Abstract
The single mitochondrion of apicomplexan protozoa is thought to be critical for all stages of the life cycle, and is a validated drug target against these important human and veterinary parasites. In contrast to other eukaryotes, replication of the mitochondrion is tightly linked to the cell cycle. A key step in mitochondrial segregation is the fission event, which in many eukaryotes occurs by the action of dynamins constricting the outer membrane of the mitochondria from the cytosolic face. To date, none of the components of the apicomplexan fission machinery have been identified and validated. We identify here a highly divergent, dynamin-related protein (TgDrpC), conserved in apicomplexans as essential for mitochondrial biogenesis and potentially for fission in Toxoplasma gondii. We show that TgDrpC is found adjacent to the mitochondrion, and is localised both at its periphery and at its basal part, where fission is expected to occur. We demonstrate that depletion or dominant negative expression of TgDrpC results in interconnected mitochondria and ultimately in drastic changes in mitochondrial morphology, as well as in parasite death. Intriguingly, we find that the canonical adaptor TgFis1 is not required for mitochondrial fission. The identification of an Apicomplexa-specific enzyme required for mitochondrial biogenesis and essential for parasite growth highlights parasite adaptation. This work paves the way for future drug development targeting TgDrpC, and for the analysis of additional partners involved in this crucial step of apicomplexan multiplication.
Author summary
Mitochondria fission is mediated by an elaborate “fission complex” of dynamin and receptor proteins, best studied in organisms such as yeast and mammalian cells. Little is known about this process in apicomplexan parasites, unicellular eukaryotes characterised by a single, lasso-shaped mitochondrion. Here, we analyse the conservation of the fission complex in Toxoplasma gondii; we find that the protein TgFis1 is the only mitochondrial adaptor that is highly conserved, but our characterisation shows that it is not essential for parasite growth or for mitochondrial dynamics. In contrast, we find that the dynamin-related protein C (TgDrpC) is a key factor of mitochondrial biogenesis in T. gondii, showing that its recruitment to the basal part of the mitochondrion is essential for the formation of mature, separate organelles. Furthermore, our analysis shows that TgDrpC GTPase domain is conserved and essential for its function, highlighting its potential as novel drug target.
Introduction
Members of the Apicomplexa phylum are unicellular obligate parasites. Being eukaryotes, they share many features of common model organisms, such as a double-membrane nuclear envelope and a conserved endomembrane system. However, they are also set apart by various novel characteristics, such as the phylum-specific apical complex, which contains specialised secretory organelles; a pellicle-like membranous compartment called the inner membrane complex (IMC); and an atypical non-photosynthetic plastid, the apicoplast.
Moreover, they present a single, ramified mitochondrion whose genome is severely reduced, encoding only three proteins [1–4]; while this organelle is usually lasso-shaped in intracellular tachyzoites, in extracellular parasites its morphology is changed drastically, reducing its proximity to the parasite periphery [5]. The mitochondrion is thought to be essential for parasite survival at all stages, as it has a central function both in energy generation and in the production of precursors of other pathways, like pyrimidine and heme biosynthesis [6–9]. Thus, it is no surprise that the mitochondrion is a validated drug target for both Plasmodium spp. and T. gondii, as reviewed in [10, 11].
Nevertheless, our knowledge about the mechanisms controlling mitochondrial behaviour in Apicomplexa is limited. In most eukaryotes studied, mitochondria form a dynamic network, whose shape depends on the opposing mechanisms of fission, fusion and motility, which occur throughout the cell cycle [12–15]. Conversely, previous research in T. gondii has shown that the duplication of the mitochondrion in tachyzoites is tightly linked to cell division (known as endodyogeny in T. gondii), similar to the single-cell red alga Cyanidioschyzon merolae [15], and spontaneous fission of mitochondria in resting intracellular parasites is not clearly observed [5, 16]. T. gondii mitochondrial duplication starts at the early stages of daughter formation, when the IMC is beginning to form, at which point the mitochondrion branches in multiple locations: these elongations are not immediately incorporated in the developing daughter cells, but stay close to the periphery of the mother until the last stage of cytokinesis. When the daughters are almost fully formed, and start emerging from the mother cell, the mitochondria migrate into them. At this point, the new mitochondria are still linked to each other, with the interconnection seen at the base of the new daughter cells. These interconnections can remain present for a variable time, but it is assumed that they eventually get cleaved leading to individual mitochondria. The mechanism underlying this putative fission step is not yet characterised.
In all eukaryotes studied to date, the key player of mitochondrial fission is a dynamin-related protein, called Drp1 or Dnm1 in humans and yeast, respectively, and DRP3A/B in plants [17–19]. This mechanochemical enzyme, characterised by a large GTPase domain, is essential for the remodelling of the mitochondria membrane, and is typically recruited to the mitochondrial outer membrane (MOM) at ER/actin preconstricted fission sites [20, 21]. Recruitment is mediated by an adaptor complex consisting of receptor/adaptor proteins, whose composition varies significantly between different eukaryotes (Table 1) [22–26]. After its recruitment, the dynamin-related protein forms spirals around the membrane. Subsequent GTP hydrolysis provokes a conformational change that results in a two-fold decrease in the diameter of the spiral, and finally in the cleavage of the membrane [27–29]. In mammalian cells this last step requires an additional constriction mediated by the classical dynamin Dyn2 [30]. Thus, dynamin-related proteins are central to mitochondrial fission.
Table 1. known factors involved in mitochondrial fission in yeast, humans and plants and their conservation in T. gondii genome.
| Protein name | Function | Toxoplasma gondii homolog |
|---|---|---|
| Drp1 (human)/Dnm1(yeast)/Drp3A-B(plants) | Dynamin related protein | TgDrpA (TGME49_267800), TgDrpB (TGME49_321620), TgDrpC (TGME49_270690) |
| Fis1 (human, yeast, plants) | Mitochondrial outer membrane adaptor | TgFis1 (TGME49_263323) |
| Mdv1/Caf4/Num1/Mdm36 (yeast) | Mitochondrial receptor for Dnm1 | - |
| Mff (Human) | Recruitment of Drp1 to the mitochondrial membrane | - |
| MiD49/50 (Human) | Drp1 receptor | - |
| ELM1 (plants) | Recruitment of Drp3 to the mitochondrial membrane | - |
Two well conserved dynamin-related proteins, TgDrpA and TgDrpB, were previously identified and functionally characterised in T. gondii, where they were shown to play essential roles in apicoplast division [31] and in the biogenesis of the unique secretory organelles [32], respectively. While a minor role for TgDrpA in mitochondrial biogenesis could not be ruled out, no clear implication in mitochondrial division was found. Bioinformatics identified a third potential member of the dynamin superfamily in these parasites [31–33]: this protein was called TgDrpC. Analysis of the phylogenetic distribution of TgDrpC identified homologs in apicomplexans and in representatives of the sister phylum Chromerida, but not in the representative of Perkinsozoa, Perkinsus marinus, or in any Ciliates representative. This suggests that TgDrpC is restricted to apicomplexan and closely related organisms. Additionally, TgDrpC was shown to be essential for parasite growth [34].
Here, we studied the putative fission mechanism in T. gondii. We showed that TgDrpC has an essential function in mitochondria biogenesis, potentially via coordinating fission during parasite division. Moreover, we investigated the presence of the adaptor complex in T. gondii. Intriguingly, we found that TgFis1, a homolog of the human Fis1, an adaptor of Drp1, is not involved in mitochondrial fission in T. gondii. Indeed, TgFis1 depletion did not result in parasite death.
Our results highlight the essentiality of mitochondrial dynamics for the parasite and demonstrate that an unconventional, Apicomplexa-specific dynamin-related protein is at the core of this process.
Materials and methods
T. gondii parasite lines, maintenance and transfections
T. gondii tachyzoites were maintained in human foreskin fibroblasts (HFF) cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 2 mM glutamine, and 25 μM gentamycin at 37°C and 5% CO2 in a humidified incubator. Transfections were carried out by electroporation using 10^7 freshly egressed or mechanically released parasites as previously described [35].
Bioinformatics analysis
Evidence for homology between fission proteins of human, yeast and plants (as reported in Fig 1B) and T. gondii was collected using reciprocal BLAST analysis. Sequences of Drp1 (Uniprot accession number: O00429), Dnm1 (P54861), Drp3 A/B (Q8S944 and Q8LFT2), Fis1 (Q9Y3D6), Mdv1 (P47025), Caf4 (P36130), Num1 (Q00402), Mdm36 (Q06820), Mff (Q9GZY8), Mid49/50 (Q96C03) and Elm1 (Q93YN4) were retrieved from UniProt database and BLASTP searches performed in ToxoDB. TgDrpC shows high score when the BLAST is with human Drp1 (e-value of 2e-06), albeit lower than TgDrpA and TgDrpB. Reciprocal BLAST with TgDrpC against NCBI identifies human Drp1 with high score (e-value of 6.5e-6). BLAST search with human Fis1 retrieves TgFis1 with an e-value of 1e-12.
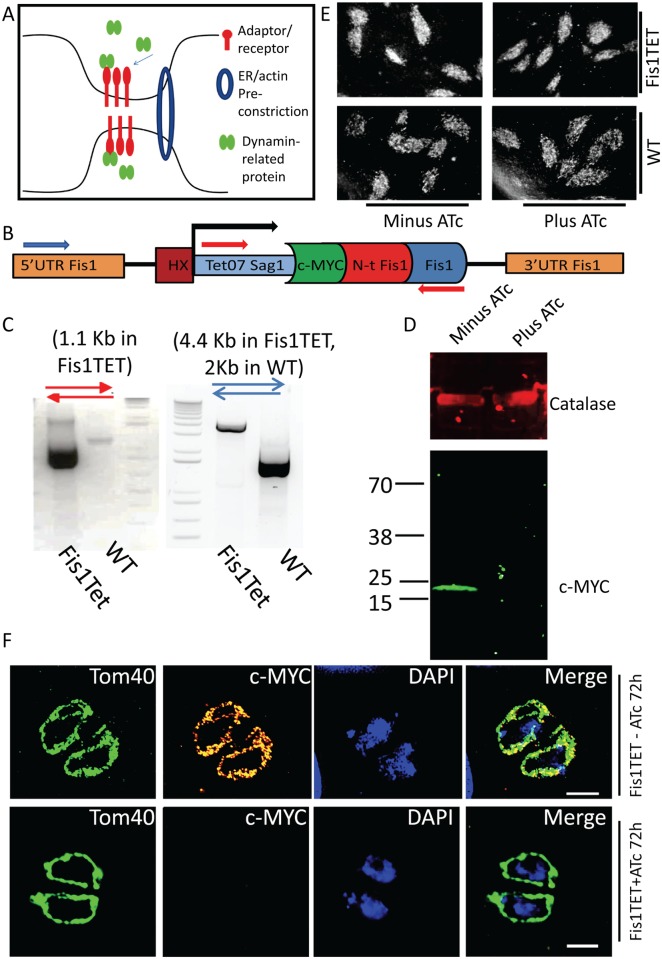
Fig 1.
