ABSTRACT
The emergence of new infectious bursal disease virus (IBDV) variants can threaten poultry health and production all over the world causing significant economic losses. Therefore, this study was performed to determine IBDV molecular epidemilogy, VP2 gene variation, and corresponding pathological lesions in IBDV infected chickens in Turkey. For this, 1855 bursa of Fabricius samples were collected from 371 vaccinated broiler flocks. Atrophia and haemorrhages were seen in the bursa Fabricius of very virulent IBDV (vvIBDV) infected chickens. Partial VP2 gene was sequenced and phylogenetic, recombination, and evolutionary analyses were performed. 1548 (83.5%) out of 1855 of bursa of Fabricius samples were IBDV positive and 1525 of those could be sequenced. The recombination analysis did not detect occurrence of any recombination event among the Turkish strains. Among 1525 sequenced samples, 1380 of them were found to be classical strains. Among 1380 classical strains, 1317 were similar to IBDV 2512, 11 to Faragher 52/70, 40 to 228 E, and 12 to Lukert strain. Out of 1525 reverse transcriptase ploymerase chain reaction positive samples, 144 of them were found to be similar to vvIBDV-VP2 gene reported to GenBank previously. The phylogenetic tree performed on a broad sequence dataset demonstrated grouping of vvIBDV Turkish strains in three different clusters, including sequences collected also from Iraq and Kuwait (Cluster 1), Indian (Cluster 2), and a distinct Turkish-only cluster (Cluster 3). The evolutionary rate estimation on branches/clades including Turkish strain mirrored the expected one for RNA viruses and no significant differences were found among different considered branches. In conclusion, results of this study indicate that vvIBDV strains similar to those circulating in various countries in the Middle East are present and undergoing evolution in chickens from Turkish broiler flocks. This point needs to be taken into account in planning adequate control strategies.
Keywords: Gumboro, virus, VP2-gene, phylogenetic, chicken
INTRODUCTION
Infectious bursal disease (IBD), also known as Gumboro disease, is an important acute, highly contagious disease of young chickens, threatening the poultry industry worldwide (Cosgrove, 1962; Dolz et al., 2005; Jackwood and Sommer-Wagner, 2007). It is caused by IBD virus (IBDV), which was first discovered in Delaware in the United States of America (USA) in 1962 (Cosgrove, 1962; Hitchner, 1970). Chickens are the primary host species for IBDV, but the antibodies and the virus are also found in wild birds including guinea fowls, ducks, quails, pheasants, and ostriches with no signs of infection (OIE, 2016). IBDV primarly infects immature B cells causing pathological changes in the bursa of Fabricius of chickens, resulting in immunosupression, which, in turn, facilitates secondary infections and poor immune response to pathogens and vaccination (Faragher et al., 1974; Jackwood et al., 2008; Aricibasi et al., 2010).
IBDV is a non-enveloped, double-stranded RNA virus that belongs to the Avibirnavirus genus in the family of Birnaviridae comprising two serotypes: serotype 1and serotype 2. Serotype 1 causes disease in chickens while serotype 2 is naturally avirulent. However, antibodies to serotype 2 are very common in turkeys, but are less frequently found in chickens and ducks (Lukert and Saif, 2003). Serotype 1 includes classical and variant antigenic subtypes whereas the classical subtypes are divided into 3 pathotypes: mild/attenuated, virulent, and very virulent IBDV (Jackwood et al., 2008; Alfonso-Morales et al., 2013). The IBDV genome is bisegmented into a linear segment A (3.4 kb) and segment B (2.8 kb). The segment A encodes four viral proteins: 2 capsid proteins VP2 (48 kDa) and VP3 (32–35 kDa), a protease VP4 (24 kDa) and a nonstructural protein VP5 (17–21 kDa).
The capsid protein VP2 has a hypervariable region which represents the major conformational and antigenic domain at amino acid positions 222 to 350 (Jackwood and Sommer-Wagner, 2007; Jackwood et al., 2008; Aricibasi et al., 2010). This region determines the antigenic and pathogenic properties of individual IBDV strains. Segment B encodes VP1 (90 kDa), which is a RNA-dependent RNA polymerase (Coulibaly et al., 2005; Durairaj et al., 2011).
Clinical and pathological lesions vary depending on the virulence of the strain, and the age of the infected chicken including hemorrhages and athrophy of the bursa of Fabriciusand lesions of kidneys, proventriculus and various muscles (Faragher et al., 1974; Rosales et al., 1989; Mazariegos et al., 1990). Very virulent IBDV (vvIBDV) strains have been shown to overcome high levels of maternal antibodies in chickens and may cause 20 to 60% mortality rates including severe pathological lesions in the above mentioned tissues (Chettle et al., 1989; Le Nouen et al., 2006).
Sequencing of the hypervariable region of the VP2 gene has been used to determine pathotype variants of the IBDV serotype 1. After the first report of vvIBDV in 1989 (Stuart, 1989), IBDV variants primarily with variations in the VP2 protein have been reported worldwide (Jackwood and Sommer-Wagner, 2007). Results of molecular epidemiology studies indicate that all vvIBDV strains described so far originated from a common ancestor although some locally circulating vvIBDV strains were found to be different (Yamaguchi et al, 1997; Rudd et al., 2002; Hon et al., 2006; Jackwood and Stoute, 2013). The molecular and pathotype characterization of IBDV is important for prevention and vaccination strategies. Until now no comprehensive study related to IBDV genetic variants in chickens in Turkey has been conducted.
