Abstract
Nicotinamide adenine dinucleotide (NAD+) is a cosubstrate for several enzymes, including the sirtuin family of NAD+-dependent protein deacylases. Beneficial effects of increased NAD+ levels and sirtuin activation on mitochondrial homeostasis, organismal metabolism and lifespan have been established across species. Here we show that α-amino-β-carboxymuconate-ε-semialdehyde decarboxylase (ACMSD), the enzyme that limits the proportion of ACMS able to undergo spontaneous cyclisation in the de novo NAD+ synthesis pathway, controls cellular NAD+ levels via an evolutionary conserved mechanism from C. elegans to the mouse. Genetic and pharmacological inhibition of ACMSD boosts de novo NAD+ synthesis and SIRT1 activity, ultimately enhancing mitochondrial function. We furthermore characterized a series of potent and selective ACMSD inhibitors, which, given the restricted ACMSD expression in kidney and liver, are of high therapeutic interest to protect these tissues from injury. ACMSD hence is a key modulator of cellular NAD+ levels, sirtuin activity, and mitochondrial homeostasis in kidney and liver.
Keywords: Tryptophan, NAD+, ACMSD, Acute Kidney Injury, NAFLD, NASH, Kynurenine, α-amino-β-carboxymuconate-ε-semialdehyde decarboxylase
Increasing NAD+ levels activates the sirtuins and positively impacts on metabolism in different model organisms1–4. Given the salutary effects of replenished NAD+ pools, there is an intense search for strategies to increase intracellular NAD+ ranging from limiting NAD+ consumption to increasing NAD+ production5. NAD+ can be produced via salvage pathways or via de novo synthesis. The de novo NAD+ synthesis starts from the amino acid tryptophan6. The formation of unstable α-amino-β-carboxymuconate-ε-semialdehyde (ACMS) constitutes a branching point of this pathway (Extended data Figure 1a). ACMS can either undergo cyclisation forming quinolinic acid (QA), the precursor for NAD+, or be directed towards total oxidation to CO2 and H2O. While the cyclisation of ACMS occurs spontaneously, the transformation of ACMS into α-amino-β-muconate-ε-semialdehyde (AMS) is catalysed by the enzyme α-amino-β-carboxymuconate-ε-semialdehyde decarboxylase (ACMSD), thereby determining the proportion of ACMS able to undergo cyclisation and produce NAD+. ACMSD is conserved across species, with mice (ACMSD), rat (ACMSD) and Caenorhabditis elegans (ACSD-1, Y71D11A.3) orthologs respectively showing 85, 85 and 48% similarity to the human protein7. Given this sequence conservation we initially characterized the function of ACSD-1 in C. elegans.
acsd-1 controls NAD+ levels in C. elegans
The newly generated C. elegans acsd-1::GFP reporter strain showed expression of acsd-1 in the majority of tissues throughout development and adulthood (Extended Data Figure 1b-c). Feeding worms with the HT115 E. coli strain expressing acsd-1 RNAi decreased acsd-1 transcripts by 46% in wild type (N2) versus 78% in rrf-3 mutant worms (pk1426), which are hypersensitive to RNAi (Extended Data Figure 1d). In rrf-3 mutants, acsd-1 RNAi furthermore resulted in the loss of function (LOF) of ACSD-1 enzymatic activity, while in N2 worms ACSD-1 activity was reduced to 30% (Extended Data Figure 1e).
It has long been postulated that C. elegans cannot de novo synthesize NAD+ and hence relies on preformed pyridine rings to produce NAD+8,9. This hypothesis, based on the fact that C. elegans do not possess a quinolinate phosphoribosyltransferase (QPRT) ortholog with obvious sequence similarities (Extended Data Figure 1a), was recently disproven as uridine monophosphate synthetase accomplishes the role of QPRT in the nematode10. We ascertained the presence of a QPRT-like enzymatic activity in both N2 and rrf-3 worms (Extended Data Figure 1f), and showed that tryptophan dose-dependently increased NAD+ levels (Figure 1a). Finally, in line with our hypothesis, acsd-1 RNAi increased NAD+ content 1.2-fold (Figure 1b).
Figure 1. acsd-1 LOF improves NAD+ levels, mitochondrial function, and lifespan through de novo synthesis in C. elegans.
a-b. NAD+ upon tryptophan supplementation (Trp 0 and 5 μM: n=4; Trp 50 μM: n=5) (a) and feeding with control (empty vector) or acsd-1 RNAi (n=14) (b), each n represents a pool of ~1000 worms.
c. Epistasis of acsd-1 and sir-2.1 RNAi. P*: ctrl vs ctrl/acsd-1 RNAi; P°: ctrl/sir-2.1 RNAi vs sir-2.1/acsd-1 RNAi.
d. GFP signal in the reporter strain, expressing a mitochondria-targeted GFP in the muscle at day 1 and 3 of adulthood (n=4, each n represents a pool of 20 worms).
e. mtDNA/nDNA ratio in wild type (N2) and sir-2.1(ok434) mutant worms (n=12 worms) upon control or acsd-1 RNAi.
f-g. mRNA levels encoding mitochondrial proteins (n=6, each n represents a pool of ~600 worms) (f) and oxygen consumption rate in basal and uncoupled conditions (n=14, each n represents a pool of 10 worms) (g), in worms fed with control or acsd-1 RNAi.
h. ATP content in rrf-3(pk1426) and N2 worms upon control or acsd-1 RNAi (n=4 and 7 respectively, each n represents a pool of ~100 worms).
i. Altered ratio between nDNA- (ATP5A) and mtDNA- (MTCO1) encoded OXPHOS subunits upon acsd-1 RNAi. Each lane represents an individual pool of ~600 worms.
j. Epistasis of acsd-1 with atfs-1 RNAi. P*: ctrl vs ctrl/acsd-1 RNAi; P°: ctrl/atfs-1 RNAi vs atfs-1/acsd-1 RNAi.
k. GFP signal in hsp-6::GFP reporter strain fed with control or acsd-1 RNAi at day 1 and 3 of adulthood (day 1: n=3; day 3: n=4, each n represents a pool of 20 worms).
l. DAF-16 nuclear translocation. Arrowheads indicate DAF-16 accumulation within nuclei. The graph represents the distribution of control and acsd-1 RNAi treated worms with DAF-16-translocated nuclei (n=25 worms).
m. Epistasis of acsd-1 with daf-16 RNAi. P*: ctrl vs ctrl/acsd-1 RNAi; P°: ctrl/daf-16 RNAi vs daf-16/acsd-1 RNAi.
n. GFP signal in control and acsd-1 RNAi treated sod-3::GFP reporter worms at day 1 and 3 of adulthood (n=4, each n represents a pool of 20 worms).
o. ROS content in worms exposed to control or acsd-1 RNAi (n=4, each n represents a pool of 20 worms).
p-q. Mobility (p) and survival (q) in N2 exposed to 4 mM paraquat starting from L4 stage, treated with control or acsd-1 RNAi throughout the entire life (n=100 worms).
All worm assays performed at 20°C and repeated at least once. Data are mean ± s.e.m. *P≤0.05, **P≤0.01, ***P≤0.001. P values calculated using one-way ANOVA (a), two-tailed t-test (b, d-h, k, n-p) or Logrank test (c, j, m, q). For gel source images, see Supplementary Fig. 1. For individual P values, see Source Data. For lifespan values, see Extended Data Table 1.
Increases in NAD+ are known to extend worm lifespan11,12. Although in basal conditions acsd-1 RNAi did not affect N2 lifespan (Extended Data Figure 1g), survival of rrf-3 mutants was significantly improved (Extended Data Figure Figure 1h). The lifespan-enhancing effect seems therefore to depend on the extent of acsd-1 downregulation. In agreement with reported data13 and with the effects of tryptophan supplementation on NAD+ content, tryptophan also extended worm lifespan, and acsd-1 RNAi did not result in a significant further extension beyond the effects of tryptophan alone (Extended Data Figure 1i). Although the effects of sir-2.1 overexpression on lifespan are debated14,15, the importance of sir-2.1 in NAD+-mediated lifespan extension has never been questioned11,12. Accordingly, sir-2.1 was required for the longevity effect of acsd-1 RNAi (Figure 1c), consolidating the importance of the NAD+/sir-2.1 axis in lifespan regulation. ACSD-1 being at the crossroad of several molecular pathways implies that the effects of its LOF might not be exclusively mediated by changes in NAD+ content, but could also partially arise from changes in the levels of products of ACSD-1 enzymatic activity, such as AMS or picolinic acid. The contribution of these downstream factors remains to be explored in the future.
Sirtuin activation is associated with enhanced mitochondrial function1,2. Feeding the myo-3::GFP(mit) and ges-1::GFP(mit) reporter strains, expressing mitochondria-targeted GFP in muscle and intestinal cells respectively, with acsd-1 RNAi robustly increased mitochondrial content in both tissues (Figure 1d, Extended Data Figure 1j). Measurement of the mitochondrial (mt) over nuclear (n) DNA ratio confirmed the increase in mitochondrial content with acsd-1 RNAi in wild type but not in sir-2.1(ok434) mutants (Figure 1e). Furthermore, acsd-1 LOF induced transcript levels of many mitochondrial proteins, basal and maximal respiration, complex II abundance upon blue native PAGE analysis of mitochondria, and ATP content (Figure 1f-h, Extended Data Figure 1k). The mitochondrial network was also more extensive and interconnected with acsd-1 RNAi (Extended Data Figure 1l).
acsd-1 LOF activates mitochondrial stress defence
Of note, acsd-1 RNAi induced nDNA-encoded OXPHOS subunits, such as H28O16.1 (ortholog of mammalian ATP5A), while mtDNA-encoded OXPHOS components, such as MTCE.26 (ortholog of mammalian MTCO1), were unchanged (Figure 1i). Such a change in the ratio in mtDNA- vs nDNA-encoded proteins is a hallmark of mitonuclear protein imbalance, that can be induced by increasing NAD+ concentrations11 and which is associated with the activation of the mitochondrial unfolded protein response (UPRmt)16. LOF of either atfs-1 or ubl-5, essential UPRmt genes17,18, attenuated the longevity seen with acsd-1 RNAi alone (Figure 1j, Extended Data Figure 1m), ascertaining the requirement of the UPRmt for the lifespan extension. By using a C. elegans hsp-6::GFP UPRmt reporter (a mitochondrial chaperone orthologous to mammalian mtHsp70), a robust activation of the UPRmt was observed upon acsd-1 RNAi (Figure 1k). This stress response was specific, since there was no activation of the UPR in the endoplasmic reticulum (UPRER) or of the cytosolic heat shock response, in the respective hsp-4::GFP and hsp-16.2::GFP reporter strains after acsd-1 LOF (Extended Data Figure 1n-o). Several UPRmt-related transcripts were induced upon acsd-1 RNAi (Extended Data Figure 1p).
In C. elegans, oxidative stress defence is launched when DAF-16, the worm Forkhead box O transcription factor (FOXO) ortholog, is translocated into the nucleus19 where it induces the mitochondrial superoxide dismutase, sod-320. Supplementing C. elegans with NAD+ was reported to activate oxidative stress defence and to improve resistance to reactive oxygen species (ROS), in a sir-2.1- and daf-16-dependent fashion12. Consistent with this mechanism, DAF-16 translocated into nuclei upon acsd-1 LOF (Figure 1l). Importantly, the improved lifespan with acsd-1 RNAi was lost upon daf-16 LOF (Figure 1m). In line with more nuclear localized DAF-16, acsd-1 LOF increased the GFP signal in sod-3::GFP reporter worms and sod-3 expression (Figure 1n, Extended Data Figure 1q). As a consequence, worms with acsd-1 knockdown had lower ROS, were more active and lived longer when exposed to paraquat, a known ROS inducer (Figure 1o-q). The better survival under paraquat was independent on the developmental stage when the worms were exposed to acsd-1 RNAi, but required daf-16 (Extended Data Figure 1r-s).
