Abstract
The permeability barrier of Gram-negative cell envelopes is the major obstacle in the discovery and development of new antibiotics. In Gram-negative bacteria, these difficulties are exacerbated by the synergistic interaction between two biochemically distinct phenomena, the low permeability of the outer membrane (OM) and active multidrug efflux. In this study, we used Pseudomonas aeruginosa and Escherichia coli strains with controllable permeability barriers, achieved through hyperporination of the OMs and varied efflux capacities, to evaluate the contributions of each of the barriers to protection from antibacterials. We analyzed antibacterial activities of β-lactams and fluoroquinolones, antibiotics that are optimized for targets in the periplasm and the cytoplasm, respectively, and performed a machine learning-based analysis to identify physicochemical descriptors that best classify their relative potencies. Our results show that the molecular properties selected by active efflux and the OM barriers are different for the two species. Antibiotic activity in P. aeruginosa was better classified by electrostatic and surface area properties, whereas topology, physical properties, and atom or bond counts best capture the behavior in E. coli. In several cases, descriptor values that correspond to active antibiotics also correspond to significant barrier effects, highlighting the synergy between the two barriers where optimizing for one barrier promotes strengthening of the other barrier. Thus, both barriers should be considered when optimizing antibiotics for favorable OM permeability, efflux evasion, or both.
Keywords: Gram-negative bacteria, outer membrane, multidrug efflux, antibiotic permeation, machine learning, physicochemical properties
Graphical Abstract
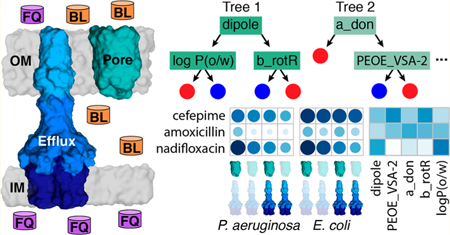
Gram-negative bacteria are notoriously more resistant to antibiotics than Gram-positive bacteria. The major reason for this resistance is that Gram-negative cell envelopes comprise two membranes of different compositions and functions.1–3 The outer membrane (OM) is an asymmetric bilayer of lipopolysaccharides (LPS) and phospholipids with nonselective porins and substrate-specific channels embedded therein.4,5 The major function of the OM is protection from toxic molecules and enzymatic attacks in a hostile environment. The inner, or cytoplasmic, membrane is a phospholipid bilayer that is responsible for diverse physiological and metabolic functions. It also contains multidrug efflux pumps that protect intracellular membrane, act synergistically and are the major factors that are responsible for the intrinsic resistance of Gram-negative bacteria to a broad range of antimicrobial agents.7,8 In addition, the sieving properties of the inner membrane, which are orthogonal to the OM, are also thought to affect the intracellular accumulation of antibiotics.2
The antibiotic resistance of Gram-negative pathogens has become particularly worrisome with the emergence of multidrug resistant strains in clinics, which often leave clinicians with no therapeutic options.9 The discovery of new antibiotics that are active against these pathogens is hindered by low hit rates in screening efforts and by the lack of practical rules to maximize functions by actively removing small, toxic molecules and peptides from the periplasm and cytoplasm.6 The two barriers, the passive, low-permeability OM and active efflux in the inner OM permeability and minimize efflux.3,10 The latter problem has been identified as a major bottleneck in addressing emerging multidrug resistance in clinics.11
To establish rules based on molecular properties that define antibiotic permeation, the two permeability barriers (OM and efflux) must be analyzed separately to define the factors contributing to each barrier.7 For this purpose, we developed a hyperporination approach that facilitates control of OM permeability in Gram-negative cells through the inducible expression of a chromosomally encoded open pore (Pore) with a 2.4 nm internal diameter.12 The expression of the Pore effectively and nonselectively allows influx of antibiotics and reduces the barrier constant, B, which is defined as the ratio of maximum attainable drug fluxes across the outer membrane into the cell and out of the cell via the efflux transporter.8 The overexpression and deletion of efflux pumps, on the other hand, allows manipulation not only of B, but also the efflux constant, KE, which measures efflux efficiency for a given antibiotic.8,13–15
The present study is focused on understanding interactions between the permeability barriers of Gram-negative bacteria and the antibacterial activities of β-lactams (BLs) and fluoroquinolones (FQs). Representatives of these antibiotic classes have been extensively developed and remain the major antibiotics administered in clinics. FQs target DNA replication by inhibiting DNA topoisomerases and, hence, to reach their targets must penetrate both the outer and inner membranes and evade efflux pumps. In contrast, transpeptidases, which are targeted by BLs, are located in the periplasm, and these antibiotics are optimized to penetrate only across the OM and to evade efflux from the periplasm. Thus, the two classes differ significantly in their structures and physicochemical properties and contain determinants that are recognized by these different barriers.
Antibacterial activities were analyzed in two Gram-negative species: P. aeruginosa and E. coli. Although the lipid compositions of the OMs overall are similar between these two species,4,16,17 they differ in the composition and structure of their major general porins. The OM of E. coli contains ∼200 000 copies per cell of OmpF/C porins, which have a molecular mass cutoff of ∼600 Da. As a result, a significant number of antibiotics are active against this species.18 In contrast, P. aeruginosa lacks such large general porins and instead utilizes substrate-specific porins of the Occ family to take up small compounds such as monosugars and amino acids.19 Nevertheless, this species is susceptible to FQs and some BLs, suggesting alternative routes of permeation across the OM. Hyperporination of the OM through the expression of large nonspecific pores negates the differences in permeabilities of the OMs in P. aeruginosa and E. coli and allows evaluation of the contributions of these barriers toward antibacterial activities.7,12
The differences between the barriers are further evident in the genetic makeup of their respective efflux pumps. In E. coli, inactivation of a single gene, tolC, leads to the complete loss of efflux across the OM because all efflux pumps capable of efflux across the OM in this species depend on TolC.20–22 In contrast, the major efflux pumps of P. aeruginosa are encoded in the same operons as specific tolC homologues, and each pump is functionally independent from the others.23 Hence, multiple pumps must be inactivated to deplete the efflux capacity of P. aeruginosa.
Identifying molecular properties that govern antibiotic activity in the presence and absence of the two barriers is expected to provide strategic guidelines for optimizing compounds against Gram-negative bacteria.2 Recently, random forest (RF) classification was used to establish a set of rules for favorable accumulation of antibiotics in E. coli.24 Molecules that are most likely to accumulate include primary or secondary amines, are rigid, have low globularity, and are amphiphilic. Although accumulation is required for antibiotic activity, it does not provide direct information about activity.
In this study, we identify molecular properties of antibiotics that are associated with their relative potencies, measured as minimum inhibitory concentrations (MICs) in P. aeruginosa and E. coli strains with controlled permeability of the OMs and variable efflux capacities. We describe the characteristics of antibiotics that display activity when both the efflux and OM barriers are removed (PΔ6-Pore and ΔTolC-Pore), when only efflux (PΔ6 and ΔTolC) or the OM barrier is removed (PAO1-Pore and WT-Pore), and in the corresponding wildtype strains (PAO1 and WT). To establish these associations, we use RF classification to extract physicochemical properties of antibiotics that separate them based on the contributions of these two barriers. Our results show that molecular properties selected by active efflux and the OM barriers are different for E. coli and P. aeruginosa.
RESULTS AND DISCUSSION
Antibiotics and Bacterial Strains Used in These Studies.
To determine how active efflux and the OM permeability barrier contribute to the activities of antibiotics, we selected 64 representatives of the BLs (cephalosporins (CEFs), penicillins (PENs), and meropenem) and FQs (Table S1). These antibiotics differ significantly in their structures and properties, ranging in molecular mass from less than 300 to 650 Da, with log D7.5 values from −3 to ∼4 and logP(o/w) values varying from −2 to 3.5 (Figure S1). In addition, we included a few representatives belonging to other classes of antibiotics that have been analyzed previously:7,12 two macrolides (azithromycin and erythromycin), the activities of which were strongly affected both by active efflux and OM permeability, chloramphenicol, which was weakly affected by both efflux and the OM barrier, and gentamicin, the activity of which was not affected by the OM and efflux.
MICs of antibiotics were measured in E. coli WT, the effluxdeficient variant ΔTolC, and the pore-producing derivatives WT-Pore and ΔTolC-Pore.12 For P. aeruginosa, four strains were also analyzed: the wild type PAO1, strain PΔ6 lacking six efflux pumps (ΔmexAB-oprM, ΔmexCD-oprJ, ΔmexXY, ΔmexJKL, ΔmexEF-oprN, ΔtriABC), and their pore-producing derivatives, PAO1-Pore and PΔ6-Pore, respectively.25 All strains were previously shown not to have significant growth defects and to differ dramatically in their susceptibilities to various classes of antibiotics.
