Summary
The rapid growth in data sharing presents new opportunities across the spectrum of biomedical research. Global efforts are underway to develop practical guidance for implementation of data sharing and open data resources. These include the recent recommendation of ‘FAIR Data Principles’, which assert that if data is to have broad scientific value, then digital representations of that data should be Findable, Accessible, Interoperable and Reusable (FAIR)1. The spinal cord injury (SCI) research field has a long history of collaborative initiatives tat include sharing of preclinical research models and outcome measures. In addition, new tools and resources are being developed by the SCI research community to enhance opportunities for data sharing and access. With this in mind, the National Institute of Neurological Disorders and Stroke (NINDS) at the National Institutes of Health (NIH) hosted a workshop on October 5–6, 2016 in Bethesda, MD, in collaboration with the Open Data Commons for Spinal Cord Injury (ODC-SCI) titled “Preclinical SCI Data: Creating a FAIR Share Community”. Workshop invitees were nominated by the workshop steering committee (co-chairs: ARF and VPL; members: AC, KDA, MSB, KF, LBJ, PGP, JMS), to bring together junior and senior level experts including preclinical and basic SCI researchers from academia and industry, data science and bioinformatics experts, investigators with expertise in other neurological disease fields, clinical researchers, members of the SCI community, and program staff representing federal and private funding agencies. The workshop and ODC-SCI efforts were sponsored by the International Spinal Research Trust (ISRT), the Rick Hansen Institute, Wings for Life, the Craig H. Neilsen Foundation and NINDS. The number of attendees was limited to ensure active participation and feedback in small groups. The goals were to examine the current landscape for data sharing in SCI research and provide a path to its future. Below are highlights from the workshop, including perspectives on the value of data sharing in SCI research, workshop participant perspectives and concerns, descriptions of existing resources and actionable directions for further engaging the SCI research community in a model that may be applicable to many other areas of neuroscience. This manuscript is intended to share these initial findings with the broader research community, and to provide talking points for continued feedback from the SCI field, as it continues to move forward in the age of data sharing.
Keywords: FAIR data principles, reproducibility, neuroscience, informatics, workshop proceedings, Open Data Commons
The culture of data sharing
Neuroscientists, including SCI researchers, have a long history of sharing data, traditionally through publications. The Institute for Scientific Information (ISI) Science Citation Index has over 46,000 publications indexed under ‘spinal cord injury’ from 2000–2016, many of which include detailed methods, results, and supplementary data that are used by other investigators in planning experiments and interpreting their own findings. Data shared in publications, however, is usually carefully selected, and represent only a fraction of the data generated by preclinical SCI researchers. Data that do not fit the ‘story’ of a discovery are often left unpublished, and most primary preclinical research data are accessible and interpretable only by individuals in a shared laboratory or collaborative group. These ‘dark data’, never made available in repositories or publications, are estimated to make up 85% of all data collected (Ferguson et al., 2014). The inability to access dark data impedes efforts to promote transparency, replication and independent validation of promising findings (Ferguson et al., 2014). Moreover, for the 15% of data that are reported in the scientific literature, inconsistent study design and statistical analysis contribute to complications and bias in interpretations (Burke et al., 2013; Watzlawick et al., 2014).
Informal data sharing occurs at meetings and symposia, where preliminary findings are presented and discussed with colleagues. At the 2016 Society for Neuroscience (SfN) meeting, for example, 2,256 presentations had the words ‘spinal cord injury’ associated with them. Only a subset of these posters and presentations will end up as publications. The informal interchange of ideas, technical approaches, and importantly, knowledge about what experiments are being done in other labs, is therefore highly valuable to the community. However, even at conferences, presenters are often careful to provide only select information to their peers. Many of us remember being admonished as students for enthusiastic sharing of not-yet-ready lab data at conferences and meetings. The free exchange of data and ideas versus ‘saving’ data for curated, peer-reviewed publications in high impact journals are competing interests in the current research landscape, in part responsible for a cultural bias against open data sharing.
In the current era of accountability and transparency, each community must consider how best to share data and seize opportunities afforded by making experimental data more widely available. The culture of sharing pre-publication findings in physics and genomics and the rapid and fruitful evolution of approaches for managing and analyzing big data in scientific research have driven discoveries in these fields. Sharing data necessitates that others can examine entire datasets from which interpretations were made. This can be seen as a challenge to the integrity of the traditional process of neuroscience research, yet it is the most transparent and useful approach to finding the ‘truth’. Recently, much attention has been paid to open data sharing as a means to increase rigor and reproducibility in neuroscience research (Ferguson et al., 2014). Effective data sharing practices can be leveraged to improve reproducibility by providing platforms for depositing published and unpublished data, enabling better meta-analyses of research studies, reducing redundancy and waste, and providing large scale resources for analytic approaches to generate new discoveries.
As a consequence, the entire biomedical research enterprise is experiencing a cultural shift in approaches to data collection and data sharing. This shift has been particularly evident in the preclinical research spectrum. In 2011, a meeting of international leaders in data science known as “The Future of Research Communications and e-Scholarship”, or FORCE 11, took on the task of creating standard recommendations for data sharing. One product of this effort was the development of “FAIR Data Principles”, which describe digital objects that hold value as those that are Findable (with sufficient explicit meata), Accessible (open and available to other researchers), Interoperable (using standard definitions and common data elements (CDEs)), and Reusable (meeting community standards, and sufficiently documented). The Office of Data Science at NIH has endorsed the FAIR Data Principles, and plans to incorporate these standards in future data sharing recommendations and programs (Wilkinson et al., 2016).
The SCI research community is well-positioned to embark on fruitful data sharing practices and lead by example. Clinical SCI researchers have joined with the International Spinal Cord Society (ISCoS), the American Spinal Injury Association (ASIA) and NINDS to develop standard definitions, case report forms, and CDEs for collection and reporting of clinical research data (Biering-Sørensen et al., 2015; Charlifue et al., 2016). In addition, basic and preclinical SCI researchers have embarked on initiatives and developed resources for data sharing over the past three decades. In the 1990s, NINDS funded a Multicenter Animal Spinal Cord Injury Study (MASCIS) as a consortium to facilitate validation of promising preclinical leads. This led to development of standard models and data collection procedures across several laboratories (Basso et al., 1996, 1995; Young, 2002).
