Abstract
Borrelia burgdorferi sensu lato, the agent of Lyme disease, exists in nature through a complex enzootic life cycle that involves both ticks and mammals. The B. burgdorferi genome encodes five Oligopeptide ABC transporters (Opp) that are predicted to be involve in transport of various nutrients. Previously, it was reported that OppA5 is important for the optimal production of OspC, a major virulence factor of B. burgdorferi. In this study, possible role of another Oligopeptide ABC transporter, OppA4 in ospC expression was investigated by construction of an oppA4 deletion mutant and the complemented strain. Inactivation of oppA4 resulted an increased production of OspC, suggesting that OppA4 has a negative impact on ospC expression. Expression of ospC is controlled by Rrp2-RpoN-RpoS, the central pathway essential for mammal infection. We showed that increased ospC expression in the oppA4 mutant was due to an increased rpoS expression. We then further investigated how OppA4 negatively regulates this pathway. Two regulators, BosR and BadR, are known to positively and negatively, respectively, regulate the Rrp2-RpoN-RpoS pathway. We found that deletion of oppA4 resulted in an increased level of BosR. Previous reports showed that bosR is mainly regulated at the post-transcriptional level by other factors. However, OppA4 appears to negatively regulate bosR expression at the transcriptional level. The finding of OppA4 involved in regulation of the Rrp2-RpoN-RpoS pathway further reinforces the importance of nutritional virulence to the enzootic cycle of B. burgdorferi.
Keywords: Borrelia burgdorferi, OppA4, OspC
INTRODUCTION
B. burgdorferi sensu lato, the Lyme disease/borreliosis pathogen, is maintained in nature in an enzootic cycle between Ixodes tick vectors and certain vertebrates, so-called reservoir hosts (Lane et al., 1991; Radolf et al., 2012). This spirochetal pathogen dramatically regulates its gene expression during its transition between ticks and the reservoir hosts (for recent reviews, see Caimano et al., 2016; Samuels, 2011; Stevenson and Seshu, 2017; Ye et al., 2016). During the process of transmission, spirochetes in the midgut of nymphal ticks begin replication triggered by tick blood meal engorgement process, activate a two-component response regulator and enhancer-binding protein Rrp2, which in turn, activates the alternative sigma factor RpoN (σN), leading to the transcription of rpoS that encodes for another alternative sigma factor and global regulator RpoS (σS). This Rrp2-RpoN-RpoS pathway controls production of a few of surface proteins that have been shown to be important for mammalian infection (e.g., OspC, DbpA/B, BBK32, BBA33, BBA64, BBA66, etc), concomitant with downregulating several factors important for spirochetal colonization in ticks (e.g., OspA, P22, lp6.6) (for review and initial discoveries of this pathway, see Caimano et al., 2016; Hübner et al., 2001; Radolf et al., 2012; Samuels, 2011; Yang et al., 2003). On the other hand, this pathway is repressed in the tick midgut during the process of tick acquisition of spirochetes (i.e., spirochetes migrate from infected vertebrate hosts to uninfected ticks), with downregulation of mammalian infection-associated proteins while upregulation of proteins important for tick colonization (Ouyang et al., 2008; Radolf et al., 2012).
Given the importance of the Rrp2-RpoN-RpoS pathway, many signals and factors have shown to regulate rpoS expression. rpoS has a σ54-type promoter, whose expression is under the control of Rrp2 and RpoN (σ54) and. In addition, a Fur/PerR-like transcription factor, BosR, is also required for rpoS and ospC expression (Hyde et al., 2009; Ouyang et al., 2011; Ouyang et al., 2009; Wang et al., 2017). BosR is capable of binding to the promoter region of rpoS in vitro, the mechanism of how BosR is involved in activating rpoS transcription from a σ54 promoter remains unclear (Hyde et al., 2009; Ouyang et al., 2011; Ouyang et al., 2009; Wang et al., 2017). Another transcription factor, BadR, directly represses rpoS expression (Miller et al., 2013; Ouyang and Zhou, 2015). Additional factors, including DsrA (Lybecker and Samuels, 2007), Rrp1 and PlzA (He et al., 2014; Rogers et al., 2009; Sze et al., 2013), BtmA (Troxell et al., 2013), SodA (Esteve-Gassent et al., 2015), BBD18 (Dulebohn et al., 2014), are also shown to influence rpoS and ospC expression (for review, see Samuels, 2011; Stevenson and Seshu, 2017). However, what signal(s) B. burgdorferi senses and how the signals feed into this pathway, are largely unknown.
B. burgdorferi has a reduced genome and lacks many biosynthetic pathways for amino acids and other essential nutrients, and thus relies on transporters to obtain nutrients from vertebrate and tick hosts (Casjens et al., 2000; Fraser et al., 1997; Groshong et al., 2017; Radolf et al., 2012). The oligopeptide permease (Opp) transporter system is an efficient system for bacteria to transport multiple amino acids (Garai et al., 2017; H., 2010). It consists of an oligopeptide-binding protein (OppA), a membrane permease (OppBC), and a nucleotide-binding domain (NBD) protein (OppDF) (Garai et al., 2017; Monnet, 2003). B. burgdorferi has five OppA proteins (OppA1 to OppA5), two OppBC permeases, and one OppDF (Bono et al., 1998; Raju et al., 2011; Wang et al., 2004). oppA1-oppA3, oppB1, oppC1, oppD, oppF are linked together and located in the chromosome, and oppB2-oppC2 are located at different loci of the chromosome. Two additional oligopeptide-binding proteins oppA4 and oppA5, encode on the endogenous plasmids cp26 and lp54, respectively (Bono et al., 1998; Fraser et al., 1997; Groshong et al., 2017). Previous studies using the Escherichia coli as a surrogate system showed that the OppA systems of B. burgdorferi can import small peptides in E. coli, and the specificity of peptide binding for OppA1, OppA2, and OppA3 were determined (Lin et al., 2001; Wang et al., 2004). All five OppA proteins have conserved residues for structure folding and peptide binding found in other OppA proteins (Lin et al., 2001; Wang et al., 2004). Sequence analysis indicates that OppA4 and OppA5 cluster together, OppA1 and OppA3 form another group, and OppA2 forms its own clade (Groshong et al., 2017). Recently, the structures of five OppA proteins of B. burgdorferi were analyzed based on the crystal structure of OppA4 (Fairman JW et al., 2012; Groshong et al., 2017), suggesting that OppA2 and OppA4 likely bind to negatively charged peptides, OppA2 and OppA5 prefer positively charged peptides, and OppA3 may bind a neutral peptides (Groshong et al., 2017).