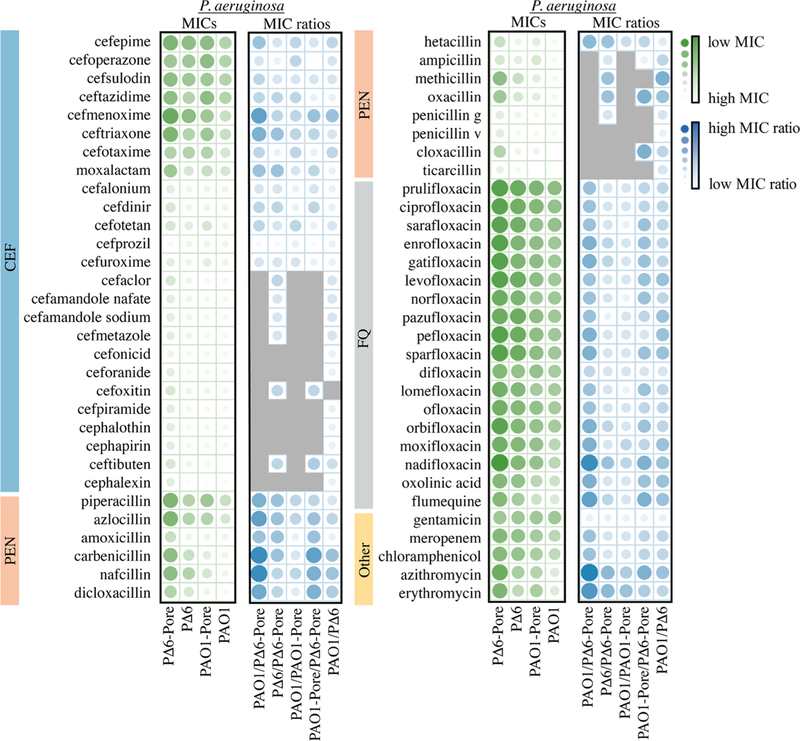
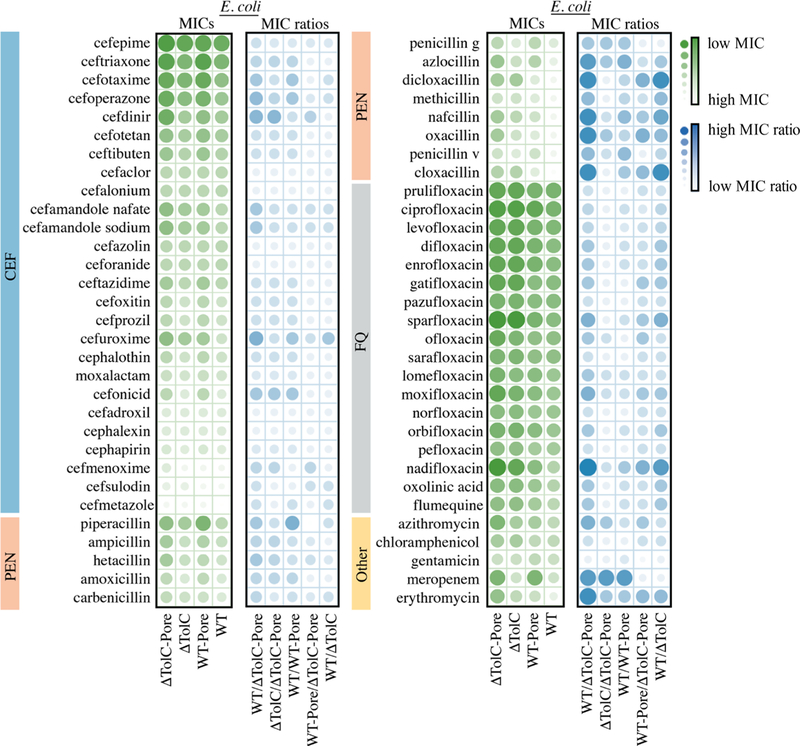
For E. coli WT cells, MICs could be measured for all tested antibiotics, whereas the MICs of ∼30% of the antibiotics were too high to be determined in P. aeruginosa PAO1 cells (Table S1 and Figures 1 and 2). However, the MICs of all antibiotics could be determined in PΔ6-Pore, highlighting the large contribution of the permeability barriers in this species toward antibiotic activities. To normalize to the differences in biochemical potency among compounds, our key measured parameters were efflux ratios and OM barrier ratios, defined as MICparent/MICmutant, for efflux mutants and hyperporinated mutants, respectively.
Figure 1.
Scaled MICs (green) and MIC ratios (blue) for antibiotics in P. aeruginosa sorted by antibiotic class and PAO1 MIC from lowest to highest within each class. Values were natural log-transformed and then scaled between 0 and 1. Gray squares indicate MIC ratios that are outside of the measurable range. MICs report on relative potency and MIC ratios report on the dependence of antibiotic activity on efflux, the OM barrier, or both.
Figure 2.
Scaled MICs (green) and MIC ratios (blue) for antibiotics in E. coli sorted by WT MIC from lowest to highest. Values were natural logtransformed and then scaled between 0 and 1. MICs report on relative potency and MIC ratios report on the dependence of antibiotic activity on efflux, the OM barrier, or both.
Cephalosporins.
These antibiotics were relatively potent against E. coli, with the lowest MICs against WT in the low nanomolar range, but not against P. aeruginosa, for which the most potent representatives had MICs in the mid to low micromolar range (Figures 1 and 2, Table S1). This gap in CEF potency can be attributed to both the species-specific differences in permeability barriers and the expression of chromosomal β-lactamases in P. aeruginosa strains.26,27 The combination of these factors resulted in about half of the analyzed CEFs lacking appreciable activities against the wildtype PAO1 strain. However, in both species, the activities of almost all CEFs were potentiated by hyperporination of the OM, inactivation of efflux, or both, albeit to different degrees. As a result, apart from the β-lactamases, all CEFs had a measurable MIC in the minimal barrier PΔ6-Pore strain with the most potent activities in the mid nanomolar range. This result suggests that in P. aeruginosa strains, the permeability barriers contribute significantly to resistance against these antibiotics.
Interestingly, in both species, CEFs were modestly (≤4-fold) affected by efflux deletions. In P. aeruginosa, the exceptions were ceftibuten (8-fold), cefotaxime and cefepime (16-fold), and cefmenoxime (64-fold), whereas in E. coli, cefuroxime was potentiated 16-fold upon efflux activation (Figure 3). In contrast to efflux, the effect of hyperporination was species-specific. With a few exceptions, the hyperporinated E. coli cells were only slightly more susceptible to CEFs (≤4-fold) than WT cells (Figures 2 and 3). Cefonicid, cefoperazone, and cefuroxime were among the most limited (16-fold) by the E. coli OM barrier (Table S1). In P. aeruginosa on the other hand, some CEFs were not significantly affected by hyperporination (e.g., cefdinir and cefalonium), whereas others were significantly limited by the OM barrier (e.g., cefotaxime and cefmenoxime).
Figure 3.
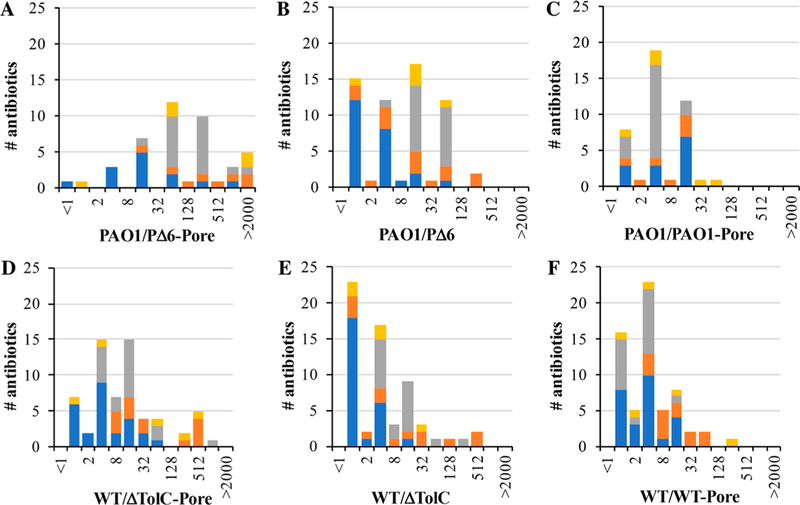
P. aeruginosa and E. coli MIC ratios colored by class (CEF = blue, PEN = orange, FQ = gray, other = gold) for (A) “barrierless” ratio (PAO1/PΔ6-Pore), (B) only efflux pump deletion (PAO1/PΔ6), and (C) only hyperporination (PAO1/PAO1-Pore), (D) “barrierless” ratio (WT/ ΔTolC-Pore), (E) only efflux pump deletion (WT/ΔTolC), (F) only hyperporination (WT/WT-Pore). Only MIC ratios with measurable values are shown. The fold changes in MICs are shown on the X-axes and the number of antibiotics with the corresponding fold changes in MICs are on Y-axes.
