Abstract
Despite its revolutionary success in hematological malignancies, chimeric antigen receptor (CAR) T cell therapy faces disappointing clinical results in solid tumors. The poor efficacy has been partially attributed to the lack of understanding in how CAR-T cells function in a solid tumor microenvironment. Hypoxia plays a critical role in cancer progression and immune editing, which potentially results in solid tumors escaping immunosurveillance and CAR-T cell-mediated cytotoxicity. Mechanistic studies of CAR-T cell biology in a physiological environment has been limited by the complexity of tumor-immune interactions in clinical and animal models, as well as by a lack of reliable in vitro models. We have engineered a microdevice platform that recapitulates a three-dimensional tumor section with a gradient of oxygen and integrates fluidic channels surrounding the tumor for CAR-T cell delivery. Our design allows for the evaluation of CAR-T cell cytotoxicity and infiltration in the heterogeneous oxygen landscape of in vivo solid tumors at a previously unachievable scale in vitro.
Keywords: Hypoxia, solid tumor, chimeric antigen receptor, immunotherapy, immune checkpoint, ovarian cancer
Introduction:
Adoptive chimeric antigen receptor T (CAR-T) cell therapy has revolutionized the treatment of hematological diseases, such as lymphoma, leukemia, and multiple myeloma. [1] In this therapy, T cells are genetically engineered with synthetic receptors (most often composed of the single chain variable fragment (scFv) of an antibody) that recognize tumor-associated antigens (TAAs) expressed on the target cell surface. The engagement activates the intracellular T cell receptor (TCR) signaling and costimulatory domains, and triggers T cell activation to elicit a TAA-specific tumor killing.[2],[3] Remarkably, two CAR-T cell therapies have been recently approved by the Food and Drug Administration (FDA), and up to 90% complete remission has been observed using anti-CD19 CAR-T cells in B cell acute lymphoblastic leukemia (B-ALL).[4] Yet, translation of CAR-T cells to solid malignancies has been met with disappointing results.[5–8] The tumor microenvironment (TME) is thought to pose a physical barrier to T cell trafficking and threaten their survival/function through immunosuppression.[9] However, there is still a lack of understanding on how the TME modulates CAR-T infiltration and function, and how to effectively overcome its adverse effects.
Solid tumors establish a unique physical and biochemical network that contributes to immune evasion. Unlike hematologic malignancies in a fluid landscape, solid tumors consist of cancer and stromal cells embedded in a dense network of extracellular matrix (ECM) with intratumoral fluid pressure.[5] In order to elicit TAA-specific cytotoxicity, CAR-T cells not only have to infiltrate the physical barriers, but also overcome additional challenges from immunosuppressive cancer- and TME-derived factors.[3] For instance, regulatory T cells (Tregs) and suppressive soluble factors and cytokines such as prostaglandin E2 (PGE2), adenosine, and TGF-β, exert a hostile environment for CAR-T cell functions.[5,10] Further, immunosuppressive molecules such as programmed death 1 (PD-1) and its ligand, PD-L1, compromise cytotoxic T cell cytokine secretion, growth, and cytotoxicity against tumor cells.[11–13] Importantly, hypoxia, a major hallmark of solid tumors, enhances chemoresistance[14] and immunosuppression in the TME,[15,16] and correlates with poor patient outcomes.[17] Cancer cells adapt to hypoxic environments by altering their biology accordingly to oxygen availability, resulting in malignant progression through proliferation, evasion of growth suppressors, resistance to cell death, replicative immortality, induction of angiogenesis, and activation of invasion and metastasis.[18] Activation of hypoxia-induced factor-1α (HIF-1α), a major regulator of hypoxia signaling, has been shown to inhibit anti-tumor cytokines such as TNF and IFN-γ,[19,20] up-regulate PD-L1,[15] and enhance secretion of immunosuppressive cytokines.[21–25] Hypoxia further alters tumor metabolism and promotes glycolysis in tumor cells.[14] The competition for glucose between cancer cells and the glycolytic anti-tumor T cells hinders the penetration and functionality of therapeutic T cells in tumors, and activates the stress responses that lead to their anergy or death.[10,26–28]
Of solid tumors, ovarian cancer is the most lethal gynecological cancer in women of the United States.[29] Despite initial response against standard treatments such as surgical resection and chemotherapy, most patients relapse with recurrent metastases, leading to poor overall survival.[30] In addition, hypoxia has been reported to increase ovarian cancer invasiveness, enhance immunosuppression through various factors such as VEGF, IL-10, and STAT3, and induce chemoresistance.[31–33] With respect to CAR-T immunotherapy, clinical trials showed no reduction in tumor burden and a lack of T cell persistence[6] despite promising preclinical results of CAR-directed T cells in ovarian cancer.[34,35] Such apparent disagreement between conventional preclinical models and clinical trials highlights the importance of engineering in vitro models that more faithfully reflect T cell-TME interactions under physiologically relevant levels of oxygen.
Recently, engineered in vitro platforms have emerged as powerful tools for evaluation of tumor immunology and immunotherapy, which allow for potential mechanistic study or high-throughput testing of immunotherapeutic regimens.[36–38] In regards to tumor hypoxia, co-cultures of tumor spheroids and lymphokine-activated killer cells revealed slower cell lysis compared to single cell suspensions.[39] Further, tumor-infiltrating lymphocytes and CD8+ T cells against human bladder and lung cancer spheroids were shown to be inefficient at cytokine release compared to traditional cell culture.[40,41] Despite their ability to induce a hypoxic gradient resembling in vivo tumors, spheroids are generally incompatible with high-content analysis.[42,43] They also lack an ECM network that would facilitate or impede cell penetration.[44] On the other hand, microfluidic tumor models that spatially isolate the tumor and immune compartments have enabled the evaluation of immune cell infiltration such as dendritic cell motility towards the tumor chamber for antigen cross-presentation.[45] Such models have also allowed for evaluation of the infiltration and cytotoxicity of T cell receptor (TCR)-engineered T cells under uniform normoxic and hypoxic conditions[46], which however does not involve the exploration of the impact of the oxygen gradients on the immune cell infiltration and cell killing. To date, there has not been a 3-D solid tumor model assessing CAR-T cell therapy under a gradient of hypoxia as seen in solid tumors.
In this study, we constructed a tumor model of human ovarian cancer cells in vitro with an oxygen gradient generated by cellular metabolism, by embedding cancer cells in a 3-D micropattern in a photo-crosslinked hydrogel and micromilled hypoxia device.[47] The platform has significant advantages over our previously reported work[47] by incorporating cell-ECM interactions in a 3-D hydrogel, allowing biomimicry of tumor masses. Further, CAR-T cells are delivered through microfluidic channels surrounding the tumor mass, and spatiotemporal examination of CAR-T cell infiltration and cytotoxicity within the hydrogel is achieved. We present the device and platform as versatile tools for gaining insights into the activities of CAR-T cells in solid tumors, and for designing more effective and personalized cancer immunotherapy.
Results:
An oxygen gradient can be engineered in a three-dimensional (3-D) tumor model
Solid tumors consist of a dense ECM network and a heterogeneous landscape of oxygen levels, which form a physical barrier to CAR-T cell infiltration as well as create an immunosuppressive network of soluble factors.[3,48] To investigate these immune-evading mechanisms and provide a fast-turnaround testing platform for CAR-T cell therapy in vitro, we introduced a 3-D micropatterned tumor model with cancer cells embedded in an ECM gel and assembled between two diffusion barriers in a hypoxia microdevice[49,47] (Figure 1A). To allow for CAR-T cell delivery to the tumor, we fabricated a polydimethylsiloxane (PDMS) chamber/channel device which is plasma-bonded to a glass slide and tightly assembled with the milled hypoxia cap (Figure 1B). Gelatin methacryloyl (GelMA), a photo-crosslinkable hydrogel derived from natural collagen, was chosen as a model ECM due to its biocompatibility, functionality and mechanical tunability.[50] Filtered ultra-violet (UV) light at a wavelength of 375 ± 14 nm was used to crosslink GelMA, with lithium phenyl-2,4,6-trimethylbenzoylphosphinate (LAP) as the photo-initiator.[49,51] Tumor tissues have a wide range of stiffness, depending on tumor grade, stage, and size.[52–54] Using a uniaxial compression test, we characterized the Young’s modulus of the hydrogel, which increased linearly with UV-curing time (R2 = 0.9909; Figure 1C). We chose 120 seconds of curing time, which yields approximately 6 kPa and is in range of multiple cancer matrices.[52,55,56] We further assessed cell viability upon UV crosslinking because our platform relies on cellular oxygen consumption for hypoxia induction. Cell viability remained above 90% after 3 days of culture in both normoxic and hypoxic conditions with no statistical significance between oxygen conditions (Figure 1D). To examine whether the ECM porosity will allow for CAR-T cell motility and infiltration, scanning electron microscopy (SEM) was used to characterize the pore size of the GelMA construct (Figure 1E). The average pore size was 8.47 µm ± 3.02 µm (Figure 1F), as measured by the diameter of randomly selected pores.
