siLATS2 transfection was validated in U-372 MG cells. The mRNA and protein expression levels of LATS2 were significantly upregulated or downregulated in cells transfected with plasmids or siRNA, respectively. Empty vector or NC siRNA were used as controls. (B) Effects of LATS2 on cell growth were determined by Cell Counting Kit-8 assay. OD450 was measured at 0, 24, 48 and 72 h post-transfection. Overexpression of LATS2 decreased cell growth (left), whereas LATS2 knockdown increased cell growth (right). (C) Effects of LATS2 on cell cycle progression were measured by flow cytometry using propidium iodide. Overexpression of LATS2 increased the percentage of cells in the G1 phase and decreased the number of cells in the S phase compared with the control, whereas knockdown of LATS2 had the opposite effects; G2/M phase was not affected. (D) Protein expression levels of cyclin D, CDK4 and CDK6 were determined by western blotting and β-actin was used as an internal reference. Expression levels of these proteins were decreased in LATS2-overexpressed cells and were increased in LATS2 knockdown cells. All data are presented as the means ± standard deviation (n=3). *P<0.05; **P<0.01; ***P<0.001 compared with control or siNC groups. CDK, cyclin-dependent kinase; LATS2, large tumor suppressor kinase 2; OD, optical density; NC, negative control; si/siRNA, small interfering RNA.
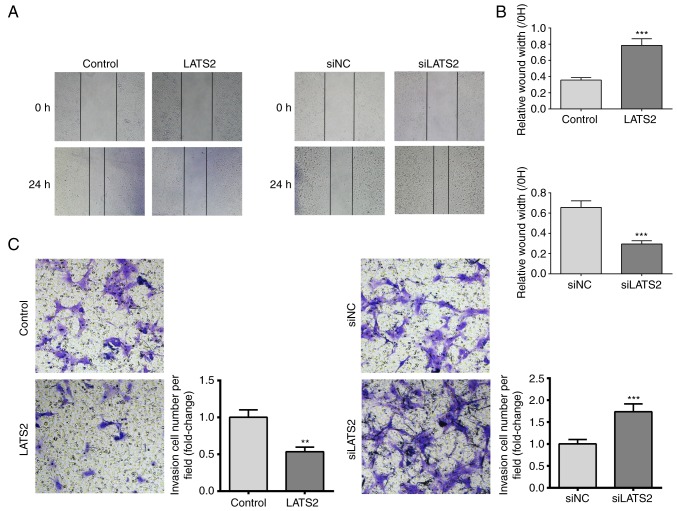
Since increased cell migration and invasion are features of glioma, the present study investigated the effects of LATS2 expression on cell migration and invasion using scratch wound healing and transwell invasion assays, respectively. A U-372 MG cell monolayer was used for the wound healing assay. The results revealed that cells with LATS2 overexpression exhibited significantly slower wound repair, whereas in cells with LATS2 knockdown, cell migration was increased compared with the control group after 24 h (Fig. 3A and B). Furthermore, transwell invasion assay revealed a reduction in the number of invasive cells when LATS2 was overexpressed compared with cells transfected with an empty vector. Consistently, LATS2 knockdown elevated the number of invasive cells (Fig. 3C). Taken together, these results indicated that LATS2 dysregulation may affect growth, migration and invasion of U-372 MG cells in vitro.
Figure 3.
LATS2 suppressed cell migration and invasion. (A) Scratch wound healing assay was performed in U-372 MG cells transfected with LATS2 (left) or siLATS2 (right) for 24 h. Wound repair rate was inversely related to the expression levels of LATS2. Representative images (magnification, ×200) of wound closure at 0 and 24 h are shown. Black dotted lines indicate initial injury at 0 h and injury closure at 24 h. (B) Cell migration was quantified as a percentage of the healed area. (C) Transwell invasion assay was performed in U-372 MG cells transfected with LATS2 (left) or siLATS2 (right). Representative images (magnification, ×400) of invasive cells are shown. The number of invasive cells was counted and was inversely related to the expression levels of LATS2. All data are presented as the means ± standard deviation (n=3). **P<0.01; ***P<0.001 compared with control or siNC groups. Each assay was performed in triplicate. LATS2, large tumor suppressor kinase 2; NC, negative control; si/siRNA, small interfering RNA.
