Abstract
At the molecular level, the evolution of new traits can be broadly divided between changes in gene expression and changes in protein-coding sequence. For proteins, the evolution of novel functions is generally thought to proceed through sequential point mutations or recombination of whole functional units. In Saccharomyces, the uptake of the sugar maltotriose into the cell is the primary limiting factor in its utilization, but maltotriose transporters are relatively rare, except in brewing strains. No known wild strains of Saccharomyces eubayanus, the cold-tolerant parent of hybrid lager-brewing yeasts (Saccharomyces cerevisiae x S. eubayanus), are able to consume maltotriose, which limits their ability to fully ferment malt extract. In one strain of S. eubayanus, we found a gene closely related to a known maltotriose transporter and were able to confer maltotriose consumption by overexpressing this gene or by passaging the strain on maltose. Even so, most wild strains of S. eubayanus lack native maltotriose transporters. To determine how this rare trait could evolve in naive genetic backgrounds, we performed an adaptive evolution experiment for maltotriose consumption, which yielded a single strain of S. eubayanus able to grow on maltotriose. We mapped the causative locus to a gene encoding a novel chimeric transporter that was formed by an ectopic recombination event between two genes encoding transporters that are unable to import maltotriose. In contrast to classic models of the evolution of novel protein functions, the recombination breakpoints occurred within a single functional domain. Thus, the ability of the new protein to carry maltotriose was likely acquired through epistatic interactions between independently evolved substitutions. By acquiring multiple mutations at once, the transporter rapidly gained a novel function, while bypassing potentially deleterious intermediate steps. This study provides an illuminating example of how recombination between paralogs can establish novel interactions among substitutions to create adaptive functions.
Author summary
Hybrids of the yeasts Saccharomyces cerevisiae and Saccharomyces eubayanus (lager-brewing yeasts) dominate the modern brewing industry. S. cerevisiae, also known as baker’s yeast, is well-known for its role in industry and scientific research. Less well recognized is S. eubayanus, which was only discovered as a pure species in 2011. While most lager-brewing yeasts rapidly and completely utilize the important brewing sugar maltotriose, no strain of S. eubayanus isolated to date is known to do so. Despite being unable to consume maltotriose, we identified one strain of S. eubayanus carrying a gene for a functional maltotriose transporter, although most strains lack this gene. During an adaptive evolution experiment, a strain of S. eubayanus without native maltotriose transporters evolved the ability to grow on maltotriose. Maltotriose consumption in the evolved strain resulted from a chimeric transporter that arose by shuffling genes encoding parent proteins that were unable to transport maltotriose. Traditionally, functional chimeric proteins are thought to evolve by shuffling discrete functional domains or modules, but the breakpoints in the chimera studied here occurred within the single functional module of the protein. These results support the less well-recognized role of shuffling duplicate gene sequences to generate novel proteins with adaptive functions.
Introduction
Proteins with novel functions can arise through a variety of mechanisms [1]. One of the best studied mechanisms is gene duplication, followed by divergence through sequential point mutations [1,2]. While this method of new protein evolution is thought to be common, evolution through stepwise point mutations can be a slow and constrained process [3]. In the mutational landscape separating the original protein from the derived protein, deleterious epistatic interactions, where multiple intermediate mutational steps interact to create fitness valleys, can make new functions difficult to access by successive point mutations. Mutational events that result in multiple amino acid changes at once can help bridge fitness valleys and speed the evolution of new functionality [3,4]. As a consequence of bypassing intermediate mutational steps, recombination can lead to intragenic reciprocal sign epistasis, where the new recombinant protein has a function not found in either parent protein [3,5]. Ectopic gene conversion, which results in chimeric protein sequences, is one such rare class of mutational events that can rapidly lead to new protein sequences with novel functions [3].
Chimeric protein-coding sequences have been found to be an important mechanism by which proteins can evolve new functions [1,6–8]. They have been implicated in the rapid radiation of multicellular animals [6] and in playing a role in both infectious and non-infectious diseases in humans [9–12]. The Drosophila gene jingwei was one of the first chimeric genes to have both its recent origin and evolution characterized in depth [1]. jingwei exemplifies many of the characteristics usually associated with chimeric proteins [1,6–8,13]. Like most other chimeric proteins that have been described in eukaryotes, jingwei is a large multidomain protein that was constructed via the movement of whole functional units (domains), facilitated by intronic sequences, a process referred to as domain or exon shuffling. In most cases, even in the absence of intronic sequences, the recombination of these modules has been considered key to the evolution of functional chimeric proteins [1,14].
The exchange of complete, independently functional units is not the only method by which functional chimeric proteins can be generated. Recombination within functional domains also has the potential to create proteins with novel characteristics. Recombination breakpoints within domains can lead to functional proteins, even between non-homologous protein sequences [4,15,16]. However, since functionally important structures are likely to be conserved between related proteins, the probability of recombination resulting in a functional protein is higher between homologous sequences where essential within-protein interactions are less likely to be disrupted [4,15,17]. Theoretical work has suggested the potential of this sort of recombination to allow proteins to rapidly bypass fitness minima in the adaptive landscape separating two protein functions [3,4]. Recombination between paralogous sequences has also been shown to be selected for in natural populations, suggesting that such sequences can indeed produce functional proteins [15,18]. In addition, recombination between paralogous sequences (DNA shuffling) has been used as an efficient way to engineer proteins with functions that are rare or difficult to evolve in natural settings (reviewed in [19–21]). For example, hexose sugar transporters in Saccharomyces cerevisiae were evolved for increased specificity to a pentose sugar, D-xylose, through DNA shuffling and selection for the ability to support growth on xylose [22].
Maltotriose, a trimer of glucose molecules, is the second most abundant fermentable sugar present in brewing wort (malt extract), but it is also the most difficult to ferment [23–26]. Among budding yeasts of the genus Saccharomyces, such as S. cerevisiae, proteins that can transport maltotriose into the cell are relatively rare [27–31]. Improving consumption of maltotriose by Saccharomyces yeasts is of general interest to the brewing community since a key consideration for any new brewing strain is its ability to rapidly and completely ferment all the sugars present in wort. Work on improving the direct uptake of maltotriose in brewing yeasts has focused on the expression of the limited set of known maltotriose transporters, either through adaptive evolution for increased expression [32,33], introducing maltotriose transporters into new strains through selective breeding [34–42], or by heterologous expression [33,40,43]. These methods all rely on the presence of functional maltotriose transporters, either natively or heterologously expressed, and are limited by the number of strains and proteins that are known to be capable of transporting maltotriose. With the focus on known transporters, how new maltotriose transporters evolve is less well studied [44,45].
Recently, special interest has been given to the development of Saccharomyces eubayanus, a distant cold-tolerant relative of S. cerevisiae, for commercial brewing [35,36,46,47]. As a hybrid with S. cerevisiae, S. eubayanus forms the industrially important lager-brewing yeasts [48], which account for more than 90% of the total beer market. So far, no strain of S. eubayanus isolated from nature has been reported to consume maltotriose [36,49–53], despite evidence for the possible presence of functional transporters in the S. eubayanus subgenome of industrial S. cerevisiae x S. eubayanus hybrids (i.e. lager-brewing yeasts) [28,54–58].
In the present study, we characterize the native MALT genes found in the taxonomic type strain of S. eubayanus for their ability to enable the transport of maltotriose. In another strain of S. eubayanus, we confirm the presence of a functional maltotriose transporter, despite that strain’s inability to consume maltotriose. We also describe a novel chimeric maltotriose transporter that resulted from the adaptive evolution of S. eubayanus for maltotriose consumption. This new maltotriose transporter was formed through a partial ectopic gene conversion event between two MALT genes. Interestingly, the parent proteins that produced the chimera were unable to transport maltotriose themselves. In addition, the breakpoints of the chimeric region do not demarcate clearly defined functional domains, suggesting that epistatic interactions between novel residue combinations, rather than domain swapping, is responsible for the new function. Overall, this study reports the first known maltotriose transporters in S. eubayanus and the first strains of this species that are able to consume maltotriose. In addition, by characterizing one of the few chimeric proteins that have been described with a novel function where recombination occurred naturally within functional modules (without being specifically targeted for engineering by DNA shuffling or mutagenesis) [59], we provide insight into how proteins can evolve novel adaptive functions through rare genetic events.
Results/Discussion
Maltotriose transporters in S. eubayanus
In a monosporic derivative of the taxonomic type strain of S. eubayanus from Patagonia (FM1318 or Pat-Seub), four genes, designated MALT1-4, have been identified as having homology to genes encoding known maltose transporters (MALT genes) [54,60]. Because MALT2 and MALT4 are predicted to encode identical amino acid sequences (see Materials and methods), we refer to these genes jointly as MALT2/4. To determine if they could enable maltotriose transport, Malt1, Malt2/4, and Malt3 were individually overexpressed using an inducible promoter in yHRVM108 (NC-Seub), a strain of S. eubayanus isolated from North Carolina that is unable to grow on maltotriose and, unlike other strains of S. eubayanus, grows sluggishly on maltose. None of these genes were able to confer growth on maltotriose when overexpressed (Table 1). These results are consistent with a recent report on the inability of these proteins to transport maltotriose [61].
Table 1. Heterologous expression of S. eubayanus MALT genes.
| Strain | Background | Transporter | Initial OD | Day 3 | Day 6 |
|---|---|---|---|---|---|
| yHRVM108* | North Carolinian strain | - | 0.16 (+/-0.05) | 0.39 (+/-0.02) | 0.48 (+/-0.01) |
| yHRVM108 | North Carolinian strain | - | 0.12 (+/-0.03) | 0.47 (+/-0.00) | 0.46 (+/-0.03) |
| yHEB1870 | yHRVM108 | MALT1 | 0.13 (+/-0.03) | 0.43 (+/-0.04) | 0.58 (+/-0.04) |
| yHEB1877 | yHRVM108 | MALT2/4 | 0.11 (+/-0.00) | 0.39 (+/-0.01) | 0.57 (+/-0.02) |
| yHEB1872 | yHRVM108 | MALT3 | 0.13 (+/-0.01) | 0.41 (+/-0.00) | 0.62 (+/-0.5) |
| yHEB1883 | yHRVM108 | ncAGT1 | 0.11 (+/-0.01) | 0.54 (+/-0.07) | 1.34 (+/-0.10) |
| yHEB1884 | yHRVM108 | lgAGT1 | 0.10 (+/-0.00) | 0.42 (+/-0.07) | 0.94 (+/-0.09) |
Growth on SC + 2% maltotriose (98% pure) of strains expressing MALT genes on a doxycycline-inducible plasmid, rather than driven by their native promoters. N = 3, standard deviation in parentheses.