The increased potency of CEFs in the barrierless strains highlights the synergistic effect of active efflux and the OM barrier. In most cases, P. aeruginosa PΔ6-Pore cells were ≥16-fold more susceptible to CEFs than the wild-type PAO1 cells (Table S1), with the exceptions of cefalonium, cefoperazone, cefprozil, and cefuroxime. Ceftriaxone and cefmenoxime were the most potentiated CEFs (≥256-fold). In contrast, most CEFs were not affected significantly (≤4-fold) by removal of the OM barrier and inactivation of efflux in E. coli (e.g., cefepime and cefalonium). Other CEFs such as cefdinir and cefuroxime had activities that were potentiated greater than 32-fold upon removal of both barriers (Figures 2 and 3).
Penicillins.
Like CEFs, these antibiotics differ significantly in their activities against E. coli and P. aeruginosa and are affected by the OM, active efflux, and β-lactamases. In general, the MICs of PENs in both species were in the micromolar and millimolar ranges. However, unlike CEFs, the effects of active efflux and hyperporination on the activity of PENs spanned orders of magnitude in both species (Table S1).
PENs were either poor substrates of P. aeruginosa efflux pumps (e.g., penicillin and amoxicillin) or excellent substrates (e.g., ampicillin and methicillin) (Figures 1 and 3). Likewise, in E. coli PENs were either poor substrates of efflux pumps (e.g., amoxicillin and ampicillin) or excellent substrates (e.g., cloxacillin and dicloxacillin), the activities of which increased by 512-fold upon the removal of efflux compared to WT (Figures 2 and 3). In both P. aeruginosa and E. coli, most PENs were limited by the OM barrier but to different degrees. Some PENs, such as piperacillin, azlocillin, and nafcillin, were minimally affected by hyperporination of PAO1-Pore. In contrast, activities of other PENs including ampicillin, methicillin, and cloxacillin were all potentiated by ≥16-fold. In E. coli, greater than four-fold increases in potentiation were observed in hyperporinated cells compared to WT cells for all PENs except ampicillin, carbenicillin, and hetacillin, with maximal increases in activity of 64-fold for piperacillin and azlocillin.
PΔ6-Pore cells were in general ≥16-times more susceptible to all PENs (Figure 3). The activities of several PENs were highly limited by both barriers in P. aeruginosa including carbenicillin, nafcillin, and azlocillin (≥103-fold). The susceptibility of ΔTolC-Pore cells increased at least eight-fold compared to WT cells for all PENs, with maximum increases of 512-fold for cloxacillin, dicloxacillin, nafcillin, and oxacillin (Table S1). Thus, the structural differences of PENs and CEFs led to dramatic effects in both the antibiotic permeation across the OM and active efflux avoidance.
Fluoroquinolones.
Unlike β-lactams, FQs are highly potent against both species, but on average P. aeruginosa PAO1 was 16-fold less susceptible to these antibiotics than E. coli WT (Table S1). Accordingly, the FQ MICs in PAO1 were in the low micromolar to high nanomolar range, whereas in E. coli, with a few exceptions, the MICs of FQ were in the submicromolar range.
FQs were relatively good substrates of efflux pumps in both species. In P aeruginosa, most FQs were potentiated by at least 16-fold upon efflux deletion, except for difloxacin (4-fold). In E. coli, FQs such as sparfloxacin and nadifloxacin were good substrates of efflux pumps, as evidenced by the potentiation of their activities in ΔTolC cells (64-fold and 256-fold, respectively) (Figure 2 and Table S1). Other FQs such as prulifloxacin and sarafloxacin only showed a four-fold increase in susceptibility upon removal of efflux capabilities. Unlike β-lactams, most FQs were not significantly affected by the removal of the OM barrier (≤4-fold) in either species. However, the susceptibility of PAO1-Pore cells toward moxifloxacin and nadifloxacin increased 16-fold compared to PAO1, whereas WT-Pore was 16-fold more susceptible than WT for nadifloxacin (Table S1).
However, even such small changes in the permeation across the OM contributed significantly to the FQ potency when synergized with active efflux. In general, PΔ6-Pore cells were ≥64-times more susceptible to FQs than WT (Table S1). Furthermore, the activities of certain FQs, such as flumequine and nadifloxacin, were potentiated by more than a thousandfold in PΔ6-Pore cells compared to PAO1 cells. In E. coli ΔTolC-Pore, most FQs displayed eight-fold or greater potentiation, although norfloxacin, pefloxacin, pazufloxacin, prulifloxacin, and sarafloxacin were potentiated by only four-fold upon removal of both barriers.
Taken together, these results show that in P. aeruginosa and coli strains the MICs of tested antibiotics ranged from millimolar to subnanomolar values, and their potencies varied dramatically depending on the presence or absence of one or both permeability barriers.
Molecular Property Fingerprints of MICs and MIC Ratios.
We used RF classification to dissect the specific effects of OM permeability and efflux that limit antibiotic activity. Specifically, we identified the most important physicochemical properties, that is, those that resulted in the largest decrease in accuracy upon removal, for classifying the relative potency of the antibiotics in mutant strains and wild-type strains (as measured by MIC) and the dependence of the antibacterial activities on efflux, the OM barrier, or both (as measured by MIC ratios). We performed RF classification on 143 descriptors for P. aeruginosa and 142 descriptors for E. coli (Table 1), and we report the top 20 descriptors for classifying the MICs or MIC ratios for each species ranked by variable importance (Tables S2, S3 and S6, S7). E. coli MICs ≤ 4 μM were classified as low, that is, active. However, because P. aeruginosa shows greater resistance to antibiotics, antibiotics classified as low had MICs ≤ 20 μM. All other MICs were classified as high (i.e., inactive). For both species, MIC ratios ≤4 were classified as low, or having no significant barrier effect, and MIC ratios >4 were considered to show a significant barrier effect.
Table 1.
Descriptor Categories and Numbers of Descriptors in Each Category
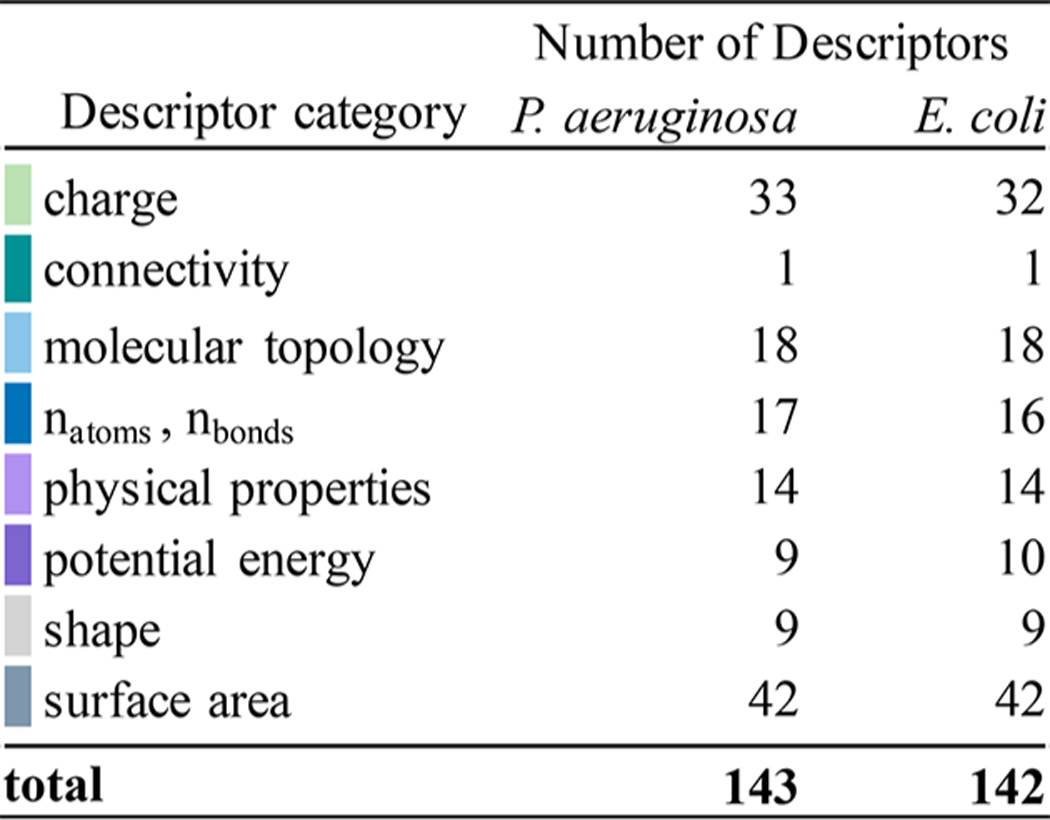 |
The most important molecular descriptors identified by RF classification provide a “fingerprint” that describes the molecular characteristics that best distinguish between high and low classifications for each set of MICs or MIC ratios (Figures S2 and S5). The descriptors belong to eight aggregate categories: charge, connectivity, molecular topology number of selected atom or bond types, physical properties, potential energy, shape, and surface area (Table 1).