The oppA genes of B. burgdorferi were differentially regulated by environmental conditions as well as during the enzootic cycle of B. burgdorferi (Troy et al., 2016; Drecktrah et al., 2015; Groshong et al., 2017; Iyer et al., 2015; Medrano et al., 2007; Wang et al., 2002). oppA1 and oppA3 are expressed in the feeding tick, and oppA2 is expressed at relatively high levels throughout the enzootic cycle (Groshong et al., 2017). The two plasmid-coded oppA genes, oppA4 and oppA5, are the most differentially expressed: oppA5 is preferably expressed in mammalian host condition, whereas oppA4 is expressed ticks during the larval and nymphal blood meals (Groshong et al., 2017; Wang et al., 2002). oppA5 expression is positively regulated by the Rrp2-RpoN-RpoS pathway, but not other oppA genes (Medrano et al., 2007). Targeted mutagenesis showed that inactivation of oppA5 impaired expression of rpoS and ospC (Raju et al., 2011), but did not affect mammalian infection. However, whether OppA4 influences virulence gene expression and whether it plays a role in the enzootic cycle of B. burgdorferi have not been determined. In this study, we report targeted inactivation of oppA4 and the complemented strain, and studied its influence on the Rrp2-RpoN-RpoS pathway and the mammalian infection.
MATERIALS AND METHODS
Bacterial strains
All strains and plasmids used in this study are described in Table 1. Low–passage, virulent B. burgdorferi strain 5A18NP1 was kindly provided by Drs. Norris and Kawabata (Kawabata et al., 2004). B. burgdorferi were cultivated in Barbour-Stoenner-Kelly (BSK-II) medium supplemented with 6% normal rabbit serum (Pel-Freez Biologicals, Rogers, AR) at 37°C with 5% CO2. Relevant antibiotics were added to the cultures at the following final concentrations: 300 μg/ml for kanamycin (Kan), 50 μg/ml for streptomycin (Strep), and 50 μg/ml for gentamicin (Gent). The constructed suicide vectors and shuttle vector were maintained in Escherichia coli strain DH5α.
Table 1.
Strains, plasmids, and primers used in this study.
| Strains, plasmids or primers | Description or sequence | Purpose or source |
|---|---|---|
| Strains | ||
| 5A18NP1 | Wild-type strain with bbeO2 gene disrupted by Kanr | (Kawabata et al., 2004) |
| OppA4-mut | oppA4 deletion mutant; 5A18NP1 transformed with pZB20 | In this study |
| OppA4-com | oppA4 complementation; OppA4-mut transformed with pZB028 | In this study |
| Plasmids | ||
| pZB020 | Suicide vector for constructing oppA4 deletion; Strr | In this study |
| pZB028 | Shuttle vector carrying oppA4 gene; Genr | In this study |
| Primers | ||
| PRBB067 (2F) | GCAGGGCCCTTAACTTTTATAATCTTTATTTTCAATCGATT CAACCATTAC | Amplify oppA4 upstream region |
| PRBB068 | GCAGTCGACACAAGCATCCTTACAAGTTTTTATAAATTTA AATTTTGCC | Same as above |
| PRBB049 | GCACCCGGGTTTAAGTAAAAATGGTTTATAGCTAGATC | Amplify oppA4 downstream region |
| PRBB050 | GCAGGATCCATGCCAAATAAGATAACAAAAGAAGCTTTA AC | Same as above |
| PRBB034 (1R) | GCACTGCAGTCATTTAATTGGTTTTATTTCAGATAAATTA AATC | Amplify oppA4 with Ndel and PstI |
| PRBB056 | GCACATATGATGTACCCATACGATGTTCCAGATTACGCTA AAATATTGATAAAAAAGTTAAAAG | Same as above |
| PRYY049 | GATCAGATCTCAGCTTTTTTTTGA | Amplify flgBp-partial aacC1 with Bg1II site |
| PRBB053 | GCGAGATCATAGATATAAATCTCACTACGCGGCTGCTC | Same as above |
| PRBB052 | GAGCAGCCGCGTAGTGAAATCTATATCTATGATCTCGC | Amplify partial aacC1 with Aatll site |
| PRYY056 | TCCGGCGACGTCATTAGGTGGCGGTACTTG | Same as above |
| PRBB033 (IF) | GCACATATGATGAAAATATTGATAAAAAAGTTAAAAGTT GTA | Amplify oppA4 for identification |
| PRBB097 (2R) | GCTGCGTAACATATATAGAAACCTCCCTC | Amplify BB_B14 for identification |
| PRBB003 (3F.4F) | GAGGAATGAATGAGGGAAGCGGTGATCGCC | Amplify aadA for identification |
| PRBB135 (3R) | ACAACAAGAATCGTTGCGGG | Amplify guaB for identification |
| PRBB009 (4R) | GCAACGCGTTATTTGCCGACTACCTTGGTGATC | Same as above |
| qPCR-flaB-F | CAGCAATAGCTTCATCTTGGTTTG | qPCR for flaB |
| qPCR-flaB-R | ACCAGCATCTTCAGGGTCTCA | qPCR for flaB |
| qhx-rpoS-F | ATTTTCAACTTATGCATCATTTTG | qPCR for rpoS |
| qhx-rpoS-R | TTTTCTTTTCTGTATGGGACTTTT | qPCR for rpoS |
| qhx-ospC-F | TAGCGGGAGCTTATGCAATATCAACC | qPCR for ospC |
| qhx-ospC-R | CATCAATTTTTTCCTTTAATCCTTCA | qPCR for ospC |
| PRBB132 | GGATACAGACCGGGACTTGC | qPCR for oppA4 |
| PRBB133 | CCTTCGGCAGTAATGGAAACTC | qPCR for oppA4 |
Construction of the oppA4 deletion mutant
To inactivate oppA4 gene in 5A18NP1, a suicide vector pZB020 was constructed for homologous recombination as following: the regions of DNA corresponding to 1.1 kb upstream and 1.5 kb downstream regions of oppA4 were PCR amplified from 5A18NP1 genomic DNA with primer pair PRBB067/PRBB068 and PRBB049/PRBB050 (Table 1), respectively. The resulting PCR fragments were cloned into pMP025 and confirmed by restriction enzyme digestion and sequencing. Firstly, pMP025 and the fragment amplified by PRBB067/PRBB068 were digested by ApaI and SalI and ligated to generate pZB020A. Then, pZB020A and the fragment amplified by PRBB049/PRBB050 were digested by XmaI and BamHI and ligated. pZB020 was constructed and confirmed by both restriction enzyme digestion and sequencing, then it is used for transformation. Transformants were selected with Kan and Strep, and twelve positive clones were obtained. PCR analysis was used to confirm correct marker insertion and inactivation of the chromosomal copy of oppA4. Endogenous plasmid profiles were performed as previously described (Bunikis et al., 2011; Xiang et al., 2017; Ye et al., 2014). The positive clones were named BBZB014.