* Control grown in SC + 0.04% glucose + doxycycline to reflect the approximate amount of growth expected from contamination with other carbon sources when using 98% pure maltotriose.
Although none of the transporters found in Pat-Seub were able to support growth on maltotriose, there is compelling evidence from lager-brewing yeasts for the existence of maltotriose transporters within the greater S. eubayanus population [28,54–57]. Of particular interest are alleles of AGT1. Two versions of AGT1 are present in the genomes of lager-brewing yeasts. One, which we call scAGT1 (S. cerevisiae-AGT1), was donated by the S. cerevisiae parent of lager yeasts, and the other, which we call lgAGT1 (lager-AGT1), has been proposed to be of S. eubayanus origin [55]. Both lgAGT1 and scAGT1, like other AGT1 alleles, can transport both maltose and maltotriose [27,28,57,62–65]. Thus far, full-length sequences closely related to this lgAGT1 have not been described in any strain of S. eubayanus [36].
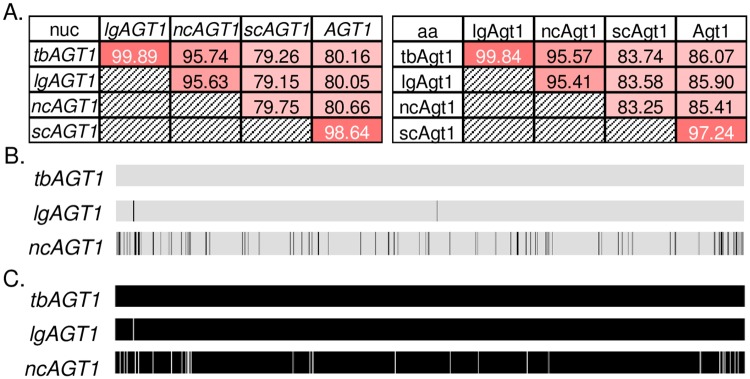
Strain CDFM21L.1 (Tb-Seub) isolated from Tibet and the closely related strain NC-Seub belong to the Holarctic subpopulation of S. eubayanus. They are close relatives of the strain(s) of S. eubayanus that hybridized with S. cerevisiae to form lager-brewing yeasts [52]. Because of their close phylogenetic relationship, Tb-Seub, NC-Seub, and the inferred S. eubayanus lager parent(s) are more likely to share strain-specific genes, such as lgAGT1 [66]. From a search of Illumina sequencing reads available for Tb-Seub and NC-Seub, we were able to assemble two full-length genes with high sequence identity to lgAGT1, which we designated tbAGT1 and ncAGT1, for Tibetan-AGT1 and North Carolinian-AGT1, respectively (Fig 1).
Fig 1. Alignment of AGT1-like genes.
A) Tables highlighting the nucleotide (nuc) and amino acid (aa) percent identities between members of the AGT1 family. Darker colors indicate greater sequence similarity. B) Multiple sequence alignment between nucleotide sequences of tbAGT1, lgAGT1, and ncAGT1. Black lines indicate nucleotide differences. C) Multiple sequence alignments between protein sequences of tbAGT1, lgAGT1, and ncAGT1. White gaps indicate amino acid differences.
Two single nucleotide polymorphisms (SNPs) separate tbAGT1 and lgAGT1. One SNP results in a synonymous substitution and the other in a nonsynonymous substitution near the N-terminus of the protein outside of any predicted transmembrane domains (Fig 1B and 1C, S1 Fig). Analyses of the predicted effect of this substitution in lgAGT1 (using STRUM and SIFT mutant protein prediction software [67,68]) suggest that it is unlikely to significantly impact protein structure or function (S1 Table). In contrast, ncAGT1 has 95% nucleotide identity with lgAGT1, with nonsynonymous differences distributed throughout the sequence (Fig 1A–1C). Despite the presence of ncAGT1, the NC-Seub wild-type strain grows poorly on maltose and is unable to grow on maltotriose.
Phylogenetic relationships among maltose and maltotriose transporters
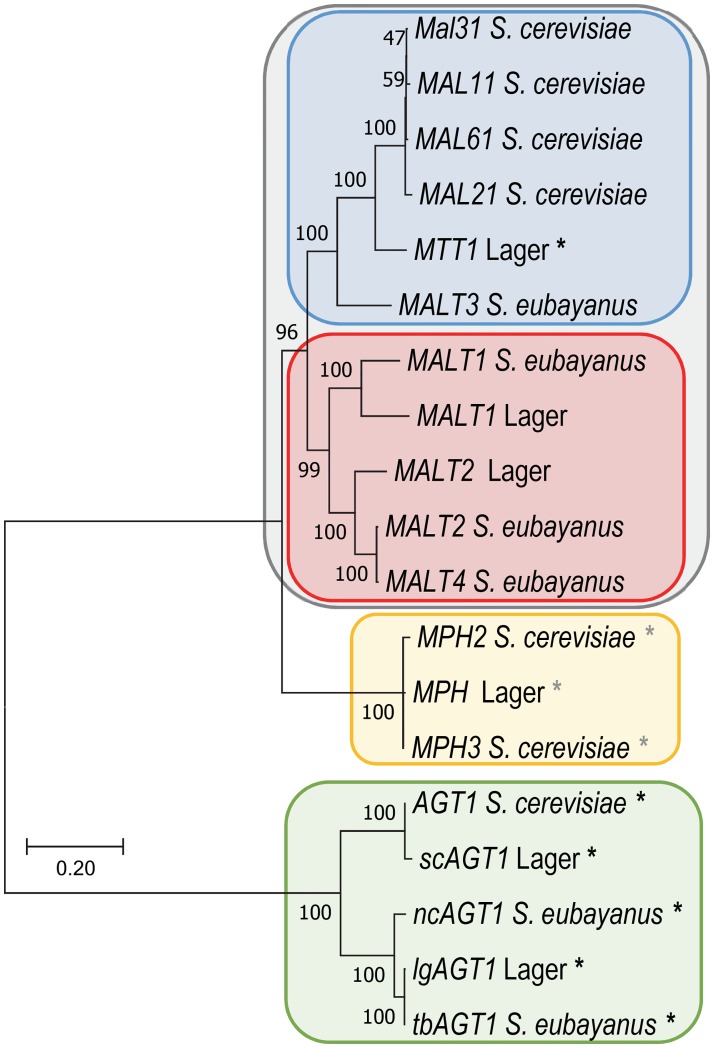
To put the relationship between S. eubayanus, S. cerevisiae, and lager MALT genes into a phylogenetic perspective, a gene tree was constructed for these three groups of genes (Fig 2). Consistent with previous analyses of MALT genes in Saccharomyces [30], the MALT genes fell into 3 major clades. The AGT1 genes formed their own group, significantly divergent from the other clades and was further split between the AGT1 genes originating from S. cerevisiae and the AGT1 genes originating from S. eubayanus. MPH genes, which are native to S. cerevisiae but also present in some lager yeasts [65,69], also formed their own clade. MPH genes are most often described as encoding maltose transporters, but their ability to transport maltotriose is ambiguous [30,65,69–71]. The final and largest clade was made up of MALT1-4 from S. eubayanus, MALx1 genes from S. cerevisiae, and the lager-specific gene MTT1 (MTY1) [28,29,54]. This clade was further subdivided into a group containing only S. eubayanus MALT genes and their close lager homologs and another group consisting of MALx1 genes, MTT1, and MALT3. Within this clade, genes encoding maltotriose transporters were rare, represented by only a single gene, MTT1 [28,29]. The phylogenetic distribution of maltotriose utilization suggests that the ability to transport maltotriose may be a difficult function for genes within this clade to evolve.
Fig 2. Phylogeny of Saccharomyces MALT genes.
ML phylogenetic tree of MALT genes described in S. cerevisiae, S. eubayanus, and lager-brewing hybrids. The scale bar equals the number of nucleotide substitutions per site. Black “*” indicate genes characterized as encoding proteins capable of transporting maltotriose. Gray “*” indicates genes encoding transporters whose ability to transport maltotriose is ambiguous.
Indirect selection for maltotriose utilization
Since NC-Seub contains a closely related homolog of a known maltotriose transporter, we anticipated that it would be simple for NC-Seub to evolve the ability to utilize maltotriose under direct selection for this trait. Because NC-Seub is unable to grow on maltotriose, we passaged the strain in 2% maltotriose medium with a small amount of added glucose to permit a limited number of cell divisions to allow mutation and selection to occur. Over the course of 100 passages under this selection regime, representing around 1,050 cell divisions across three experimental replicates, no maltotriose-utilizing lineage of NC-Seub arose.
While evolving NC-Seub directly for maltotriose consumption was not successful, we found that an alternative, indirect selection regime was effective at evolving maltotriose utilization in this background. Concurrent with the maltotriose selection regime, we also began selecting for increased growth of NC-Seub on maltose to improve this strain’s sluggish growth on this carbon source. All three replicates of this experiment eventually evolved the ability to grow rapidly on maltose. Interestingly, in addition to growing on maltose four times more rapidly over two days (S2 Table, S1 Dataset), single-colony isolates from the first two replicates that evolved rapid maltose utilization also gained the ability to utilize maltotriose (S3 Table, S2 Dataset), despite never being exposed to maltotriose during the course of the adaptive evolution experiment. Based on the presence of ncAGT1 in the genome of NC-Seub, we hypothesized that increased expression of this gene could have provided maltotriose utilization in the evolved strain. We confirmed that overexpression of ncAGT1 in NC-Seub conferred growth on maltotriose, similar to the known maltotriose transporter gene lgAGT1 (Table 1).
Though we found the difficulty of directly selecting for expression of a functional transporter surprising, such a result is not unprecedented. In a long-term evolution experiment in Escherichia coli, a functioning citrate transporter was present in the founding strain. Though expression of this gene would have been highly favored in the citrate-rich experimental environment, it took thousands of generations, even after the necessary potentiating mutations had appeared, before a gene amplification and rearrangement event joined the citrate transporter gene to a new promoter, resulting in a novel expression pattern [72]. These results show how an organism’s preexisting genetic architecture, interacting with the selective environment, can facilitate or impede evolution along a particular path [73–76]. Understanding why direct selection for maltotriose consumption may have impeded the evolution of this trait, compared to selection on maltose, will require further dissection of the genetic and molecular basis of the concomitant increase in maltose and maltotriose utilization by the evolved strains of NC-Seub.