Conservation of the fission machinery in T. gondii: TgFis1 is not essential for mitochondrial fission A) Illustration of the main events that lead to mitochondrial fission in higher eukaryotes. ER and actin enwrap the mitochondrial membrane at future sites of fission, and an “adaptor complex” recruits Drp1/Dnm1, which forms spirals around the MOM and constricts it. (B) Scheme of the promoter replacement strategy adopted for the conditional KD of TgFis1; red and blue arrows indicate oligonucleotides designed to verify promoter exchange via genomic PCR. (C) Genomic PCR confirms the replacement of the endogenous promoter of TgFis1 with the inducible promoter Tet07-Sag1. A clonal parasite line was used for genomic PCR; primer positions and amplicon length are specified. (D) Confirmation of protein depletion upon addition of ATc for 48h in Fis1KD. In absence of ATc, western blot analysis shows the expected protein size of 17 kDa, tagged with c-MYC; the induction of TgFis1 knockdown with ATc shows that at 48 hours the protein is no longer detectable by western blot. (E) Plaque assay on parasites grown on HFF cells for 7 days in presence and absence of ATc shows that depletion of TgFis1 is not deleterious for parasite fitness. The images shown are representative of three experiments. (F) Immunofluorescence analysis shows that TgFis1 is evenly distributed in the mitochondrial outer membrane. In presence of ATc, the signal of Fis1 is no longer detectable, but mitochondria morphology is not affected. The experiment was performed in triplicate; representative images are shown. Scale bar: 2 μm.
BLAST searches using human Drp1 finds an additional putative gene (TGME49_326100) which seem to encode a fragment of a dynamin-like protein. However, consultation with the ToxoDB team suggested that this is likely a duplicate of the same sequence as that assembled into chromosome 1b (Omar Harb, personal communication).
Homology modelling and molecular dynamics simulations
To construct a reliable homology for TgDrpC, BLAST+ [2] was used to identify related sequences in the UniProt data base (The UniProt Consortium, 2016). The conserved N-terminal GTPase domain (300 residues) was then used to construct the homology model using SWISSMODEL [36, 37]. All potential models were evaluated manually with the model based on PDB:3L43 determined at a resolution of 2.3 Å, which shares a sequence identity of approximately 22%, being the most reliable (30% over the GTPase domain). To further assess the validity of the active site, GDP was included into the putative binding of the homology model and the GTPase-GDP complex model was optimized by energy minimization followed by a short molecular dynamics simulation using GROMACS [38]. GDP parameters were obtained in the Gromos54a7 forcefield from the ATP Repository (molID:35650) [39]. The simulation converged quickly resulting in an excellent binding pose for GDP consistent with its propose catalytic activity. Visual inspections, structural comparisons, least-squares superpositions and figures were prepared using Pymol [40].
Plasmid construction
The plasmid TgFis1TET was generated as described in [41]. Fis1 5’UTR was amplified with primers Fis5’-fw and fis5’-rev, and the N-terminus of Fis1 was amplified with primers FisNterminus-fw and FisNterminus-rev. All fragments were cloned into the parental vector Tet07-Sag1-HX [41], which was transfected in the strain TATIΔKu80ΔHX and selected by HX selection [42]. Integration was confirmed using the primers FisTET integration fw 1 and FisTET integration rev, and FisTET integration fw 2 and FisTET integration rev.
To generate the plasmid TgDrpC-YFP LIC, the C-terminal part of the gene was amplified using the primers DrpC-LIC fw and DrpC-LIC rev; the resulting PCR product was inserted into the plasmid pLIC-YFP by ligation independent cloning [43, 44]. Following transfection into the Δku80 strain [43], transfected parasites were selected using pyrimethamine. Integration was confirmed using the primers DrpCYFP integration fw and DrpCYFP integration rev.
To generate the plasmids DD-GFP-DrpCwt and DD-GFP-DrpCtruncated, full length and truncated cDNA encoding TgDrpC was amplified using oligonucleotides DD-DrpCwt-fw and DD-DrpCwt-rev, and DD-DrpCtruncated-fw and DD-DrpCtruncated-rev, respectively (Table 2). Subsequently, ligation through NsiI-NotI restriction into the vector p5RT70DDmycGFP-HX [32] was performed. The vector DD-DrpCwt was subsequently modified with Q5 modification kit (New England BioLabs, Catalog number E0554S) to delete the DD cassette, and obtain the plasmid GFP-DrpCwt, using primers Q5wt-fw and Q5wt-rev. Similarly, the vectors DD-GFP-DrpCDN and DD-GFP-DrpCGTPase only were obtained through modification of the DD-GFP-DrpCwt plasmid, using the primers Q5DN-fw and Q5DN-rev, and Q5GTPase only-fw and Q5GTPase only–rev, respectively. Transfections were made in the RH parental strain.
Table 2. Oligonucleotides used.
| Oligo name | Sequence |
|---|---|
| Fis5’fw | gggTCATGAGTCTCTTTTGAAGACGTGCACCG |
| Fis5’rev | ggGGATCCTCTCGTACAGTGCTCACAAAAAACG |
| FisNterminusfw | cccAGATCTATGGAAGACTCCAACTTCAGTC |
| FisNterminusrev | gggACTAGTGGGCGAAACACGCAAGTAAC |
| FisTET integration fw 1 | GCTGCACCACTTCATTATTTCTTCTGG |
| FisTET integration fw 2 | TCATGAGTCTCTTTTGAAGACGTGCACCG |
| FisTET integration rev | GCTATAAACACAGCCGAGGCGAC |
| DrpC-LIC fw | TACTTCCAATCCAATTTAATGCCCGAG |
| DrpC-LIC rev | TCCTCCACTTCCAATTTTAGCAGCC |
| DrpCYFP integration fw | GCGCCACTCACGACGAAG |
| DrpCYFP integration rev | ATGGGCACCACCCCGG |
| DrpCYFP integration fw 2 | TTTGTGATGCTCGTCAGG |
| DrpCYFP integration rev 2 | CTGGAACCCTTCCATACTG |
| DD-DrpCwt | CCATGCATCGAACGCGCTGCCGCGTC |
| DD-DrpCw | CCTTAATTAATTACGCCCCATTCAACGGTGACGGAAGC |
| Q5wt-fw | TTTAGATCTAAAAGGGAATTCAAGAAAAAATG |
| Q5wt-rev | GCCATGGCCATGCATAGT |
| Q5DN-fw | GAGCATGGGCgcgACGACCCTTCTC |
| Q5DN-rev | TGCTGCCCGAAGACGACA |
| Q5GTPase only-fw | TAATCACCGTTGTGCTCAC |
| Q5GTPase only–rev | CTCACTCAAGAGGCTCTG |
| DD-DrpCtruncated-fw | CCTTAATTAATTACGCCCCATTCAACGGTGACGGAAGC |
| DD-DrpCtruncated-rev | CCATGCATGGAGCCTTCGAGAGTTCATTCTCTCTGCACCTCC |
Immunoblot analysis
Parasites were cultivated for the indicated times in the following conditions: the line Fis1TET was grown in presence or absence of 0.5 μg/mL ATc for 72 hours; the strain TgDrpC-YFP was grown in normal medium; parasites TgDrpC-U1 were grown for 24, 48 and 72 hours in presence of 50 nM of Rapamycin; the line DD-GFP-DrpCDN was grown with 0.5 μM of Shield-1 for 0, 4, 8 or 24 hours; the strains DD-GFP-DrpCwt, DD-GFP-DrpCGTPase only and DD-GFP-DrpCwt were cultivated in presence or absence of 1 μM Shield-1 for 24 hours. Parasites were subsequently harvested and western blot analysis of total parasite lysates were performed as described previously [45], using the antibodies indicated in the figures and figure legends.
Plaque assay
For plaque assays, the clonal lines Fis1TET and DD-GFP-DrpCDN were grown for 7 days in the presence or absence of 0.5 μg/mL ATc or 1 μM of Shield-1, respectively. Fixation, staining and visualization of plaques were performed as previously described [32].
Immunofluorescence analysis
Immunofluorescence analysis was carried out on cover slips. Cells were fixed with 4% paraformaldehyde for 20 minutes, then a permeabilisation step using 0.2% Triton X-100 in PBS was performed for 20 minutes, followed by a blocking step using 2% bovine serum albumin in PBS for 20 minutes, then stained for 60 minutes with the indicated antibodies. Primary antibodies used were: mouse anti c-MYC (Sigma, M-4439), rabbit anti-catalase, rabbit anti-Mic4 and rabbit anti-GAP45 (gifts from Dominique Soldati), mouse anti-HSP60 (gifts from Boris Striepen), rabbit anti-Tom40 (gifts from Giel Van Dooren), rat anti-DrpB, rabbit anti-Rop13 (gift from Peter Bradley). Subsequently, secondary stain was performed for 60 minutes. Secondary antibodies used were goat anti-mouse, goat anti-rabbit or goat anti-rat AlexaFluor 350, AlexaFluor 488, AlexaFluor 594 or AlexaFluor 633-conjugated antibodies (Life Technologies). These reactions were carried out at room temperature. Co-localisation analysis was performed using FIJI plugin “JACoP” to determine Manders’ coefficient, counting at least 20 images per condition [46].
Microscopy
Widefield images were acquired in z-stacks of 2 μm increments and were collected using an Olympus UPLSAPO 100x oil (1.40NA) objective on a Deltavision Core microscope (Applied Precision, GE) attached to a CoolSNAP HQ2 CCD camera. Deconvolution was performed using SoftWoRx Suite 2.0 (Applied Precision, GE). Video microscopy was conducted with the DeltaVision Core microscope as above. Normal growth conditions were maintained throughout the experiment (37°C; 5% CO2). Further image processing was performed using ImageJ64 software.
Moreover, an ELYRA PS.1 microscope (Zeiss) was used for super-resolution microscopy (SR-SIM), as described in [47].
Results
T. gondii does not possess a conserved mitochondrial fission complex
To identify candidates involved in T. gondii mitochondrial fission, we performed bioinformatics analysis to assess the conservation of known fission factors from yeast, human and plants in the genome of the parasite (illustrated in Fig 1A). We could only identify putative homologs for the dynamin component (TgDrpA, TgDrpB and TgDrpC) and for the fission component (TgFis1) (Fig 1B). We ruled out TgDrpA and TgDrpB, since earlier analyses indicated no major involvement of these dynamin-related proteins in mitochondria function. Therefore, we hypothesised that TgFis1 (TGME49_263323) and TgDrpC (TGME49_270690) could be involved in mitochondrial fission in T. gondii, and set out to examine these two proteins in detail.