Construction of the oppA4 complemented strain.
To complement oppA4 in BBZB014, we constructed a shuttle vector, pZB028, that harbors a wild-type oppA4 gene (with a human influenza virus hemagglutinin [HA] tag at the 5’ end of the coding region) driven by flaB promoter. Wide-type oppA4 with a HA-tag was amplified by PRBB034/PRBB056 and digested by NdeI and PstI, the resulting fragment was ligated into pYY025, which harbors a flgB promoter (flaBp)-aadA cascade (streptomycin-resistant gene), resulting in pZB028A. The positive clones were identified by restriction enzyme digestion and sequencing. Then pZB028A was digested by BglII and AatII to remove flgBp-aadA (streptomycin-resistant gene) and ligated with flgBp-aacC1 (gentamicin-resistant gene) digested by BglII and AatII. To get rid of BglII cutting site in aacC1, we introduced a point mutation to change BglII cutting site from AGATCT to AAATCT without change of amino acid sequence by overlap PCR. To make this, we use pYY020 as template and amplify flgBp-partial aacC1 and the rest partial aacC1 by PRYY049/PRBB053, PRBB052/PRYY056, respectively. And the whole flgBp-aacC1 without BglII was amplified by PRYY049 and PRYY056 using the two fragments amplified above. The new flgBp-aacC1 without BglII was ligated into pZB028A, resulting in pZB028. Positive clones were confirmed by restriction enzyme digestion and sequencing. Then pZB028 was transformed to BBZB014 for complementation. Complemented strains were selected by Kanamycin, Streptomycin and Gentamicin. Positive clones were analyzed by PCR, SDS-PAGE and immunoblotting and named as BBZB018.
SDS-PAGE and immunoblotting.
SDS-PAGE and immunoblotting were performed as previously described (Troxell et al., 2013). The images were developed using chemiluminescent method (Pierce ECL Western Blotting Substrate, Thermo Scientific, IL). All proteins were separated on 12.5% SDS-PAGE. Spirochetes were collected at both early phase and late-log phase for immunoblotting. Antibodies directed against BosR, RpoS, BadR, OspC and FlaB have been described previously (Xu et al., 2010).
RNA isolation and quantitative RT-PCR
Total RNA was isolated using the TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. To reduce trace amounts of DNA contamination, samples were further digested with RNase-free DNaseI (Qiagen), and purified using the RNeasy kit (Qiagen). DNA-free RNA was confirmed by PCR. cDNA was generated using the SuperScript III First-Strand Synthesis System according to the manufacturer’s protocol (Invitrogen). Real-time quantitative PCR was performed using the RT2 SYBR Green ROX qPCR Mastermix (Qiagen) on an ABI 7000 sequence detection system. As previously described (Yang et al., 2009), changes in gene expression level were analyzed through relative quantification method (ΔΔCT) using flaB of B. burgdorferi for normalization. Statistical analysis of the data were performed using an unpaired, two-tailed Student’s t-test, in which statistical significance was determined when P < 0.05.
Mouse infection.
All mouse experiments were approved by the IACUC committee of Indiana University School of Medicine under the protocol number # 11339 MD/R/USDA/HZ/AR. Four-week-old C3H/HeN mice (Harlan, Indianapolis, IN) were subcutaneously inoculated with 1×105 spirochetes. Ear punch was done 4 weeks after infection and cultivated in 2 ml of the BSK-II medium (Sigma-Aldrich, St. Louis, MO) containing an antibiotic mixture of phosphomycin (2 mg/ml), rifampin (5 mg/ml), and amphotericin B (250 mg/ml) (Sigma-Aldrich). All cultures were maintained at 37°C and examined for the presence of spirochetes by dark-field microscopy beginning from 5 days after inoculation. Mouse infection was determined by positive growth of ear punch culture.
RESULTS
Construction of the oppA4 mutant and complemented strain
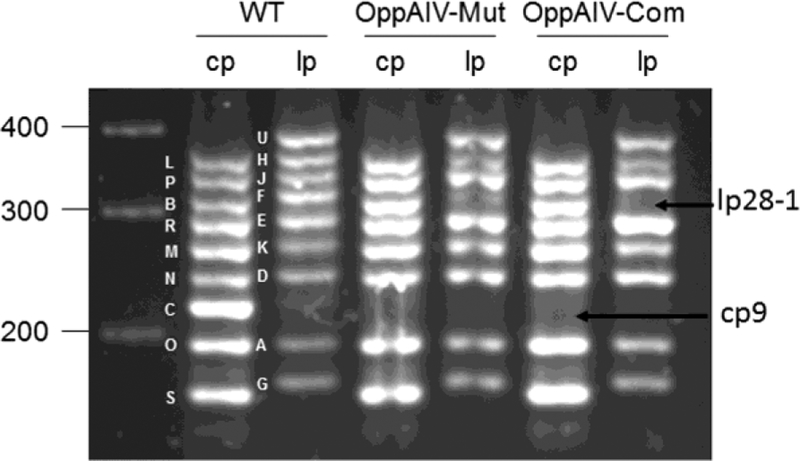
The oppA4 mutant was generated by allele exchange via homologous recombination by transforming a suicide vector (pZB020) carrying aadA (Strep-resistant cassette) flanked by 1.1 kb upstream and 1.5 kb downstream regions of oppA4 into an infectious B. burgdorferi strain B31–5A18NP1 (Fig. 1A). Twelve positive clones were obtained and PCR analysis was performed to confirm that oppA4 was replaced by the aadA marker (Fig. 1C). The oppA4 mutant was designated OppA4-Mut. For complementation, a shuttle vector containing a flaB-driven oppA4 gene (pZB028, Fig. 1B) was transformed with the oppA4 mutant. Positive clones were confirmed by PCR (Fig. 1C), and the complemented strain was designated OppA4-Com. qRT-PCR analysis showed that the oppA4 mutant lost oppA4 expression (Fig. 1D). The complemented strain showed much higher oppA4 expression level, as the result of the flaB promoter (Fig. 1D). Endogenous plasmid profiles were examined, and one clone of the oppA4 mutant and the complemented strain with identical plasmid contents with the parental strain was chosen for further study (Fig. 2).