LATS2 regulates phosphorylation of YAP and TAZ, and their subcellular localization
Since YAP and TAZ are well recognized as downstream effectors of the Hippo pathway, the present study aimed to determine whether LATS2 functions through phosphorylating YAP and TAZ in U-372 MG cells. The phosphorylated and total protein expression levels of YAP and TAZ were detected in cells transfected with LATS2 plasmids or LATS2 siRNA. Notably, although alterations in LATS2 expression had no effect on the total expression levels of YAP and TAZ, p-YAP and p-TAZ were increased in cells transfected with LATS2 plasmids and decreased in cells transfected with LATS2 siRNA (Fig. 4A). These findings indicated that expression of LATS2 may regulate the phosphorylation of YAP and TAZ. Since it has been reported that YAP and TAZ phosphorylation promotes their translocation from the nucleus to the cytoplasm (13), this study also investigated whether LATS2-induced YAP and TAZ phosphorylation altered their subcellular localization. Immunostaining revealed that overexpression of LATS2 induced nuclear to cytoplasm translocation of p-YAP and p-TAZ (Fig. 4B).
Figure 4.
LATS2 regulates YAP and TAZ by phosphorylation and subcellular localization. (A) Levels of p- and total YAP and TAZ were determined by western blotting; β-actin was used as an internal reference. Overexpression (left) increased the phosphorylation of YAP and TAZ, whereas knockdown (right) of LATS2 decreased the phosphorylation of YAP and TAZ; no effects were detected on total protein levels. (B) Subcellular localization of YAP and TAZ was determined by immunofluorescence (magnification, ×800). YAP and TAZ [Alexa Fluor® 488-conjugated secondary antibody (top)], nuclei [DAPI (middle)] and merged images (bottom) are presented. YAP and TAZ were translocated from the nucleus to the cytoplasm in cells with LATS2 overexpression. LATS2, large tumor suppressor kinase 2; NC, negative control; p, phosphorylated; si/siRNA, small interfering RNA; TAZ, tafazzin; YAP, yes-associated protein.
YAP S127A mutant abolishes the inhibitory effects of LATS2 on cell proliferation, migration and invasion
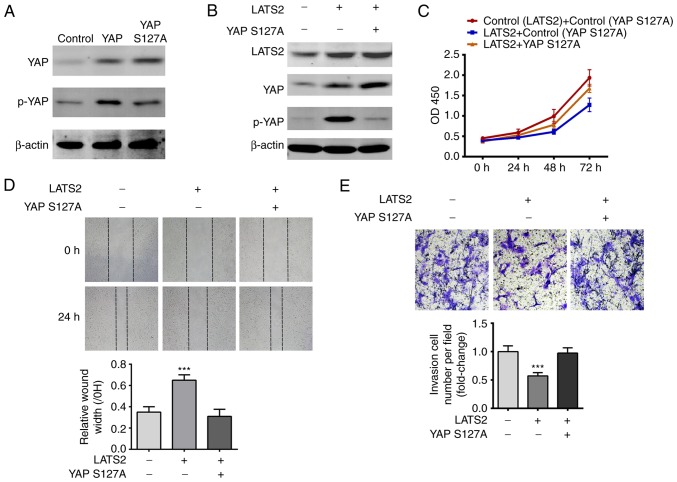
Although YAP and TAZ are the main effectors of the Hippo pathway, LATS2 can operate through other signaling pathways, such as Wnt signaling (14). In order to confirm that LATS2 inhibited cell proliferation, migration and invasion through the YAP/TAZ signaling pathway, a YAP S127A mutant, which lacks the phosphorylation site recognized by LATS2 (15), was transfected into cells, which were analyzed by CCK-8, scratch wound healing and transwell invasion assays. These studies aimed to investigate whether disrupting YAP phosphorylation abolished the inhibitory effects of LATS2 on cell proliferation, migration and invasion. Initially, the present study validated that YAP S127A transfection increased the total protein expression levels of YAP, but not p-YAP in U-372 MG cells (Fig. 5A). Compared with transfection with LATS2 alone, cotransfection with YAP S127A and LATS2 decreased the expression levels of p-YAP (Fig. 5B). The results of CCK-8, scratch wound healing and transwell invasion assays consistently revealed that YAP S127A relieved the suppressive effects of LATS2 on cell proliferation, migration and invasion (Fig. 5C-E). These findings are consistent with the hypothesis that LATS2 regulated cell proliferation, migration and invasion via YAP/TAZ signaling.
Figure 5.