Conditional knock-down of Fis1 does not impair mitochondria biogenesis
The fission-protein 1 (TgFis1)—a tetratricopeptide repeat domain-containing protein that is essential for mitochondrial fission in yeast—is highly conserved in T. gondii, as shown by Padgett et al. [48], who identified it as a tail-anchored protein, and showed that when ectopically expressed it localises to the mitochondrion.
To investigate the role of TgFis1 in T. gondii, we employed the tetracyclin inducible transactivator system [49] to generate a knockdown parasite line for TgFis1. Briefly, the endogenous promoter of TgFis1 was replaced via homologous recombination, as previously described [41, 42], resulting in a parasite line where TgFis1 is N-terminally tagged with c-MYC epitope tag and is under the control of the conditional promoter T7Sag1 (Fig 1C) [49]. Promoter replacement was confirmed by PCR and the regulation is shown by western blot analysis (Fig 1D and 1E). As shown in Fig 1G, in absence of ATc the TgFis1-Myc signal overlaps with the signal of the mitochondrial marker TgTOM40 [50]. Immunoblot and immunofluorescence analysis demonstrate that the signal of TgFis1 is undetectable at 72 hours post induction with ATc. Plaque assay measuring parasite growth for 7days shows that depletion of TgFis1 does not have a deleterious effect on parasite proliferation (Fig 1F); furthermore, no defect in mitochondria morphogenesis is observable by immunofluorescence analysis (IFA) (Fig 1G). These results are confirmed by a CRISPR/Cas9 genome-wide screen, where TgFis1 is classified as “dispensable”, with a phenotypic score of +0.94 [51]. We conclude that TgFis1 is not required for the growth of T. gondii tachyzoites in cell culture, and that it does not play a central role in the control of mitochondrial morphology under normal growth conditions.
TgDrpC is an apicomplexan dynamin-related protein and in silico studies predict its GTPase domain to be active
To understand TgDrpC function in T. gondii, we first performed a detailed in silico analysis. TgDrpC is a highly divergent, potential dynamin-related protein, characterised by a GTPase domain at its N-terminus, and by a tail domain which is largely disordered. While the first 118 residues of TgDrpC, as well as the C-terminal tail, share no detectable sequence similarity with any known structures, the putative GTP binding domain shows low yet marked sequence identities with a range of other GTP binding proteins.
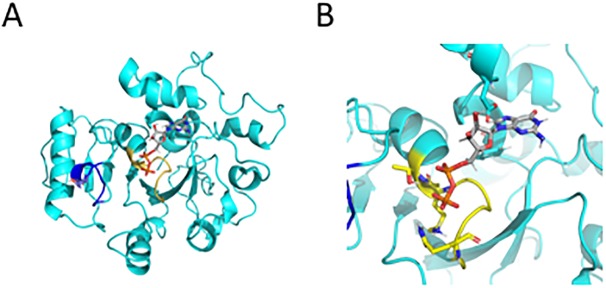
To provide molecular insight into the putative GTPase activity, a homology model was constructed using SWISSmodel [36]: the closest model in the protein database as identified by BLAST [52] is the crystal structure of the human dynamin 3 GTPase domain (PDB: 3L43), determined by the Toronto Structural Genomics Consortium at a resolution of 2.3 Å. The GTPase domain of human dynamin 1 and dynamin 3 are highly conserved, with a sequence similarity of over 80% for the GTPase domain (identity 56%) and more importantly, all GTP contacting residues are fully conserved. As TgDrpC and human dynamin 3 share a sequence identity of 31% over the GTPase domain (amino-acids 118–401), a reliable homology model for the overall structure of this domain can be calculated for TgDrpC; however, as shown in S1A Fig, there are a number of insertions and deletions, in particular in the C-terminal side of TgDrpC GTPase domain. We predict that while these differences will lead to significant local structural changes, the overall fold will be conserved. To further validate GDP/GTP binding, GDP was included in the putative binding pocket and the structure of the resulting GTPase-GDP complex was optimized by molecular dynamics simulations.
As shown in Fig 2A and 2B, the GTP binding site is mainly conserved, with the diphosphate-contacting P-loop (shown in yellow) displaying only small changes in sequence with its characteristic GX4GKT motif and structure. In this region, the residues Ser41 and Lys44 (numbering corresponding to the human dynamin 3), which form a salt bridge to the β-phosphate group, are conserved, while the following Ser44 and Ser45 residues–which in human dynamin 3 form H-bonds to the β-phosphate and α-phosphate, respectively—correspond to threonine residues able to form the same interactions in TgDrpC. Importantly, the switch I region shown in blue, contains the conserved threonine residues that may serve in the GTP hydrolysis by interacting with the Mg2+ and the γ-phosphate [53].
Fig 2.
Modelling of the GTP binding domain of TgDrpC (A) Monomer model for TgDrpC GTPase domain. The P-loop responsible for tri/diphosphate binding is indicated in yellow, while the switch I region with the putative MG2+ coordinating threonine is in blue. (B) close-up of the GDP binding site after molecular dynamics simulation with key residue interacting to the co-substrate shown in ball-and-stick representation, yellow for residues in the P-loop, cyan for residues interacting with the ribose and the guanine.
In addition, the TgDrpC equivalent to the Switch II region, which in other GTPases contains either a conserved glutamine or histidine residue that can activate the water molecule for GTP hydrolysis, harbours a lysine residue close-by, which could fulfil the same function. In contrast, the guanine-contacting residues (Lys206, Asp208, Asp211 and Asn236 in the human dynamin 3 GTPase structure, shown in blue in S1A Fig) are not conserved in TgDrpC, and as a result, some reorganisation of the loop regions were required in the molecular dynamics simulation to accommodate GDP/GTP binding. The 3D homology model, shown in Fig 2b, clearly supports the notion of GTPase activity; however, no further conclusion about which residue acts as water-activating residue to perform the nucleophilic attack should be drawn, as in this part of the model the sequence similarity with human dynamin 3 is significantly lower.
Based on the homodimeric human dynamin 3 structure [37], the TgDrpC homology model adopts a similar homodimer (Fig 2C), though the oligomeric structure of the full-length TgDrpC remains to be elucidated. Finally, the C-terminus of the protein is highly disordered. Clustal-Omega alignment of TgDrpC homologs in Apicomplexa (Table 3, S1B Fig) shows that there are two highly conserved regions in the tail (highlighted in red). These regions, however, do not correspond to any canonical GTPase effector domain (GED) or middle domain, which are commonly found in dynamin-related proteins.
Table 3. accession numbers of sequences used for alignment.
| Sequence | UniProt accession number |
|---|---|
| Tg DrpC | S8GIW4_TOXGM |
| Ta DrpC | Q4UHJ0_THEAN |
| Bb DrpC | A7AS48_BABBO |
| Pf DrpC | Q8I5M3_PLAF7 |
| Pv DrpC | A5JZZ2_PLAVS |
| Pb DrpC | A0A077XK90_PLABA |
| Cp DrpC | Q5CW16_CRYPI |
TgDrpC forms foci at the mitochondrial periphery and basal end
To assess the localization of TgDrpC, we generated a knock-in strain where TgDrpC is endogenously tagged at its C-terminus with YFP, as confirmed by PCR (Fig 3A and 3B). Western blot analysis performed with the obtained clonal line TgDrpC-YFP confirms the expression of the fusion protein, which has the predicted mass of ≈160 kDa (Fig 3C).
Fig 3. Endogenous tagging of TgDrpC reveals a mitochondria localisation.
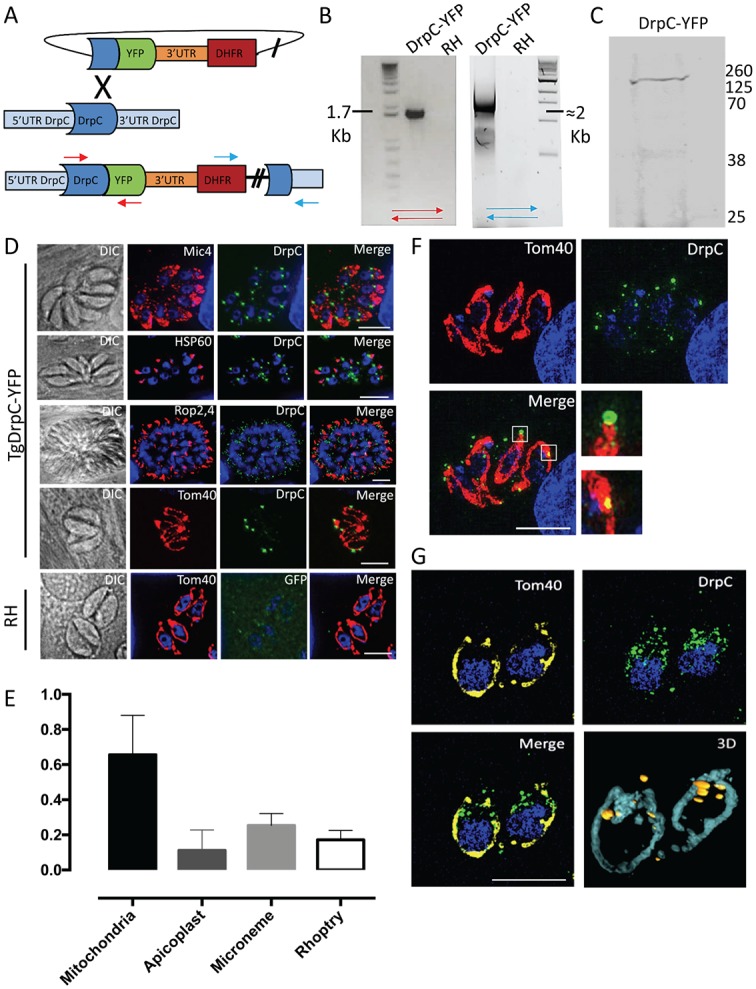
(A) Strategy for the endogenous tagging of TgDrpC with YFP in RHΔku80 parasite strain: The ligation-independent cloning vector LIC-YFP was integrated at the 3’ end of TgDrpC, through a single-crossover mechanism. (B) PCR analyses using the primers indicated in (A) confirms the integration of the tagging construct at 5’ and 3’ ends. (C) Western blot analysis using α-GFP antibody on the TgDrpC-YFP clonal line verifies the expected protein size of ≈160 kDa. (D) Immunofluorescence images showing the co-localisation between TgDrpC and the indicated organelles (Mic4, micronemes; HSP60, apicoplast; Rop2-4, rhoptries; Tom40, mitochondrion). (E) Quantification of colocalisation between TgDrpC and the indicated organelles; the Manders’ coefficient (average of n>20 values) is reported in the y axis. (F) TgDrpC puncta (green) have different shapes and sizes, varying from spirals to smaller dots on the membrane; 3-D reconstruction (G) confirms that the vast majority of TgDrpC aggregates are in close proximity with mitochondria. Scale bar: 5 μm.