Fig. 1. Construction of the oppA4 mutant and the complemented strain.
(A) Strategy for constructing the oppA4 mutant. WT: wild type. (B) Diagram of the shuttle vector used for complementation. (C) PCR analysis of the oppA4 mutant (Mut) and the complemented strain (Com). Primers used are labeled on top and their locations are indicated in (A). (D) qRT-PCR analysis of oppA4 mRNA levels in wild-type strain (WT), the oppA4 mutant (OppA4-mut), and the complemented strain (OppA4-com). The values of the oppA4 mRNA were normalized with the flaB mRNA level.
Fig. 2. Endogenous plasmid profile analysis.
WT, wild-type strain 5A18NP1. OppA4-Mut, the oppA4 mutant. OppA4-Com, the oppA4 mutant complemented with a shuttle vector carrying a wild-type copy of oppA4. cp, circular plasmid; lp, linear plasmid.
Inactivation of oppA4 increased the OspC level
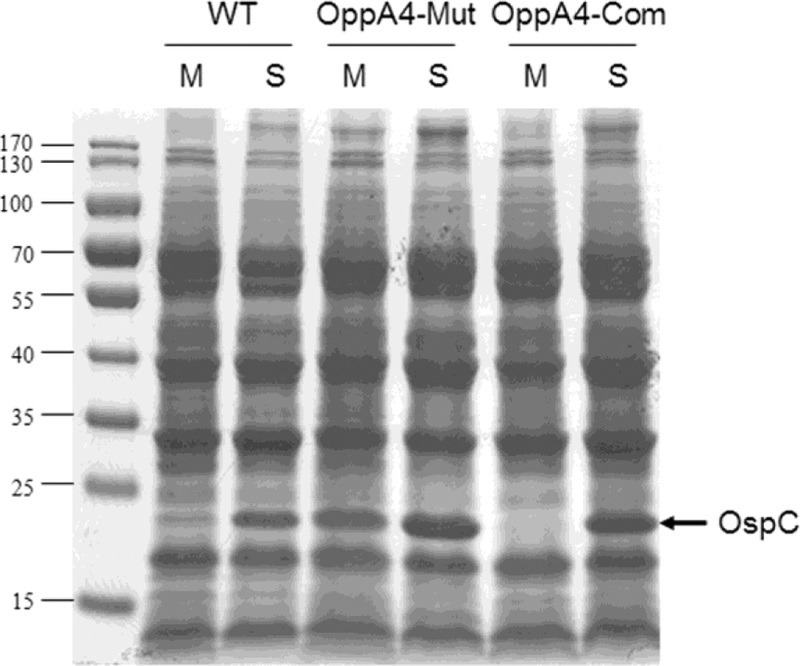
The oppA4 mutant had similar growth rate in vitro as that of parental strain (data not shown). To examine the effect of oppA4 deletion on protein profile of B. burgdorferi, spirochetes were grown in the BSK-II medium at 37°C, and harvested at the mid-logarithmic phase (M, 1 X 107 cells/ml, M) and stationary phase (S, 5 X 108 cells/ml) (Fig. 3). As expected, wild-type B. burgdorferi did not have visible OspC production on SDS-PAGE gel at the mid-log phase, but readily detected at the stationary phase (Fig. 3). In contract, the oppA4 mutant showed readily visible OspC production even at the mid-log phase, and the level of OspC was further increased at the stationary phase (Fig. 3). Enhanced OspC levels in the oppA4 mutant in both mid-log and stationary phase spirochetes were more apparent in the immunoblot assay using monoclonal antibody against OspC (Fig. 4). Noted that 50-fold less samples were loaded for immunoblotting than that used for SDS-PAGE gel because of high levels of OspC. Further qRT-PCR analysis demonstrated that the enhanced OspC production in the oppA4 mutant occurred at the transcription level (Fig. 5). Complementation restored the OspC pattern close that in wild-type spirochetes (Fig. 4 & 5). These data suggest that OppA4 negatively regulates ospC expression.
Fig. 3. Increased level of OspC production in the oppA4 mutant.
Strains of WT, OppA4-Mut, and OppA4-Com were cultured in standard BSK-II medium at 37°C. Spirochetes were collected at mid-log (M, 107 spirochetes/ml) or stationary phase (S, 5 X 108 spirochetes/ml) and subjected to SDS-PAGE analysis. The band corresponding to OspC was labeled and indicated by arrow.
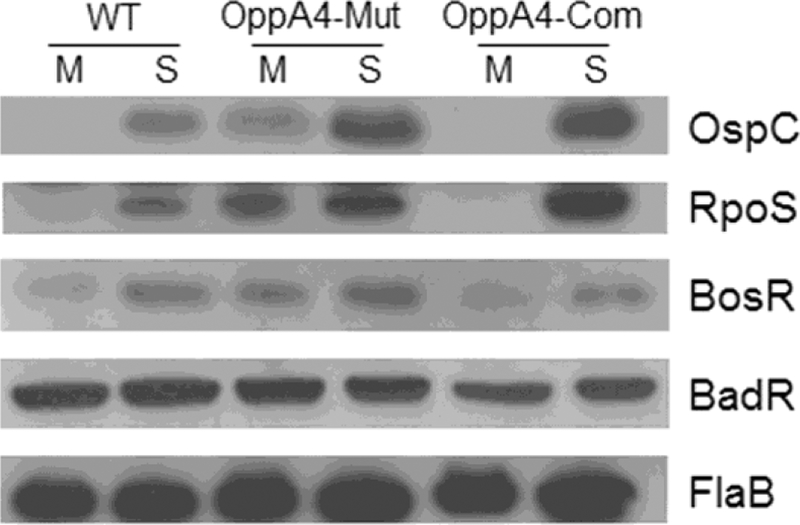
Fig. 4. Increased RpoS and BosR levels of the oppA4 mutant.
