Although reactive oxygen species (ROS) are required for spermatogonial stem cell (SSC) self-renewal, the mechanism has remained unknown. We show that SSC self-renewal signals activate MAPK14/MAPK7 pathway to induce nuclear translocation of BCL6B and activation of NOX1.
Abstract
Reactive oxygen species (ROS) play critical roles in self-renewal division for various stem cell types. However, it remains unclear how ROS signals are integrated with self-renewal machinery. Here, we report that the MAPK14/MAPK7/BCL6B pathway creates a positive feedback loop to drive spermatogonial stem cell (SSC) self-renewal via ROS amplification. The activation of MAPK14 induced MAPK7 phosphorylation in cultured SSCs, and targeted deletion of Mapk14 or Mapk7 resulted in significant SSC deficiency after spermatogonial transplantation. The activation of this signaling pathway not only induced Nox1 but also increased ROS levels. Chemical screening of MAPK7 targets revealed many ROS-dependent spermatogonial transcription factors, of which BCL6B was found to initiate ROS production by increasing Nox1 expression via ETV5-induced nuclear translocation. Because hydrogen peroxide or Nox1 transfection also induced BCL6B nuclear translocation, our results suggest that BCL6B initiates and amplifies ROS signals to activate ROS-dependent spermatogonial transcription factors by forming a positive feedback loop.
Introduction
Spermatogonial stem cells (SSCs) undergo continuous self-renewal and produce numerous progenitors that ultimately give rise to spermatozoa (Meistrich & van Beek, 1993; de Rooij & Russel, 2000). Although the frequency of SSCs in the testis is very low (0.02–0.03%) (Meistrich & van Beek, 1993; Tegelenbosch & de Rooij, 1993), these cells produce sperm throughout the life span of male animals. SSCs have a unique mode of self-renewal because they do not undergo asymmetric division; a single SSC produces two stem cells by self-renewal division or two differentiated cells by differentiating division. These two types of divisions are considered to occur at the same frequency to maintain a constant population size (Meistrich & van Beek, 1993; de Rooij & Russel, 2000). Because excessive self-renewal division leads to the accumulation of SSCs and increased differentiating division depletes SSCs, imbalances between the two types of divisions can result in male infertility. Therefore, the regulation of these two types of divisions in SSCs requires sophisticated control, but the molecular factors that regulate self-renewal division remain largely unknown.
Studies within the last decade suggest that reactive oxygen species (ROS) influence various stem cells. For example, hematopoietic stem cells are sensitive to ROS, and increased ROS levels induce senescence and compromise stem cell function (Ito et al, 2006). Embryonic stem (ES) cells are sensitive to hydrogen peroxide–induced apoptosis but are resistant to oxidative stress–induced senescence, entering a transient cell cycle arrest state (Guo et al, 2010). However, ROS are not necessarily harmful for self-renewal because proliferative neural stem cells (NSCs) have high endogenous ROS levels (Le Belle et al, 2011). Likewise, transient generation of ROS activates hair follicle stem cells, thereby promoting hair growth, and accelerates burn healing (Carrasco et al, 2015). Thus, ROS can also promote self-renewal in some tissues. Whereas ROS-induced senescence and damage have been well characterized, little is known about how ROS promote self-renewal machinery.
ROS have important influences on SSCs. We recently found that constitutive active Hras transfection induces SSC self-renewal without the need for self-renewal factors as well as increases ROS (Morimoto et al, 2013). The addition of ROS inhibitors suppressed self-renewal division, whereas hydrogen peroxide increased cell recovery. These results suggest that self-renewal division is positively regulated by ROS in SSCs. Consistent with this notion, testes of mice deficient in Nox1, which generates ROS, are smaller than those of WT mice, and SSCs in Nox1 KO mice have reduced self-renewal activity upon serial transplantation. Depletion of Nox1 in vitro by shRNA significantly suppressed self-renewal. These results suggest that ROS generated by Nox1 are necessary for self-renewal. This conclusion was unexpected because Nox1 expression is relatively low in germ cells and ROS are thought to be harmful to spermatogenesis. In fact, ROS suppression is a commonly accepted treatment for male infertility.
Although these studies demonstrated the critical roles of ROS generated by Nox genes, they are only weakly expressed in germ cells, and the link between ROS generation and self-renewal has not been elucidated. SSC self-renewal is based on the complex interplay between stably expressed transcription factors and cytokine-induced transcriptional activators (Kanatsu-Shinohara & Shinohara, 2013). The p38 MAPK appears to be involved in this process because 1) the addition of self-renewal factors to cultured SSCs induces sustained phosphorylation of p38 MAPK and 2) inhibition of p38 MAPK by a chemical inhibitor SB203580 suppresses self-renewal and down-regulates Nox1 (Morimoto et al, 2013). However, there are several genes responsible for p38 MAPK activity, and the critical target of p38 MAPK activity in SSCs has not been identified. Most importantly, it remains unclear how ROS integrate with the self-renewal machinery in general. In this respect, SSCs are uniquely favorable for research because both stem cell culture and transplantation techniques are available (Brinster & Zimmermann, 1994; Kanatsu-Shinohara et al, 2003). In this study, we aimed to identify the gene responsible for p38 MAPK activity and investigate its effect in SSCs. Using KO mice, we carried out a functional analysis of p38 MAPK and found that ROS create a positive feedback loop to sustain self-renewal by activating the MAPK14/MAPK7/BCL6B pathway. This process of ROS amplification by an SSC transcription factor may explain why ROS play important roles despite relatively low expression levels of Nox1.
Results
Essential role of Mapk14 in SSC self-renewal
FGF2 and GDNF play critical roles in deriving germline stem (GS) cells, which are cultured SSCs with enriched SSC activity (Kanatsu-Shinohara et al, 2003). Addition of FGF2 and GDNF to GS cells that had been starved for cytokines induced ROS (Fig S1A), which suggested close relationship between ROS and self-renewal. We previously showed that the p38 MAPK inhibitor SB203580 inhibits GS cell proliferation and reduces Nox1 expression (Morimoto et al, 2013). Depletion of Nox1 by shRNA suppressed ROS (Fig S1B). Although there are four p38 MAPK isoforms, it has not been determined which isoform is involved in GS cell proliferation. We carried out real-time PCR analysis and found that Mapk14 is expressed in many organs, including testis (Fig S2), and that GS cells express Mapk14 most strongly among the four p38 MAPK isoforms (Fig 1A). However, GS cells also expressed Mapk11. Because both MAPK11 and MAPK14 are sensitive to SB203580, we used another inhibitor VX-745, which is more specific to MAPK14. We added VX-745 to GS cells cultured in the presence of self-renewal factors. As expected, VX-745 suppressed GS cell proliferation in a dose-dependent manner (Fig 1B and C). Moreover, flow cytometric analysis showed that GS cells treated with VX-745 generated significantly lower levels of ROS in a similar manner to Nox1 knockdown (KD) (Fig 1D and Table S1). MAPK14 activation level was closely related to ROS levels: although MAPK14 was phosphorylated by hydrogen peroxide treatment (Fig 1E and Table S2), addition of N-acetyl cysteine (NAC), a ROS scavenger, decreased MAPK14 phosphorylation (Fig 1F and Table S2). These results suggested that ROS generated by Nox1 regulates MAPK14 and that MAPK14 plays some role in both GS cell proliferation and in ROS production.
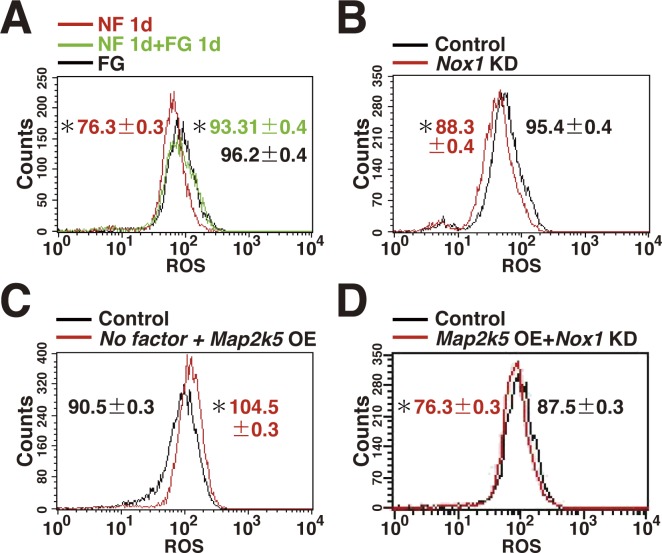
Figure S1. Generation of ROS in GS cells.
(A) ROS generation after cytokine stimulation. GS cells were cultured without cytokine (No factor, NF) for 1 d, and restimulated with FGF2 and GDNF. FG culture indicates exponentially growing GS cells cultured with FGF2 and GDNF (n = 3 cultures). (B) Decreased ROS generation by Nox1 KD in GS cells (n = 3 cultures; moi = 4). The cells were recovered on the next day after transfection. (C) ROS generation after Map2k5 OE in GS cells cultured without cytokines (n = 3 cultures; moi = 4). The cells were recovered 2 d after transfection. (D) ROS generation 2 d after Map2k5 KD and Nox1 OE in GS cells (n = 3 cultures; moi = 4). Data information: in (A–D), data are presented as mean ± SEM. *P < 0.05 (linear regression).
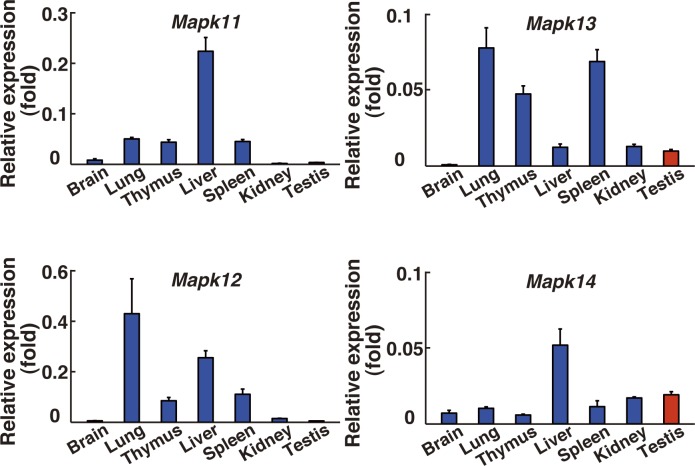
Figure S2. Real-time PCR analysis of four p38 MAPK isoform gene expression in various organs of adult male mice (n = 3).
The values are normalized to Hprt expression levels. Data information: data are presented as mean ± SEM.
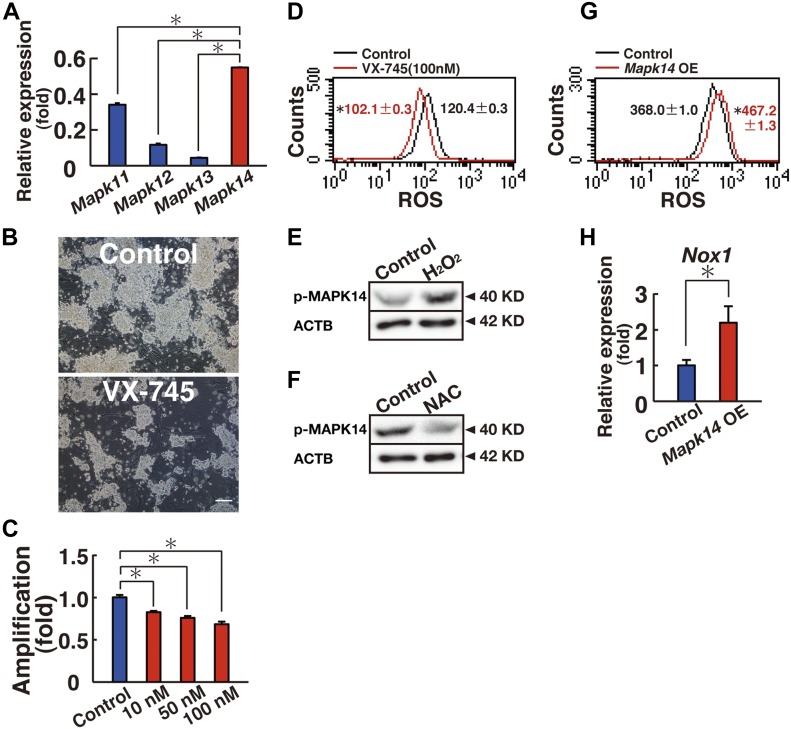
Figure 1. MAPK14 regulates ROS generation in GS cells.