YAP S127A mutant abolished the effects of LATS2 on cell proliferation, migration and invasion. (A) Expression levels of p- and total YAP were determined in wild-type YAP- and YAP S127A-transfected cells by western blotting. β-actin was used as an internal reference. Total levels of YAP were increased in both wild-type YAP- and YAP S127A-transfected cells compared with the control; however, p-YAP was only increased in wild-type YAP-transfected cells. (B) YAP S127A abolished LATS2-induced p-YAP upregulation. Expression levels of LATS2, total YAP and p-YAP were determined by western blotting. β-actin was used as an internal reference. (C) YAP S127A abolished LATS2-induced suppression of cell proliferation. Growth curves were plotted with the results of the Cell Counting Kit-8 assay, and OD450 was measured at 24, 48 and 72 h. Cell growth was restored by cotransfection of cells with LATS2 and YAP S127A. (D) YAP S127A restored the migration of the LATS2-transfected cells. Cell migration in each group was determined using the scratch wound healing assay; representative images (magnification, ×200) are shown. Following cotransfection with YAP S127A and LATS2, cells had a similar wound repair rate to the control group, whereas transfection with LATS2 alone resulted in a significantly lower repair rate compared with cells cotransfected with LATS2 and YAP S127A. (E) Overexpression of YAP S127A restored the invasive ability of LATS2-transfected cells. Cell invasion in each group was determined by transwell invasion assay and the number of invasive cells was counted (magnification, ×800). Cells cotransfected with YAP S127A and LATS2 had a similar number of invasive cells to the control group, whereas transfection with LATS2 alone resulted in significantly fewer invasive cells compared with the control group. All data are presented as the means ± standard deviation (n=3). ***P<0.001 compared with cells cotransfected with LATS2 and YAP S127A. Each assay was performed in triplicate. LATS2, large tumor suppressor kinase 2; p, phosphorylated; si/siRNA, small interfering RNA; YAP, yes-associated protein.
Discussion
The present study revealed the LATS2 was downregulated in glioma samples compared with in normal tissue samples, as determined by RT-qPCR and immunohistochemistry. Subsequently, the effects of LATS2 on cell proliferation, migration and invasion were detected in vitro using U-372 MG glioma cells. The results indicated that LATS2 suppressed cell proliferation, likely by decreasing the expression levels of cyclin D1, CDK4 and CDK6, which are necessary for G1/S transition. This result supported previous findings by Li et al, which reported that LATS2 suppressed cell proliferation by inhibiting G1/S transition through downregulating cyclin E/CDK2 kinase in NIH3T3/v-ras cells (16). Notably, dysregulation of the aforementioned proteins have also been implicated in other types of human cancer (17), thus suggesting that LATS2 may inhibit cell proliferation through several pathways. Increased cell migration and invasion are two important features in development and oncogenic transformation, which may be associated with the high invasive ability of gliomas. The present study demonstrated using scratch wound healing and transwell invasion assays, that LATS2 also suppressed cell migration and invasion of U-372 MG cells. Therefore, reduced LATS2 expression in glioma samples may explain the increased cell migration and invasiveness.
The present study aimed to determine the underlying pathway that mediates the effects of LATS2 on cell proliferation, migration and invasion. The Hippo pathway has been well recognized as a key player in cellular proliferation, transformation and tumorigenesis (18), and YAP/TAZ act as downstream effectors. The present results revealed that LATS2 knockdown decreased the expression levels of p-YAP/TAZ. This finding is consistent with other findings, which reported that downregulation of LATS2 alters phosphorylation of YAP/TAZ, and in turn increases YAP and TAZ nuclear accumulation; these proteins are pervasively activated in human malignancies (6). These findings suggested that LATS2 may act as a tumor suppressor in glioma, as well as in other cancers (14,19).