ACMSD function is conserved in mammals
We next explored whether the molecular mechanisms observed in C. elegans upon acsd-1 LOF are evolutionary conserved. In humans, ACMSD was mainly detected in liver and kidney, with some Acmsd transcripts present in brain, even though they were respectively 1300- and 30-fold lower than in kidney or liver21; this is in line with data in the Human Protein Atlas (http://www.proteinatlas.org/ENSG00000153086-ACMSD/tissue). Mouse primary hepatocytes expressed the highest levels of Acmsd amongst different mouse (AML-12, Hepa 1-6) and human (HepG2, HuH-7, HEK 293, HK-2) hepatic and renal cell lines (Extended Data Figure 2a) and were hence selected for further study.
Acmsd expression was reduced by >98% in mouse primary hepatocytes transduced with an adenovirus encoding an shRNA targeting Acmsd (Extended Data Figure 2b). Consistent with our C. elegans data, total cellular NAD+ was 1.4-fold higher compared to hepatocytes transduced with scramble shRNA (Figure 2a), whereas no effect was evident on mitochondrial NAD+ content (Extended Data Figure 2c). Acmsd downregulation also enhanced mitochondrial function in hepatocytes, as evidenced by increased mtDNA/nDNA ratio, citrate synthase activity and expression of OXPHOS genes both at the transcript and protein levels (Figure 2b-e). In-gel activity for CI and CIV showed an overall increase in Acmsd shRNA infected cells, including the high molecular weight supercomplexes (Extended Data Figure 2d-e). UPRmt transcripts were also increased by Acmsd shRNA (Figure 2d). Furthermore, Acmsd shRNA increased basal and maximal oxygen consumption rate and ATP content in murine primary hepatocytes grown both in high and low glucose (Figure 2f-g).
Figure 2. Pathways activated by Acmsd knockdown in worms are conserved in mammalian cells.
a-f. Mouse primary hepatocytes obtained from C57BL/6J mice were transduced with an adenovirus encoding either control shRNA or shRNA against Acmsd for 48h. NAD+ levels (n=6) (a), mtDNA/nDNA ratio (n=4) (b), citrate synthase activity (n=3) (c), expression of OXPHOS and UPRmt genes (n=3) (d), protein expression of the respiratory complex subunits (e), oxygen consumption in basal and uncoupled (FCCP 2 μM) conditions (n=15) (f).
g. ATP content in control vs Acmsd shRNA conditions, in mouse primary hepatocytes grown in both medium with high glucose (HG) and low glucose (LG) supplemented with 1% oleate (n=15).
h. Apoptosis evaluated by caspase 3/7 activity after 24h exposure of mouse primary hepatocytes to 0.75 mM palmitate (palmitate- condition: n=3, palmitate+ condition: n=6).
i. Triglyceride (TGs) content after 24h exposure to 0.5 mM oleate in AML12 cells transduced with an adenovirus encoding either control or Acmsd shRNA (n=9).
j-k. Primary hepatocytes extracted from a Sirt1L2/L2 mouse transduced with an adenovirus either encoding GFP (WT condition) or the CRE-recombinase to generate Sirt1 KO primary hepatocytes. (j) Steatosis quantification in hepatocytes treated with control or Acmsd shRNA after 24h exposure to 0.75 mM palmitate (palmitate- condition: n=3; palmitate+ condition: n=6). (k) ROS content in hepatocytes exposed to control shRNA versus Acmsd shRNA. The positive control treated with 550 μM of H2O2 (H2O2- conditions: n=3; H2O2+ condition: n=6).
Data are mean ± s.e.m.; each n represents a biologically independent sample. All experiments performed independently at least twice. *P≤0.05, **P≤0.01, ***P≤0.001. P values calculated using two-tailed t-test (a-d, f-i) or two-way ANOVA (j-k). For gel source images, see Supplementary Fig. 1. For individual P values, see Source Data.
In combination these changes indicate that hepatocyte mitochondrial function was improved by Acmsd LOF, which incited us to test whether knocking down Acmsd could render hepatocytes more resilient to steatosis and apoptosis inflicted by high concentrations of fatty acids in the medium. LOF of Acmsd protected primary murine hepatocytes from fatty acid-induced apoptosis (Figure 2h) and attenuated triglyceride (TGs) accumulation (Figure 2i); this effect was SIRT1-dependent (Figure 2j, Extended Data Figure 2f). Acmsd shRNA also reduced levels of ROS in primary hepatocytes in a SIRT1-dependent fashion (Figure 2k) and lowered the acetylation levels of FOXO1, a deacetylation target of SIRT1 (Extended Data Figure 2g).
Characterisation of ACMSD inhibitors
The first indications that inhibiting ACMSD activity could enhance de novo NAD+ synthesis came from studies on phthalate esters and pyrazinamide; however, these compounds have pleiotropic effects22,23. We therefore developed selective and potent ACMSD inhibitors, TES-991 and TES-102524, and explored their therapeutic potential. Similar to genetic Acmsd LOF, pharmacological inhibition of ACMSD with both compounds dose-dependently increased NAD+ levels and resulted in SIRT1 activation in primary hepatocytes (Figure 3a-c). Both ACMSD inhibitors induced mitochondrial transcripts and enhanced both basal and maximal oxygen consumption rate and mitochondrial SOD2 activity in primary mouse hepatocytes (Figure 3d-e, Extended Data Figure 3a-c). Like the genetic Acmsd LOF, ACMSD inhibition protected primary hepatocytes from apoptosis induced by high doses of fatty acids (Figure 3f) and promoted fatty acid oxidation in SIRT1-dependent way, confirmed by both the induction of β-oxidation genes and functional assays (Figure 3g-i and Extended Data Figure 3d).
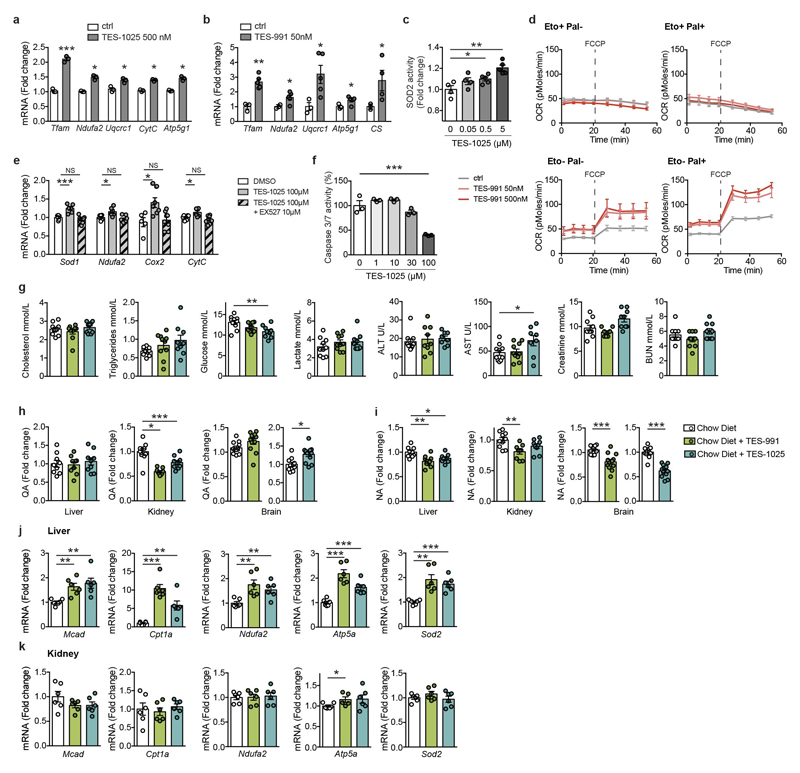
Figure 3. Pharmacological inhibition of ACMSD has similar effects as its genetic downregulation.
a-b. NAD+ levels in mouse primary hepatocytes (PH) treated for 24h with vehicle (DMSO), TES-1025 (n=3) (a) or TES-991 (n=6) (b).
c. SIRT1 activity in PH treated for 24h with vehicle or the ACMSD inhibitors (500nM) (n=3).
d-f. SOD2 activity (n=3) (d), oxygen consumption in basal and uncoupled conditions (ctrl: n=6; TES-991: n=7) (e), and apoptosis rate after 36h exposure to 0.75 mM palmitate (ctrl: n=6; TES-991: n=5) (f) in PH treated for 24h with vehicle or TES-991.
g. mRNA levels of fatty acid oxidation (FAO) genes in PH treated with vehicle or TES-991 (500 nM) after 6h exposure to 0.33 mM palmitate and 0.66 mM oleate (n=6).
h-i. PH from a Sirt1L2/L2 mouse transduced with an adenovirus encoding GFP (Sirt1 WT) or CRE-recombinase (Sirt1 KO). FAO after 24h treatment with vehicle or TES-991 under basal (h) and uncoupled (i) conditions (n=5).
j-m. Changes in NAD+ (n=4) (j), mRNA levels of mitochondrial and oxidative stress defence genes (n=6) (k), OXPHOS subunits (l) and ATP content (ctrl: n=12; TES-1025: n=13) (m) in HK-2 cells upon treatment with TES-1025 (100 μM) and EX527 (10 μM), for 24h.
n. Apoptosis in HK-2 cells 16h after addition of cisplatin (50 μM). TES-1025 added 1h prior to cisplatin (n=3).
o. NAD+ in livers (ctrl: n=6; TES-991: n=7; TES-1025: n=9 mice), kidneys (ctrl, TES-1025: n=8; TES-991: n=9 mice) and brains (ctrl: n=11; TES-991, TES-1025: n=12 mice) of mice fed with normal chow diet or supplemented with ACMSD inhibitors (15mg/kg body weight/day).
Data are mean ± s.e.m.; each n represents a biologically independent sample. All experiments performed independently at least twice (a-n). *P≤0.05, **P≤0.01, ***P≤0.001. P values calculated using two-tailed t-test (a-c, f-g, j-k, m-o), one-way (d-e) or two-way ANOVA (h-i). For gel source images, see Supplementary Fig. 1. For individual P values, see Source Data.
TES-1025, the compound with the best exposure profile for the kidney24, also increased NAD+ content in human HK-2 kidney cells (Figure 3j). TES-1025 induced transcript levels of mitochondrial and ROS defence genes, as well as protein levels of the OXPHOS subunits in a SIRT1-dependent fashion (Figure 3k-l, Extended Data Figure 3e). Treatment of HK-2 cells with TES-1025 also increased their ATP content (Figure 3m). Cisplatin-induced apoptosis was attenuated when HK-2 cells were either pre-treated (Figure 3n) or exposed to TES-1025 simultaneously with cisplatin (Extended Data Figure 3f).
We then characterized the effect of both compounds in the mouse. Feeding 9-week old male C57BL/6J mice for 10 days with chow diet supplemented with TES-991 or TES-1025 (15 mg/kg body weight) did not result in any pronounced effects on metabolic homeostasis and did not affect liver or kidney function (Extended Data Figure 3g). Both TES-991 and TES-1025 increased NAD+ content in liver, kidneys and brain (Figure 3o), whereas NAD+ levels in heart and skeletal muscle seemed unaffected (not shown). Changes in QA did not show a consistent pattern after pharmacological inhibition of ACMSD. While in brain QA tended to increase (statistically significant for TES-1025, but to concentrations too low to provoke neurotoxicity25), its kidney levels were reduced and liver levels remained unchanged (Extended Data Figure 3h). Intriguingly, both ACMSD inhibitors led to a consistent depletion of nicotinic acid (NA) in liver, kidney and brain, which could serve as another readout for ACMSD inhibition (Extended Data Figure 3i). Besides this, administration of ACMSD inhibitors induced transcript levels of mitochondrial genes in the liver, while the expression of the same genes in the kidneys was unaffected (Extended Data Figure 3j-k).