(A) Real-time PCR analysis of Mapk11, Mapk12, Mapk13, and Mapk14 in GS cells (n = 3 cultures). *P < 0.05 (ANOVA). (B) Appearance of GS cells after culturing with VX-745 for 6 d. Scale bar: 100 μm. (C) Cell recovery (n = 9 cultures). GS cells were cultured with VX-745 for 6 d. *P < 0.05 (ANOVA). (D) Flow cytometric analysis of ROS generation 5 d after VX-745 supplementation. (n = 3 cultures). *P < 0.05 (linear regression). (E) Western blot analysis of phosphorylated MAPK14 10 min after hydrogen peroxide (30 μM) supplementation. (F) Western blot analysis of phosphorylated MAPK14 2 d after NAC (0.5 mM) supplementation. (G) Flow cytometric analysis of ROS generation 1 d after transfection with constitutively active Mapk14 (n = 3 cultures; multiplicity of infection [moi] = 4). *P < 0.05 (linear regression). (H) Real-time PCR analysis of Nox1 expression 1 d after transfection with constitutively active Mapk14 (moi = 4; n = 5 cultures). *P < 0.05 (t test). Data information: in (A, C, D, G, and H), data are presented as mean ± SEM.
Table S1 Statistical analysis of ROS levels. (14.7KB, docx)
Table S2 Quantification of Western blot analysis. (22KB, docx)
MAPK14 phosphorylation is induced after cytokine supplementation (Morimoto et al, 2013). However, it was not clear whether MAPK14 plays any role in ROS generation. We transfected constitutively active Mapk14 in GS cells to determine whether MAPK14 activation is sufficient for ROS generation. As expected, GS cells transfected with constitutively active Mapk14 exhibited increased ROS generation by flow cytometry even in the presence of self-renewal factors (Fig 1G and Table S1). Because SB203580 inhibited Nox1 expression in GS cells (Morimoto et al, 2013), we examined its expression level after transfection of constitutively active Mapk14. As expected, real-time PCR analysis showed up-regulation of Nox1 after transfection (Fig 1H). These results showed a close correlation between MAPK14 activation and ROS.
We previously showed that the number of undifferentiated spermatogonia is significantly reduced in Nox1 KO mouse testes (Morimoto et al, 2013). Using this KO mouse strain, we examined whether reduced ROS production can influence MAPK14 activity in vivo. To test this hypothesis, we performed immunohistochemistry for Nox1 KO testes using antibodies against GFRA1 (a component of GDNF receptor; marker of Asingle [As], Apaired [Apr] spermatogonia, and some Aaligned [Aal] spermatogonia), a component of GDNF receptor, to assess the phosphorylation levels of MAPK14 in these cells (Fig S3A) (Grasso et al, 2012). Enumeration of GFRA1+ cells positive for phosphorylated MAPK14 revealed that the number of GFRA1+ spermatogonia positive for phosphorylated MAPK14 was significantly reduced (Fig S3B), suggesting that NOX1-mediated ROS production is involved in MAPK14 phosphorylation in undifferentiated spermatogonia, possibly including SSCs. However, no changes were found in GATA4+ Sertoli cells, suggesting that ROS generation has a greater impact on germ cells (Fig S3C).
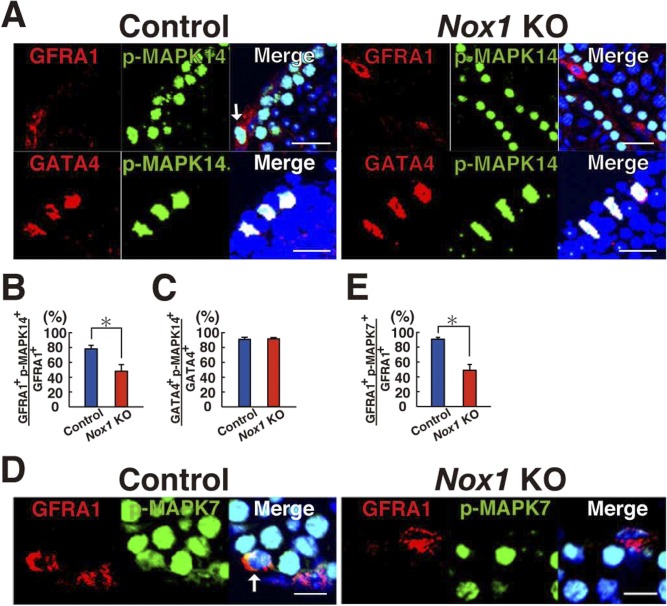
Figure S3. Reduced GFRA1+ cells in Nox1 KO testes.
(A) Immunohistochemical analysis of Nox1 KO testes using antibodies against phosphorylated MAPK14 (p-MAPK14) and markers for undifferentiated spermatogonia (GFRA1) or Sertoli cells (GATA4). Arrow indicates a cell expressing both antigens. Scale bar: 20 μm. (B, C) Quantification of cells expressing GFRA1 (B) or GATA4 (C). *P < 0.05 (t test). (D) Immunohistochemical analysis of Nox1 KO testes using antibodies against phosphorylated MAPK7 (p-MAPK7) and an undifferentiated spermatogonia marker (GFRA1). Arrow indicates a cell expressing both antigens. Scale bar: 10 μm. (E) Quantification of cells expressing GFRA1. At least 20 tubules were counted. Counterstaining using Hoechst 33342. *P < 0.05 (t test). Data information: in (B, C, E), data are presented as mean ± SEM.
Mapk14 gene targeting in SSCs
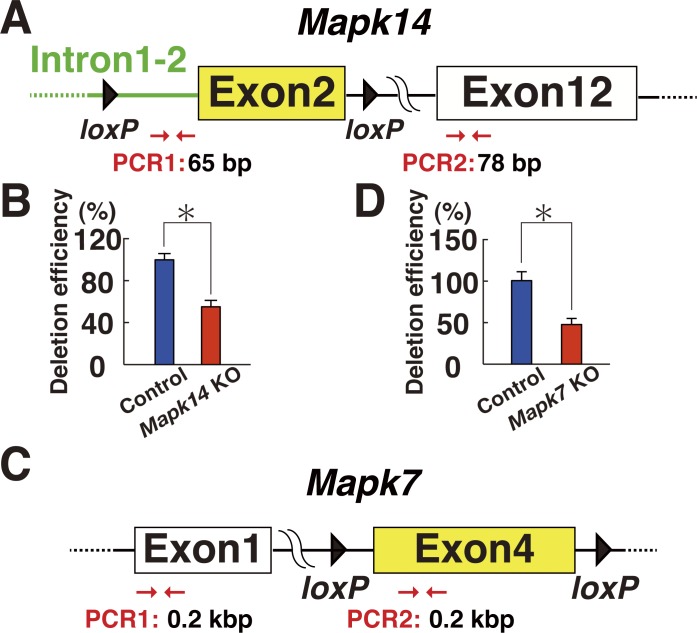
Based on these observations, we evaluated the function of MAPK14 by gene targeting. For this purpose, we used a floxed mutant mouse strain that possess loxP sites flanking exon 2 of the Mapk14 gene (Mapk14f/f mice) (Fig S4A). The cells were collected from 5- to 10-d-old pup testes, which are enriched with SSCs because of lack of differentiating germ cells. The cells were then exposed to a Cre-expressing adenovirus (AxCANCre) overnight (Takehashi et al, 2007). Testes from littermate mouse were used as controls. After trypsinization, an average of 67.1% and 59.0% of the input Mapk14f/f and WT cells were collected, respectively, with no statistically significant difference (n = 7). Real-time PCR analysis confirmed the deletion of the target gene at the time of transplantation (Fig S4B).
Figure S4. Production of Mapk14 and Mapk7 KO cells.
(A) Mapk14 conditional KO mice used in the experiment. Exon 2 of the Mapk14 gene was deleted by CRE-mediated recombination (moi = 2). The indicated primers were used for PCR analysis. (B) Deletion efficiency of the target gene at the time of transplantation by real-time PCR of genomic DNA (n = 4 testes). Exon 12 was used as a control. *P < 0.05 (t test). (C) Mapk7 conditional KO mice used in the experiment. Exon 4 of the Mapk7 gene was deleted by CRE-mediated recombination (moi = 2). (D) Deletion efficiency of the target gene at the time of transplantation by real-time PCR of genomic DNA (n = 12 cultures). Exon 1 was used as a control. *P < 0.05 (t test).
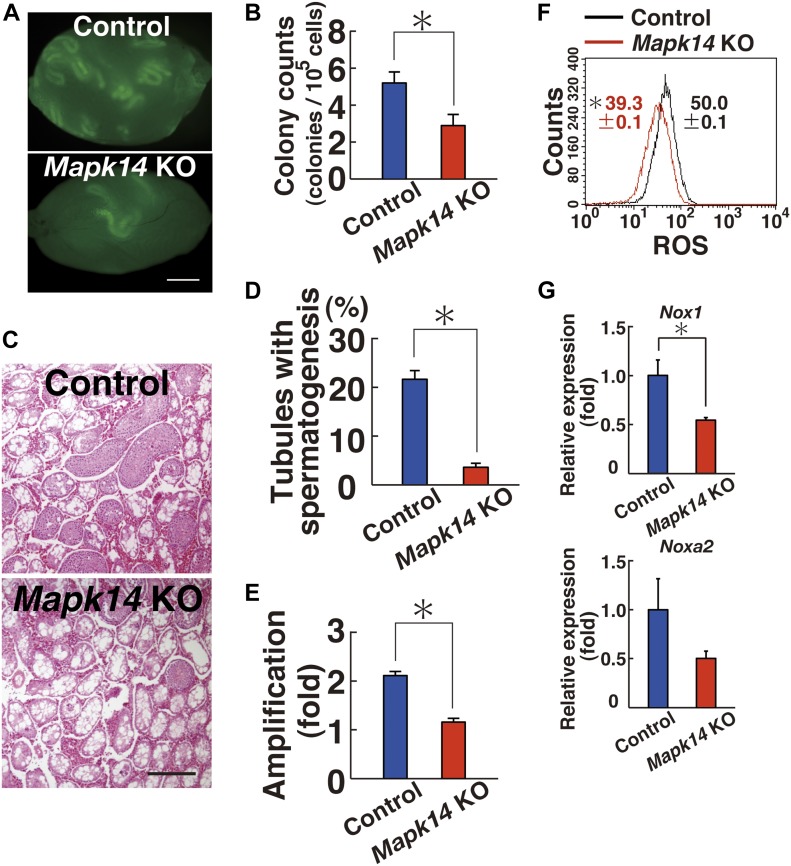
Analysis of the recipients showed significantly reduced donor cell colonization after Mapk14 deletion (Fig 2A). The numbers of colonies generated by Mapk14 KO and control testis cells were 2.9 and 5.2 per 105 transplanted cells, respectively (Fig 2B). This difference was statistically significant. We also examined the colonization efficiency by counting the number of tubules exhibiting spermatogenesis by histology (Fig 2C). Whereas testes with mutant donor cells exhibited spermatogenesis in 3.6% of tubules, those that received control transplants exhibited spermatogenesis in 21.7% of tubules (n = 10). This difference was also statistically significant (Fig 2D).
Figure 2. Functional analysis of Mapk14 in SSCs by spermatogonial transplantation.
(A) Macroscopic appearance of a recipient testis that underwent transplantation of AxCANCre-treated Mapk14f/f testis cells. Scale bar: 1 mm. (B) Colony counts (n = 25 testes for Mapk14f/f; n = 20 testes for control). *P < 0.05 (t test). (C) Histological appearance of a recipient testis. Scale bar: 250 μm. (D) Quantification of tubules exhibiting spermatogenesis. At least 1,139 tubules were counted. *P < 0.05 (t test). (E) Defective proliferation of Mapk14f/f GS cells exposed to AxCANCre (n = 14 cultures; moi = 2). The cells were recovered 6 d after transfection. *P < 0.05 (t test). (F) Flow cytometric analysis of ROS generation in AxCANCre-treated Mapk14f/f GS cells 1 d after transfection (moi = 2; n = 3 cultures). *P < 0.05 (linear regression). (G) Real-time PCR analysis of Nox1 and Noxa2 expression in Mapk14f/f GS cells exposed to AxCANCre 4 d after infection (n = 3 cultures; moi = 2). *P < 0.05 (t test). Data information: in (B, D–G), data are presented as mean ± SEM.