From 2003–2013, NINDS executed contract agreements as Facilities of Research Excellence in Spinal Cord Injury (FORE-SCI), which led to additional outcome measures in mice and rats (Aguilar and Steward, 2010; Anderson et al., 2009), established a research training course for investigators new to the field, and completed 18 controlled replication studies in order to identify leads for translation (Steward et al., 2012). The FORE-SCI investment enriched the field with a highly-trained workforce, highlighted the challenges in replication attempts, and contributed to a larger effort across the NIH to enhance transparency, rigor and data quality for all preclinical research (Landis et al., 2012).
Since 2013, four projects have added data resources and tools for the SCI preclinical research community: (1) the VISION-SCI data repository with source data contributed by multiple research laboratories (Nielson et al., 2015a, 2014), (2) a consensus guideline of minimal reporting expectations for preclinical SCI research (MIASCI) (Lemmon et al., 2014), (3) a knowledge base and ontology for integration of SCI research data that is compatible with domain wide terminology standards (RegenBase) (Callahan et al., 2016), and (4) a rapidly-developing open data commons for SCI research. Each of these efforts has been a product of wide collaboration with dozens of contributing SCI scientists and multiple authors and is described in more detail below.
Given the state of readiness of the SCI research community and the availability of these unique resources, NINDS hosted the FAIR Share Workshop to engage stakeholders in discussion of the new challenges and opportunities for data sharing (Figure 1). The goals of the workshop were to (1) bring together researchers and data science experts with policy/program staff, (2) get feedback from the community about perceived barriers and incentives for data sharing and reuse, (3) study the lessons learned and best practices from other preclinical research fields, (4) identify opportunities for expanding the data sharing community and (5) identify solutions and a path for moving SCI preclinical data sharing efforts forward.
Figure 1.
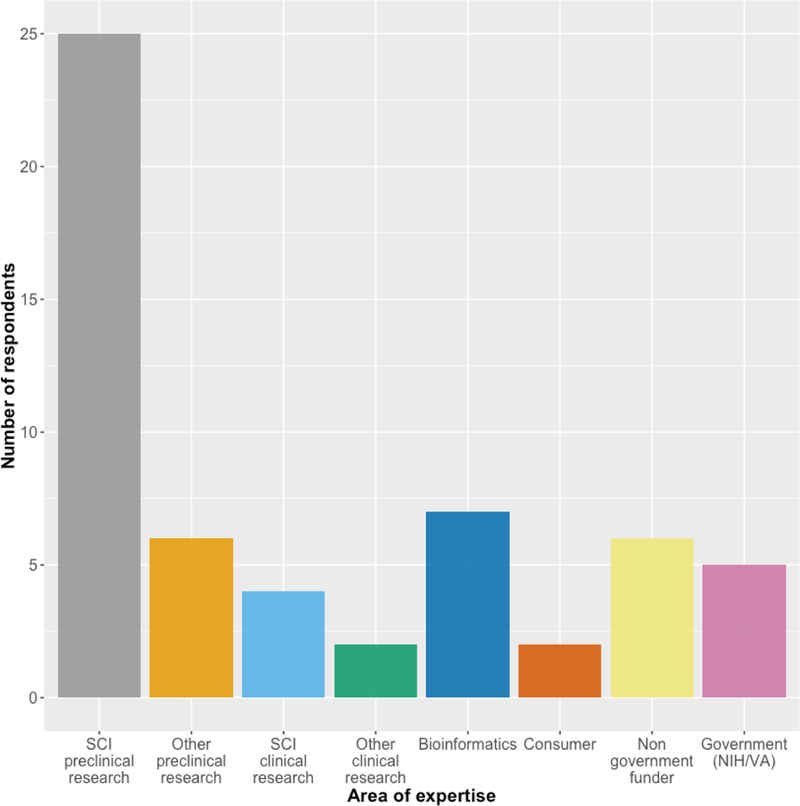
Workshop participant areas of expertise. Some participants indicated more than one.
Challenges and incentives
In theory, the rationale for public data sharing seems obvious. A large proportion of academic research is funded by taxpayer dollars, and most stakeholders would benefit from knowing whether specific experiments have already been tried and whether the experiments were successful or not. Regardless of whether the results of a study are positive or negative, the availability of that study’s data would increase research efficiency and reduce research costs by eliminating unnecessary repetition of experiments for which data are already available. However, among the participants at the FAIR Share Workshop, only 2 of 19 indicated that they currently share their data publicly with no access restrictions (Figure 2). Nearly half of respondents share their data upon request, which is a fairly open approach, but still restrictive in that a researcher has to initiate the data sharing process via direct communication with the lab generating the data. Why is it that so few scientists publicly share their data? The reality is that even though the concept of data sharing is appealing, the logistics of and cultural barriers to data sharing are daunting.
Figure 2.
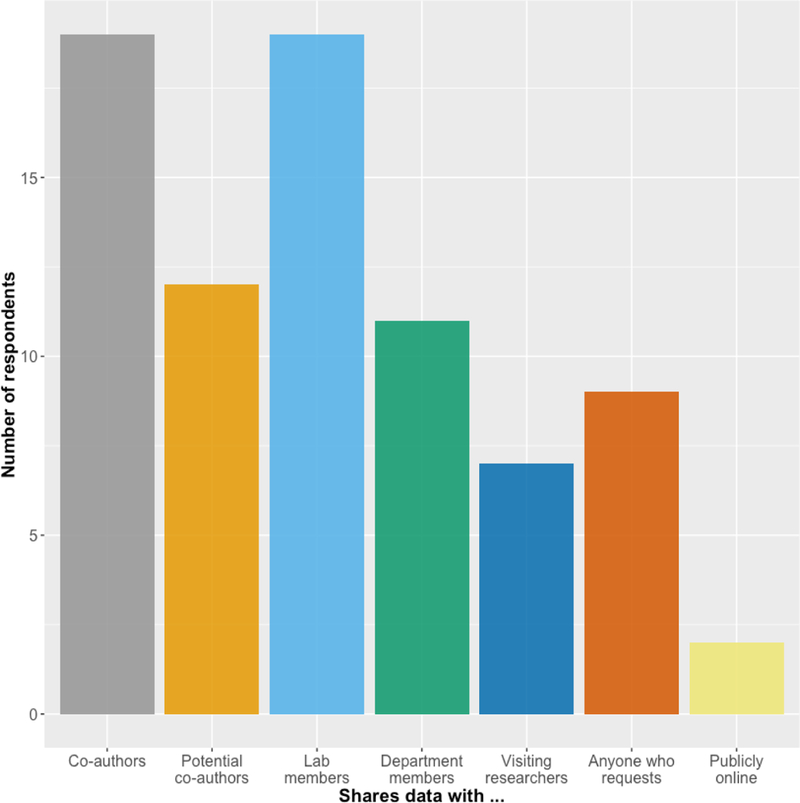
Workshop survey responses to the question “Who do you share experimental data generated by your lab with?”. All respondents share data with co-authors and lab members; fewer than half share their data with anyone who requests it, and only 2 of 19 respondents share any data publicly online.