Evolution of maltotriose utilization through a chimeric transporter by direct selection
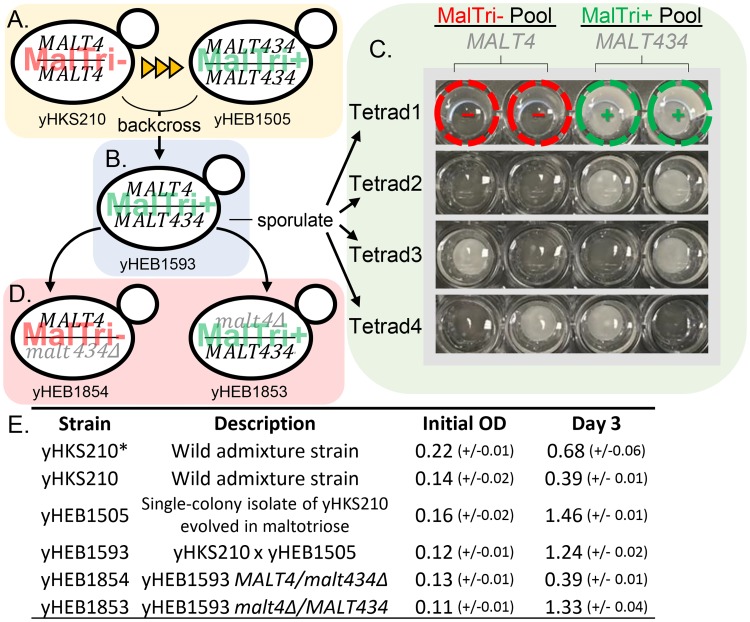
To determine how strains lacking any maltotriose transporters could evolve them de novo, we experimentally evolved Pat-Seub [48] and yHKS210 (WI-Seub) [53] for maltotriose utilization. Unlike NC-Seub, previous reports have confirmed that the taxonomic type strain of S. eubayanus, from which Pat-Seub is derived, is able to utilize maltose [36,48,49,58,77], and we determined that WI-Seub can robustly grow on maltose as well (S3 Table, S2 Dataset). However, like NC-Seub, neither Pat-Seub nor WI-Seub could grow on maltotriose. A search of the available genome sequence reads for Pat-Seub [54,60] and WI-Seub [52] confirmed that neither of these strains contain sequences that are closely related to AGT1-like genes or other known maltotriose transporters [28,29]. Based on our analysis of the available whole-genome sequencing data, these strains only contain the four MALT transporter genes previously identified in Pat-Seub [54], which are unable to confer maltotriose utilization even when overexpressed (Table 1) [61]. Since neither strain could grow on maltotriose, a small amount of glucose was also added to the medium to permit a limited number of cell divisions for mutation and selection. Over the course of 100 passages, representing approximately 2,100 cell divisions in total between the two strains and their replicates, a single replicate, derived from WI-Seub, evolved the ability to grow on maltotriose. Two single-colony isolates (yHEB1505-6) from this replicate were isolated and confirmed to be able to grow on maltotriose without added glucose (Fig 3A, S3 Table, S2 Dataset).
Fig 3. Evolution and validation of the chimeric maltotriose transporter Malt434.
A) After continuous culturing on maltotriose with a small amount of added glucose, WI-Seub (yHKS210), which was originally unable to use maltotriose (MalTri-), evolved the ability to consume maltotriose (MalTri+). B) Strain yHEB1593, which is a backcross between yHKS210 and yHEB1505, was also MalTri+. C) To test the inheritance of maltotriose utilization, yHEB1593 was sporulated. The panel shows a subset of tetrads screened growing on SC + 2% maltotriose. Examples of MalTri- spores in Tetrad 1 are circled in red, and MalTri+ examples are circled in green. Whole genome sequencing of MalTri+ and MalTri- pools showed that maltotriose utilization perfectly correlated with the presence/absence of MALT434. D) Reciprocal hemizygosity test [81] of the MALT4/MALT434 locus in the backcross strain yHEB1593. E) Table of initial and day-three OD600 (OD) readings of yHKS210, yHEB1505, yHEB1593, yHEB1853, and yHEB1854 on SC + 2% maltotriose as the sole carbon source. N = 3, standard deviation in parentheses. * Control grown in SC + 0.04% glucose to reflect the approximate amount of growth expected from contamination with other carbon sources when using 98% pure maltotriose.
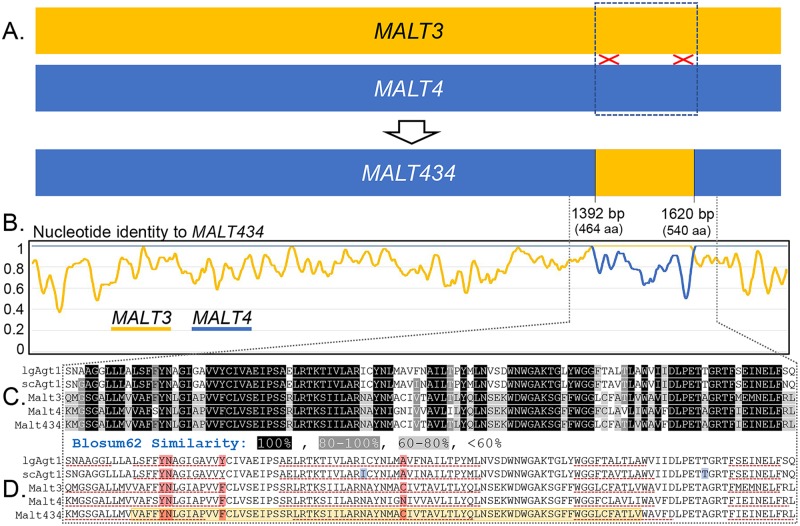
To determine the genetic architecture of maltotriose utilization in the replicate of WI-Seub that evolved the ability to grow on maltotriose, we set up an F1 backcross between the evolved maltotriose-utilizing isolate yHEB1505 and the parent strain, WI-Seub (yHKS210), producing strain yHEB1593, a putative heterozygote capable of growth on maltotriose (Fig 3B and 3E). In a test of 15 fully viable F2 tetrads, maltotriose utilization segregated in a perfect 2:2 manner (Fig 3C). These results suggest that the ability of the evolved strain to utilize maltotriose is conferred by a dominant mutation at a single genetic locus. We performed bulk segregant analysis [78–80] using strains derived from the F2 spores, dividing them between those that could (MalTri+) and those that could not (MalTri-) utilize maltotriose (Fig 3C), with a total of 30 strains in each category. Twelve 1-kb regions were identified as potentially containing fixed differences between the MalTri+ and MalTri- strains. Of these regions, eight mapped to genes encoding ribosomal proteins and most likely represent assembly artefacts due to the presence of many closely related paralogs and/or their absence from the MalTri- de novo assembly that was used for comparisons. Three other regions contained fixed differences between the MalTri+ and MalTri- groups but had no clear relationship to carbon metabolism. The final 1-kb region mapped to the MALT4 locus of S. eubayanus genome [54,60]. The coding sequence of MALT4 from the MalTri+ group contained 52 SNPs relative to the MALT4 allele found in WI-Seub, all of which occurred within a single 230-bp region. Of these, 11 were predicted to lead to non-synonymous changes. Closer inspection revealed that the changes within the 230-bp region were the result of a recombination event between MALT4 and MALT3, creating a chimeric gene (Fig 4), likely through ectopic gene conversion. We call this chimeric MALT4 allele MALT434 after the arrangement of sequences from its parent genes and confirmed by Sanger sequencing that the other maltotriose utilizing strain we isolated (yHEB1506) also carried this chimeric gene at the MALT4 locus. The sequence of MALT3 was not impacted by this mutational event.
Fig 4. Sequence architecture of MALT434.
A) Schematic of the origin of MALT434. B) Line graphs representing the identity between nucleotide sequences of MALT3 and MALT4 from WI-Seub (yHKS210) to MALT434 over 10-bp sliding windows. C-D) Segment of the alignment of the chimeric region between Malt3, Malt4, Malt434, scAgt1, and lgAgt1. The region highlighted in yellow in the Malt434 sequence indicates the chimeric region. The regions underlined with a red dashed line are predicted transmembrane domains. The amino acids highlighted in red are predicted maltose-binding residues. The residues highlighted in blue were experimentally found to be important for maltotriose transport by Smit et al. 2008.
To confirm that MALT434 was the causative locus of maltotriose utilization, we performed a reciprocal hemizygosity test [81] in the heterozygous F1 backcross strain (Fig 3D). Removal of MALT434 eliminated the F1 backcross strain’s ability to utilize maltotriose (Fig 3E), demonstrating that MALT434 is required for maltotriose utilization. Conversely, removing the parental, non-chimeric allele of MALT4 in the heterozygous F1 backcross strain had no impact on maltotriose utilization. Furthermore, overexpression of Malt434 in both the unevolved parent, WI-Seub, and in the NC-Seub background (Fig 5) supported growth on maltotriose, demonstrating that overexpression of Malt434 is sufficient to confer maltotriose utilization. The fact that maltotriose utilization maps to a derivative of maltose transporter genes strongly suggests that the mutant MALT434 gene encodes a functional maltotriose transporter.
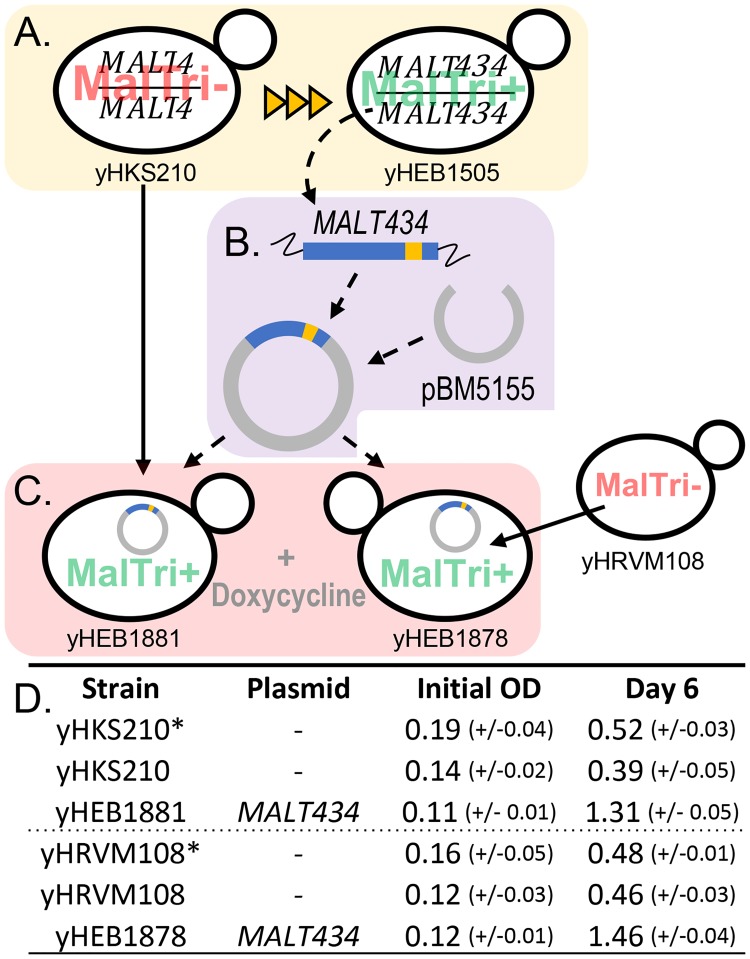
Fig 5. Heterologous expression of MALT434.