Charge Properties.
These descriptors quantify electrostatic properties in a molecule or portion of a molecule. Partial charges calculated with the partial equalization of orbital electronegativity (PEOE) method28 are the most prevalent charge descriptors identified from the RF analysis. Several of these descriptors map charges to specific van der Waals surface area (VSA) regions (e.g., fractional negative or total negative VSA). This category is prominent for both P. aeruginosa and E. coli MICs and MIC ratios. Charge properties are abundant in the top descriptors for P. aeruginosa, comprising 8–10 of the top 20 descriptors for each strain (Table S2). However, for E. coli MICs, charge descriptors are less abundant (≤7 of the 20) (Table S3). Compared to MICs, the MIC ratios feature fewer charge descriptors (4–8 of the top 20 for P. aeruginosa MIC ratios and 2–5 for E. coli) (Tables S6 and S7). For both E. coli and P. aeruginosa, efflux ratios (PAO1/PΔ6, PAO1-Pore/PΔ6-Pore, WT/ΔTolC, and WT-Pore/ΔTolC-Pore) include the most charge descriptors, suggesting that electrostatic properties may help in distinguishing between antibiotics that are significantly limited by the efflux barrier and those that are not.
Atom Connectivity, Shape, and Molecular Topology.
Connectivity and topological descriptors represent molecules as graphs in which vertices correspond to atoms, and edges correspond to bonds. These descriptors are not abundant in the fingerprints of either species (Figures S2 and S5). ΔTolC-Pore is the only strain that contains a connectivity descriptor, along with the MIC ratios WT-Pore/ΔTolC-Pore and PAO1/PΔ6-Pore. Shape descriptors are present in the fingerprints of ΔTolC/ΔTolC-Pore, PAO1/PΔ6-Pore, PAO1/PAO1-Pore, and PAO1-Pore/PΔ6-Pore ratios. In addition, most MIC or MIC ratio fingerprints contain topological descriptors. Five of the top 20 descriptors for WT-Pore are topological, incorporating measures of the partition coefficient, partial charges, or polarizability. This category is also highly abundant in the E. coli ratios WT/ΔTolC-Pore and WT/WT-Pore, (8 and 7 of the top 20, respectively), indicating that molecular topology may be relevant for distinguishing antibiotics that are severely limited by the OM barrier in the presence and absence of efflux compared to WT.
Atom and Bond Counts.
Examples of these descriptors include the numbers of hydrogen bond donor atoms (a_don), aromatic rings, and oxygen atoms as well as measures of flexibility in the form of total and fractional rotatable bond counts. The fingerprints for all strains of P. aeruginosa contain a single atom or bond count descriptor, and all E. coli strains include 2–4 descriptors in this category (Figures S2 and S5). These descriptors are present in the fingerprints of all ratios except for WT/WT-Pore and PAO1/PΔ6-Pore. The top descriptor lists for ΔTolC-Pore and WT-Pore/ΔTolC-Pore each include four atom or bond count descriptors. Thus, certain descriptors in this category may be useful for classifying antibiotic activity in the absence of efflux in E. coli.
Physical Properties.
Physical properties such as molecular weight, solubility coefficient, and partition coefficient are commonly considered in drug design. Descriptors of this type are more abundant in MICs and MICs ratios for E. coli than for P. aeruginosa (Tables S2 and S3). E. coli MIC ratios all list 2–4 physical property descriptors, and the ratios WT/ΔTolC-Pore and WT/WT-Pore have the most physical descriptors (Table S7). The trend is similar for P. aeruginosa, with PAO1/PΔ6-Pore and PAO1/PAO1-Pore having the most physical descriptors among P. aeruginosa MIC ratios (Table S6). Thus, common metrics used in rational drug design classify antibiotics in the OM ratios better for E. coli than for P. aeruginosa.
Potential Energy Descriptors.
Potential energy descriptors quantify energetic contributions from, for example, van der Waals (VDW) effects or solvation. Similar to physical properties, these descriptors appear in the fingerprints for all MICs and MIC ratios in both species at least once (Figures S2 and S5). The wild-type and hyperporinated strains contain two potential energy descriptors in both species, but the effluxdeficient strains contain only a single descriptor in this category.
Surface Area Properties.
Surface area (SA) descriptors are the second most abundant descriptor category, with all MIC and ratio fingerprints including 2–8 occurrences (Tables S2, S3 and S6, S7). Many of these descriptors are based on either the total or subdivided VSA of a molecule combined with another property such as lipophilicity (SlogP and logP(o/w)), hydrophobicity, shape, or connectivity. Compared to E. coli, the respective ratios in P. aeruginosa have more descriptors in this category, highlighting the differences between the two species. The descriptor fingerprints for hyperporinated strains in both species contained five SA descriptors, suggesting that these descriptors can distinguish active from inactive antibiotics in the presence of only the efflux barrier.
P. aeruginosa and E. coli Permeability Barriers Select for Different Molecular Properties.
To provide a set of descriptor guidelines that are favorable for antibiotic potency in the presence and absence of efflux and OM barriers, we selected from among the top descriptors those that trend with MICs or MIC ratios (Figure S8). To identify the “optimal” descriptor values for active antibiotics, we determined the average best split value or threshold (Tavg) that separates high and low MICs or ratios in the RF analysis (Tables S4, S5 and S8, S9). As in previous studies, antibiotics were grouped on the basis of how their antibacterial activities were affected by the OM barrier and active efflux barriers.7
OM Barrier.
Antibiotic properties that are favorable for OM permeation were captured in the MIC fingerprints of the effluxdeficient PΔ6 and ΔTolC strains (Figure S2) and in the fingerprints of the MIC ratios for cells with hyperporinated and intact OM barriers, that is, PΔ6/PΔ6-Pore, PAO1/PAO1-Pore, ΔTolC/ΔTolC-Pore, and WT/WT-Pore (Figure S5). Antibiotics with high OM barrier ratios were classified as being strongly affected by permeation across the OM. This group includes BLs, predominantly CEFs.
The properties that trend with changes in MICs in both ΔTolC and PΔ6 strains are rigidity and VDW potential energy (E_vdw) (Table 2). The most potent antibiotics have high values for E_vdw, with average threshold values of 4.1 kcal mol−1 for ΔTolC and 4.6 kcal mol−1 for PΔ6. Molecular rigidity was quantified here as the fraction of rotatable bonds (b_rotR = brotatable/btotal). Rigid antibiotics (i.e., b_rotR below Tavg of 0.2) were more active against both E. coli and P. aeruginosa in the absence of efflux, suggesting that less flexible molecules more readily overcome the OM barrier in both species. Rigidity was previously found to be important for increasing accumulation in wild-type E. coli.24
Table 2.
Top Descriptors That Trend with MICs and MIC Ratios and Their Average Threshold Values.
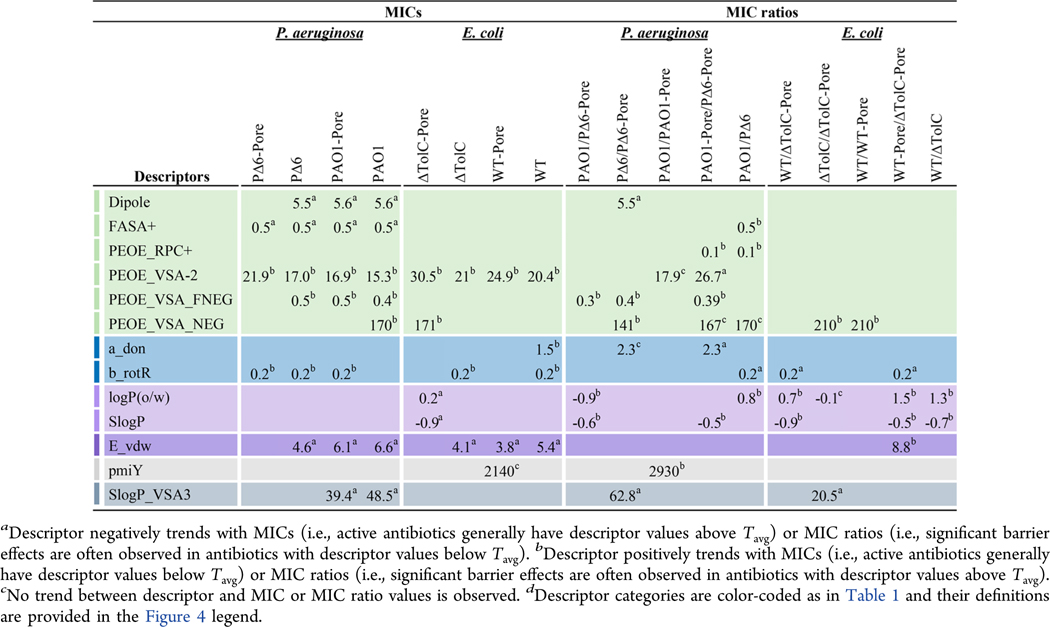 |
As mentioned previously, charge properties are more abundant in the P. aeruginosa fingerprints. Several charge descriptors trend with antibiotic potencies in PΔ6 but not in ΔTolC cells. In general, the most effective antibiotics have larger dipole moments (Tavg = 5.5 D), suggesting that molecules with greater charge separation more readily permeate the OM in P. aeruginosa. Both fractional positive water accessible SA (FASA+) and fractional negative VSA (PEOE_VSA_FNEG) provide information about favorable charge distributions in these molecules. Antibiotics with a high FASA+ (Tavg = 0.5) and a low PEOE_VSA_FNEG (Tavg = 0.5) generally have low MICs. However, these descriptors trend with MICs in other P. aeruginosa strains as well (Table 2), suggesting their importance in antibiotic permeation across both barriers.