Figure 1.
Physical characteristics of the 3-D tumor model that recapitulates an oxygen gradient and matrix microenvironment for T cell infiltration. (A) Illustration of the working principle. (B) Schematics of the microdevice assembly. A PDMS fluidic component is plasma-bonded to a glass slide before assembling with a milled polycarbonate (PC) cap to form a cell-laden GelMA layer under the oxygen barrier pillar. (C) Stiffness of the GelMA hydrogels in relation to UV-curing time (R2 = 0.9909, linear regression). (D) Viability of GelMA-encapsulated SKOV3 human ovarian cancer cells under hypoxic or normoxic incubation in device (n = 3; n.s.: not significant, p < 0.05 by Student’s t-test). (E) Scanning electron microscopy image (scale bar = 10 µm), and (F) the average pore size of the 120-second UV-crosslinked GelMA hydrogel.
An oxygen gradient is established in the 3-D tumor model over time
The spatial and temporal profiles of expected oxygen in the device were first simulated with COMSOL Multiphysics® during the device design phase. At steady state, a 100 µm-thick tumor bulk in the device had a hypoxic core and a gradient that rapidly approached above physioxic levels[57] at the periphery (Figure 2A). Three points of interest were selected along the base of the tumor section: the edge (A), the intermediate region (B), and the center (C) in relation to the circular pillar geometry (Figure 2A). Temporal analysis demonstrated that oxygen levels dropped quickly within the first 10 minutes of the device assembly, resembling those at 24 hours (or 1440 minutes) (Figure 2B). Next, we simulated the effect of cell density on the oxygen gradient at 24 hours after device assembly. As expected, the spatial gradient of oxygen evolves gradually at lower cell density (e.g. 1 million cells/mL), whereas higher cell densities show a steeper slope from point A to C (Figure 2C). At the densities of 10 million cells/mL and above, oxygen level reaches near-zero levels at the center of the device. Interestingly, at these densities, the oxygen gradient is almost absent in the core (within 1/3 of the radius) while it rapidly approaches normoxic levels from 2/3 of the radius to the edge, indicating that the samples may eventually develop a necrotic core upon longer culture time (Figure 2C). As our microdevice is milled with a < 28 µm precision[58] which may lead to variations in the hydrogel thickness controlled by the difference of heights between the barrier and reference pillars, we further evaluated the effect of hydrogel thickness on the oxygen gradient through COMSOL simulations. For a wide range of gap sizes (50 – 150 µm), the oxygen gradients are strikingly similar, with slightly greater variability towards the micropattern periphery (Figure 2D).
Figure 2.
Characterization of the oxygen gradient. (A) Heat map illustrating the oxygen levels at the steady state in the hypoxic microdevice. (B) Evolvement of oxygen concentration in the microdevice within 24 hours of device assembly. Oxygen gradient profiles in relations to (C) the cell density and (D) the thickness of the cell-laden hydrogel at the 10 million/mL density. (E) Fluorescent images (scale bar = 1,000 µm) and (F) measurements with microparticle-based oxygen sensors without or with cells in the device (n = 4 with cells, n = 2 without cells). (G) Measured/calculated vs. simulated oxygen profiles. (H) Expression of Glut-1, a hypoxia marker, in the 3-D tumor model incubated without or with the hypoxic microdevice (scale bar = 1,000 µm), and (I) a radial analysis of high Glut-1 expression across cells (n = 190, 185, 249 for normoxic center, intermediate, and edge, respectively, and n = 163, 183, 231 for hypoxic center, intermediate, and edge, respectively; n.s.: not significant, *: p < 0.05 by Student’s t-test).
To further characterize and validate the hypoxia gradient under cell culture conditions, we embedded fluorescence-based oxygen sensors into PDMS, which was coated onto the oxygen barrier pillar.[47,59] The ruthenium compound absorbed in the silica microparticle sensors showed enhanced fluorescent signal only when incubated with cell-laden GelMA, suggesting a low oxygen environment induced by cellular oxygen consumption (Figure 2E). In addition, the enhanced signal followed a radial distribution across the oxygen barrier pillar (Figure 2F), which corresponded to a gradient of oxygen concentrations that closely resembled our simulation results (Pearson’s correlation coefficient, r = 0.8895; Figure 2G). Next, glucose transporter-1 (Glut-1), an intrinsic cellular marker of hypoxia,[60] was studied to confirm that the suspended cells in the hydrogel can create and respond to the oxygen gradient in the device. Upon immunostaining, Glut-1 was up-regulated in hypoxia device-incubated samples, with signals from the tumor cores (corresponding to the inner 2000 µm radius of the micropattern) significantly different between the normoxia and hypoxia samples (Figure 2H, I).
CAR-T cell-induced cytotoxicity is spatially modulated in the hypoxic tumor model
Next, we investigated whether CAR-T cells have altered therapeutic behavior under a hypoxic gradient compared to normoxia. We used HER2-targeting 4D5 scFv CAR-T cells (CAR-T), where the CAR is derived from a clinically-used anti-HER2 antibody, trastuzumab, and has been previously characterized for their activation with cancer cell lines of differing HER2 expression[61]. We confirmed the induction of antigen-specific cytotoxicity by the CAR-T cells using a degranulation assay and IFN-γ secretion assay (Figure S1). 3-D tumor sections were incubated under normoxic (no cap) or hypoxic (cap) conditions for 24 hours, where the normoxic devices are essentially partially “disassembled” hypoxic devices after GelMA crosslinking, with media retained within the channel through surface tension (Figure 3A). Then, CAR-T and NT-T in fresh media were loaded at a 20:1 effector:target (E:T) ratio through microfluidic channels and co-incubated with the 3-D micropatterned tumor consisting of HER2+ SKOV3 ovarian cancer cells for 24 hours. After 24 hours of co-culture, embedded SKOV3 cancer cells were assessed for viability with a live-dead stain (with calcein AM and propidium iodide, respectively). The staining revealed sporadic cell death without T cells in both normoxic and hypoxic samples (top row; Figure 3B), consistent with cellular viability data in Figure 1D. Treatment with NT-T cells did not yield any additional increase in cell death (middle row; Figure 3B). On the other hand, CAR-T cells increased cell killing in both oxygenation conditions at the same E:T ratio, while an up-regulation of cytotoxicity was observed in the periphery of hypoxic micropatterns (bottom row; Figure 3B). Analysis of the number of PI-positive (dead) cells within concentric rings with radius Δr from the center of each micropattern confirmed this trend, although not statistically significant (blue curves; Figure 3C,D). To modulate the hypoxic gradient, similar analysis was carried out in tumor sections micropatterned at 1 million cells/mL. the core region now corresponds to 9.52 – 10% oxygen within a 1000 µm radii, and 13.65% to 19.93% oxygen at the edges (red line; Figure 2C). Cytotoxicity was minimal throughout the construct, with no preferential cell killing at the edge or at the core (Figure S2). Next, we aimed to compare CAR-T cell-induced cytotoxicity in our 3-D platform against conventional multi-well plate co-culture assays as well as at different E:T ratios. 2-D multi-well plate assays of CAR-T efficiency show significantly higher cytotoxicity at each E:T ratio compared to the dead cell proportion in our 3-D tumor bulks, except the 1:1 ratio in hypoxic condition (Figure. 3E). Intriguingly, the proportion of dead cells in 3-D micropatterns is not affected by increasing E:T ratios, while 2-D results suggest a positive correlation that saturates at 10:1 E:T ratio.
Figure 3.
T cell mediated killing in the 3-D tumor models. (A) Schematic of normoxic and hypoxic conditions in the platform. Upon crosslinking of cell-laden GelMA, media was flushed through the PDMS fluidics channels. The cap was then disassembled for normoxic conditons (top), while the construct remained assembled for hypoxic conditions (bottom). (B) Live/dead staining of the 3-D tumor models after 24 hours of culture without or with microfluidic channel-delivered T cells in normoxia and hypoxia devices (scale bar = 1,000 µm). Radial analysis of dead cell counts in (C) normoxia and (D) hypoxia devices (n = 9 for no treatment, n = 5 for NT-T and CAR-T). (E) Comparison of non-transduced and CAR-T cell-mediated cytotoxicity under 2-D vs. 3-D, normoxia vs. hypoxia, and three effector:target (E:T) ratios (n = 3; n.s.: not significant, ‡: p < 0.05 between compared conditions, and *: p < 0.05 compared to the corresponding 2-D culture under the same E:T ratio conditions; all comparisons done by one-way ANOVA).