A) Evolution of non-maltotriose utilizing strain (MalTri-), WI-Seub (yHKS210), to maltotriose utilizing (MalTri+) strain, yHEB1505, by serial passing on maltotriose containing media (same as Fig 3A). B) Insertion of MALT434 into vector pBM5155 for doxycycline-inducible heterologous expression in MalTri- strains. C) Transformation of MALT434 expression plasmid in MalTri- S. eubayanus strains yHKS210 and NC-Seub (yHRVM108). D) Table of initial and day-six OD600 (OD) measurements of parent strains and strains carrying the MALT434 expression plasmid grown in SC media with maltotriose as the sole carbon and doxycycline to induce plasmid expression. N = 3, standard deviation in parentheses. * Control grown in SC + 0.04% glucose + doxycycline to reflect the approximate amount of growth expected from contamination with other carbon sources when using 98% pure maltotriose.
Potential structural impact of Malt434 chimerism
It was surprising that sequences from MALT3 enabled MALT4 to encode a maltotriose transporter because neither MALT3 nor MALT4 (cloned from Pat-Seub) supported maltotriose utilization on their own (Table 1). The change in function of the MALT434 chimera cannot be the result of differences between the Malt3 and Malt4 proteins encoded by WI-Seub and Pat-Seub. MALT3 in WI-Seub is identical to MALT3 in Pat-Seub, while MALT4 in WI-Seub has several base pair differences relative to MALT4 from Pat-Seub, but none result in amino acid substitutions. Malt3 and Malt4 share about 80% amino acid sequence identity overall and 85% amino acid sequence identity in the chimeric region specifically (Fig 4B). Most residues in the chimeric region had high similarity between Malt3 and Malt4, as measured by Blosum62 similarity matrix (Fig 4C) [82], but there were a handful of low-similarity amino acids as well. To gain insight into what changes in protein structure may be driving the new functionality of Malt434, we used I-TASSER [83–85] to predict the protein structure of Malt3, Malt4, and Malt434. I-TASSER predicts a protein’s structure based on its homology to proteins whose structures have already been solved. Consistent with other studies on the structure of maltose transporters in Saccharomyces [27,86–88], I-TASSER predicted that Malt3, Malt4, and Malt434 were similar to members of the Major Facilitator Superfamily (MFS) of transporters, specifically the sugar porter family [88]. Protein structure is predicted to be conserved between Malt3 and Malt4, including within the chimeric region, which encompasses one full transmembrane domain and parts of two other transmembrane domains (Fig 4D). Four maltose-binding sites were also predicted in the chimeric region. These same domains and predicted binding residues were predicted for Malt434 as well. Interestingly, I-TASSER predicted several of the alpha helices to be shorter in the chimera relative to the parent proteins: two alpha helices in the chimeric region and two towards the N-terminal end of the protein (Fig 4D, S1 Fig). The regions covered by these alpha helices were otherwise predicted to be conserved, out to phylogenetically distantly related Malt proteins lgAgt1 and scAgt1 (Fig 2, Fig 4D, S1 Fig). The predicted shortening of some alpha helices suggests that recombining the MALT3 region into MALT4 may have decreased the overall rigidity of the encoded chimeric protein, allowing it to accommodate bulkier substrates, such as maltotriose. Mutations that increase structural flexibility have been recognized in protein engineering as an important step in accommodating new substrates [89,90].
Besides increasing overall flexibility, the specific location of the chimeric region could have also played a role in supporting maltotriose transport. A previous study found that two residues were important for scAgt1’s ability to transport maltotriose, while not affecting its ability to transport maltose [44]. One of these residues lies within the chimeric region of Malt434, and the other is 10 amino acids downstream (Fig 4D, S1 Fig). Since the overall structure of maltose/maltotriose transporters is conserved [27,86–88], the area in and around the chimeric region of Malt434 may itself be important for substrate specificity.
Thus, the chimeric structure of Malt434 may have facilitated maltotriose transport in two ways. First, it may have increased the overall flexibility of the protein, allowing it to accommodate the larger maltotriose molecule. Second, it could also have specifically altered an important substrate interface to facilitate a better interaction with maltotriose, possibly also by making this region more flexible. Testing these biophysical and structural models will require future experiments, such as solving the crystal structures for Malt3, Malt4, and Malt434 as complexes with maltose and/or maltotriose.
A non-modular chimeric path to novel substrate utilization
Most of the work on functional innovations by chimeric proteins has focused on the rearrangement of discrete functional domains, with or without the benefit of intronic sequences [6,7,14,91–94]. However, Malt434 does not fit easily into the framework of new protein creation by the reordering or exchanging of domains. While residues important for sugar specificity probably exist in Malt3 and Malt4 [44,87], with respect to maltotriose, Malt3 and Malt4 do not seem to have different functions or sugar specificities in their native backgrounds. In Malt3 and Malt4, there are no specific “maltotriose-transporting” residues to be swapped. Instead, the ability of the residues from Malt3 to facilitate maltotriose transport must rely on their interaction with one or more residues in Malt4, not simply on their independent ability to interact with maltotriose.
Rather than the modular framework of novel protein formation, Malt434 exemplifies another framework for how recombination can lead to the evolution of novel functions. Theoretical and experimental work has demonstrated the important role that recombination between related proteins can play in facilitating the evolution of new functions [3,4,15,95]. Indeed, protein engineering has utilized the technique of DNA shuffling since the mid-1990’s to recombine closely related coding sequences to efficiently generate proteins with novel or improved functions [19]. More recently, experimental work has begun to demonstrate the importance of recombination between closely related proteins in nature for the evolution of new functions [15,18,95]. In this model, two duplicate proteins neutrally accumulate the multiple amino acid changes needed for a new function independently. All of the mutations that are needed for the new function are then brought together at once, en masse, through recombination. This molecular mechanism allows proteins to “tunnel” to new functions, bypassing potentially deleterious intermediates that would be encountered through a series of amino acid substitutions [3,4].
While MALT3 and MALT4 are not recent duplicates, they are distant paralogs (Fig 2). In addition, as members of the sugar porter family of proteins, they share a highly conserved protein structure [27,86–88]. The conservative nature of sugar porter family proteins means that recombination events like the one that formed Malt434, which do not fall between clear domains, probably have a relatively high likelihood of creating functional transporters [96], albeit ones of unpredictable specificity. In the case of Malt434, we do not yet know which specific amino acid interactions were important for the gain of maltotriose utilization in the chimera, let alone the function or history of the residues in the background of their native protein sequences. It may be that they represent neutral changes in their parental backgrounds, but they also could have been selected for other specificities. Nevertheless, the independent accumulation of these changes in a common ancestral protein background eventually allowed these sequences to recombine and create a novel function. One intriguing hypothesis proposes that other maltotriose transporters, such as AGT1 and MTT1, and indeed other diverse MFS superfamily genes might also be chimeras [97]. The placement of MTT1 in our phylogenetic analysis is consistent with its origin as a chimera between MALT3 and a MALx1 gene (Fig 2), but rigorous phylogenetic analyses will be required to evaluate its origin and the generality of this model.
Conclusions
Our findings suggest that the evolution of maltotriose utilization by Saccharomyces spp. is not a straightforward process. Even when a functioning maltotriose transporter is available in the parent genome, the regulatory changes necessary to support atypical expression may be difficult to evolve under certain experimental conditions. Conversely, when a maltotriose transporter is not already present, single point mutations may be insufficient to switch or expand the specificity of available Malt proteins. Recombination between paralogous proteins can rapidly do what a single point mutation cannot and, in a single rare mutational event, introduce the multiple residue changes needed to perform a new function. Our report on the evolution of a chimeric maltotriose transporter from parental proteins that could not transport maltotriose supports the role of recombination, beyond the simple swapping of functional protein domains and discrete peptide motifs, in the formation of proteins with novel functions. Our results establish that strains of Saccharomyces without known maltotriose transporters are capable of evolving novel transporters during experimental evolution for maltotriose consumption. Future, larger scale experiments could establish whether chimeric transporters, similar to the one we observed, are a common mechanism to gain this new function or if other routes to maltotriose utilization are open to some Saccharomyces lineages.
Note added in proof
While we were revising this manuscript in response to peer review, we became aware of a manuscript posted on the preprint server bioRxiv that also recovered a chimeric maltotriose transporter from the recombination of maltose transporters in S. eubayanus [97]. The similarity of these results further suggests that the evolutionary trajectories leading to maltotriose utilization are limited in some genetic backgrounds.
Materials and Methods
Strains
All strains discussed in this paper are listed in S4 Table. Briefly, FM1318 (Pat-Seub) is a monosporic derivative of the taxonomic type strain of S. eubayanus, which was isolated from Patagonia [48]. yHRVM108 (NC-Seub) was isolated from Durham, North Carolina, and is closely related to the S. eubayanus strains that hybridized with S. cerevisiae to give rise to lager-brewing yeasts [52]. yHKS210 (WI-Seub) was isolated from Sheboygan, Wisconsin, and is the result of admixture between populations A and B of S. eubayanus. WI-Seub is nearly homozygous due to selfing after the initial admixture event [53]. Of these strains, Pat-Seub and WI-Seub grew well on maltose, but they did not grow on maltotriose. NC-Seub grew sluggishly on maltose and did not grow on maltotriose. yHAB47 is a copy of Weihenstephan 34/70 [52], a representative of the Frohberg or Group II [77] lineage of lager-brewing hybrids (S. cerevisiae (2n) x S. eubayanus (2n) [58]). CDFM21L.1 (Tb-Seub) is a strain of S. eubayanus isolated from Tibet [51] and is closely related to NC-Seub. Of known S. eubayanus strains, Tb-Seub is the most genetically similar to the S. eubayanus parents of lager-brewing hybrids [51,52].