To further investigate the role of MAPK14 in SSCs, we derived GS cells from control and Mapk14f/f mice. Both types of GS cells were exposed to AxCANCre to remove the target gene. The virus-containing medium was removed the next day, and cells were recovered 6 d after culture initiation by trypsinization. Comparison of cell recovery showed that Mapk14f/f GS cells proliferated significantly slower after AxCANCre exposure compared with control GS cells (Fig 2E). Moreover, these cells showed reduced ROS levels upon AxCANCre-mediated Mapk14 deletion (Fig 2F and Table S1). As expected, Mapk14 deletion resulted in reduced Nox1 expression (Fig 2G). These results showed that activation of Mapk14 is involved in GS cell proliferation.
MAPK7 regulation by MAPK14
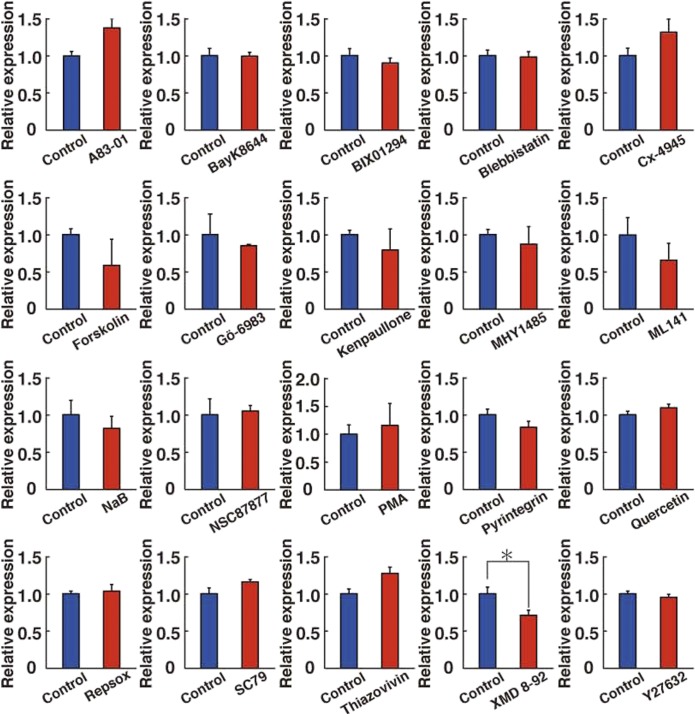
To further explore the signaling pathway involved in Nox1 regulation, we screened GS cells for chemical activators/inhibitors that are involved in Nox1 expression. We selected small chemicals that are involved in cell morphology (blebbistatin, pyrintegrin, thiazovivin, XMD 8-92, and Y27632), signal transduction (A83-010, BayK8644, Cx-4945, forskolin, Gö-6983, kenpaullone, MHY1485, NSC87877, PMA, quercetin, RepSox, and SC79), or epigenetic regulation (BIX01294, ML141, and NaB). Because many of these chemical inhibitors have known targets, screening would help identify candidate molecules that might regulate Nox1 expression. Logarithmically growing GS cells were cultured for 3 d in the presence of candidate chemical compounds, and Nox1 expression was analyzed by real-time PCR. Of the compounds evaluated, we found that XMD 8-92 efficiently reduced Nox1 expression (Figs 3A and S5).
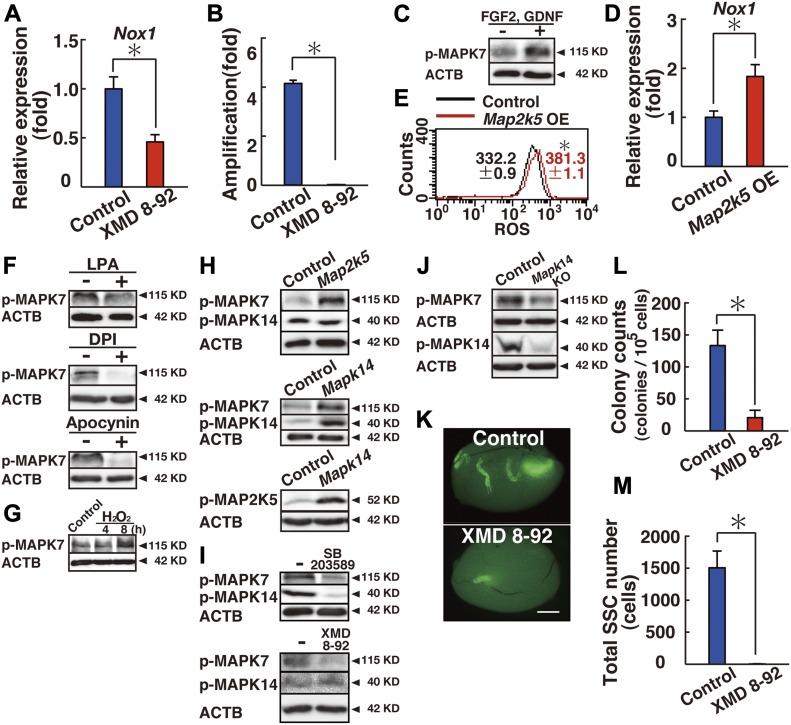
Figure 3. Regulation of Nox1 expression by MAPK7 in GS cells.
(A) Real-time PCR analysis of Nox1 expression 1 d after supplementation of XMD 8-92 (n = 6 cultures). *P < 0.05 (t test). (B) Defective proliferation of GS cells exposed to XMD 8-92 (n = 9 cultures). GS cells were cultured with XMD 8-92 for 5 d. *P < 0.05 (t test). (C) Western blot analysis of MAPK7 in GS cells treated with self-renewal factors. GS cells were cultured without cytokines for 3 d and restimulated with GDNF and FGF2 for 3 h. (D) Real-time PCR analysis of Nox1 expression 1 d after transfection of constitutively active Map2k5 (n = 6 cultures; moi = 4). *P < 0.05 (t test). (E) Flow cytometric analysis of ROS generation 2 d after transfection of constitutively active Map2k5 (n = 3 cultures; moi = 4). *P < 0.05 (linear regression). (F) Western blot analysis of MAPK7 in GS cells 2 d after supplementation with the indicated ROS inhibitors. (G) Western blot analysis of MAPK7 phosphorylation. The cells were incubated with hydrogen peroxide (30 μM) for indicated time points before sample collection. (H) Western blot analysis of GS cells 2 d after transfection with constitutively active Map2k5 or Mapk14 (moi = 10). (I) Western blot analysis of GS cells 1 d or 4 d after supplementation with a MAPK14 or MAPK7 inhibitor, respectively. (J) Western blot analysis of Mapk14f/f GS cells 3 d after AxCANCre treatment (moi = 2). (K) Macroscopic appearance of a recipient testis that underwent transplantation of GS cells cultured with XMD 8-92 for 6 d. Scale bar: 1 mm. (L) Colony counts (n = 12 testes). *P < 0.05 (t test). (M) Total number of SSCs recovered from culture (n = 12 cultures). *P < 0.05 (t test). Data information: In (A, B, D, E, L, M), data are presented as mean ± SEM. *P < 0.05 (t test).
Figure S5. Real-time PCR analysis of Nox1 expression in GS cells exposed to the indicated chemicals for 3 d (n = 3).
*P < 0.05 (t test). Data information: data are presented as mean ± SEM.
XMD 8-92 is a potent and selective inhibitor of MAPK7. GS cells treated with XMD 8-92 showed significantly reduced proliferation in a manner similar to that associated with MAPK14 inhibitors (Fig 3B). Western blot analysis showed that MAPK7 is activated upon cytokine stimulation (Fig 3C and Table S2). When GS cells were starved for 3 d and restimulated with self-renewal factors, MAPK7 phosphorylation was significantly increased, which suggests that MAPK7 plays a role in driving GS cell proliferation.
To determine whether MAPK7 is involved in ROS generation, we transfected GS cells with constitutively active Map2k5, which can activate MAPK7 (Terasawa et al, 2003). Real-time PCR analysis showed that transfection of constitutively active Map2k5 induced Nox1 expression (Fig 3D). In addition, flow cytometric analysis showed that exponentially growing GS cells produce a significant amount of ROS in constitutively active Map2k5-transfected cells (Fig 3E and Table S1). ROS generation by Map2k5 also occurred in cells cultured without cytokines (Fig S1C). This was mediated by Nox1 because Nox1 suppression down-regulated ROS after Map2k5 transfection (Fig S1D). The patterns of Nox1 induction and ROS generation induced by Mapk7 were similar to those induced by constitutively active Mapk14. These results suggest that MAPK7 is involved in the regulation of Nox1 expression.
We then examined the effect of ROS inhibitors on MAPK7 activation in stably growing GS cells. Western blot analysis showed that phosphorylation of MAPK7 is significantly suppressed by several Nox inhibitors, including diphenyleneiodonium (DPI), and apocynin (Fig 3F and Table S2). Similar results were found when we used α-lipoic acid, which is an antioxidant (Fig 3F). Because adding hydrogen peroxide increased MAPK7 phosphorylation (Fig 3G and Table S2), we examined the status of MAPK7 phosphorylation in vivo using Nox1 KO mice (Morimoto et al, 2013). Immunohistochemistry of Nox1 KO testes using antibodies against GFRA1 showed a significant reduction in MAPK7 phosphorylation in these cells, in a manner similar to MAPK14 (Fig S3D and E). These results suggest that ROS generation by Nox1 induces MAPK7 and MAPK14 activation in GFRA1+ undifferentiated spermatogonia in vivo.
Because MAPK7 and MAPK14 acted similarly on Nox1 expression and ROS generation, we examined the relationship between these molecules. Because MAPK7 phosphorylation is mediated by its upstream kinase MAP2K5, we first confirmed the effect of MAP2K5 on MAPK7 by transfecting constitutively active Map2k5. Western blot analysis at 2 d after transfection showed increased levels of phosphorylated MAPK7, which suggests that MAPK7 is indeed activated by MAP2K5 (Fig 3H and Table S2). However, we could not detect significant changes in MAPK14 phosphorylation levels in these cells. In contrast, when we transfected constitutively active Mapk14, we observed increased phosphorylation levels of both MAP2K5 and MAPK7 (Fig 3H). These results suggested that MAPK14 acts upstream of MAPK7 via MAP2K5.
To further confirm this relationship based on overexpression (OE) experiments, we used chemical inhibitors. SB203589 not only inhibited MAPK14 phosphorylation but also reduced MAPK7 phosphorylation, suggesting that MAPK14 activation is necessary for MAPK7 activation (Fig 3I and Table S2). In contrast, XMD 8-92 supplementation did not have any apparent effects on MAPK14 phosphorylation. The relationship between MAPK14 and MAPK7 was finally confirmed by Mapk14f/f GS cells, which showed decreased phosphorylation of MAPK7 upon AxCANCre treatment (Fig 3J and Table S2). These results suggested that MAPK14 and MAPK7 phosphorylation levels are closely related to each other.
Although XMD 8-92 inhibits GS cell proliferation, it was possible that XMD 8-92 suppressed proliferation of progenitors and may not have effect on SSCs. This is because the frequency of SSCs is relatively low (∼1–2%) in GS cell cultures (Kanatsu-Shinohara & Shinohara, 2013). Therefore, functional analysis of SSCs is required to demonstrate the effect of XMD 8-92 on GS cells. For this purpose, we carried out germ cell transplantation. We transplanted the cells at the beginning and end of the culture period and measured the increase in SSC number. GS cells treated with XMD 8-92 stopped proliferating and generated a significantly smaller number of colonies after spermatogonial transplantation (Fig 3K and L). The total increase in SSC number was also significantly attenuated during culture (Fig 3M). These results confirmed that XMD 8-92 inhibits the self-renewal division of SSCs in GS cell cultures.
