Abstract
Piwi-interacting RNAs (piRNAs) comprise the largest class of non-coding RNAs. They represent a molecular feature shared by all non-aging biological systems, including germline and somatic cancer stem cells, which display an indefinite capacity of renewal and proliferation and are potentially immortal. They have been identified in animal stomachs, but their relationship with human gastric cancers remains largely unclear. The present study aimed to identify the piRNAs associated with human gastric cancers across the whole transcriptome. Fresh tumor tissues and adjacent non-tumorous tissues from stomachs were examined using a piRNA microarray (23,677 piRNAs) that was then validated by qPCR. The differential expression of piRNAs between cases and controls was analyzed. The transposable elements (TEs) that are potentially targeted by the risk piRNAs were searched. The expression of the nearest genes that are complementary to the sequences of the piRNAs was examined in the stomach tissue. The regulatory effects of genome-wide significant and replicated cancer-risk DNA variants on the piRNA expression in stomach were tested. Based on the findings, we identified a total of 8,759 piRNAs in human stomachs. Of all, 50 were significantly (P<0.05) and differentially (>2-fold change) expressed between the cases and controls, and 64.7% of the protein-coding genes potentially regulated by the gastric cancer-associated piRNAs were expressed in the human stomach. The expression of many cancer-associated piRNAs was correlated with the genome-wide and replicated cancer-risk SNPs. In conclusion, we conclude that piRNAs are abundant in human stomachs and may play important roles in the etiological processes of gastric cancers.
Keywords: piRNA, gastric cancer, transposable element, transcriptome-wide, microarray
Introduction
Gastric cancer (GC) is the fourth most common diagnosed cancer worldwide and ranks second as the cause of cancer-related death (1). The etiology of GC is complicated and remains an active focus of research. In addition to environmental risk factors, genetics may play a role in the development and progression of the disease (2,3). Many studies have explored the genetic mechanisms underlying GC, including the direct effects of protein-coding genes, e.g. TP53, HOXB5, TGF-β, SK and THBS1 (4–8), and the indirect effects of non-coding RNAs (ncRNAs), e.g., let-7a, miR-21 and SNGH8 (9,10).
Recently, a new and largest class (11) of non-coding small RNAs, i.e., piwi-interacting RNAs (piRNAs), which interact with a subset of Argonaute proteins related to Piwi, has become a focus of intense research (12). piRNAs are non-coding RNAs with 24–32 nucleotides (nt). Although structurally only slightly longer than microRNAs (miRNAs; 19–25 nt), piRNAs are different in expression pattern and quantity, as well as in genomic organization (13–15). piRNAs are initially discovered in germline cells. Germline and certain ‘lower’ eukaryotic organisms are ‘non-aging’ biological systems that display an indefinite capacity for renewal and a lifespan-stable genome integrity, and are potentially immortal (16). piRNA is a shared feature of all non-aging biological systems (16). Interestingly, cancer stem cells also display an indefinite capacity of renewal and proliferation and are potentially immortal (16). An increasing number of studies have shown aberrant piRNA expression as a molecular signature across multiple tumor types (17,18), including renal cancer (19), breast cancer (14,20), multiple myeloma (21), and hepatocellular carcinoma (22), suggesting that piRNAs may be involved in the pathogenesis of cancer (12). For instance, upregulated expression of piR-36743, piR-20365, piR-20485, piR-4987 and piR-932 was observed in breast cancer as compared to matched non-cancerous tissues (23,24); and the expression of piRABC was increased in bladder cancer, as compared to their corresponding adjacent non-cancerous tissues (25). These findings also suggest that piRNAs are a shared feature of this non-aging biological system, cancers. In an earlier work, we reported associations between piRNAs and Alzheimer's disease (26), a clinical condition strongly related to aging. Cancers are just opposite to the aging diseases in renewal and proliferation capacity, which also suggests a potential role of piRNAs in cancers. Recently, a study reported that 156 piRNAs were significantly deregulated in GC (27).
The sequences of piRNAs primarily complement the transposable elements (TEs). The most widely recognized and well-characterized function of piRNAs is to suppress the activities of TEs at genomic and epigenetic levels (17). TEs are capable of moving from one genomic locus to another, thereby causing insertional mutations (28,29). There are two major classes of TEs: RNA transposons (or retrotransposons) and DNA transposons, as distinguished by their mechanism of transposition (28). Retrotransposons include long terminal repeats (LTRs), long interspersed nuclear element (LINE), and short interspersed nuclear elements (SINE). As the primary determinants of genomic instability (16), TEs exert pathological effects via controlling the transcription of neighboring genes (30). The downregulation of piRNA pathways would increase the repeats of retrotransposon and cause DNA damage, which represents the primary genetic determinant of aging. The upregulation of piRNAs suppresses the activity of TEs, which protects against TE-mediated mutagenesis, maintains genomic integrity, and self-renewal and proliferation capacity, and thereby preserves the non-aging features of cancer cells (16). Additionally, piRNA may also be involved in a TE-independent mechanism in cancer risk; that is, it may regulate the transcription of certain key cancer-related genes via non-TE sequence complementarity (16,31) or modulation of chromatin organization (16). In the present study, we aimed to identify the piRNAs in association with GC across the transcriptome using a piRNA profile. Further, we explored whether these GC-associated piRNAs are expressed in other tissues, and whether their nearest protein-coding genes were expressed in the stomach and perhaps even related to gastrointestinal cancers.
For many years investigators have scanned the whole genome to search for risk DNA variants for GC. In reviewing all published genome-wide association studies (GWASs) and whole genome/exome sequencing studies of GC, we found that only three variants were genome-wide significant in association with GC and replicated across at least two independent studies at single-point level. The three variants are rs4072037 at MUC1 (on Chr1) (32–35), rs13361707 at PRKAA1 (on Chr5) (34,36) and rs2294008 at PSCA (on Chr8) (34,35). Numerous candidate gene studies supported these GWAS findings. However, the mechanisms underlying these SNP-GC associations are not clear. Here we examined whether the GC-related piRNAs might mediate the SNP-GC associations, in order to explore the potential roles of piRNAs in the pathogenesis of GC.
Materials and methods
Subjects
The primary cohort including eight gastric cancer(GC) tissues and adjacent normal tissues extracted from eight male patients (aged 49–79 years old) was examined in a piRNA microarray study. The clinical characteristics of the GC patients are shown in Table I. Fresh gastric tissue samples were collected from GC patients who underwent surgical resection at the Fujian Provincial Cancer Hospital. A second cohort of 20 paired GC and adjacent normal tissues extracted from 15 male and 5 female patients (aged 32–78 years old) was also obtained from the same hospital for validating the findings obtained from the first cohort. A third cohort of 181 paired GC and adjacent normal tissues extracted from 134 male and 47 female patients (aged 28–73 years old) from the same hospital was tested for piwi protein expression in the stomach. These cohorts were recruited between June 2015 and February 2018. None of the patients received chemotherapy before surgery and tissues were obtained immediately after tumor resection. Medical diagnoses were confirmed histopathologically, and clinical pathological data were obtained. Histological classification of GC was assessed according to the Lauren's criteria (37). Patients were staged according to the 7th Edition of the TNM staging system of the American Joint Commission on Cancer (AJCC) (38). The study was approved by the Research Ethics Committee of the Fujian Provincial Cancer Hospital, China. Informed consents from all patients were obtained prior to the study. All experiments were performed in accordance with relevant guidelines and regulations.
Table I.