Despite the availability and application of IBDV vaccines in poultry worldwide, the emergence of new IBDV variants can threaten poultry health and production all over the world causing significant economic losses. In view of the genetic variability of IBDVs’ reported in many studies, it is important to determine genetic evolution and changes in virulence in circulating strains of IBDV in order to reduce the economic impact of highly virulent vvIBDV. Based on such studies, effective national and/or regional mitigation strategies, which include improved diagnostics and vaccination, can be established. Therefore, this study was performed to determine VP2 gene variation and corresponding pathological lesions in IBDV infected chickens in Turkey to understand phylogeographic dynamics of IBDV, viral population expansion, and IBDV evolution in Turkey.
MATERIALS AND METHODS
Study Population and Sampling
Commercial broiler flocks (n = 371) were selected from 3 geographic regions (Egean, Marmara and Western Black Sea) of Turkey, and sampled from 1st of February to 31st December 2017. Biosecurity at the farms was good to moderate. All flocks were vaccinated against IBDV either in the hatchery (Winterfield 2515 or Vector vaccine) or in the farms (Winterfield 2512, Faragher 52/70, 228E, Lukert or MB strain) between 12 to 16 d of age. The rationale of sampling of vaccinated flocks was mainly for monitoring the vaccine uptake, applicatian afficacy, and also for the diagnosis of IBDV suspected birds. In order to monitor vaccine intake and diagnosis of IBDV suspected birds. Two criteria were considered in selection of flocks for sampling: 1. Flocks showing clinical signs of IBD; 2. Randomly selected flocks not showing IBD signs. Five humanely euthanized birds at the age of 27 to 40 d from each broiler flock were necropsied and sampled for bursa of Fabricius (1855 bursae in total).
Histopathology
Samples of bursa of Fabricius of 28 broiler flocks detected vvIBDV were analysed histopathologically. Only the bursa of Fabricius showing severe necropsi findings (edema, heamorrhages and atrophy or enlargement) were examined histopathologically. For this, samples were fixed in 10% neutral buffered formalin, embedded in paraffin blocks, cut into 4 to 5 μm sections, stained with hematoxylin and eosin, and blindly examined in the Department of Pathology.
RNA Extraction and Rreverse Transcription
Total RNA was extracted from bursa of Fabricius tissues using the RNeasy RNA extraction kit as described by the manufacturer (Qiagen, Cat No:74,106). The amount of RNA in the extracted material (50 μl) was measured using a NanoDrop spectrophotometer (NanoDrop 1000c, Thermo Scientific, Waltham, USA). Reverse transcription and generation of cDNA was performed by using High Capacity cDNA reverse transcription kit (Applied Biosystems, Cat No: 4,368,814) as described by the manufacturer.
Partial VP2-Gene Sequencing
In order to differentiate vaccine and field IBDV strains, the cDNA generated from tissues as described above was subjected to PCR by using IBDV-specific primers as described previously (Toroghi et al., 2001). The partial IBDV VP2 gene amplicons were partially sequenced. The partial VP2 gene sequences consisted of a 552 bp PCR product from nucleotide positions 651 to 1202 in the IBDV VP2 gene Toroghi et al. 2001). For this purpose, our optimized PCR reaction consisted of a total volume of 50 μl reaction mixture containing 3 μl (10 μM) of each forward and reverse primers, 25 μl Maxima Hot Start PCR Master Mix (Termo Scientific, K1052), 15 μl nuclease-free water, 1 μl MgCl2 (25 mM), and 3 μl of cDNA. The mixture was placed in a thermal cycler (Biorad, Chromo-4), and the polymerase was then activated by incubation at 95°C for 10 min. Cycling conditions were 94°C for 1 min, 60°C for 1 min, and 72°C for 1 min for 35 cycles. Final extension was performed at 72°C for 5 min. For all PCR reactions, nuclease-free water was used as negative control in place of template. After the PCR, the presence of the 552 bp product was confirmed by agarose gel (1.5%) electrophoresis.
Positive controls were obtained from samples submitted to the Department of Virology, Veterinary Faculty of Istanbul, and previously confirmed to be IBDV positive by reverse transcriptase ploymerase chain reaction (RT-PCR) and subsequent sequencing. For PCR efficiency, the avian glyceraldehyde 3-phosphate dehydrogenase gene was used as internal control. Products obtained by PCR using the primers specific for the partial IBDV VP2 gene were sequenced by a commercial company (MedSanTek, Istanbul, Turkey).
Phylogenetic, Recombination and Evolutionary Analyses
Multiple alignments of partial VP2 gene region sequences were made using the MUSCLE algorithm implemented in MEGA-7 software. Phylogenetic analyses were carried out using the criterion of neighbor-joining trees based on genetic distance model by Tamura and Nei (1993). The reference sequence set for IBDV classification (Michel and Jackwood, 2017) was used to reconstruct the topology of the VP2 gene sequences generated in this study. The partial VP2 gene sequences obtained in this study were submitted to GenBank (Accession Number: MH137942-MH137957).
To investigate the occurence of recombination events, a full collection of partial IBDV VP2 gene sequences was downloaded from GenBank, aligned to the one obtained in the present study using MAFFT (Katoh and Standley, 2013) and analyzed using RDP4 (Martin et al., 2015). The RDP4 settings for each method were adjusted to account for the dataset features according to the RDP manual recommendations. Only recombination events detected by more than 2 methods with a significance value lower than 0.05 (P-value < 0.05) and Bonferroni correction were accepted (Moreno et al. 2017). To investigate the relationship between Turkish strains and worldwide collected ones, a phylogentic tree was reconstructed on the non-recombinant sequence dataset using the Maximum likelihood (ML) approach implemented in PhyML (Guindon et al, 2010). The best substitution model was selected based on the Akaike information criteria, calculated using JmodelTest (Darriba et al., 2012). The branch support was evaluated using the fast non-parametric version of the aLRT (Shimodaira–Hasegawa [SH]-aLRT), developed and implemented in PhyML 3.0 (Anisimova and Gascuel, 2006). For clarity reason, only sequences whose collection date and country were available were included in the analysis. The evolutionary rates featuring the monophyletic Turkish clades were estimated using BEAST, similarly to what performed by Franzo and others (32).