Identification of MALT genes
Previously, we identified four genes with homology to genes encoding maltose transporters in S. cerevisiae and lager-brewing hybrids in the genome assembly of FM1318 (Pat-Seub) published by Baker et al. 2015 [54]. These genes were previously designated MALT1-4. Only a partial contig was available for MALT4 in this assembly, but a BLAST [98] search of the Okuno et al. 2016 [60] assembly of the taxonomic type strain of S. eubayanus (of which FM1318 is a monosporic derivative) allowed us to annotate the full-length sequence of MALT4. MALT4 has 99.7% identity to MALT2 at the nucleotide level and is predicted to have 100% identity at the amino acid level. The regions from 900 bp downstream of MALT2 and MALT4 and upstream to the ends of chromosomes V and XVI (regions of approximately 12 kb in the Okuno et al. 2016 [60] assembly), respectively, share 99.1% nucleotide identity. The 10 kb outside of this region only share 49.8% nucleotide identity. Thus, MALT2 and MALT4 are close paralogs that are likely related by a recent subtelomeric duplication and/or translocation event.
Reads for homologs of AGT1 were retrieved using the functional AGT1 sequence from lager yeast (lgAGT1) as the query sequence [55] in an SRA-BLAST search of the SRA databases of NCBI for yHRVM108 (SRR2586159) and CDFM21L.1 (SRR1507225). All reads identified in the BLAST searches were downloaded and assembled using the de novo assembler in Geneious v. 9.0.3 (http://www.geneious.com) [99]. The homologs identified in yHRVM108 and CDFM21L.1 were designated ncAGT1 (for North Carolinian AGT1) and tbAGT1 (for Tibetan AGT1), respectively. The presence and sequence of ncAGT1 in yHRVM108 was further verified by PCR amplification and Sanger sequencing (S5 Table). CDFM21L.1 was not available at the time of this work for further verification of the presence of tbAGT1.
Adaptive evolution
Design of the adaptive evolution experiments was based on Parreiras et al. 2014 [100]. The highest available purities of carbon sources were used: 98% pure maltotriose, ≥99% pure maltose, and 100.0% pure glucose. Adaptive evolution was initiated by growing parent strains overnight in liquid YPD medium (1% yeast extract, 2% peptone, 2% glucose). One mL of maltotriose or maltose medium was inoculated with enough overnight culture to give an OD600 reading of ~0.1, as measured with an IMPLEN OD600 DiluPhotometer. Evolution on maltotriose was conducted in synthetic complete (SC) medium (0.17% yeast nitrogen base, 0.5% ammonium sulfate, 0.2% complete drop out mix) with 2% maltotriose and 0.1% glucose. The addition of 0.1% glucose ensured enough growth that mutations could occur and be selected for through the ensuing generations. Adaptive evolution of yHRVM108 (NC-Seub) on maltose was carried out in SC with 2% maltose. Because NC-Seub grew so poorly on maltose alone, an additional 0.1% glucose was supplemented into its medium; after increased growth was observed around generation 110 for replicate A (from which strains yHEB1585-1587 were derived), around generation 80 for replicate B (from which strains yHEB1588-90 were derived), and around generation 155 for replicate C (from which strains yHEB1778-80 were derived), subsequent generations of NC-Seub adaptive evolution on maltose for these replicates were conducted with 2% maltose only. Adaptive evolution experiments of each strain were carried out in triplicate. Samples were grown on a culture wheel at room temperature (22°C) and diluted 1:10 into fresh media every 3–4 days. At the beginning of the experiment, before consumption of the primary carbon sources had evolved, 1/10 of the population at the time of passaging contained approximately 10 million cells. Samples of each experimental evolution replicate were taken every other passage and placed into long-term storage by mixing 700uL of culture with 300uL of 50% glycerol in a cryotube and storing it at -80°C. The number of cells passaged and of doublings between passages were estimated from cell counts during the second and third passages. Experimental evolution was carried out for a total of 100 passages. Strains that could not use the primary carbon source in the adaptive evolution medium underwent approximately one cell division per day on average.
Sporulation and backcrossing
To induce sporulation, strains were grown to saturation, washed twice, and then resuspended in 200μL liquid sporulation (spo) medium (1% potassium acetate, 0.5% zinc acetate). 30μL of this suspension was added to 1.5mL of spo medium and incubated on a culture wheel at room temperature. Cultures were checked for sporulation after 2–5 days. Tetrads were dissected using a Singer SporePlay. For backcrossing, tetrads of the strains to be crossed were dissected on a single YPD plate. A spore from one parent was placed in close proximity to a spore from the other parent, and they were observed over several hours for mating and zygote formation. Transformations of the diploid F1 backcross strain for gene knockouts were carried out as described below in the section describing the construction of gene expression plasmids.
Construction of gene expression plasmids
Genes encoding transporters of interest were cloned via gap repair into the NotI site of plasmid pBM5155 (GenBank KT725394.1), which contains the complete machinery necessary for doxycycline-based induction of genes cloned into this site [101]. Transformation was carried out using standard lithium acetate transformation [102] with modifications to optimize transformation in S. eubayanus. Specifically, transformation reactions were heat-shocked at 34°C. After 55 minutes, 100% ethanol was added to 10% total volume, and the reactions heat shocked for another 5 minutes before they were allowed to recover overnight and plated to selective media the next day. When necessary, plasmids were recovered and amplified in E. coli for transformation into multiple strains. The sequences of genes encoding transporters cloned into pBM5155 were verified by Sanger sequencing. S. eubayanus MALT1, MALT3, and MALT4 were amplified from Pat-Seub, lgAGT was amplified from yHAB47, and ncAGT1 was amplified from NC-Seub. Primers used for plasmid construction and sequence verification are listed in S5 Table.
Growth assays
Growth was measured in liquid media in 96-well plates using OD600 measurements on a FLUOstar Omega microplate reader. Strains were first grown to saturation in liquid YPD medium, then washed twice and diluted in SC without added carbon to OD600 = 1.9 +/- 0.05 to ensure that all cultures had approximately the same starting concentration. 15μL of each diluted culture was added to 235μL of the test medium. Three technical replicates, randomly distributed on a 96-well plate to control for position effects, were carried out for each strain. Single-colony isolates of WI-Seub evolved on maltotriose and single-colony isolates of NC-Seub evolved on maltose were tested in SC medium + 2% maltotriose. Single-colony isolates of NC-Seub evolved on maltose were also tested on SC medium + 2% maltose. Strains carrying MALT genes expressed on an inducible plasmid were tested in SC medium + 2% maltotriose and 5 ng/mL doxycycline to induce plasmid gene expression. To control for growth from the small amount of non-maltotriose sugar in 98% pure maltotriose, the parent strains of NC-Seub and WI-Seub were also tested in SC medium + 0.04% glucose, reflecting the approximate amount of other carbon sources expected in SC medium + 2% maltotriose.
Bulk segregant analysis
60 spores from 15 fully viable tetrads of strain yHEB1593 (F1 of yHKS210 x yHEB1505) were dissected and individually screened for their ability to grow in SC + 2% maltotriose. F2 segregants that could grow on maltotriose were classified as MalTri+, and those that could not were classified as MalTri-. Each F2 segregant was then individually grown to saturation in liquid YPD. The saturated cultures were spun down, the supernatant removed, and enough cells resuspended in liquid SC medium to give an OD600 measurement of between 1.9 and 1.95, as measured with an IMPLEN OD600 DiluPhotometer. Strains were pooled based on their ability to grow on maltotriose, forming a MalTri+ pool and a MalTri- pool. To pool, 1mL of each strain dilution was added to the appropriate pool of cells and vortexed to mix. Phenol-chloroform extraction and ethanol precipitation was used to isolate gDNA from the segregant pools. The gDNA was sonicated and ligated to Illumina TruSeq-style dual adapters and index sequencing primers using the NEBNext DNA Library Prep Master Mix Set for Illumina kit following the manufacturer’s instructions. The paired-end libraries were sequenced on an Illumina MiSeq instrument, conducting a 2 x 250bp run.
Analysis of bulk segregant sequencing reads
To identify fixed differences between the meiotic segregant pools, de novo assemblies were made for the MalTri- group of segregants using the meta-assembler iWGS with default settings [103]. The final genome assembly of the MalTri- pool was made by DISCOVAR [104] in iWGS. This assembly was used for reference-based genome assembly and variant calling using reads from the MalTri+ pool following the protocol described in Peris and Langdon et al. 2016 [52]. Assemblies of the putative chimeric maltotriose transporter were retrieved from the MalTri+ pool of reads using the program HybPiper [105]. Briefly, HybPiper uses a BLAST search of read sequences to find reads that map to a query sequence; it then uses the programs Exonerate [106] and SPAdes [107] to assemble the reads into contigs. The sequence and genomic location of the chimeric transporter were further verified by PCR amplification and Sanger sequencing (S5 Table), as was the sequence of MALT4 from yHKS210.
Phylogenetic analyses and computational predictions of protein structures and functions
Multiple sequence alignments between the proteins encoded by the MALT genes were carried out using MUSCLE [108], as implemented in Geneious v.9.0.3 [99] (http://www.geneious.com). Phylogenetic relationships were determined using codon alignments. Codon alignments were made using PAL2NAL [109] (http://www.bork.embl.de/pal2nal/) to convert the MUSCLE alignments of amino acid sequences to nucleotide alignments. A phylogenetic tree of nineteen MALT genes from S. eubayanus and S. cerevisiae and lager-brewing yeasts was constructed as described in Baker et al. 2015 [54] using MEGA v.6. Most genes used in the phylogenetic analysis were retrieved as previously described in Baker et al. 2015 [54] as follows: MAL21, MAL31, and MAL61 from S. cerevisiae; MALT1 and MALT3 from S. eubayanus; MALT1, MALT2, and MPH from lager-brewing yeast; MPH2 and MPH3 from S. cerevisiae; AGT1 (MAL11 in Baker et al. 2015 [54]) from S. cerevisiae; scAGT1 (WeihenMAL11-CB in Baker et al. 2015 [54]); and lgAGT1 (WeihenMAL11-CA in Baker et al. 2015 [54]). Sequences for MALT2 and MALT4 were retrieved from the genome assembly of CBS 12357T from Okuno et al. 2016 [60]. MAL11 was retrieved from the genome assembly of S. cerevisiae strain YJM456 [110]. Sequences for tbAGT1 and ncAGT1 were retrieved as described above. MAL11 and AGT1 both encode α-glucoside transporters located at the MAL1 locus in S. cerevisiae and, as such, are considered alleles of each other [27,111]. Their shared genomic location notwithstanding, MAL11 and AGT1 are not phylogenetically closely related, with MAL11 clustering with other MALx1 type transporters (Fig 2). In addition, while AGT1 can support maltotriose transport, MAL11, like other known MALx1 genes, cannot [27,30]. Despite their dissimilarity, AGT1 is recorded in the Saccharomyces Genome Database (yeastgenome.org) as MAL11 since the reference strain carries the AGT1 allele at the MAL1 locus [62,65]. For this reason, MAL11 is often used to refer to AGT1 [30,32,54]. For clarity, here we use MAL11 to only refer to the MALx1-like allele and AGT1 to refer to the distinct maltotriose-transporting allele.