Pathological features of the subjects with gastric carcinoma.
| Parameters | Discovery samples (n) (for sequencing) | Validation samples (n) (for qPCR) |
|---|---|---|
| Sex | ||
| Male | 8 | 15 |
| Female | 0 | 5 |
| Age (years) | ||
| <60 | 4 | 9 |
| ≥60 | 4 | 11 |
| Location | ||
| EGJ | 4 | 6 |
| Non-EGJ | 4 | 14 |
| Depth of invasion | ||
| <T2 | 0 | 3 |
| ≥T2 | 8 | 17 |
| Lauren's type | ||
| Intestinal-type | 4 | 6 |
| Diffuse-type | 4 | 14 |
| Tumor size (cm) | ||
| <5 | 4 | 10 |
| ≥5 | 4 | 10 |
| TNM stage | ||
| I+II | 3 | 6 |
| III+IV | 5 | 14 |
| LN metastasis | ||
| Absent | 2 | 6 |
| Present | 6 | 14 |
n, sample size; EGJ, esophagogastric junction; T2, tumor invades the muscularis propria; TNM, Tumor Node Metastasis; LN, lymph node.
piRNA microarray experiment
The ArrayStar HG19 piRNA array was used to profile piRNA expression in humans (ArrayStar; Agilent Technologies, Santa Clara, CA, USA). This array contains probes for 23,677 piRNAs obtained from the National Center for Biotechnology Information (NCBI) database (https://www.ncbi.nlm.nih.gov/nuccore/?term=piRNA) and landmark publications (15,39). Human piRNAs were mapped to the HG19 genome sequence using UCSC Blat (https://genome.ucsc.edu/cgi-bin/hgBlat?command=start). Total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to manufacturer's instructions. Sample labeling was performed using an RNA ligase method (40). The labeled samples were hybridized onto Arraystar Human piRNA Array in Agilent's SureHyb Hybridization Chambers according to manufacturer's standard protocols (Agilent Technologies). After slides were scanned, data were extracted using Agilent Feature Extraction software. Raw signal intensities were normalized in quantiles by GeneSpring GX v11.5.1 (Agilent Technologies), and low intensity piRNAs were filtered. Agilent Feature Extraction software (version 11.0.1.1) was used to analyze the acquired array images (40). Normalized intensity values were then log2-transformed.
Quality assessment of piRNA data after normalization and filtering
We used a Box Plot to visualize the distributions of a dataset. We compared the distributions of the intensities from all samples and showed that. after normalization, the distributions of log2-ratios among the samples were nearly the same (Fig. S1). We used a Scatter Plot to assess the piRNA expression variation (or reproducibility) between arrays (Fig. S2). This plot showed good reproducibility of our array data.
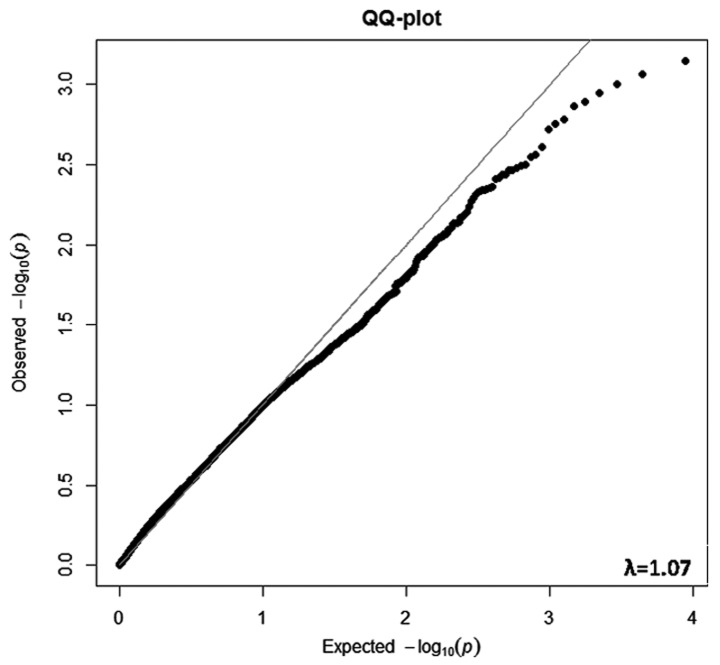
The distribution of all of the observed P-values from the differential expression analysis (see below) was fitted to the expected P-values using a QQ plot. A transcriptomic inflation factor (λ) was computed from these P-values and defined as the ratio of the median of the empirically observed distribution of the test statistic to the expected median, thus quantifying the extent of the bulk inflation and the excess false positive rate. It was computed for the genomic control analysis to reflect the maximum possible inflation factor if the associations were affected by population stratification. A λ with a departure of less than 0.1 from a value of 1.0 is considered an indicator of very good quality and robust associations.
Differential expression analysis
The normalized and log2-transformed intensities were compared between GC cases and normal controls by paired Student's t-test. Fold-changes (FCs) between the two groups were calculated by ‘normalized intensity values in Group 1 divided by Group 2’. Significantly differentially expressed piRNAs, which were associated with GC, were identified through Volcano Plot filtering (FC≥1.5 and P≤0.05). The expression level of each piRNA in each individual was also shown in a Heat Map, and clustered using Hierarchical clustering analysis to show the relationships among the expression levels of samples.
Quantitative reverse transcription-polymerase chain reaction (qRT-PCR) experiment
We selected some candidate piRNAs from the top list of the significantly differentially expressed piRNAs to be examined by qPCR in an independent cohort (20 cases vs. 20 controls), in order to control the quality of microarray experiment and validate piRNA-GC associations as derived from the differential expression analysis on the array data. The selection criteria for these candidate piRNAs include that: i) their expression in gastric cancer tissues should have top intensities >1,500 in the array data; ii) their expression in gastric cancer tissues should be higher than adjacent tissues with top FCs ≥2.4; and iii) they are located at or close to those genes that have been reported by literature to be cancer-related. After this selection, only two piRNAs, i.e., DQ579739 and DQ600515, were included (Table II). Both piRNAs were abundantly expressed in gastric cancer tissues [log2(normalized intensity) >10], and differentially expressed between cases and controls in the array data (P<0.05).
Table II.
Top piRNAs significantly differentially expressed between the cases and controls.