Isolation of 3 Different Clusters of vvIBDV Strains
IBDV strains, which belong to 3 different clusters of vvIBDV identified in this study by the phylogentic analyses, were inoculated in to chorio-allontoic membrane of 11 d old embryonated specific pathogen free chicken eggs (Bornova Veterinary Control Institute, Izmir, Turkey) by using conventional methods (OIE, 2016). They were tested by RT-PCR and sequenced if the corresponding bands present as explained previously in the Materials and Methods of this study.
RESULTS
Clinical Observations
Mortality rates in clinically affected broiler flocks (detected vvIBDV) ranged from 8 to 14%. The clinical signs observed in the infected flocks were depression, ruffled feathers, lethargy, poor feed intake, poor growth, and white watery diarrhea. The feathers around the cloaca were stained with feces.
Necropsy and Histopathological Findings
At necropsy, various gross changes were seen in bursa of Fabricius in 28 broiler flocks detected vvIBDV. Atrophy was remarkable in some of them. They were firm and almost receded to one third of their original sizes. There were distinct edema, hemorrhage, atrophy, and enlargement in some bursae of Fabricius. In addition, petechial and stripe-like hemorrhages were seen on the pectoral and leg muscles of some IBDV infected birds.
Histopathologically, different sizes of vacuolizations were prominent in the plicae of lamina epithelialis in the bursa of Fabricius of IBDV infected chickens. On the other hand, in some of them, invaginations in lamina epithelialis were observed and correspondingly gland like structures both in the follicules and lamina epithelialis had formed (Figure 1a). Edema and enlargement due to IBDV infection were detected in the interfollicules areas (Figure 1b). In some bursa of Fabricius, hemorrhages in the inter and intrafollicular areas and lympho-mononuclear cell infiltration interfollicules areas were also seen (Figure 1c). In some follicules, vacuolizations and necrosis in the medullary areas were noted and in some of them infiltration of endothelial macrophages, diffuse vacuolizations, and atrophy were also evident. In some bursa of Fabricius, there was an excess formation of fibrous connective tissue and follicular atrophy (Figure 1d).
Figure 1.
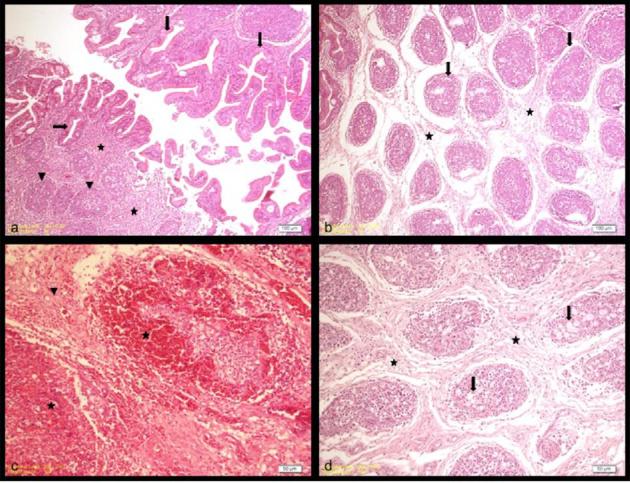
Histopathological lesions in the bursa of Fabricius in chickens infected with very virulent IBDV (H.E.). a: gland like structures both in the follicules and lamina epithelialis (arrow) and lympho-mononuclear cell infiltration in the interfollicular areas (star) and follicular atrophy (arrow head), bar: 100 μm; b: Edema and enlargement in the interfollicular areas (star) and follicular atrophy (arrow), bar: 100 μm; c: Lympho-mononuclear cell infiltration in the interfollicular areas (arrow head) and hemorrhages (star) bar: 50 μm; d: In some follicules, vacuolizations and necrosis in medullary areas and in some of them infiltration of endothelial macrophages (arrow) and excess formation of fibrous connectivetissue (star) and follicular atrophy, bar: 50μm.
Overall Detection of Partial IBDV VP2-Gene in Broiler Flocks by RT-PCR
Out of 1855 bursa of Fabricius samples analysed, IBDV was detected by RT-PCR in 1548 (83.5%). On gel electrophoresis, 552 bp amplicon corresponding to the parital IBDV VP2 gene was seen. All of these flocks had been previously vaccinated against IBDV by using commercial vaccines containing either Winterfield 2512, Faragher 52/70, 228 E, Lukert or MB strain.