Efficacy of ACMSD inhibitors in NAFLD and AKI models
The observed differences in exposure profiles of the two ACMSD inhibitors, with TES-991 being enriched in liver and TES-1025 in the kidneys24, instructed us to assess the translational potential of TES-991 in the setting of liver disease, whereas we oriented in vivo studies with TES-1025 to the kidney. Non-alcoholic fatty liver disease (NAFLD) constitutes the most common liver disease worldwide, often leading to progressive liver damage, termed non-alcoholic steatohepatitis26. Given the efficacy of NAD+ boosting to attenuate NAFLD in mouse models27,28, we tested the efficacy of ACMSD inhibition in a mouse model of NAFLD, induced by feeding 13-week old C57BL/6J mice with a methionine-choline deficient (MCD) diet for 2.5 weeks28. Supplementing the MCD diet with TES-991 (15 mg/kg/day prophylactically) attenuated hepatic steatosis (Figure 4a-b) and plasma ALT and AST levels (Figure 4c, Extended Data Figure 4a). As expected, the MCD diet depleted the hepatic NAD+ stock; an effect reverted by TES-991 (Figure 4d). TES-991 also protected against hepatic lipid accumulation and attenuated inflammation (Extended Data Figure 4b-c). Hepatic SOD2 activity and ATP content, which were reduced with the MCD diet, also partially recovered with TES-991 (Figure 4e, Extended Data Figure 4d). Furthermore, ACMSD inhibition reversed changes in transcripts of genes involved in ROS protection, β-oxidation, inflammation, and mitochondrial function, known to be modulated in NAFLD (Figure 4f). Importantly, while supplementation of the MCD diet with TES-991 was still able to increase hepatic NAD+ content in Sirt1 KO (Sirt1hep-/-) mice (Extended Data Figure 4e), TES-991 no longer protected the Sirt1hep-/- livers from MCD diet-induced NAFLD, as reflected by the hepatic lipid content, plasma markers of liver damage, SOD2 activity, and changes in liver transcript levels (Extended Data Figure 4f-k).
Figure 4. ACMSD inhibitors protect hepatic function from MCD diet-induced NAFLD.
a. Comparison of gross liver morphology in representative 16-week old C57BL/6J male mice fed for 2.5 weeks with control diet, MCD diet, and MCD diet supplemented with 15 mg/kg/day of TES-991. In vivo MCD diet study was performed once.
b-e. Liver TGs (b), plasma ALT (c), liver NAD+ (d) and ATP (e) levels in the mouse cohorts described in (a) (CD, MCD: n=8; MCD+TES-991: n=7 mice).
f. mRNA levels of oxidative stress defence, mitochondrial, β-oxidation, inflammatory and fibrosis genes in livers of the mice described in (a) (n=8 mice).
Data are mean ± s.e.m. *P≤0.05, **P≤0.01, ***P≤0.001. P values calculated using two-tailed t-test. For individual P values, see Source Data.
Acute kidney injury (AKI) affects 3-7% of all hospitalized patients and has a high mortality rate, especially when occurring in the setting of intensive care29. Given the recent study reporting that increased NAD+ levels can provide protection against AKI30 and the observed protective effect of ACMSD inhibitors in cisplatin-challenged HK-2 cells, we explored the impact of ACMSD inhibition in a mouse model of AKI, induced by application of single intraperitoneal dose of cisplatin to 12-week old male C57BL/6J mice (Extended Data Figure 5a). Supplementation of mouse chow diet with TES-1025 (15 mg/kg/day prophylactically), protected against AKI, as reflected by the normalization of blood creatinine, blood urea nitrogen (BUN) levels and KIM1 levels (Figure 5a-c). Glomerular filtration rate (GFR) was severely compromised in vehicle-treated animals after cisplatin challenge, but was preserved in mice receiving TES-1025 (Figure 5d). While the administration of cisplatin depleted renal NAD+ levels, this drop in NAD+ was less pronounced in TES-1025 treated mice (Figure 5e). Consistent with these biochemical improvements, the increase in cumulative histopathological score, that evaluates tubular necrosis, tubular dilation, inflammation, edema and cast formation, was also reduced by ACMSD inhibition (Figure 5f, Extended Data Figure 5b-d). TES-1025 also restored the cisplatin-induced changes in expression of the OXPHOS complexes in the kidney (Figure 5g).
Figure 5. ACMSD inhibitors protect renal function in two different models of AKI.
a-c. Plasma creatinine (a), BUN (b) and renal KIM1 protein levels (c) in 12-week old C57BL/6J male mice 72h post cisplatin administration. AKI was induced at day 10 after the beginning of the study by a single intraperitoneal injection of cisplatin (20 mg/kg body weight). Sham controls injected with saline solution. TES-1025 administered at 15 mg/kg/day (n=6 mice).
d. Glomerular filtration rate (GFR) in mice 52h after cisplatin or saline administration (Sham ctrl: n=7; Cis-AKI: n=6; Cis-AKI+TES-1025: n=5 mice).
e. NAD+ levels in kidneys of the cisplatin-AKI cohorts (Sham ctrl: n=7; Cis-AKI, Cis-AKI+TES-1025: n=5 mice).
f. Histopathological scoring of H&E stained kidney sections from mice described in (a), evaluating tubular necrosis, tubular dilation, inflammation, edema and cast (Sham ctrl, Cis-AKI+TES-1025: n=6; Cis-AKI: n=5 mice). The scoring performed by two pathologists in a blinded and independent way.
g. Protein expression of the respiratory complex subunits in kidneys from mouse cohorts described in (a). The experiment was performed twice.
h-i. AKI was induced in 12-week old C57BL/6J male mice by a dorsal surgical incision and bilateral occlusion of the renal pedicles for 25 min; a simple dorsal incision was performed for the Sham controls. TES-1025 administered at 15 mg/kg/day. Plasma creatinine (h) and BUN (i) in mice 48h post-surgery (n=5 mice).
Data are mean ± s.e.m. *P≤0.05, **P≤0.01, ***P≤0.001. P values calculated using two-tailed t-test. For gel source images, see Supplementary Fig. 1. For individual P values, see Source Data.
We further confirmed the protective effects of TES-1025 (15 mg/kg body weight/day) in a distinct AKI model, i.e. ischemia-reperfusion AKI (IR-AKI). Renal pedicles of 12-week old male C57BL/6J mice were clamped for 25 minutes to induce mild to moderate AKI (Extended Data Figure 5e). Similar to the cisplatin-induced AKI model, administration of TES-1025 protected from structural and functional renal damage inflicted by IR-AKI (Figure 5h-i, Extended Data Figure 5f-j). GSH depletion, caused by IR-AKI, was rescued and MPO activity, reflecting neutrophil infiltration, was attenuated by TES-1025 (Extended Data Figure 5k-l). Higher NAD+ levels were furthermore detected in kidneys of mice receiving TES-1025 (Extended Data Figure 5m). TES-1025 also normalized the renal expression of OXPHOS complexes, which was impaired by IR (Extended Data Figure 5n).
Conclusions
Accumulating evidence that supports the importance of a robust NAD+ homeostasis to protect against aging and numerous diseases, stimulated the interest in approaches to maintain and/or raise tissue NAD+ content5. While the translational potential of NAD+ salvaging from precursor molecules has been extensively studied, the possibility to increase NAD+ levels via the de novo NAD+ synthesis pathway has been largely disregarded by the scientific community. However, a few recent reports emphasising the vital role of de novo NAD+ biosynthesis for the maintenance of the whole-body NAD+ homeostasis have brought this pathway onto centre stage and made it bona fide target for the efforts to boost NAD+ content31,32.
Our data position ACMSD, an evolutionary conserved enzyme, as a key checkpoint of the de novo NAD+ synthesis pathway. We show that ACMSD acts as a tissue-selective modulator of cellular NAD+ levels, sirtuin activity, and mitochondrial homeostasis across species, from C. elegans to the mouse. The enriched expression of ACMSD in the kidney and liver in mammals underscored the therapeutic potential of ACMSD inhibition for diseases compromising hepatic and renal function, such as NAFLD, non-alcoholic steatohepatitis, AKI, and chronic kidney disease, which, despite their high prevalence, represent large unmet medical needs. We show here rather striking beneficial effects of two potent and selective ACMSD inhibitors in animal models of NAFLD and AKI. The beneficial effects of in vivo ACMSD inhibition were associated with increases in tissue NAD+ content and were dependent on the activity of SIRT1. Furthermore, the administration of ACMSD inhibitors was well tolerated and devoid of systemic side effects, such as the accumulation of the potential neurotoxic metabolite, QA25. In combination our data hence warrant further studies to explore the long-term benefits of ACMSD inhibition in a broader range of disease models and bode well for potential future translation of ACMSD inhibition in a clinical setting.
Methods
C. elegans experiments
C. elegans strains were provided by the Caenorhabditis Genetics Center (University of Minnesota). Worms were maintained on Nematode Growth Medium (NGM) agar plates seeded with E.coli OP50 bacteria at 20°C, unless stated otherwise. The strains used for the experiments are the following: Bristol N2, NL2099 (rrf-3(pk1426)II), SJ4143 (zcIs17[ges-1::GFP(mit)]), SJ4103 (zcIs14[myo-3::GFP(mit)]), KN259 (huIs33[sod-3::GFP+pRF4(rol-6(su1006))]), SJ4005 (zcIs4[hsp-4::GFP]), TJ356 (zIs356[daf-16p::daf-16a/b::GFP+rol-6(su1006)], SJ4100 (zcIs13[hsp-6::GFP]), VC199 (sir-2.1(ok434)IV). If not indicated explicitly in the text the strain used for the experiments was NL2099 (rrf-3(pk1426)II).
Bacterial feeding RNAi experiments were carried out as described34. Clones used were acsd-1 (Y71D11A.3), sir-2.1 (R11A8.4), and daf-16 (R13H8.1), ubl-5 (F46F11.4), atfs-1 (ZC376.7). Clones were purchased from GeneService and their identity was confirmed by sequencing. For the double RNAi experiments bacterial cultures were mixed before seeding on NGM plates. The control RNAi in this kind of experiments was 50% diluted with control empty vector RNAi bacteria.
Lifespan assays
C. elegans lifespan assays were carried at 20°C as described35. 100 worms were used per condition and scored every 2 days. The reasons for censoring were the «exploded vulva» phenotype or worms that crawled off the plate and were pre-established before the beginning of the experiment. Where indicated, paraquat dichloride (Sigma-Aldrich) was added on top of the agar plates to obtain the indicated final concentration. Once the paraquat solution was absorbed completely by the agar, L4 worms were transferred to these agar plates and monitored for 5-6 days every day. The tests were performed in non-blinded manner and repeated twice, unless indicated otherwise.
Mobility assessment
The movement of worms was recorded for 45 seconds at days 1, 3, and 5 of adulthood using a Nikon DS-L2 / DS-Fi1 camera and controller setup, attached to both a computer and a standard bright field microscope. For each condition 5 plates of worms, with 10 worms per plate was used. The movement of worms was calculated by taking an integral of the speed value, which was assessed by following the worm centroids with a modified version of the freely-available for the Parallel Worm Tracker for MATLAB36. The experiments were non-blinded and repeated twice.
Generation of transgenic C. elegans strains
acsd-1p::gfp expression vector was created by amplifying 1425 bp sequence upstream from the transcription start site of acsd-1 coding region by using C. elegans genomic DNA and 5’-gtaACATGTcagtcgacgcaaaaattgtt-3’ and 5’-gtaCCCGGGtttgattcaggaaaattataaaaattaaatg-3’ primers. The PCR product was digested with PciI and XmaI and ligated into the PciI and AgeI digested pPD30.38 expression vector containing gfp coding sequence cloned between inserted AgeI and NotI restriction sites. Two independent lines carrying acsd-1p::gfp transgene as extrachromosomal array were analyzed in the study.