Time and workflow conflicts were perceived by workshop participants as some of the primary obstacles to data sharing. Every day researchers are faced with time-intensive tasks including grant and manuscript writing, complying with administrative requirements from funding organizations and universities, graduate student and postdoctoral researcher training, teaching, and a wide range of service activities. Participants were concerned about the additional time required to collect, format, and prepare research data for sharing.
Consider how a laboratory researcher currently manages data. Trainees typically collect and present data in various formats (presentation slides, spreadsheets, hard copy printouts, lab notebooks etc.). Together over months to years, lab members and collaborators work to interpret, reanalyze, repeat, and finally publish some of those data in manuscripts. The mechanics of the process of collecting data for publication varies greatly among laboratories and even among researchers in the same laboratory. Workshop participants collect data using many different modalities, including notebooks, spreadsheets, electronic lab notebooks, as well as experiment-specific software and other formats (Figure 3). Moreover, different types of data (consider histological data versus cell culture data versus molecular data such as PCR readouts) are collected and stored in different ways.
Figure 3.
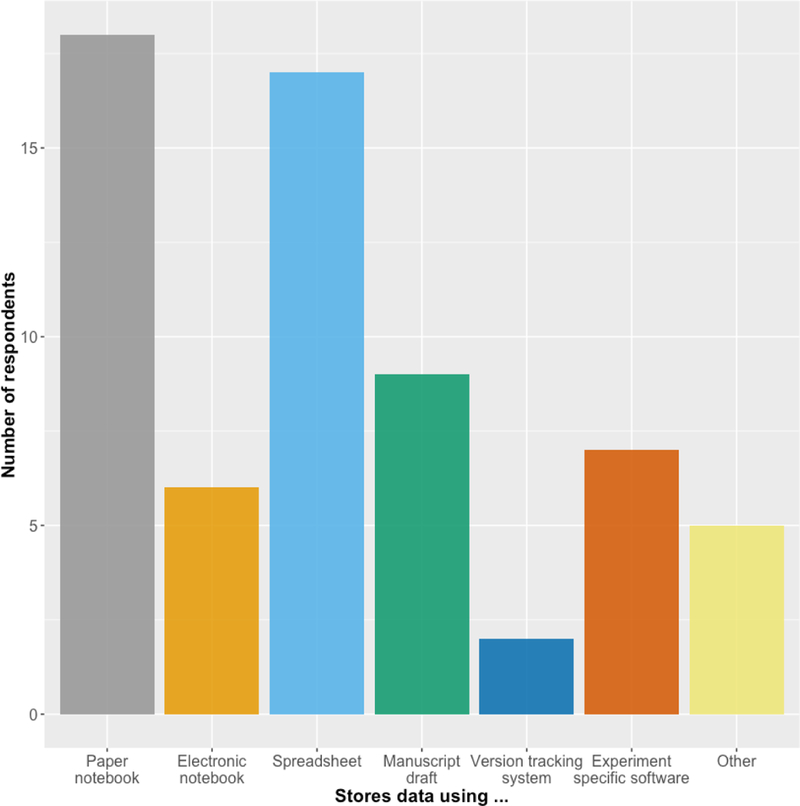
Workshop survey responses to the question “How do you store experimental data generated in your lab?”. Most respondents use paper notebooks and spreadsheets to store data; close to half use manuscript drafts, and few use electronic notebooks and version tracking systems.
Regardless of research field, challenges to data sharing broadly (Table 1) include developing community consensus around terms and definitions, identifying data stewardship policies and infrastructure, enabling researchers to participate in sharing and reuse of datasets, implementing practices that protect intellectual property and allow appropriate citation of data, and establishing models that are sustainable (Briggs, 2016; McKiernan et al., 2016; Steckler et al., 2015; Zinner et al., 2016). While efforts are underway to address these issues, specific solutions will differ widely across research domains. International data science experts and leaders at the NIH agree that best practices must be developed by individual research communities in order to be accepted and appropriate for their needs and the types of data generated.
Table 1.
Summary of challenges in data sharing discussed at the FAIR Share Workshop
| •Data collection and organization schemes vary across laboratories - a ‘one-size-fits-all’ approach to data collection and storage may not work for every laboratory |
| •Training laboratory members in best practices for data collection and storage requires additional time and expertise for lab managers and PIs |
| •Infrastructure for data storage and upload to repositories is expensive and not available in all labs |
| •Data repository security is essential to protecting researchers and research subjects |
| •Repositories must have metrics of data quality to ensure that data shared are accurate and sufficiently described - variety in data quality checks will exist across labs and study types |
| •Shared data must be attributable and citable |
Raw data collected and stored using different modalities are unlikely to be easily understood and used by others in the scientific community. In preclinical research, nearly every dataset is unique in the variables it captures and would likely require extensive annotation to be compatible with FAIR principles. Methods of collecting and organizing each lab’s data will likely require developing an infrastructure and standard operating procedure (SOP) that can be optimized for upload to a data server in order to make the data useful to others, or ‘Interoperable’. Some workshop participants also expressed concern that standardization of collection and archiving methods could be incompatible with creativity and optimal scientific training in their laboratories. For most investigators, open data sharing is therefore not likely to be an easy transition and the time and expense required to create the necessary infrastructure could be prohibitive. Most labs will not have the expertise or resources to do this, so it will be up to institutions to develop solutions to support their researchers. Additional issues of data security, quality control and intellectual property must also be addressed before many are willing to share data outside conventional formats. However, it seems clear that data and resource sharing initiatives are already in motion, with both valuable incentives and policy directives.
What are the incentives for data sharing? For the SCI research community, compliance with journal requirements, scientific discovery and compliance with funder requirements were the incentives ranked most important on average by 23 workshop participants (Figure 4). This suggests that the tools and platforms supporting data sharing must easily integrate into the paper submission and funding application processes used by neuroscientists. The perceived importance of the role of data sharing in scientific discovery means that data must be shared in a way that is compatible with analysis workflows, and that shared data must be accompanied by metadata that describes the experimental conditions under which the data were collected, and any data transformations that were applied.