| Normalized intensity | Cancer vs. control | |||||||
|---|---|---|---|---|---|---|---|---|
| piRNA | Length (nt) | Chr | Cancer | Control | FC | P-value | Gene | |
| With top intensities in cancer (intensities >1,500) | ||||||||
| DQ588779a | 29 | chr1 | 14,194 | 6,844 | 2.1 | 0.037 | SKI | |
| DQ595533a | 29 | chr3 | 13,314 | 6,148 | 2.2 | 0.035 | COLQ | |
| DQ571813 | 31 | chr16 | 12,123 | 5,710 | 2.1 | 0.046 | VAC14 | |
| DQ590386 | 29 | chr16 | 11,953 | 5,766 | 2.1 | 0.046 | VAC14 | |
| DQ595534 | 30 | chr16 | 11,307 | 5,429 | 2.1 | 0.038 | VAC14 | |
| DQ600670a | 30 | chr11 | 4,410 | 1,805 | 2.4 | 0.032 | EED | |
| DQ600515a | 29 | chr1 | 4,071 | 1,668 | 2.4 | 0.041 | ATAD3B | |
| DQ580665a | 26 | chr7 | 2,349 | 5,133 | 2.2 | 0.041 | POLR2J | |
| DQ579739a | 30 | chr19 | 1,844 | 774 | 2.4 | 0.029 | CXCL17 | |
| DQ578739 | 31 | chr2 | 1,709 | 3,422 | 2.0 | 0.007 | to LOC100507334 | |
| DQ583443 | 30 | chr9 | 1,582 | 3,683 | 2.3 | 0.015 | FAM225A | |
| With top FC between cases and controls (FC≥2.4) | ||||||||
| DQ590922a | 29 | chr9 | 32 | 10 | 3.1 | 0.036 | to TLE4 | |
| DQ572848a | 30 | chr9 | 22 | 8 | 2.7 | 0.024 | to TLE4 | |
| DQ600670a | 30 | chr11 | 4,410 | 1,805 | 2.4 | 0.032 | EED | |
| DQ600515a | 29 | chr1 | 4,071 | 1,668 | 2.4 | 0.041 | ATAD3B | |
| DQ579739a | 30 | chr19 | 1,844 | 774 | 2.4 | 0.029 | CXCL17 | |
| DQ571419 | 28 | chr1 | 1,443 | 605 | 2.4 | 0.032 | RPS8 | |
| With top FC ↓ between cases and controls (FC≥3.8) | ||||||||
| DQ574148 | 30 | chr22 | 25 | 110 | 4.4 | 0.012 | IGLL1 | |
| DQ583043 | 30 | chr19 | 32 | 135 | 4.2 | 0.015 | to LOC100652909 | |
| DQ574145 | 30 | chr22 | 18 | 75 | 4.2 | 0.01 | IGLL1 | |
| DQ574146 | 30 | chr22 | 49 | 197 | 4.1 | 0.012 | IGLL1 | |
| DQ598137 | 28 | chr10 | 15 | 58 | 4.0 | 0.017 | NUTM2A | |
| DQ570015 | 30 | chr15 | 429 | 1,642 | 3.8 | 0.036 | GOLGA8A | |
| DQ574147 | 31 | chr22 | 55 | 208 | 3.8 | 0.010 | IGLL1 | |
| With lowest P-values between cases and controls and FC ↑ (P≤0.025) | ||||||||
| DQ570687a | 28 | chr22 | 910 | 448 | 2.0 | 0.012 | RPL3 | |
| DQ580689 | 28 | chr1 | 180 | 90 | 2.0 | 0.016 | NBPF3 | |
| DQ575884a | 32 | chr2 | 1,393 | 691 | 2.0 | 0.017 | to EPCAM | |
| DQ572848a | 30 | chr9 | 22 | 8 | 2.7 | 0.024 | to TLE4 | |
| With lowest P-values between cases and controls and FC ↓ (P≤0.005) | ||||||||
| DQ577345 | 31 | chr19 | 383 | 969 | 2.5 | 0.003 | to LOC400685 | |
| DQ587262 | 31 | chr6 | 97 | 233 | 2.4 | 0.003 | to LOC100507584 | |
| DQ575557 | 29 | chr3 | 25 | 83 | 3.4 | 0.003 | to IQSEC1 | |
| DQ594511 | 29 | chr8 | 22 | 45 | 2.0 | 0.004 | to LOC100287846 | |
| DQ594440 | 31 | chr3 | 60 | 164 | 2.7 | 0.004 | to IQSEC1 | |
| DQ576821 | 28 | chr19 | 28 | 77 | 2.7 | 0.004 | ZNF490 | |
| DQ570858 | 31 | chr15 | 30 | 106 | 3.5 | 0.005 | HERC2P7 | |
| With locations at or close to cancer-related genes | ||||||||
| DQ600515a | 29 | chr1 | 4,071 | 1,668 | 2.4 | 0.041 | ATAD3B | |
| DQ594797 | 26 | chr2 | 14 | 7 | 2.1 | 0.042 | to TEKT4 | |
| DQ580529 | 30 | chr22 | 13 | 30 | 2.3 | 0.022 | to WBP2NL | |
| DQ579739a | 30 | chr19 | 1,844 | 774 | 2.4 | 0.029 | CXCL17 | |
| DQ570687a | 28 | chr22 | 910 | 448 | 2.0 | 0.012 | RPL3 | |
| DQ600670a | 30 | chr11 | 4,410 | 1,805 | 2.4 | 0.032 | EED | |
| DQ590922a | 29 | chr9 | 32 | 10 | 3.1 | 0.036 | to TLE4 | |
| DQ595533a | 29 | chr3 | 13,314 | 6,148 | 2.2 | 0.035 | COLQ | |
| DQ588779a | 29 | chr1 | 14,194 | 6,844 | 2.1 | 0.037 | SKI | |
| DQ588880 | 30 | chr17 | 30 | 67 | 2.2 | 0.032 | to HOXB5 | |
| DQ580665a | 26 | chr7 | 2,349 | 5,133 | 2.2 | 0.041 | POLR2J | |
| DQ590827 | 30 | chr16 | 179 | 367 | 2.0 | 0.037 | DHODH | |
| DQ575884a | 32 | chr2 | 1,393 | 691 | 2.0 | 0.017 | to EPCAM | |
| DQ580779 | 28 | chr9 | 61 | 184 | 3.0 | 0.035 | TMOD1 | |
| DQ600254 | 29 | chr14 | 144 | 354 | 2.5 | 0.027 | PNMA1 | |
chr, chromosome; ↑, upregulated; ↓, downregulated. FC, fold-change; ‘to’, proximate to.
Appears at least twice in this table. P-value from t-test.
To measure the piRNA expression levels, a miScript Reverse Transcription (RT) kit (Guangzhou RiboBio Co., Ltd., Guangzhou, China) was used to generate cDNA, according to the manufacturer's instructions (40). Next, qRT-PCR was performed in a 20 µl reaction volume with the following reagents (Guangzhou RiboBio Co., Ltd): SYBR-Green Mix [10 µl of SYBR-Green I mix, 0.5 µl of miDETECTATrack™ miRNA forward and reverse primers (10 µM), and 2 µl of cDNA template] on the ABI7500 system (Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA) with the following protocol: 95°C for 3 min and 40 cycles of 95°C for 10 sec, 60°C for 20 sec, and 70°C for 1 sec.
Using the comparative 2−ΔΔCq method (41) and an example gastric carcinoma sample as a calibrator, the relative expression levels in all gastric carcinoma samples and adjacent non-tumorous stomach tissues were quantified. Hs_5S_miScript expression was used to normalize piRNA expression levels between different samples.
Risk gene expression in the stomach
The primary function of piRNAs is to suppress TEs that will then control the transcription of nearby genes. piRNAs affect the phenotypes primarily via TE suppression. The target TEs that are potentially regulated by the risk piRNAs were searched from UCSC Genome Browser (http://genome.ucsc.edu; genome assembly, hg19; track, RepeatMasker). We hypothesized that, without prior knowledge, the nearest genes to which the sequences of the piRNAs are complementary should be the most likely targets for TE regulation (30,31). These genes were thus carefully examined in this study. The relationship between the genes and GC or related cancers was searched in PubMed (Table SI). The mRNA expression of these genes in stomach was examined in two independent cohorts without cancers, including an American cohort (n=262) (42), and a UK European cohort (n=176) (43). The density of the proteins encoded by these genes was also examined in stomach of a Germany cohort (44). If a gene had a normalized intensity >36 (by RNA microarray), a TPM >1 (by RNA-Seq) or a value >10 ppm (by mass spectrometry-based proteomics microarray) in the expression levels in human stomach of these three cohorts or had been reported to be expressed in human stomach in the literature, it was marked ‘E: Expressed’ (Table SI). The detailed demographic data of these three cohorts and the expression values of the top four genes are shown in Table SII. Overall, the genes expressed in stomach are more likely to be implicated in stomach diseases than non-stomach genes. Of note, the expression of these genes was not examined in the primary cohort, which represented a limitation of the study.
The piwi protein expression in the stomach
piRNAs regulate cellular activities by interacting with piwi proteins to form a complex (17). The golden criterion to confirm the presence of piRNAs is the existence of piwi proteins in the same tissue. To test the expression of piwi in stomach, the mRNA and protein expression of piwi, including PIWIL1, PIWIL2, PIWIL3 and PIWIL4 was examined in the above three independent cohorts, including the American (42), UK European (43) and Germany cohorts (44). The most abundant piwi protein (here, PIWIL4) was further tested in our own samples (181 GC and 181 adjacent tissues), using microarray and immunohistochemistry (IHC) technologies (see Supplementary Methods).