Phylogenetic Analysis of the Partial IBDV VP2-Gene Sequences and Vaccine Monitoring
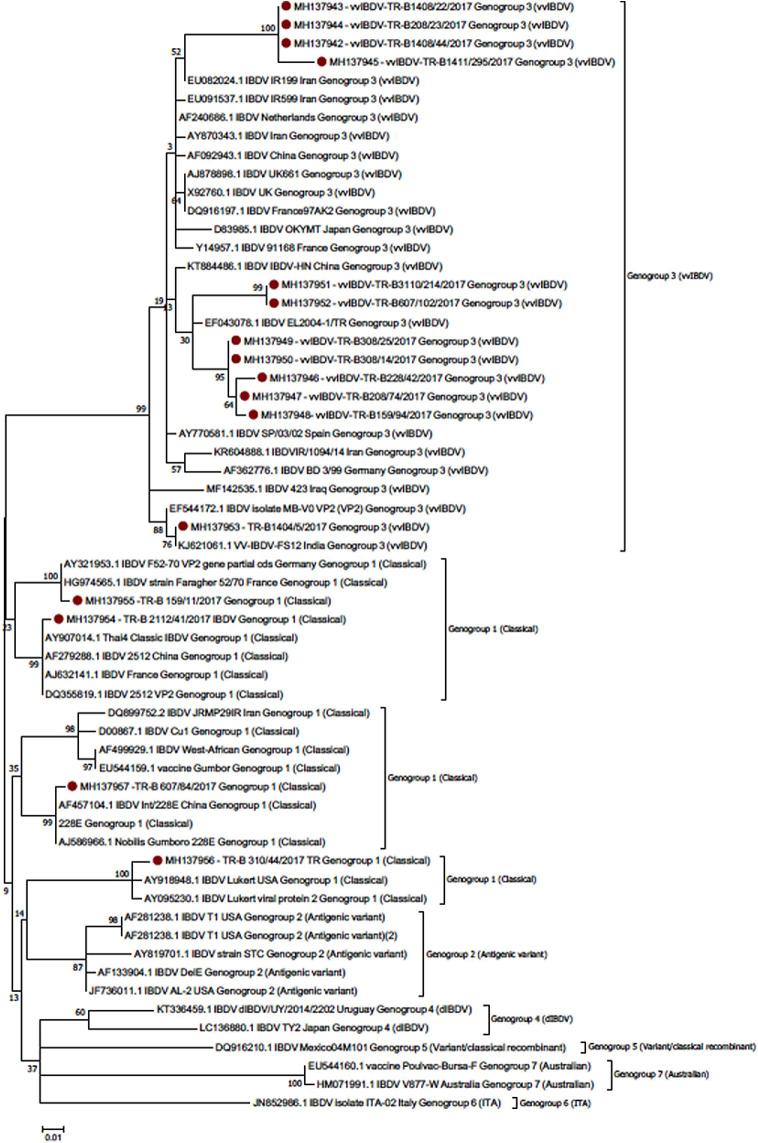
Amongst 1548 RT-PCR positive samples, 1525 samples could be sequenced. Out of 1525 samples, 144 (9.5%) of them were found to be similar to vvIBDV VP2 gene reported in other countries and submitted to GenBank previously. These vvIBDV strains formed 3 different clusters, all part of the Genogroup 3 (Michel and Jackwood, 2017). 79 of the vvIBDV strains (representative sequences: MH137942, MH137943, MH137944, MH137945) formed different cluster amongst all vvIBDV strains reported so far in the world but were related to strains reported from China (AF092943) and Iran (AY870343, EU082024, EU091537), the Netherlands AF240686) UK (AJ 878,898), and France (Y14957). 32 of the 144 vvIBDV strains (representative sequences: MH137946, MH137947, MH137948, MH137949, MH137950, MH137951, MH137952) were similar but slightly different from each other. They were closely related to the vvIBDV strain (EF043078) previously identified in Turkey as well as strains reported from China (KT884486) (Figure 2). 33 of the vvIBDV strains (representative sequence: MH137953) was found to be closely related to the vvIBDV strains reported from India (KJ621061), Iraq (MF142535) as well as MB strain (EF544172). It is most likely that these strains might be vaccine strain since this vaccine used in the field where the samples were collected. Because of the genetic identity among most of the considered vvIBDV strains, only 12 sequences, representative of the 3 detected clusters (Figure 2), were submitted to Genbank
Figure 2.
Neighbour-joining phylogentic tree based on partial IBDV VP2 sequences. Strain classification has been performed using the reference sequences described in Michel and Jackwood, (2017). Circular dots indicate strains detected in this study. Bootstrap supports reported near to the corresponding tree node.
In addition, 1380 (90.5%) classical strains were detected. These strains were similar to classical strains reported from other countries as well as to vaccine strains. These strains formed 4 different clusters, classified in the Genogroup 1 according to Michel and Jackwood, (2017). Amongst classical strains, 1317 strains (95%) (representative sequence: MH137954) were similar to IBDV 2512 (AF251298, DQ355819, EU544157). 11 strains (representative sequence: MH137955) to Faragher 52/70 strain (HG974565), 40 strains (representative sequence: MH137957) to 228E (AF457104, AJ586966) and 12 strains (representative sequence: MH137956) to Lukert strain (AY918948, AY095230) (Figure 2). Because of the genetic identity among most of the considered classical IBDV strains, only 4 sequences, representative of the four detected clusters (Figure 2), were submitted to Genbank.
Recombination and Evolutionary Analyses
The recombination analysis did not detect occurrence of any recombination event in the considered region among the Turkish strains. However, presence of recombination breakpoints affecting other regions cannot be excluded. The phylogenetic tree performed on a broad sequence dataset demonstrated grouping of Turkish strains in 3 different clusters (Supplementary material Figure S1). The first one (Cluster 1), likely introduced in Turkey about 2010 based on the time to most recent ancestor estimation, includes sequences collected also from Iraq and Kuwait (B3110/214, B607/102, B308/25, B308/14, B228/42, B208/74 and B159/94), the second (Cluster 2)(sequence B1404/5), introduced approximately in 2010, is closely related to Indian strains while the last one (Cluster 3) (1408/22, 1408/44, 1408/23 and 1408/295) formed a distinct Turkish-only cluster potentially originating in 2014. The evolutionary rate estimation on branches/clades including Turkish strain mirrored the expected one for RNA viruses (i.e., mean 4,53*10−3; Range 2,2*10−3; -6,2*10−3; ) and no significant differences were found among different considered branches (Supplementary material Figure S1)
Isolation of 3 Different Clusters of vvIBDV Strains and Pathological Changes in IBDV Inoculated Embryonated Eggs
Harvests from the embryonated eggs were found to be positive for 552 bp product analysed by RT-PCR at first and/or second passages. When these products were sequenced, they were similar to strains which form 3 different clusters as explained in the section “Phylogenetic analysis of the partial IBDV VP2 gene sequences and vaccine monitoring”.