Figure 4.
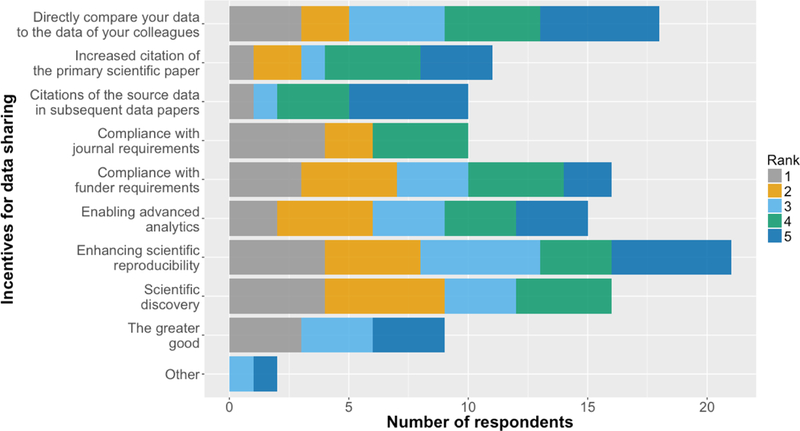
Workshop survey responses to the question “How do you rank possible incentives for data sharing?”. ‘Compliance with journal requirements’, ‘scientific discovery’ and ‘compliance with funder requirements’ were the incentives ranked most important on average. Citations were ranked least important on average.
Interestingly, ‘increased citation of the primary scientific paper’ and ‘citations of the source data’ were ranked least important on average. This may be evidence of the perceived lack of reward associated with the additional effort required to make one’s data FAIR. Indeed, evaluation criteria for scientists at academic institutions and the current criteria used when reviewing grant applications do not reward data sharing. This is critical since data sharing is likely to be time consuming, at least until standardized methods of collection, storage and sharing are streamlined for a given type of research. In other words, before FAIR data sharing practices become widely adopted, a cultural change is required for researchers, academic institutions and funding organizations.
Supporting data sharing and reuse
Informatics is the study of information, its structure and use (Stead, 1998). Neuroscience as a field is exploiting diverse technologies, including functional magnetic resonance imaging (fMRI), electrophysiology, RNA-sequencing, and optogenetics, that produce big data (Sejnowski et al., 2014). To make sense of this flood of data, especially across sub-disciplines, informatics has an important role to play alongside biology, statistics and information technology.
The life cycle of a dataset starts in pilot experiments, scrawled observations and gel photos taped and kept securely in lab notebooks (only recently in electronic form), where successes reside along with (usually) many failures - due to technical problems, suboptimal reagents, and just plain mistakes. The successes (some signed and dated by the experimenter in case they might lead to patentable intellectual property) are moved ‘up the food chain’ to the lab meeting and the principal investigator. When that scientist and their colleagues collect many findings, and combine them, it has moved into the next phase – some details about the original data may be lost, but it is part of a larger collection from which new information can be extracted in the form of trends and associations. Those data become the figures in conference posters, and, highly filtered, in grants and publications. Only at this point do peers and public see the data, or rather the interpretation of the data given by the authors, in collaboration with reviewers and editors whose job is to ensure transparency and validity of the results. The power and significance of these findings can be evaluated using statistical analyses, which are also usually described in a publication and shared with the scientific community, and this summary of data itself becomes a new data point for meta-analyses and further research.
Building on advances in information technology, informatics can facilitate data sharing at each point in this data life cycle by providing easy-to-use-interfaces for data upload to repositories, describing data provenance and quality control. On the data organization and analysis side, ontologies and data science methods can inform best practices for data sharing by providing actionable definitions of concepts and associated logic, and means to study their effects on interpretation and analysis. Informatics tools also have the potential to make data sharing easy and rewarding for both data donors and data (re)users. User-friendly interfaces for data upload, data download, data citation, author credit attribution, and analytics will further motivate data sharing. The ultimate goal of data sharing efforts is data reuse, allowing researchers and the public to link existing knowledge and new data together to make new discoveries.
Informatics research in itself will benefit from the availability of more data across all scientific fields. Informatics methods such as multidimensional analytics and machine learning are ‘data-hungry’, meaning that the more enriched datasets there are, the more potential they create for improving methods and for knowledge discovery. This is true in neuroscience as well – the public availability of more data, and the methods and infrastructure to analyze those data, will move the entire field forward.
For SCI and axon regeneration research, several efforts across multiple institutions are underway to enable data sharing and reuse. Beginning in 2004, Adam Ferguson and collaborators began curating archived data donated from different SCI preclinical research groups, with the goal of enabling re-analysis and data-driven analytical discovery (Ferguson et al., 2013, 2011, 2004). This data repository, now known as VISION-SCI, contains data from over 3000 animal subjects and approximately 2700 curated variables including the MASCIS preclinical trials from the 1990s, and donations from 13 laboratories (Nielson et al., 2014). Application of analytics and machine learning tools to these pooled data have contributed insights into outcome scaling, anatomical substrates of recovery, and acute critical care predictors of long term recovery (Ferguson et al., 2013, 2004; Friedli et al., 2015; Irvine et al., 2014; Nielson et al., 2015b). This provides proof-of-concept for the potential value of data sharing within SCI, and illustrates the willingness of the SCI research community to share data.
In 2014, Vance Lemmon and collaborators published the Minimum Information About a Spinal Cord Injury (MIASCI) reporting guideline, to capture the methodological details of SCI experiments using animal models. The MIASCI team has since developed a publicly available online tool for literature curation, MIASCI Online (http://regenbase.org/miasci-online/), that uses MIASCI as a backbone. MIASCI Online allows researchers to curate in vivo SCI experiments from the published literature or unpublished experiments, and produces a structured representation of experimental details (metadata) and a summary of experimental findings that can be shared. Examples of metadata include animal housing conditions and anaesthetics, experimental treatments like drugs or stem cell growth conditions. A challenge for scientists is that in vivo experiments are very complex and it is well known that seemingly innocuous details can have a significant impact on experimental outcomes. Documenting all the details about reagents, surgical practice and outcome measures is difficult but is best done at the time of the experiment, following a structured plan. Using a standard spreadsheet or an online tool like MIASCI Online can allow this documentation and reduce some of the pain.