Dynamin-related proteins studied in other systems form a diffuse cytoplasmic pool until recruited to the target membrane, where they oligomerize and form spirals [13, 54]. Similarly, our immunofluorescence analysis of TgDrpC-YFP detected a faint but specific cytoplasmic signal, alongside a distinct pattern of punctate structures that appear associated with the mitochondrial membrane (Fig 3D and 3E). The number and size of these structures are highly variable: when viewed from above, it is possible to visualise spiral-like structures of TgDrpC around the mitochondrial tubule (Fig 3F, upper inset), while in other cases the puncta are small and form dots (Fig 3F, lower inset). Evaluation of images from 3D reconstruction confirm that most puncta are in close proximity to the mitochondrion (Fig 3G). Quantification indicates that the mitochondrion is the only organelle that shows a strong co-localisation with TgDrpC (Manders’ coefficient ≈0.7); in particular, while the foci at the periphery are not always in contact with the organelle, the ones at the basal end of the parasite consistently co-localise with the mitochondrion (Fig 3E).
TgDrpC is recruited to the basal end of mitochondria during endodyogeny
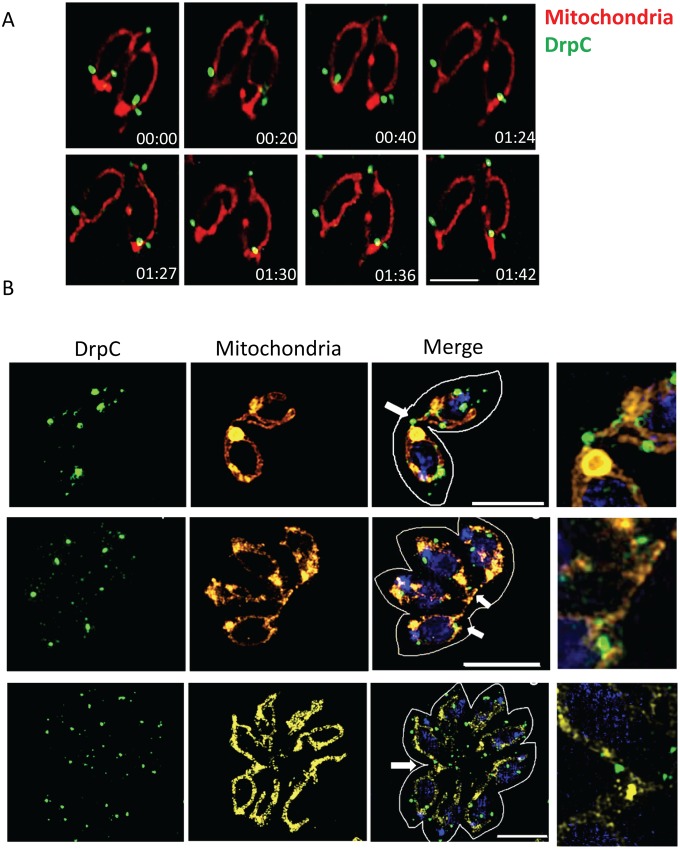
In the last stages of endodyogeny, newly-formed mitochondria migrate into the two developing daughter cells [16]; they remain attached for an unknown period at the basal part, where they are eventually separated. To follow the localization of TgDrpC during endodyogeny, we performed live cell imaging on parasites expressing the outer mitochondrial marker TGME49_215430 fused to tdTomato [5] in the strain TgDrpC-YFP. We focused our analysis on parasites at the late stages of replication, when mitochondria are inside the newly formed daughter, but still connected at the basal end (S1 Video). As shown in Fig 4A, the puncta of TgDrpC are both at the periphery and at the basal connection between the two mitochondria. While the TgDrpC puncta found at the periphery of the mitochondria remain static, the signal at the basal connection of the mitochondria is highly dynamic. In the example shown in Fig 4A, at time point 00:20 two distinct TgDrpC foci are at the basal interconnection; at 00:40, a constriction is noticeable in one of the foci, and at 1:27 TgDrpC seems to be at the terminal end of the mitochondria tubule, and the connection is no longer detectable. Interestingly, one of the TgDrpC foci persisted at the basal end of the mitochondria, even twenty minutes after the cleavage step occurred. These behaviours are in support of a TgDrpC role in mitochondrial fission.
Fig 4. Recruitment of TgDrpC at interconnected mitochondria during replication.
(A) Time-lapse analysis of parasites TgDrpC-YFP/TGME49_215430-tdTomato with interconnected mitochondria (in red, TGME49_215430-tdTomato; in green, TgDrpC-YFP). TgDrpC is at the mitochondria periphery and at the basal interconnection; while the puncta at the periphery are more static, the signal at the basal end is highly dynamic. Time is indicated in minutes. Scale bar: 5 μm. (B) SIM microscopy shows that the puncta at the basal end coincide with sites of constriction of the MOM (white arrows). Scale bar: 5 μm.
To provide further support for this model, we performed structured illumination microscopy (SIM) on the TgDrpC-YFP/TGME49_215430-tdTomato strain, choosing vacuoles containing two, four or eight parasites which show mitochondria that are still interconnected. Multiple puncta of TgDrpC are visible at the interconnections of the mitochondria at the basal end (Fig 4B). Importantly, in some cases an invagination of the mitochondrial tubule is seen and what seems like constriction of the membrane colocalises with TgDrpC signal.
Together, our results from live and fixed samples indicate that TgDrpC is present at the basal end of mitochondria during the last stages of endodyogeny, and that TgDrpC may constrict the interconnected mitochondria, mirroring the behaviour of Drp1 and Dnm1 in humans and yeast [55].
Conditional knock-down of TgDrpC results in interconnected mitochondria
We reasoned that if involved in fission, TgDrpC depletion would inhibit mitochondrial separation after division [13, 54]. We analysed the phenotypic consequences of TgDrpC depletion using the U1 system [34] to obtain a TgDrpC-U1 inducible mutant, where TgDrpC is tagged with an HA epitope tag, and addition of rapamycin results in TgDrpC mRNA degradation (Fig 5A). In accordance to our previous findings (also shown here, S2A Fig), we observed that TgDrpC is essential for parasite survival.
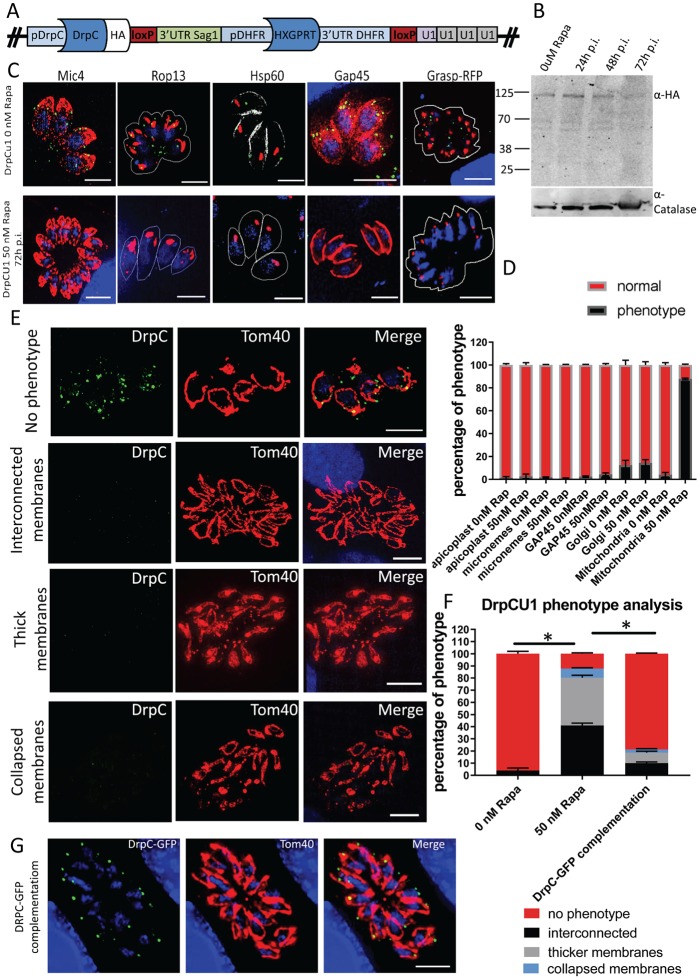
Fig 5. Inducible Knock-down of DrpC has a specific mitochondrial effect.
(A) Scheme of the genomic locus after integration of the DrpC-U1 construct, as detailed in Pieperhoff et al., 2015. (B) Western Blot analysis showing efficient downregulation of TgDrpC 72 hours post Rapamycin induction. The membrane was probed with α-HA and α-Catalase antibodies. (C) Immunofluorescence analysis shows that TgDrpC signal is no longer observable at 72 hours post induction; at this time, the distribution of micronemes and rhoptries, probed with Mic4 and Rop13, was not affected in Rapamycin-treated parasites; similarly, the morphology of apicoplast (αHSP60), IMC (αGAP45) and Golgi (GRASP-RFP) appeared normal, as quantified in (D). (D) Organelle phenotypes were scored in the following way: for micronemes and rhoptries, an apical distribution was counted as “normal”; for the apicoplast, both location (at the apical part of resting parasites) and number (one per parasite in a vacuole) were assessed; for the Golgi, correct distribution above the nucleus and morphology (i.e., not fragmented) were checked; the IMC stain forming the “banana shape” typical of tachyzoites was scored as normal. For each data set, 100 parasitophorous vacuoles were counted. This was performed in triplicate. In contrast, as shown by Immunofluorescence analysis in (E) and quantified in (F), induced parasites showed severe morphology defects in the mitochondrion, classified as “interconnected mitochondria” (41%), “thick mitochondrial membranes” (39.2%) and “collapsed mitochondria” (7%) Scale bar: 5μm. (G) Complementation experiments through transient transfection of plasmid p5RT70-GFP-DrpC in Rapamycin-treated parasites led to rescue of the mitochondrial phenotype, as quantified in (F). Quantification in (F) was obtained through comparison of three independent experiments, each with at least 300 vacuoles. Error bars show SD. Asterisks indicate significant difference (P<0.001 multiple t-test).