After primary inoculation, no death of chicken embryo was observed in all inoculated embryonated eggs. However, dwarfed embryos with noticeable congestion, haemorrhages, and edema were observed at day 5 after inoculation. Similarly, chorio-allantoic membranes were also edematous, congested, and hemorrhagic. These findings were not seen in control eggs.
DISCUSSION
Virulent IBDV strains have been detected in different countries with an economic impact on poultry production worldwide (Dolz et al., 2005; Jackwood and Sommer-Wagner, 2007; Alfonso-Morales et al., 2013). Poor protection due to a low titer of maternally derived antibodies (MAbs) is of a concern in controlling IBDV in the field as well as poor sanitation and hygiene (Jackwood et al., 2008). The vaccines used and vaccination strategy applied are also important factor in prevention and control of IBDV infection. In order to apply good preventive and control strategies, field IBDV strains as well as vaccine strains need to be monitored. There is no comprehensive study on current IBDV strains circulating in broilers at present in Turkey. Therefore, this study targeted monitoring of circulating virulent and vaccine IBDV strains of in broilers in this country. The current study was designed to determine VP2 gene variation and corresponding lesions in IBDV infected chickens in Turkey to understand phylogeographic dynamics of IBDV, viral population expansion, and IBDV evolution in Turkey.
The lesions associated with IBDVs’ in Turkey were similar to those reported previously (Khenenou et al., 2017). The predominant necropsy and histopathologic findings included atrophy, enlargement, edema, hemorrhages, and congestions in the bursa of Fabricius and petechial and stripe-like hemorrhages in the pectoral and leg muscles, lympho-mononuclear cell infiltration of interfollicular areas, vacuolizations, and necrosis, which are consistent with previous findings reported from the field studies (Khenenou et al., 2017).
Depending on geographic origin, previous reports indicated that phylogenetic analyses of the hypervariable region or complete VP2 gene sequences of vvIBDV strains clustered in one major monophyletic lineage (Cortey et al., 2012; Silva et al., 2013). However, recent reports from People's Republic of China on vvIBDV showed divergent VP2 phylogenetic clusters. Despite similarity in antigenic characteristics with previous isolates of vvIBDV, the recent isolates of vvIBDVs have mutations in both segments and increased virulence compared to the European vvIBDV (Li et al., 2015; Alkie and Rautenschlein, 2016). Similar findings have been reported in the USA since numerous VP2 gene sequences of IBDV field isolates were different from any known IBDV VP2 gene sequence (Durairaj et al., 2011). All of these reports indicate that evolution of the VP2 gene is continuous and diverse vvIBDVs are circulating in the fields posing a risk to poultry health. In addition, it is important to emphasize that the existence of a worldwide-spread genetic lineage of IBDVs designated as distinct IBDVs has been described, which cause only immunosuppression (Hernandez et al, 2011; Alkie and Rautenschlein, 2016).
After the detection of vvIBDV strains in Europe, these viruses have been reported in many countries. In Europe and in the USA, various prevalence of virulent strains of IBDV has been reported. Many studies reported existence of vvIBDV strains in different geographic areas. In a study performed by Jackwood and Sommer-Wagner [3], phylogenetic relationship of 113 IBDVs from 18 countries on 4 continents was investigated by sequencing hypervariable region of the VP2 gene. Results indicated that 2 viruses from South Africa were genetically similar to USA variant viruses. Of the 113 viruses sequenced, 71 had the amino acid alanine at the position 222 and 67 of these vvIBDV suspects also had amino acids I242, I256, I294, and S299, which are highly conserved among vvIBDV strains. In the USA, an outbreak of IBD in 2 California layer flocks have occurred and two IBD viruses designated rA and rB were isolated. Sequence analysis has shown that their amino acid sequences in the hypervariable region of the VP2 gene were similar to the vvIBDV strains UK 661, OKYM and Harbin (Jackwood et al., 2009). In a study performed in Europe, phylogenetic analysis of the hypervariable region of the VP2 gene demonstrated that all eight viruses were more similar to USA variant viruses (Jackwood et al., 2006). In Italy, characterization of IBDV strains detected between 2013 and 2014 showed that the majority of the genotyped strains (28 out of 41) showed the highest similarity (>99%) to a peculiar Italian strain named ITA strains. Four very virulent strains (DV86) and one classical strain (HPR2), together with eight vaccine strains, were also detected (Lupini et al., 2016). In Slovenia, phylogenetic analyses of VP2 gene has indicated that recent vvIBDV isolates were closely related to those from outbreaks reported in the 1990s (Rojs et al., 2008). In France, the phylogenetic analyses of the (IBDV) segments A and B of 50 natural or vaccine IBDV strains that were isolated or produced between 1972 and 2002 in 17 countries from four continents, with phenotypes ranging from attenuated to very virulent have shown that 26% of these strains exhibited a significantly different phylogenetic relationship depending on which segment was analysed (Le Nouen et al., 2006).