Glioma is a frequent primary CNS tumor, with an average annual age-adjusted incidence rate of 6.0/100,000 in the United States (20). Due to its highly invasive ability, it is considered an intractable disease in the clinic with a very short median survival time (20). Therefore, there is an urgent requirement to further study the mechanism of tumorigenesis and develop novel treatment strategies. Targeted therapy that inhibits oncogenic signaling pathways and tumor-associated angiogenesis exerts impressive effects in the clinic (21). It has previously been reported that dysregulation of the Hippo pathway is heavily involved in the initiation and progression of various types of cancer (22). The Hippo pathway is an evolutionarily conserved mechanism, which serves a crucial role in cell proliferation, apoptosis, differentiation and development. The core component of the mammalian Hippo pathway is the MST1/2 and LATS1/2 kinase cascade. Activation of the kinase cascade induces translocation of YAP and TAZ from the nucleus to the cytoplasm and degradation, which suppresses the expression of genes that promote proliferation and inhibit apoptosis. Therefore, LATS2 is a potential tumor suppressor that may be able to regulate several oncogenes. Low expression of LATS2, or LATS2 copy number loss has been identified in breast, ovarian, hepatocellular and lung cancer (23). The present study confirmed the downregulation of LATS2 expression in glioma. The functions and downregulation of LATS2 in various cancers indicates that it may serve an important role in glioma formation and progression. Notably, studies also revealed that low expression of LATS2 predicts a better outcome for patients with cancer (24,25). One possible reason for the contradictory function of LATS2 may be the crosstalk of LATS2 with other signaling pathways. The roles of LATS2 in tumorigenesis remain unclear; LATS2 may potentially function as a tumor suppressor, not only by regulating YAP and TAZ phosphorylation as indicated in this study, but also by suppressing oncogenic Wnt signaling through disrupting β-catenin/B cell CLL/lymphoma 9 interaction, as shown in a previous study (12); conversely, LATS2 has been reported to promote the stability of p53 via inactivation of MDM2 proto-oncogene (26). However, p53 mutations have been detected in the majority of cancers; in this case, lower expression of LATS2 may decrease the transcriptional activity of mutant p53 and further decrease the proliferation, invasion and migration of the cancer cells. Therefore, heterogeneity in cytology and gene expression should be taken into account when exploring tumorigenic mechanisms.
The present results confirmed that LATS2 was downregulated in glioma samples. It was also revealed that overexpression of LATS2 suppressed proliferation, migration and invasion of U-372 MG cells by altering YAP/TAZ phosphorylation and nuclear translocation. These results highlight the potential function of LATS2 as a tumor suppressor in glioma, and its reduced expression in glioma may be a contributing factor to cancer progression.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
CG and CL were involved in conception and design of the study. JY acquired the data. HH, XL and BF analyzed the data and drafted the manuscript. CL gave final approval of the version to be published. All authors revised, read and approved the final version of the manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of The Second Affiliated Hospital of Hebei Medical University, and all patients provided written informed consent.
Patient consent for publication
All patients provided written informed consent.
Competing interests
There is no conflict of interest to declare.
References
- 1.Dellaretti M, Reyns N, Touzet G, Dubois F, Gusmao S, Pereira JL, Blond S. Diffuse brainstem glioma: Prognostic factors. J Neurosurg. 2012;117:810–814. doi: 10.3171/2012.7.JNS111992. [DOI] [PubMed] [Google Scholar]
- 2.Grossman SA, Ye X, Piantadosi S, Desideri S, Nabors LB, Rosenfeld M, Fisher J, NABTT CNS Consortium Survival of patients with newly diagnosed glioblastoma treated with radiation and temozolomide in research studies in the United States. Clin Cancer Res. 2010;16:2443–2449. doi: 10.1158/1078-0432.CCR-09-3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Y, Jiang T. Understanding high grade glioma: Molecular mechanism, therapy and comprehensive management. Cancer Lett. 2013;331:139–146. doi: 10.1016/j.canlet.2012.12.024. [DOI] [PubMed] [Google Scholar]