Western blot analysis confirmed that at 72 hours post induction TgDrpC-HA is highly reduced at the protein level (Fig 5B). Similarly, we observe no puncta of TgDrpC by IFA at 72 hours post-induction (Fig 5C, lower panel). In agreement with TgDrpC involvement in mitochondrial biogenesis, at this time point 90% of mitochondria show abnormal morphology, while in non-induced TgDrpC-HA-U1 96% of the mitochondria show the typical lasso-shape morphology (Fig 5D and 5E). We identified three different mitochondrial phenotypes in the induced parasites: 41% of the vacuoles contain mitochondria that remain interconnected with each other; 39.2% of the vacuoles show mitochondria with what seem to be swollen membranes, a phenotype we refer to as “thick membranes”; finally, ~7% of the vacuoles show small mitochondria shaped like closed circles (Fig 5E and 5F). In contrast, at the same time point we found that TgDrpC down-regulation has no significant effect on other organelles such as the apicomplexan-specific secretory organelles (micronemes and rhoptries) and apicoplast; likewise, rosette organisation is not significantly affected by TgDrpC knock-down, and Golgi morphology, as assessed with the signal of transiently expressed GRASP-RFP marker [56], shows no significant differences in induced and non-induced parasites 72 hours after induction (Fig 5C and 5D). When TgDrpC knock-down was analysed at later time points, a drastic morphological change was observed, where mitochondria completely lost their shape. At this late time point some parasites also showed additional defects, such as disruption of the IMC, which most likely represents a non-specific secondary effect caused by depletion of TgDrpC (S2B Fig).
Finally, to correlate the observed mitochondria phenotype to TgDrpC depletion, we carried out a complementation experiment by transiently expressing a second copy of TgDrpC tagged with GFP (GFP-DrpC). GFP-DrpC localises to the MOM in rapamycin treated and non-treated parasites, and fully rescues the mitochondrial morphological defect when expressed in induced TgDrpC-U1 parasites (Fig 5F and 5G). Taken together, these data support a role for TgDrpC in mitochondria morphogenesis.
The expression of a dominant-negative form of DrpC impairs correct mitochondria segregation
The variable morphological phenotypes observed upon prolonged TgDrpC depletion (S2 Fig) raise the possibility that this protein could contribute to additional functions during parasite replication; an alternative explanation is that some of the morphologies represent a secondary outcome occurring in response to DrpC depletion. We reasoned that a rapid induction of protein depletion would allow us to identify the primary function of TgDrpC.
To achieve this, we employed the ddFKBP-system [57] to rapidly express a dominant-negative version of TgDrpC. This strategy is based on previous research showing that expression of dynamins with mutations in the GTPase domain has a dominant-negative effect. In these mutants, loss of the GTPase activity impairs the dynamin function, but does not affect membrane recruitment. As a result, competition between the endogenous and the ectopic mutant over membrane binding leads to functional impairment of endogenous TgDrpC [17, 58, 59].
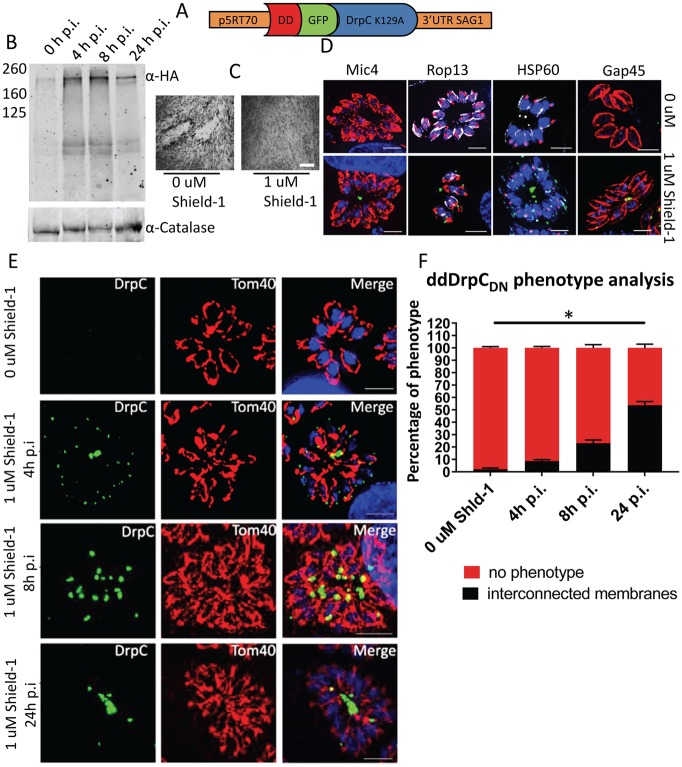
We generated a line (DD-GFP-DrpCDN) whereby ddFKBP-DrpC(K129A) is randomly integrated in the genome of RH parasites; efficient regulation was demonstrated by western blot analysis following 0 to 24 hours induction with 0.5 μM Shield-1 (Fig 6B). We detected very low levels of DD-GFP-DrpCDN in the absence of Shield-1, and 4 hours after induction, full expression levels are reached, with no significant change after 8–24 hours.
Fig 6. Over-expression of a dominant-negative form of TgDrpC leads to interconnected mitochondria.
A) Scheme of the plasmid DD-GFP-DrpCDN. (B) Western blot analysis with the indicated antibodies shows that Shield-1 induction efficiently regulates the expression of DD-GFP-DrpCDN as soon as 4 hours post induction. (C) Plaque assay shows that induction of DD-GFP-DrpCDN causes parasite death. Parasites were grown for 7 days on HFF cells in presence and absence of 1μM Shield-1. The experiment was performed in triplicate; representative images are shown. (D) The effects of DD-GFP-DrpCDN expression were assessed 24 hours after Shield-1 induction; immunofluorescence analysis of the specific markers Mic4, Rop13, HSP60 and Gap45 shows no effect on secretory organelles, apicoplast or IMC morphology. (E and F) Eight hours after stabilisation, DD-GFP-DrpCDN shows an accumulation at the basal part of the mitochondria, which increasingly appear interconnected (23% of the total); 24 hours after induction, more than half of the parasites show abnormally interconnected mitochondria, and DD-GFP-DrpCDN accumulates at the interconnections (scale bar: 5 μm). At least 100 parasitophorous vacuoles were counted in triplicate. Error bars represent SD from the three independent experiments. Asterisks indicate significant difference (P<0.001 multiple t-test).
In agreement with our observations for TgDrpC depletion, plaque assays showed impaired growth in response to Shield-1 induced expression of dominant-negative TgDrpC (Fig 6C). Likewise, immunofluorescence analysis of DD-GFP-DrpCDN parasites grown in presence of Shield-1 for 24 hours shows that, while no effect is observed on the biogenesis of the secretory organelles, the apicoplast and the IMC (Fig 6D), a mitochondrial defect is evident, as more than half of the vacuoles show an “interconnected mitochondria” phenotype (Fig 6E and 6F). Interestingly, DD-GFP-DrpCDN is both at the periphery and at the basal part of the mitochondrion at 4 hours post-induction, when only 9% of the vacuoles show interconnected mitochondria. At 8 hours post-induction, almost all the signal is accumulated at the basal part of the mitochondrion, and the proportion of interconnected mitochondria rises to 23%; at 24 hours post-induction, 53.6% of the vacuoles have this phenotype, and show an accumulation of DD-GFP-DrpCDN signal at the interconnection regions. Importantly, at these time points neither of the other mitochondrial phenotypes seen upon U1-knockdown of TgDrpC (thicker membrane or closed-circled mitochondria) was observed. These data support the function of TgDrpC in mitochondrial fission and suggest that the additional phenotypes observed upon extended knockdown may result from downstream effects.
Notably, the over-expression of the wild-type copy of TgDrpC (DD-GFP-DrpCwt) does not influence parasite fitness; Shield-1 mediated stabilisation of DD-GFP-DrpCwt causes the protein to form puncta at the mitochondrion, and no mitochondrial phenotype is observed upon induction (S3C Fig). The stronger fluorescence signal obtained by overexpression of DD-GFP-DrpCwt allowed us to follow TgDrpC localisation over longer times in using time lapse analysis and confirmed the data obtained for endogenous tagged TgDrpC, supporting the proposed role of TgDrpC in mitochondrial fission (S2D and S2E Fig).
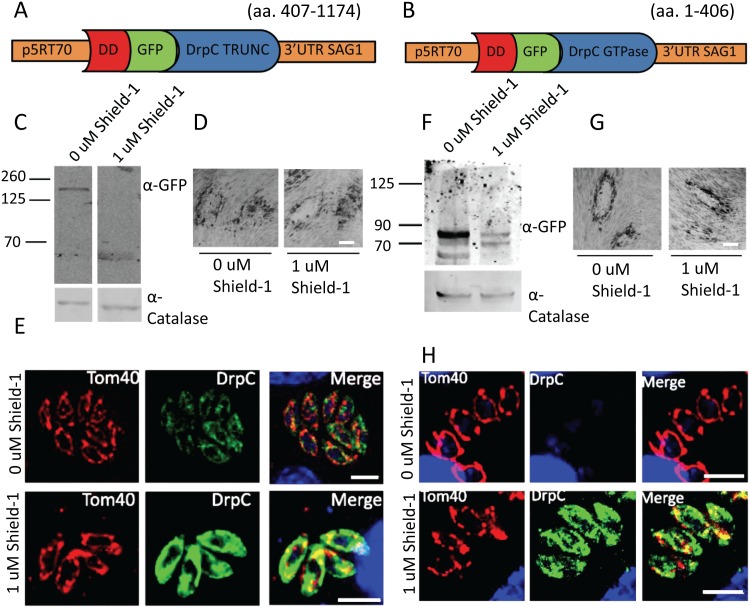
Since our data provides support for TgDrpC involvement in mitochondrial morphogenesis, we were interested to better understand its potential interaction with the mitochondrion. To gain some information about the minimal mitochondrial targeting domain of TgDrpC, we obtained the parasite strains DD-GFP-DrpCtruncated and DD-GFP-DrpCGtpase only, which express only the GTPase or the C-terminus domain of TgDrpC, respectively (Fig 7A and 7B); western blot analysis confirmed inducible expression of the truncated proteins (Fig 7C and 7F). We found that the over-expression of DD-GFP-DrpCtruncated and of DD-GFP-DrpCGtpase only does not impair parasite fitness (Fig 7D and 7G); these truncated versions of TgDrpC do not associate with the mitochondrion, but localise diffusely in the cytosol of the parasite (Fig 7E and 7H). Moreover, our attempts to complement TgDrpC knockdown with these truncated forms of the protein were unsuccessful. Since a dominant negative version of TgDrpC localises correctly to the mitochondrial periphery, we conclude that the full-length protein is required for association with the mitochondria, though no GTPase activity is required.
Fig 7. Truncated TgDrpC does not localise to the mitochondrion.