Expression quantitative trait locus (eQTL) analysis
The 3 genome-wide significant (P<5×10−8) risk variants for GC were genotyped in the primary cohort of 16 subjects (8 cases and 8 controls). The associations between these risk variants and GC have been replicated by at least two independent genome-wide association studies (GWASs) (Table SIII). To examine whether the GC-associated piRNA expression was controlled by any of these GC-risk SNPs, we performed eQTL analysis. Usually, a piRNA controlled by DNA variants is more likely to be functional than those uncontrolled. Associations between the genotypes and expression level of each GC-associated piRNA were analyzed using t-test or one-way ANOVA. This correlation is assumed to potentially underlie SNP-disease associations. The design of the whole study was based on a regulation pathway as illustrated in Fig. S3.
Results
Differential expression of piRNAs between cases and controls (Figs. S4, 1 and 2, and Tables II and SI)
Among the 23,677 piRNAs, 8,759 (37.0%) were detected in the human stomach. The expression levels of these 8,759 piRNAs in each individual are shown in the HeatMap (Fig. S4). A total of 8,759 P-values were obtained from the 8,759 differential expression analyses. The observed and expected P-values fitted very well (see QQ plot in Fig. 1). The λ was 1.07, indicating that the array data were of high quality, the differential expression analysis was robust, and transcriptome-wide inflation and excess false positive rates in the differential expression analysis were very low. That is, the associations were less likely to be affected by population stratification.
Figure 1.
QQ plot of P-values for the differential expression between cases and controls across the transcriptome. [x-axis: Expected-log(P) values; y-axis: Observed-log(P) values; P-values correspond to associations between piRNAs and gastric cancer; λ, transcriptomic inflation factor].
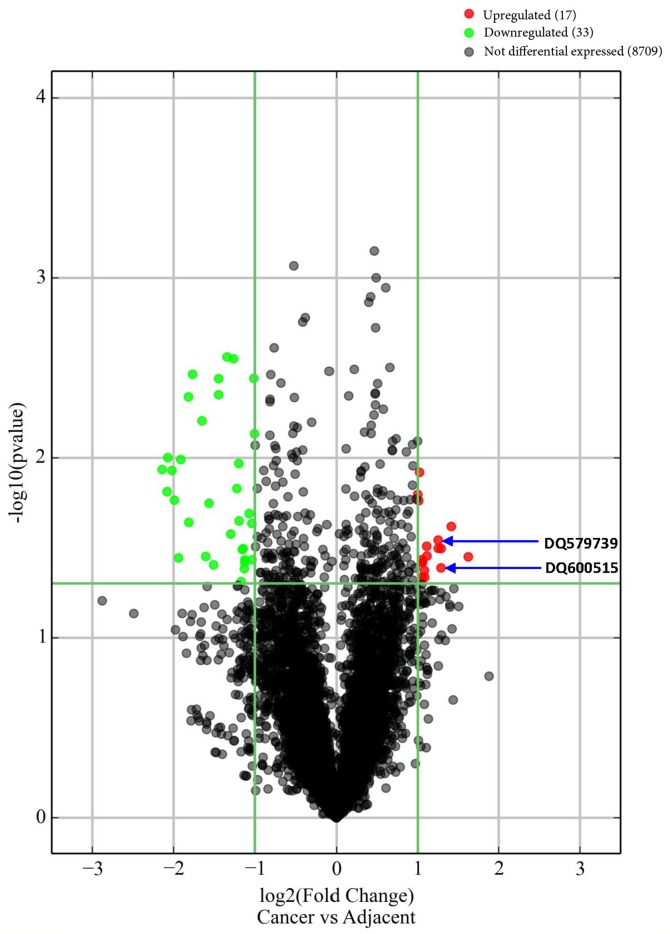
Fifty piRNAs with length of 26–32 nt were nominally differentially expressed between cases and controls (FC>2.0; P<0.05; without Bonferroni correction) (Fig. 2; Table SI). The mean log2-transformed normalized intensity of these 50 piRNAs was 7.46±3.34 (3.21–13.79) in GC cases, and 8.02±2.97 (2.75–12.74) in controls. Among the 50 risk/protective piRNAs, 17 were upregulated and 33 were downregulated in cases in contrast to controls. Among the 50 piRNAs, 41 risk piRNAs potentially targeted at 41 TEs, including 1 DNA transposon, 20 SINEs, 12 LINEs and 8 LTRs (Table SI). Thirty piRNAs mapped within 23 protein-coding genes, 13 mapped close to 11 protein-coding genes, and the other 7 mapped close to 5 ncRNAs. Twenty-two of these 34 (23+11) protein-coding genes (64.7%) were expressed in stomach, and 18 piRNAs were located in 15 piRNA clusters, 10 of which were located in intergenic regions. Seventeen (77.2%) of these 22 protein-coding stomach genes have been related to various human cancers in the literature, including gastric cancer (6–8), colon cancer (45–50), colorectal cancer (51–53), pancreatic cancer (54–56), hepatocellular carcinoma (57–59) and bladder cancer (60) (Table SI).
Figure 2.
The differential expression between cases and controls. [x-axis, fold-change; y-axis, -log10(p); red/green points, the differentially expressed piRNAs with 2-fold change and P<0.05].
Thirteen piRNAs had log2-transformed normalized intensities >10 (i.e., >1,000 before transformation). The top five piRNAs with highest intensities in cases were DQ588779 at SKI (SKI proto-oncogene) (on chr1), DQ595533 at COLQ (on chr3), and DQ571813, DQ590386 and DQ595534 at VAC14 (on chr16) (Table II).
Two piRNAs, i.e., DQ590922 and DQ572848 close to TLE4 (on chr9), were expressed with >2.5 FCs in cases compared to controls. Nine piRNAs were expressed with >3.5 FCs in controls compared to cases; and the top five of them were DQ574148, DQ574145 and DQ574146 at IGLL1 (on chr22), DQ598137 at NUTM2A (on chr10), and DQ583043 close to LOC100652909 (on chr19) (Table II).
Ten piRNAs were significantly differentially expressed between cases and controls with P<0.01. The four most significant ones with higher FCs in cases were DQ570687 at RPL3 (on chr22), DQ580689 at NBPF3 (on chr1), DQ575884 close to EPCAM (on chr2) and DQ572848 close to TLE4 (on chr9) (0.012≤P≤0.024). The five most significant ones with lower FCs in cases were all intergenic, including DQ577345 close to LOC400685 (chr19), DQ587262 close to LOC100507584 (chr6), DQ594511 close to LOC100287846 (chr8), and DQ575557 and DQ594440 close to IQSEC1 (chr3) (0.003≤P≤0.004) (Table II).