Imaging and image processing
Confocal images were acquired with Zeiss LSM 700 Upright confocal microscope (Carl Zeiss AG) under non-saturating exposure conditions. The worms were prepared for imaging as described16. Briefly, nematodes were immobilized with 7.5 mM solution of tetramisole hydrochloride (Sigma-Aldrich) in M9 and mounted on 6% agarose pads on glass slides. Image processing was performed with the Fiji software (http://imagej.nih.gov/ij; version 1.47b). Each experiment was repeated at least once.
GFP quantification
Fluorescence intensity in worm strains expressing GFP-reporter proteins was quantified using Victor X4 plate reader (Perkin Elmer). The animals were prepared in the following way: eighty worms per condition (at the corresponding ages) were transferred into wells filled with M9 medium (20 worms per well of a black-walled 96-well plate). Each experiment was repeated at least twice.
MitoSox ROS quantification
MitoSox staining was performed as previously described11. Briefly, a population of 100 worms was recovered in 1 ml of M9 buffer, washed five times to remove residual bacteria, and resuspended in 200 μl of 1:200 MitoSox (Life technologies) stock solution (initial stock solution was dissolved at 1 mM in DMSO). After 20 min of treatment, worms were washed five times in 1 ml of M9 buffer to eliminate the MitoSox reagent, and then transferred in a black-walled 96-well plate for reading using Victor X4 plate reader (Perkin Elmer).
Enzymatic assays in C. elegans
Worms were collected in M9 and washed 8 times to get rid of all the residual bacteria. After the last washing the maximal possible amount of M9 was aspirated and the remaining worm pellet was snap frozen in liquid nitrogen. Activity of QPRT was assessed as described in37. Briefly, we coupled the QPRT reaction to the conversion of the reaction product NAMN to NAD+ with the help of the ancillary enzymes NadD and NadE, followed by a quantification of NAD+ with fluorometric cycling assay (Extended Data Figure 1c). The buffer of the assay mixture consisted of 30 mM potassium phosphate buffer, pH 7.0, 0.5 mM MgCl2. ACMSD activity was assayed as described in21. Briefly, formation of ACMS was monitored at 360 nm, at 37°C, in a pre-assay mixture consisting of 30 µM hydroxyanthranilic acid and an excess quantity of recombinant R. metallidurans 3-hydroxyanthranilic acid dioxygenase. After the reaction was complete, an appropriate aliquot of worm extract was added and ACMS consumption was calculated by the decrease in absorbance. The activity value was corrected for the spontaneous decrease in absorbance due to the non-enzymatic cyclization of ACMS to QA. To this end a control mixture containing all reagents except the extract (replaced by an equal volume of a BSA solution at the same concentration of the extract) was monitored in parallel.
Tryptophan supplementation experiments in C. elegans
L-tryptophan (Sigma-Aldrich) was dissolved in water to obtain a 50 mM solution. This freshly prepared solution was added into the NGM agar at the moment of plate preparation to obtain the concentrations stated in the main text. Worms were maintained on the tryptophan-supplemented plates seeded with live bacteria (E.coli OP50) through their larval stages to allow development. At L4 larvae stage, worms were collected in M9 solution, washed 7 times to remove all the residual bacteria in their intestines and transferred to tryptophan-supplemented plates seeded with heat-inactivated bacteria (E.coli OP50) to avoid metabolism of L-tryptophan by the bacteria. 500 ml culture of OP50 bacteria was incubated at 37°C overnight, the bacteria were then centrifuged at 3320g for 30 min at 4°C and the resulting bacterial pellet was dissolved in 50 ml of fresh LB medium. Subsequently, the heat inactivation was performed by incubating bacteria at 95°C for 30 min.
NAD+ measurements
NAD+ was extracted using acidic extraction method and analyzed by HPLC-mass-spectrometry as described38. Briefly, ~10 mg of frozen tissue samples were used for NAD+ extraction in 10% perchloric acid and neutralized in 3M K2CO3 on ice. After final centrifugation, the supernatant was filtered and the internal standard (NAD+-C13) was added and loaded onto a column (150 Å~ 2.1 mm; Kinetex EVO C18, 100 Å). HPLC was run for 1 min at a flow rate of 300 ml/min with 100% buffer A (Methanol/H2O, 80/20% v/v). Then, a linear gradient to 100% buffer B [H2O + 5mM ammonium acetate] was performed (at 1 to 6min). Buffer B (100%) was maintained for 3 min (at 6 to 9 min), and then a linear gradient back to 100% buffer A (at 9 to 13 min) started. Buffer A was then maintained at 100% until the end (at 13 to 18 min). NAD+ eluted as a sharp peak at 3.3 min and was quantified on the basis of the peak area ratio between NAD+ and the internal standard and normalized to tissue weight and protein content.
Cell culture
The mouse hepatocytes cell line AML-12 (alpha mouse liver 12) was obtained from ATCC and grown at 37°C in a humidified atmosphere of 5% CO2/95% air in Dulbecco’s Modified Eagle Medium / Nutrient Mixture F-12 (DMEM / F-12) supplemented with 0.005 mg/ml insulin, 0.005 mg/ml transferrin, 5 ng/ml selenium, 40 ng/ml dexamethasone, 0.5 mM tryptophan and 1% gentamycin.
Proximal tubular cell line HK-2 (human kidney 2) was purchased from ATCC and grown at 37°C in a humidified atmosphere of 5% CO2/95% air in normal DMEM medium (Gibco) including 10% FCS (Gibco), 10 units per ml penicillin, 0.5 mM tryptophan and HEPES for buffering. ACMSD inhibitor was initially diluted from powder in DMSO until the stock concentration of 1 mM. This stock was further diluted with water until the 100 μM solution, which was used for the cell treatments.
All the cell lines purchased from ATCC have been authenticated by morphology, karyotyping and PCR-based approaches. All the used cell lines have been routinely checked in the laboratory for mycoplasma contamination with the MycoProbe detection kit (R&D systems). Only cells negative for mycoplasma contamination were used.
Primary hepatocytes culture
Primary hepatocytes were prepared from 8–12-week-old C57BL/6 or Sirt1L2/L2 mice (males and females) by liberase perfusion method as described in39 with minor modifications. Isolated primary hepatocytes were plated with DMEM medium (Gibco) including 10% FCS (Gibco), 10 units per ml penicillin, 0.5 mM tryptophan and HEPES for buffering. After 6-8 h of attachment, this medium was replaced with media containing either different concentrations of ACMSD inhibitor or the corresponding concentration of DMSO, or transduced with adenovirus encoding either control shRNA or shRNA against Acmsd. Primary hepatocytes extracted from Sirt1L2/L2 mice were transduced either with adenovirus encoding CRE-recombinase (to generate Sirt1 KO) or GFP. Primary hepatocytes were harvested 24h later in the case of pharmacological treatment and 48h post adenoviral transduction. When indicated, cell culture medium was supplemented with palmitate-BSA and/or oleate-BSA.
Genetic knockdown of Acmsd
Five different pairs of single-stranded DNA oligonucleotides were designed and tested for Acmsd knockdown.
“Top strand” oligos:
-
1)
5’-caccggaagctcttcagagtgatcccgaaggatcactctgaagagcttcc-3’
-
2)
5’-caccggagatggagcgttgtgttaacgaattaacacaacgctccatctcc-3’
-
3)
5’-caccgctattgacagatgtcataggcgaacctatggacatctgtcaatagc-3’
-
4)
5’-caccggaagctgatagagtccatggcgaaccatggactctatcagcttcc -3’
-
5)
5’-caccgcagagtttgatgaagaaacacgaatgtttcttcatcaaactctgc -3’
“Bottom strand” oligos (respectively):
-
1)
5’-aaaaggaagctcttcagagtgatccttcgggatcactctgaagagcttcc-3’
-
2)
5’-aaaaggagatggagcgttgtgttaattcgttaacacaacgctccatctcc-3’
-
3)
5’-aaaagctattgacagatgtcataggttcgcctatgacatctgtcaatagc-3’
-
4)
5’-aaaaggaagctgatagagtccatggttcgccatggactctatcagcttcc-3’
-
5)
5’-aaaagcagagtttgatgaagaaacattcgtgtttcttcatcaaactctgc-3’
BLOCK-iT U6 RNAi Entry Vector Kit (Intvitrogen) was used for production of an entry clone. BLOCK-iT™ Adenoviral RNAi Expression System (Invitrogen) was used afterwards to produce adenoviral expression clone. The shRNA N°3 showed the highest knockdown efficacy and was used for all the experiments in this manuscript.
Apoptosis assessment
Caspase 3/7 activities were measured with the Caspase-Glo assay (Promega) according to the manufacturer’s instructions. HK-2 cells were plated at 5x103 cells/well in white ViewPlate-384 microplate (PerkinElmer). The cells were treated with 50 μM cisplatin (Sigma Aldrich) for 16h. ACMSD inhibitor was either added simultaneously with cisplatin or 1h prior to cisplatin addition (preventive). The data were graphed considering the cisplatin treatment as 100% and the vehicle treatment as 0% of caspase activity. Apoptosis in primary hepatocytes was induced by exposing the cells to 0.75 mM palmitate for 36h.
SOD2 activity assay
Primary hepatocytes, AML-12 cells or pieces of frozen liver were lysed in a 20 mM HEPES buffer (Gibco), pH 7.2, containing 1 mM EGTA (Sigma-Aldrich), 210 mM mannitol (Sigma-Aldrich), and 70 mM sucrose (AMRESCO). Total protein concentration was determined using the Bradford assay (BioRad). SOD2 activity was determined at indicated times after ACMSD inhibitor treatment by the SOD Assay Kit (Cayman Chemical) according to the manufacturer’s instructions. In order to specifically detect the SOD2 activity 2 mM potassium cyanide was added to the assay, which inhibited both Cu/Zn-SOD and extracellular SOD, resulting in the detection of only Mn-SOD (SOD2) activity. Absorbance was determined with a Victor X4 multilabel plate reader (Perkin-Elmer) at 450 nm. Results are expressed in U/ml/mg of protein according to the standard curve and measured protein concentration.
Oxygen consumption
Oxygen consumption was measured with the Seahorse XF96 instrument (Seahorse Bioscience) according to the manufacturer’s protocol. FCCP at the indicated concentrations was used as an uncoupler to reach maximal respiration.
Fatty acid oxidation
Fatty acid oxidation was measured in primary murine hepatocytes with the Seahorse XF96 instrument (Agilent Seahorse), following the manufacturer’s protocol. This protocol quantified the oxidation of both exogenous and endogenous fatty acids (FAs). Cells were cultured in conditions that would stimulate the depletion of endogenous substrates to prime the cells for oxidation of exogenous FAs. Four different conditions were used for each of the treatment groups. These conditions included Etomoxir+ Palmitate-, Etomoxir+ Palmitate+, Etomoxir- Palmitate- and Etomoxir- Palmitate+. In order to determine endogenous fatty acid utilization etomoxir+ conditions were used, as cells treated with etomoxir were unable to import exogenous fatty acids into the mitochondria.
Steatosis assessment in cells
Extent of steatosis and triglycerides (TGs) accumulation within cells were assessed with Steatosis Colorimetric Assay Kit (Cayman) and Triglyceride Colorimetric Assay Kit (Cayman) according to manufacturer’s instructions. TGs were normalized by protein content, which was determined using the Bradford assay (BioRad).
ROS quantification in cells
ROS levels were measured by staining cells with 20 μM 2’,7’-dichlorofluorescin (DCFDA) reagent (Abcam) for 45-60 min at 37°C. The DCFDA solution was then removed and cells were washed twice with PBS. The quantification of the signal was performed with a Victor X4 multilabel plate reader (Perkin-Elmer) with maximum excitation and emission spectra of 495 nm and 529 nm respectively.