(A) and (B) Schematics of the plasmids DD-GFP-DrpCtruncated (amino-acids 407–1174) and DD-GFP-DrpCGTPase only (amino-acids 1–406). (C) Western blot analysis shows efficient DD-GFP-DrpCtruncated stabilisation 24 hours after Shield-1 induction. (D) Plaque assay analysis shows that overexpression of DD-GFP-DrpCtruncated does not impair parasite fitness. Parasites were grown for 7 days on HFF cells in presence and absence of 1μM Shield-1. (E) Immunofluorescence analysis shows that DD-GFP-DrpCtruncated localises to the cytoplasm and does not form puncta. (F) Immunoblot analysis of clonal DD-GFP-DrpCGTPase only parasites in presence and absence of Shield-1 using the indicated antibodies. (G) DD-GFP-DrpCGTPase only parasites grown in presence of 1 μM of Shield-1 for 7 days do not show any growth defect compared to non-induced controls. (H) Immunofluorescence analysis shows that DD-GFP-DrpCGTPase only localise to the cytoplasm. Scale bar: 5 μm.
Discussion
Mitochondrial fission is crucial for the control of organelle morphology and function. Here, we investigated the role in T. gondii of conserved components of the fission machinery.
We show for the first time that a dynamin-related protein, termed TgDrpC, is essential for mitochondrial biogenesis; we propose that it plays a role in mitochondrial fission at the end of cytokinesis. TgDrpC localises in puncta, and its recruitment at the basal end of the mitochondrion is evident in the last steps of endodyogeny. It has been suggested that, after the entry of the new mitochondria into the budding daughter cells, the mitochondrial connection between the two branches is cleaved [16]. We show by time-lapse and fixed microscopy that TgDrpC is at the interconnections until they are no longer visible. In addition to its localization, a potential role in fission is supported also by functional studies. Protein ablation through a genetic knock-down and functional inhibition via dominant negative strategy show that in the absence of active TgDrpC the mitochondria of daughter cells remain permanently interconnected. Strikingly, when using a dominant-negative form of TgDrpC, which has an inactive GTPase domain and is tagged with YFP, we can see that the interconnections between mitochondria are decorated with the inactive TgDrpC.
In both strategies, we observe a growth defect, suggesting that inhibition of TgDrpC function is lethal in T. gondii. Similarly, defects in mitochondrial fission are lethal during embryo development in mice and humans [60, 61], and Drp1 has been involved in cancer development, as reviewed in [62]. In contrast, Dnm1 depletion is not lethal in yeast: in absence of fission, the mitochondrial network is only shaped by fusion, and as a result it becomes a long, tubular structure, which can still be transferred to buds during mitosis [17]. Although our analysis suggests that TgDrpC is essential for mitochondrial fission, which subsequently leads to non-specific secondary defects while the parasite dies, we cannot exclude that TgDrpC has additional functions during parasite replication, as suggested in a recent study [63].
In this context, it is worth noting that TgDrpC puncta are not only localised at mitochondria interconnections, but also at its periphery. This is in good accordance with previous observations showing that only a fraction of dynamin-related protein puncta on the MOM participates in mitochondrial division in yeast [64]. It was also observed that a different population of Dnm1 puncta, which is less dynamic than the one involved in mitochondrial fission, co-localises with the mitochondrial-ER-cortex anchor (MECA), a key element for the tethering of mitochondria, but the functional significance of this colocalisation is not yet clear [65, 66]. Previous work in Apicomplexa parasites has shown that the mitochondrion is in close juxtaposition with the IMC and suggested that the two organelles are tethered [5, 67]. It could be speculated that the puncta of TgDrpC at the mitochondrial periphery are likewise involved in the formation of such tethers.
Another component of the fission machinery is the “adaptor complex” that brings the dynamin-related proteins Drp1/Dnm1 to the mitochondrial membrane. The composition of this complex varies in different organisms; in T. gondii, only one of the proteins involved is identifiable by BLAST, the transmembrane protein TgFis1, also conserved in P. falciparum [68]. We show here that this protein is not required for mitochondrial fission and is altogether dispensable for tachyzoite growth in culture. These findings are consistent to what observed in mammals, where Fis1 has only a minor role in mitochondrial fission, while other receptors, most notably Mff, Mid49 and MiD50, are required for recruitment of Drp1 and modulation of its GTPase activity [69]. Homologs of these proteins are not found in the T. gondii genome via BLAST, thus the question of how TgDrpC is recruited to the mitochondrial membrane remains open.
Moreover, research has shown that Drp1/Dnm1 role in mitochondrial fission is facilitated by a “pre-constriction” step, which is mediated by two main actors: ER and actin. The ER wraps around mitochondria in sites of future fission [70] and in those sites actin polymerization occurs, mediated by the ER-localised formin 2 (INF2) and by the mitochondrial protein Spire1C [20, 71]. It is proposed that myosin II is subsequently recruited to these sites to help pre-constrict the mitochondrial tubules [72]. In contrast, no direct role of actin and myosins in mitochondrial fission has been observed in T. gondii, and TgAct1 depletion does not affect mitochondria morphology [73]. Moreover, though ER and mitochondria seem to be strictly associated in T. gondii, we were not able to prove a direct interaction between their membranes during endodyogeny; better tools for the analysis of ER localisation and behaviour are required to further explore the role of the ER in T. gondii mitochondrial fission. It was recently shown that ER-mediated preconstriction is not the only mechanism that induces mitochondrial fission in mammalian cells, as other types of mechanical force can promote the same response [21]; thus, we cannot exclude that in T. gondii the preconstriction step is due to different mechanisms, such as the extension of the daughter bud’s IMC or the action of the basal body during cytokinesis, which have been proposed to be important for the fission of the apicoplast in this parasite [31, 74, 75].
In conclusion, we show here that the Apicomplexa-specific TgDrpC is highly unusual among dynamin-related proteins, has only a conserved GTPase domain and is essential for mitochondrial biogenesis and for parasite survival. It therefore holds great potential as a drug target against apicomplexan parasites: future research on this point will benefit from the recent discovery of a novel class of compounds that directly inhibit Drp1 GTPase activity in vitro [76].
Supporting information
A) Sequence alignment of the N-terminus of TgDrpC (aa 1–465) and the GTPase domain of human dynamin 3 (aa 16–304). (B) Clustal-Omega alignment of the indicated dynamin-related proteins. Comparison with orthologues in the Apicomplexa family shows that other 2 regions (red) are conserved in this group, but they do not correspond to canonical GED or Middle domain. Tg, Toxoplasma gondii; Ta, Theileria annulata; Bb, Babesia bovis; Pv, Plasmodium vivax; Pb, Plasmodium berghei; Cp, Cryptosporidium parvum. Black letters indicate identical and grey letters similar amino acids.
(TIF)
(A) Plaque assay shows that, upon induction of rapamycin, the line TgDrpC-U1 shows a severe growth phenotype leading to collapse of parasites within the PV (upper lane and inset). Parasites were grown for 7 days on HFF cells in presence or absence of 50 nM Rapamycin. The experiment was performed in triplicate; representative images are shown. RH parasites were used as control. Scale bars = 200 μm. (B) Analysis of TgDrpC-U1 after 96 hours of induction with rapamycin. Most vacuoles present “collapsed” mitochondria (αTom40); moreover, some of the bigger vacuoles look misshapen (as shown here with αGap45 staining) and in few cases the apicoplast (αHSP60) is not present in every parasite. Scale bars = 5 μm.
(TIF)
(A) Schematics of the plasmid DD-GFP-DrpCwt. (B) Western blot of clonal DD-GFP-DrpCwt parasites in presence and absence of Shield-1 using the indicated antibodies. (C) Plaque assay for RH and DD-GFP-DrpCwt lines in presence and absence of Shield-1. The experiment was performed in triplicate; representative images are shown. Scale bars = 200 μm. (D) Immunoflorescence analysis showing DD-GFP-DrpCwt signal at 24 hours post-induction with Shield-1. (E) Time lapse analysis of parasites DD-GFP-DrpCwt/HSP60-RFP undergoing endodyogeny. (Scale bar = 5 μm. Red: HSP60-RFP; green: DD-GFP-DrpCwt).
(TIF)
The time lapse follows a vacuole with two parasites that just completed their cytokinesis but still show interconnected mitochondria until the connection is no longer detectable.