Mapk7 gene targeting in SSCs
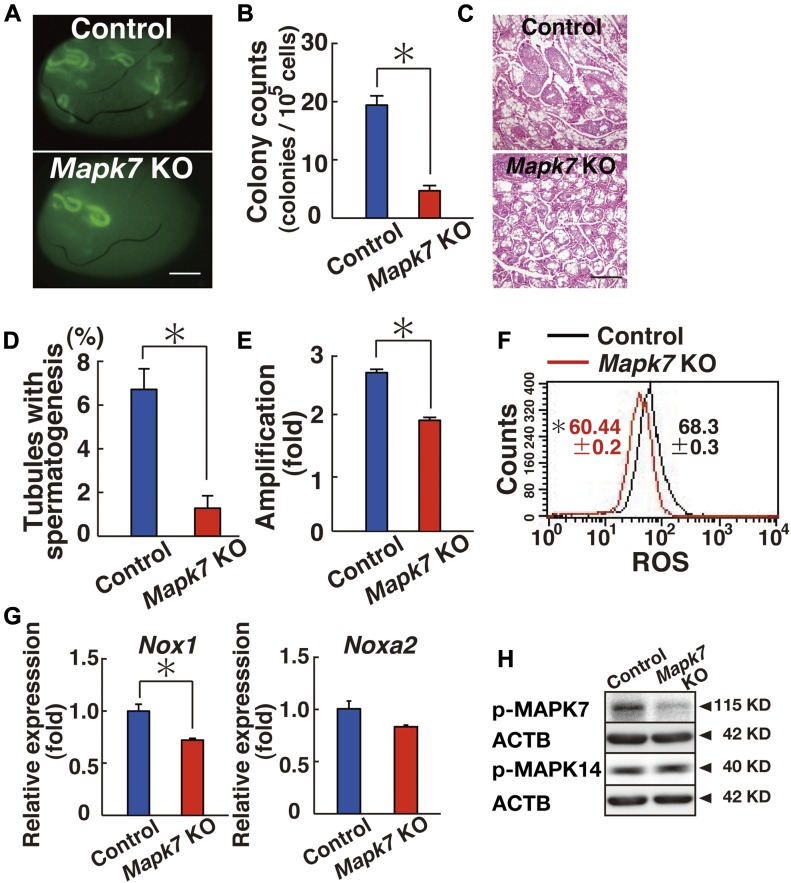
In the next set of experiments, we used a floxed mutant mouse strain that possess loxP sites flanking exon 4 of the Mapk7 gene (Mapk7f/f floxed mice) to confirm the effects of XMD 8-92 (Fig S4C) (Wang et al, 2006). The cells were collected from 5- to 10-d-old pup testes, and exposed to AxCANCre overnight. Testes from littermate mouse were used as controls. The transfected cells were recovered for transplantation the next day. After transplantation, an average of 57.5% and 65.5% of the input Mapk7f/f and control cells were collected by trypsinization, respectively, and the difference was not statistically significant (n = 4). Real-time PCR analysis confirmed deletion of the target gene at the time of transplantation (Fig S4D). Three experiments were performed.
The analysis of recipient mice revealed significantly decreased colony numbers after CRE-mediated deletion of Mapk7 (Fig 4A). Testis cells exposed to AxCANCre generated 4.7 colonies per 105 transplanted cells. In contrast, control testis cells generated 19.4 colonies per 105 cells (Fig 4B). The difference between the mutant and control samples was statistically significant. Histological analysis of the recipient testes also showed poor colonization of the Mapk7-deficient testis cells (Fig 4C). Spermatogenesis was detected in an average of 1.3% of the tubules that received mutant testis cells, whereas it was detected in an average of 6.7% of the seminiferous tubules that received control transplants (n = 10), and this difference was also statistically significant (Fig 4D).
Figure 4. Functional analysis of Mapk7 in SSCs using spermatogonial transplantation.
(A) Macroscopic appearance of a recipient testis that underwent transplantation of AxCANCre-treated Mapk7f/f testis cells. Scale bar: 1 mm. (B) Colony counts (n = 32 testes for Mapk7; n = 36 testes for control). *P < 0.05 (t test). (C) Histological appearance of a recipient testis. Scale bar: 250 μm. (D) Quantification of tubules with spermatogenesis. At least 1,247 tubules were counted. *P < 0.05 (t test). (E) Defective proliferation of Mapk7f/f KO GS cells exposed to AxCANCre (n = 16 cultures; moi = 2). The cells were recovered 6 d after transfection. *P < 0.05 (t test). (F) Flow cytometric analysis of ROS generation in AxCANCre-treated Mapk7f/f GS cells 3–4 d after transfection (n = 3 cultures; moi = 2). *P < 0.05 (linear regression). (G) Real-time PCR analysis of Nox1 and Noxa2 expression in Mapk7f/f GS cells exposed to AxCANCre 2 d after infection (n = 3 cultures; moi = 2). *P < 0.05 (t test). (H) Western blot analysis of Mapk7f/f GS cells 2 d after AxCANCre treatment (moi = 2). Data information: in (B, D–G), data are presented as mean ± SEM.
To examine the role of MAPK7 in GS cell proliferation, we derived GS cells from Mapk7 conditional KO mouse testes and exposed the cells to AxCANCre. In a manner similar to XMD 8-92, GS cells exposed to AxCANCre also proliferated slowly; whereas control cells increased 2.7-fold, Mapk7 KO cells increased only 1.9-fold, and this difference was significant (Fig 4E). Moreover, ROS generation was significantly reduced when AxCANCre was added to Mapk7f/f GS cells (Fig 4F and Table S1).
Because only chemical inhibitors were used to study the mechanism of Nox1 expression, we confirmed this relationship using Mapk14f/f and Mapk7f/f GS cells. As expected from the inhibitor experiments, real-time PCR showed down-regulation of Nox1 expression levels in both GS cell types after treatment with AxCANCre (Figs 2G and 4G). AxCANCre treatment of Mapk7f/f GS cells also did not influence MAPK14 phosphorylation, which also confirmed that MAPK7 acts downstream of MAPK14 (Fig 4H and Table S2). Considering that MAPK7 phosphorylation is decreased in Mapk14f/f GS cells after AxCANCre treatment (Fig 3J), the results of these experiments showed that MAPK7 acts downstream of MAPK14.
ROS generation by Mapk14 and Mapk7 activation
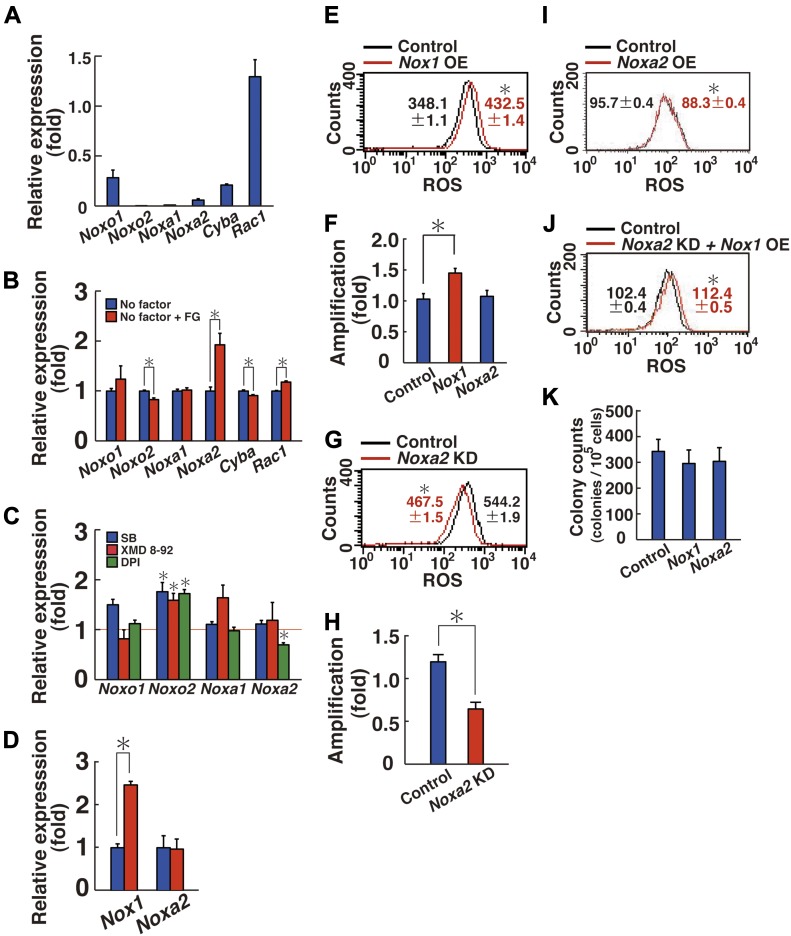
The generation of ROS by NOX1 requires additional components, including CYBA (p22phox), NOXO1 (or NOXO2 [p47phox]), NOXA1, and RAC1 (Bedard & Krause, 2007). Therefore, we searched for the genes responsible for ROS generation that allow ROS generation by co-transfection with Nox1. Real-time PCR analysis of stably growing GS cells showed that the cells express all of these genes except for Noxo2 (Fig 5A). When we examined the cytokine response by adding GDNF and FGF2 to starved GS cells, the expression of Noxa2 was most significantly up-regulated by cytokine restimulation (Fig 5B). This suggested that NOXA2 (p67phox) collaborates with NOX1 for ROS generation. In contrast, the addition of self-renewal factors only modestly changed the expression of other component genes. Therefore, these results suggested that Noxa2 is an important regulator of ROS generation in GS cells and that Nox1 and Noxa2 are regulated in a similar manner.
Figure 5. ROS generation and enhanced proliferation of GS cells by Nox1 transfection.
(A) Real-time PCR analysis of Nox1-related components in GS cells (n = 3 cultures). (B) Real-time PCR analysis of Nox1-related components by FGF2 and GDNF (FG) supplementation (n = 3 cultures). The cells were starved for 3 d, and FGF2 and GDNF were added for 6 h before sample collection. *P < 0.05 (t test). (C) Real-time PCR analysis of Nox1-related components 2 (DPI) or 3 d (SB203589 and XMD 8-92) after supplementation with the indicated inhibitors (n = 3 cultures). Mean values for control experiment are indicated by the horizontal line. *P < 0.05 (t test). (D) Real-time PCR analysis of indicated genes 10 min after hydrogen peroxide (90 μM) supplementation (n = 3 cultures). *P < 0.05 (t test). (E) Flow cytometric analysis of ROS generation 3 h after Nox1 transfection (n = 3 cultures; moi = 10). (F) Cell recovery 5 d after Nox1 or Noxa2 transfection (n = 6 cultures; moi = 10). *P < 0.05 (ANOVA). (G) Flow cytometric analysis of ROS generation 6 d after Noxa2 depletion (n = 3 cultures; moi = 4). *P < 0.05 (linear regression). (H) Cell recovery 7 d after Noxa2 depletion (n = 6 cultures; moi = 4). *P < 0.05 (t test). (I) Flow cytometric analysis of ROS generation 3 h after Noxa2 transfection (n = 3 cultures; moi = 4). (J) Flow cytometric analysis of ROS generation 3 h after Nox1 transfection into GS cells, in which Noxa2 was depleted 6 d before (n = 3 cultures; moi = 4). Eyfp was transfected into Noxa2-depleted cells as a control. (K) Colony counts after spermatogonial transplantation of GS cells transfected with the indicated genes (n = 12 testes). *P < 0.05 (t test). Data information: in (A–K), data are presented as mean ± SEM.
To test this hypothesis, we first used chemical inhibitors. When GS cells are proliferating by GDNF and FGF2 stimulation, the expression of Noxa2 was suppressed only by DPI (Fig 5C). Moreover, adding hydrogen peroxide induced the expression of Nox1 but not Noxa2 (Fig 5D). To confirm the impact of these genes, we checked its expression using Mapk14 and Mapk7 KO GS cells (Figs 2G and 4G). However, deletion of these genes did not show significant effects on Noxa2 expression. These results suggested that Nox1 and Noxa2 are regulated in a different manner.
We also compared the effect of Noxa2 in ROS generation and cell proliferation. Transfection experiments revealed that Nox1 transfection alone is sufficient to generate ROS and enhances cell proliferation (Fig 5E and F, and Table S1). Although Noxa2 depletion decreased ROS and impaired cell proliferation (Figs 5G, H, and S6A), neither ROS increase nor enhanced proliferation occurred after Noxa2 transfection (Fig 5F and I, and Table S1). As expected, Nox1 transfection generated ROS even when Noxa2 was inhibited by shRNA (Fig 5J and Table S1). Spermatogonial transfection showed that the colony counts did not change after Nox1 or Noxa2 transfection (Fig 5K), which suggested that ROS levels did not influence SSC concentration in GS cell cultures. Taken together, these results suggest that NOX1 is primarily important for ROS generation.
Figure S6. KD efficiency of target gene expression.