In a study performed in Spain, sequences of the hypervariable region of the VP2 gene from IBDV strains isolated from diverse geographic locations were obtained from the GenBank database as well as the results of Cuban sequencing study (Alfonso-Morales et al., 2013). The phylogeographic association-trait analysis indicated that viruses analysed from individual countries tend to cluster in the same phylogeny. However, spatial analysis showed that strains having sequences that were related to increased virulence of IBDV appeared in Iran in 1981 and spread to Western Europe (Belgium) in 1987, Africa (Egypt) around 1990, East Asia (China and Japan) in 1993, the Caribbean Region (Cuba) by 1995, and South America (Brazil) around 2000 (Alfonso-Morales et al., 2013).
vvIBD Vs were also reported in neighboring countries to Turkey. Phylogenetic analyses of the hypervariable region of the VP2 gene of IBDV in 10 pooled bursa Fabricius samples collected from broiler farms in Iraq have demonstrated that 5 of these viruses were vvIBDV (Amin and Jackwood, 2014). In Iran, a total of 10 vvIBDVs were analysed for the diversity of the VP2 gene. Amongst these viruses, 97.6 to 100% similarities were found indicating the presence of a common ancestor virus circulating in Iran. However, they were partially different from previous Iranian and neighboring countries' isolates (96.2 to 97.3% similarity to Shiraz isolate and 95.7 to 96.7% to Iraq and Turkey isolates) (Norouzian et al., 2017). In a previous study performed in Turkey, classical and variant IBDVs were found by RT-PCR-RFLP method (Ture et al., 1998). In another study in Turkey, 280 bursa of Fabricius samples were analysed for IBDV by using RFLP method. The results indicated existence of field isolates with new molecular patterns different than those previously found, which may be specific to Turkey (Sareyyüpoğlu and Akan, 2006). In this study, 1548 (83.5%) out of 1855 of bursa of Fabricius samples were IBDV positive. Among 1525 sequenced samples, 1380 (90.5%) of them were found to be classical strains. These strains were similar to strains reported from other countries as well as to vaccine strains. These strains formed 4 different clusters. Among the 1380 classical strains, 1317 were similar to IBDV 2512, 11 to Faragher 52/70, 40 to 228 E, and 12 to Lukert strain. Out of 1525 RT-PCR positive samples, 144 (9.5%) of them were found to be similar to vvIBDV VP2 gene reported to GenBank previously.
In Turkey, chickens are routinely vaccinated with various IBDV vaccines, representing heterogeneous genotypes, including recent introduction of virulent (intermediate plus) vaccine strains to control vvIBDV in poultry. The presence of these recently introduced vaccine strains and other used vaccine strains were identified in samples taken from poultry flocks as part of this study. Overall, these results demonstrate existence of different genotypes of IBDV affecting poultry, and in Turkey in particular, it suggests the co-persistence of different IBDV strains in poultry flocks, which could favor the emergence of new IBDV variants and escape mutants.
Turkey has a large and increasing poultry production industry and serves as a bridge between Europe and Asia with eight neighboring poultry-producing countries. The vvIBDV strains detected in this study were found to belong to three independent clusters, which suggest the occurrence of multiple introduction events. Particularly, 2 out of 3 clusters comprised sequences from India, Iraq, and Kuwait. The present scenario suggests both a widespread vvIBDV circulation of some IBDV strains in the Middle East region and the presence of long distance importation events. However, the limited sequence availability could hide a more complex spreading network among the considered countries. Remarkably, relevant within-group variability was evidenced in Cluster 1 and Cluster 3, suggesting a prolonged circulation of imported strains in Turkey and independent evolution. Especially, a cluster of vvIBDVs was different compared to all previously reported strains from other countries, suggesting the presence of distinctive IBDV Turkish variants.
Regardless of the considered cluster, the vvIBDV strains identified in this study displayed an evolutionary rate within the range expected for RNA viruses. Further studies will be required to understand if the observed evolution is attributable to vaccine-induced selection or to a stochastic genetic drift. On the other hand, no recombination event was observed, suggesting that this mechanism did not play a relevant role in the genesis of the Turkish vvIBDV genetic variability, at least in the considered region. However, presence of recombination breakpoints affecting other regions cannot be excluded.
Presence and spread of the vvIBDV strains in broiler farms in Turkey might be associated with improper vaccination strategy, biosecurity, trade between Middle East countries, backyard poultry farming which recently increased in Turkey and wildlife birds. In Finland, amongst 51 backyard chicken farms, antibodies to IBDV were detected in 20% of non-vaccinated farms (Pohjola et al., 2017). This point should also be investigated in Turkish backyard poultry.
Vaccines for IBDV should induce a good humoral and cellular immune response in order to protect against IBD. In Turkey, IBDV vaccination mainly follows the recommendations of vaccine suppliers who usually recommend drinking water vaccination between 12 to 16 d of age. As the breeder gets older, the vaccination is performed earlier. Measurement of MAbs in day old broilers to determine the proper vaccination time is not routinely performed in Turkey. This may cause vaccination failure in vaccinated flocks as has been observed in this study. One of the main reasons for IBDV vaccination failure is most likely that chicks with heterogeneous MAbs antibody levels are vaccinated with the above mentioned consequences. Another reason for IBDV vaccination failure is the quality of vaccine application. As reported in this study, presence of vvIBDV in the IBDV vaccinated flocks can be attributed to the vaccination failures due to bad quality of vaccine applications and improper vaccination regimes. Therefore, in order to overcome vaccination failure, new strategies like “day old chick vaccination” are becoming more popular to provide a better protection.
CONCLUSIONS
vvIBDVs’ similar to those circulating in various countries in the Middle East were identified in chickens from Turkish broiler flocks. Pathological changes were detected in the bursa of Fabricius of vvIBDV infected chickens. The results of phylogenetic analysis indicate that very virulent IBDV circulating in Turkey is undergoing evolution. Care should be taken for quality of vaccine application and proper vaccination time. These points need to be taken into account in planning adequate control strategies.
SUPPLEMENTARY DATA
Supplementary figure S1. Maximum likelihood phylogenetic tree based on non recombinant strains. Nodes displaying a bootstrap support higher that 70% are represented as black circles.It is possible to zoom in to appreciate further details and tip labels. Turkish strains have been colored in red
ACKNOWLEDGMENTS
We would like to thank to the University of Istanbul (Teknokent NO: BSTB 049628) for funding this study.
FUNDING
University of Istanbul (Teknokent NO: BSTB 04,9628).