Protein structure predictions for MALT3, MALT4, lgAGT1, and scAGT1 were carried out using the I-TASSER server, and the structure prediction of MALT434 was carried out using the command line version of I-TASSER [83–85] (https://zhanglab.ccmb.med.umich.edu/I-TASSER/, accessed between 2-7-2018 and 2-28-2018). The potential impact of the single residue difference between lgAGT1 and tbAGT1 was analyzed by two different methods. Prediction of the change in free energy (ΔΔG) was carried out using the STRUM server (https://zhanglab.ccmb.med.umich.edu/STRUM/, accessed 3-21-18) [67]. A ΔΔG score of < +/- 0.5 was considered to be unlikely to affect function [112]. Homology-based predictions were made using SIFT at http://sift.jcvi.org/ (accessed 3-30-18) [68,113–116]. The SIFT Related Sequences analysis was done using the amino acid sequences of MALT genes in the phylogenetic analysis above. Several SIFT analyses were also carried out using the SIFT Sequence analysis program. This analysis operates using the same principle as the SIFT Related Sequences analysis, but rather than being supplied by the user, homologous sequences were provided by a PSI-BLAST search of the indicated protein database. The SIFT Sequence analyses were carried out using default settings and the following databases available on http://sift.jcvi.org/ (accessed 3-30-18): NCBI nonredundant 2011 Mar, UniRef90 2011 Apr, UniProt-SwissProt 57.15 2011 Apr.
Supporting information
Protein structural alignment between Malt3, Malt4, Malt434, scAgt1, and lgAgt1. The purple blocks represent predicted alpha helices, and the orange lines represent predicted beta strands. Red ticks mark predicted maltose-binding sites. Blue ticks mark residues found to be important for maltotriose transport by Smit et al. 2008. A green tick marks the location of the single non-synonymous substitution between lgAGT1 and tbAGT1. Arrows point to alpha helices in Malt434 whose predicted sizes are reduced compared to other transporters in the alignment.
(TIF)
Results of mutation prediction analyses for E18V, the sole amino acid substitution in the lgAgt1 protein sequence, relative to tbAgt1.
(XLSX)
Growth on maltose of single-colony isolates. Isolated from adaptive evolution of yHRVM108 on 2% maltose + 0.1% glucose. N = 3. * Control grown in SC + 0.04% glucose to reflect the approximate amount of growth expected from contamination with other carbon sources when using 98% pure maltotriose.
(XLSX)
Growth on maltotriose of single-colony isolates from adaptive evolution experiments. Strains were evolved with either maltotriose or maltose as the primary carbon source (2%) with 0.1% added glucose. N = 3. * Control grown in SC + 0.04% glucose to reflect the approximate amount of growth expected from contamination with other carbon sources when using 98% pure maltotriose.
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
We thank Dana A. Opulente, Quinn K. Langdon, Ryan V. Moriarty, and Kelly V. Buh for their assistance maintaining the experimental evolution lines described in this manuscript; Diego Libkind, José Paulo Sampaio, and the Portuguese Yeast Culture Collection for the taxonomic type strain of S. eubayanus; and Russell L. Wrobel for assistance with allele confirmation reactions.
Data Availability
All Illumina sequencing data have been uploaded to GenBank as BioProject PRJNA507896. All other relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the USDA National Institute of Food and Agriculture, Hatch project 1003258, the National Science Foundation (grant numbers DEB-1253634); and funded in part by the DOE Great Lakes Bioenergy Research Center (DOE BER Office of Science DE-SC0018409 and DE-FC02-07ER64494). EB is supported by a Louis and Elsa Thomsen Wisconsin Distinguished Graduate Fellowship. CTH is a Pew Scholar in the Biomedical Sciences and a Vilas Faculty Early Career Investigator, supported by the Pew Charitable Trusts and the Vilas Trust Estate. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Long M, Betrán E, Thornton K, Wang W. The origin of new genes: glimpses from the young and old. Nat Rev Genet. 2003;4: 865–875. 10.1038/nrg1204 [DOI] [PubMed] [Google Scholar]
- 2.Ohno S. Evolution by Gene Duplication. Berlin, Heidelberg: Springer Berlin Heidelberg; 1970. [Google Scholar]
- 3.Bittihn P, Tsimring LS. Gene Conversion Facilitates Adaptive Evolution on Rugged Fitness Landscapes. Genetics. 2017;207: 1577–1589. 10.1534/genetics.117.300350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cui Y, Wong WH, Bornberg-Bauer E, Chan HS. Recombinatoric exploration of novel folded structures: a heteropolymer-based model of protein evolutionary landscapes. Proc Natl Acad Sci U S A. 2002;99: 809–14. 10.1073/pnas.022240299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poelwijk FJ, Kiviet DJ, Weinreich DM, Tans SJ. Empirical fitness landscapes reveal accessible evolutionary paths. Nature. 2007;445: 383–386. 10.1038/nature05451 [DOI] [PubMed] [Google Scholar]
- 6.Patthy L. Genome evolution and the evolution of exon-shuffling—a review. Gene. 1999;238: 103–114. [DOI] [PubMed] [Google Scholar]
- 7.Patthy L. Modular Assembly of Genes and the Evolution of New Functions. Genetica. 2003;118: 217–231. [PubMed] [Google Scholar]
- 8.Henikoff S, Greene EA, Pietrokovski S, Bork P, Attwood TK, Hood L. Gene families: the taxonomy of protein paralogs and chimeras. Science. 1997;278: 609–14. [DOI] [PubMed] [Google Scholar]
- 9.Leffler EM, Band G, Busby GBJ, Kivinen K, Le QS, Clarke GM, et al. Resistance to malaria through structural variation of red blood cell invasion receptors. Science. 2017;356: eaam6393 10.1126/science.aam6393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mitelman F, Johansson B, Mertens F. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer. 2007;7: 233–245. 10.1038/nrc2091 [DOI] [PubMed] [Google Scholar]
- 11.Rippey C, Walsh T, Gulsuner S, Brodsky M, Nord AS, Gasperini M, et al. Formation of Chimeric Genes by Copy-Number Variation as a Mutational Mechanism in Schizophrenia. Am J Hum Genet. 2013;93: 697–710. 10.1016/j.ajhg.2013.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Malfavon-Borja R, Wu LI, Emerman M, Malik HS. Birth, decay, and reconstruction of an ancient TRIMCyp gene fusion in primate genomes. Proc Natl Acad Sci U S A. 2013;110: E583–92. 10.1073/pnas.1216542110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang W, Zhang J, Alvarez C, Llopart A, Long M. The Origin of the Jingwei Gene and the Complex Modular Structure of Its Parental Gene, Yellow Emperor, in Drosophila melanogaster. Mol Biol Evol. 2000;17: 1294–1301. 10.1093/oxfordjournals.molbev.a026413 [DOI] [PubMed] [Google Scholar]
- 14.De Chateau M, Bjorck L, Smith JM. Identification of interdomain sequences promoting the intronless evolution of a bacterial protein family. Proc Natl Acad Sci. 1996;93: 8490–8495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rogers RL, Hartl DL. Chimeric Genes as a Source of Rapid Evolution in Drosophila melanogaster. Mol Biol Evol. 2012;29: 517–529. 10.1093/molbev/msr184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bogarad LD, Deem MW. A hierarchical approach to protein molecular evolution. Proc Natl Acad Sci U S A. 1999;96: 2591–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Voigt CA, Martinez C, Wang Z-G, Mayo SL, Arnold FH. Protein building blocks preserved by recombination. Nat Struct Biol. 2002;9: 553–8. 10.1038/nsb805 [DOI] [PubMed] [Google Scholar]
- 18.Thomas JH. Concerted evolution of two novel protein families in Caenorhabditis species. Genetics. 2006;172: 2269–81. 10.1534/genetics.105.052746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giver L, Arnold FH. Combinatorial protein design by in vitro recombination. Curr Opin Chem Biol. 1998;2: 335–338. [DOI] [PubMed] [Google Scholar]
- 20.Cole MF, Gaucher EA. Utilizing natural diversity to evolve protein function: applications towards thermostability. Curr Opin Chem Biol. 2011;15: 399–406. 10.1016/j.cbpa.2011.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Minshull J, Willem Stemmer PC. Protein evolution by molecular breeding. Curr Opin Chem Biol. 1999;3: 284–290. [DOI] [PubMed] [Google Scholar]
- 22.Nijland JG, Shin HY, de Waal PP, Klaassen P, Driessen AJM. Increased xylose affinity of Hxt2 through gene shuffling of hexose transporters in Saccharomyces cerevisiae. J Appl Microbiol. 2018;124: 503–510. 10.1111/jam.13670 [DOI] [PubMed] [Google Scholar]
- 23.Eßlinger HM. Fermentation, Maturation and Storage In: Eßlinger HM, editor. Handbook of brewing processes, technology, markets. Weinheim (Germany): Wiley-VCH; 2009. p. 209. [Google Scholar]
- 24.Briggs DE, Brookes PA, Stevens R, Boulton CA. Metabolism of wort by yeast Brewing: science and practice. Abington Hall, Abington Cambridge CB1 6AH, England: Woodhead Publishing Limited; 2004. p. 413. [Google Scholar]
- 25.Briggs D.E., Brookes P.A., Stevens R. BCA. The science of mashing Brewing: science and practice. Abington Hall, Abington Cambridge CB1 6AH, England: Woodhead Publishing Limited; 2004. p. 108, 109,, 123,, 125. [Google Scholar]
- 26.Meussdoerffer F, Zarnkow M. Starchy Raw Materials In: Eßlinger HM, editor. Handbook of brewing processes, technology, markets. Weinheim (Germany): Wiley-VCH; 2009. p. 58. [Google Scholar]
- 27.Han E, Cotty F, Sottas C, Jiang H, Michels CA. Characterization of AGT1 encoding a general α‐glucoside transporter from Saccharomyces. Mol Microbiol. 1995;17: 1093–1107. [DOI] [PubMed] [Google Scholar]
- 28.Dietvorst J, Londesborough J, Steensma HY. Maltotriose utilization in lager yeast strains: MTT1 encodes a maltotriose transporter. Yeast. 2005;22: 775–788. 10.1002/yea.1279 [DOI] [PubMed] [Google Scholar]
- 29.Salema-Oom M, Valadão Pinto V, Gonçalves P, Spencer-Martins I. Maltotriose utilization by industrial Saccharomyces strains: characterization of a new member of the alpha-glucoside transporter family. Appl Environ Microbiol. 2005;71: 5044–9. 10.1128/AEM.71.9.5044-5049.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brown CA, Murray AW, Verstrepen KJ. Rapid Expanion and Functional Divergence of Subtelomeric Gene Families in Yeasts. Curr Biol. 2010;20: 895–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Horák J. Regulations of sugar transporters: insights from yeast. Curr Genet. 2013;59: 1–31. 10.1007/s00294-013-0388-8 [DOI] [PubMed] [Google Scholar]