WT, OppA4-Mut and OppA4-Com were cultured in standard BSK-II medium at 37°C. Spirochetes were collected at mid-log (M, 107 spirochetes/ml) or stationary phase (S, 5 X 108 spirochetes/ml) and subjected to immunoblot analysis using antibodies specific for FlaB, RpoS, OspC, BosR or BadR. For immunoblotting against OspC, 50-fold less amount of samples were loaded.
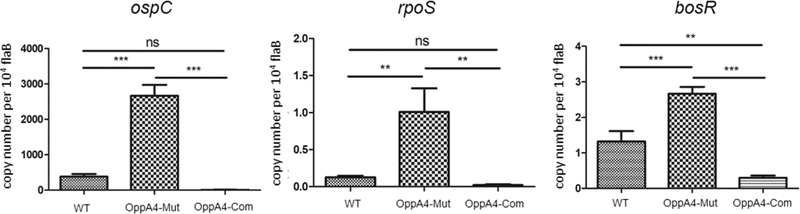
Fig 5. Increased levels of rpoS, bosR and ospC mRNA in the oppA4 mutant.
WT, OppA4-Mut and OppA4-Com were cultured in standard BSK-II medium at 37°C, and were collected at mid-log phase (M, 107 spirochetes/ml). RNAs were isolated and then subjected to qRT-PCR analyses. Levels of each gene transcript were normalized with flaB. The bars represent the mean values of three independent experiments, and the error bars represent the SD. *, P < 0.05.
Inactivation of oppA4 increased the RpoS and BosR levels
ospC has an RpoS-type promoter and its expression is under the direct control of RpoS (Hübner et al., 2001; Lybecker and Samuels, 2007; Yang et al., 2005). Accordingly, expression of rpoS at mRNA and protein levels were examined. The result showed that the oppA4 mutant had increased levels of rpoS expression at both mRNA (~8 folds) and protein levels (~50 folds) (Fig. 4 & 5). As an initial attempt to understand how OppA4 negatively regulates rpoS and subsequently ospC expression, we investigated the known factors involved in the Rrp2-RpoN-RpoS pathway. Specifically, we focused on two major factors BosR and BadR. BosR positively regulates rpoS expression, while BadR negatively regulates rpoS expression. The result showed that the BosR level, not BadR, was upregulated in the oppA4 mutant in spirochetes harvested at both mid-log and stationary phases (Fig. 4), suggesting that OppA4 down-regulates rpoS and ospC expression via BosR.
OppA4 negatively regulates bosR at the transcriptional level
The BosR level is often regulated at the post-transcription level (Hyde et al., 2007; Troxell et al., 2013). Transcriptional regulation of bosR was also reported previously (He et al., 2014; Ouyang and Zhou, 2015). Thus, levels of bosR mRNA were determined in the oppA4 mutant. As shown in Fig. 5, there was a significant increase in bosR mRNA levels in the oppA4 mutant in comparison to wild-type and the complemented strains. This result indicates that OppA4 negatively regulates bosR at transcriptional level.
OppA4 is not essential for mammalian infection.
To examine the oppA4 mutant’s ability to infect mice, groups of C3H/HeN mice were needle inoculated with wild-type, the oppA4 mutant, or the complemented strain with a dose of 1×105 spirochetes per mouse. Ear punch biopsies were collected 4 weeks post-infection and then cultured for presence of spirochetes. As shown in Table 2, all mice inoculated with wild-type B. burgdorferi were culture-positive. Four out of five mice infected with the oppA4 mutant showed culture positive. Since the complemented strain also had the same number of mice infected, this minor difference in infection was not related to oppA4 inactivation. These data suggested that OppA4 is not essential for B. burgdorferi to establish infection in mice.
Table 2.
Mouse infection of the OppA4-mut and OppA4-com
C3H/HeN mice were needle infected with 105 spirochetes/mouse.
Fisher Exact Test statistic value between WT and OppA4-mut is 1.
DISCUSSION
In this study, we successfully constructed an oppA4 deletion mutant and the complemented strain in B. burgdorferi. These results showed that the oppA4 mutant showed increased ospC expression via influencing BosR and RpoS levels. These data are different from previous findings for OppA5, another substrate-binding protein, in which deletion of oppA5 resulted in an impaired activation of the Rrp2-RpoN-RpoS pathway (Raju et al., 2011). These observations indicate that although they all involve in acquiring amino acids from either tick or vertebrate hosts, effects of OppA1 to OppA5 on the Rrp2-RpoNRpoS pathway are different. This is consistent with the observations that expressions of each oppA genes are different in the enzootic cycle of B. burgdorferi (Groshong et al., 2017; Wang et al., 2002).
The findings of Opp transporter systems influencing virulence gene expression of B. burgdorferi from this and previous studies highlight the important role of nutrients in regulating bacterial virulence, which represents one aspect of the concept of “nutritional virulence” (Garai et al., 2017; Kwaik and Bumann, 2013). During tick feeding, a flux of blood meals provides rich nutrient conditions that allows the most robust replication of B. burgdorferi in the tick midgut (de Silva and Fikrig, 1995; Schwan and Piesman, 2000). Thus, it is the spirochete’s advantage to sense such change in nutritional conditions as a signal for triggering virulence gene expression. In this regard, we and other showed that metals are important signals for regulating B. burgdorferi virulence gene expression (Troxell and Yang, 2013; Troxell et al., 2013; Wang et al., 2017). Pyruvate, a key molecule in the central metabolic pathway could help B. burgdorferi, an organism that lacks catalase, defensing against H2O2 (Troxell et al., 2014). More importantly, we and others showed that acetyl phosphate, an intermediate molecule of AckA-Pta pathway that B. burgdorferi employs to synthesize the essential molecule, acetyl-coA, from acetate, contributes to the activation of the Rrp2-RpoN-RpoS pathway (Raju et al., 2011; Van Laar et al., 2012; Xu et al., 2010). Recent data showed that during in vitro cultivation conditions, the AckA-Pta pathway is not essential for the activation of the Rrp2-RpoN-RpoS pathway (Richards et al., 2016). Such finding suggests that multiple mechanisms are present for Rrp2 phosphorylation, but this study does not dispute the data from earlier studies showing that acetate and acetyl phosphate play an important role in the activation of the Rrp2-RpoN-RpoS pathway during tick feeding (Xu et al., 2010). All these data support the notion that nutritional factor is one of the key player in regulating virulence gene expression of B. burgdorferi.