In parallel to MIASCI and MIASCI Online, researchers at the University of Miami and Stanford University collaborated to develop RegenBase (http://regenbase.org), a knowledge base of SCI biology and experimental data (Callahan et al 2016). RegenBase integrates literature-sourced facts and experimental details from publications curated using MIASCI Online, raw assay data profiling the effect of tens of thousands of compounds on enzyme activity and cell growth, and gene expression data for more than 40,000 rat and mouse genes and gene probes. RegenBase consists of an ontology to capture knowledge about the biological entities studied in SCI research and the relationships between them and a triplestore (a database for storing Resource Description Framework statements called “triples”) to collect and publish linked data about those biological entities as collected during experiments. The knowledge base has been used to identify potential gene and protein targets for SCI drug therapies and to identify drugs that improve behavioral outcomes following SCI across studies. RegenBase uses standard languages for its ontology, triplestore, and for querying, and supports reasoning and inference based on formal semantics. MIASCI Online records can also be deposited into RegenBase, and current work focuses on automatically extracting statements from published literature and annotating them with ontologies to augment high quality statements generated by expert curators using MIASCI Online.
Common to each of these efforts are templates for structuring metadata describing experiments and a digital home for data captured during experiments. It is notable that the experience of VISION-SCI suggested that SCI researchers were willing to share their data in full if they were provided with templates. The VISION-SCI repository also had dedicated funding for data entry personnel to limit the burden on data donors, and several of the initial donors had sufficient resources to absorb the added burden of data sharing. VISION-SCI, MIASCI, RegenBase and prior efforts like MASCIS demonstrate the fundamental potential for data sharing in SCI and have set the stage for an expanded community repository and data-sharing platform currently being developed for the ODC-SCI (https://scicrunch.org/odc-sci). However, in the long term no data-sharing effort will succeed without well-developed standards to support assigning unique identifiers to data and datasets, registration across laboratories and detailed metadata to allow responsible data reuse. Such resources are essential for neuroscientists sharing research data with others, as are associated online repositories and software resources. Future work must focus on making these valuable resources interoperable through the use of standard knowledge representation languages and data capture formats, and accessible to researchers for data deposition, access and search.
Ideas for community data sharing can be gleaned from other fields where preclinical researchers are working to enhance transparency and reproducibility. Representatives from preclinical research communities in stroke, epilepsy, and traumatic brain injury (TBI) attended the FAIR Share Workshop to share their experiences to date. The stroke research community met in November of 2016 to address barriers to translation and are working on developing community standards specifically for translational research studies. The American Epilepsy Society (AES) and International League Against Epilepsy (ILAE) had a similar gathering in 2012 and published consensus papers with recommendations for use of standard data and outcomes (Galanopoulou et al., 2013). They have since produced case report forms for researchers to use for preclinical models of epilepsy. Researchers studying TBI published an initial set of CDEs for preclinical research (Smith et al., 2015) and are currently working to develop CDEs for specific outcome domains. The TBI CDE effort has included the NINDS and participants from the SCI and epilepsy research communities to maximize harmonization of these tools and resources where possible. This communication across research areas is exciting and can be harnessed to inform data sharing practices for the SCI research community and enhance the value of published datasets.
The NIH Data Science Office is also developing approaches for making publicly funded data readily available, while managing the prohibitive costs of creating and curating data archives. The recent growth of data generated across the NIH is astounding. In 2012, the data archived in the entire US Library of Congress amounted to 3 PB (1 petabyte=1015 bytes or 1 million gigabytes), while the total data from NIH-funded research is currently estimated at 650 PB. Maintaining existing data archives cost the NIH ~$1.2 billion from 2007–2014, and this cost is cited with the knowledge that only 12% of data described in published papers is available in recognized archives (Read et al., 2015). The NIH and other government agencies alone cannot conceivably support the storage and archiving of all funded research data. At the same time, NIH recognizes the numerous examples where data sharing has led to new discoveries and treatments for disease. A proposed solution is for NIH to support the development of an open platform that would enable accessible datasets and analytical tools to be maintained and housed with commercial cloud providers (Bourne et al., 2015). In this model, proposed as the NIH Data Commons (https://datascience.nih.gov/commons), investigators might receive credits to support the use of the shared resources. The NIH Commons is currently undergoing pilot testing at selected data centers in the NIH Big Data To Knowledge (BD2K; https://datascience.nih.gov/bd2k) network. If successful, this model may provide a feasible solution for enabling sustainability of preclinical SCI research data for use by the community and public.
The future of data sharing for SCI research
Today, SCI research is characterized by a low number of randomized controlled trials (RCTs) compared with other neurological diseases (Lammertse, 2013); this is evidence of little success in translating preclinical findings to clinical studies. Publication bias and errors in experimental design result in biased effect estimates for treatments investigated in animal model studies (Macleod et al., 2009; Watzlawick et al., 2014). Despite these challenges, and differences between human and experimental SCI models (Courtine et al., 2007) there have been several lines of evidence supporting the translational value of findings in preclinical animal models for understanding human neurological disorders and treatment (Dirnagl and Endres, 2014). Data sharing initiatives therefore have the potential to bolster future translational efforts in SCI, by making a wealth of data available for comparison and inclusion in meta-analyses. One of the main scientific products of data sharing efforts in the SCI research community will be more accurate estimates of interventional effect size achieved in preclinical studies.
Variability in SCI research findings also stems from differences in experimental procedures (Simard et al., 2012). In most cases, published preclinical findings are based on observations derived from data collected at a single laboratory. Data sharing efforts will allow for multi-center cohort studies, which will take the variability between different centers (the “center effect”) into account, resulting in a more reliable effect size for clinical translation. This will more accurately mirror the conditions of a clinical multicenter trial, and will therefore both enhance the translational value of preclinical studies and save significant resources. The power of this approach has been recently demonstrated in a re-analysis of data generated during preclinical multi-center testing of one of the interventions tested in a clinical Phase III RCT (Nielson et al., 2015b). Data sharing methods and their integration into the workflow of neuroscience researchers stemming from the FAIR Share Workshop represent an unparalleled effort to tackle a prospective challenge in a timely and resource-sparing manner, equipped to improve the chance of translation success and research quality across the field.