From the high PAO1/PAO1-Pore ratios, the OM barrier of P. aeruginosa counterselects for antibiotics with a high principal moment of inertia in the Y direction (pmiY, Tavg = 2930) (Table 2). PmiY is not present in the WT/WT-Pore fingerprint and, therefore, is specific for the P. aeruginosa OM barrier. Antibiotics that show a significant barrier effect in PΔ6/PΔ6-Pore generally have high negative and fractional negative VSA (PEOE_VSA_NEG, Tavg = 141 Å2 and PEOE_VSA_FNEG, Tavg = 0.4), and low VSA with SlogP values in the range of 0 to 0.1 (SlogP_VSA3, Tavg = 62.8 Å2). The same trends are evident for PEOE_VSA_NEG (Tavg = 210 Å2) and SlogP_VSA3 (Tavg = 20.5) in the corresponding E. coli ΔTolC/ΔTolC-Pore ratio.
Thus, several descriptors that trend with MICs or MIC ratios are common for the OM barriers of both E. coli and P. aeruginosa. For example, rigid molecules more readily permeate the OM in both species. On the other hand, charge and shape descriptors are characteristic only for the OM barrier in P. aeruginosa.
Active Efflux Barrier.
Descriptors that quantify the effect of the active efflux barrier are present in the fingerprints for PAO1-Pore and WT-Pore MICs. They are also present in MIC ratios PAO1-Pore/PΔ6-Pore, PAO1/PΔ6 in P. aeruginosa, and WT-Pore/ΔTolC-Pore and WT/ΔTolC in E. coli (Figures S2 and S5). In both species, FQs are the dominant antibiotics limited by this barrier. Meropenem is known to cross the OM of P. aeruginosa using the amino acid-specific channel OprD.29 This carbapenem is a substrate of the P. aeruginosa efflux pumps. In contrast, in E. coli meropenem potency is strongly limited by OM permeation but not by efflux (Table S1).
For PAO1-Pore, active antibiotics have a balance of fractional positive and negative SA, with high FASA+ (Tavg = 0.5) and low PEOE_VSA_FNEG (Tavg = 0.5) and a high dipole moment (Tavg = 5.6 D) (Table S6). As observed with the OM barrier, antibiotics with the greatest activity against PAO1-Pore are more rigid (b_rotR Tavg = 0.2).
The top descriptors that trend with the PAO1/PΔ6 ratio are primarily charge descriptors. Interestingly, relative positive partial charges (PEOE_RPC+, Tavg = 0.1) trend with both the PAO1/PΔ6 and PAO1-Pore/PΔ6-Pore efflux ratios, but not with the OM ratios. Thus, P. aeruginosa efflux pumps may select for antibiotics with high positive partial charges. In addition, rigid antibiotics have higher PAO1/PΔ6 ratios, suggesting that P. aeruginosa efflux pumps may favor rigid molecules. Lipophilicity trends with PAO1-Pore/PΔ6-Pore ratios. The more lipophilic molecules tend to have higher values for this ratio. Thus, a metric such as SlogP (Tavg = −0.53), which is often considered for maximizing membrane permeability, may also promote efflux in P. aeruginosa.
In E. coli, active antibiotics in WT-Pore have E_vdw values above ∼4 kcal mol−1. However, charge descriptors are mostly absent for this strain and do not show any notable trends with MICs or ratios in E. coli (Table 2). Antibiotics that are significantly limited by active efflux in the presence of the OM barrier in E. coli (WT/ΔTolC) have logP(o/w) and SlogP values greater than 1.3 and −0.7, respectively, indicating that lipophilicity plays a role in efflux pump specificity. Similarly, antibiotics with high WT-Pore/ΔTolC-Pore ratios have logP(o/w) and SlogP values greater than the average thresholds of 1.5 and −0.5, respectively.
Thus, the lipophilic properties identified by RF for active efflux are similar in both species. These descriptors may be useful for guiding the prediction of antibiotic potencies and the effects of efflux. In addition, partial positive charges in antibiotics are selected by active efflux in P. aeruginosa.
OM and Active Efflux Synergy.
Some antibiotics are strongly affected both by hyperporination and efflux inactivation. In both species, FQs, macrolides, and BLs are all included in this group. Antibiotics with activities that were significantly affected by the removal of both barriers (PAO1/PΔ6-Pore and WT/ΔTolC-Pore) generally have positive logP(o/w) values (Table 2). Interestingly, the average threshold for logP(o/w) in WT/ΔTolC-Pore (Tavg = 0.7) is higher than for PAO1/PΔ6-Pore (Tavg = −0.9). Lipophilicity (SlogP) values greater than the average threshold are associated with high PAO1/PΔ6-Pore and WT/ΔTolC-Pore ratios with values of −0.6 and −0.9, respectively. In E. coli, b_rotR is an important feature for describing the differences in antibiotic effectiveness between the maximal and minimal barrier strains. In general, rigid molecules have lower WT/ΔTolC-Pore ratios (Tavg = 0.2), and thus antibiotics below this threshold value were often limited by both barriers.
PΔ6-Pore and ΔTolC-Pore are minimal-barrier strains in which both barriers have been removed. The positive trends of MICs in these strains with the lipophilic descriptors logP(o/w) and SlogP and the rigidity descriptor b_rotR are consistent with the trends in the respective MIC ratios described above (Table 2). In addition, FASA+ is also a top descriptor for PΔ6-Pore, with active antibiotics having higher values than Tavg = 0.5.
The synergy between the OM and active efflux barriers is also captured by descriptors that trend with MICs in more than one strain (Table 2). High values of SlogP_VSA3 characterize antibiotics with low MICs in the efflux-proficient strains of P. aeruginosa (Tavg = 40–50). E_vdw is present in the fingerprints of all strains except ΔTolC-Pore and PΔ6-Pore, suggesting the importance of this descriptor for both the OM barrier and active efflux in the two species. In general, higher values of E_vdw correspond to effective antibiotics (Tavg = 3.8–6.6 kcal mol−1). Some descriptors appear in the MIC fingerprints of all strains, suggesting they are important features both for permeation and target inhibition (Table 2).
Taken together, these results show that several top descriptors identified by RF classification trend with MICs or MIC ratios in P. aeruginosa and E. coli and that these descriptors vary between species and barriers. In several cases, optimizing for one barrier promotes strengthening of the other barrier, suggesting a synergistic relationship between the OM and efflux barriers. For example, rigid antibiotics (e.g., FQs) are more active against PΔ6 and thus more readily permeate the OM in P. aeruginosa, but these antibiotics are also often excellent substrates of efflux pumps. The activity of antibiotics in P. aeruginosa is primarily captured by charge and surface area descriptors, whereas properties identified for the E. coli barriers point to the role of topology, physical properties, and atom or bond counts. The calculated threshold values of these descriptors provide guidelines that may be useful for selecting or designing antibiotics with favorable properties to overcome these barriers.
Chemical Structure and Descriptor Relationships.