The Rrp2-RpoN-RpoS pathway is repressed when spirochetes migrate to the tick midgut from the vertebrate host (for review, see Radolf et al., 2012; Samuels, 2011). The signaling pathway that leads to this repression remains largely unknown. BadR is the repressor of rpoS, and it was proposed that phosphorylated sugar may be one of the signals (Miller et al., 2013). We previously showed that the metal ion manganese (Mn2+) represses rpoS expression while zinc (Zn2+) enhances rpoS expression. Recently, Wang et al., reported that another transition metal, copper (Cu2+) also plays a represses rpoS expression via inhibiting BosR binding to DNA. Given that oppA4 is expressed ticks during the larval and nymphal blood meals (Groshong et al., 2017; Wang et al., 2002), and inactivation of oppA4 leads to increased activation of the Rrp2-RpoN-RpoS pathway, we postulate that the ligand that OppA4 transports may be a cue to downregulates the Rrp2-RpoN-RpoS pathway during the acquisition phase of the enzootic cycle of B. burgdorferi.
Several regulators were known to regulate rpoS and subsequently, ospC expression in B. burgdorferi (Radolf et al., 2012; Samuels, 2011; Stevenson and Seshu, 2017). In this study, we only examined two major transcription factors, BosR and BadR, and uncovered that the OppA4 negatively regulates the rpoS and ospC levels via alteration of bosR expression. Note that the complemented strain had lower levels of bosR than that of wild-type strain (Fig. 5). This is likely due to the high level of constitutive expression of oppA4 driven by the flaB promoter in the complemented strain. We did not examine the level of rrp2 and rpoN in the oppA4 mutant, as activation of rpoS by Rrp2 and RpoN is controlled by phosphorylation of Rrp2. We and others recently showed that under the in vitro cultivation conditions, Rrp2 phosphorylation appears to be constitutive, as Rrp2 phosphorylation is vital for B. burgdorferi replication (Groshong et al., 2012; Ouyang and Zhou, 2016; Yin et al., 2016). How OppA4 negatively regulates bosR expression warrants further investigation.
In this study, we showed that the oppA4 mutant is capable of infecting mice via needle infection, indicating that OppA4 is not essential for mammalian infection. It remains possible that the oppA4 mutant has reduced infectivity. Performing ID50 experiment or competition assay by co-infection wild-type B. burgdorferi are needed to determine whether OppA4 contributes partially to mammalian infection of B. burgdorferi. Furthermore, whether OppA4 is required for mammalian infection via tick bite remains to be determined. If OppA4 plays a role in the tick vector, the outcomes for infectivity via tick infection would be different from that of needle inoculation. One caveat of this study is that the complete enzootic cycle of the oppA4 mutant was not performed. The parental strain of B. burgdorferi used in this study is 5A18NP1, which does not contain the endogenous plasmids lp56 and lp28–4: both plasmids were reported to contribute to colonize ticks (Jacobs et al., 2006; Strother et al., 2005). Given that expression of oppA4 is preferably expressed in ticks, we postulate that OppA4 plays a more prominent role in the tick part of the cycle of B. burgdorferi. Generating an oppA4 mutant in a B. burgdorferi strain with complete endogenous plasmids is warranted to examine the exact role of OppA4 in the enzootic cycle of B. burgdorferi.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Bono JL, Tilly K, Stevenson B, Hogan D, Rosa P, 1998. Oligopeptide permease in Borrelia burgdorferi: putative peptide-binding components encoded by both chromosomal and plasmid loci. Microbiology. 144, 1033–1044. [DOI] [PubMed] [Google Scholar]
- Bunikis I, Kutschan-Bunikis S, Bonde M, Bergström S, 2011. Multiplex PCR as a tool for validating plasmid content of Borrelia burgdorferi. J Microbiol Methods. 86, 243–247. [DOI] [PubMed] [Google Scholar]
- Caimano MJ, Drecktrah D, Kung F, Samuels DS, 2016. Interaction of the Lyme disease spirochete with its tick vector. Cellular microbiol. 18, 919–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casjens S, Palmer N, van Vugt R, Huang WM, Stevenson B, Rosa P, Lathigra R, Sutton g., Peterson J, Dodson RJ, Haft D, Hickey E, Gwinn M, White O, Fraser CM, 2000. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol Microbiol. 35, 490–516. [DOI] [PubMed] [Google Scholar]
- de Silva AM, Fikrig E, 1995. Growth and migration of Borrelia burgdorferi in Ixodes ticks during blood feeding. Am J Trop Med Hyg. 53, 397–404. [DOI] [PubMed] [Google Scholar]
- Drecktrah D, Lybecker M, Popitsch N, Rescheneder P, Hall LS, Samuels DS, 2015. The Borrelia burgdorferi RelA/SpoT homolog and stringent response regulate survival in the tick vector and global gene expression during starvation. PLoS Pathog. 11, e1005160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulebohn DP, Hayes BM, Rosa PA, 2014. Global Repression of host-associated genes of the Lyme Disease spirochete through post-transcriptional modulation of the alternative sigma factor RpoS. PLoS ONE. 9, e93141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteve-Gassent MD, Smith TC II, Small CM, Thomas DP, Seshu J, 2015. Absence of sodA increases the levels of oxidation of key metabolic determinants of Borrelia burgdorferi. PLoS ONE. 10, e0136707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairman JW, Abendroth J, Sankaran B, Staker BL, Structural, S. (SSGCID), G.C.f.I.D., 2012. X-ray crystal structure of a periplasmic oligopeptide-binding protein/oligopeptide ABC transporter (OppAIV) from Borrelia burgdorferi. https://proteindiffraction.org/project/4gl8/.