RNA isolation and RT-PCR
Total RNA was extracted using TRIzol (Invitrogen) according to the manufacturer’s instructions. RNA was treated with DNase, and 2 μg of RNA was used for reverse transcription (RT). 15X or 20X diluted cDNA was used for RT-quantitative PCR (RT-qPCR) reactions. The RT-qPCR reactions were performed using the Light-Cycler system (Roche Applied Science) and a qPCR Supermix (QIAGEN) with the indicated primers. The average of at least three technical repeats was used for each biological data point. The list of primer used and their sequences are available upon request. The experiments were repeated at least twice (starting from RNA isolation). In case of contradictions in the results between the two repetitions, the experiment has been repeated for the third time.
Protein isolation and Western blot
Proteins were extracted with RIPA buffer containing protease and phosphatase inhibitors and analyzed by SDS-PAGE/western blot. Proteins were detected using the following specific antibodies: ACMSD (Abcam, ab96081), Actin (Sigma, A5441), acetyl-FKHR (Santa Cruz, sc-49437), FOXO1 (Cell Signaling, 2880), HSP90 (BD transduction laboratories, 610418), Total OXPHOS Rodent Antibody Cocktail (Abcam, ab110413), Tubulin (Santa Cruz, sc-5286), Vinculin (Abcam, ab129002). All the antibodies were validated by the manufacturer.
Mitochondrial isolation, Blue Native PolyAcrylamide Gel Electrophoresis (BN-PAGE) and In-gel activity assays
The mitochondrial isolation was performed as described in details in40. In brief, ~30 mg of liver tissue or a pool of 10’000 worms was used for mitochondrial isolation. The samples were homogenized in ice-cold sucrose-containing isolation buffer with Glass/Teflon Potter Elvehjem homogenizer (Wheaton, cat. No. 358029) and mitochondria were pelleted through multiple rounds of centrifugation at different speeds. For BN-PAGE, 50 μg of mitochondria from liver was solubilized in digitonin and sample buffer (Invitrogen, BN2008). Electrophoresis was performed using Native PAGE Novex Bis-Tris Gel System (3 to 12%), as per manufacturer’s instructions with minor modifications. Gel transfer was performed using Invitrogen iBlot gel transfer system. For detection of the complexes, anti-OXPHOS cocktail (Invitrogen, 457999) and WesternBreeze Chromogenic Western Blot Immunodetection Kit (Invitogen, WB7103) were used. In the final detection step, the membrane was incubated with the chromogenic substrate for 8 min for all the gels.
For in-gel activity assays, electrophoresis was performed for 3h (30 min at 150 V and 2.5h at 250 V). Complex I activity was performed by incubating the gels for 15 to 30 min in the substrate composed of 2 mM tris-HCl pH 7.4; 0.1 mg/mL NADH, and 2.5 mg/mL nitrotetrazolium blue. CIV activity was performed by incubating the gels for 30 to 40 min in the substrate composed of 25 mg of 3,3′-diamidobenzidine tetrahydrochloride; 50 mg cytochrome c; 45 mL of 50 mM phosphate buffer pH 7.4, and 5 mL water. CIV+CI activity was performed by subsequently incubating the gels in the substrate for CIV followed by incubation in CI. CII activity was checked on a separate gel, which was incubated in the substrate composed of 20 mM sodium succinate, 2.5 mg/ml of nitrotetrazolium blue, 0.2 mM phenazine methosulfate and 5 mM tris-HCl buffer. All reactions were stopped with 10% acetic acid.
SIRT1 activity
SIRT1 activity was assessed with SIRT1 Direct Fluorescent Screening Assay Kit (Cayman Chemical), according to the manufacturer’s instructions. The absorbance was read at 450 nm.
Mitochondrial DNA copy number
Mitochondrial number was assessed by using mitochondrial DNA – nuclear DNA ratio. Mitochondrial DNA was quantified as described with slight modifications41. Briefly, total DNA was extracted with the Nucleospin tissue kit (Macherey Nagel). 40 ng of total DNA was assessed by real-time PCR using a Light Cycler 480 (Roche). The reaction was performed in a final volume of 8 μl with 1× SYBR green master mix (Roche) and 1.25 μM of the reverse and forward primers. UCP2 primers were used as endogenous control for nuclear DNA and 16s as marker for mitochondrial DNA.
Statistics
Survival analyses were performed using the Kaplan Meier method and the significance of differences between survival curves calculated using the Logrank test. Differences between two groups were assessed using two-tailed t-tests. To compare the interaction between age and genotype, two-way ANOVA tests were performed. Analysis of variance, assessed by Tukey’s or Dunnett’s multiple comparison test, was used when comparing more than two groups. We used GraphPad Prism 6 (GraphPad Software, Inc.) for all other statistical analyses. Sample sizes were chosen without performing statistical tests, but based on studies with similar experimental design and on the known variability of the assay. All p values < 0.05 were considered significant. *P < 0.05; **P ≤ 0.01; ***P ≤ 0.001 unless stated otherwise.
Mouse experiments
Mice were maintained in a controlled environment with 22°C temperature, 50% humidity, 15-20 fresh air changes/hour and light/dark cycle of 12h. They were housed 3-4 animals per cage with ad libitum access to fresh water and food and with appropriate environmental enrichment within the cage. All animal experiments were performed according to Swiss ethical guidelines and approved by the Service de la consommation et des affaires vétérinaires (SCAV) of the Canton de Vaud. Animals that showed signs of severity, predefined by the animal authorizations, were euthanized and removed from the calculations. All experiments on mice were done in a non-blinded way.
Generation of Sirt1hep-/- mice
Liver-specific Sirt1 KO (Sirt1hep-/-) mice were generated as previously described28, by breeding Sirt1L2/L2 mice11 with Alb-Cre mice, expressing Cre Recombinase under the control of the Albumin promoter.
MCD diet-induced NAFLD
12 weeks-old, C57BL/6J males were acclimatized for a period of 7 days before initiation of the experiment. After acclimatization period the animals were randomized based on their body weight and separated into three groups, one receiving methionine-choline deficient (MCD) diet (Harlan Teklad TD.90262) containing vehicle (DMSO 5% + polyethylene glycol (PEG) 20%), the second receiving MCD diet supplemented with ACMSD inhibitor (TES-991) at concentration of 15 mg/kg body weight/day and the third receiving the matched control diet (Harlan Teklad TF.94149) with the vehicle (DMSO 5% + PEG 20%). 2.5 weeks after the beginning of the special diets, animals fasted for 4h were subjected to isoflurane anesthesia and blood was sampled via cardiac puncture. Plasma was collected by centrifugation at 4000 rpm for 10 min at 4°C. Collected tissues and plasma were snap frozen in liquid nitrogen. The experiment was performed once.
Histopathology on MCD diet liver tissues
A piece of liver tissue (always from the same lobe) was fixed with 10% neutral buffered formaldehyde, processed and embedded in paraffin. After sectioning, tissue was stained with hematoxylin and eosin (H&E). Another liver piece (always from the same lobe) was embedded in Shandon™ Cryomatrix™ resin (Thermo Scientific Scientific) and snap-frozen in isopentane cooled in liquid nitrogen, before being placed on dry ice. Oil red O and CD45 stainings were performed on cryosections as described previously28.
Hepatic triglyceride content measurement
Hepatic lipids were extracted as described previously42. Triglyceride content in hepatic lipid fraction was quantified with enzymatic assays (Roche).
Ischemia/reperfusion-induced AKI
9 weeks-old, C57BL/6J males were acclimatized for a period of 7 days before initiation of the experiment. After acclimatization period the animals were randomized based on their body weight and separated into three groups, two of them receiving receiving normal chow diet (Harlan Teklad 2916) containing vehicle (DMSO 5% + polyethylene glycol (PEG) 20%), the third group receiving normal chow diet supplemented with ACMSD inhibitor (TES-1025) at concentration of 15 mg/kg body weight/day. On day 10 after the beginning of the diet mice were anesthetized with isoflurane and placed on a surgical platform in a dorsal position. Both kidneys were exposed through flank incisions and renal pedicles were occluded using vascular clamps for 25 min. The clamp was then removed and the surgical site was sutured. 1 ml of physiological saline was administered intraperitoneally after closing the wound to prevent dehydration. The Sham-operated group was subjected to similar surgical procedures, except that the occluding clamp was not applied. A suitable analgesic (Dafalgan) was administered post operatively to all animals. Animals were monitored for recovery from anaesthesia and then housed singly in their home cage with appropriate environmental enrichment. 48h after the surgery animals were subjected to isoflurane anesthesia and blood was sampled via cardiac puncture, followed by organ collection. Plasma was collected by centrifugation at 4000 rpm for 10 min at 4°C. Plasma was separated into a fresh tube and stored at -80°C. Collected tissues were snap frozen in liquid nitrogen. The experiment was performed once.
Cisplatin-induced AKI
Mice were maintained in a controlled environment with 22°C temperature, 50% humidity, 15-20 fresh air changes/hour and light/dark cycle of 12h. They were housed 3-4 animals per cage with free access to fresh water and food. 8 weeks-old, C57BL/6J males were acclimatized for a period of 5-7 days before initiation of the experiment. Animals were randomized into different treatment groups based on their body weight and basal measured blood urea nitrogen (BUN) levels. After randomization the groups were maintained on specified diet, either normal chow diet (Teklad global 16% protein rodent diet; Harlan Laboratories 2016S) or chow diet supplemented with ACMSD inhibitor, for 10 days. Body weight and food consumption were monitored during this period. Cisplatin was injected intraperitoneally with 1 ml syringe with 26G needle at a dose of 20 mg/kg of body weight. The Sham control group was administered with the same volume of 0.9% saline. Animals were monitored every day for bodyweight loss, general health conditions and signs of pain, distress and mortality. 72h after cisplatin injection animals were subjected to isoflurane anesthesia and blood was sampled via cardiac puncture, followed by organ collection. Plasma was collected by centrifugation at 4000 rpm for 10 min at 4°C. Plasma was separated into a fresh tube and stored at -80°C. Collected tissues were snap frozen in liquid nitrogen. The experiment was performed twice.
Glomerular filtration rate measurement
Determination of glomerular function rate was performed in conscious mice using transcutaneous measurement of FITC-sinistrin (Mannheim Pharma and diagnostic, Mannheim, Germany) elimination as previously described, using miniaturized devices containing an optical component and a microprocessor (Medibeacon, Mannheim, Germany)43,44. Briefly, 55h post-cisplatin administration mice were subjected to a light (1.5-2% Isoflurane) anesthesia to affix the device and a rechargeable battery on a depilated region of the back using a double-sided sticky patch. The device was further held in place with a piece of Transpore tape (3M, Minnesota, USA) surrounding the animal body. After a resting period of 2-5 min, a bolus of FITC-sinistrin (35 mg/ 100g, dissolved in saline) was injected through the tail vein. The excretion kinetics of FITC-sinistrin was recorded in conscious animals for an average of 60 min as a decay of subcutaneous fluorescence of FITC-sinistrin. Elimination half-life (t1/2) was determined from the single exponential phase of the excretion curve and then converted into GFR using a semi-empirical conversion factor developed and validated for mice as previously described44.
Histopathology on AKI kidneys
Half of a kidney was fixed with 10% neutral buffered formaldehyde, processed and embedded in paraffin. After sectioning, tissue was stained with hematoxylin and eosin (H&E). Histopathological scoring was performed by two different pathologists in a blinded and independent way. The scoring system was adapted from45 with slight modifications. Tubular cell necrosis, tubular dilatation, inflammatory cell infiltration, edema and cast formation were scored.
Quantification of intermediates in tryptophan pathway
Snap frozen brain, liver and kidney samples were grinded into powder with liquid nitrogen, using mortar and pestle. Each sample was then pre-weighed (~50 mg) into lysis tubes (soft tissue homogenizing CK 14 tubes) and stored at -80°C prior to metabolite extraction. Frozen tissue powders were extracted by the addition of an ice-cold methanol-H2O 4:1 mixture (100 μl for every 10 mg of tissue) and homogenized using ceramic beads in the Cryolys Precellys 24 sample Homogenizer (2x20 sec at 10000 rpm, Bertin Technologies, Rockville, MD, US), which was air-cooled by a flow rate at 110 L/min at 6 bar. Homogenized extracts were centrifuged for 15 min at 4000 g at 4°C and the supernatant (tissue extract) was removed and used in further preparation for liquid chromatography – tandem mass spectrometry (LC-MS/MS) analysis.