Some genes within or near which the GC-associated piRNAs are located have been reported to be associated with gastrointestinal cancers. They are CXCL17, EPCAM, EED, TLE4, SKI, RPL3 and HOXB5 (Table SI).
qPCR validation
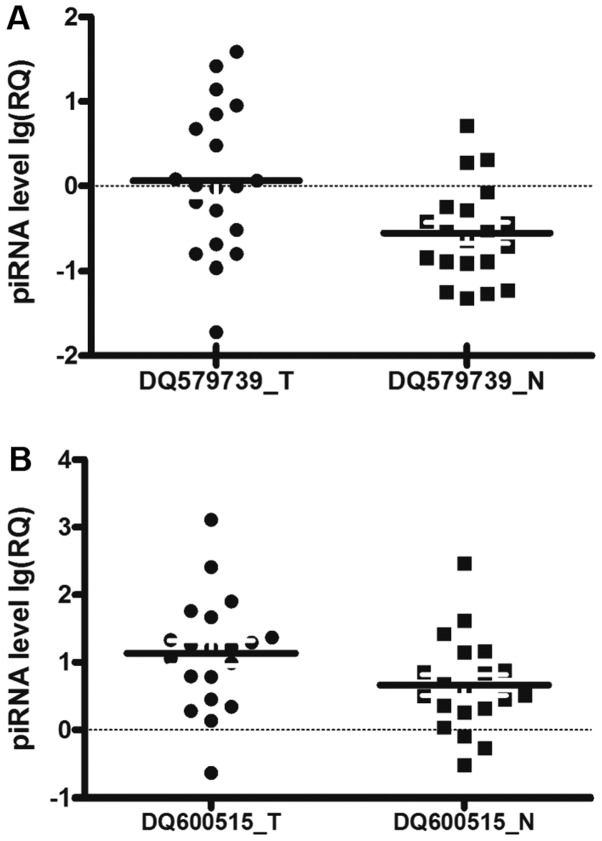
In a second cohort of 20 patients with gastric cancer, qPCR analysis on two piRNAs selected from the top list verified the quality of microarray experiment and validated the piRNA-GC associations in the primary cohort. DQ579739 and DQ600515 expression in the second cohort was concordant with the microarray data in the primary cohort. Notably, DQ579739 (P=0.001) and DQ600515 (P=0.005) expression in GC in the second cohort was significantly higher than in adjacent tissues (Fig. 3), consistent with the differential analysis on the array data in the primary cohort (Fig. 2) and in the same direction of association (i.e., upregulated).
Figure 3.
Distribution of piRNA expression levels in cases (T) and controls (N) [bold lines represent the mean values of expression levels; T, tumor tissue group; N, normal tissue group; RQ=2−ΔΔCq, where Cq values were generated from qPCR. (A) DQ579739; (B) DQ600515].
Gene expression in the stomach
Among the 50 GC-associated piRNAs, 43 were located within or close to 34 protein-coding genes. Twenty-two (64.7%) of the 34 genes within or close to which the 26 GC-associated piRNAs (60.5%) were located were expressed in human stomach (Table SI). The top four risk genes most abundant in stomach were CXCL17, EPCAM, RPL3 and RPS8 (Table SII).
piRNA expression is correlated with SNPs
eQTL analysis showed that many GC-associated piRNAs were nominally correlated with the genome-wide significant risk SNPs (1.6×10−4≤P<0.05; Table III). After Bonferroni correction (α=0.001), the correlations of rs4072037 at MUC1 with DQ595533, DQ590386, DQ595534 and DQ571813 in the whole sample (N+T) (P=3.6×10−4, 2.6×10−4, 1.6×10−4 and 2.6×10−4, respectively) and with DQ573237 in control group (P=9.3×10−4) remained significant. The results are also illustrated in Fig. S3.
Table III.
The risk piRNAs correlated with the genome-wide significant replicated risk SNPs.
| Chr1: MUC1 rs4072037 | Chr5: PRKAA1 rs13361707 | Chr8: PSCA rs2294008 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| piRNA | Chr | Gene | N | T | N+T | N | T | N+T | N | N+T |
| DQ575884 | chr2 | to EPCAM | 0.026 | 0.048 | 0.006 | 0.044 | 0.023 | |||
| DQ578739 | chr2 | to LOC100507334 | 0.038 | |||||||
| DQ571556 | chr2 | to SPATA31C1 | 0.027 | |||||||
| DQ595533 | chr3 | COLQ | 0.003 | 0.023 | 3.6×10−4 | 0.033 | 0.005 | |||
| DQ593293 | chr5 | to C5orf52 | 0.018 | |||||||
| DQ573237 | chr6 | SAYSD1 | 9.3×10−4 | 0.039 | 0.005 | 0.016 | 0.035 | 0.003 | ||
| DQ580665 | chr7 | POLR2J | 0.044 | 0.039 | 0.025 | |||||
| DQ583443 | chr9 | FAM225A | 0.042 | |||||||
| DQ571073 | chr12 | to CLLU1 | 0.039 | |||||||
| DQ600254 | chr14 | PNMA1 | 0.008 | 0.045 | 0.002 | 0.032 | 0.010 | |||
| DQ572562 | chr15 | DNM1P46 | 0.016 | |||||||
| DQ590386 | chr16 | VAC14 | 0.003 | 0.022 | 2.6×10−4 | 0.032 | 0.004 | |||
| DQ595534 | chr16 | VAC14 | 0.002 | 0.015 | 1.6×10−4 | 0.026 | 0.003 | |||
| DQ571813 | chr16 | VAC14 | 0.002 | 0.025 | 2.6×10−4 | 0.024 | 0.004 | |||
| DQ577343 | chr19 | to LOC400685 | 0.002 | 0.023 | ||||||
| DQ577344 | chr19 | to LOC400685 | 0.003 | 0.015 | ||||||
| DQ577345 | chr19 | to LOC400685 | 0.005 | 0.013 | ||||||
| DQ583043 | chr19 | to LOC100652909 | 0.029 | |||||||
| DQ574146 | chr22 | IGLL1 | 0.045 | |||||||
| DQ574147 | chr22 | IGLL1 | 0.042 | |||||||
| DQ580529 | chr22 | to WBP2NL | 0.004 | |||||||
SNP, single nucleotide polymorphism; N, controls; T, cases; ‘to’, proximate to. Correlations with P<α=0.001 (=0.05/50 piRNAs) are in bold print.
Piwi proteins are expressed in the stomach
Among the four piwi proteins, PIWIL4 was most abundant in stomach in the American cohort (TPM=3.9; GTEx). It was also expressed in our stomach samples of both 80 GC and 127 adjacent tissues (all expression scores ≥2+). There was significant difference in the weighted expression scores between GC and adjacent tissues (P=2.3×10−7; Table IV; Fig. S5).
Table IV.
Piwi protein expression in 181 gastric cancer and adjacent tissues.
| Weighted expression scores | |||||
|---|---|---|---|---|---|
| − | 1+ | 2+ | 3+ | P-value | |
| Cancer | 49 | 52 | 57 | 23 | |
| Normal | 21 | 33 | 110 | 17 | 2.3×10−7 |
Weighted expression scores: -, scores 0–1; 1+, scores 1–2; 2+, scores 3–4; 3+, scores 6–12 (see detailed scoring in Supplementary Methods).
Discussion
In the present study, we found that piRNAs are abundant in the human stomach, with 50 of them significantly associated with GC. Many GC-associated piRNAs were correlated with the genome-wide significant risk SNPs for GC and potentially regulated the transcription of cancer-related genes in the stomach via TE-suppression or TE-independent mechanisms. These findings support the proposition that piRNAs might play a role in determining GC risk.
These 50 piRNAs might impact GC risk via targeting the TEs and then regulating the expression of the proximate protein-coding genes, by sequence complementarity. Of them, 17 piRNAs, were upregulated in GC. This upregulation might suppress the activity of TEs, which maintains the self-renewal and proliferation capacity of cancer cells. The other 33 piRNAs were significantly downregulated in GC. This downregulation would increase the repeats of retrotransposon and cause DNA damage. We illustrate these potential mechanisms underlying the pathogenicity of piRNAs in Fig. S3.
In the present study, 8,759 piRNAs were detected in the human stomach. Across the transcriptome, an average of 5% of the piRNAs mapped within the protein-coding genes (61). However, in the present study, of the 50 differentially expressed piRNAs, 30 (60%) mapped within protein-coding genes, suggesting a strong relationship between these piRNAs and GC. Among the 34 protein-coding genes where the GC-associated piRNAs are located, 50% were expressed in stomach and associated with cancers, again in support of a strong relationship between the piRNAs and GC. Thus, the GC-associated piRNAs contribute to the pathogenesis of GC possibly via regulating their nearest genes based on sequence complementarity (31).