PD-L1 expression is altered by hypoxia, but its blockade does not enhance CAR-T cytotoxicity
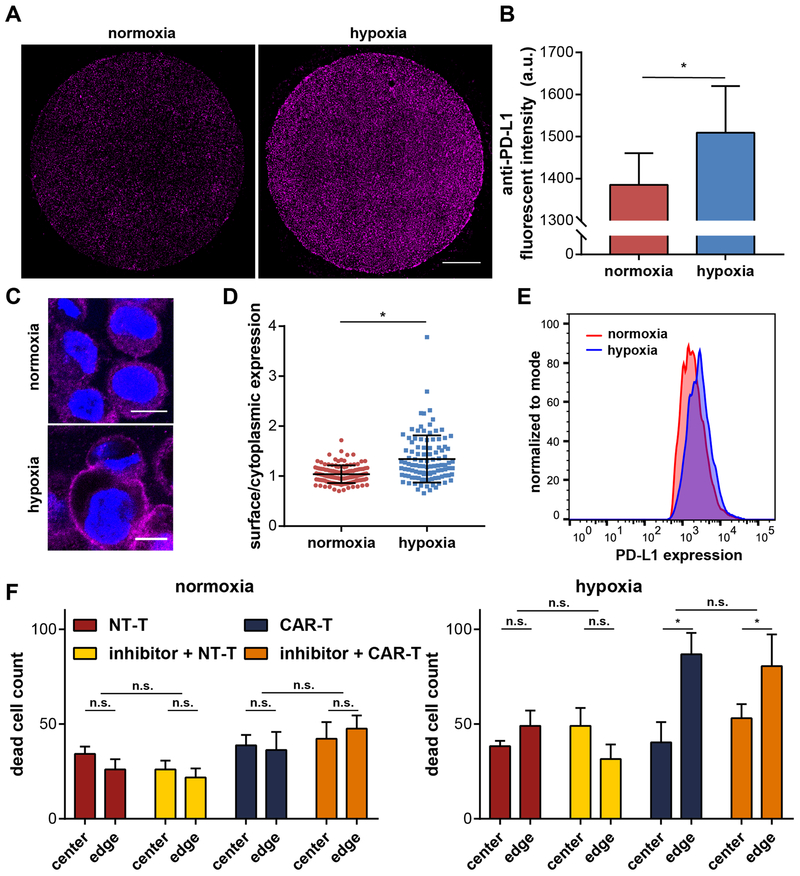
To understand the immunosuppression and the change in CAR-T induced cell killing under hypoxia, we investigated PD-L1 expression and blockade in our 3-D tumor model. PD-L1 is an immune suppressive molecule that is expressed by many cancer cells and regulates the cell cycle, survival, and proliferation of tumor-infiltrating T cells.[13,62–64] Fluorescence microscopy of our 3-D tumor models immunostained for PD-L1 reveal an overall up-regulation of the molecule under hypoxic conditions (Figure 4A, B). Under confocal microscopy, we further observe a difference in the sub-cellular distribution of PD-L1 between normoxia and hypoxia. At 24 hours, cells under hypoxia increased their surface expression of PD-L1, as compared to a more cytoplasmic profile under the normoxic condition (Figure 4C). We confirmed the difference by quantifying the ratio of the surface vs. cytoplasmic levels of PD-L1 in single cells (p < 0.0001 with Mann-Whitney test; Figure 4D). To validate our finding on PD-L1 surface expression, cells cultured in incubators with uniform normoxic (21% oxygen) and hypoxic (1% oxygen) environments for 24 hours were live-immunostained and assessed with flow cytometry. Indeed, cells up-regulated their surface PD-L1 expression upon hypoxia induction (Figure 4E). PD-L1 expression was evaluated over 72 hours in order to assess the temporal evolution of this immunosuppressive marker. Induction of surface PD-L1 expression was observed up to 72 hours with confocal imaging (Figure S3).
Figure 4.
PD-L1 characterization and inhibition in the 3-D tumor models. (A) PD-L1 immunostaining showing an up-regulation in the hypoxia microdevice (scale bar = 1,000 µm). (B) Quantification of the fluorescence intensity of the PD-L1 staining (n = 100, *: p < 0.05 by Mann-Whitney test). (C) Confocal images of single cancer cells in the tumor models under normoxia and hypoxia (scale bar = 10 µm). (D) Up-regulation of PD-L1 surface/cytoplasmic ratio by hypoxia (n = 129 for normoxia, n =105 for hypoxia; *: p < 0.05 by Mann-Whitney test). (E) Confirmation of PD-L1 surface up-regulation by flow cytometric analysis with live-stained cancer cells. (F) PD-L1 inhibition has negligible effect on NT-T and CAR-T killing behavior and efficacy in the 3-D tumor models (n = 6; n.s.: not significant, *: p < 0.05 by one-way ANOVA).
Due to its immunosuppressive role, we reasoned that PD-L1 inhibition may rescue the low CAR-T induced cytotoxicity in the 3-D tumor models. Cancer cells were micropatterned into 3-D bulks 24 hours before combination treatment of T cell co-incubation and PD-L1 inhibition. Combination treatment of PD-L1 inhibition and CAR-T cells for 24 hours did not yield any significant therapeutic benefit under normoxic or hypoxic conditions (blue versus orange bars; Figure 4F). We still observed elevated levels of CAR-specific cytotoxicity at the edges of hypoxic micropatterns, although this trend was also not enhanced by the addition of PD-L1 inhibition. We then assessed the inhibitor in traditional 2-D cytotoxicity assays incubated at normoxic or hypoxic incubators. 2-D assays also confirmed the lack of enhanced cytotoxicity upon PD-L1 inhibition (Figure S4).
Oxygen condition regulates temporal evolution of CAR-T cell function
To evaluate the longer term functionality of CAR-T cells in the TME, we extended the co-culture study to 72 hours in the 3-D tumor models. After 24 hours, CAR-T cells exhibited increased cytotoxicity against cancer cells at the periphery of the hypoxic tumors (Figure 3B, D; Figure 5A). Extending incubation time to 48 hours enhanced this trend in hypoxic samples, while also improving peripheral cytotoxicity in the normoxic samples. Notably, hypoxic samples treated with CAR-T cells showed elevated cytotoxicity in the hypoxic core, which is absent in the samples treated with non-transduced T cells (Figure 5D, E) nor in control samples without T cells (Figure 5F, G). At 72 hours, extensive CAR-T cell-induced cytotoxicity is observed at the periphery of the hypoxic samples, while it is uniformly enhanced across the normoxic samples (Figure 5A-C). Importantly, the degree of overall cell killing in normoxic and hypoxic counterparts become comparable after 72 hours (blue lines; Figure 5B, C; Figure S5A-C) and between CAR-T and NT-T (Figure 5B-E). In addition, we averaged the number of dead cells within concentric circles with Δr of 500 µm at the inner, intermediate, and outer regions, which correspond to oxygen levels of 0.2 – 0.3%, 1.5 – 4%, and 6 – 17%, respectively, and assessed statistical significance. Significant difference at the edge was observed between normoxic and hypoxic conditions at 48 hours and 72 hours, and at the intermediate zone at 72 hours (Figure S5D-F). As a possible mechanism of CAR-T cell-induced cytotoxicity across the tumor section, we immunostained the samples for granzyme B, a serine proteinase that is secreted by cytotoxic leukocytes to induce apoptosis in cancer cells[65]. Confocal imaging of cells at the hydrogel center and edge revealed that cells at the edge consistently express granzyme B at 24- and 72-hour, while such staining is absent at the center (Figure S6). To evaluate the origin of granzyme B, we co-stained samples with CD45. Interestingly, we observed granzyme B signal from both CD45- cancer cells and CD45+ immune cells at the hydrogel edge. The intensity of granzyme B staining also appeared to be higher in CAR-T than NT-T, and the pattern was consistent regardless of oxygenation conditions, implying that granzyme B is not influenced by the oxygen levels.
Figure 5.