- 32.Brickwedde A, van den Broek M, Geertman J-MA, Magalhães F, Kuijpers NGA, Gibson B, et al. Evolutionary Engineering in Chemostat Cultures for Improved Maltotriose Fermentation Kinetics in Saccharomyces pastorianus Lager Brewing Yeast. Front Microbiol. 2017;8: 1690 10.3389/fmicb.2017.01690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jansen MLA, Daran-Lapujade P, de Winde JH, Piper MDW, Pronk JT. Prolonged maltose-limited cultivation of Saccharomyces cerevisiae selects for cells with improved maltose affinity and hypersensitivity. Appl Environ Microbiol. 2004;70: 1956–63. 10.1128/AEM.70.4.1956-1963.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krogerus K, Seppänen-Laakso T, Castillo S, Gibson B. Inheritance of brewing-relevant phenotypes in constructed Saccharomyces cerevisiae × Saccharomyces eubayanus hybrids. Microb Cell Fact. 2017;16: 66 10.1186/s12934-017-0679-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krogerus K, Magalhães F, Vidgren V, Gibson B. New lager yeast strains generated by interspecific hybridization. J Ind Microbiol Biotechnol. 2015;42: 769–778. 10.1007/s10295-015-1597-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hebly M, Brickwedde A, Bolat I, Driessen MRM, de Hulster EAF, van den Broek M, et al. S. cerevisiae × S. eubayanus interspecific hybrid, the best of both worlds and beyond. FEMS Yeast Res. 2015;15: fov005 10.1093/femsyr/fov005 [DOI] [PubMed] [Google Scholar]
- 37.Krogerus K, Arvas M, De Chiara M, Magalhães F, Mattinen L, Oja M, et al. Ploidy influences the functional attributes of de novo lager yeast hybrids. Appl Microbiol Biotechnol. 2016;100: 7203–22. 10.1007/s00253-016-7588-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mertens S, Steensels J, Saels V, De Rouck G, Aerts G, Verstrepen KJ. A large set of newly created interspecific Saccharomyces hybrids increases aromatic diversity in lager beers. Appl Environ Microbiol. 2015;81: 8202–14. 10.1128/AEM.02464-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nikulin J, Krogerus K, Gibson B. Alternative Saccharomyces interspecies hybrid combinations and their potential for low-temperature wort fermentation. Yeast. 2018;35: 113–127. 10.1002/yea.3246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stewart GG. The genetic manipulation of industrial yeast strains. Can J Microbiol. 1981;27: 973–990. [Google Scholar]
- 41.Bilinski CA, Casey GP. Developments in sporulation and breeding of brewer’s yeast. Yeast. Wiley Online Library; 1989;5: 429–438. [Google Scholar]
- 42.Mukai N, Nishimori C, Fujishige W, Mizuno A, Takahashi T, Sato K. Beer Brewing Using a Fusant between a Sake Yeast and a Brewer’s Yeast. J Biosci Bioeng. 2001;91: 482–486. [DOI] [PubMed] [Google Scholar]
- 43.Yamakawa S, Yamada R, Tanaka T, Ogino C, Kondo A. Repeated batch fermentation from raw starch using a maltose transporter and amylase expressing diploid yeast strain. Appl Microbiol Biotechnol. 2010;87: 109–115. 10.1007/s00253-010-2487-5 [DOI] [PubMed] [Google Scholar]
- 44.Smit A, Moses SG, Pretorius IS, Cordero Otero RR. The Thr505 and Ser557 residues of the AGT1-encoded alpha-glucoside transporter are critical for maltotriose transport in Saccharomyces cerevisiae. J Appl Microbiol. 2008;104: 1103–11. 10.1111/j.1365-2672.2007.03671.x [DOI] [PubMed] [Google Scholar]
- 45.Smit A, Dissertation. Maltotriose Transport in Yeast [Internet]. Stellenbosch University. 2007. http://scholar.sun.ac.za/bitstream/handle/10019.1/21760/Smit_Maltotriose_1007.pdf?sequence=1&isAllowed=y
- 46.Krogerus K, Magalhães F, Vidgren V, Gibson B. Novel brewing yeast hybrids: creation and application. Appl Microbiol Biotechnol. 2017;101: 65–78. 10.1007/s00253-016-8007-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hittinger CT, Steele JL, Ryder DS. Diverse yeasts for diverse fermented beverages and foods. Curr Opin Biotechnol. 2018;49: 199–206. 10.1016/j.copbio.2017.10.004 [DOI] [PubMed] [Google Scholar]
- 48.Libkind D, Hittinger CT, Valério E, Gonçalves C, Dover J, Johnston M, et al. Microbe domestication and the identification of the wild genetic stock of lager-brewing yeast. Proc Natl Acad Sci U S A. 2011;108: 14539–14544. 10.1073/pnas.1105430108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gibson BR, Storgårds E, Krogerus K, Vidgren V. Comparative physiology and fermentation performance of Saaz and Frohberg lager yeast strains and the parental species Saccharomyces eubayanus. Yeast. 2013;30: 255–266. 10.1002/yea.2960 [DOI] [PubMed] [Google Scholar]
- 50.Gibson B, Geertman J-MA, Hittinger CT, Krogerus K, Libkind D, Louis EJ, et al. New yeasts—new brews: modern approaches to brewing yeast design and development. FEMS Yeast Res. 2017;17. [DOI] [PubMed] [Google Scholar]
- 51.Bing J, Han P-J, Liu W-Q, Wang Q-M, Bai F-Y. Evidence for a Far East Asian origin of lager beer yeast. Curr Biol. 2014;24: R380–1. 10.1016/j.cub.2014.04.031 [DOI] [PubMed] [Google Scholar]
- 52.Peris D, Langdon QK, Moriarty R V., Sylvester K, Bontrager M, Charron G, et al. Complex Ancestries of Lager-Brewing Hybrids Were Shaped by Standing Variation in the Wild Yeast Saccharomyces eubayanus. PLOS Genet. 2016;12: e1006155 10.1371/journal.pgen.1006155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peris D, Sylvester K, Libkind D, Gonçalves P, Sampaio JP, Alexander WG, et al. Population structure and reticulate evolution of Saccharomyces eubayanus and its lager-brewing hybrids. Mol Ecol. 2014;23: 2031–2045. 10.1111/mec.12702 [DOI] [PubMed] [Google Scholar]
- 54.Baker E, Wang B, Bellora N, Peris D, Hulfachor AB, Koshalek JA, et al. The Genome Sequence of Saccharomyces eubayanus and the Domestication of Lager-Brewing Yeasts. Mol Biol Evol. 2015;32: 2818–2831. 10.1093/molbev/msv168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nakao Y, Kanamori T, Itoh T, Kodama Y, Rainieri S, Nakamura N, et al. Genome sequence of the lager brewing yeast, an interspecies hybrid. DNA Res. 2009;16: 115–129. 10.1093/dnares/dsp003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vidgren V, Multanen J-P, Ruohonen L, Londesborough J. The temperature dependence of maltose transport in ale and lager strains of brewer’s yeast. FEMS Yeast Res. 2010;10: 402–411. 10.1111/j.1567-1364.2010.00627.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cousseau FEM, Alves SL, Trichez D, Stambuk BU. Characterization of maltotriose transporters from the Saccharomyces eubayanus subgenome of the hybrid Saccharomyces pastorianus lager brewing yeast strain Weihenstephan 34/70. Lett Appl Microbiol. 2013;56: 21–29. 10.1111/lam.12011 [DOI] [PubMed] [Google Scholar]
- 58.Nguyen H-V, Boekhout T. Characterization of Saccharomyces uvarum (Beijerinck, 1898) and related hybrids: assessment of molecular markers that predict the parent and hybrid genomes and a proposal to name yeast hybrids. FEMS Yeast Res. 2017;17. [DOI] [PubMed] [Google Scholar]
- 59.Nijland JG, Shin HY, de Jong RM, de Waal PP, Klaassen P, Driessen AJ. Engineering of an endogenous hexose transporter into a specific D-xylose transporter facilitates glucose-xylose co-consumption in Saccharomyces cerevisiae Biotechnol Biofuels. 2014;7: 168 10.1186/s13068-014-0168-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Okuno M, Kajitani R, Ryusui R, Morimoto H, Kodama Y, Itoh T. Next-generation sequencing analysis of lager brewing yeast strains reveals the evolutionary history of interspecies hybridization. DNA Res. 2016;23: 67–80. 10.1093/dnares/dsv037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brickwedde A, Brouwers N, van den Broek M, Gallego Murillo JS, Fraiture JL, Pronk JT, et al. Structural, Physiological and Regulatory Analysis of Maltose Transporter Genes in Saccharomyces eubayanus CBS 12357T. Front Microbiol. 2018;9: 1786 10.3389/fmicb.2018.01786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vidgren V, Huuskonen A, Virtanen H, Ruohonen L, Londesborough J. Improved fermentation performance of a lager yeast after repair of its AGT1 maltose and maltotriose transporter genes. Appl Environ Microbiol. 2009;75: 2333–45. 10.1128/AEM.01558-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vidgren V, Londesborough J. Characterization of the Saccharomyces bayanus-type AGT1 transporter of lager yeast. J Inst Brew. 2012;118: 148–151. [Google Scholar]
- 64.Day RE, Rogers PJ, Dawes IW, Higgins VJ. Molecular analysis of maltotriose transport and utilization by Saccharomyces cerevisiae. Appl Environ Microbiol. 2002;68: 5326–35. 10.1128/AEM.68.11.5326-5335.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vidgren V, Ruohonen L, Londesborough J. Characterization and functional analysis of the MAL and MPH Loci for maltose utilization in some ale and lager yeast strains. Appl Environ Microbiol. 2005;71: 7846–57. 10.1128/AEM.71.12.7846-7857.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bergström A, Simpson JT, Salinas F, Barré B, Parts L, Zia A, et al. A High-Definition View of Functional Genetic Variation from Natural Yeast Genomes. Mol Biol Evol. 2014;31: 872–888. 10.1093/molbev/msu037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Quan L, Lv Q, Zhang Y. STRUM: structure-based prediction of protein stability changes upon single-point mutation. Bioinformatics. 2016;32: 2936–2946. 10.1093/bioinformatics/btw361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11: 863–74. 10.1101/gr.176601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Day RE, Higgins VJ, Rogers PJ, Dawes IW. Characterization of the putative maltose transporters encoded by YDL247w and YJR160c. Yeast. 2002;19: 1015–1027. 10.1002/yea.894 [DOI] [PubMed] [Google Scholar]
- 70.Stambuk BU, Alves SL, Hollatz C, Zastrow CR. Improvement of maltotriose fermentation by Saccharomyces cerevisiae. Lett Appl Microbiol. 2006;43: 370–376. 10.1111/j.1472-765X.2006.01982.x [DOI] [PubMed] [Google Scholar]
- 71.Alves SL, Herberts RA, Hollatz C, Trichez D, Miletti LC, de Araujo PS, et al. Molecular analysis of maltotriose active transport and fermentation by Saccharomyces cerevisiae reveals a determinant role for the AGT1 permease. Appl Environ Microbiol. 2008;74: 1494–501. 10.1128/AEM.02570-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Blount ZD, Barrick JE, Davidson CJ, Lenski RE. Genomic analysis of a key innovation in an experimental Escherichia coli population. Nature. 2012;489: 513–518. 10.1038/nature11514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fedoroff N V. Presidential address. Transposable elements, epigenetics, and genome evolution. Science. American Association for the Advancement of Science; 2012;338: 758–67. [DOI] [PubMed] [Google Scholar]