For the analysis of tryptophan pathway intermediates and NAD+, in negative ionization mode, the sample was prepared by mixing an aliquot of tissue extract (50 μl) with 250 μl of the ice-cold internal standard solution (in 100% acetonitrile). For the analysis of end products of kynurenine pathway (i.e. picolinic, quinolinic and nicotinic acid), in positive ionization mode, 300 μl of the tissue extract was evaporated to dryness (using a CentriVap vacuum concentrator, LabConco), and reconstituted with 75 μl 0.2% FA in H2O. Samples were centrifuged and supernatant was injected for LC-MS/MS analysis.
LC-MS/MS analyses were performed using a 6495 triple quadrupole mass spectrometer (QqQ) interfaced with a 1290 UHPLC system (Agilent Technologies, Basel, CH) and operated in the dynamic Multiple Reaction Monitoring (dMRM) mode.
Quinolinic, picolinic and nicotinic acid were measured in positive electrospray ionization (ESI + MS) mode, using an Acquity HSS T3 column (2.1x100mm, 1.8µm, Waters, Montreux-Chailly, CH). An 11-minute gradient was applied starting at 0% B (0.2% FA in methanol, 0-2 min) and increasing to 50% (2-4 min), and further to 90% (4-5 min), before returning to starting conditions (5-7 min) and re-equilibrating for 4 min (7-11 min). Mobile phase A was 0.2% FA in H2O, the flow rate for this method was 400 μl/min and the sample injection volume was 5μl. ESI source conditions were set as follows: dry gas temperature 250°C, nebulizer 35 psi and flow 15 L/min, sheath gas temperature 250°C and flow 8 L/min, nozzle voltage 1000 V, and capillary voltage +3000 V. The cycle time in dMRM mode was 500 ms. Standard calibration curves ranging from 0-50 μM were used for quantification for all three organic acids.
NAD+ was measured in negative electrospray ionization (ESI - MS) mode, using a SeQuant ZIC-pHILIC column (10x2.1 mm I.D., 5 μm) with a SeQuant ZIC-pHILIC guard column (20x2.1 mm I.D., 5 μm (Merck, Darmstadt, Germany)) at an operation temperature of 30°C. Mobile phase A was composed of 20 mM ammonium acetate and 20 mM ammonium hydroxide in H2O (pH 9.3) and mobile phase B was 100% acetonitrile, and the sample injection volume was 2 µl. The linear gradient elution started at 90% B (0-1.5 min), decreased to 50% B (8-11 min), decreased further to 45% B (12-15 min), restored to 90% (15-16 min) and subsequently re-equilibrated for 9 min at a flow rate of 300 μl/min. ESI source conditions were set as follows: dry gas temperature 290°C, nebulizer 35 psi and flow 14 L/min, sheath gas temperature 350°C and flow 12 L/min, nozzle voltage 0 V, and capillary voltage -2000 V. Cycle time in dMRM mode was 600 ms. The following calibration curve range was used for NAD+ quantification: 0-316 µM. NAD+ concentrations were reported with an external calibration curve only (IS not available). Collision energies and product ions (MS2 or quantifier and qualifier ion transitions) were pre-optimized for each metabolite of interest.
Data processing, including the peak integration and concentration calculation was performed in Masshunter Quantitative Analysis (for QqQ, version B.07.01/ Build 7.1.524.0, Agilent Technologies). Linearity of the standard curves was evaluated for each metabolite using a 6-point range; in addition, peak area integration was manually curated and corrected where necessary. Concentrations of the compounds for which the IS were available were corrected for the ratio of MS response (peak area) between the analyte and the IS, to account for matrix effects.
GSH assay
Glutathione quantities in the kidney were assayed by the ELISA kit (Cayman Chemicals) according to the manufacturer instructions.
MPO assay
Myeloperoxidase in kidney tissue was quantified by ELISA kit (Abcam) according to the manufacturer instructions.
Extended Data
Extended Data Figure 1. acsd-1 LOF improves NAD+ levels, mitochondrial function, and lifespan through de novo synthesis in C. elegans.
a. De novo synthesis of NAD+ from tryptophan. Names of the worm’s orthologs are in blue.
b. acsd-1 expression pattern across different developmental stages in wild-type worms expressing extrachromosomal array of acsd-1::gfp transgene. Scale bar=100 μm.
c. acsd-1 expression pattern in adult wild-type worms expressing extrachromosomal array of acsd-1::gfp transgene; i = intestine, m = muscle, p = pharynx, v = vulva.
d. acsd-1 mRNA levels in wild type and rrf-3(pk1426) mutants (n=6, each n represents a pool of ~600 worms).
e. ACSD-1 activity in control (empty vector) vs acsd-1 RNAi fed worms quantified both wild type and rrf-3 mutants (n=3, where each n represents a pool of ~3600 worms) with compensation for negative controls.
f. QPRT-like activity can be detected in both wild type and rrf-3 mutants (n=3, each n represents a pool of ~3600 worms).
g-h. Effects of acsd-1 knockdown throughout the entire life on N2 (g) and rrf-3 mutant (h) worm lifespan.
i. Lifespan of rrf-3(pk1426) mutants exposed to control or acsd-1 RNAi upon tryptophan supplementation. P*: ctrl vs ctrl+Trp 50 μM; P^: ctrl vs acsd-1 RNAi; P°: ctrl+Trp 50 μM vs acsd-1 RNAi+Trp 50 μM.
j. Quantification of GFP signal in ges-1::mito::gfp reporter strain, expressing mitochondria-targeted GFP in the intestine at day 1 and 3 of adulthood (n=4, each n represents a pool of 20 worms).
k. Blue native PAGE on mitochondria extracted from rrf-3 mutant worms fed with either empty vector or acsd-1 RNAi bacteria at day 2 of adulthood (n=3, each n represents mitochondria extracted from a pool of ~10’000 worms).
l. Mitochondrial morphology in the pmyo-3::mito::GFP reporter strain fed with control or acsd-1 RNAi. Stars represent nuclei. Scoring includes the total perimeter of the mitochondrial network, its total area, the area occupied by the mitochondria within the cell and the circularity assessment, where 1=perfect circle and 0=line (n=6 worms).
m. Epistasis between RNAi for acsd-1 and the UPRmt regulator, ubl-5. P*: ctrl vs ctrl/acsd-1 RNAi; P°: ctrl/ubl-5 RNAi vs ubl-5/acsd-1 RNAi.
n. Quantification of the GFP signal in hsp-4::gfp reporter strain (n=4, each n represents a pool of 20 worms) at day 1 and 3 of adulthood.
o. Quantification of the GFP signal in hsp-16.2::gfp reporter strain (n=4, each n represents a pool of 20 worms). After the first time-point sampled at 20°C, worms were exposed to 37°C, and the measurement was repeated every hour for 6 hours.
p. Expression of UPRmt genes in worms at day 2 of adulthood fed with control or acsd-1 RNAi (n=6, each n represents a pool of ~600 worms).
q. Expression of sod-3 mRNA at day 1 of adulthood in control or acsd-1 RNAi fed worms (n=3, each n represents a pool of ~600 worms).
r. Survival of wild type (N2) worms exposed to 4 mM paraquat starting from the L4 stage, in which the knockdown of acsd-1 was performed at different life stages,. P*: ctrl vs acsd-1 RNAi whole life; P^: ctrl vs acsd-1 RNAi development; P°: ctrl vs acsd-1 RNAi adulthood.
s. Epistasis between RNAi for acsd-1 and daf-16 in wild type (N2) worms exposed to 4 mM paraquat. P*: ctrl vs ctrl/acsd-1 RNAi; P^: ctrl/daf-16 RNAi vs daf-16/acsd-1 RNAi.
All worm assays, except for hsp-16.2::gfp reporter strain, performed at 20°C and repeated at least once. Data are mean ± s.e.m. *P≤0.05, **P≤0.01, ***P≤0.001. P values calculated using two-tailed t-test (d, e, j, l, n-q) or Logrank test (g-i, m, r, s). For individual P values, see Source Data. For lifespan values, see Extended Data Table 1.
Extended Data Figure 2. Pathways activated by Acmsd knockdown in worms are conserved in mammalian cells.
a. Acmsd transcript levels reflected by the Ct values in different hepatic and renal cells and cell lines (n=4).
b. Efficiency of Acmsd shRNA in mouse primary hepatocytes 48h post adenoviral transduction (n=6).
c. NAD+ levels in mitochondria of AML-12 cells transduced with either shRNA control or shRNA against Acmsd (n=5).
d-e. Blue native PAGE followed by in-gel activity for Complex II (blue) (d) and Complex I (purple) + IV (brown) (e) on mitochondria extracted from mouse primary hepatocytes transduced with either shRNA control or shRNA against Acmsd for 48h. The experiment was performed once.
f. Primary hepatocytes extracted from a Sirt1L2/L2 mouse were transduced either with an adenovirus encoding GFP (WT condition) or the CRE-recombinase to generate Sirt1 KO primary hepatocytes. These hepatocytes were exposed to an shRNA targeting a random sequence or shRNA targeting Acmsd. Transcript levels of Acmsd and Sirt1 (n=3).
g. FOXO1 acetylation levels in mouse primary hepatocytes transduced with either shRNA control or shRNA against Acmsd for 48h. The experiment was independently performed twice.
Data are mean ± s.e.m.; each n represents a biologically independent sample. *P≤0.05, **P≤0.01, ***P≤0.001. P values calculated using two-tailed t-test. For gel source images see Supplementary Fig. 1. For individual P values, see Source Data.
Extended Data Figure 3. Pharmacological inhibition of ACMSD has similar effects as its genetic downregulation.
a-b. mRNA levels of mitochondrial genes in mouse primary hepatocytes treated for 24h with DMSO or TES-1025 (a) or TES-991 (b), at the indicated concentrations (n=3).
c. SOD2 activity in mouse primary hepatocytes treated for 24h with DMSO or TES-1025, at indicated concentrations (n=4).
d. Fatty acid oxidation assessed in mouse primary hepatocytes treated with DMSO or TES-991 for 24h at the indicated concentrations (n=5). FCCP (2 μM) was used as an uncoupler to reach maximal respiration.
e. mRNA levels of mitochondrial genes in HK-2 cells after 24h of treatment with TES-1025 or TES-1025 in combination with SIRT1 inhibitor, EX527, at the indicated concentrations (n=5-8).
f. Apoptosis rate in HK-2 cells assessed 16h after addition of 50 μM cisplatin by caspase 3/7 activity. TES-1025 was added simultaneously with the cisplatin.
g. Biochemical analysis of plasma from mice fed with chow diet or chow diet supplemented with TES-991 or TES-1025 at the dose of 15 mg/kg body weight/day (ctrl and TES-991: n=10; TES-1025: n=9 mice).
h-i. Quinolinic acid (QA) (h) and Nicotinic acid (NA) (i) levels in livers (n=9), kidneys (ctrl: n=10; TES-991 and TES-1025: n=9 mice) and brains (ctrl: n=11; TES-991 and TES-1025: n=12 mice). from mice fed with control chow diet or chow diet supplemented with TES-991 or TES-1025 at the dose of 15 mg/kg body weight/day.
j-k. mRNA levels of β-oxidation, mitochondrial and oxidative stress defence genes in livers (j) and kidneys (k) of mice described in (g) (n=6 mice).
Data are mean ± s.e.m. *P≤0.05, **P≤0.01, ***P≤0.001. P values calculated using two-tailed t-test (a-d, g-k) or one-way ANOVA (e, f). For individual P values, see Source Data.