Temporal analysis of CAR-T cell-induced cytotoxicity. (A) Live/dead staining of the 3-D tumor models incubated in normoxia or hypoxia with NT-T or CAR-T cells at a 20:1 E:T ratio for 24, 48, and 72 hours (scale bar = 1,000 µm). The number of dead cells from CAR-T cell-treated conditions were then radially quantified for the (B) normoxic and (C) hypoxic microdevice conditions. Control samples were treated with NT-T cells under (D) normoxia and (E) hypoxia or no T cells under (F) normoxia and (G) hypoxia (n = 4 for all conditions).
CAR-T infiltration is limited in the 3-D tumor bulk, with killing associated with cell-cell contact
The degree of tumor infiltrating T cells has been correlated with prognosis.[66–68] Therefore, we sought to quantify the degree of T cell infiltration in the 3-D tumor models. We carried out immunostaining and analysis on CD45, a pan-leukocyte surface marker, to investigate the degree of infiltrating T cells into cell-laden GelMA. Interestingly, we observe negligible infiltration throughout the micropattern, regardless of the oxygen levels after 24 hours (Figure 6A). Next, cancer-immune cell interactions were assessed under confocal microscopy (Figure 6B). The distance between the centroids of a CD45+ cell (T cell) and the nearest GelMA-embedded cancer cell (distinguished by CD45- and DAPI nuclei staining) was calculated as an indicator of the level of interaction between the two cell types, after 24 hours (Figure 6C) and 72 hours (Figure 6D) in culture. Compared to NT-T cells, CAR-T cells were significantly closer to SKOV3 cancer cells. CAR-T cells tightly associated with cancer cells in the hypoxic microdevice as soon as 24 hours, whereas their level of interaction was not statistically different from the NT-T cells under normoxic conditions, which explains the enhanced cytotoxicity pattern observed at the periphery of hypoxic micropatterns earlier (Figure 3B). CAR-T cells bind to cancer cells regardless of oxygen level by 72 hours of treatment, with increasingly visible infiltration into the GelMA hydrogel. In addition, we investigated the infiltration distance into the hydrogel bulk by quantifying the distance from the hydrogel boundary to the centroids of T cells. Positive distance was chosen to indicate infiltration into GelMA, while negative values indicate immune cells remaining outside of GelMA. CAR-T cells infiltrate into the periphery of the hydrogel as soon as 24 hours, but only in hypoxic conditions (Figure 6E). By 72 hours, CAR-T cells have infiltrated regardless of oxygenation condition (Figure 6F).
Figure 6.
Assessment of T cell infiltration into the tumor bulk. (A) 3-D micropatterns after treatment fixed and immuno-stained against CD45 for identification of T cells within the tumor bulk (scale bar = 1,000 µm). (B) Confocal images of NT-T and CAR-T cells in the device near the tumor-channel boundary after 72 hours of treatment (scale bar = 50 µm). Quantification of the distance between CD45+ cells and the nearest GelMA-embedded cancer cells at (C) 24 hours and (D) 72 hours (*: p < 0.05, n.s.: not significant, by one-way ANOVA). Quantification of the infiltration distance into or outside of GelMA was depicted as a positive or negative value, respectively, and depicted at (E) 24 hours and (F) 72 hours (*: p < 0.05, n.s.: not significant, by one-way ANOVA). (C, E) n = 8 for normoxia NT-T and CAR-T, n = 7 for hypoxia NT-T, n = 12 for hypoxia CAR-T. (D, F) n = 29 for normoxia NT-T, n = 20 for normoxia CAR-T, n = 13 for hypoxia NT-T, n = 25 for hypoxia CAR-T.
Discussion:
In this study, we have introduced a novel platform in which 3-D immune-tumor interactions can be studied under a hypoxic gradient. The tumor compartment can accurately generate and control oxygen gradients, while eliminating complex microfluidic handling of external oxygen control. Despite studying a 3-D tissue bulk, our methodology allows for high-content imaging-based analysis and has the potential for live cell tracking. Moreover, GelMA is a highly configurable scaffold hydrogel for 3-D cell cultures with mechanically tunable properties, allowing for investigation of matrix stiffness and its effect on tumor biology and therapeutic outcome. In our study, we crosslinked GelMA under approximately 2 mW/cm2 UV light for 120 seconds. It is important to note that varying matrix stiffness, most often correlated with ECM density and pore size, has been suggested to affect immune cell trafficking and functionality[69]. For example, the degrees of tumor-associated macrophages (TAMs) and cytotoxic T cell infiltrates are correlated and inversely correlated, respectively, with ECM rigidity[70,71]. Lymphocytes also migrated along collagen fibers through low-affinity interactions[72] and T cells were found localized in tumor sections with low collagen/fibronectin content, suggesting that extremely dense ECM regions exclude T cell infiltrates through a physical barrier[71]. However, the exact role of matrix stiffness on T cell infiltration remains an open question. To apply similar studies against different types of solid tumors at varying stages,[73,74] the elastic moduli of the resulting matrix can be easily modulated by changing the UV power intensity, curing time, concentration of GelMA used, and degree of methacrylation.[51] Similarly, the porosity of GelMA can be modulated by the concentration of photo-initiator to match the dense ECM network seen in tissues.[75] Additionally, tumor microenvironments also involve different types of ECM proteins. For instance, the dominant ECM types in ovarian cancer include fibrous proteins such as collagens I, IX and fibronectin, as well as proteoglycans such as hyaluronic acid (HA)[76]. We are currently investigating the effect of substituting/mixing GelMA with other types of natural or chemically modified ECM to evaluate their individual roles in T cell infiltration in our model. Compared to traditional 2-D models, 3-D cell culture models have been proven to provide insights that are better translated into clinically observed phenomena.[77–79] By extending our hypoxic model into a 3-D micropattern of cancer cells, the microdevice platform can potentially offer unique insights to evaluate CAR-T cell therapy in solid tumors, which would otherwise have not been captured in traditional 2-D models.
In our reported tumor model, we established a gradient of oxygen across a 3-D micropattern of cancer cells without the need for external sources of oxygen control. Oxygen has been revealed as a main driving factor for metabolic changes, which leads to gradients of metabolites such as glucose and lactate as well as different cell phenotypes and interactions in the TME[14,80]. While the induction of hypoxia may also be accompanied by a multitude of downstream changes, we observed enhanced cell killing by CAR-T cells in these physiologically relevant oxygen levels despite the evidence of increasing immunosuppression under a hypoxic microenvironment. This region of enhanced killing, however, falls in physiologically relevant tissue oxygen levels.[57] Immune cells, and notably CD8+ cytotoxic T cells, travel through the bloodstream and into tissues, which may experience oxygen levels varying from 14.5% to lower than 1% oxygen depending on the location.[81] On the other hand, normoxia, or 21% oxygen, is never experienced within the tissues. In lieu of this, we indeed observed no selective up-regulation of CAR-T cell functionality at the hyperoxic, physiologically irrelevant oxygen concentration at the micropattern edge upon hypoxia modulation (Figure S2). In this study, we capture enhanced antitumor cytotoxicity at more physioxic microenvironments, with a unique cytotoxicity trend affected by oxygen availability over time, which is in agreement with a recent report on hypoxia enhancing anti-tumor functionality of CD8+ T cells.[82] Strikingly, we further demonstrated the sensitivity of CAR-T cells function to various oxygen tensions by revealing the differential killing under the oxygen gradient. Our study highlights the importance of studying the interconnectivity of tumor biology and immunology in physiologically appropriate oxygen levels.