- 74.Gresham D, Desai MM, Tucker CM, Jenq HT, Pai DA, Ward A, et al. The Repertoire and Dynamics of Evolutionary Adaptations to Controlled Nutrient-Limited Environments in Yeast. Snyder M, editor. PLoS Genet. 2008;4: e1000303 10.1371/journal.pgen.1000303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dunham MJ, Badrane H, Ferea T, Adams J, Brown PO, Rosenzweig F, et al. Characteristic genome rearrangements in experimental evolution of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2002;99: 16144–16149. 10.1073/pnas.242624799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liti G, Louis EJ. Yeast evolution and comparative genomics. Annu Rev Microbiol. 2005;59: 135–153. 10.1146/annurev.micro.59.030804.121400 [DOI] [PubMed] [Google Scholar]
- 77.Magalhães F, Vidgren V, Ruohonen L, Gibson B. Maltose and maltotriose utilisation by group I strains of the hybrid lager yeast Saccharomyces pastorianus. FEMS Yeast Res. 2016;16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ehrenreich IM, Torabi N, Jia Y, Kent J, Martis S, Shapiro JA, et al. Dissection of genetically complex traits with extremely large pools of yeast segregants. Nature. 2010;464: 1039–1042. 10.1038/nature08923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Brauer MJ, Christianson CM, Pai DA, Dunham MJ. Mapping novel traits by array-assisted bulk segregant analysis in Saccharomyces cerevisiae. Genetics. 2006;173: 1813–6. 10.1534/genetics.106.057927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Segrè A V, Murray AW, Leu J-Y. High-Resolution Mutation Mapping Reveals Parallel Experimental Evolution in Yeast. PLoS Biol. 2006;4: e256 10.1371/journal.pbio.0040256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Stern DL. Identification of loci that cause phenotypic variation in diverse species with the reciprocal hemizygosity test. Trends Genet. 2014;30: 547–554. 10.1016/j.tig.2014.09.006 [DOI] [PubMed] [Google Scholar]
- 82.Henikoff S, Henikoff JG. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci U S A. 1992;89: 10915–10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5: 725–738. 10.1038/nprot.2010.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. The I-TASSER Suite: protein structure and function prediction. Nat Methods. 2015;12: 7–8. 10.1038/nmeth.3213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9: 40 10.1186/1471-2105-9-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cheng Q, Michels CA. The maltose permease encoded by the MAL61 gene of Saccharomyces cerevisiae exhibits both sequence and structural homology to other sugar transporters. Genetics. 1989;123: 477–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Barrett MP, Walmsleyt AR, Gould GW. Structure and function of facultative sugar transporters. Curr Opin Cell Biol. 1999;11: 496–502. [DOI] [PubMed] [Google Scholar]
- 88.Yan N. Structural Biology of the Major Facilitator Superfamily Transporters. Annu Rev Biophys. Annual Reviews; 2015;44: 257–283. [DOI] [PubMed] [Google Scholar]
- 89.Olga Khersonsky, Tawfik DS. Enzyme Promiscuity: A Mechanistic and Evolutionary Perspective. Annu Rev Biochem. 2010;79: 471–505. 10.1146/annurev-biochem-030409-143718 [DOI] [PubMed] [Google Scholar]
- 90.Mannige R V. Dynamic New World: Refining Our View of Protein Structure, Function and Evolution. Proteomes. 2014;2: 128–153. 10.3390/proteomes2010128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bashton M, Chothia C. The Generation of New Protein Functions by the Combination of Domains. Structure. 2007;15: 85–99. 10.1016/j.str.2006.11.009 [DOI] [PubMed] [Google Scholar]
- 92.Vogel C, Bashton M, Kerrison ND, Chothia C, Teichmann SA. Structure, function and evolution of multidomain proteins. Curr Opin Struct Biol. 2004;14: 208–216. 10.1016/j.sbi.2004.03.011 [DOI] [PubMed] [Google Scholar]
- 93.Furuta Y, Kobayashi I. Movement of DNA sequence recognition domains between non-orthologous proteins. Nucleic Acids Res. 2012;40: 9218–9232. 10.1093/nar/gks681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Doolittle RF. The multiplicity of domains in proteins. Annu Rev Biochem. 1995;64: 287–314. 10.1146/annurev.bi.64.070195.001443 [DOI] [PubMed] [Google Scholar]
- 95.Mody A, Weiner J, Ramanathan S. Modularity of MAP kinases allows deformation of their signalling pathways. Nat Cell Biol. 2009;11: 484–491. 10.1038/ncb1856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Drummond DA, Silberg JJ, Meyer MM, Wilke CO, Arnold FH. On the conservative nature of intragenic recombination. Proc Natl Acad Sci U S A. 2005;102: 5380–5. 10.1073/pnas.0500729102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Brouwers N, Vries de ARG, Broek van den M, Weening SM, Schuurman TDE, Kuijpers NGA, et al. In vivo recombination of Saccharomyces eubayanus maltose-transporter genes yields a chimeric transporter that enables maltotriose fermentation. PLOS Genetics. 2019; 15(4): e1007853 10.1371/journal.pgen.1007853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28: 1647–1649. 10.1093/bioinformatics/bts199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Parreiras LS, Breuer RJ, Avanasi Narasimhan R, Higbee AJ, La Reau A, Tremaine M, et al. Engineering and Two-Stage Evolution of a Lignocellulosic Hydrolysate-Tolerant Saccharomyces cerevisiae Strain for Anaerobic Fermentation of Xylose from AFEX Pretreated Corn Stover. PLoS One. 2014;9: e107499 10.1371/journal.pone.0107499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Alexander WG, Peris D, Pfannenstiel BT, Opulente DA, Kuang M, Hittinger CT. Efficient engineering of marker-free synthetic allotetraploids of Saccharomyces. Fungal Genet Biol. 2016;89: 10–17. 10.1016/j.fgb.2015.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gietz DR, Woods RA. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 2002;350: 87–96. [DOI] [PubMed] [Google Scholar]
- 103.Zhou X, Peris D, Kominek J, Kurtzman CP, Hittinger CT, Rokas A. in silico Whole Genome Sequencer and Analyzer (iWGS): A Computational Pipeline to Guide the Design and Analysis of de novo Genome Sequencing Studies. G3. 2016;6: 3655–3662. 10.1534/g3.116.034249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Weisenfeld NI, Yin S, Sharpe T, Lau B, Hegarty R, Holmes L, et al. Comprehensive variation discovery in single human genomes. Nat Genet. 2014;46: 1350–5. 10.1038/ng.3121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Johnson MG, Gardner EM, Liu Y, Medina R, Goffinet B, Shaw AJ, et al. HybPiper: Extracting coding sequence and introns for phylogenetics from high-throughput sequencing reads using target enrichment. Appl Plant Sci. 2016;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Slater G, Birney E. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics. 2005;6: 31 10.1186/1471-2105-6-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19: 455–477. 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32: 1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Suyama M, Torrents D, Bork P. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006;34: W609–W612. 10.1093/nar/gkl315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Strope PK, Skelly DA, Kozmin SG, Mahadevan G, Stone EA, Magwene PM, et al. The 100-genomes strains, an S. cerevisiae resource that illuminates its natural phenotypic and genotypic variation and emergence as an opportunistic pathogen. Genome Res. 2015;25: 762–74. 10.1101/gr.185538.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Charron MJ, Michels CA. The naturally occurring alleles of MAL1 in Saccharomyces species evolved by various mutagenic processes including chromosomal rearrangement. Genetics. 1988;120: 83–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bromberg Y, Rost B. Correlating protein function and stability through the analysis of single amino acid substitutions. BMC Bioinformatics. 2009;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ng PC, Henikoff S. Accounting for human polymorphisms predicted to affect protein function. Genome Res. 2002;12: 436–46. 10.1101/gr.212802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31: 3812–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ng PC, Henikoff S. Predicting the Effects of Amino Acid Substitutions on Protein Function. Annu Rev Genomics Hum Genet. 2006;7: 61–80. 10.1146/annurev.genom.7.080505.115630 [DOI] [PubMed] [Google Scholar]
- 116.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4: 1073–1081. 10.1038/nprot.2009.86 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Protein structural alignment between Malt3, Malt4, Malt434, scAgt1, and lgAgt1. The purple blocks represent predicted alpha helices, and the orange lines represent predicted beta strands. Red ticks mark predicted maltose-binding sites. Blue ticks mark residues found to be important for maltotriose transport by Smit et al. 2008. A green tick marks the location of the single non-synonymous substitution between lgAGT1 and tbAGT1. Arrows point to alpha helices in Malt434 whose predicted sizes are reduced compared to other transporters in the alignment.
(TIF)
Results of mutation prediction analyses for E18V, the sole amino acid substitution in the lgAgt1 protein sequence, relative to tbAgt1.
(XLSX)
Growth on maltose of single-colony isolates. Isolated from adaptive evolution of yHRVM108 on 2% maltose + 0.1% glucose. N = 3. * Control grown in SC + 0.04% glucose to reflect the approximate amount of growth expected from contamination with other carbon sources when using 98% pure maltotriose.
(XLSX)
Growth on maltotriose of single-colony isolates from adaptive evolution experiments. Strains were evolved with either maltotriose or maltose as the primary carbon source (2%) with 0.1% added glucose. N = 3. * Control grown in SC + 0.04% glucose to reflect the approximate amount of growth expected from contamination with other carbon sources when using 98% pure maltotriose.
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
All Illumina sequencing data have been uploaded to GenBank as BioProject PRJNA507896. All other relevant data are within the paper and its Supporting Information files.