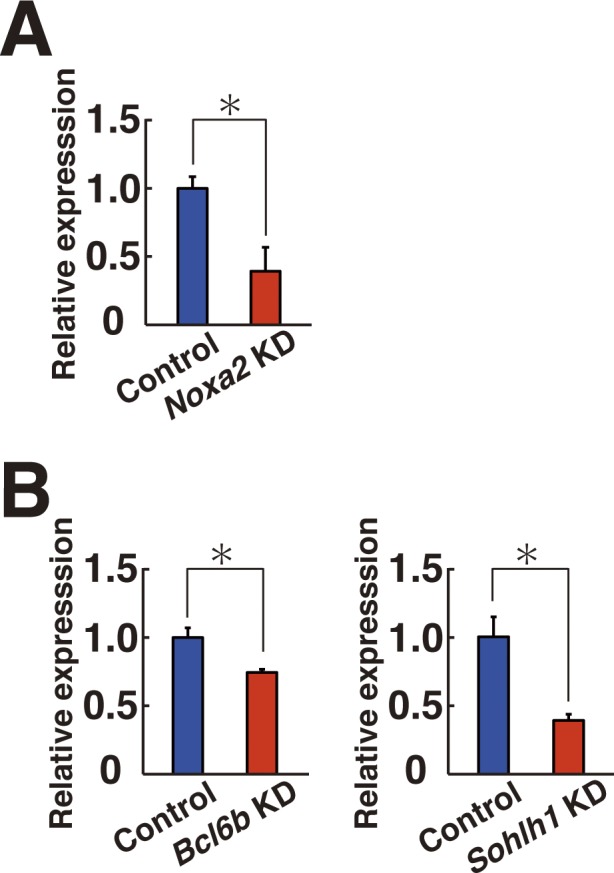
(A, B) Real-time PCR analysis of Noxa2 (A), Bcl6b (B), and Sohlh1 (B) expression after KD (n = 3 cultures; moi = 4). The cells were collected at 2 (Bcl6b) or 5 d (Noxa2 or Sohlh1) after transfection. *P < 0.05 (t test). Data information: in (A, B), data are presented as mean ± SEM.
Involvement of Bcl6b in ROS generation in GS cells
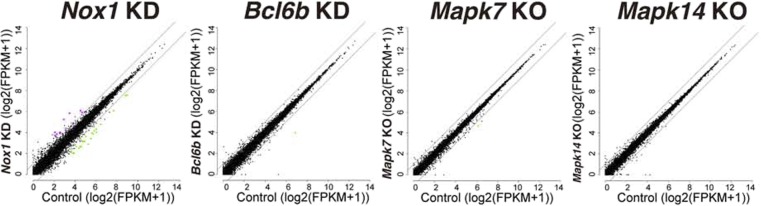
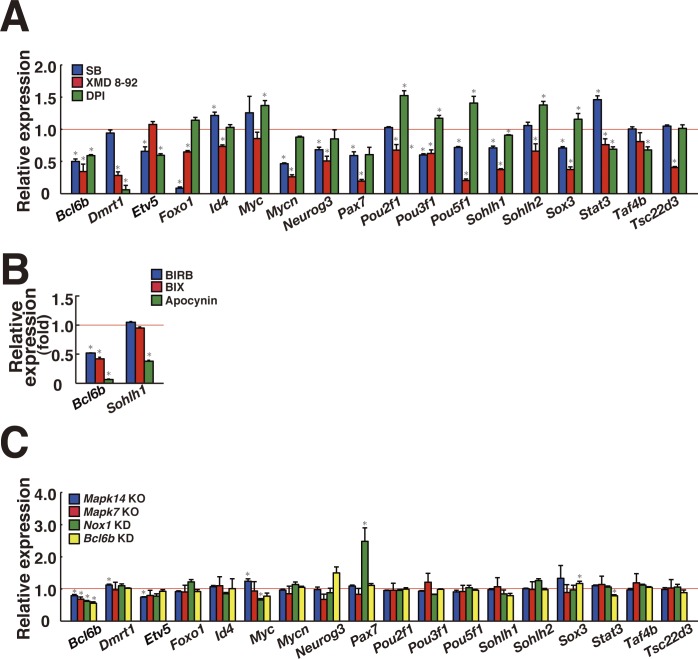
Because the induction of ROS by Nox1 transfection increased GS cell proliferation, we reasoned that a subset of transcription factors acts downstream of ROS to drive self-renewal division. To identify such genes, we initially carried out microarray and RNA sequencing (RNA-seq) using Mapk14f/f GS cells, Mapk7f/f GS cells, or Nox1 KD GS cells. However, we were not able to identify genes that are commonly involved in ROS regulation (Fig S7, Tables S3–S6). Because it was possible that lack of commonly expressed genes could be due to relatively low sensitivity of the assays (Mehta et al, 2016), we screened for spermatogonial transcription factor genes down-regulated by SB203589, XMD 8-92, and DPI by real-time PCR. Triplicate analysis of GS cells revealed down-regulation of many transcription factors whose deficiency causes defective spermatogenesis or self-renewal (Fig S8A), suggesting that ROS-dependent transcription factors have important functions in SSCs and spermatogenesis. In particular, the Bcl6b and Sohlh1 genes were commonly down-regulated by the three inhibitors (Table S6).
Figure S7. Scatter plots represent log2 (FPKM (average) +1) values for genes in control and KO/KD samples.
The magenta and green dots indicate up-regulated and down-regulated genes (>2-folds, q-values < 0.05), respectively, in each KO/KD sample. The diagonal lines are shown as twofold changes. The cells were collected at 7 d after the treatment (moi = 4). For Mapk14f/f, the cells were collected between 5 and 6 d after AxCANCre treatment (moi = 2).
Figure S8. Screening of candidate transcription factors that are sensitive to ROS levels.
(A) Real-time PCR analysis of transcription factors in GS cells 3 d after SB203589 or XMD 8-92 supplementation (n = 3). For DPI, the cells were recovered on the next day (n = 3). Mean values for control experiment are indicated by horizontal line. *P < 0.05 (t test). (B) Real-time PCR analysis of Bcl6b and Sohlh1 in GS cells 5 d after BIRB796 and BIX02189 (n = 3). For apocynin, the cells were recovered 2 d after the treatment (n = 3). *P < 0.05 (t test). (C) Real-time PCR analysis of spermatogonial transcription factors in GS cells. Mapk14f/f and Mapk7f/f GS cells were recovered 7 and 10 d after AxCANCre treatment, respectively (n = 6). For Nox1 or Bcl6b KD, the cells were recovered 8 and 3 d after transfection, respectively (n = 3 for Nox1; n = 6 for Bcl6b). *P < 0.05 (t test). Data information: in (A–C), data are presented as mean ± SEM.
Table S3 Microarray analysis. (45.8KB, docx)
Table S4 RNA sequence analysis. (1.8MB, docx)
Table S5 Raw values for candidate transcription factors determined by microarray. (34.5KB, docx)
Table S6 Raw values for candidate transcription factors determined by RNA-seq. (19.6KB, docx)
To confirm this result, the effect of individual chemical inhibitors on these target genes was further examined by another set of inhibitors (BIRB796, BIX02189, and apocynin) against the same molecules. Although Bcl6b was down-regulated by all of these inhibitors, only apocynin down-regulated Sohlh1 (Fig S8B). Moreover, when we checked the expression of all candidate genes in Mapk14 KO, Mapk7 KO, and Nox1 KD GS cells by real-time PCR, only Bcl6b showed consistent down-regulation (Fig S8C). These results suggested that Bcl6b is the strongest candidate in ROS regulation.
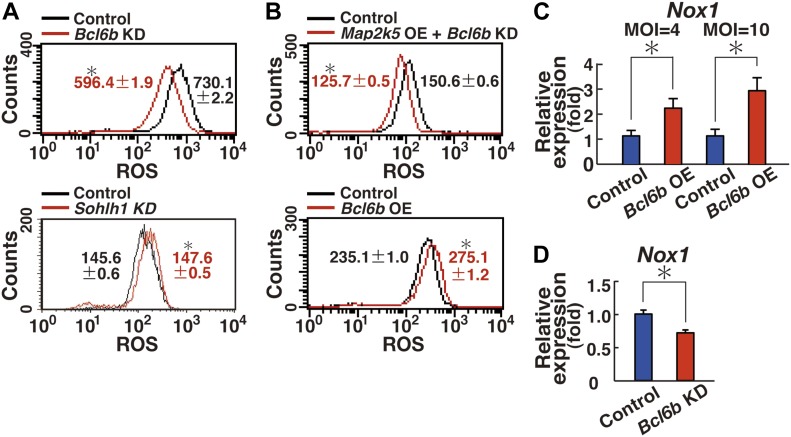
To determine whether the candidate genes are involved in ROS generation, GS cells were transfected with shRNA and ROS levels were examined by flow cytometry 7 d after transfection. Whereas depletion of Sohlh1 did not show reduced ROS, GS cells transfected with shRNA against Bcl6b showed significantly reduced ROS (Figs 6A and B, and Table S1). As expected, Bcl6b depletion also inhibited the ROS caused by transfection of constitutively active Map2k5 (Fig 6B and Table S1). We then examined whether Bcl6b is sufficient for ROS generation and found that Bcl6b OE could indeed induced ROS (Fig 6B and Table S1). Consistent with this observation, real-time PCR analysis revealed that Bcl6b transfection induced Nox1 in a dose-dependent manner (Fig 6C). Real-time PCR analysis also showed Bcl6b KD significantly decreased Nox1 expression (Fig 6D). These results suggest that Bcl6b is responsible for ROS generation.
Figure 6. Nox1 up-regulation by Bcl6b.
(A) Flow cytometric analysis of ROS generation 7 d after depletion of the indicated genes by shRNA transfection (n = 3 cultures; moi = 4). *P < 0.05 (linear regression). (B) Flow cytometric analysis of ROS generation after transfection of the indicated genes (n = 3 cultures; moi = 4). The cells were recovered 2 (Bcl6b OE) or 4 d (Map2k5 OE + Bcl6b KD) after transfection. *P < 0.05 (linear regression). (C) Real-time PCR analysis of Nox1 expression 1 d after transfection with the indicated genes, respectively (n = 6 cultures for moi = 4; n = 4 cultures for moi = 10). *P < 0.05 (t test). (D) Real-time PCR analysis of Nox1 expression 8 d after Bcl6b depletion (n = 6 cultures; moi = 4). *P < 0.05 (t test). Data information: in (A–D), data are presented as mean ± SEM.
ETV5-mediated nuclear translocation of BCL6B
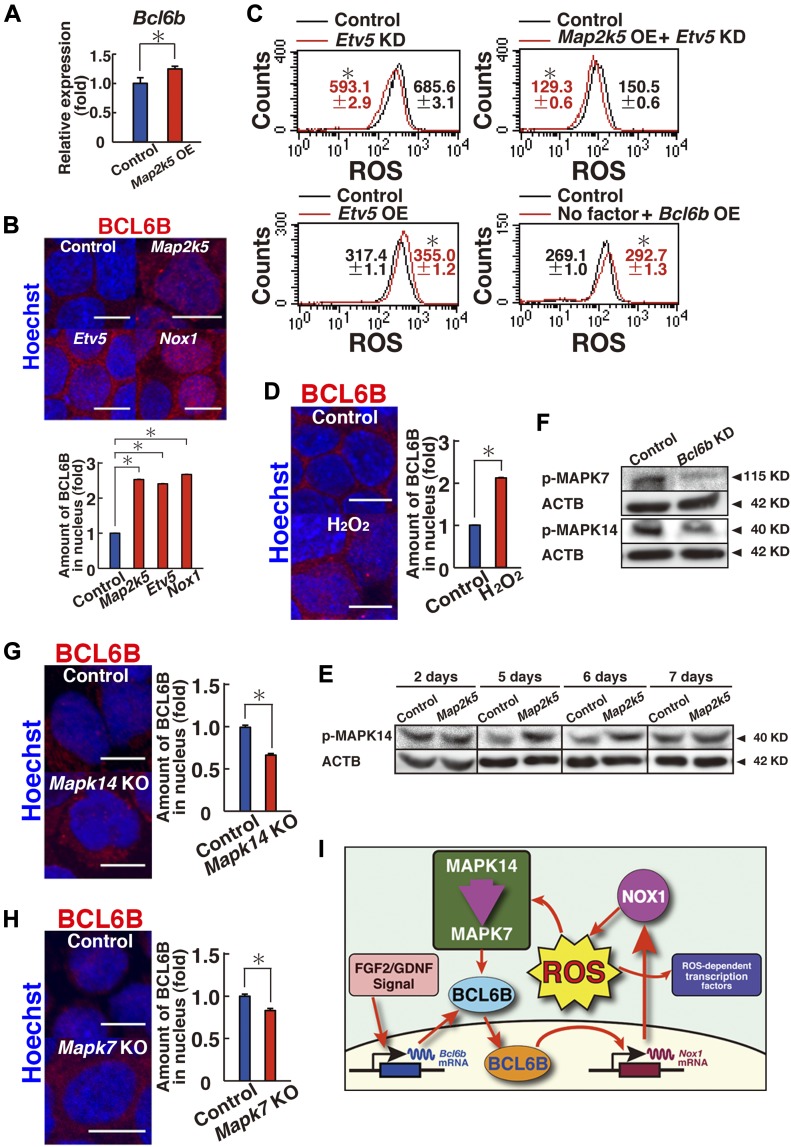
To clarify the relationship between BCL6B and the MAPK14/MAPK7 signaling pathway, we performed real-time PCR to examine changes in Bcl6b expression levels. Although GS cells transfected with constitutively active Map2k5 showed a modest but significant increase in Bcl6b expression (Fig 7A), we reasoned that the modest induction of Bcl6b could not explain the significant increase in ROS generation. We then performed immunostaining of the transfected cells to assess the changes in BCL6B localization. Although BCL6B is a transcription factor, BCL6B was predominantly found in the cytoplasm of GS cells, whereas it exhibited only weak signals in the nucleus (Figs 7B and S9A). However, when GS cells were transfected with constitutively active Map2k5, the density of BCL6B increased by 2.5-fold in the nucleus (Fig 7B), which suggests that Map2k5 regulates the nuclear translocation of BCL6B.