Immunosuppressive molecules expressed or secreted from cancer cells create a hostile microenvironment for effector immune cells, potentially rendering CAR-T cell therapy less effective against solid tumors[3]. We studied the expression of PD-L1, an immune checkpoint protein, under a hypoxic gradient and its role in CAR-T cell-mediated cytotoxicity. Increased surface expression of PD-L1 was verified in our 3-D tumor bulk induced with hypoxia.[11,16,83] Interestingly, pharmacological inhibition of PD-L1 did not enhance the CAR-T mediated cancer cell killing within the time scale evaluated. Our results agree with outcomes from clinical trials, where checkpoint inhibition fell short of expectations in multiple cancer types (non-small cell lung cancer, melanoma, colorectal cancer, renal cell cancer, ovarian cancer, pancreatic cancer, gastric cancer, and breast cancer) despite promising preclinical results.[84,85] Further investigation of temporal effects, as well as immunosuppressive molecules such as A2AR and TGF-β will help elucidate the role of hypoxic immunosuppression against CAR-T cell therapy.[21–23,86]
We found negligible infiltration by CAR-T cells into tumor bulks, although long-term hypoxic culture seemed to slightly increase infiltration. Nonetheless, evidence of CAR-T cell-induced cytotoxicity in these 3-D sections suggest a non-contact mediated, bystander killing mechanism. Cytotoxic T cells under 1% oxygen have been reported to increase the amount of granzyme-B released, resulting in enhanced cytolytic activity, while the penetration in vivo is largely limited to the periphery of the tumor.[82] In agreement, we also observed enhanced cytotoxicity in the hypoxic core after 48 hours of treatment, with insignificant CAR-T cell infiltration in this region. Analysis of granzyme B immunostaining, however, did not correlate with oxygenation level or the unique cytotoxicity trend over time. CAR-T cells at the edge of our hypoxic micropatterns are exposed to approximately 15% oxygen, which may explain the lack of increased granzyme B secretion. Interestingly, we observed low levels of granzyme B at the edges of tumor sections without immune cells. This observation is in agreement with several other studies that report endogenous expression of granzyme B in some cancer cells[87–89]. The absence of granzyme B and CD45+ infiltrating cells at the center of our 3-D micropatterns suggests that the non-contact mediated CAR-T cell cytotoxicity may not be dependent on granzyme B. Instead, we speculate that it is mediated by metabolic competition for metabolites such as glucose between cancer and immune cells. For example, engagement of CAR-T cells with cancer cells in the periphery leads to their activation and enhancement of aerobic glycolysis[90,91], which may lower the availability of glucose in the tumor bulk. Under a hypoxic gradient, this competition exacerbates the metabolic stress experienced by the cancer cells near the core that rely solely on glycolysis, while cells in the intermediate zone may survive with a higher supply of glucose. In normoxic samples, on the other hand, the uniformly higher metabolic rate may have accelerated glucose consumption and/or increased oxidative stress across the tumor bulk, leading to relatively uniform, enhanced cancer cell death. We are further exploring these killing mechanisms. Our study further sheds insight into a contact independent mechanism of CAR-T mediated killing in hypoxic tumors, as well as the timescale at which CAR-T cells target cancer cells adapted to a range of oxygen availability.
With our platform, we can easily incorporate additional components of the TME. Angiogenic factors such as VEGF have been implicated to play a role in tumor immunity.[92] To investigate the modulatory role of VEGF on immune cells, as well as to depict infiltration through the vasculature from the bloodstream into tumors, endothelial cells can be incorporated as a 2-D layer surrounding the fluidics channels.[93] 3-D tumor bulks can be further modified to include multiple fibroblastic cells. Stromal cell types such as cancer-associated fibroblasts (CAFs) and bone marrow stromal cells (BMSCs) have been reported to confer resistance against therapeutic T cells.[94,95] Additionally, innate immune cells such as macrophages,[96] natural killer (NK) cells,[97] neutrophils,[98] and dendritic cells[99] can be assessed for their anti-tumor activity and migration upon activation[45,97] or as cancer vaccines.[100] In our particular platform, regulatory mechanisms that give rise to immunosuppression can be studied as well, as tumor hypoxia attracts M2-like TAMs and Treg cells, both of which function against cytotoxic T cells.[82] Combination immunotherapy with adjuvant agents to activate both innate and adaptive immune components[101] can also be evaluated for maximal antitumor effects. Incorporation of additional TME factors will allow for a systemic evaluation of on-target, off-tumor effects from targeted CAR-T cell therapy, providing a wholesome insight into expected outcome and potential side effects.
In summary, we have established a 3-D cell-laden hydrogel construct in a hypoxia microdevice, which offers unique insights into CAR-T cell anti-tumor mechanisms in the solid TME. This platform is a powerful tool that can depict CAR-T cell cytotoxicity with spatial and temporal resolution, which may provide a new avenue for the assessment of CAR constructs against various solid tumors before in vivo models.
Materials and Methods:
Cell culture
SKOV3 human epithelial ovarian cancer cells were obtained from ATCC and maintained in Dulbecco’s Modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS; Omega Scientific), 100 U/mL penicillin and 100 µg/mL streptomycin (P/S, Thermo Fisher), 4mM sodium pyruvate, and 2mM GlutaMAX (Thermo Fisher). Cells from 75–90% confluent monolayers were passaged using 1% trypsin in phosphate buffered saline (PBS, Thermo Fisher).
Preparation of T cells, CAR-T cells and 2-D cytotoxicity
Primary blood mononuclear cells (PBMCs) and CAR constructs with scFv 4D5 (anti-HER2) were prepared as previously described.[61] T cells were thawed 24 hours prior to co-culturing with cancer cells. T cells and all co-cultures were maintained in T cell medium (TCM) containing X-VIVO™ 15 serum-free medium (Lonza), 5% GemCell human serum antibody AB (Gemini Bio-Products), 2mM GlutaMAX (Thermo Fisher), 10nM HEPES buffer (Corning), P/S, 12.25 mM N-Acetyl-L-cysteine (Sigma), and 10 ng/mL human IL-2 (Peprotech). For the degranulation assay, 500,000 T cells were co-cultured with target cells at a 1:1 E:T ratio for 4 hours with GolgiStop (BD Biosciences) and FITC anti-CD107a antibody. PerCP/Cy5.5 anti-CD4 (BioLegend) and Pacific Blue-conjugated anti-CD8 (BioLegend) antibodies were used to identify T cells. For the cytokine release assay, 100,000 T cells were co-cultured with target cells at a 1:1 ratio for 6 hours with Brefeldin-A (Sigma Aldrich), which prevents protein transport. Cells were then permeabilized using the CytoFix/CytoPerm kit (BD Biosciences), intracellularly stained for CD8 and interferon gamma (IFN-γ) with Pacific Blue-conjugated anti-human CD8 (BioLegend) and PE-conjugated anti-human IFN-γ (BioLegend), and analyzed via flow cytometry. Anti-human CD3/CD28 were used as a positive control. For conventional 2-D cytotoxicity assays, target cells (20,000 cells/well) were labeled with 1 µM of carboxyfluorescein succinimidyl ester (CFSE; Life Technologies) and co-incubated with T cells at various ratios in round-bottom 96-well plate for 24 hours. Cells were then stained for 7-AAD (Life Technologies) in PBS (1:1,000 dilution) for 15 minutes at room temperature and analyzed via flow cytometry as previously described.[61]
3-D micropatterning and hypoxic tumor model
The design and toolpaths for the hypoxia microdevice were created in Autodesk Fusion360 (Autodesk, Inc.). The hypoxia cap consists of an oxygen barrier pillar (6 mm diameter) 100 µm shorter than the three reference pillars, which determine the gap size for oxygen diffusion and hydrogel height.[47] A master mold was milled to produce a PDMS fluidic chamber and channel through replica molding, which fit with the hypoxia cap (Fig. 2A). The PDMS chamber and channel device is plasma-bonded onto a clean glass slide, which is then assembled with the hypoxia cap (Fig. 1B). Both the hypoxia cap and the master mold for the PDMS device were milled in polycarbonate (PC). SKOV3 cells were resuspended in gelatin methacryloyl (GelMA) and 10x DMEM (prepared from powder DMEM, Gibco) to achieve a cell density of 10 million cells/mL in 10% GelMA and 1x DMEM. The final pH of the cell-laden solution was adjusted to approximately 7, as determined by a pH indicator strip. This mixture was pipetted onto the oxygen barrier pillar, before it was assembled into the PDMS chamber/channel device, to form the tumor layer by the surface tension between the glass slide and the oxygen barrier pillar. Next, the platform was exposed to UV light for 120 seconds to crosslink the GelMA. The UV power was measured as 2.34 ± 0.26 mW/cm2 by a handheld radiometer (Solarmeter Model 5.0, Solar Light). Upon crosslinking, the 3-D device was either kept assembled (hypoxic) or disassembled from the PC cap and replaced with a PDMS cap (normoxic), and incubated for up to 72 hours. For live/dead analysis, cells were stained for calcein-AM (Sigma Aldrich) and propidium iodide (PI; Thermo Fisher) for 30 minutes at room temperature.
Mechanical properties
The Young’s elastic moduli of bulk GelMA hydrogels were quantified using a uniaxial mechanical compression test (Instron Model 5942 MicroTester, Precision Instrument). Briefly, GelMA was crosslinked under a polycarbonate pillar of 1 mm height and 3 mm diameter. Upon incubation in 1x PBS for 24 hours, the hydrogel was placed on the tester and subjected to up to 40% strain, with a compression rate of 0.5 mm/min. The stress-strain relation was used to derive the elastic modulus. Porosity was examined by scanning electron microscopy (SEM). Briefly, crosslinked hydrogel samples were placed in 1x PBS for 24 hours. Next, samples were flash-frozen in liquid nitrogen, immediately followed by lyophilization. Lyophilized samples were sputter coated with a thin layer of platinum and palladium, and imaged on a scanning electron microscope (JEOL JSM-7001).