- Fraser CM, Casjens S, Huang WM, Sutton GG, Clayton R, Lathigra R, White O, Ketchum KA, Dodson R, Hickey EK, Gwinn M, Dougherty B, Tomb JF, Fleischmann RD, Richardson D, Peterson J, Kerlavage AR, Quackenbush J, Salzberg S, Hanson M, van Vugt R, Palmer N, Adams MD, Gocayne J, Weidman J, Utterback T, Watthey L, McDonald L, Artiach P, Bowman C, Garland S, Fujii C, Cotton MD, Horst K, Roberts K, Hatch B, Smith HO, Venter JC, 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature. 390:580–586. [DOI] [PubMed] [Google Scholar]
- Garai P, Chandra K, Chakravortty D, 2017. Bacterial peptide transporters: Messengers of nutrition to virulence. Virulence. 8, 297–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groshong AM, Dey A, Bezsonova I, Caimano MJ, Radolf JD, 2017. Peptide uptake is essential for Borrelia burgdorferi viability and involves structural and regulatory complexity of its oligopeptide transporter. mBio. 8, pii: e02047–17. doi: 10.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groshong AM, Gibbons NE, Yang XF, Blevins JS, 2012. Rrp2, a prokaryotic enhancer-like binding protein, is essential for viability of Borrelia burgdorferi. J Bacteriol. 194, 3336–3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M, Zhang J-J, Ye M, Lou Y, Yang XF, 2014. The cyclic di-GMP receptor PlzA controls virulence gene expression through RpoS in Borrelia burgdorferi. Infect Immun. 82, 445–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hübner A, Yang X, Nolen DM, Popova TG, Cabello FC, Norgard MV, 2001. Expression of Borrelia burgdorferi OspC and DbpA is controlled by a RpoN-RpoS regulatory pathway. Proc Natl Acad Sci U S A. 98, 12724–12729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde JA, Shaw DK, Smith Iii R, Trzeciakowski JP, Skare JT, 2009. The BosR regulatory protein of Borrelia burgdorferi interfaces with the RpoS regulatory pathway and modulates both the oxidative stress response and pathogenic properties of the Lyme disease spirochete. Mol Microbiol. 74, 1344–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde JA, Trzeciakowski JP, Skare JT, 2007. Borrelia burgdorferi alters its gene expression and antigenic profile in response to CO2 levels. J. Bacteriol 189, 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer R, Caimano MJ, Luthra A, Axline D, Corona A, Iacobas DA, Radolf JD, Schwartz I, 2015. Stage-specific global alterations in the transcriptomes of Lyme disease spirochetes during tick feeding and following mammalian host adaptation. Mol Microbiol. 95, 509–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs MB, Norris SJ, Phillippi-Falkenstein KM, Philipp MT, 2006. Infectivity of the Highly Transformable BBE02- lp56- Mutant of Borrelia burgdorferi, the Lyme Disease Spirochete, via Ticks. Infect Immun. 74, 3678–3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabata H, Norris SJ, Watanabe H, 2004. BBE02 disruption mutants of Borrelia burgdorferi B31 have a highly transformable, infectious phenotype. Infect Immun. 72, 7147–7154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwaik YA, Bumann D, 2013. Microbial quest for food in vivo: ‘Nutritional virulence’ as an emerging paradigm. Cell Microbiol. 15, 882–890. [DOI] [PubMed] [Google Scholar]
- Lane RS, Piesman J, Burgdorfer W, 1991. Lyme borreliosis: relation of its causative agent to its vectors and hosts in North America and Europe. Annu Rev Entomol. 36, 587–609. [DOI] [PubMed] [Google Scholar]
- Lin B, Short SA, Eskildsen M, Klempner MS, Hu LT, 2001. Functional testing of putative oligopeptide permease (Opp) proteins of Borrelia burgdorferi: a complementation model in opp(−) Escherichia coli. Biochim Biophys Acta. 1499, 222–31. [DOI] [PubMed] [Google Scholar]
- Lybecker MC, Samuels DS, 2007. Temperature-induced regulation of RpoS by a small RNA in Borrelia burgdorferi. Mol Microbiol. 64, 1075–1089. [DOI] [PubMed] [Google Scholar]
- Medrano MS, Ding Y, Wang XG, Lu P, Coburn J, Hu LT, 2007. Regulators of expression of the oligopeptide permease A proteins of Borrelia burgdorferi. J Bacteriol. 189, 2653–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CL, Karna SLR, Seshu J, 2013. Borrelia host adaptation regulator (BadR) regulates rpoS to modulate host adaptation and virulence factors in Borrelia burgdorferi. Mol Microbiol. 88, 105–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monnet V, 2003. Bacterial oligopeptide-binding proteins. CMLS, Cell. Mol. Life Sci. 60, 2100–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang Z, Blevins JS, Norgard MV, 2008. Transcriptional interplay among the regulators Rrp2, RpoN, and RpoS in Borrelia burgdorferi. Microbiology. 154, 2641–2658. [DOI] [PubMed] [Google Scholar]
- Ouyang Z, Deka RK, Norgard MV, 2011. BosR (BB0647) controls the RpoN-RpoS regulatory pathway and virulence expression in Borrelia burgdorferi by a novel DNA-binding mechanism. PLoS Pathog. 7, e1001272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang Z, Kumar M, Kariu T, Haq S, Goldberg M, Pal U, Norgard MV, 2009. BosR (BB0647) governs virulence expression in Borrelia burgdorferi. Mol Microbiol. 74, 1331–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang Z, Zhou J, 2015. BadR (BB0693) controls growth phase-dependent induction of rpoS and bosR in Borrelia burgdorferi via recognizing TAAAATAT motifs. Mol Microbiol. 98, 1147–1167. [DOI] [PubMed] [Google Scholar]