(AVI)
Acknowledgments
We are grateful to Elena Jimenez Ruiz, Jana Ovciarikova, Javier Periz, Natalie Tatum and Mariana Serpeloni for their help and useful advice; Gurman Pall and Mattew Gow for their support in the group. We thank Peter Bradley, Dominique Soldati-Favre, Boris Striepen and Giel Van Dooren for sharing of reagents; Volodymyr Nechyporuk-Zloy for support of the imaging facility at the WCMP during the initial phase of this project.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
Markus Meissner is funded by an ERC-Starting grant (ERC-2012-StG 309255-EndoTox) and by a Wellcome Trust 087582/Z/08/Z Senior Fellowship Lilach Sheiner is funded by a Royal society of Edinburg Personal Research Fellowship and by a Biotechnology and Biological Sciences Research Council grant BB/N003675/1 The work also was supported by BBSRC’s grant BB/M024156/1 to Ehmke Pohl. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Melo EJ, Attias M, De Souza W. The single mitochondrion of tachyzoites of Toxoplasma gondii. J Struct Biol. 2000;130(1):27–33. Epub 2000/05/12. 10.1006/jsbi.2000.4228 . [DOI] [PubMed] [Google Scholar]
- 2.de Souza W, Attias M, Rodrigues JC. Particularities of mitochondrial structure in parasitic protists (Apicomplexa and Kinetoplastida). Int J Biochem Cell Biol. 2009;41(10):2069–80. Epub 2009/04/22. 10.1016/j.biocel.2009.04.007 . [DOI] [PubMed] [Google Scholar]
- 3.Sheiner L, Vaidya AB, McFadden GI. The metabolic roles of the endosymbiotic organelles of Toxoplasma and Plasmodium spp. Curr Opin Microbiol. 2013;16(4):452–8. Epub 2013/08/10. 10.1016/j.mib.2013.07.003 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Dooren GG, Stimmler LM, McFadden GI. Metabolic maps and functions of the Plasmodium mitochondrion. FEMS Microbiol Rev. 2006;30(4):596–630. 10.1111/j.1574-6976.2006.00027.x . [DOI] [PubMed] [Google Scholar]
- 5.Ovciarikova J, Lemgruber L, Stilger KL, Sullivan WJ, Sheiner L. Mitochondrial behaviour throughout the lytic cycle of Toxoplasma gondii. Sci Rep. 2017;7:42746 Epub 2017/02/17. 10.1038/srep42746 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jacot D, Waller RF, Soldati-Favre D, MacPherson DA, MacRae JI. Apicomplexan Energy Metabolism: Carbon Source Promiscuity and the Quiescence Hyperbole. Trends Parasitol. 2016;32(1):56–70. Epub 2015/10/17. 10.1016/j.pt.2015.09.001 . [DOI] [PubMed] [Google Scholar]
- 7.Seeber F, Limenitakis J, Soldati-Favre D. Apicomplexan mitochondrial metabolism: a story of gains, losses and retentions. Trends Parasitol. 2008;24(10):468–78. Epub 2008/09/09. 10.1016/j.pt.2008.07.004 . [DOI] [PubMed] [Google Scholar]
- 8.Vaidya AB, Mather MW. Mitochondrial evolution and functions in malaria parasites. Annu Rev Microbiol. 2009;63:249–67. Epub 2009/07/07. 10.1146/annurev.micro.091208.073424 . [DOI] [PubMed] [Google Scholar]
- 9.MacRae JI, Sheiner L, Nahid A, Tonkin C, Striepen B, McConville MJ. Mitochondrial metabolism of glucose and glutamine is required for intracellular growth of Toxoplasma gondii. Cell Host Microbe. 2012;12(5):682–92. Epub 2012/11/20. 10.1016/j.chom.2012.09.013 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mather MW, Vaidya AB. Mitochondria in malaria and related parasites: ancient, diverse and streamlined. J Bioenerg Biomembr. 2008;40(5):425–33. Epub 2008/09/25. 10.1007/s10863-008-9176-4 . [DOI] [PubMed] [Google Scholar]
- 11.Goodman CD, Buchanan HD, McFadden GI. Is the Mitochondrion a Good Malaria Drug Target? Trends in parasitology. 2017;33(3):185–93. 10.1016/j.pt.2016.10.002 [DOI] [PubMed] [Google Scholar]
- 12.Meeusen S, Nunnari J. Evidence for a two membrane-spanning autonomous mitochondrial DNA replisome. J Cell Biol. 2003;163(3):503–10. Epub 2003/11/05. 10.1083/jcb.200304040 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smirnova E, Griparic L, Shurland DL, van der Bliek AM. Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol Biol Cell. 2001;12(8):2245–56. Epub 2001/08/22. 10.1091/mbc.12.8.2245 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van der Bliek AM. A mitochondrial division apparatus takes shape. J Cell Biol. 2000;151(2):F1–4. Epub 2000/10/19. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuroiwa T, Nishida K, Yoshida Y, Fujiwara T, Mori T, Kuroiwa H, et al. Structure, function and evolution of the mitochondrial division apparatus. Biochimica et Biophysica Acta (BBA)—Molecular Cell Research. 2006;1763(5):510–21. 10.1016/j.bbamcr.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 16.Nishi M, Hu K, Murray JM, Roos DS. Organellar dynamics during the cell cycle of Toxoplasma gondii. J Cell Sci. 2008;121(Pt 9):1559–68. Epub 2008/04/16. 10.1242/jcs.021089 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Otsuga D, Keegan BR, Brisch E, Thatcher JW, Hermann GJ, Bleazard W, et al. The dynamin-related GTPase, Dnm1p, controls mitochondrial morphology in yeast. J Cell Biol. 1998;143(2):333–49. Epub 1998/10/24. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smirnova E, Shurland DL, Ryazantsev SN, van der Bliek AM. A human dynamin-related protein controls the distribution of mitochondria. J Cell Biol. 1998;143(2):351–8. Epub 1998/10/24. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arimura S-i, Tsutsumi N. A dynamin-like protein (ADL2b), rather than FtsZ, is involved in Arabidopsis mitochondrial division. Proceedings of the National Academy of Sciences. 2002;99(8):5727–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Korobova F, Ramabhadran V, Higgs HN. An actin-dependent step in mitochondrial fission mediated by the ER-associated formin INF2. Science. 2013;339(6118):464–7. Epub 2013/01/26. 10.1126/science.1228360 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Helle SCJ, Feng Q, Aebersold MJ, Hirt L, Grüter RR, Vahid A, et al. Mechanical force induces mitochondrial fission. eLife. 2017;6:e30292 10.7554/eLife.30292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mozdy AD, McCaffery JM, Shaw JM. Dnm1p GTPase-mediated mitochondrial fission is a multi-step process requiring the novel integral membrane component Fis1p. J Cell Biol. 2000;151(2):367–80. Epub 2000/10/19. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koirala S, Guo Q, Kalia R, Bui HT, Eckert DM, Frost A, et al. Interchangeable adaptors regulate mitochondrial dynamin assembly for membrane scission. Proc Natl Acad Sci U S A. 2013;110(15):E1342–51. Epub 2013/03/27. 10.1073/pnas.1300855110 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Palmer CS, Osellame LD, Laine D, Koutsopoulos OS, Frazier AE, Ryan MT. MiD49 and MiD51, new components of the mitochondrial fission machinery. EMBO Rep. 2011;12(6):565–73. Epub 2011/04/22. 10.1038/embor.2011.54 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gandre-Babbe S, van der Bliek AM. The novel tail-anchored membrane protein Mff controls mitochondrial and peroxisomal fission in mammalian cells. Mol Biol Cell. 2008;19(6):2402–12. Epub 2008/03/21. 10.1091/mbc.E07-12-1287 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Otera H, Wang C, Cleland MM, Setoguchi K, Yokota S, Youle RJ, et al. Mff is an essential factor for mitochondrial recruitment of Drp1 during mitochondrial fission in mammalian cells. J Cell Biol. 2010;191(6):1141–58. Epub 2010/12/15. 10.1083/jcb.201007152 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frohlich C, Grabiger S, Schwefel D, Faelber K, Rosenbaum E, Mears J, et al. Structural insights into oligomerization and mitochondrial remodelling of dynamin 1-like protein. Embo j. 2013;32(9):1280–92. Epub 2013/04/16. 10.1038/emboj.2013.74 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ingerman E, Perkins EM, Marino M, Mears JA, McCaffery JM, Hinshaw JE, et al. Dnm1 forms spirals that are structurally tailored to fit mitochondria. J Cell Biol. 2005;170(7):1021–7. Epub 2005/09/28. 10.1083/jcb.200506078 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mears JA, Lackner LL, Fang S, Ingerman E, Nunnari J, Hinshaw JE. Conformational changes in Dnm1 support a contractile mechanism for mitochondrial fission. Nat Struct Mol Biol. 2011;18(1):20–6. Epub 2010/12/21. 10.1038/nsmb.1949 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee JE, Westrate LM, Wu H, Page C, Voeltz GK. Multiple dynamin family members collaborate to drive mitochondrial division. Nature. 2016;540(7631):139–43. Epub 2016/11/01. 10.1038/nature20555 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Dooren GG, Reiff SB, Tomova C, Meissner M, Humbel BM, Striepen B. A novel dynamin-related protein has been recruited for apicoplast fission in Toxoplasma gondii. Curr Biol. 2009;19(4):267–76. Epub 2009/02/17. 10.1016/j.cub.2008.12.048 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Breinich MS, Ferguson DJ, Foth BJ, van Dooren GG, Lebrun M, Quon DV, et al. A dynamin is required for the biogenesis of secretory organelles in Toxoplasma gondii. Curr Biol. 2009;19(4):277–86. Epub 2009/02/17. 10.1016/j.cub.2009.01.039 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Purkanti R, Thattai M. Ancient dynamin segments capture early stages of host-mitochondrial integration. Proc Natl Acad Sci U S A. 2015;112(9):2800–5. Epub 2015/02/19. 10.1073/pnas.1407163112 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pieperhoff MS, Pall GS, Jimenez-Ruiz E, Das S, Melatti C, Gow M, et al. Conditional U1 Gene Silencing in Toxoplasma gondii. PLoS One. 2015;10(6):e0130356 Epub 2015/06/20. 10.1371/journal.pone.0130356 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roos DS, Donald RG, Morrissette NS, Moulton AL. Molecular tools for genetic dissection of the protozoan parasite Toxoplasma gondii. Methods Cell Biol. 1994;45:27–63. Epub 1994/01/01. . [DOI] [PubMed] [Google Scholar]
- 36.Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42(Web Server issue):W252–8. Epub 2014/05/02. 10.1093/nar/gku340 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bertoni M, Kiefer F, Biasini M, Bordoli L, Schwede T. Modeling protein quaternary structure of homo- and hetero-oligomers beyond binary interactions by homology. Sci Rep. 2017;7(1):10480 Epub 2017/09/07. 10.1038/s41598-017-09654-8 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berendsen HJC vdS D.; van Drunen R. GROMACS: A message-passing parallel molecular dynamics implementation. Computer Physics Communications. 1995;91(1–3): 43–56. 10.1016/0010-4655(95)00042-e [DOI] [Google Scholar]
- 39.Schmid N, Eichenberger AP, Choutko A, Riniker S, Winger M, Mark AE, et al. Definition and testing of the GROMOS force-field versions 54A7 and 54B7. Eur Biophys J. 2011;40(7):843–56. Epub 2011/05/03. 10.1007/s00249-011-0700-9 . [DOI] [PubMed] [Google Scholar]
- 40.DeLano WL. Pymol: An open-source molecular graphics tool. CCP4 Newsletter On Protein Crystallography. 2002;40:82–92. [Google Scholar]
- 41.Salamun J, Kallio JP, Daher W, Soldati-Favre D, Kursula I. Structure of Toxoplasma gondii coronin, an actin-binding protein that relocalizes to the posterior pole of invasive parasites and contributes to invasion and egress. Faseb j. 2014;28(11):4729–47. Epub 2014/08/13. 10.1096/fj.14-252569 . [DOI] [PubMed] [Google Scholar]