COMSOL Multiphysics® modeling
Expected oxygen gradients across the 3-D cell bulk were simulated with COMSOL Multiphysics modeling software (COMSOL). Passive oxygen diffusion within the media was assumed to be governed by the generic diffusion equation of gas in water,[102] with a diffusion coefficient of 3×10−9 m2/s. Boundary conditions were approximated so that the microdevice was impermeable to oxygen, the media surface in direct contact with atmospheric media had a fixed concentration of oxygen corresponding to normoxic levels (0.2 mol/m3), and the interface between GelMA and media had a fixed oxygen flux rate of 6×10−8 m2/s. Cellular density was assumed to be homogeneous throughout the hydrogel. Cellular oxygen consumption was assumed to follow Michaelis-Menten kinetics with a logistic function constraining consumption below a critical oxygen level:
| Equation (1) |
where Rmax is the maximum oxygen consumption rate of cancer cells adjusted for their average cell volume (0.02 mol s−1 m−3),[102,103] kMM,O2 is the Michaelis-Menten constant corresponding to the oxygen concentration where consumption is half maximal, Ccr is the critical oxygen concentration below which necrosis is assumed to happen and cells cease oxygen consumption, and δ is the step-down function accounting for the termination of oxygen consumption.[102] The step-down function was COMSOL’s smoothed Heaviside function with a continuous first derivative and no overshoot (flc1hs in COMSOL Multiphysics®). Cell number in the 3-D micropattern was then incorporated into the total oxygen consumption rate of the 3-D bulk. All geometries in the model were defined with an extremely fine mesh in COMSOL Multiphysics.
Measurement of oxygen concentration with fluorescent probe
Oxygen levels were measured using fluorophore-based microparticle sensors as described earlier.[47,59] Briefly, 10–14µm silica gel particles were absorbed with 0.5 mM tris(4,7-diphenyl-1,10-phenanthroline) ruthenium (II) dichloride (Thermo Fisher) and 0.5mM Nile blue chloride (Sigma Aldrich). The microparticle sensors were immobilized onto the surface of the oxygen barrier pillar by curing in a PDMS thin layer. After 24 hours of incubation with cell micropatterns, fluorescent intensity from tris(4,7-diphenyl-1,10-phenanthroline) ruthenium(II) dichloride was divided by those from Nile blue chloride to obtain a ratio of differential quenching in the oxygen-sensitive and -insensitive fluorophores. This data was then related to physiological oxygen concentration following a conventional Stern-Volmer model:[47,59,104]
| Equation (2) |
where IR,0 and IR are the fluorescence ratios of the two fluorophores in the absence and presence of oxygen, respectively, and KSV is the Stern-Volmer quenching constant. Anoxic water was prepared by mixing 1 g sodium sulfite (Na2SO3), 50 µL of 0.5 mol/L cobalt nitrate (Co(NO3)2) dissolved in nitric acid (HNO3), and 100 mL water. Upon confirmation of the oxygen levels in anoxic water using a commercial oxygen sensor (Presens Precision Sensing GmbH), IR,0 and IR were obtained from sensor particles that have been calibrated in anoxic water or in regular water, respectively. Derived oxygen concentrations for each bin were plotted against pillar radii.
T cell cytotoxicity assay in 3-D models
After selected timepoints of co-culture incubation, the fluidics channels were rinsed with fresh media and stained for calcein-AM (Sigma Aldrich) and propidium iodide (PI) (Thermo Fisher) for 30 minutes at room temperature. Cell death was quantified by the number of identified PI-positive objects in the radial analysis and by the fraction of cells expressing PI signal in the fractional analysis. For PD-L1 inhibition, cells were treated with 10 nM PD-L1/PD-1 Inhibitor 3 (Selleck Chemicals, IC50 = 5.6 nM) for 24 hours concurrently with T cell co-incubation.
Immunostaining
After 24, 48, or 72 hours of hypoxia or normoxia incubation, samples were disassembled and fixed in 4% paraformaldehyde (PFA), permeabilized with 0.1% Triton X-100 (Fisher Scientific), blocked for 2 hours with 4% bovine serum albumin (GE Healthcare Bio-Sciences), incubated in primary (overnight for anti-CD45 and 2 hours for all others) and secondary antibody (1 hour), and mounted with FluoroGel II containing 4′,6-diamidino-2-phenylindole (DAPI) (Electron Microscopy Sciences) on glass slides. Primary antibodies used were anti-Glucose Transporter 1 (Glut-1) (ab15309, 1:200) (Abcam) and anti-PD-L1 (D8T4X, 1:200) (Cell Signaling Technology). For T cell infiltration studies, micropatterns were prepared similarly after co-incubation for selected time points and stained for CD45 (HI30, 1:200) (Thermo Fisher Scientific). A Nikon inverted fluorescent microscope/confocal microscope and the ImageJ/Matlab/Python software were used to image and analyze immunostained samples, respectively.
Image analysis
Images were analyzed using the ImageJ, Python, and MATLAB software. For oxygen measurements, fluorescent intensity from sensor microparticles was quantified in each fluorophore’s corresponding fluorescence channel (Acridine Orange for ruthenium compound and Cy5 for Nile blue chloride). Raw, pixel-by-pixel fluorescence from tris(4,7-diphenyl-1,10-phenanthroline ruthenium (II) dichloride was divided by those from Nile blue chloride to obtain a ratio of differential quenching in the oxygen -sensitive and -insensitive fluorophores depending on oxygen levels. This data was then binned into concentric circles from the measured centroid of each pillar and translated to oxygen concentration following a conventional Stern-Volmer model[59,104] (Eq. 2). For immunostained samples, the fraction of micropattern area with fluorescence above a pre-defined threshold value was measured. This fraction was also binned into 100 radially evolving concentric circles and plotted against micropattern radii. For cytotoxicity in 3-D samples, PI-positive (dead) cells were identified and counted. These cells’ locations were referenced to the centroid of each micropattern and binned into 100 radially evolving concentric circles and plotted against micropattern radii. Temporal evolution of 3-D cytotoxicity was evaluated in a similar fashion, except each micropattern was divided into equal halves before radially analyzed. To determine the fraction of dead cells in 3-D micropatterns, the area of dead cells was compared to the total area of cells (areal sum of dead cells and calcein-AM-positive, living cells). For the surface/cytoplasmic ratio of PD-L1 expression, fluorescent intensities from a pre-defined cell border and cytoplasm was compared in a custom Python code. In T cell infiltration analysis, the distance from the centroid of CD45+ cells to the centroid of the nearest GelMA-embedded SKOV3 cancer cell or the nearest GelMA boundary was obtained from confocal images.
Statistical analysis
All data are presented in mean ± S.D and all statistical analyses were performed using GraphPad Prism 7. Pearson’s correlation coefficient (r) was used to depict correlation between oxygen sensors and simulation results, linear regression (R2) was used between hydrogel stiffness and curing time, and a goodness to fit (R2) to a Gaussian distribution for pore size. Statistics for cell viability and Glut-1 were assessed using the Student’s t-test (unpaired, multiple), while cytotoxicity and infiltration were assessed using one-way ANOVA. Since PD-L1 expression in 3-D tumor models and surface/cytoplasmic expression in single cells were not normally distributed, data were analyzed with the Mann-Whitney test. In all statistical analysis, p < 0.05 was considered significant.
Supplementary Material
Acknowledgements
This work was supported by NIH NIBIB Trailblazer Award R21EB024748, USC Viterbi School of Engineering, STOP CANCER Marni Levine Memorial Research Career Development Award, Phi Beta Psi Charity Trust, Rose Hills Fellowship, USC Provost’s PhD Fellowship, USC Women in Science and Engineering (WiSE) Fellowship, and USC Undergraduate Research Associates Program. It is also in part supported through shared resources by award number P30CA014089 from the National Cancer Institute at the NIH.
Footnotes
Conflict of Interest
K.S., Y.A., and H.P.T. are listed as inventors for a US patent application disclosing the hypoxia device, which includes part of the data described in the manuscript.