Figure 7. Nuclear translocation of BCL6B.
(A) Real-time PCR analysis of Bcl6b expression 1 d after transfection of Map2k5 (n = 6 cultures; moi = 4). *P < 0.05 (t test). (B) Immunostaining and quantification of nuclear BCL6B staining in GS cells 2 d after transfection of the indicated genes (moi = 4). At least 119 cells were counted for quantification. Scale bar: 5 μm. *P < 0.05 (ANOVA). (C) Flow cytometric analysis of ROS generation following transfection of the indicated genes (n = 3; moi = 4). The cells were recovered on 3 (Etv5 OE), 4 (Map2k5 OE + Etv5 KD), or 5 d (Etv5 KD) after transfection. For cytokine starvation, the cells were cultured without cytokines for 2 d, and recovered 1 d after transfection. *P < 0.05 (linear regression). (D) Immunostaining and quantification of nuclear BCL6B staining of GS cells 2 d after hydrogen peroxide treatment. At least 133 cells were counted for quantification. Scale bar: 5 μm. *P < 0.05 (t test). (E) Western blot analysis of MAPK14 in GS cells at indicated time pointes after transfection with constitutively active Map2k5 (moi = 4). (F) Western blot analysis of MAPK7 and MAPK14 in GS cells 9 d after Bcl6b KD (moi = 4). (G, H) Immunostaining and quantification of nuclear BCL6B staining of Mapk14f/f (G) or Mapk7f/f (H) GS cells 1 d after AxCANCre treatment (moi = 4). At least 205 cells were counted for quantification. Scale bar: 5 μm. *P < 0.05 (t test). (I) Summary of results. MAPK14 induces MAPK7 phosphorylation, which promotes BCL6B entry into the nucleus via ETV5. BCL6B induces expression of Nox1. ROS generated by NOX1 activate MAPK14, thereby creating a positive feedback loop. ROS also activate many transcription factors. Data information: in (A–D, G, H), data are presented as mean ± SEM.
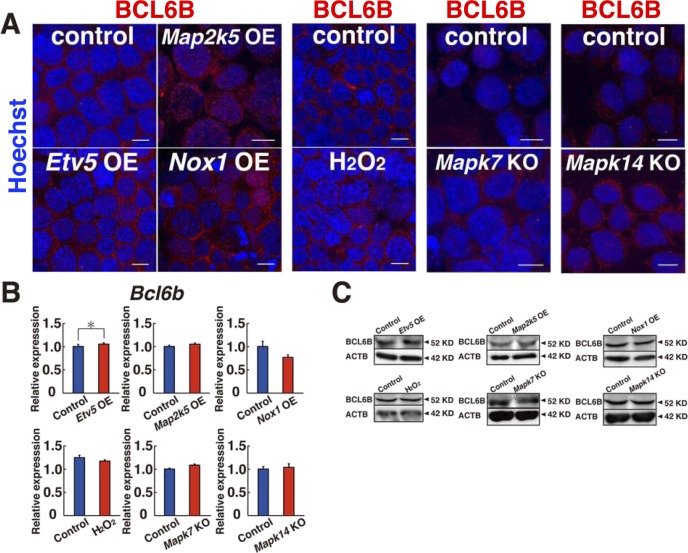
Figure S9. Expression of BCL6B in GS cells after indicated treatment.
The cells were analyzed 2 d after the treatment (moi = 4). For Mapk7 KO and Mapk14 KO cells, immunostaining was performed 1 d after AxCANCre treatment (moi = 2). (A) Immunostaining. (B, C) Real-time PCR (B) and Western blot (C) (n = 3 cultures). Scale bar: 10 μm. *P < 0.05 (t test). Data information: in (B), data are presented as mean ± SEM.
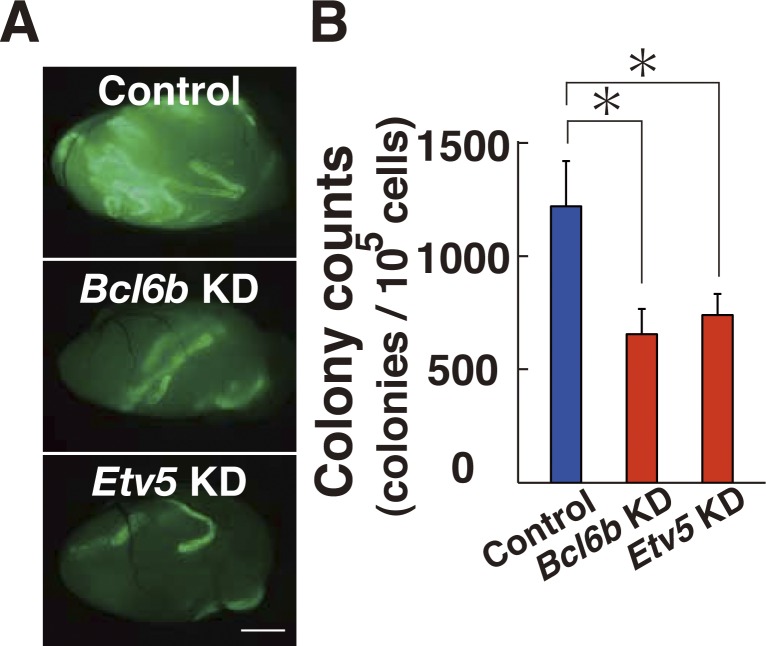
To gain insight into the underlying mechanism, we focused on the role of Etv5. Etv5 is essential for SSC self-renewal, and Etv5 KO mice results in loss of spermatogenesis (Chen et al, 2005). It also regulates CXCR4 expression and may regulate SSC homing (Wu et al, 2011). Although Etv5 expression was not suppressed by XMD 8-92 or Bcl6b KD (Fig S8A and C), it was down-regulated in Mapk14 KO GS cells (Fig S8C). Because Etv5 transfection induces Bcl6b up-regulation in stable transfectants (Ishii et al, 2012), we hypothesized that ETV5 might play a role in BCL6B translocation into the nucleus. Consistent with this idea, when we depleted Etv5 by shRNA, GS cells showed a significant decrease in ROS (Fig 7C and Table S1). The same treatment also blocked ROS induced by constitutively active Map2k5 OE (Fig 7C). On the other hand, transfection of constitutively active Etv5 generated ROS (Fig 7C), suggesting that ETV5 acts downstream of MAPK14/MAPK7 signaling pathway for ROS. As expected from these observations, transfection of constitutively active Etv5 increased the number of cells expressing that contain BCL6B in the nucleus (Fig 7B). The density of BCL6B staining in the nucleus was increased 2.4-fold by constitutively active Etv5 transfection, suggesting that Etv5 regulates BCL6B translocation into the nucleus. GS cells that had been depleted of Etv5 or Bcl6b by shRNA showed significantly reduced SSC activity (Fig S10A and B), when constitutive active Map2k5 was transfected, which confirmed the critical role of this signaling pathway in SSC self-renewal.
Figure S10. Spermatogonial transplantation of GS cells transfected with constitutive active Map2k5 and shRNA for Bcl6b or Etv5.
The cells were transplanted 2 d after transfection. (A) Macroscopic appearance of recipient testes 2 mo after transplantation of active Map2k5 and shRNA for Bcl6b or Etv5. The cells were transplanted 2 d after transfection. (B) Colony counts (n = 15–18 testes). Scale bar: 1 mm. *P < 0.05 (ANOVA). Data information: in (B), data are presented as mean ± SEM.
Because the addition of hydrogen peroxide promoted GS cell proliferation and increased Nox1 expression (Fig 5D) (Morimoto et al, 2013), we reasoned that the ROS generated by Nox1 also promote nuclear translocation of BCL6B, thereby creating a positive feedback loop. To test this hypothesis, we transfected Nox1 or added hydrogen peroxide, both of which also increased BCL6B nuclear localization 2.7- or 2.1-fold, respectively (Figs 7B, D, and S9A). Although these experiments were performed using stably growing GS cells, ROS induction occurred even when Bcl6b was transfected into cytokine-starved GS cells (Fig 7C and Table S1), indicating that Bcl6b is primarily responsible for ROS generation even after cytokine deprivation. Although Etv5 transfection induced modest increase in Bcl6b mRNA expression, none of the treatments that increase ROS levels did not influence protein levels of BCL6B (Fig S9B and C). Consistent with this idea, we also noted delayed phosphorylation of MAPK14 after Map2k5 transfection (Fig 7E and Table S2). Map2k5 transfection did not influence MAPK14 2 d after transfection, but it induced phosphorylation at 5–6 d after transfection. Blc6b expression was important for MAPK14 and MAPK7 activation because Blc6b KD down-regulated phosphorylated MAPK14 and MAPK7 levels 9 d after KD (Fig 7F and Table S2). Considering that MAPK14 activation induces MAPK7 phosphorylation and the fact that MAPK7 activation causes delayed MAPK14 activation (Figs 3H and 7E), these results suggested that a positive feedback loop operates between MAPK14 and MAPK7 (Fig 7I).
Finally, we examined the contribution of BCL6B to the defective growth of Mapk14 KO and Mapk7 KO GS cells. Immunostaining of both cell types showed that intranuclear BCL6B expression is decreased upon Cre treatment (Figs 7G, H, and S9A). Because Cre treatment of both cell types also decreased Nox1 expression (Figs 2G and 4G), these results suggest that ETV5 drives the ROS generation by regulating the nuclear translocation of BCL6B, which subsequently up-regulates Nox1 to induce ROS.
Discussion
Mapk14 has context-dependent roles in different tissues. It is involved in the proliferation of stem cells in the lungs and muscle (Ventura et al, 2007; Bernet et al, 2014), whereas its activation leads to senescence in hematopoietic stem cells (Ito et al, 2006). In this respect, SSCs are similar to stem cells in the lungs and muscle in that Mapk14 positively contributes to self-renewal division. Mapk14 activation can be caused by increased ROS levels. Previous studies have suggested a link between Mapk14 and Nox1, but this relationship is not clear and appears to depend on the tissue type. In the present study, we showed that ROS and MAPK14 create a positive feedback loop to promote SSC self-renewal. To our knowledge, this is the first demonstration of ROS-mediated positive feedback in sustaining stem cell self-renewal.
Our chemical screening identified MAPK7 as a critical regulator of ROS generation and Nox1 expression. Although MAPK7 and MAPK1/3 share the Thr-Glu-Tyr activation motif, MAPK7 is different from MAPK1/3 in that MAPK7 has a long C-terminal domain with transcriptional activity. In addition, Mapk7 KO mice show a phenotype distinct from that of Mapk1/3 KO mice (Hayashi & Lee, 2004). Therefore, despite its similarity to MAPK1/3, MAPK7 plays a distinct role. In mouse ES cells, MAPK7 phosphorylation occurred downstream of BMP4 via MAP2K5, which is critical for mediating BMP4-induced self-renewal (Morikawa et al, 2016). In GS cells, we found a critical role for MAPK7 in self-renewal, but it was apparently different because MAPK7 expression was accompanied by Nox1 induction and ROS generation. Because Mapk7 KO SSCs showed significantly reduced self-renewal activity and MAPK7 phosphorylation was induced by constitutively active Mapk14 OE, we initially thought that MAPK7 acts downstream of MAPK14 and is similarly involved in ROS generation.