- 42.Sheiner L, Demerly JL, Poulsen N, Beatty WL, Lucas O, Behnke MS, et al. A systematic screen to discover and analyze apicoplast proteins identifies a conserved and essential protein import factor. PLoS Pathog. 2011;7(12):e1002392 Epub 2011/12/07. 10.1371/journal.ppat.1002392 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huynh MH, Carruthers VB. Tagging of endogenous genes in a Toxoplasma gondii strain lacking Ku80. Eukaryot Cell. 2009;8(4):530–9. Epub 2009/02/17. 10.1128/EC.00358-08 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fox BA, Ristuccia JG, Gigley JP, Bzik DJ. Efficient gene replacements in Toxoplasma gondii strains deficient for nonhomologous end joining. Eukaryot Cell. 2009;8(4):520–9. Epub 2009/02/17. 10.1128/EC.00357-08 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hettmann C, Herm A, Geiter A, Frank B, Schwarz E, Soldati T, et al. A dibasic motif in the tail of a class XIV apicomplexan myosin is an essential determinant of plasma membrane localization. Mol Biol Cell. 2000;11(4):1385–400. Epub 2000/04/06. 10.1091/mbc.11.4.1385 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bolte S, Cordelieres FP. A guided tour into subcellular colocalization analysis in light microscopy. Journal of microscopy. 2006;224(Pt 3):213–32. Epub 2007/01/11. 10.1111/j.1365-2818.2006.01706.x . [DOI] [PubMed] [Google Scholar]
- 47.Harding CR, Egarter S, Gow M, Jimenez-Ruiz E, Ferguson DJ, Meissner M. Gliding Associated Proteins Play Essential Roles during the Formation of the Inner Membrane Complex of Toxoplasma gondii. PLoS Pathog. 2016;12(2):e1005403 Epub 2016/02/06. 10.1371/journal.ppat.1005403 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Padgett LR, Arrizabalaga G, Sullivan WJ Jr,. Targeting of tail-anchored membrane proteins to subcellular organelles in Toxoplasma gondii. Traffic. 2017;18(3):149–58. Epub 2016/12/20. 10.1111/tra.12464 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meissner M, Schluter D, Soldati D. Role of Toxoplasma gondii myosin A in powering parasite gliding and host cell invasion. Science. 2002;298(5594):837–40. Epub 2002/10/26. 10.1126/science.1074553 . [DOI] [PubMed] [Google Scholar]
- 50.van Dooren GG, Yeoh LM, Striepen B, McFadden GI. The Import of Proteins into the Mitochondrion of Toxoplasma gondii. J Biol Chem. 2016. Epub 2016/07/28. 10.1074/jbc.M116.725069 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sidik SM, Huet D, Ganesan SM, Huynh MH, Wang T, Nasamu AS, et al. A Genome-wide CRISPR Screen in Toxoplasma Identifies Essential Apicomplexan Genes. Cell. 2016;166(6):1423–35.e12. Epub 2016/09/07. 10.1016/j.cell.2016.08.019 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, et al. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421 Epub 2009/12/17. 10.1186/1471-2105-10-421 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Carvalho AT, Szeler K, Vavitsas K, Aqvist J, Kamerlin SC. Modeling the mechanisms of biological GTP hydrolysis. Arch Biochem Biophys. 2015;582:80–90. Epub 2015/03/04. 10.1016/j.abb.2015.02.027 . [DOI] [PubMed] [Google Scholar]
- 54.Bleazard W, McCaffery JM, King EJ, Bale S, Mozdy A, Tieu Q, et al. The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat Cell Biol. 1999;1(5):298–304. Epub 1999/11/13. 10.1038/13014 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rosenbloom AB, Lee SH, To M, Lee A, Shin JY, Bustamante C. Optimized two-color super resolution imaging of Drp1 during mitochondrial fission with a slow-switching Dronpa variant. Proc Natl Acad Sci U S A. 2014;111(36):13093–8. 10.1073/pnas.1320044111 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pelletier L, Stern CA, Pypaert M, Sheff D, Ngo HM, Roper N, et al. Golgi biogenesis in Toxoplasma gondii. Nature. 2002;418(6897):548–52. Epub 2002/08/02. 10.1038/nature00946 . [DOI] [PubMed] [Google Scholar]
- 57.Herm-Gotz A, Agop-Nersesian C, Munter S, Grimley JS, Wandless TJ, Frischknecht F, et al. Rapid control of protein level in the apicomplexan Toxoplasma gondii. Nat Methods. 2007;4(12):1003–5. Epub 2007/11/13. 10.1038/nmeth1134 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van der Bliek AM, Redelmeier TE, Damke H, Tisdale EJ, Meyerowitz EM, Schmid SL. Mutations in human dynamin block an intermediate stage in coated vesicle formation. J Cell Biol. 1993;122(3):553–63. Epub 1993/08/01. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Herskovits JS, Burgess CC, Obar RA, Vallee RB. Effects of mutant rat dynamin on endocytosis. J Cell Biol. 1993;122(3):565–78. Epub 1993/08/01. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ishihara N, Nomura M, Jofuku A, Kato H, Suzuki SO, Masuda K, et al. Mitochondrial fission factor Drp1 is essential for embryonic development and synapse formation in mice. Nat Cell Biol. 2009;11(8):958–66. Epub 2009/07/07. 10.1038/ncb1907 . [DOI] [PubMed] [Google Scholar]
- 61.Waterham HR, Koster J, van Roermund CW, Mooyer PA, Wanders RJ, Leonard JV. A lethal defect of mitochondrial and peroxisomal fission. N Engl J Med. 2007;356(17):1736–41. Epub 2007/04/27. 10.1056/NEJMoa064436 . [DOI] [PubMed] [Google Scholar]
- 62.Lima AR, Santos L, Correia M, Soares P, Sobrinho-Simoes M, Melo M, et al. Dynamin-Related Protein 1 at the Crossroads of Cancer. Genes (Basel). 2018;9(2). Epub 2018/02/22. 10.3390/genes9020115 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Heredero-Bermejo I, Varberg JM, Charvat R, Jacobs K, Garbuz T, Sullivan WJ Jr., et al. TgDrpC, an atypical dynamin-related protein in Toxoplasma gondii, is associated with vesicular transport factors and parasite division. Mol Microbiol. 2018. Epub 2018/10/27. 10.1111/mmi.14138 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schauss AC, Bewersdorf J, Jakobs S. Fis1p and Caf4p, but not Mdv1p, determine the polar localization of Dnm1p clusters on the mitochondrial surface. J Cell Sci. 2006;119(Pt 15):3098–106. Epub 2006/07/13. 10.1242/jcs.03026 . [DOI] [PubMed] [Google Scholar]
- 65.Cerveny KL, Studer SL, Jensen RE, Sesaki H. Yeast mitochondrial division and distribution require the cortical num1 protein. Dev Cell. 2007;12(3):363–75. Epub 2007/03/06. 10.1016/j.devcel.2007.01.017 . [DOI] [PubMed] [Google Scholar]
- 66.Lackner LL, Ping H, Graef M, Murley A, Nunnari J. Endoplasmic reticulum-associated mitochondria-cortex tether functions in the distribution and inheritance of mitochondria. Proc Natl Acad Sci U S A. 2013;110(6):E458–67. Epub 2013/01/24. 10.1073/pnas.1215232110 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kudryashev M, Lepper S, Stanway R, Bohn S, Baumeister W, Cyrklaff M, et al. Positioning of large organelles by a membrane-associated cytoskeleton in Plasmodium sporozoites. Cellular microbiology. 2010;12(3):362–71. 10.1111/j.1462-5822.2009.01399.x [DOI] [PubMed] [Google Scholar]
- 68.Scarpelli P, Almeida GT, Vicoso KL, Lima WR, Pereira LB, Meissner KA, et al. Melatonin activate FIS1, DYN1 and DYN2 Plasmodium falciparum related-genes for mitochondria fission: mitoemerald-GFP as a tool to visualize mitochondria structure. J Pineal Res. 2018. Epub 2018/02/27. 10.1111/jpi.12484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pagliuso A, Cossart P, Stavru F. The ever-growing complexity of the mitochondrial fission machinery. Cell Mol Life Sci. 2018;75(3):355–74. Epub 2017/08/06. 10.1007/s00018-017-2603-0 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Friedman JR, Lackner LL, West M, DiBenedetto JR, Nunnari J, Voeltz GK. ER tubules mark sites of mitochondrial division. Science. 2011;334(6054):358–62. Epub 2011/09/03. 10.1126/science.1207385 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Manor U, Bartholomew S, Golani G, Christenson E, Kozlov M, Higgs H, et al. A mitochondria-anchored isoform of the actin-nucleating spire protein regulates mitochondrial division. Elife. 2015;4 Epub 2015/08/26. 10.7554/eLife.08828 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Korobova F, Gauvin TJ, Higgs HN. A role for myosin II in mammalian mitochondrial fission. Curr Biol. 2014;24(4):409–14. Epub 2014/02/04. 10.1016/j.cub.2013.12.032 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Periz J, Whitelaw J, Harding C, Gras S, Del Rosario Minina MI, Latorre-Barragan F, et al. Toxoplasma gondii F-actin forms an extensive filamentous network required for material exchange and parasite maturation. Elife. 2017;6 Epub 2017/03/23. 10.7554/eLife.24119 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Heaslip AT, Dzierszinski F, Stein B, Hu K. TgMORN1 is a key organizer for the basal complex of Toxoplasma gondii. PLoS Pathog. 2010;6(2):e1000754 Epub 2010/02/09. 10.1371/journal.ppat.1000754 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lorestani A, Sheiner L, Yang K, Robertson SD, Sahoo N, Brooks CF, et al. A Toxoplasma MORN1 null mutant undergoes repeated divisions but is defective in basal assembly, apicoplast division and cytokinesis. PLoS One. 2010;5(8):e12302 Epub 2010/09/03. 10.1371/journal.pone.0012302 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mallat A, Uchiyama LF, Lewis SC, Fredenburg RA, Terada Y, Ji N, et al. Discovery and characterization of selective small molecule inhibitors of the mammalian mitochondrial division dynamin, DRP1. Biochem Biophys Res Commun. 2018;499(3):556–62. Epub 2018/03/31. 10.1016/j.bbrc.2018.03.189 . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A) Sequence alignment of the N-terminus of TgDrpC (aa 1–465) and the GTPase domain of human dynamin 3 (aa 16–304). (B) Clustal-Omega alignment of the indicated dynamin-related proteins. Comparison with orthologues in the Apicomplexa family shows that other 2 regions (red) are conserved in this group, but they do not correspond to canonical GED or Middle domain. Tg, Toxoplasma gondii; Ta, Theileria annulata; Bb, Babesia bovis; Pv, Plasmodium vivax; Pb, Plasmodium berghei; Cp, Cryptosporidium parvum. Black letters indicate identical and grey letters similar amino acids.
(TIF)
(A) Plaque assay shows that, upon induction of rapamycin, the line TgDrpC-U1 shows a severe growth phenotype leading to collapse of parasites within the PV (upper lane and inset). Parasites were grown for 7 days on HFF cells in presence or absence of 50 nM Rapamycin. The experiment was performed in triplicate; representative images are shown. RH parasites were used as control. Scale bars = 200 μm. (B) Analysis of TgDrpC-U1 after 96 hours of induction with rapamycin. Most vacuoles present “collapsed” mitochondria (αTom40); moreover, some of the bigger vacuoles look misshapen (as shown here with αGap45 staining) and in few cases the apicoplast (αHSP60) is not present in every parasite. Scale bars = 5 μm.
(TIF)
(A) Schematics of the plasmid DD-GFP-DrpCwt. (B) Western blot of clonal DD-GFP-DrpCwt parasites in presence and absence of Shield-1 using the indicated antibodies. (C) Plaque assay for RH and DD-GFP-DrpCwt lines in presence and absence of Shield-1. The experiment was performed in triplicate; representative images are shown. Scale bars = 200 μm. (D) Immunoflorescence analysis showing DD-GFP-DrpCwt signal at 24 hours post-induction with Shield-1. (E) Time lapse analysis of parasites DD-GFP-DrpCwt/HSP60-RFP undergoing endodyogeny. (Scale bar = 5 μm. Red: HSP60-RFP; green: DD-GFP-DrpCwt).
(TIF)
The time lapse follows a vacuole with two parasites that just completed their cytokinesis but still show interconnected mitochondria until the connection is no longer detectable.
(AVI)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.