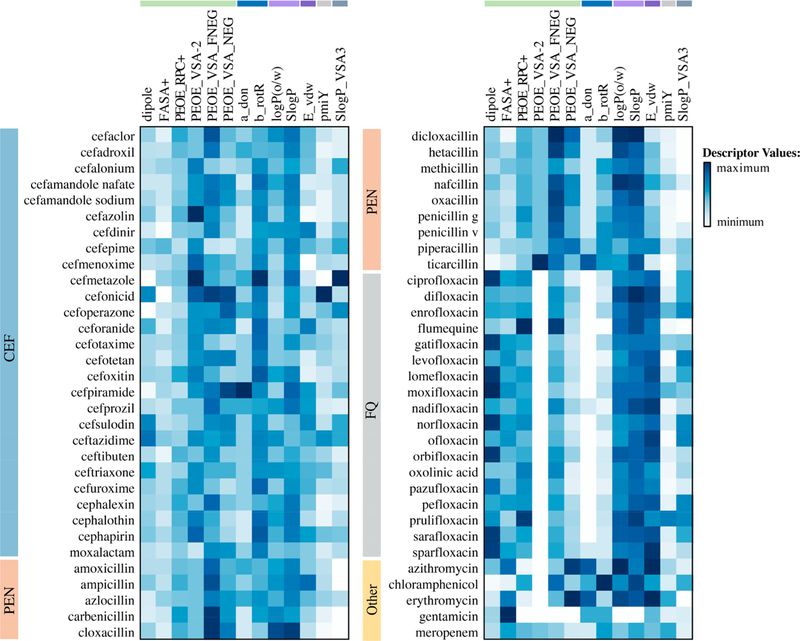
The top descriptors described above are sensitive to small changes in the chemical structures of antibiotics. For example, the CEFs ceftriaxone and cefepime (Table 3) differ by the cyclic substitutions at position 3 of the cephalosporin nucleus. These two antibiotics were active against both E. coli and P. aeruginosa but differed significantly in the effects of efflux and the OM barrier (Figures 1 and 2). Cefepime, with a methylpyrrolidine substitution at position 3, was only weakly sensitive to efflux or the OM barrier in E. coli but was affected strongly by both barriers in P. aeruginosa. In the case of P. aeruginosa, cefmenoxime, which has a methyltetrazole group at this position, was further affected by both barriers with a thousandfold decrease in activity compared to WT. Although cefepime and cefmenoxime have similar values for FASA+, PEOE_RPC+, a_don, and b_rotR, cefmenoxime has a higher fractional and total negative SA, whereas cefepime has a higher dipole moment, E_vdw, and lipophilicity (Figure 4 and Table S1).
Table 3.
Example CEFs, PENs, and FQs Highlighting How Small Changes in Antibiotic Structure Contribute to Differences in MIC Ratios and Molecular Descriptors
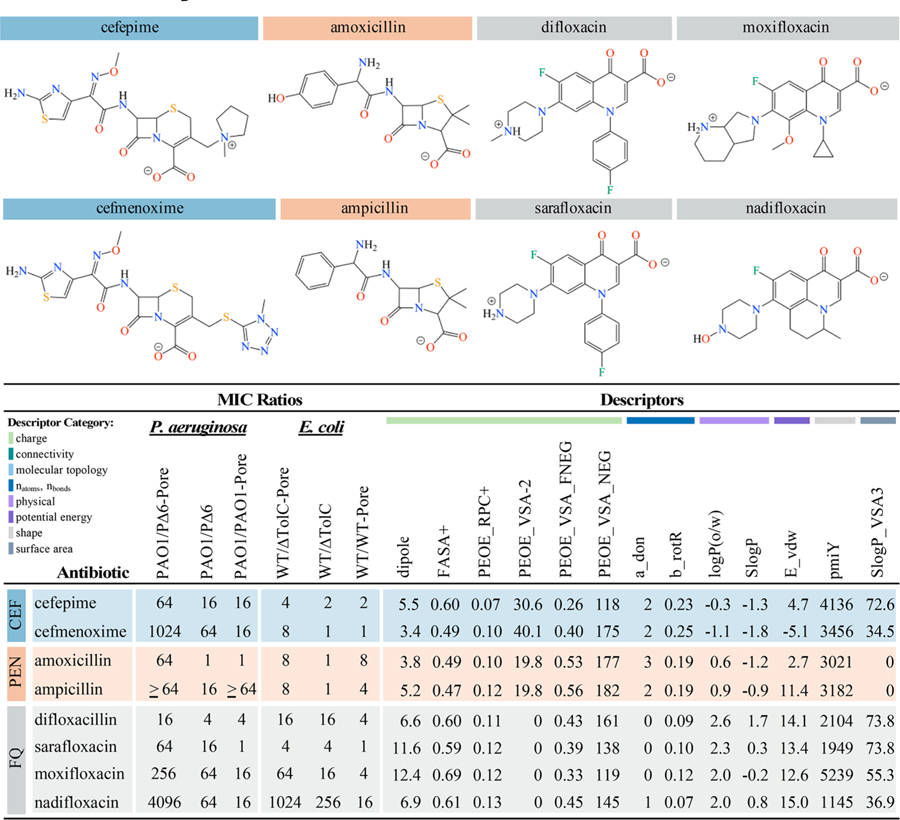 |
Figure 4.
Selected top molecular descriptors from RF classification of antibiotics with activity against P. aeruginosa and E. coli. Descriptor values were scaled between 0 and 1 and colored blue, with darker blue indicating higher descriptor values. Minimal values for a given descriptor are shown in white. Descriptors: dipole (dipole moment calculated from partial charges of the molecule), FASA+ (fractional positive water accessible surface area), PEOE_RPC+ (Relative positive partial charge), PEOE_VSA-2 (amount of van der Waals surface area with partial charge between −0.15 and −0.10), PEOE_VSA_FNEG (fractional negative van der Waals surface area), PEOE_VSA_NEG (total negative van der Waals surface area), a_don (number of hydrogen bond donor atoms), b_rotR (fraction of rotatable bonds), logP(o/w) (predicted log of the octanol/water partition coefficient), SlogP (predicted log of the octanol/water partition coefficient calculated as described in38), E_vdw (van Der Waals potential energy), pmiY (Y component of the principal moment of inertia), SlogP_VSA3 (VDW surface area with SlogP values ranging from 0 to 0.1). Descriptor categories are color-coded as in Table 1.
The aminopenicillins amoxicillin and ampicillin differ by the presence of a hydroxy group at C4 in the phenyl ring (Table 3). These two penicillins had high MICs in PAO1, but both are relatively active against WT (Figures 1 and 2). In E. coli, the activity of these antibiotics was mainly limited by the OM barrier. For P. aeruginosa, either inactivation of efflux or hyperporination was required to obtain a measurable MIC for ampicillin. In contrast, both inactivation of efflux and hyperporination were needed to obtain a measurable MIC for amoxicillin. These two antibiotics have similar descriptor values for several properties such as b_rotR, SlogP_VSA3, and FASA +, but ampicillin has a higher dipole moment, lipophilicity, E_vdw, and pmiY (Figure 4). Amoxicillin has a higher a_don as a result of its additional hydroxyl group, which may decrease the effect of the OM barrier, at least in E. coli.
For FQs, difloxacin differs from sarafloxacin by the presence of a methyl group at N4 in the piperazinyl ring and sarafloxacin is a stronger base and is more hydrophilic (Table 3). Difloxacin has a higher total and fractional negative SA, SlogP, and pmiY, and a lower dipole moment than sarafloxacin (Figure 4 and Table S1). In P. aeruginosa, both of these antibiotics were affected by the removal of both barriers, but only sarafloxacin was significantly limited by efflux alone. In both species, hyperporination did not greatly limit antibiotic activity (≤4-fold change) (Figure 1). However, in E. coli active efflux provided a significant barrier to overcome for difloxacin but not sarafloxacin (Figure 2).
The activities of most FQs were mainly limited by active efflux. The exceptions are moxifloxacin and nadifloxacin, which also showed OM barrier limitations and an even greater increase in activity in the minimal-barrier strains, reflecting the synergy of these barriers (Table 3). Several of the top descriptors differ between these two antibiotics. For example, moxifloxacin has a higher dipole moment, pmiY, and SlogP_VSA3, but nadifloxacin has a higher negative SA and an additional hydrogen bond donor. Moxifloxacin, a fourthgeneration FQ belonging to the 6-hydrogenquinolones, bears a cyclopropyl group at N1 coupled with a C8 methoxy and a pyrrolopyridine at C7 (Table 3). The bulky heterocyclic group and overall lipophilicity strongly affect both the permeation across the OM and the effect of active efflux. Chemically, nadifloxacin has a lipophilic tricyclic benzoquinolizine core, with a 4-hydroxypiperidino moiety at the C8 position (Table 3). This singular moiety lacks a distal basic functionality, which is unusual for a side chain of a quinolone, as all marketed quinolones bear side chains with a basic functionality, thereby providing two or three ionizable groups compared with only one for nadifloxacin (pKa = 6.8). Thus, even small changes in the chemical structures of antibiotics such as a single substituent change can significantly affect one or more descriptors that capture the behavior of the OM barrier, active efflux, or both.