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
REFERENCES
- Alfonso-Morales A., Martínez-Pérez O., Dolz R., Valle R., Perera C. L., Bertran K., Frías M. T., Majó N., Ganges L., Pérez L. J.. 2013. Spatiotemporal phylogenetic analysis and molecular characterisation of infectious bursal disease viruses based on the VP2 hyper-variable region. PLoS One 8:e65999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkie T. N., Rautenschlein S.. 2016. Infectious bursal disease virus in poultry: current status and future prospects. Vet. Med. 7:9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin O. G., Jackwood D. J.. 2014. Identification and molecular analysis of infectious bursal disease in broiler farms in the Kurdistan Regional Government of Iraq. Trop. Anim. Health Prod. 46:1297–1301. [DOI] [PubMed] [Google Scholar]
- Anisimova M., Gascuel O.. 2006. Approximate likelihood-ratio test for branches: A fast, accurate, and powerful alternative. Syst. Biol. 55:539–552. [DOI] [PubMed] [Google Scholar]
- Aricibasi M., Jung A., Heller E. D., Rautenschlein S.. 2010. Differences in genetic background influence the induction of innate and acquired immune responses in chickens depending on the virulence of the infecting infectious bursal disease virus (IBDV) strain. Vet. Immunol. Immunopathol. 135:79–92. [DOI] [PubMed] [Google Scholar]
- Chettle N., Stuart J. C., Wyeth P. J.. 1989. Outbreak of virulent infectious bursal disease in East Anglia. Vet. Rec. 125:271–272. [DOI] [PubMed] [Google Scholar]
- Cortey M., Bertran K., Toskano J., Majó N., Dolz R.. 2012. Phylogeographic distribution of very virulent infectious bursal disease virus isolates in the Iberian Peninsula. Avian Pathol. 41:277–284. [DOI] [PubMed] [Google Scholar]
- Cosgrove A. S. 1962. An apparently new disease of chickens: Avian nephrosis. Avian Dis. 6:385–389. [Google Scholar]
- Coulibaly F., Chevalier C., Gutsche I., Pous J., Navaza J., Bressanelli S., Delmas B., Rey F. A.. 2005. The birnavirus crystal structure reveals structural relationships among icosahedral viruses. Cell 120:761–772. [DOI] [PubMed] [Google Scholar]
- Darriba D., Taboada G. L., Doallo R., Posada D.. 2012. jModelTest 2: More models, new heuristics and parallel computing. Nat Methods 9:772–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolz R., Majó N., Ordóñez G., Porta R.. 2005. Viral genotyping of infectious bursal disease viruses isolated from the 2002 acute outbreak in Spain and comparison with previous isolates. Avian Dis. 49:332–339. [DOI] [PubMed] [Google Scholar]
- Durairaj V., Sellers H. S., Linnemann E. G., Icard A. H., Mundt E.. 2011. Investigation of the antigenic evolution of field isolates using the reverse genetics system of infectious bursal disease virus (IBDV). Arch. Virol. 156:1717–1728. [DOI] [PubMed] [Google Scholar]
- Faragher J. T., Allan W. H., Wyeth P. J.. 1974. Immunosuppressive effect of infectious bursal agent on vaccination against Newcastle disease. Vet. Rec. 95:385–388. [DOI] [PubMed] [Google Scholar]
- Guindon S., Dufayard J.-F., Lefort V., Anisimova M., Hordijk W., Gascuel O.. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59:307–321. [DOI] [PubMed] [Google Scholar]
- Hernández M., Tomás G., Hernández D., Villegas P., Banda A., Maya L., Panzera Y., Pérez R.. 2011. Novel multiplex RT-PCR/RFLP diagnostic test to differentiate low- from high-pathogenic strains and to detect reassortant infectious bursal disease virus. Avian Dis. 55:368–374. [DOI] [PubMed] [Google Scholar]
- Hitchner S. B. 1970. Infectivity of infectious bursal disease virus for embryonating eggs. Poult. Sci. 49:511–516. [DOI] [PubMed] [Google Scholar]
- Hon C. C., Lam T.-Y., Drummond A., Rambaut A., Lee Y. F., Yip C.-W., Zeng F., Lam P.-Y., Ng P. T. W., Leung F. C. C.. 2006. Phylogenetic analysis reveals a correlation between the expansion of very virulent infectious bursal disease virus and reassortment of its genome segment B. J. Virol. 80:8503–8509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackwood D. J., Sommer-Wagner S.. 2007. Genetic characteristics of infectious bursal disease viruses from four continents. Virology 365:369–375. [DOI] [PubMed] [Google Scholar]
- Jackwood D. J., Stoute S. T.. 2013. Molecular evidence for a geographically restricted population of infectious bursal disease viruses. Avian Dis. 57:57–64. [DOI] [PubMed] [Google Scholar]
- Jackwood D. J., Sreedevi B., LeFever L. J., Sommer-Wagner S. E.. 2008. Studies on naturally occurring infectious bursal disease viruses suggest that a single amino acid substitution at position 253 in VP2 increases pathogenicity. Virology 377:110–116. [DOI] [PubMed] [Google Scholar]
- Jackwood D. J., Cookson K. C., Sommer-Wagner S. E., Le Galludec H., de Wit J. J.. 2006. Molecular characteristics of infectious bursal disease viruses from asymptomatic broiler flocks in Europe. Avian Dis. 50:532–536. [DOI] [PubMed] [Google Scholar]
- Jackwood D. J., Sommer-Wagner S. E., Stoute A. S., Woolcock P. R., Crossley B. M., Hietala S. K., Charlton B. R.. 2009. Characteristics of a very virulent infectious bursal disease virus from California. Avian Dis. 53:592–600. [DOI] [PubMed] [Google Scholar]