Through the ODC-SCI initiative, next steps to foster and support data sharing in the SCI research community include developing guidelines and training resources for researchers to enable FAIR sharing practices, implementing tools for data collection, and creating mechanisms for dataset citation, quality evaluation and annotation. NINDS, other NIH Institutes and the Department of Defense are devoting substantial resources to data sharing infrastructure, including the development of CDEs for both clinical and preclinical research. Such common vocabularies are ideal for collecting data and metadata for preclinical research, and can be integrated into existing repositories to enable standardization and data integration. Importantly, they can also be used by informaticians, ontologists and software engineers to develop new tools to make experimental data FAIR. Institutions such as university libraries can provide repositories and registry services for datasets that are not deposited in specialty databases like RegenBase, VISION-SCI or the Federal Interagency Traumatic Brain Injury Research (FITBIR) system (Thompson et al., 2015). Feedback we received from workshop participants make it clear that the community must be engaged broadly to facilitate essential cultural changes around data sharing, while the NIH and publishers will be the institutions that incentivize researchers to make their data FAIR and publicly available. Now is the time for SCI researchers promoting open data sharing to work together with these institutions to move SCI data accessibility forward in ways that encourage participation, while balancing the needs for resources to do science and to support the infrastructure for sharing, curating and preserving scientific data. To this end, the broad SCI research community must be involved in the next steps to develop ambitious and realistic expectations, as well as to create and test the tools and resources that are needed to make SCI data both FAIR and widely shared.
Appendix.
FAIR Share Workshop Participants
Michele Basso, EdD, PT
School of Health and Rehabilitation Sciences Ohio State University
Patrick S. F. Bellgowan, PhD
National Institute of Neurological Disorders and Stroke The National Institutes of Health
Fiona P. Brabazon, PhD
Kentucky Spinal Cord Injury Research Center University of Louisville
Sarah A. Busch, PhD
Athersys, Inc.
Aristea S Galanopoulou, MD, PhDSaul R. Korey
Department of Neurology Dominick P. Purpura Department of Neuroscience Albert Einstein College of Medicine
Ryan Gilbert, PhD
Biomedical Engineering Rensselaer Polytechnic Institute
Jeffrey S. Grethe, PhD
Center for Research in Biological Systems University of California, San Diego
James W. Grau, PhD
Behavioral and Cellular Neuroscience Institute for Neuroscience Texas A & M University
Jenny Haefeli, PhD
Weill Institute for Neurosciences Brain and Spinal Injury Center (BASIC) Department of Neurological Surgery University of California, San Francisco
Linda Jones, PT, MS
Spinal Cord Injury Research on the Translational Spectrum (SCIRTS)
Craig H. Neilsen Foundation
Naomi Kleitman, PhD
Craig H. Neilsen Foundation
Audrey Kusiak, PhD
Rehabilitation Research and Development Service
Office of Research and Development
Department of Veterans Affairs
Rosi Lederer, MD
Wings for Life InternationalSpinal Cord Research Foundation
David S. K. Magnuson, PhD
Kentucky Spinal Cord Injury Research CenterUniversity of Louisville
Maryann Martone, PhD
Center for Research in Biological Systems
University of California San Diego
Verena May, PhD
Wings for Life InternationalSpinal Cord Research Foundation
Lawrence Moon, PhD
Wolfson Centre for Age-Related Diseases
King’s College London
Alexander G. Rabchevsky, PhD
Spinal Cord & Brain Injury Research Center (SCoBIRC)
University of Kentucky
Carol Taylor-Burds
National Institute of Neurological Disorders and Stroke
The National Institutes of Health
Wolfram Tetzlaff, MD, PhD
University of British ColumbiaVancouver Coastal Health Blusson Spinal Cord Centre
Veronica J. Tom, PhD
Department of Neurobiology and AnatomyCollege of MedicineDrexel University
Abel Torres-Espín, PhD
Department of Physical Therapy Faculty of Rehabilitation Medicine University of Alberta
Vicky Whittemore, PhD
National Institute of Neurological Disorders and Stroke The National Institutes of Health
Xiao-Ming Xu, PhD
Stark Neurosciences Research Institute Indiana University School of Medicine
Binhai Zheng, PhD
Department of Neurosciences University of California San Diego
Highlights.
Preclinical spinal cord injury researchers are open to data sharing, yet just over 10% of those attending a data sharing workshop currently share their data online
Challenges in data sharing for this community include logistics of data collection and storage requirements, and personnel training
Researchers consider enabling scientific discovery and complying with journal and funder requirements more important incentives for data sharing than the potential for increased citations of papers and datasets
To promote data sharing broadly, a cultural shift is required not only for researchers but institutions and funding agencies
Next steps to support data sharing include developing guidelines and training resources, implementing tools for data collection, and creating mechanisms for dataset citation, quality evaluation and annotation
Acknowledgements
The FAIR Share workshop was co-sponsored by the NINDS and the University of Alberta, with contributions from the International Spinal Research Trust (ISRT) (KF), the Rick Hansen Institute (KF), and Wings for Life (KF). The ODC-SCI project is supported by a grant from the Craig H. Neilsen Foundation (ARF). Representatives from each of the funding agencies participated in the workshop and provided individual opinions that do not represent the official position or opinions of the funding agencies, their Boards, or the governments of the United States or Canada.