The importance of Nox1 in ROS generation in GS cells was clearly demonstrated by transfection experiments. Although ROS can be induced in cultured cells by transfection of Nox genes, transfection of additional components, such as Noxa or Noxo genes is usually required (Bedard & Krause, 2007). In GS cells, however, many of these components are already expressed without induction by cytokines. Although self-renewal factor stimulation also increased Noxa2 expression and Noxa2 depletion decreased cell recovery, transfection of Nox1 alone was sufficient for ROS production, suggesting that the level of Nox1 expression is the most critical factor in ROS generation. Because Nox1 OE increased ROS and adding hydrogen peroxide induced Nox1 expression, these results led us to speculate that ROS-dependent transcription factors drive a positive feedback loop to sustain self-renewal.
Based on this notion, we screened transcription factors that respond to ROS signals and identified that Bcl6b is the most critical transcription factor for ROS generation. Although we failed to identify commonly regulated genes by microarray and RNA-seq, we think that this was caused by the low sensitivity of RNA-seq (Mehta et al, 2016). Moreover, the timing of RNA analysis is complicated by the positive feedback loop because each analyzed molecule contributes to the regulation of BCL6B localization in a different manner (i.e., phosphorylation, ROS generation, and transcription repression). The involvement of Bcl6b in SSC self-renewal was initially discovered by determining the genes induced by GDNF supplementation (Oatley et al, 2006). However, FGF2 also induces Bcl6b via Etv5 induction (Ishii et al, 2012). Although Bcl6b was thought to drive self-renewal genes, our results suggest that the role of Bcl6b is not so simple. Our analyses suggest that Nox1 is one of the critical effectors of BCL6B. Moreover, delayed MAPK14 phosphorylation by Map2k5 transfection strongly suggested that ROS generated by NOX1 creates a positive feedback loop because ROS also induces BCL6B translocation and Nox1 up-regulation. We think that this phase of self-renewal may be considered as maintenance phase, which should be analyzed separately from initial Bcl6b up-regulation by self-renewal signal. Therefore, BCL6B translocation is regulated by a complex manner involving MAPK14/MAPK7 signaling and ETV5. This link between BCL6B and NOX1 to amplify ROS may explain why Nox1 plays such important roles in SSCs despite its relatively low expression levels.
At least three questions arose from this study. The first concerns the effect of ROS on transcription factors. Although only Bcl6b was able to induce ROS, several other transcription factors also showed significant changes in expression after ROS depletion by DPI and their significance in SSC self-renewal is unknown. Because chemicals can have off-targets, we still cannot totally exclude the possibility that other pathways may be involved. It has been known that GDNF and FGF2 induce significant changes in the gene expression profiles. However, the degree and the impact of ROS-dependent and ROS-independent pathways on these changes in gene expression have been poorly addressed. The second question concerns understanding the mechanism of ETV5-mediated increase in nuclear BCL6B. We speculate that ETV5 may up-regulate molecules that help nuclear transport. BCL6, which is a close counterpart of BCL6B, interact with many proteins, including NCOR2 or BCOR (Huynh et al, 2000). It is possible that ETV5 may regulate such additional targets, which may help transport BCL6B into the nucleus. The last concerns the mechanism of ROS-based MAPK14 activation. ROS can activate MAPK by oxidative modification of MAPK signaling proteins (e.g., receptor tyrosine kinase or MAPK kinase; MKP) or inactivation of MAPK phosphatases. For example, ROS can activate the EGF receptor in the absence of its ligand. It has been also demonstrated that ROS-induced MKP inactivation causes sustained activation of JNK pathway (Hou et al, 2008). However, we still do not know such regulatory molecules responsible for feedback regulation in SSCs. Future studies are required to resolve these problems.
In many self-renewing tissues, ROS are harmful to stem cells, but some tissues require ROS for self-renewal. However, the positive role of ROS in self-renewal division has only recently begun to be analyzed. Our study shows that in SSCs, ROS amplification plays a critical role to drive self-renewal division. Because ROS can be toxic to germ cells and cause spermatogenic defect, success of spermatogenesis appears to depend on the delicate control of ROS levels. Identification and functional analysis of such ROS regulatory molecules will enhance our understanding of the molecular mechanisms of spermatogenesis and ROS-mediated self-renewal in tissue-specific stem cells.
Materials and Methods
Animals and cell culture
Nox1 KO, Mapk14f/f, and Mapk7f/f conditional KO mice were generated as described previously (Nishida et al, 2004; Matsuno et al, 2005; Wang et al, 2006). These mice were crossed with transgenic Gt (ROSA)26Sortm1(EYFP)Cos mice (designated R26R-Eyfp; a gift from Dr. F Costantini, Columbia University Medical Center, New York, NY) to introduce a donor cell marker for transplantation experiments. GS cells from a transgenic mouse line of B6-TgR(ROSA26)26Sor mice (designated ROSA) (Jackson Laboratory) were previously described (Kanatsu-Shinohara et al, 2003), and GS cells were derived from Mapk14f/f and Mapk7f/f conditional KO mice using culture medium based on Iscove’s modified MEM (Invitrogen), which was supplemented with 10 ng/ml human FGF2, 15 ng/ml recombinant rat GDNF (both from Peprotech), and 1% FBS, as described previously (Kanatsu-Shinohara et al, 2014). Cultures were maintained on mitomycin C–treated mouse embryonic fibroblasts (MEFs). Where indicated, hydrogen peroxide (30 μM) or NAC (0.5 mM; Sigma) was added to the cultures. The chemicals used in the study are listed in Table S7. FGF2 and GDNF are supplemented in the medium unless stated otherwise.
Table S7 Chemicals used in this study. (13.8KB, docx)
Transplantation
Spermatogonial transplantation was carried out as described previously using busulfan (44 mg/ml)-treated C57BL/6 (B6) × DBA/2 F1 (BDF1) mice (Ogawa et al, 1997). Recipients were used at least 4 wk after busulfan treatment. For histological analysis to evaluate donor-derived spermatogenesis levels, we used 4–8-wk-old WBB6F1-W/Wv (W) mice (Japan SLC) because they lack endogenous spermatogenesis. Approximately 10 or 4 μl of cell suspension was microinjected through the efferent duct of busulfan-treated or W mice, respectively. Each injection filled 75–85% of all seminiferous tubules. The Institutional Animal Care and Use Committee of Kyoto University approved all of our animal experimentation protocols.
Viral transfection
For OE experiments, mouse Mapk14 D176A-F327S (a gift from Dr. D Engelberg, National University of Singapore, Singapore) and mouse Map2k5 cDNA (a gift from Dr. E Nishida, Kyoto University, Kyoto, Japan) were cloned into the CSII-EF-IRES2-Puro lentivirus vector. We also produced a mutant mouse Etv5 (T135D/T139D/S142D) and introduced into CSII-EF-IRES2-Puro. CSII-EF-IRES2-Puro expressing Bcl6b was described previously (Ishii et al, 2012). CSII-EF-Efyp-IRES2-Puro was used as a control. For shRNA-mediated gene KD, all vectors were purchased from Open Biosystems. A mixture of lentiviral particles was used to transfect GS cells or testis cells. pLKO1-Scramble shRNA (Addgene) was used as a control (Open Biosystems). All KD vectors are listed in Table S8. Lentiviral transfection was carried out as described previously using polyethylenimine MAX (Polysciences) (Morimoto et al, 2015). The virus culture supernatant was concentrated by ultracentrifugation at 50,000 g for 2 h. The virus titer was determined by infection of 293T cells, using the qPCR Lentivirus titration (Titer) kit (Abcam). We used AxCANCre and AxCANLacZ (RIKEN BRC) for adenoviral infection. Virus particles preparation and titer determination were carried out as described previously (Takehashi et al, 2007).
Table S8 KD vectors used in this study. (18.1KB, docx)
Analysis of recipient mice
Recipients were euthanized 2 mo after transplantation, and donor cell colonies were examined under UV light. Donor cell clusters were defined as colonies when the entire basal surface of the tubule was occupied and was at least 0.1 mm in length (Nagano et al, 1999).
For the evaluation of donor cell colonization levels by histology, W recipient testes were embedded in paraffin, and histological sections were stained with hematoxylin and eosin. The number of tubules with spermatogenesis, defined based on the presence of multiple layers of germ cells in the entire circumference of the tubules, was recorded for one section from each testis.
Immunostaining
For immunohistochemistry of Nox1 KO mouse testes, the samples were fixed in 4% paraformaldehyde for 2 h. They were then embedded in Tissue-Tek OCT Compound (Sakura Finetek) for cryosectioning. Immunostaining of cryosections was performed by treating samples with 0.1% Triton-X in PBS. For staining GS cells, single-cell suspensions were concentrated on glass slides by centrifugation using a Cytospin 4 unit (Thermo Electron Corp.). After immersing the slides in blocking buffer (0.1% Tween 20, 1% bovine serum albumin, and 10% donkey serum in PBS) for >1 h, the samples were incubated with antibodies against the indicated antigens. Secondary antibodies were incubated for 1 h at room temperature. The samples were counterstained with Hoechst 33342 (Sigma). The images were collected using confocal microscope (Fluoview FV1000D; Olympus). Images were analyzed by Photoshop software (Adobe Systems). Quantification of BCL6B levels was performed using MetaMorph software (Molecular Devices). All antibodies are listed in Table S9.
Table S9 Antibodies used in this study. (18.8KB, docx)
Flow cytometry
To measure the ROS levels in GS cells, we used CellROX Deep Red Reagent (Thermo Fisher Scientific) following the manufacturer’s instructions (Morimoto et al, 2015). In brief, the cells were dissociated with trypsin and incubated for 30 min at 37°C with 5 μM CellROX Deep Red in GS cell culture medium containing 1% FBS (Kanatsu-Shinohara et al, 2014). After washing with PBS containing 1% FBS (PBS/FBS), the stained cells were analyzed using the FACSCalibur (BD Biosciences). Three separate experiments using at least two different cell lines were carried out for each staining. Flow cytometric analysis of the same experimental data set was analyzed under the same conditions. All antibodies are listed in Table S9.
Microarray
To examine genome-wide gene expression profiles, total RNA samples were extracted with TRIzol reagent (Invitrogen) and purified using the RNeasy cleanup system (QIAGEN). Microarray data collection and analysis were carried out as described previously (Tanaka et al, 2016).
RNA-seq
For library preparation, total RNA was extracted using RNeasy Mini kit (QIAGEN) following the manufacturer’s instructions. RNA-seq libraries were generated from 200 ng total RNA with the TruSeq Stranded mRNA LT Sample Prep kit according to the manufacturer’s protocol (Illumina). The obtained RNA-seq libraries were sequenced on NextSeq 500 (86-bp single-end reads, Illumina).
The sequenced reads were mapped to the mm10 mouse reference genome using HISAT2 (version 2.1.0) (Kim et al, 2015) with the GENCODE annotation gtf file (version M17) after trimming adaptor sequences and low-quality bases by cutadapt-1.16 (Martin, 2011). The uniquely mapped reads (mapping quality [MAPQ] ≥ 20) were used for further analyses. The differentially expressed genes (>2-fold changes and q-values <0.05 between each pair of samples) were identified using Cuffdiff within Cufflinks version 2.2.1 package and GENCODE annotation gtf file (version M17, protein coding genes) (Trapnell et al, 2013). Fragments per kilobase million (FPKM) were calculated as the expression values by Cuffdiff, and the low expressed genes (<10 FPKM) both in control and KD/KO samples were filtered out from the differentially expressed genes.
PCR analyses
Total RNA was isolated using TRIzol (Invitrogen), and first-strand cDNA was synthesized using the Verso cDNA Synthesis kit (Thermo Fisher Scientific). For real-time PCR, the StepOnePlus Real-Time PCR System and Power SYBR Green PCR Master Mix were used according to the manufacturer’s protocol (Applied Biosystems). Transcript levels were normalized relative to those of Hprt. PCR conditions were 95°C for 10 min, followed by 40 cycles of 95°C for 15 s, and 60°C for 1 min. Each reaction was performed in duplicate. PCR primer sequences are listed in Table S10.
Table S10 PCR Primers used in this study. (20.7KB, docx)
Western blot analysis
Samples were separated by SDS–PAGE, transferred onto Hybond P membranes (Amersham Biosciences), and incubated with primary antibodies. All antibodies are listed in Table S9.