Two piRNAs, i.e., DQ579739 and DQ588779, are particularly worthy of attention (1). In our study, DQ579739 expression in GC was higher than in adjacent tissues in both microarray and qPCR validation data across two independent cohorts. The sequence of DQ579739 is complementary to AluSc8, a SINE-type TE, within the CXCL17 gene. CXCL17 mRNA was the most abundant in human stomach among all protein-coding genes examined in our study. As a novel acid CXC chemokine, CXCL17 is significantly upregulated in colon cancer (45), hepatocellular carcinoma (57), pancreatic carcinogenesis (56) and lung macrophages (62), and is associated with carcinogenesis, tumor proliferation, tumor progression (63), and angiogenesis (64,65). CXCL17 might act as a chemokine that accelerates tumor progression. The changes in individual piRNA level could influence both auto-regulatory gene expression and the expression of the gene to which the piRNA is complementary (66). Our finding suggests that this piRNA may be involved in GC development by regulating its host gene and its interacting proteins via AluSc8 (2). Among all piRNAs examined, the most abundant one in human stomach is DQ588779 at SKI (SKI proto-oncogene) (on chr1). This SKI proto-oncogene encodes the nuclear proto-oncogene protein homolog of avian sarcoma viral (v-ski) oncogene. It is expressed abundantly in the stomach, and has been associated with many types of cancers including GC (6,7). DQ588779 might play important roles in the development of GC by targeting AluSz6, a SINE-type TE, which may regulate the expression of stomach SKI gene.
Many GC-associated piRNAs were nominally correlated with the genome-wide significant risk SNPs, suggesting that these GC-associated piRNAs may be biologically functional. Several piRNAs complementary to COLQ, SAYSD1 and VAC14 were significantly correlated with rs4072037 on MUC1, suggesting a potential trans-acting regulation that may underlie SNP-GC associations.
To conclude, piRNAs are abundant in the human stomach and may play important roles in the pathogenesis of gastric cancer, and underlie the SNP-cancer associations as have been identified by GWASs. Overall, the current findings support piRNAs as potential biomarkers for GC diagnosis and prognosis. It is warranted to replicate these findings in larger sample in future work.
Supplementary Material
Acknowledgements
We thank Dr Chiang-Shan Li of Yale University for thorough editing of the manuscript.
Funding
The present study was supported by the National Clinical Key Specialty Construction Program of China, research grants from the Ministry of Health, China (WKJ2016-2-05), the Fujian Provincial Health Technology Project (2015-CX-7, 2013-ZQN-JC-8, 2015-ZQN-JC-7), the Natural Science Foundation of Fujian Province (nos. 2016J01513, 2016J01508 and 2019J01196), the Joint Funds for the Innovation of Science and Technology, Fujian province (no. 2017Y9082), the Science and Technology Program of Fujian Province (no. 2018Y2003) and the National Institute on Alcohol Abuse and Alcoholism (NIAAA) grants R21 AA021380, R21 AA020319 and R21 AA023237. To note, the study sponsors only provided the funds, but did not participated in the study.
Availability of data and materials
The datasets used during the present study are available from the corresponding author upon reasonable request.
Authors' contributions
XLi, XZ, JX, QM, HHC and XLu contributed to the formulation of the research goals and aims and the writing, reviewing, and editing of the article. XLi, YX, DH, HeZ, ZY, CL, GC, FL, WZ, YS, HuZ, JZ, TS, JX, XZ and XLu provided the patient resources and searched and collected the data. XLi, YX, DH, QM, HeZ, ZY, CL, GC, FL WZ, YS, HuZ, JZ, TS, HHC and XLu conducted the study and performed the analysis. XLi, XZ and XLu secured funding for the present study. XLi and XLu are responsible for the overall content as guarantors. All authors read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
The study was approved by the Research Ethics Committee of the Fujian Provincial Cancer Hospital, China. Informed consents from all patients were obtained prior to the study. All experiments were performed in accordance with relevant guidelines and regulations.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Lin X, Hu D, Chen G, Shi Y, Zhang H, Wang X, Guo X, Lu L, Black D, Zheng XW, Luo X. Associations of THBS2 and THBS4 polymorphisms to gastric cancer in a Southeast Chinese population. Cancer Genet. 2016;209:215–222. doi: 10.1016/j.cancergen.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 3.Berger H, Marques MS, Zietlow R, Meyer TF, Machado JC, Figueiredo C. Gastric cancer pathogenesis. Helicobacter. 2016;21(Suppl 1):S34–S38. doi: 10.1111/hel.12338. [DOI] [PubMed] [Google Scholar]
- 4.Kim J, Yum S, Kang C, Kang SJ. Gene-gene interactions in gastrointestinal cancer susceptibility. Oncotarget. 2016;7:67612–67625. doi: 10.18632/oncotarget.11701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin XD, Chen SQ, Qi YL, Zhu JW, Tang Y, Lin JY. Polymorphism of THBS1 rs1478604 A>G in 5-untranslated region is associated with lymph node metastasis of gastric cancer in a Southeast Chinese population. DNA Cell Biol. 2012;31:511–519. doi: 10.1089/dna.2011.1344. [DOI] [PubMed] [Google Scholar]
- 6.Nakao T, Kurita N, Komatsu M, Yoshikawa K, Iwata T, Utsunomiya T, Shimada M. Expression of thrombospondin-1 and Ski are prognostic factors in advanced gastric cancer. Int J Clin Oncol. 2011;16:145–152. doi: 10.1007/s10147-010-0147-5. [DOI] [PubMed] [Google Scholar]
- 7.Takahata M, Inoue Y, Tsuda H, Imoto I, Koinuma D, Hayashi M, Ichikura T, Yamori T, Nagasaki K, Yoshida M, et al. SKI and MEL1 cooperate to inhibit transforming growth factor-beta signal in gastric cancer cells. J Biol Chem. 2009;284:3334–3344. doi: 10.1074/jbc.M808989200. [DOI] [PubMed] [Google Scholar]
- 8.Hong CS, Jeong O, Piao Z, Guo C, Jung MR, Choi C, Park YK. HOXB5 induces invasion and migration through direct transcriptional up-regulation of β-catenin in human gastric carcinoma. Biochem J. 2015;472:393–403. doi: 10.1042/BJ20150213. [DOI] [PubMed] [Google Scholar]