1Abbreviations:
- AES
American Epilepsy Society
- ASIA
American Spinal Injury Association
- CDE
Common Data Element
- BD2K
Big Data To Knowledge
- FORE-SCI
Facilities of Research Excellence in Spinal Cord Injury
- FAIR
Findable, Accessible, Interoperable, Reusable
- FITBIR
Federal Interagency Traumatic Brain Injury Research
- fMRI
functional magnetic resonance imaging
- FORCE 11
Future of Research Communications and e-Scholarship
- ISI
Institute for Scientific Information
- ILAE
International League Against Epilepsy
- ISCoS
International Spinal Cord Society
- ISRT
International Spinal Research Trust
- MIASCI
Minimum Information About a Spinal Cord Injury
- MASCIS
Multicenter Animal Spinal Cord Injury Study
- NINDS
National Institute of Neurological Disorders and Stroke
- ODC-SCI
Open Data Commons for Spinal Cord Injury
- PB
petabyte
- PCR
polymerase chain reaction
- RCT
randomized controlled trial
- RDF
resource description framework
- RNA
ribonucleic acid
- SCI
Spinal Cord Injury
- SfN
Society for Neuroscience
- SOP
Standard Operating Procedure
References
- Aguilar RM, Steward O, 2010. A bilateral cervical contusion injury model in mice: assessment of gripping strength as a measure of forelimb motor function. Exp. Neurol. 221, 38–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson KD, Sharp KG, Hofstadter M, Irvine K-A, Murray M, Steward O, 2009. Forelimb locomotor assessment scale (FLAS): novel assessment of forelimb dysfunction after cervical spinal cord injury. Exp. Neurol. 220, 23–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basso DM, Beattie MS, Bresnahan JC, 1995. A sensitive and reliable locomotor rating scale for open field testing in rats. J. Neurotrauma 12, 1–21. [DOI] [PubMed] [Google Scholar]
- Basso DM, Beattie MS, Bresnahan JC, Anderson DK, Faden AI, Gruner JA, Holford TR, Hsu CY, Noble LJ, Nockels R, Perot PL, Salzman SK, Young W, 1996. MASCIS evaluation of open field locomotor scores: effects of experience and teamwork on reliability. Multicenter Animal Spinal Cord Injury Study. J. Neurotrauma 13, 343–359. [DOI] [PubMed] [Google Scholar]
- Biering-Sørensen F, Alai S, Anderson K, Charlifue S, Chen Y, DeVivo M, Flanders AE, Jones L, Kleitman N, Lans A, Noonan VK, Odenkirchen J, Steeves J, Tansey K, Widerström-Noga E, Jakeman LB, 2015. Common data elements for spinal cord injury clinical research: a National Institute for Neurological Disorders and Stroke project. Spinal Cord 53, 265–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne PE, Lorsch JR, Green ED, 2015. Perspective: Sustaining the big-data ecosystem. Nature 527, S16–7. [DOI] [PubMed] [Google Scholar]
- Briggs KA, 2016. Is preclinical data sharing the new norm? Drug Discov. Today. doi: 10.1016/j.drudis.2016.05.003 [DOI] [PubMed] [Google Scholar]
- Burke DA, Whittemore SR, Magnuson DSK, 2013. Consequences of common data analysis inaccuracies in CNS trauma injury basic research. J. Neurotrauma 30, 797–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan A, Abeyruwan SW, Al-Ali H, Sakurai K, Ferguson AR, Popovich PG, Shah NH, Visser U, Bixby JL, Lemmon VP, 2016. RegenBase: a knowledge base of spinal cord injury biology for translational research. Database 2016 . doi: 10.1093/database/baw040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlifue S, Tate D, Biering-Sorensen F, Burns S, Chen Y, Chun S, Jakeman LB, Kowalski RG, Noonan VK, Ullrich P, 2016. Harmonization of Databases: A Step for Advancing the Knowledge About Spinal Cord Injury. Arch. Phys. Med. Rehabil. 97, 1805–1818. [DOI] [PubMed] [Google Scholar]
- Courtine G, Bunge MB, Fawcett JW, Grossman RG, Kaas JH, Lemon R, Maier I, Martin J, Nudo RJ, Ramon-Cueto A, Rouiller EM, Schnell L, Wannier T, Schwab ME, Edgerton VR, 2007. Can experiments in nonhuman primates expedite the translation of treatments for spinal cord injury in humans? Nat. Med. 13, 561–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirnagl U, Endres M, 2014. Found in translation: preclinical stroke research predicts human pathophysiology, clinical phenotypes, and therapeutic outcomes. Stroke 45, 1510–1518. [DOI] [PubMed] [Google Scholar]
- Ferguson AR, Hook MA, Garcia G, Bresnahan JC, Beattie MS, Grau JW, 2004. A simple post hoc transformation that improves the metric properties of the BBB scale for rats with moderate to severe spinal cord injury. J. Neurotrauma 21, 1601–1613. [DOI] [PubMed] [Google Scholar]
- Ferguson AR, Irvine K-A, Gensel JC, Nielson JL, Lin A, Ly J, Segal MR, Ratan RR, Bresnahan JC, Beattie MS, 2013. Derivation of multivariate syndromic outcome metrics for consistent testing across multiple models of cervical spinal cord injury in rats. PLoS One 8, e59712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson AR, Nielson JL, Cragin MH, Bandrowski AE, Martone ME, 2014. Big data from small data: data-sharing in the “long tail” of neuroscience. Nat. Neurosci. 17, 1442–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson AR, Stück ED, Nielson JL, 2011. Syndromics: a bioinformatics approach for neurotrauma research. Transl. Stroke Res. 2, 438–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedli L, Rosenzweig ES, Barraud Q, Schubert M, Dominici N, Awai L, Nielson JL, Musienko P, Nout-Lomas Y, Zhong H, Zdunowski S, Roy RR, Strand SC, van den Brand R, Havton LA, Beattie MS, Bresnahan JC, Bézard E, Bloch J, Edgerton VR, Ferguson AR, Curt A, Tuszynski MH, Courtine G, 2015. Pronounced species divergence in corticospinal tract reorganization and functional recovery after lateralized spinal cord injury favors primates. Sci. Transl. Med. 7, 302ra134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galanopoulou AS, Simonato M, French JA, O’Brien TJ, 2013. Joint AES/ILAE translational workshop to optimize preclinical epilepsy research. Epilepsia 54 Suppl 4, 1–2. [DOI] [PubMed] [Google Scholar]
- Irvine K-A, Ferguson AR, Mitchell KD, Beattie SB, Lin A, Stuck ED, Huie JR, Nielson JL, Talbott JF, Inoue T, Beattie MS, Bresnahan JC, 2014. The Irvine, Beatties, and Bresnahan (IBB) Forelimb Recovery Scale: An Assessment of Reliability and Validity. Front. Neurol. 5, 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammertse DP, 2013. Clinical trials in spinal cord injury: lessons learned on the path to translation. The 2011 International Spinal Cord Society Sir Ludwig Guttmann Lecture. Spinal Cord 51, 2–9. [DOI] [PubMed] [Google Scholar]