- Katoh K., Standley D. M.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khenenou T., Bougherara M., Melizi M., Lamraoui R.. 2017. Histomorphological study of the bursae of fabricius of broiler chickens during Gumboro disease in algeria area. Global Vet 18:132–136. [Google Scholar]
- Le Nouën C., Rivallan G., Toquin D., Darlu P., Morin Y., Beven V., de Boisseson C., Cazaban C., Comte S., Gardin Y., Eterradossi N.. 2006. Very virulent infectious bursal disease virus: reduced pathogenicity in a rare natural segment-B-reassorted isolate. J. Gen. Virol. 87:209–216. [DOI] [PubMed] [Google Scholar]
- Li Z., Qi X., Ren X., Cui L., Wang X., Zhu P.. 2015. Molecular characteristics and evolutionary analysis of a very virulent infectious bursal disease virus. Sci. China Life Sci. 58:731–738. [DOI] [PubMed] [Google Scholar]
- Lukert P. D, Saif Y. M.. 2003. Diseases of Poultry, 11th ed. by Y. M. Saif, J. R. Glisson, A. M. Fadly, L. R. McDougald, and D. E. Swayne. Iowa State Press. Ames. pp. 161–180. [Google Scholar]
- Lupini C., Giovanardi D., Pesente P., Bonci M., Felice V., Rossi G., Morandini E., Cecchinato M., Catelli E.. 2016. A molecular epidemiology study based on VP2 gene sequences reveals that a new genotype of infectious bursal disease virus is dominantly prevalent in Italy. Avian Pathol. 45:458–464. [DOI] [PubMed] [Google Scholar]
- Martin D. P., Murrell B., Golden M., Khoosal A., Muhire B.. 2015. RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evol. 1:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazariegos L. A., Lukert P. D., Brown J.. 1990. Pathogenicity and immunosuppressive properties of infectious bursal disease ``Intermediate" strains. Avian Dis. 34:203–208. [PubMed] [Google Scholar]
- Michel L. O., Jackwood D. J.. 2017. Classification of infectious bursal disease virus into genogroups. Arch. Virol. 162:3661–3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno A., Franzo G., Massi P., Tosi G., Blanco A., Antilles N., Biarnes M., Majó N., Nofrarías M., Dolz R., Lelli D., Sozzi E., Lavazza A., Cecchinato M.. 2017. A novel variant of the infectious bronchitis virus resulting from recombination events in Italy and Spain. Avian Pathol. 46:28–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norouzian H., Farjanikish G., Hosseini H.. 2017. Genetic and pathologic characteristics of infectious bursal disease viruses isolated from broiler chickens in Iran during 2014–2015. av. 61:191–196. [DOI] [PubMed] [Google Scholar]
- OIE, Terrestrial Manual. 2016. Chapter 2.3.12. http://www.oie.int/fileadmin/Home/eng/Health_standards/tahm/2.03.12_IBD. [Google Scholar]
- Pohjola L., Tammiranta N., Ek-Kommonen C., Soveri T., Hänninen M. L.. 2017. A survey for selected avian viral pathogens in backyard chicken farms in Finland. Avian Pathol. 46:166–172. [DOI] [PubMed] [Google Scholar]
- Rojs O. Z., Krapez U., Slavec B., Mankoc S., Juriric-Cizerl R., Barlic-Maganja D.. 2008. Molecular characterisation of infectious bursal disease viruses isolated in recent acute outbreaks in Solvenia. Acta Vet. Hung. 56:255–264. [DOI] [PubMed] [Google Scholar]
- Rosales A. G., Villegas P., Lukert P. D., Fletcher O. J., Mohamed M. A., Brown J.. 1989. Isolation, identification, and pathogenicity of two field strains of infectious bursal disease virus. Avian Dis. 33:35–41. [PubMed] [Google Scholar]
- Rudd M. F., Heine H. G., Sapats S. I., Parede L., Ignjatovic J.. 2002. Characterisation of an Indonesian very virulent strain of infectious bursal disease virus. Arch. Virol. 147:1303–1322. [DOI] [PubMed] [Google Scholar]
- Sareyyüpoğlu B., Akan M.. 2006. Restriction fragment length polymorphism typing of infectious bursal disease virus field strains in Turkey. Avian Dis. 50:545–549. [DOI] [PubMed] [Google Scholar]
- Silva F. M. F., Vidigal P. M. P., Myrrha L. W., Fietto J. L. R., Silva A., Almeida M. R.. 2013. Tracking the molecular epidemiology of Brazilian Infectious bursal disease virus (IBDV) isolates. Infect. Genet. Evol. 13:18–26. [DOI] [PubMed] [Google Scholar]
- Stuart J. C. 1989. Acute infectious bursal disease in poultry. Vet. Rec. 125:281–281. [DOI] [PubMed] [Google Scholar]
- Tamura K., Nei M.. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10:512–526. [DOI] [PubMed] [Google Scholar]
- Toroghi R., Kataria J. M., Verma K. C., Kataria R. S., Tiwari A. K.. 2001. Amino acid changes in the variable region of VP2 in three infectious bursal disease viruses with different virulence, originating from a common ancestor. Avian Pathol. 30:667–673. [DOI] [PubMed] [Google Scholar]
- Ture O., Saif Y. M., Jackwood D. J.. 1998. Restriction fragment length polymorphism analysis of highly virulent strains of infectious bursal disease viruses from Holland, Turkey, and Taiwan. Avian Dis. 42:470–479. [PubMed] [Google Scholar]
- Yamaguchi T., Ogawa M., Miyoshi M., Inoshima Y., Fukushi H., Hirai K.. 1997. Sequence and phylogenetic analyses of highly virulent infectious bursal disease virus. Arch. Virol. 142:1441–1458. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.