References
- [1].Beatty GL, O’Hara M, Pharmacol. Ther 2016, 166, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Yu S, Li A, Liu Q, Li T, Yuan X, Han X, Wu K, J. Hematol. Oncol.J Hematol Oncol 2017, 10, 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Scarfò I, Maus MV, Immunother J. Cancer 2017, 5, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Maude SL, Frey N, Shaw PA, Aplenc R, Barrett DM, Bunin NJ, Chew A, Gonzalez VE, Zheng Z, Lacey SF, et al. , N. Engl. J. Med 2014, 371, 1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Newick K, Moon E, Albelda SM, Mol. Ther. - Oncolytics 2016, 3, 16006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Kershaw MH, Westwood JA, Parker LL, Wang G, Eshhar Z, Mavroukakis SA, White DE, Wunderlich JR, Canevari S, Rogers-Freezer L, et al. , Clin. Cancer Res 2006, 12, 6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Beatty GL, O’Hara MH, Nelson AM, McGarvey M, Torigian DA, Lacey SF, Melenhorst JJ, Levine B, Plesa G, June CH, J. Clin. Oncol 2015, 33, 3007. [Google Scholar]
- [8].Zhang C, Wang Z, Yang Z, Wang M, Li S, Li Y, Zhang R, Xiong Z, Wei Z, Shen J, et al. , Mol. Ther 2017, 25, 1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Li S, Siriwon N, Zhang X, Yang S, Jin T, He F, Kim YJ, Mac J, Lu Z, Wang S, et al. , Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res 2017, 23, 6982. [DOI] [PubMed] [Google Scholar]
- [10].Xia A-L, Wang X-C, Lu Y-J, Lu X-J, Sun B, Oncotarget 2017, 8, 90521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Qu Q-X, Xie F, Huang Q, Zhang X-G, Cell. Physiol. Biochem 2017, 43, 1893. [DOI] [PubMed] [Google Scholar]
- [12].Wang X, Teng F, Kong L, Yu J, OncoTargets Ther 2016, 9, 5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Hamanishi J, Mandai M, Iwasaki M, Okazaki T, Tanaka Y, Yamaguchi K, Higuchi T, Yagi H, Takakura K, Minato N, et al. , Proc. Natl. Acad. Sci 2007, 104, 3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Eales KL, Hollinshead KER, Tennant DA, Oncogenesis 2016, 5, e190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Barsoum IB, Smallwood CA, Siemens DR, Graham CH, Cancer Res 2014, 74, 665. [DOI] [PubMed] [Google Scholar]
- [16].Noman MZ, Desantis G, Janji B, Hasmim M, Karray S, Dessen P, Bronte V, Chouaib S, J. Exp. Med 2014, 211, 781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Harris AL, Nat. Rev. Cancer 2002, 2, 38. [DOI] [PubMed] [Google Scholar]
- [18].Hanahan D, Weinberg RA, Cell 2011, 144, 646. [DOI] [PubMed] [Google Scholar]
- [19].Nizet V, Johnson RS, Nat. Rev. Immunol 2009, 9, 609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Lukashev D, Klebanov B, Kojima H, Grinberg A, Ohta A, Berenfeld L, Wenger RH, Ohta A, Sitkovsky M, J. Immunol 2006, 177, 4962. [DOI] [PubMed] [Google Scholar]
- [21].Curran CS, Keely PJ, Matrix Biol. J. Int. Soc. Matrix Biol 2013, 32, 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Furuta C, Miyamoto T, Takagi T, Noguchi Y, Kaneko J, Itoh S, Watanabe T, Itoh F, Cancer Sci 2015, 106, 1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Vaupel P, Mayer A, in Oxyg. Transp. Tissue XXXVII (Eds: Elwell CE, Leung TS, Harrison DK), Springer; New York, 2016, pp. 177–183. [Google Scholar]
- [24].Blay J, White TD, Hoskin DW, Cancer Res 1997, 57, 2602. [PubMed] [Google Scholar]
- [25].Sitkovsky MV, Ohta A, Trends Immunol 2005, 26, 299. [DOI] [PubMed] [Google Scholar]
- [26].Fischer K, Hoffmann P, Voelkl S, Meidenbauer N, Ammer J, Edinger M, Gottfried E, Schwarz S, Rothe G, Hoves S, et al. , Blood 2007, 109, 3812. [DOI] [PubMed] [Google Scholar]
- [27].Jacobs SR, Herman CE, MacIver NJ, Wofford JA, Wieman HL, Hammen JJ, Rathmell JC, J. Immunol 2008, 180, 4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Howie D, Waldmann H, Cobbold S, Front. Immunol 2014, 5, DOI 10.3389/fimmu.2014.00409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Ghoneum A, Afify H, Salih Z, Kelly M, Said N, Oncotarget 2018, 9, 22832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].von Strandmann EP, Reinartz S, Wager U, Müller R, Trends Cancer 2017, 3, 137. [DOI] [PubMed] [Google Scholar]
- [31].Horiuchi A, Hayashi T, Kikuchi N, Hayashi A, Fuseya C, Shiozawa T, Konishi I, Int. J. Cancer 2012, 131, 1755. [DOI] [PubMed] [Google Scholar]
- [32].Duechler M, Peczek L, Szubert M, Suzin J, Anticancer Res 2014, 34, 2811. [PubMed] [Google Scholar]
- [33].Selvendiran K, Bratasz A, Kuppusamy ML, Tazi MF, Rivera BK, Kuppusamy P, Int. J. Cancer J. Int. Cancer 2009, 125, 2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Hwu P, Shafer GE, Treisman J, Schindler DG, Gross G, Cowherd R, Rosenberg SA, Eshhar Z, J. Exp. Med 1993, 178, 361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Hwu P, Yang JC, Cowherd R, Treisman J, Shafer GE, Eshhar Z, Rosenberg SA, Cancer Res 1995, 55, 3369. [PubMed] [Google Scholar]
- [36].Cui X, Morales R-TT, Qian W, Wang H, Gagner J-P, Dolgalev I, Placantonakis D, Zagzag D, Cimmino L, Snuderl M, et al. , Biomaterials 2018, 161, 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Sherman H, Gitschier HJ, Rossi AE, Front. Immunol 2018, 9, DOI 10.3389/fimmu.2018.00857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Hirt C, Papadimitropoulos A, Mele V, Muraro MG, Mengus C, Iezzi G, Terracciano L, Martin I, Spagnoli GC, Adv. Drug Deliv. Rev 2014, 79–80, 145. [DOI] [PubMed] [Google Scholar]
- [39].Sacks PG, Taylor DL, Racz T, Vasey T, Oke V, Schantz SP, Cancer Immunol. Immunother 1990, 32, 195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Dangles V, Validire P, Wertheimer M, Richon S, Bovin C, Zeliszewski D, Vallancien G, Bellet D, Int. J. Cancer 2002, 98, 51. [DOI] [PubMed] [Google Scholar]
- [41].Dangles-Marie V, Richon S, El Behi M, Echchakir H, Dorothée G, Thiery J, Validire P, Vergnon I, Menez J, Ladjimi M, et al. , Cancer Res 2003, 63, 3682. [PubMed] [Google Scholar]
- [42].Weiswald L-B, Bellet D, Dangles-Marie V, Neoplasia 2015, 17, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Herter S, Morra L, Schlenker R, Sulcova J, Fahrni L, Waldhauer I, Lehmann S, Reisländer T, Agarkova I, Kelm JM, et al. , Cancer Immunol. Immunother 2017, 66, 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Hallmann R, Zhang X, Di Russo J, Li L, Song J, Hannocks M-J, Sorokin L, Curr. Opin. Cell Biol 2015, 36, 54. [DOI] [PubMed] [Google Scholar]
- [45].Parlato S, Ninno AD, Molfetta R, Toschi E, Salerno D, Mencattini A, Romagnoli G, Fragale A, Roccazzello L, Buoncervello M, et al. , Sci. Rep 2017, 7, 1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Pavesi A, Tan AT, Koh S, Chia A, Colombo M, Antonecchia E, Miccolis C, Ceccarello E, Adriani G, Raimondi MT, et al. , JCI Insight 2017, 2, DOI 10.1172/jci.insight.89762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Ando Y, Ta HP, Yen DP, Lee S-S, Raola S, Shen K, Sci. Rep 2017, 7, 15233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Lu P, Weaver VM, Werb Z, J. Cell Biol 2012, 196, 395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Nichol JW, Koshy ST, Bae H, Hwang CM, Yamanlar S, Khademhosseini A, Biomaterials 2010, 31, 5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Klotz BJ, Gawlitta D, Rosenberg AJWP, Malda J, Melchels FPW, Trends Biotechnol 2016, 34, 394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Yue K, Trujillo-de Santiago G, Alvarez MM, Tamayol A, Annabi N, Khademhosseini A, Biomaterials 2015, 73, 254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Samani A, Zubovits J, Plewes D, Phys. Med. Biol 2007, 52, 1565. [DOI] [PubMed] [Google Scholar]