- Ouyang Z, Zhou J, 2016. The putative Walker A and Walker B motifs of Rrp2 are required for the growth of Borrelia burgdorferi. Mol Microbiol. 103, 86–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radolf JD, Caimano MJ, Stevenson B, Hu LT, 2012. Of ticks, mice and men: understanding the dual-host lifestyle of Lyme disease spirochaetes. Nat Rev Micro. 10, 87–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raju BV, Esteve-Gassent MD, Karna SL, Miller CL, Van Laar TA, Seshu J, 2011. Oligopeptide permease A5 modulates vertebrate host-specific adaptation of Borrelia burgdorferi. Infect Immun. 79, 3407–3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards CL, Lawrence KA, Su H, Yang Y, Yang XF, Dulebohn DP, Gherardini FC, 2015. Acetyl-phosphate is not a global regulatory bridge between virulence and central metabolism in Borrelia burgdorferi. PLoS One. 10, e0144472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers EA, Terekhova D, Zhang H, Hovis KM, Schwartz I, Marconi RT, 2009. Rrp1, a cyclic-di-GMP-producing response regulator, is an important regulator of Borrelia burgdorferi core cellular functions. Mol Microbiol. 71, 1551–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels DS, 2011. Gene regulation in Borrelia burgdorferi. Annu Rev Microbiol 65, 479–499. [DOI] [PubMed] [Google Scholar]
- Schwan TG, Piesman J, 2000. Temporal changes in outer surface proteins A and C of the Lyme disease-associated spirochete, Borrelia burgdorferi, during the chain of infection in ticks and mice. J Clin Microbiol. 38, 382–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson B, Seshu J, 2017. Regulation of Gene and Protein Expression in the Lyme Disease Spirochete In: Curr Top Microbiol Immunol. Springer, Berlin, Heidelberg: 10.1007/82_2017_49. [DOI] [PubMed] [Google Scholar]
- Strother KO, Broadwater A, de Silva A, 2005. Plasmid requirements for infection of ticks by Borrelia burgdorferi. Vector Borne Zoonotic Dis. 5, 237–245. [DOI] [PubMed] [Google Scholar]
- Sze CW, Smith A, Choi YH, Yang X, Pal U, Yu A, Li C, 2013. Study of the response regulator Rrp1 reveals its regulatory role in chitobiose utilization and virulence of Borrelia burgdorferi. Infect Immun. 81, 1775–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas GH, 2010. Homes for the orphans: utilization of multiple substrate-binding proteins by ABC transporters. Mol Microbiol. 75, 6–9. [DOI] [PubMed] [Google Scholar]
- Troxell B, Yang XF, 2013. Metal-dependent gene regulation in the causative agent of Lyme disease. Front Cell Infect Microbiol. 3, 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troxell B, Ye M, Yang Y, Carrasco SE, Lou Y, Yang XF, 2013. Manganese and zinc regulate virulence determinants in Borrelia burgdorferi. Infect Immun. 81, 2743–2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troxell B, Zhang J-J, Bourret TJ, Zeng MY, Blum J, Gherardini F, Hassan HM, Yang XF, 2014. Pyruvate protects pathogenic spirochetes from H2O2 killing. PLoS ONE. 9, e84625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troy EB, Lin T, Gao L, Lazinski DW, Lundt M, Camilli A, Norris S,J, Hu L, 2016. Global Tnseq analysis of carbohydrate utilization and vertebrate infectivity of Borrelia burgdorferi. Mol Microbiol. 101, 1003–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Laar TA, Lin Y-H, Miller CL, Karna SLR, Chambers JP, Seshu J, 2012. Effect of levels of acetate on the mevalonate pathway of Borrelia burgdorferi. PLoS One. 7, e38171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Yu Z, Santangelo TJ, Olesik J, Wang Y, Heldwein E, Li X, 2017. BosR is a novel Fur family member responsive to copper and regulating copper homeostasis in Borrelia burgdorferi. J Bacteriol. 199:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XG, Kidder JM, Scagliotti JP, Klempner MS, Noring R, Hu LT, 2004. Analysis of differences in the functional properties of the substrate binding proteins of the Borrelia burgdorferi oligopeptide permease (opp) operon. J Bacteriol. 186, 51–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XG, Lin B, Kidder JM, Telford S, Hu LT, 2002. Effects of environmental changes on expression of the oligopeptide permease (opp) genes of Borrelia burgdorferi. J Bacteriol. 184, 6198–6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang X, Yang Y, Du J, Lin T, Chen T, Yang XF, Lou Y, 2017. Investigation of ospC expression variation among Borrelia burgdorferi strains. Front Cell Infect Microbiol. 7, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Caimano MJ, Lin T, He M, Radolf JD, Norris SJ, Gheradini F, Wolfe AJ, Yang XF, 2010. Role of acetyl-phosphate in activation of the Rrp2-RpoN-RpoS pathway in Borrelia burgdorferi. PLoS Pathog. 6, e1001104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Coleman AS, Anguita J, Pal U, 2009. A chromosomally encoded virulence factor protects the Lyme disease pathogen against host-adaptive immunity. PLoS pathogens. 5, e1000326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XF, Alani SM, Norgard MV, 2003. The response regulator Rrp2 is essential for the expression of major membrane lipoproteins in Borrelia burgdorferi. Proc Natl Acad Sci U S A. 100, 11001–11006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XF, Lybecker MC, Pal U, Alani SM, Blevins J, Revel AT, Samuels DS, Norgard MV, 2005. Analysis of the ospC regulatory element controlled by the RpoN-RpoS regulatory pathway in Borrelia burgdorferi. J Bacteriol. 187, 4822–4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye M, Zhang J-J, Fang X, Lawlis G, Troxell B, Zhou Y, Gomelsky M, Lou Y, Yang X, 2014. DhhP, a c-di-AMP phosphodiesterase of Borrelia burgdorferi, is essential for cell growth and virulence. Infect Immun. 82, 1840–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye M, Zhou Y, Lou Y, Yang XF, 2016. Genome reduction of Borrelia burgdorferi: two TCS signaling pathways for two distinct host habitats. SCI CHINA LIFE SCI. 59, 19–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Youyun Y, Xiang X, Wang Q, Yang Z-N, Blevins J, Lou Y, Yang XF, 2016. Insight into the dual functions of bacterial enhancer-binding protein Rrp2 of Borrelia burgdorferi. J Bacteriol. 198, 1543–52. [DOI] [PMC free article] [PubMed] [Google Scholar]