CONCLUSIONS
The OM barrier and efflux synergistically limit antibiotic activity in Gram-negative bacteria. In the present study, we have used P. aeruginosa and E. coli strains with controlled OM permeability and varying efflux capacity to identify trends in the antibacterial activities of CEFs, PENs, and FQs. In addition to high-affinity target binding, OM permeation should be maximized and efflux should be minimized to obtain optimal antibacterial activity. Using RF classification, we have identified properties that distinguish between antibiotics with high and low antibiotic activities (MICs) in the various strains and highlight the properties related to the effect of altering the OM permeability and efflux barriers (MIC ratios).
The top descriptors for P. aeruginosa are dominated by electrostatic properties. Active antibiotics have a higher dipole moment and antibiotics with low dipole moments are significantly limited by the OM barrier. On the other hand, partial positive charges trend specifically with the efflux ratios. Rigid antibiotics and antibiotics with high VDW energies (E_vdw) were more active against both E. coli and P. aeruginosa. Lipophilicity (logP(o/w) and SlogP) trends positively with efflux ratios in both species and classify antibiotics in the OM ratios better for E. coli than for P. aeruginosa, likely the result of the more complex barriers in P. aeruginosa.
The descriptors identified here reflect chemical bias toward CEFs, PENs, and FQs. However, performing a similar predictive analysis on diverse molecular libraries is expected to be beneficial for identifying new antibiotics. Our results suggest that different properties need to be targeted for optimization of antibiotics against P. aeruginosa and E. coli species. Furthermore, we have shown that the properties selected as important for the OM barrier ratios differ significantly from those for the efflux ratios and that these two barriers work together to protect cells from antibiotics. Therefore, antibiotics should be optimized to evade efflux and enhance OM permeability simultaneously.
METHODS
P. aeruginosa and E. coli strains used in this study are listed in Table 4. All strains were grown in Luria–Bertani broth at 37 °C with shaking. Susceptibilities of cells to different classes of antibiotics were determined by 2-and 4-fold dilutions as described previously.7,12 Therefore, MIC ratios of 2–4-fold changes are within error of the assay. For RF classification of MICs and MIC ratios, the lowest MIC in the range was used. Sixty-two antibiotics were included in the analysis for P. aeruginosa and E. coli. Antibiotics were purchased from MicroSource Discovery Systems, Inc. and Sigma-Aldrich. All minimal inhibitory concentration (MICs) determinations were done at least twice.
Table 4.
Strains Used in This Study
| strains | description | source |
|---|---|---|
| E. coli | ||
| BW 25113 | Wild-type strain Δ(araD-araB)567 Δ(rhaD-rhaB)568 ΔlacZ4787 (::rrnB- 3) hsdR514 rph-1 | 39 |
| GD102 | BW25113 ΔtolC-ygiBC | 40 |
| GKCW101 (WT-Pore) | BW25113 attTn7::mini-Tn7T-Kanr- araC-ParaBAD-fhuAΔCΔ4L | 12 |
| GKCW102 (WT) | BW25113 attTn7::mini-Tn7T -Kanr -araC-ParaBAD-MCS | 12 |
| GKCW103 (ΔTolC- Pore) | GD102 attTn7::mini-Tn7T-Kanr -araCParaBAD-fhuAΔCΔ4L | 12 |
| GKCW104 (ΔTolC) | GD102 attTn7::mini-Tn7T -Kanr -araCParaBAD-MCS | 12 |
| P. aeruginosa | ||
| PAO1 | Wild-type strain | O. Lomovskaya |
| PAO1116 | PAO1 ΔmexAB-oprM ΔmexCD-oprJ ΔmexEF-oprN ΔmexJK ΔmexXY ΔtriABC | H. Schweizer |
| GKCW111 (PAO1) | PAO1 attTn7::mini-Tn7T-Gm-lacIq-pLAC-MCS | |
| GKCW120 (PΔ6) | PAO1116 attTn7::mini-Tn7T-Gm-lacIq-pLAC-MCS | |
| GKCW115 (PAO1- Pore) | PAO1 attTn7::mini-Tn7T- Gmr-lacIq-pLAC- fhuAΔCΔ4L | |
| GKCW122 (PΔ6-Pore) | PAO1116 attTn7::mini-Tn7T-Tpr -araC-ParaBAD-fhuAΔCΔ4L | 7 |
Three-dimensional structures of antibiotics used in RF classification were obtained from the ZINC database.30,31 Marvin calculator plugins32 were used to calculate the most likely tautomeric and protonation states at pH 7.4. Geometries were optimized with the Amber12:EHT molecular mechanics force field33 implemented in MOE version 2015.34 MOE was then used to calculate >300 2D and 3D molecular descriptors,35 and the resulting descriptor values were analyzed with respect to the MIC and MIC ratio data. Descriptors with standard deviations equal to zero were discarded. Redundant descriptors (i.e., correlation coefficients >0.85) were identified and removed using the findCorrelation function in the R package caret.36 A total of 143 descriptors were used for P. aeruginosa and 142 descriptors for E. coli (Table 1). Prior to analysis, the descriptor values were scaled and centered so they all had the same variance.
RF combines the information from numerous decision trees to obtain a consensus classification of “high” or “low” activity (MICs) or barrier effects (MIC ratios) from molecular descriptor values. At each node of a tree, RF determines which descriptor from a randomly selected subset of descriptors best separates the antibiotics in the training set classified as “high” from those that are classified as “low”. Each time a descriptor is selected as the best splitter, a best split value, or threshold (T), is obtained based on the descriptor values for that subset of antibiotics. The threshold values for each descriptor were averaged over all occurrences in each model (Tavg) to obtain general guidelines for desirable descriptor values.
R version 3.3.237 was used to perform the RF analysis of molecular descriptors. RF scripts were adapted from Richter et al.,24 and classification was performed with caret using 10-fold cross validation repeated ten times on a set of 2000 decision trees. The RF classification models were assessed with receiver operating characteristic (ROC) curves and confusion matrices. The top 20 descriptors for each set of response variables (MICs or MIC ratios) were determined by quantifying the overall variable importance of the machine learning model using the out-of-bag error, that is, the decrease in classification accuracy when a single descriptor is removed. Scatter plots containing the Tavg, Tmin, and Tmax values were generated with ggplot2 for the top 20 descriptors with natural log-transformed MICs and MIC ratios. All data files and R scripts are provided in the Supporting Information.
Supplementary Material
ACKNOWLEDGMENTS
This study was sponsored by the Department of Defense, Defense Threat Reduction Agency (HDTRA1-14-1-0019) and by NIH/NIAID Grant No. RO1AI132836. The content of the information does not necessarily reflect the position or the policy of the federal government, and no official endorsement should be inferred. S.J.C. was supported by NIH/NIGMSIMSD Grant No. R25GM086761 and a National Science Foundation Graduate Research Fellowship under Grant No. 2017219379. We thank Dr. James Aggen and Rosemarie O’Shea for their help at earlier stages of the project.
ABBREVIATIONS
- OM
outer membrane
- LPS
lipopolysaccharide
- BL
beta-lactams
- FQ
fluoroquinolones
- RF
Random Forest
- MIC
minimal inhibitory concentration
- PENs
penicillins
- CEFs
cephalosporins
- Tavg
average best split value or threshold (Tavg) that separates high and low MICs or ratios in RF analysis
- FASA+
fractional positive water accessible surface area
- PEOE_RPC+
relative positive partial charge
- PEOE_VSA-2
amount of van der Waals surface area with partial charge between −0.15 and −0.10
- PEOE_VSA_FNEG
fractional negative van der Waals surface area
- PEOE_VSA_NEG
total negative van der Waals surface area
- a_don
number of hydrogen bond donor atoms
- b_rotR
fraction of rotatable bonds
- logP(o/w)
predicted log of the octanol/water partition coefficient
- SlogP
predicted log of the octanol/water partition coefficient
- E_vdw
van Der Waals potential energy
- pmiY
Y component of the principal moment of inertia
- SlogP_VSA3
VDW surface area with slogp values ranging from 0 to 0.1
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsinfecdis.8b00036.
Descriptor fingerprints, variable importance of top 20 descriptors for MICs and MIC ratios in P. aeruginosa and E. coli strains, representative correlations of natural logtransformed MICs with top 20 descriptors (PDF) Tables with MICs, MIC ratios, and molecular descriptor values; top subsets of descriptors for MICs and MIC ratios in P. aeruginosa and E. coli strains; maximum, minimum, and average threshold values of top descriptors for MICs and MIC ratios in P. aeruginosa and E. coli strains, all data files and R scripts (ZIP)
Notes
The authors declare no competing financial interest.