Statistical analyses and data visualization
Microsoft Excel was used for statistical evaluation using unpaired two-tailed t tests. Multiple comparison analyses were performed using ANOVA followed by Tukey’s honest significant difference test. For analysis of flow cytometry, fluorescence values obtained from FlowJo software (TreeStar) were used to compare control and experimental samples by linear regression model adjusting for the timing of experiments. Adobe illustrator (CS5) was used for compiling figures and designing schematic figures. All variance is reported as SEM, and P-values for all statistically analyzed experiments are listed in Table S1.
Data availability
The accession numbers for the microarray and RNA-seq data are GSE106103 and GSE118191, respectively.
Supplementary Material
Acknowledgements
We thank Ms. Y Ogata and S Ikeda for the technical assistance. This research was supported by Grants-in-aid for MEXT (18H05281, 25112003).
Author Contributions
H Morimoto: conceptualization, formal analysis, investigation, and writing—original draft.
M Kanatsu-shinohara: conceptualization, investigation, and writing—original draft, review, and editing.
N Ogonuki: investigation.
S Kamimura: investigation.
A Ogura: investigation.
C Yabe-Nishimura: resources.
Y Mori: formal analysis.
T Morimoto: formal analysis.
S Watanbe: formal analysis.
K Otsu: resources.
T Yamamoto: data curation and investigation.
T Shinohara: conceptualization, formal analysis, funding acquisition, investigation, and writing—original draft, review, and editing.
Conflict of Interest Statement
The authors declare that they have no conflict of interest.
References
- Bedard K, Krause KH (2007) The NOX family of ROS-generating NADPH oxidases: Physiology and pathophysiology. Physiol Rev 87: 245–313. 10.1152/physrev.00044.2005 [DOI] [PubMed] [Google Scholar]
- Bernet JD, Doles JD, Hall JK, Tanaka KK, Carter TA, Olwin BB (2014) p38 MAPK signaling underlies a cell-autonomous loss of stem cell self-renewal in skeletal muscle of aged mice. Nat Med 20: 265–271. 10.1038/nm.3465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinster RL, Zimmermann JW (1994) Spermatogenesis following male germ-cell transplantation. Proc Natl Acad Sci USA 91: 11298–11302. 10.1073/pnas.91.24.11298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasco E, Calvo MI, Blázquez-Castro A, Vecchio D, Zamarrón A, de Almeida IJ, Stockert JC, Hamblin MR, Juarranz Á, Espada J (2015) Photoactivation of ROS production in situ transiently activates cell proliferation in mouse skin in the hair follicle stem cell niche promoting hair growth and wound healing. J Invest Dermatol 135: 2611–2622. 10.1038/jid.2015.248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Ouyang W, Grigura V, Zhou Q, Cames K, Lim H, Zhao GQ, Arber S, Kurpios N, Murphy TL, et al. (2005) ERM is required for transcriptional control of the spermatogonial stem cell niche. Nature 436: 1030–1034. 10.1038/nature03894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Rooij DG, Russell LD (2000) All you wanted to know about spermatogonia but were afraid to ask. J Androl 21: 776–798. [PubMed] [Google Scholar]
- Grasso M, Fuso A, Dovere L, de Rooij DG, Stefanini M, Boitani C, Vicini E (2012) Distribution of GFRA1-expressing spermatogonia in adult mouse testis. Reproduction 143: 325–332. 10.1530/rep-11-0385 [DOI] [PubMed] [Google Scholar]
- Guo Y-L, Chakraborty S, Rajan SS, Wang R, Huang F (2010) Effects of oxidative stress on mouse embryonic stem cell proliferation, apoptosiss, senescence, and self-renewal. Stem Cell Dev 19: 1321–1331. 10.1089/scd.2009.0313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M, Lee JD (2004) Role of the BMK1/ERK5 signaling pathway: Lessons from knockout mice. J Mol Med 82: 800–808. 10.1007/s00109-004-0602-8 [DOI] [PubMed] [Google Scholar]
- Hou N, Torii S, Saito N, Hosaka M, Takeuchi T (2008) Reactive oxygen species-mediated pancreatic beta-cell death is regulated by interactions between stress-activated protein kinases, p38 and c-Jun N-terminal kinase, and mitogen-activated protein kinase phosphatases. Endocrinolgy 149: 1654–1665. 10.1210/en.2007-0988 [DOI] [PubMed] [Google Scholar]
- Huynh KD, Fischle W, Verdin E, Bardwell VJ (2000) BCoR, a novel corepressor involved in BCL-6 repression. Genes Dev 14: 1810–1823. [PMC free article] [PubMed] [Google Scholar]
- Ishii K, Kanatsu-Shinohara M, Toyokuni S, Shinohara T (2012) FGF2 mediates mouse spermatogonial stem cell self-renewal via upregulation of Etv5 and Bcl6b through MAP2K1 activation. Development 139: 1734–1743. 10.1242/dev.076539 [DOI] [PubMed] [Google Scholar]
- Ito K, Hirao A, Arai F, Takubo K, Matsuoka S, Miyamoto K, Ohmura M, Naka K, Hosokawa K, Ikeda Y, et al. (2006) Reactive oxygen species act through p38 MAPK to limit the lifespan of hametopoietic stem cells. Nat Med 12: 446–451. 10.1038/nm1388 [DOI] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M, Ogonuki N, Inoue K, Miki H, Ogura A, Toyokuni S, Shinohara T (2003) Long-term proliferation in culture and germline transmission of mouse male germline stem cells. Biol Reprod 69: 612–616. 10.1095/biolreprod.103.017012 [DOI] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M, Shinohara T (2013) Spermatogonial stem cell self-renewal and development. Annu Rev Cell Dev Biol 29: 163–187. 10.1146/annurev-cellbio-101512-122353 [DOI] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M, Ogonuki N, Matoba S, Morimoto H, Ogura A, Shinohara T (2014) Improved serum- and feeder-free culture of mouse germline stem cells. Biol Reprod 91: 88 10.1095/biolreprod.114.122317 [DOI] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL (2015) HISAT: A fast spliced aligner with low memory requirements. Nat Methods 12: 357–360. 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Belle JE, Orozco NM, Paucar AA, Saxe JP, Mottahedeh J, Pyle AD, Wu H, Kornblum HI (2011) Proliferative neural stem cells have high endogenous ROS levels that regulate self-renewal and neurogenesis in a PI3K/Akt-dependent manner. Cell Stem Cell 8: 59–71. 10.1016/j.stem.2010.11.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuno K, Yamada H, Iwata K, Jin D, Katsuyama M, Matsuki M, Takai S, Yamanishi K, Miyazaki M, Matsubara H, et al. (2005) Nox1 is involved in angiotensin II-mediated hypertension: A study in Nox1-deficient mice. Circulation 112: 2677–2685. 10.1161/circulationaha.105.573709 [DOI] [PubMed] [Google Scholar]
- Martin M. (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet Journal 17: 10–12. 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- Mehta S, Tsai P, Lasham A, Campbell H, Reddel R, Braithwaite A, Print C (2016) A study of TP53 RNA splicing illustrates pitfalls of RNA-seq methodology. Cancer Res 76: 7151–7159. 10.1158/0008-5472.can-16-1624 [DOI] [PubMed] [Google Scholar]
- Meistrich ML, van Beek MEAB (1993) Spermatogonial stem cells In Cell and Molecular Biology of the Testis. Desjardins C, Ewing LL (eds). pp 266–295. New York: Oxford University Press. [Google Scholar]
- Morikawa M, Koinuma D, Mizutani A, Kawasaki N, Holmborn K, Sundqvist A, Tsutsumi S, Watabe T, Aburatani H, Heldin CH, et al. (2016) BMP4 sustains embryonic stem cell self-renewal through distinct functions of different Krüppel-like factors. Stem Cell Rep 6: 64–73. 10.1016/j.stemcr.2015.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto H, Iwata K, Ogonuki N, Inoue K, Ogura A, Kanatsu-Shinohara M, Morimoto T, Yabe-Nishimura C, Shinohara T (2013) ROS are required for mouse spermatogonial stem cell self-renewal. Cell Stem Cell 12: 774–786. 10.1016/j.stem.2013.04.001 [DOI] [PubMed] [Google Scholar]
- Morimoto H, Kanatsu-Shinohara M, Shinohara T (2015) ROS-generating oxidase Nox3 regulates the self-renewal of mouse spermatogonial stem cells. Biol Reprod 92: 147 10.1095/biolreprod.114.127647 [DOI] [PubMed] [Google Scholar]
- Nagano M, Avarbock MR, Brinster RL (1999) Pattern and kinetics of mouse donor spermatogonial stem cell colonization in recipient testes. Biol Reprod 60: 1429–1436. 10.1095/biolreprod60.6.1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida K, Yamaguchi O, Hirotani S, Hikoso S, Higuchi Y, Watanabe T, Takeda T, Osuka S, Morita T, Kondoh G, et al. (2004) p38alpha mitogen-activated protein kinase plays a critical role in cardiomyocyte survival but not in cardiac hypertrophic growth in response to pressure overload. Mol Cell Biol 24: 10611–10620. 10.1128/mcb.24.24.10611-10620.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oatley JM, Avarbock MR, Telaranta AI, Fearon DT, Brinster RL (2006) Identifying genes important for spermatogonial stem cell self-renewal and survival. Proc Natl Acad Sci USA 103: 9524–9529. 10.1073/pnas.0603332103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa T, Aréchaga JM, Avarbock MR, Brinster RL (1997) Transplantation of testis germinal cells into mouse seminiferous tubules. Int J Dev Biol 41: 111–122. [PubMed] [Google Scholar]
- Takehashi M, Kanatsu-Shinohara M, Inoue K, Ogonuki N, Miki H, Toyokuni S, Ogura A, Shinohara T (2007) Adenovirus-mediated gene delivery into mouse spermatogonial stem cells. Proc Natl Acad Sci USA 104: 2596–2601. 10.1073/pnas.0609282104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Kanatsu-Shinohara M, Lei Z, Rao CV, Shinohara T (2016) The luteinizing hormone-testosterone pathway regulates mouse spermatogonial stem cell self-renewal by suppressing WNT5A expression in Sertoli cells. Stem Cell Rep 7: 279–291. 10.1016/j.stemcr.2016.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegelenbosch RAJ, de Rooij DG (1993) A quantitative study of spermatogonial multiplication and stem cell renewal in the C3H/101 F1 hybrid mouse. Mutat Res 290: 193–200. [DOI] [PubMed] [Google Scholar]
- Terasawa K, Okazaki K, Nishida E (2003) Regulation of c-Fox and Fra-1 by the MEK5-ERK5 pathway. Genes Cells 8: 263–273. 10.1046/j.1365-2443.2003.00631.x [DOI] [PubMed] [Google Scholar]
- Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L (2013) Differential analysis of gene regulation at transcript resoluteion with RNA-seq. Nat Biotechnol 31: 46–53. 10.1038/nbt.2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura JJ, Tenbaum S, Perdiguero E, Huth M, Guerra C, Barbacid M, Pasparakis M, Nebreda AR (2007) P38alpha MAP kinase is essential in lung stem and progenitor cell proliferation and differentiation. Nat Genet 39: 750–758. 10.1038/ng2037 [DOI] [PubMed] [Google Scholar]
- Wang X, Finegan KG, Robinson AC, Knowles L, Khosravi-Far R, Hinchliffe KA, Boot-Handford RP, Tournier C (2006) Activation of extracellular signal-regulated protein kinase 5 down-regulates FasL upon osmotic stress. Cell Death Differ 13: 2099–2108. 10.1038/sj.cdd.4401969 [DOI] [PubMed] [Google Scholar]
- Wu X, Goodyear SM, Tobias JW, Avarbock MR, Brinster RL (2011) Spermatogonial stem cell self-renewal requires ETV5-mediated downstream activation of Brachyury in mice. Biol Reprod 85: 1114–1123. 10.1095/biolreprod.111.091793 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Statistical analysis of ROS levels. (14.7KB, docx)
Table S2 Quantification of Western blot analysis. (22KB, docx)
Table S3 Microarray analysis. (45.8KB, docx)
Table S4 RNA sequence analysis. (1.8MB, docx)
Table S5 Raw values for candidate transcription factors determined by microarray. (34.5KB, docx)
Table S6 Raw values for candidate transcription factors determined by RNA-seq. (19.6KB, docx)
Table S7 Chemicals used in this study. (13.8KB, docx)
Table S8 KD vectors used in this study. (18.1KB, docx)
Table S9 Antibodies used in this study. (18.8KB, docx)
Table S10 PCR Primers used in this study. (20.7KB, docx)
Data Availability Statement
The accession numbers for the microarray and RNA-seq data are GSE106103 and GSE118191, respectively.