- Landis SC, Amara SG, Asadullah K, Austin CP, Blumenstein R, Bradley EW, Crystal RG, Darnell RB, Ferrante RJ, Fillit H, Finkelstein R, Fisher M, Gendelman HE, Golub RM, Goudreau JL, Gross RA, Gubitz AK, Hesterlee SE, Howells DW, Huguenard J, Kelner K, Koroshetz W, Krainc D, Lazic SE, Levine MS, Macleod MR, McCall JM, Moxley RT 3rd, Narasimhan K, Noble LJ, Perrin S, Porter JD, Steward O, Unger E, Utz U, Silberberg SD, 2012. A call for transparent reporting to optimize the predictive value of preclinical research. Nature 490, 187–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon VP, Ferguson AR, Popovich PG, Xu X-M, Snow DM, Igarashi M, Beattie CE, Bixby JL, MIASCI Consortium, 2014. Minimum information about a spinal cord injury experiment: a proposed reporting standard for spinal cord injury experiments. J. Neurotrauma 31, 1354–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macleod MR, Fisher M, O’Collins V, Sena ES, Dirnagl U, Bath PMW, Buchan A, van der Worp HB, Traystman R, Minematsu K, Donnan GA, Howells DW, 2009. Good laboratory practice: preventing introduction of bias at the bench. Stroke 40, e50–2. [DOI] [PubMed] [Google Scholar]
- McKiernan EC, Bourne PE, Brown CT, Buck S, Kenall A, Lin J, McDougall D, Nosek BA, Ram K, Soderberg CK, Spies JR, Thaney K, Updegrove A, Woo KH, Yarkoni T, 2016. How open science helps researchers succeed. Elife 5. doi: 10.7554/eLife.16800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielson JL, Guandique CF, Liu AW, Burke DA, Lash AT, Moseanko R, Hawbecker S, Strand SC, Zdunowski S, Irvine K-A, Brock JH, Nout-Lomas YS, Gensel JC, Anderson KD, Segal MR, Rosenzweig ES, Magnuson DSK, Whittemore SR, McTigue DM, Popovich PG, Rabchevsky AG, Scheff SW, Steward O, Courtine G, Edgerton VR, Tuszynski MH, Beattie MS, Bresnahan JC, Ferguson AR, 2014. Development of a database for translational spinal cord injury research. J. Neurotrauma 31, 1789–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielson JL, Haefeli J, Salegio EA, Liu AW, Guandique CF, Stück ED, Hawbecker S, Moseanko R, Strand SC, Zdunowski S, Brock JH, Roy RR, Rosenzweig ES, Nout-Lomas YS, Courtine G, Havton LA, Steward O, Reggie Edgerton V, Tuszynski MH, Beattie MS, Bresnahan JC, Ferguson AR, 2015a. Leveraging biomedical informatics for assessing plasticity and repair in primate spinal cord injury. Brain Res. 1619, 124–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielson JL, Paquette J, Liu AW, Guandique CF, Tovar CA, Inoue T, Irvine K-A, Gensel JC, Kloke J, Petrossian TC, Lum PY, Carlsson GE, Manley GT, Young W, Beattie MS, Bresnahan JC, Ferguson AR, 2015b. Topological data analysis for discovery in preclinical spinal cord injury and traumatic brain injury. Nat. Commun. 6, 8581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read KB, Sheehan JR, Huerta MF, Knecht LS, Mork JG, Humphreys BL, NIH Big Data Annotator Group, 2015. Sizing the Problem of Improving Discovery and Access to NIH-Funded Data: A Preliminary Study. PLoS One 10, e0132735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sejnowski TJ, Churchland PS, Movshon JA, 2014. Putting big data to good use in neuroscience. Nat. Neurosci. 17, 1440–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simard JM, Popovich PG, Tsymbalyuk O, Gerzanich V, 2012. Spinal cord injury with unilateral versus bilateral primary hemorrhage--effects of glibenclamide. Exp. Neurol. 233, 829–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DH, Hicks RR, Johnson VE, Bergstrom DA, Cummings DM, Noble LJ, Hovda D, Whalen M, Ahlers ST, LaPlaca M, Tortella FC, Duhaime A-C, Dixon CE, 2015. Pre-Clinical Traumatic Brain Injury Common Data Elements: Toward a Common Language Across Laboratories. J. Neurotrauma 32, 1725–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stead WW, 1998. Medical informatics--on the path toward universal truths. J. Am. Med. Inform. Assoc. 5, 583–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steckler T, Brose K, Haas M, Kas MJ, Koustova E, Bespalov A, ECNP Preclinical Data Forum Network, 2015. The preclinical data forum network: A new ECNP initiative to improve data quality and robustness for (preclinical) neuroscience. Eur. Neuropsychopharmacol. 25, 1803–1807. [DOI] [PubMed] [Google Scholar]
- Steward O, Popovich PG, Dietrich WD, Kleitman N, 2012. Replication and reproducibility in spinal cord injury research. Exp. Neurol. 233, 597–605. [DOI] [PubMed] [Google Scholar]
- Thompson HJ, Vavilala MS, Rivara FP, 2015. Chapter 1 Common Data Elements and Federal Interagency Traumatic Brain Injury Research Informatics System for TBI Research. Annu. Rev. Nurs. Res. 33, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watzlawick R, Sena ES, Dirnagl U, Brommer B, Kopp MA, Macleod MR, Howells DW, Schwab JM, 2014. Effect and reporting bias of RhoA/ROCK-blockade intervention on locomotor recovery after spinal cord injury: a systematic review and meta-analysis. JAMA Neurol. 71, 91–99. [DOI] [PubMed] [Google Scholar]
- Wilkinson MD, Dumontier M, Aalbersberg IJJ, Appleton G, Axton M, Baak A, Blomberg N, Boiten J-W, da Silva Santos LB, Bourne PE, Bouwman J, Brookes AJ, Clark T, Crosas M, Dillo I, Dumon O, Edmunds S, Evelo CT, Finkers R, Gonzalez-Beltran A, Gray AJG, Groth P, Goble C, Grethe JS, Heringa J, ‘t Hoen PAC, Hooft R, Kuhn T, Kok R, Kok J, Lusher SJ, Martone ME, Mons A, Packer AL, Persson B, Rocca-Serra P, Roos M, van Schaik R, Sansone S-A, Schultes E, Sengstag T, Slater T, Strawn G, Swertz MA, Thompson M, van der Lei J, van Mulligen E, Velterop J, Waagmeester A, Wittenburg P, Wolstencroft K, Zhao J, Mons B, 2016. The FAIR Guiding Principles for scientific data management and stewardship. Sci Data 3, 160018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young W, 2002. Spinal cord contusion models. Prog. Brain Res. 137, 231–255. [DOI] [PubMed] [Google Scholar]
- Zinner DE, Pham-Kanter G, Campbell EG, 2016. The Changing Nature of Scientific Sharing and Withholding in Academic Life Sciences Research: Trends From National Surveys in 2000 and 2013. Acad. Med 91, 433–440. [DOI] [PMC free article] [PubMed] [Google Scholar]