Extended Data Figure 4. ACMSD inhibitors protect hepatic function from MCD diet-induced NAFLD.
a. Plasma AST levels in 16-week old C57BL/6J male mice fed for 2.5 weeks with control diet, MCD diet, and MCD diet supplemented with 15 mg/kg/day of TES-991 (n=8 mice).
b. Representative photomicrographs of liver tissues stained with hematoxylin and eosin (H&E) and Oil red O from the mouse cohorts described in (a). The experiment was performed twice in an independent way.
c. Representative photomicrographs of liver tissues from the mouse cohorts described in (a) stained with CD45 and the corresponding negative control. The experiment was performed twice in an independent way.
d. Hepatic SOD2 activity in mouse cohorts described in (a) (CD: n=8; MCD: n=7; MCD+TES-991: n=6 mice).
e-h. Liver NAD+ (e), triglyceride (TGs) content (f), plasma ALT (g), AST (h) levels in congenic C57BL/6J Sirt1hep-/- mice that match the mouse cohorts described in (a) related to age, gender and treatment duration (CD: n=8; MCD and MCD+TES-991: n=10 mice).
i. Representative photomicrographs of liver tissues stained with hematoxylin and eosin (H&E) from the Sirt1hep-/- mice described in (e-h). The experiment was performed once.
j. Hepatic SOD2 activity in congenic C57BL/6J Sirt1hep-/- mice described in (e-h) (CD: n=8; MCD: and MCD+TES-991: n=9 mice).
k. mRNA levels of oxidative stress defence, mitochondrial, β-oxidation, inflammatory and fibrosis genes in livers of Sirt1hep-/- mice (n=8 mice).
Data are mean ± s.e.m. *P≤0.05, **P≤0.01, ***P≤0.001. P values calculated using two-tailed t-test. For individual P values, see Source Data.
Extended Data Figure 5. ACMSD inhibitors protect renal function in two different models of AKI.
a. Schematic timeline of the cisplatin-induced AKI study. AKI was induced at day 10 after the beginning of the study in male C57BL/6J mice by a single intraperitoneal dose of cisplatin (20mg/kg body weight). Mice in the sham control group were injected with a saline solution. GFR was measured non-invasively 52h post-cisplatin administration.
b-d. Representative photomicrographs of H&E stained kidney sections and the histopathological scoring for tubular necrosis (c), and inflammatory cell infiltration (d) of mouse cohorts described in (a) (Sham ctrl, Cis-AKI+TES-1025: n=6; Cis-AKI: n=5 mice).
e. Schematic timeline of the IR-induced AKI study. AKI was induced at day 10 after the beginning of the study in anesthetised male C57BL/6J mice by a dorsal surgical incision and bilateral occlusion of the renal pedicles for 25 min. Mice in the sham control group underwent the same surgical procedure without application of the occluding clamp on the renal pedicles.
f-j. Representative photomicrographs of H&E stained kidney sections (f) and histopathological scoring for cumulative score (g), tubular necrosis (h), tubular dilation (i), and cast formation (j) of mouse cohorts described in (e) (n=5 mice). Tubular cell necrosis (arrows), tubular dilation and casts (asterisk) and interstitial edema (crescent moon) are indicated on the pictures by the corresponding symbols in brackets.
k-m. GSH protein levels (k), MPO activity (l) and NAD+ content (m) in kidneys of the IR-AKI cohorts described in (e) (n=5 mice).
n. Protein expression of the respiratory complex subunits in kidneys from the mouse cohorts described in (e). The experiment was independently performed twice.
Data are mean ± s.e.m. *P≤0.05, **P≤0.01, ***P≤0.001. P values calculated using two-tailed t-test. For gel source images see Supplementary Fig. 1. For individual P values, see Source Data. The histopathological scoring performed by two pathologists in a blinded and independent way (b-d, f-j).
Extended Data Table 1. Summary of C. elegans lifespan experiments.
$ vs rrf-3 (pk1426) sir-2.1 ½, # vs rrf-3 (pk1426) atfs-1 ½, * vs rrf-3 (pk1426) ubl-5 ½, + vs rrf-3 (pk1426) daf-16 ½, § vs rrf-3 (pk1426) ev/Trp 50 μM
Errors are represented as s.e.m. Survival analyses were performed using the Kaplan Meier method and the significance of differences between survival curves calculated using the Logrank test (two-sided).
| Conditions | Cumulative statistics | Statistics of individual exprts | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Strain | Treatment/RNAi | No. of expts. | Mean lifespan [days] (treatment/ctrl) |
variation [%] compared to control | P-value | No. of animals (treatment/ctrl) |
Mean lifespan [days] (treatment/ctrl) |
variation [%] compared to control | P-value | No. of animals (treatment/ctrl) |
| N2 | acsd-1 | 2 | 20.5/20.5 (±0.47/0.41) |
0 | 0.3 | 116/124 | 20.2/20.9 | -3.35 | 0.303 | 43/44 |
| 20.6/20.2 | +1.98 | 0.582 | 73/80 | |||||||
|
rrf-3 (pk1426) |
acsd-1 | 2 | 21.4/18.5 (±0.56/0.56) |
+15.7 | 0.00034 | 137/118 | 22.0/18.6 | +18.3 | 0.004 | 70/66 |
| 20.9/18.4 | +13.6 | 0.042 | 67/52 | |||||||
|
rrf-3 (pk1426) |
acsd-1 1/2 | 4 | 20.2/18.5 (±0.35/0.32) |
+9.2 | <0.0001 | 262/246 | 18.6/17.0 | +9.4 | 0.047 | 61/57 |
| 19.6/17.6 | +11.4 | 0.03 | 59/64 | |||||||
| 20.2/18.2 | +11.0 | 0.009 | 63/61 | |||||||
| 21.4/20.6 | +3.9 | 0.036 | 79/64 | |||||||
|
rrf-3 (pk1426) |
sir-2.1 1/2 | 1 | 17.4/18.1 (±0.51/0.62) |
-3.9 | 0.32 | 61/61 | 17.4/18.1 | -3.9 | 0.32 | 61/61 |
|
rrf-3 (pk1426) |
sir-2.1/acsd-1 | 1 | 17.6/18.1 (±0.56/0.62)17.6/17.4$ (±0.56/0.51)$ |
-2.8 +1.1$ |
0.67 0.57$ |
66/61 66/61$ |
17.6/18.1 | -2.8 | 0.67 | 66/61 |
|
rrf-3 (pk1426) |
atfs-1 1/2 | 2 | 17.1/17.9 (±0.45/0.46) |
-4.5 | 0.42 | 135/125 | 16.8/17.6 | -4.5 | 0.42 | 69/64 |
| 17.4/18.2 | -4.4 | 0.67 | 66/61 | |||||||
|
rrf-3 (pk1426) |
atfs-1/acsd-1 | 2 | 18.0/17.9 (±0.48/0.46) 18.0/17.1# (±0.48/0.45)# |
+0.6 +5.2# |
0.72 0.25# |
117/125 117/135# |
18.2/17.6 | +3.4 | 0.58 | 55/64 |
| 17.9/18.2 | -1.6 | 0.94 | 62/61 | |||||||
|
rrf-3 (pk1426) |
ubl-5 1/2 | 1 | 17.0/20.6 (±0.3/0.45) |
-17.5 | <0.0001 | 68/64 | 17.0/20.6 | -17.5 | <0.0001 | 68/64 |
|
rrf-3 (pk1426) |
ubl-5/ acsd-1 | 1 | 17.2/20.6 (±0.34/045) 17.2/17.0*(±0.34/0.3)* |
-16.5 +1.2* |
<0.0001 0.36* |
74/64 74/68* |
17.2/20.6 | -16.5 | <0.0001 | 74/64 |
|
rrf-3 (pk1426) |
daf-16 1/2 | 2 | 15.0/17.8 (±0.41/0.52) |
-15.7 | <0.0001 | 80/126 | 13.4/17.0 | -21.2 | <0.0001 | 39/60 |
| 16.4/18.6 | -11.8 | 0.01+ | 41/66 | |||||||
|
rrf-3 (pk1426) |
daf-16/acsd-1 | 2 | 14.1/17.8 (±0.34/0.52) 14.1/15.0+ (±0.34/0.4)+ |
-20.8 -6.0+ |
<0.0001 0.06+ |
99/126 99/80+ |
13.0/17.0 | -23.5 | <0.0001 | 49/60 |
| 15.1/18.6 | -18.8 | <0.0001 | 50/66 | |||||||
| Paraquat experiments | ||||||||||
|
rrf-3 (pk1426) |
acsd-1 (whole life) | 3 | 7.45/6.85 (±0.14/0.17) |
+8.0 | 0.0034 | 110/137 | 8.97/8.57 | +4.7 | 0.014 | 42/64 |
| 6.42/5.71 | +12.4 | 0.028 | 47/48 | |||||||
| 6.11/5.33 | +14.7 | 0.025 | 24/35 | |||||||
|
rrf-3 (pk1426) |
acsd-1 1/2 | 2 | 6.19/5.84 (±0.13/0.15) |
+6.48 | 0.0142 | 38/44 | 3.9/3.73 | +4.56 | 0.334 | 30/32 |
| 6.97/6.68 | +4.34 | 0.043 | 8/12 | |||||||
|
rrf-3 (pk1426) |
daf-16/acsd-1 | 1 | 3.07/3.73 (±0.14/0.15) |
-21.49 | 0.006 | 47/32 | 3.07/3.73 | -21.49 | 0.006 | 47/32 |
|
rrf-3 (pk1426) |
acsd-1 (develop) | 2 | 3.55/3.28 (±0.13/0.15) |
+8.2 | 0.0149 | 73/83 | 3.43/3.08 | +11.4 | 0.0175 | 53/52 |
| 3.68/3.48 | +5.74 | 0.0279 | 20/31 | |||||||
|
rrf-3 (pk1426) |
acsd-1 (adult) | 2 | 3.68/3.28 (±0.13/0.15) |
+12.2 | 0.0013 | 69/83 | 3.7/3.1 | +21.4 | 0.00082 | 51/52 |
| 3.66/3.48 | +5.17 | 0.005 | 18/31 | |||||||
| Tryptophan supplementation experiments | ||||||||||
|
rrf-3 (pk1426) |
evl Trp 50μM | 2 | 22.3/20.1 (±0.51/0.40) |
+10.9 | <0.0001 | 129/173 | 22.4/20.6 | +8.73 | 0.025 | 71/91 |
| 22.2/20.4 | +8.82 | 0.001 | 58/78 | |||||||
|
rrf-3 (pk1426) |
acsd-1/Trp 50μM | 2 | 22.7/20.1 (±0.50/0.40) 22.7/22.3§ (±0.50/0.51)§ |
+12.9 +1.79§ |
<0.0001 0.81§ |
143/173 129/173§ |
23.2/20.6 | +12.6 | 0.001 | 62/91 |
| 22.2/20.4 | +8.82 | 0.06 | 81/78 | |||||||
Supplementary Material
Acknowledgements
We thank P. Gönczy and the Caenorhabditis Genetics Center for providing reagents, the Bioimaging and Optics Core Facility and the Phenotyping Unit of EPFL, Norman Moullan, Thibaud Clerc and Sabrina Bichet for technical assistance. EK was supported by Fondation Romande pour la Recherche sur le Diabète. MZ was supported by the KNOW consortium 'Healthy Animal—Safe Food' MS&HE No. 05-1/KNOW2/2015 and the Foundation for Polish Science. The work was supported by funds from EPFL and Swiss National Science Foundation (grant 310030B).
Footnotes
Data availability
The authors declare that all the data supporting the findings of this study are available from the corresponding author upon request.
Materials availability
Materials can be requested from JA (admin.auwerx@epfl.ch). ACMSD inhibitors used in this study are described previously in a manuscript24 and a patent application that was licensed to TES Pharma (US9708272 B2).