- 9.Xie SS, Jin J, Xu X, Zhuo W, Zhou TH. Emerging roles of non-coding RNAs in gastric cancer: Pathogenesis and clinical implications. World J Gastroenterol. 2016;22:1213–1223. doi: 10.3748/wjg.v22.i3.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang T, Ji Y, Hu D, Chen B, Zhang H, Li C, Chen G, Luo X, Zheng XW, Lin X. SNHG8 is identified as a key regulator of epstein-barr virus(EBV)-associated gastric cancer by an integrative analysis of lncRNA and mRNA expression. Oncotarget. 2016;7:80990–81002. doi: 10.18632/oncotarget.13167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fu A, Jacobs DI, Hoffman AE, Zheng T, Zhu Y. PIWI-interacting RNA 021285 is involved in breast tumorigenesis possibly by remodeling the cancer epigenome. Carcinogenesis. 2015;36:1094–1102. doi: 10.1093/carcin/bgv105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng J, Guo JM, Xiao BX, Miao Y, Jiang Z, Zhou H, Li QN. piRNA, the new non-coding RNA, is aberrantly expressed in human cancer cells. Clin Chim Acta. 2011;412:1621–1625. doi: 10.1016/j.cca.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 13.Zuo L, Wang Z, Tan Y, Chen X, Luo X. piRNAs and their functions in the brain. Int J Hum Genet. 2016;16:53–60. doi: 10.1080/09723757.2016.11886278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ross RJ, Weiner MM, Lin H. PIWI proteins and PIWI-interacting RNAs in the soma. Nature. 2014;505:353–359. doi: 10.1038/nature12987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- 16.Sturm Á, Perczel A, Ivics Z, Vellai T. The Piwi-piRNA pathway: Road to immortality. Aging Cell. 2017;16:906–911. doi: 10.1111/acel.12630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mei Y, Clark D, Mao L. Novel dimensions of piRNAs in cancer. Cancer Lett. 2013;336:46–52. doi: 10.1016/j.canlet.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ng KW, Anderson C, Marshall EA, Minatel BC, Enfield KS, Saprunoff HL, Lam WL, Martinez VD. Piwi-interacting RNAs in cancer: Emerging functions and clinical utility. Mol Cancer. 2016;15:5. doi: 10.1186/s12943-016-0491-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Busch J, Ralla B, Jung M, Wotschofsky Z, Trujillo-Arribas E, Schwabe P, Kilic E, Fendler A, Jung K. Piwi-interacting RNAs as novel prognostic markers in clear cell renal cell carcinomas. J Exp Clin Cancer Res. 2015;34:61. doi: 10.1186/s13046-015-0180-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hashim A, Rizzo F, Marchese G, Ravo M, Tarallo R, Nassa G, Giurato G, Santamaria G, Cordella A, Cantarella C, Weisz A. RNA sequencing identifies specific PIWI-interacting small non-coding RNA expression patterns in breast cancer. Oncotarget. 2014;5:9901–9910. doi: 10.18632/oncotarget.2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yan H, Wu QL, Sun CY, Ai LS, Deng J, Zhang L, Chen L, Chu ZB, Tang B, Wang K, et al. piRNA-823 contributes to tumorigenesis by regulating de novo DNA methylation and angiogenesis in multiple myeloma. Leukemia. 2015;29:196–206. doi: 10.1038/leu.2014.135. [DOI] [PubMed] [Google Scholar]
- 22.Law PT, Qin H, Ching AK, Lai KP, Co NN, He M, Lung RW, Chan AW, Chan TF, Wong N. Deep sequencing of small RNA transcriptome reveals novel non-coding RNAs in hepatocellular carcinoma. J Hepatol. 2013;58:1165–1173. doi: 10.1016/j.jhep.2013.01.032. [DOI] [PubMed] [Google Scholar]
- 23.Huang G, Hu H, Xue X, Shen S, Gao E, Guo G, Shen X, Zhang X. Altered expression of piRNAs and their relation with clinicopathologic features of breast cancer. Clin Transl Oncol. 2013;15:563–568. doi: 10.1007/s12094-012-0966-0. [DOI] [PubMed] [Google Scholar]
- 24.Zhang H, Ren Y, Xu H, Pang D, Duan C, Liu C. The expression of stem cell protein Piwil2 and piR-932 in breast cancer. Surg Oncol. 2013;22:217–223. doi: 10.1016/j.suronc.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 25.Chu H, Hui G, Yuan L, Shi D, Wang Y, Du M, Zhong D, Ma L, Tong N, Qin C, et al. Identification of novel piRNAs in bladder cancer. Cancer Lett. 2015;356:561–567. doi: 10.1016/j.canlet.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 26.Qiu W, Guo X, Lin X, Yang Q, Zhang W, Zhang Y, Zuo L, Zhu Y, Li CR, Ma C, Luo X. Transcriptome-wide piRNA profiling in human brains of Alzheimer's disease. Neurobiol Aging. 2017;57:170–177. doi: 10.1016/j.neurobiolaging.2017.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martinez VD, Enfield KSS, Rowbotham DA, Lam WL. An atlas of gastric PIWI-interacting RNA transcriptomes and their utility for identifying signatures of gastric cancer recurrence. Gastric Cancer. 2016;19:660–665. doi: 10.1007/s10120-015-0487-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levin HL, Moran JV. Dynamic interactions between transposable elements and their hosts. Nat Rev Genet. 2011;12:615–627. doi: 10.1038/nrg3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malone CD, Hannon GJ. Small RNAs as guardians of the genome. Cell. 2009;136:656–668. doi: 10.1016/j.cell.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reilly MT, Faulkner GJ, Dubnau J, Ponomarev I, Gage FH. The role of transposable elements in health and diseases of the central nervous system. J Neurosci. 2013;33:17577–17586. doi: 10.1523/JNEUROSCI.3369-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roy J, Sarkar A, Parida S, Ghosh Z, Mallick B. Small RNA sequencing revealed dysregulated piRNAs in Alzheimer's disease and their probable role in pathogenesis. Mol Biosyst. 2017;13:565–576. doi: 10.1039/C6MB00699J. [DOI] [PubMed] [Google Scholar]
- 32.Hu N, Wang Z, Song X, Wei L, Kim BS, Freedman ND, Baek J, Burdette L, Chang J, Chung C, et al. Genome-wide association study of gastric adenocarcinoma in Asia: A comparison of associations between cardia and non-cardia tumours. Gut. 2016;65:1611–1618. doi: 10.1136/gutjnl-2015-309340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abnet CC, Freedman ND, Hu N, Wang Z, Yu K, Shu XO, Yuan JM, Zheng W, Dawsey SM, Dong LM, et al. A shared susceptibility locus in PLCE1 at 10q23 for gastric adenocarcinoma and esophageal squamous cell carcinoma. Nat Genet. 2010;42:764–767. doi: 10.1038/ng.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Z, Dai J, Hu N, Miao X, Abnet CC, Yang M, Freedman ND, Chen J, Burdette L, Zhu X, et al. Identification of new susceptibility loci for gastric non-cardia adenocarcinoma: Pooled results from two Chinese genome-wide association studies. Gut. 2017;66:581–587. doi: 10.1136/gutjnl-2015-310612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Helgason H, Rafnar T, Olafsdottir HS, Jonasson JG, Sigurdsson A, Stacey SN, Jonasdottir A, Tryggvadottir L, Alexiusdottir K, Haraldsson A, et al. Loss-of-function variants in ATM confer risk of gastric cancer. Nat Genet. 2015;47:906–910. doi: 10.1038/ng.3342. [DOI] [PubMed] [Google Scholar]
- 36.Shi Y, Hu Z, Wu C, Dai J, Li H, Dong J, Wang M, Miao X, Zhou Y, Lu F, et al. A genome-wide association study identifies new susceptibility loci for non-cardia gastric cancer at 3q13.31 and 5p13.1. Nat Genet. 2011;43:1215–1218. doi: 10.1038/ng.978. [DOI] [PubMed] [Google Scholar]
- 37.Chen YC, Fang WL, Wang RF, Liu CA, Yang MH, Lo SS, Wu CW, Li AF, Shyr YM, Huang KH. Clinicopathological variation of lauren classification in gastric cancer. Pathol Oncol Res. 2016;22:197–202. doi: 10.1007/s12253-015-9996-6. [DOI] [PubMed] [Google Scholar]