- [53].Broders-Bondon F, Ho-Bouldoires THN, Fernandez-Sanchez M-E, Farge E, J Cell Biol 2018, jcb. 201701039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Gkretsi V, Stylianopoulos T, Front. Oncol 2018, 8, DOI 10.3389/fonc.2018.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Levental KR, Yu H, Kass L, Lakins JN, Egeblad M, Erler JT, Fong SFT, Csiszar K, Giaccia A, Weninger W, et al. , Cell 2009, 139, 891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Kawano S, Kojima M, Higuchi Y, Sugimoto M, Ikeda K, Sakuyama N, Takahashi S, Hayashi R, Ochiai A, Saito N, Cancer Sci 2015, 106, 1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Hammond EM, Asselin M-C, Forster D, O’Connor JPB, Senra JM, Williams KJ, Clin. Oncol 2014, 26, 277. [DOI] [PubMed] [Google Scholar]
- [58].Yen DP, Ando Y, Shen K, TECHNOLOGY 2016, 04, 234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Acosta MA, Ymele-Leki P, Kostov YV, Leach JB, Biomaterials 2009, 30, 3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Airley RE, Loncaster J, Raleigh JA, Harris AL, Davidson SE, Hunter RD, West CML, Stratford IJ, Int. J. Cancer 2003, 104, 85. [DOI] [PubMed] [Google Scholar]
- [61].Siegler E, Li S, Kim YJ, Wang P, Hum. Gene Ther 2017, 28, 726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Clark CA, Gupta HB, Sareddy G, Pandeswara S, Lao S, Yuan B, Drerup JM, Padron A, Conejo-Garcia J, Murthy K, et al. , Cancer Res 2016, 76, 6964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Dong H, Strome SE, Salomao DR, Tamura H, Hirano F, Flies DB, Roche PC, Lu J, Zhu G, Tamada K, et al. , Nat. Med 2002, 8, 793. [DOI] [PubMed] [Google Scholar]
- [64].Chatterjee J, Dai W, Aziz NHA, Teo PY, Wahba J, Phelps DL, Maine CJ, Whilding LM, Dina R, Trevisan G, et al. , Clin. Cancer Res 2017, 23, 3453. [DOI] [PubMed] [Google Scholar]
- [65].Rousalova I, Krepela E, Int. J. Oncol 2010, 37, 1361. [DOI] [PubMed] [Google Scholar]
- [66].Barnes TA, Amir E, Br. J. Cancer 2017, 117, 451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].de Ruiter EJ, Ooft ML, Devriese LA, Willems SM, Oncoimmunology 2017, 6, e1356148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Li J, Wang J, Chen R, Bai Y, Lu X, Oncotarget 2017, 8, 15621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Cohen IJ, Blasberg R, Breast Cancer Basic Clin. Res 2017, 11, DOI 10.1177/1178223417731565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Acerbi I, Cassereau L, Dean I, Shi Q, Au A, Park C, Chen YY, Liphardt J, Hwang ES, Weaver VM, Integr. Biol 2015, 7, 1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Salmon H, Franciszkiewicz K, Damotte D, Dieu-Nosjean M-C, Validire P, Trautmann A, Mami-Chouaib F, Donnadieu E, J. Clin. Invest 2012, 122, 899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Friedl P, Entschladen F, Conrad C, Niggemann B, Zänker KS, Eur. J. Immunol 1998, 28, 2331. [DOI] [PubMed] [Google Scholar]
- [73].Wang Y, Yao B, Li H, Zhang Y, Gao H, Gao Y, Peng R, Tang J, J. Ultrasound Med 2017, 36, 955. [DOI] [PubMed] [Google Scholar]
- [74].McKenzie AJ, Hicks SR, Svec KV, Naughton H, Edmunds ZL, Howe AK, Sci. Rep 2018, 8, 7228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Benton JA, DeForest CA, Vivekanandan V, Anseth KS, Tissue Eng. Part A 2009, 15, 3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Cho A, Howell VM, Colvin EK, Front. Oncol 2015, 5, DOI 10.3389/fonc.2015.00245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Duval K, Grover H, Han L-H, Mou Y, Pegoraro AF, Fredberg J, Chen Z, Physiology 2017, 32, 266. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- [78].Hoarau-Véchot J, Rafii A, Touboul C, Pasquier J, Hoarau-Véchot J, Rafii A, Touboul C, Pasquier J, Int. J. Mol. Sci 2018, 19, 181. [Google Scholar]
- [79].Riedl A, Schlederer M, Pudelko K, Stadler M, Walter S, Unterleuthner D, Unger C, Kramer N, Hengstschläger M, Kenner L, et al. , J Cell Sci 2017, 130, 203. [DOI] [PubMed] [Google Scholar]
- [80].Petrova V, Annicchiarico-Petruzzelli M, Melino G, Amelio I, Oncogenesis 2018, 7, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].de Silly RV, Dietrich P-Y, Walker PR, OncoImmunology 2016, 5, e1232236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Gropper Y, Feferman T, Shalit T, Salame T-M, Porat Z, Shakhar G, Cell Rep 2017, 20, 2547. [DOI] [PubMed] [Google Scholar]
- [83].Tatli Dogan H, Kiran M, Bilgin B, Kiliçarslan A, Sendur MAN, Yalçin B, Ardiçoglu A, Atmaca AF, Gumuskaya B, Diagn. Pathol 2018, 13, 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Brahmer JR, Tykodi SS, Chow LQM, Hwu W-J, Topalian SL, Hwu P, Drake CG, Camacho LH, Kauh J, Odunsi K, et al. , N. Engl. J. Med 2012, 366, 2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Li HY, McSharry M, Bullock B, Nguyen TT, Kwak J, Poczobutt JM, Sippel TR, Heasley LE, Weiser-Evans MC, Clambey ET, et al. , Cancer Immunol. Res 2017, 5, 767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Soleimani A, Bahreyni A, Roshan MK, Soltani A, Ryzhikov M, Shafiee M, Soukhtanloo M, Jaafari MR, Mashkani B, Hassanian SM, J. Cell. Physiol n.d., 0, DOI 10.1002/jcp.27249. [DOI] [PubMed] [Google Scholar]
- [87].Kontani K, Sawai S, Hanaoka J, Tezuka N, Inoue S, Fujino S, Eur. J. Surg. Oncol 2001, 27, 180. [DOI] [PubMed] [Google Scholar]
- [88].Hu SX, Wang S, Wang JP, Mills GB, Zhou Y, Xu H-J, Br. J. Cancer 2003, 89, 135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].D’Eliseo D, Pisu P, Romano C, Tubaro A, De Nunzio C, Morrone S, Santoni A, Stoppacciaro A, Velotti F, Int. J. Cancer 2010, 127, 1283. [DOI] [PubMed] [Google Scholar]
- [90].van der Windt GJW, Pearce EL, Immunol. Rev 2012, 249, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Menk AV, Scharping NE, Moreci RS, Zeng X, Guy C, Salvatore S, Bae H, Xie J, Young HA, Wendell SG, et al. , Cell Rep 2018, 22, 1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Li Y-L, Zhao H, Ren X-B, Cancer Biol. Med 2016, 13, 206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Myers DR, Sakurai Y, Tran R, Ahn B, Hardy ET, Mannino R, Kita A, Tsai M, Lam WA, JoVE J Vis. Exp 2012, e3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Lakins MA, Ghorani E, Munir H, Martins CP, Shields JD, Nat. Commun 2018, 9, 948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Li M, Sun X, Kuang X, Liao Y, Li H, Luo D, Clin. Exp. Immunol 2014, 178, 516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Cassetta L, Kitamura T, Immunology 2018, DOI 10.1111/imm.12976. [DOI] [Google Scholar]
- [97].Li Y, Sun R, Chin. J. Cancer Res 2018, 30, 173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Ocana A, Nieto-Jiménez C, Pandiella A, Templeton AJ, Mol. Cancer 2017, 16, 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Sabado RL, Balan S, Bhardwaj N, Cell Res 2017, 27, 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Pizzurro GA, Barrio MM, Front. Immunol 2015, 6, DOI 10.3389/fimmu.2015.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Moynihan KD, Opel CF, Szeto GL, Tzeng A, Zhu EF, Engreitz JM, Williams RT, Rakhra K, Zhang MH, Rothschilds AM, et al. , Nat. Med 2016, 22, 1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Buchwald P, Theor. Biol. Med. Model 2009, 6, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Wagner BA, Venkataraman S, Buettner GR, Free Radic. Biol. Med 2011, 51, 700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Wang X, Wolfbeis OS, Chem. Soc. Rev 2014, 43, 3666. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.