Author contributions
EK, JA and RP conceived and designed the project. EK, AM, MZ, FDF, KG, DR, NG performed experiments. PL synthesised ACMSD inhibitors. OM made the acsd-1::gfp C. elegans reporter strain. NR and LC performed activity assays for the enzymes in de novo NAD+ biosynthesis. VvdH and JI performed the metabolomics and NS-R the histopathology. SDS and DL assisted with GFR measurements. EK, KS and JA wrote the manuscript with the contributions from all other authors.
Author information
Reprints and permissions information is available at www.nature.com/reprints.
Competing Financial Interests
JA, RP and NR are inventors on a patent application describing ACMSD inhibitors (application No: 14/839,209; applicant - TES Pharma S.r.l., Corciano, Italy). Granted US patent 9,708,272 (18 July, 2017). The patent application covers the results obtained with the compounds TES-991 and TES-1025 described in Figures 3-5 RP, FDF, NG and PL are employed by TES Pharma.
References
- 1.Houtkooper RH, Pirinen E, Auwerx J. Sirtuins as regulators of metabolism and healthspan. Nat Rev Mol Cell Biol. 2012;13:225–238. doi: 10.1038/nrm3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Imai S, Guarente L. It takes two to tango: NAD+ and sirtuins in aging/longevity control. npj Aging and Mechanisms of Disease. 2016;2 doi: 10.1038/npjamd.2016.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belenky P, Bogan KL, Brenner C. NAD+ metabolism in health and disease. Trends in biochemical sciences. 2007;32:12–19. doi: 10.1016/j.tibs.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 4.Yang Y, Sauve AA. NAD+ metabolism: Bioenergetics, signaling and manipulation for therapy. Biochim Biophys Acta. 2016;1864:1787–1800. doi: 10.1016/j.bbapap.2016.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Katsyuba E, Auwerx J. Modulating NAD(+) metabolism, from bench to bedside. The EMBO journal. 2017;36:2670–2683. doi: 10.15252/embj.201797135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bender DA. Biochemistry of tryptophan in health and disease. Mol Aspects Med. 1983;6:101–197. doi: 10.1016/0098-2997(83)90005-5. 0098-2997 83)90005-5 [pii] [DOI] [PubMed] [Google Scholar]
- 7.Fukuoka SI. Identification and Expression of a cDNA Encoding Human alpha -Amino-beta -carboxymuconate-epsilon -semialdehyde Decarboxylase (ACMSD). A KEY ENZYME FOR THE TRYPTOPHAN-NIACINE PATHWAY AND "QUINOLINATE HYPOTHESIS". Journal of Biological Chemistry. 2002;277:35162–35167. doi: 10.1074/jbc.M200819200. [DOI] [PubMed] [Google Scholar]
- 8.Vrablik TL, Huang L, Lange SE, Hanna-Rose W. Nicotinamidase modulation of NAD+ biosynthesis and nicotinamide levels separately affect reproductive development and cell survival in C. elegans. Development. 2009;136:3637–3646. doi: 10.1242/dev.028431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rongvaux A, Andris F, Van Gool F, Leo O. Reconstructing eukaryotic NAD metabolism. BioEssays : news and reviews in molecular, cellular and developmental biology. 2003;25:683–690. doi: 10.1002/bies.10297. [DOI] [PubMed] [Google Scholar]
- 10.McReynolds MR, Wang W, Holleran LM, Hanna-Rose W. Uridine monophosphate synthetase enables eukaryotic de novo NAD+ biosynthesis from quinolinic acid. The Journal of biological chemistry. 2017 doi: 10.1074/jbc.C117.795344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mouchiroud L, et al. The NAD(+)/Sirtuin Pathway Modulates Longevity through Activation of Mitochondrial UPR and FOXO Signaling. Cell. 2013;154:430–441. doi: 10.1016/j.cell.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hashimoto T, Horikawa M, Nomura T, Sakamoto K. Nicotinamide adenine dinucleotide extends the lifespan of Caenorhabditis elegans mediated by sir-2.1 and daf-16. Biogerontology. 2010;11:31–43. doi: 10.1007/s10522-009-9225-3. [DOI] [PubMed] [Google Scholar]
- 13.Gebauer J, et al. A Genome-Scale Database and Reconstruction of Caenorhabditis elegans Metabolism. Cell systems. 2016;2:312–322. doi: 10.1016/j.cels.2016.04.017. [DOI] [PubMed] [Google Scholar]
- 14.Burnett C, et al. Absence of effects of Sir2 overexpression on lifespan in C. elegans and Drosophila. Nature. 2011;477:482–485. doi: 10.1038/nature10296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Viswanathan M, Guarente L. Regulation of Caenorhabditis elegans lifespan by sir-2.1 transgenes. Nature. 2011;477:E1–2. doi: 10.1038/nature10440. [DOI] [PubMed] [Google Scholar]
- 16.Houtkooper RH, et al. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature. 2013;497:451–457. doi: 10.1038/nature12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benedetti C, Haynes CM, Yang Y, Harding HP, Ron D. Ubiquitin-like protein 5 positively regulates chaperone gene expression in the mitochondrial unfolded protein response. Genetics. 2006;174:229–239. doi: 10.1534/genetics.106.061580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haynes CM, Yang Y, Blais SP, Neubert TA, Ron D. The matrix peptide exporter HAF-1 signals a mitochondrial UPR by activating the transcription factor ZC376.7 in C. elegans. Molecular cell. 2010;37:529–540. doi: 10.1016/j.molcel.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berdichevsky A, Viswanathan M, Horvitz HR, Guarente L. C. elegans SIR-2.1 interacts with 14-3-3 proteins to activate DAF-16 and extend life span. Cell. 2006;125:1165–1177. doi: 10.1016/j.cell.2006.04.036. [DOI] [PubMed] [Google Scholar]
- 20.Honda Y, Honda S. The daf-2 gene network for longevity regulates oxidative stress resistance and Mn-superoxide dismutase gene expression in Caenorhabditis elegans. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 1999;13:1385–1393. [PubMed] [Google Scholar]
- 21.Pucci L, Perozzi S, Cimadamore F, Orsomando G, Raffaelli N. Tissue expression and biochemical characterization of human 2-amino 3-carboxymuconate 6-semialdehyde decarboxylase, a key enzyme in tryptophan catabolism. FEBS Journal. 2007;274:827–840. doi: 10.1111/j.1742-4658.2007.05635.x. [DOI] [PubMed] [Google Scholar]
- 22.Fukuwatari T. Phthalate Esters Enhance Quinolinate Production by Inhibiting -Amino- -Carboxymuconate- -Semialdehyde Decarboxylase (ACMSD), a Key Enzyme of the Tryptophan Pathway. Toxicological Sciences. 2004;81:302–308. doi: 10.1093/toxsci/kfh204. [DOI] [PubMed] [Google Scholar]
- 23.Saito K, et al. Mechanism of increases in L-kynurenine and quinolinic acid in renal insufficiency. Am J Physiol Renal Physiol. 2000;279:F565–572. doi: 10.1152/ajprenal.2000.279.3.F565. [DOI] [PubMed] [Google Scholar]
- 24.Pellicciari R, et al. alpha-Amino-beta-carboxymuconate-epsilon-semialdehyde Decarboxylase (ACMSD) Inhibitors as Novel Modulators of De Novo Nicotinamide Adenine Dinucleotide (NAD(+)) Biosynthesis. Journal of medicinal chemistry. 2018;61:745–759. doi: 10.1021/acs.jmedchem.7b01254. [DOI] [PubMed] [Google Scholar]
- 25.Chen Y, Guillemin GJ. Kynurenine pathway metabolites in humans: disease and healthy States. International journal of tryptophan research : IJTR. 2009;2:1–19. doi: 10.4137/ijtr.s2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Michelotti GA, Machado MV, Diehl AM. NAFLD, NASH and liver cancer. Nature reviews Gastroenterology & hepatology. 2013;10:656–665. doi: 10.1038/nrgastro.2013.183. [DOI] [PubMed] [Google Scholar]
- 27.Gariani K, et al. Eliciting the mitochondrial unfolded protein response by nicotinamide adenine dinucleotide repletion reverses fatty liver disease in mice. Hepatology. 2016;63:1190–1204. doi: 10.1002/hep.28245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gariani K, et al. Inhibiting poly-ADP ribosylation increases fatty acid oxidation and protects against fatty liver disease. Journal of hepatology. 2016 doi: 10.1016/j.jhep.2016.08.024. [DOI] [PubMed] [Google Scholar]
- 29.Lewington AJ, Cerda J, Mehta RL. Raising awareness of acute kidney injury: a global perspective of a silent killer. Kidney international. 2013;84:457–467. doi: 10.1038/ki.2013.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tran MT, et al. PGC1alpha drives NAD biosynthesis linking oxidative metabolism to renal protection. Nature. 2016;531:528–532. doi: 10.1038/nature17184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu L, et al. Quantitative Analysis of NAD Synthesis-Breakdown Fluxes. Cell metabolism. 2018 doi: 10.1016/j.cmet.2018.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi H, et al. NAD Deficiency, Congenital Malformations, and Niacin Supplementation. The New England journal of medicine. 2017;377:544–552. doi: 10.1056/NEJMoa1616361. [DOI] [PubMed] [Google Scholar]
- 34.Kamath RS, Martinez-Campos M, Zipperlen P, Fraser AG, Ahringer J. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2001;2 doi: 10.1186/gb-2000-2-1-research0002. RESEARCH0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mouchiroud L, et al. Pyruvate imbalance mediates metabolic reprogramming and mimics lifespan extension by dietary restriction in Caenorhabditis elegans. Aging Cell. 2011;10:39–54. doi: 10.1111/j.1474-9726.2010.00640.x. [DOI] [PubMed] [Google Scholar]
- 36.Mouchiroud L, et al. The Movement Tracker: A Flexible System for Automated Movement Analysis in Invertebrate Model Organisms. Current protocols in neuroscience. 2016;77 doi: 10.1002/cpns.17. 8 37 31-38 37 21. [DOI] [PubMed] [Google Scholar]
- 37.Zamporlini F, et al. Novel assay for simultaneous measurement of pyridine mononucleotides synthesizing activities allows dissection of the NAD(+) biosynthetic machinery in mammalian cells. The FEBS journal. 2014;281:5104–5119. doi: 10.1111/febs.13050. [DOI] [PubMed] [Google Scholar]
- 38.Yang T, Sauve AA. NAD metabolism and sirtuins: metabolic regulation of protein deacetylation in stress and toxicity. AAPS J. 2006;8:E632–643. doi: 10.1208/aapsj080472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oosterveer MH, et al. LRH-1-dependent glucose sensing determines intermediary metabolism in liver. The Journal of clinical investigation. 2012;122:2817–2826. doi: 10.1172/JCI62368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jha P, Wang X, Auwerx J. Analysis of Mitochondrial Respiratory Chain Supercomplexes Using Blue Native Polyacrylamide Gel Electrophoresis (BN-PAGE) Current protocols in mouse biology. 2016;6:1–14. doi: 10.1002/9780470942390.mo150182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lagouge M, et al. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 2006;127:1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 42.Jha P, et al. Role of adipose tissue in methionine-choline-deficient model of non-alcoholic steatohepatitis (NASH) Biochim Biophys Acta. 2014;1842:959–970. doi: 10.1016/j.bbadis.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schock-Kusch D, et al. Transcutaneous measurement of glomerular filtration rate using FITC-sinistrin in rats. Nephrol Dial Transplant. 2009;24:2997–3001. doi: 10.1093/ndt/gfp225. [DOI] [PubMed] [Google Scholar]
- 44.Schreiber A, et al. Transcutaneous measurement of renal function in conscious mice. Am J Physiol Renal Physiol. 2012;303:F783–788. doi: 10.1152/ajprenal.00279.2012. [DOI] [PubMed] [Google Scholar]
- 45.Melnikov VY, et al. Neutrophil-independent mechanisms of caspase-1- and IL-18-mediated ischemic acute tubular necrosis in mice. The Journal of clinical investigation. 2002;110:1083–1091. doi: 10.1172/JCI15623. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.