- 4.Baker GJ, Yadav VN, Motsch S, Koschmann C, Calinescu AA, Mineharu Y, Camelopiragua S, Orringer D, Bannykh S, Nichols WS, et al. Mechanisms of glioma formation: Iterative perivascular glioma growth and invasion leads to tumor progression, VEGF-independent vascularization, and resistance to antiangiogenic therapy. Neoplasia. 2014;16:543–561. doi: 10.1016/j.neo.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meng Z, Moroishi T, Guan KL. Mechanisms of Hippo pathway regulation. Genes Dev. 2016;30:1–17. doi: 10.1101/gad.274027.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zanconato F, Cordenonsi M, Piccolo S. YAP/TAZ at the roots of cancer. Cancer Cell. 2016;29:783–803. doi: 10.1016/j.ccell.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mo J, Park HW, Guan KL. The Hippo signaling pathway in stem cell biology and cancer. EMBO Rep. 2014;15:642–656. doi: 10.15252/embr.201438638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y, Pan P, Wang Z, Zhang Y, Xie P, Geng D, Jiang Y, Yu R, Zhou X. β-catenin-mediated YAP signaling promotes human glioma growth. J Exp Clin Cancer Res. 2017;36:136. doi: 10.1186/s13046-017-0606-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhat KP, Salazar KL, Balasubramaniyan V, Wani K, Heathcock L, Hollingsworth F, James J, Gumin J, Diefes K, Kin SH, et al. The transcriptional coactivator TAZ regulates mesenchymal differentiation in malignant glioma. Genes Dev. 2011;25:2594–2609. doi: 10.1101/gad.176800.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grzmil M, Morin P, Jr, Lino MM, Merlo A, Frank S, Wang Y, Moncayo G, Hemmings BA. MAP kinase-interacting kinase 1 regulates SMAD2-dependent TGF-β signaling pathway in human glioblastoma. Cancer Res. 2011;71:2392–2402. doi: 10.1158/0008-5472.CAN-10-3112. [DOI] [PubMed] [Google Scholar]
- 11.Guo C, Wang X, Liang L. LATS2-mediated YAP1 phosphorylation is involved in HCC tumorigenesis. Int J Clin Exp Pathol. 2015;8:1690–1697. [PMC free article] [PubMed] [Google Scholar]
- 12.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 13.Piccolo S, Dupont S, Cordenonsi M. The Biology of YAP/TAZ: Hippo signaling and beyond. Physiol Rev. 2014;94:1287–1312. doi: 10.1152/physrev.00005.2014. [DOI] [PubMed] [Google Scholar]
- 14.Li J, Chen X, Ding X, Cheng Y, Zhao B, Lai ZC, Al-Hezaimi K, Hakem R, Guan KL, Wang CY. LATS2 suppresses oncogenic Wnt signaling by disrupting β-catenin/BCL9 interaction. Cell Rep. 2013;5:1650–1663. doi: 10.1016/j.celrep.2013.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, Li L, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Y, Pei J, Xia H, Ke H, Wang H, Tao W. Lats2, a putative tumor suppressor, inhibits G1/S transition. Oncogene. 2003;22:4398–4405. doi: 10.1038/sj.onc.1206603. [DOI] [PubMed] [Google Scholar]
- 17.Bonelli P, Tuccillo FM, Borrelli A, Schiattarella A, Buonaguro FM. CDK/CCN and CDKI alterations for cancer prognosis and therapeutic predictivity. Biomed Res Int. 2014;2014:361020. doi: 10.1155/2014/361020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu FX, Zhao B, Guan KL. Hippo pathway in organ size control, tissue homeostasis, and cancer. Cell. 2015;163:811–828. doi: 10.1016/j.cell.2015.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Visser S, Yang X. LATS tumor suppressor: A new governor of cellular homeostasis. Cell Cycle. 2010;9:3892–3903. doi: 10.4161/cc.9.19.13386. [DOI] [PubMed] [Google Scholar]
- 20.Ostrom QT, Kinnersley B, Wrensch MR, Eckel-Passow JE, Armstrong G, Rice T, Chen YW, Wiencke J, McCoy LS, Hansen HM, et al. Sex-specific glioma genome-wide association study identifies new risk locus at 3p21.31 in females, and finds sex-differences in risk at 8q24.21. Sci Rep. 2018;8:7352. doi: 10.1038/s41598-018-24580-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hughes PE, Caenepeel S, Wu LC. Targeted therapy and checkpoint immunotherapy combinations for the treatment of cancer. Trends Immunol. 2016;37:462–476. doi: 10.1016/j.it.2016.04.010. [DOI] [PubMed] [Google Scholar]
- 22.Moroishi T, Hansen CG, Guan KL. The emerging roles of YAP and TAZ in cancer. Nat Rev Cancer. 2015;15:73–79. doi: 10.1038/nrc3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Furth N, Aylon Y. The LATS1 and LATS2 tumor suppressors: Beyond the Hippo pathway. Cell Death Differ. 2017;24:1488–1501. doi: 10.1038/cdd.2017.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao Y, Yi J, Zhang K, Bai F, Feng B, Wang R, Chu X, Chen L, Song H. Downregulation of MiR-31 stimulates expression of LATS2 via the hippo pathway and promotes epithelial-mesenchymal transition in esophageal squamous cell carcinoma. J Exp Clin Cancer Res. 2017;36:161. doi: 10.1186/s13046-017-0622-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y, Hu CF, Chen J, Yan LX, Zeng YX, Shao JY. LATS2 is de-methylated and overexpressed in nasopharyngeal carcinoma and predicts poor prognosis. BMC Cancer. 2010;10:538. doi: 10.1186/1471-2407-10-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aylon Y, Michael D, Shmueli A, Yabuta N, Nojima H, Oren M. A positive feedback loop between the p53 and Lats2 tumor suppressors prevents tetraploidization. Genes Dev. 2006;20:2687–2700. doi: 10.1101/gad.1447006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.