REFERENCES
- (1).Nikaido H (2009) Multidrug resistance in bacteria. Annu. Rev. Biochem 78, 119–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Silver LL (2016) A Gestalt approach to Gram-negative entry. Bioorg. Med. Chem 24 (24), 6379–6389. [DOI] [PubMed] [Google Scholar]
- (3).Zgurskaya HI, Lopez CA, and Gnanakaran S (2015) Permeability Barrier of Gram-Negative Cell Envelopes and Approaches To Bypass It. ACS Infect. Dis 1 (11), 512–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Tamber S, and Hancock RE (2004) The outer membranes of Pseudomonads, in Pseudomonas (Ramos J-L, Ed.) Vol. 1, pp 575–601, Kluwer Academic/Plenum Publishers, New York. [Google Scholar]
- (5).Pages JM, James CE, and Winterhalter M (2008) The porin and the permeating antibiotic: a selective diffusion barrier in Gramnegative bacteria. Nat. Rev. Microbiol 6 (12), 893–903. [DOI] [PubMed] [Google Scholar]
- (6).Zgurskaya HI, Krishnamoorthy G, Tikhonova EB, Lau SY, and Stratton KL (2003) Mechanism of antibiotic efflux in Gramnegative bacteria. Front. Biosci., Landmark Ed 8, s862–73. [DOI] [PubMed] [Google Scholar]
- (7).Krishnamoorthy G, Leus IV, Weeks JW, Wolloscheck D, Rybenkov VV, and Zgurskaya HI (2017) Synergy between Active Efflux and Outer Membrane Diffusion Defines Rules of Antibiotic Permeation into Gram-Negative Bacteria. mBio 8 (5), e01172–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Westfall DA, Krishnamoorthy G, Wolloscheck D, Sarkar R, Zgurskaya HI, and Rybenkov VV (2017) Bifurcation kinetics of drug uptake by Gram-negative bacteria. PLoS One 12 (9), e0184671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Current Epidemiology and Growing Resistance of GramNegative Pathogens FAU: Livermore, D. M. (2012) Korean J. Intern. Med 27 (2), 128–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Lewis K (2012) Antibiotics: Recover the lost art of drug discovery. Nature 485 (7399), 439–440. [DOI] [PubMed] [Google Scholar]
- (11).A Scientific Roadmap for Antibiotic Discovery: A sustainable and robust pipeline of new antibacterial drugs and therapies is critical to preserve public health, in Antibiotic Resistance Project, The PEW Charitable Trusts; http://www.pewtrusts.org/en/projects/antibioticresistance-project (accessed June 21, 2016). [Google Scholar]
- (12).Krishnamoorthy G, Wolloscheck D, Weeks JW, Croft C, Rybenkov VV, and Zgurskaya HI (2016) Breaking the Permeability Barrier of Escherichia coli by Controlled Hyperporination of the Outer Membrane. Antimicrob. Agents Chemother 60 (12), 7372–7381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Lomovskaya O, Lee A, Hoshino K, Ishida H, Mistry A, Warren MS, Boyer E, Chamberland S, and Lee VJ (1999) Use of a genetic approach to evaluate the consequences of inhibition of efflux pumps in Pseudomonas aeruginosa. Antimicrob. Agents Chemother 43 (6), 1340–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Cao L, Srikumar R, and Poole K (2004) MexAB-OprM hyperexpression in NalC-type multidrug-resistant Pseudomonas aeruginosa: identification and characterization of the nalC gene encoding a repressor of PA3720-PA3719. Mol. Microbiol 53 (5), 1423–36. [DOI] [PubMed] [Google Scholar]
- (15).De Kievit TR, Parkins MD, Gillis RJ, Srikumar R, Ceri H, Poole K, Iglewski BH, and Storey DG (2001) Multidrug Efflux Pumps: Expression Patterns and Contribution to Antibiotic Resistance in Pseudomonas aeruginosa Biofilms. Antimicrob. Agents Chemother 45 (6), 1761–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Bystrova OV, Lindner B, Moll H, Kocharova NA, Knirel YA, Zahringer U, and Pier GB (2004) Full structure of the lipopolysaccharide of Pseudomonas aeruginosa immunotype 5. Biochemistry (Moscow) 69 (2), 170–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Nikaido H (2003) Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol. Biol. Rev 67 (4), 593–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Nikaido H (1994) Porins and specific diffusion channels in bacterial outer membranes. J. Biol. Chem 269 (6), 3905–8. [PubMed] [Google Scholar]
- (19).Chevalier S, Bouffartigues E, Bodilis J, Maillot O, Lesouhaitier O, Feuilloley MGJ, Orange N, Dufour A, and Cornelis P (2017) Structure, function and regulation of Pseudomonas aeruginosa porins. FEMS Microbiol Rev 41 (5), 698–722. [DOI] [PubMed] [Google Scholar]
- (20).Fralick JA (1996) Evidence that TolC is required for functioning of the Mar/AcrAB efflux pump of Escherichia coli. J. Bacteriol 178 (19), 5803–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Morona R, Manning PA, and Reeves P (1983) Identification and characterization of the TolC protein, an outer membrane protein from Escherichia coli. J. Bacteriol 153 (2), 693–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Zgurskaya HI, Krishnamoorthy G, Ntreh A, and Lu S (2011) Mechanism and Function of the Outer Membrane Channel TolC in Multidrug Resistance and Physiology of Enterobacteria. Front. Microbiol 2, 189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Poole K, and Srikumar R (2001) Multidrug efflux in Pseudomonas aeruginosa: components, mechanisms and clinical significance. Curr. Top. Med. Chem 1 (1), 59–71. [DOI] [PubMed] [Google Scholar]
- (24).Richter MF, Drown BS, Riley AP, Garcia A, Shirai T, Svec RL, and Hergenrother PJ (2017) Predictive compound accumulation rules yield a broad-spectrum antibiotic. Nature 545, 299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Wolloscheck D, Krishnamoorthy G, Nguyen J, and Zgurskaya HI (2018) Kinetic control of quorum sensing in Pseudomonas aeruginosa by multidrug efflux pumps. ACS Infect. Dis 4, 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Zhang Y, Bao Q, Gagnon LA, Huletsky A, Oliver A, Jin S, and Langaee T (2010) ampG Gene of Pseudomonas aeruginosa and Its Role in β-Lactamase Expression. Antimicrob. Agents Chemother 54 (11), 4772–4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Juan C, Macia MD, Gutieŕrez O, Vidal C, Peŕez JL, and Oliver A (2005) Molecular Mechanisms of β-Lactam Resistance Mediated by AmpC Hyperproduction in Pseudomonas aeruginosa Clinical Strains. Antimicrob. Agents Chemother 49 (11), 4733–4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Gasteiger J, and Marsili M (1980) Iterative partial equalization of orbital electronegativity a rapid access to atomic charges. Tetrahedron 36 (22), 3219–3228. [Google Scholar]
- (29).Ochs MM, McCusker MP, Bains M, and Hancock REW (1999) Negative Regulation of the Pseudomonas aeruginosa Outer Membrane Porin OprD Selective for Imipenem and Basic Amino Acids. Antimicrob. Agents Chemother 43 (5), 1085–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Irwin JJ, and Shoichet BK (2005) ZINC – A Free Database of Commercially Available Compounds for Virtual Screening. J. Chem. Inf. Model 45 (1), 177–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Irwin JJ, Sterling T, Mysinger MM, Bolstad ES, and Coleman RG (2012) ZINC: A Free Tool to Discover Chemistry for Biology. J. Chem. Inf. Model 52 (7), 1757–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).(2017) Marvin 17.9.0, ChemAxon, Cambridge, MA. [Google Scholar]
- (33).Gerber PR, and Moeller K (1995) MAB, a generally applicable molecular force field for structure modelling in medicinal chemistry. J. Comput.-Aided Mol. Des 9, 251–268. [DOI] [PubMed] [Google Scholar]
- (34).(2015) Molecular Operating Environment (MOE), 2012.10, Chemical Computing Group Inc., Montreal, QC, Canada. [Google Scholar]
- (35).Leach AR, and Gillet VJ (2007) An Introduction to Chemoinformatics, Springer Publishing Company, Inc.. [Google Scholar]
- (36).Kuhn M, Wing J, Weston S, Williams A, Keefer C, Engelhardt A, Cooper T, Mayer Z, and Kenkel B (2016) caret: Classification and Regression Training. R package version 6.0-73, R Project, https://CRAN.R-project.org/package=caret.
- (37).R Development Core Team (2011) R: A language and environment for statistical computing, R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- (38).Wildman SA, and Crippen GM (1999) Prediction of Physicochemical Parameters by Atomic Contributions. J. Chem. Inf. Comput. Sci 39, 868–873. [Google Scholar]
- (39).Datsenko KA, and Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A 97 (12), 6640–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Dhamdhere G, Krishnamoorthy G, and Zgurskaya HI (2010) Interplay between drug efflux and antioxidants in Escherichia coli resistance to antibiotics. Antimicrob. Agents Chemother 54 (12), 5366–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.