- 38.Edge SB, Compton CC. The American joint committee on cancer: The 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17:1471–1474. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 39.Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, Bartel DP, Kingston RE. Characterization of the piRNA complex from rat testes. Science. 2006;313:363–367. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- 40.He QQ, Xiong LL, Liu F, He X, Feng GY, Shang FF, Xia QJ, Wang YC, Qiu DL, Luo CZ, et al. MicroRNA-127 targeting of mitoNEET inhibits neurite outgrowth, induces cell apoptosis and contributes to physiological dysfunction after spinal cord transection. Sci Rep. 2016;6:35205. doi: 10.1038/srep35205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yuan JS, Reed A, Chen F, Stewart CN., Jr Statistical analysis of real-time PCR data. BMC Bioinformatics. 2006;7:85. doi: 10.1186/1471-2105-7-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.GTEx Consortium. The genotype-tissue expression (GTEx) project. Nat Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S, Hodge CL, Haase J, Janes J, Huss JW, III, Su AI. BioGPS: An extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009;10:R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wilhelm M, Schlegl J, Hahne H, Gholami AM, Lieberenz M, Savitski MM, Ziegler E, Butzmann L, Gessulat S, Marx H, et al. Mass-spectrometry-based draft of the human proteome. Nature. 2014;509:582–587. doi: 10.1038/nature13319. [DOI] [PubMed] [Google Scholar]
- 45.Ohlsson L, Hammarström ML, Lindmark G, Hammarström S, Sitohy B. Ectopic expression of the chemokine CXCL17 in colon cancer cells. Br J Cancer. 2016;114:697–703. doi: 10.1038/bjc.2016.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oosterhoff D, Overmeer RM, de Graaf M, van der Meulen IH, Giaccone G, van Beusechem VW, Haisma HJ, Pinedo HM, Gerritsen WR. Adenoviral vector-mediated expression of a gene encoding secreted, EpCAM-targeted carboxylesterase-2 sensitises colon cancer spheroids to CPT-11. Br J Cancer. 2005;92:882–887. doi: 10.1038/sj.bjc.6602362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xie X, Li F, Zhang H, Lu Y, Lian S, Lin H, Gao Y, Jia L. EpCAM aptamer-functionalized mesoporous silica nanoparticles for efficient colon cancer cell-targeted drug delivery. Eur J Pharm Sci. 2016;83:28–35. doi: 10.1016/j.ejps.2015.12.014. [DOI] [PubMed] [Google Scholar]
- 48.Russo A, Maiolino S, Pagliara V, Ungaro F, Tatangelo F, Leone A, Scalia G, Budillon A, Quaglia F, Russo G. Enhancement of 5-FU sensitivity by the proapoptotic rpL3 gene in p53 null colon cancer cells through combined polymer nanoparticles. Oncotarget. 2016;7:79670–79687. doi: 10.18632/oncotarget.13216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pagliara V, Saide A, Mitidieri E, d'Emmanuele di Villa Bianca R, Sorrentino R, Russo G, Russo A. 5-FU targets rpL3 to induce mitochondrial apoptosis via cystathionine-β-synthase in colon cancer cells lacking p53. Oncotarget. 2016;7:50333–50348. doi: 10.18632/oncotarget.10385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Russo A, Pagliara V, Albano F, Esposito D, Sagar V, Loreni F, Irace C, Santamaria R, Russo G. Regulatory role of rpL3 in cell response to nucleolar stress induced by Act D in tumor cells lacking functional p53. Cell Cycle. 2016;15:41–51. doi: 10.1080/15384101.2015.1120926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang SY, Gao K, Deng DL, Cai JJ, Xiao ZY, He LQ, Jiao HL, Ye YP, Yang RW, Li TT, et al. TLE4 promotes colorectal cancer progression through activation of JNK/c-Jun signaling pathway. Oncotarget. 2016;7:2878–2888. doi: 10.18632/oncotarget.6694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu YL, Gao X, Jiang Y, Zhang G, Sun ZC, Cui BB, Yang YM. Expression and clinicopathological significance of EED, SUZ12 and EZH2 mRNA in colorectal cancer. J Cancer Res Clin Oncol. 2015;141:661–669. doi: 10.1007/s00432-014-1854-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seo GS, Yu JI, Chae SC, Park WC, Shin SR, Yoo ST, Choi SC, Lee SH. EED gene polymorphism in patients with colorectal cancer. Int J Biol Markers. 2013;28:274–279. doi: 10.5301/JBM.5000024. [DOI] [PubMed] [Google Scholar]
- 54.Song L, Chen X, Gao S, Zhang C, Qu C, Wang P, Liu L. Ski modulate the characteristics of pancreatic cancer stem cells via regulating sonic hedgehog signaling pathway. Tumour Biol. 2016 Oct 12; doi: 10.1007/s13277-016-5461-8. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 55.Jiang SH, He P, Ma MZ, Wang Y, Li RK, Fang F, Fu Y, Tian GA, Qin WX, Zhang ZG. PNMA1 promotes cell growth in human pancreatic ductal adenocarcinoma. Int J Clin Exp Pathol. 2014;7:3827–3835. [PMC free article] [PubMed] [Google Scholar]
- 56.Hiraoka N, Yamazaki-Itoh R, Ino Y, Mizuguchi Y, Yamada T, Hirohashi S, Kanai Y. CXCL17 and ICAM2 are associated with a potential anti-tumor immune response in early intraepithelial stages of human pancreatic carcinogenesis. Gastroenterology. 2011;140:310–321. doi: 10.1053/j.gastro.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 57.Li L, Yan J, Xu J, Liu CQ, Zhen ZJ, Chen HW, Ji Y, Wu ZP, Hu JY, Zheng L, Lau WY. CXCL17 expression predicts poor prognosis and correlates with adverse immune infiltration in hepatocellular carcinoma. PLoS One. 2014;9:e110064. doi: 10.1371/journal.pone.0110064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chan AW, Tong JH, Chan SL, Lai PB, To KF. Expression of stemness markers (CD133 and EpCAM) in prognostication of hepatocellular carcinoma. Histopathology. 2014;64:935–950. doi: 10.1111/his.12342. [DOI] [PubMed] [Google Scholar]
- 59.Kang JS. Increased expression of epithelial cell adhesion molecule (EpCAM) in rat hepatic tumors induced by diethylnitrosamine. Asian Pac J Cancer Prev. 2012;13:3627–3630. doi: 10.7314/APJCP.2012.13.8.3627. [DOI] [PubMed] [Google Scholar]
- 60.Luo J, Cai Q, Wang W, Huang H, Zeng H, He W, Deng W, Yu H, Chan E, Ng CF, et al. A microRNA-7 binding site polymorphism in HOXB5 leads to differential gene expression in bladder cancer. PLoS One. 2012;7:e40127. doi: 10.1371/journal.pone.0040127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 62.Burkhardt AM, Maravillas-Montero JL, Carnevale CD, Vilches-Cisneros N, Flores JP, Hevezi PA, Zlotnik A. CXCL17 is a major chemotactic factor for lung macrophages. J Immunol. 2014;193:1468–1474. doi: 10.4049/jimmunol.1400551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Matsui A, Yokoo H, Negishi Y, Endo-Takahashi Y, Chun NA, Kadouchi I, Suzuki R, Maruyama K, Aramaki Y, Semba K, et al. CXCL17 expression by tumor cells recruits CD11b+Gr1 high F4/80- cells and promotes tumor progression. PLoS One. 2012;7:e44080. doi: 10.1371/journal.pone.0044080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Weinstein EJ, Head R, Griggs DW, Sun D, Evans RJ, Swearingen ML, Westlin MM, Mazzarella R. VCC-1, a novel chemokine, promotes tumor growth. Biochem Biophys Res Commun. 2006;350:74–81. doi: 10.1016/j.bbrc.2006.08.194. [DOI] [PubMed] [Google Scholar]
- 65.Mu X, Chen Y, Wang S, Huang X, Pan H, Li M. Overexpression of VCC-1 gene in human hepatocellular carcinoma cells promotes cell proliferation and invasion. Acta Biochim Biophys Sin (Shanghai) 2009;41:631–637. doi: 10.1093/abbs/gmp051. [DOI] [PubMed] [Google Scholar]
- 66.Esposito T, Magliocca S, Formicola D, Gianfrancesco F. piR_015520 belongs to Piwi-associated RNAs regulates expression of the human melatonin receptor 1A gene. PLoS One. 2011;6:e22727. doi: 10.1371/journal.pone.0022727. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used during the present study are available from the corresponding author upon reasonable request.