Summary
Adult neural stem cells and multiciliated ependymal cells are glial cells essential for neurological functions. Together, they make up the adult neurogenic niche. Using both high-throughput clonal analysis and single-cell resolution of progenitor division patterns and fate, we show that these two components of the neurogenic niche are lineally related: adult neural stem cells are sister cells to ependymal cells, whereas most ependymal cells arise from the terminal symmetric divisions of the lineage. Unexpectedly, we found that the antagonist regulators of DNA replication, GemC1 and Geminin, can tune the proportion of neural stem cells and ependymal cells. Our findings reveal the controlled dynamic of the neurogenic niche ontogeny and identify the Geminin family members as key regulators of the initial pool of adult neural stem cells.
Keywords: neural stem cells, multiciliated ependymal cells, radial glial cells, cell lineage, symmetric division, geminin family, GemC1, multiciliogenesis, cell division
Highlights
-
•
High-throughput clonal analysis shows a common origin of ependymal and B1 stem cells
-
•
Ependymal cells are mostly produced through symmetric divisions
-
•
B1 cells are generated through asymmetric divisions with ependymal cells
-
•
Geminin family members tune the proportion of ependymal and B1 stem cells in clones
Adult neurogenesis persists in mammals. Ortiz-Álvarez et al. analyze the clonal relationship among ependymal cells and B1 astrocytes, which together compose the adult neurogenic niche. They show that their proportion is tuned by the DNA replication regulators Geminin and GemC1.
Introduction
Neurons and glial cells are continuously produced throughout life. In the adult, a subpopulation of astrocytes (type B1) located in the ventricular-subventricular zone (V-SVZ) region of the lateral ventricles (LVs) retain stem cell properties; i.e., self-renewal and multilineage differentiation (Doetsch et al., 1999). These cells have a multipolar shape, contact both the LV and the blood vessels, and are surrounded by multiciliated ependymal cells (Shen et al., 2008, Tavazoie et al., 2008, Mirzadeh et al., 2008). The coordinated beating of ependymal cilia contributes to cerebrospinal fluid (CSF) dynamics, which is crucial for the exposure of type B1 cells to trophic and metabolic signals and to clear toxins and waste from the brain (Spassky and Meunier, 2017). Proper functioning of adult neurogenesis thus depends on the production and positioning of the controlled number of ependymal cells and type B1 astrocytes composing the neurogenic niche.
Type B1 astrocytes and ependymal cells are both derived from radial glial cells (RGCs) between embryonic day 13.5 (E13.5) and E15.5 and progressively acquire identical phenotypic markers (Sox2, Sox9, Nestin, and CD133) (Ferri et al., 2004, Mirzadeh et al., 2008, Sun et al., 2017). However, in the adult, these cells have very different morphologies and fulfill different functions: e.g., although B1 astrocytes are reactivable quiescent neuronal progenitors, multiciliated ependymal cells are postmitotic throughout life (Fuentealba et al., 2015, Furutachi et al., 2015, Shah et al., 2018, Spassky et al., 2005). It is totally unknown how these cells are allocated to the neurogenic niche and how they acquire their common characteristics and distinct identities and functions.
Recent studies have demonstrated that GemC1 and Mcidas are early regulators of multiciliogenesis in different organs (Arbi et al., 2016, Boon et al., 2014, Kyrousi et al., 2015, Ma et al., 2014, Stubbs et al., 2012, Terré et al., 2016, Zhou et al., 2015). Interestingly, these coiled-coil proteins, together with their antagonist Geminin, are part of the Geminin superfamily, which was initially characterized for its role in DNA replication control (Balestrini et al., 2010, McGarry and Kirschner, 1998, Pefani et al., 2011). More recently, Geminin was found to regulate neural cell fate and to be highly expressed in cycling type B1 cells in the adult SVZ (Khatri et al., 2014, Sankar et al., 2016).
Here we exploited high-resolution lineage-tracing techniques—multi-addressable genome-integrative color (MAGIC) markers (Loulier et al., 2014) and mosaic analysis with double markers (MADM) strategies (Gao et al., 2014)—in the mouse brain to show that type B1 astrocytes and ependymal cells share a common RGC progenitor. These RGCs first produce type B1 astrocytes through both symmetric and asymmetric divisions. Ultimately, ependymal cells are produced through terminal symmetric division. We also examined the role of antagonist regulators of DNA replication (GemC1 and Geminin) in lineage progression. We show that GemC1 promotes premature symmetric division of RGCs producing ependymal cells at the expense of astrocytes, whereas Geminin favors symmetric divisions producing type B1 astrocytes. Altogether, we show that ependymal cells and type B1 astrocytes share a common lineage in which type B1 cells are produced first, followed by a majority of ependymal cells. This dynamic can be modulated by the Geminin family members.
Results
Ependymal Cells Originate from Locally Differentiated RGCs
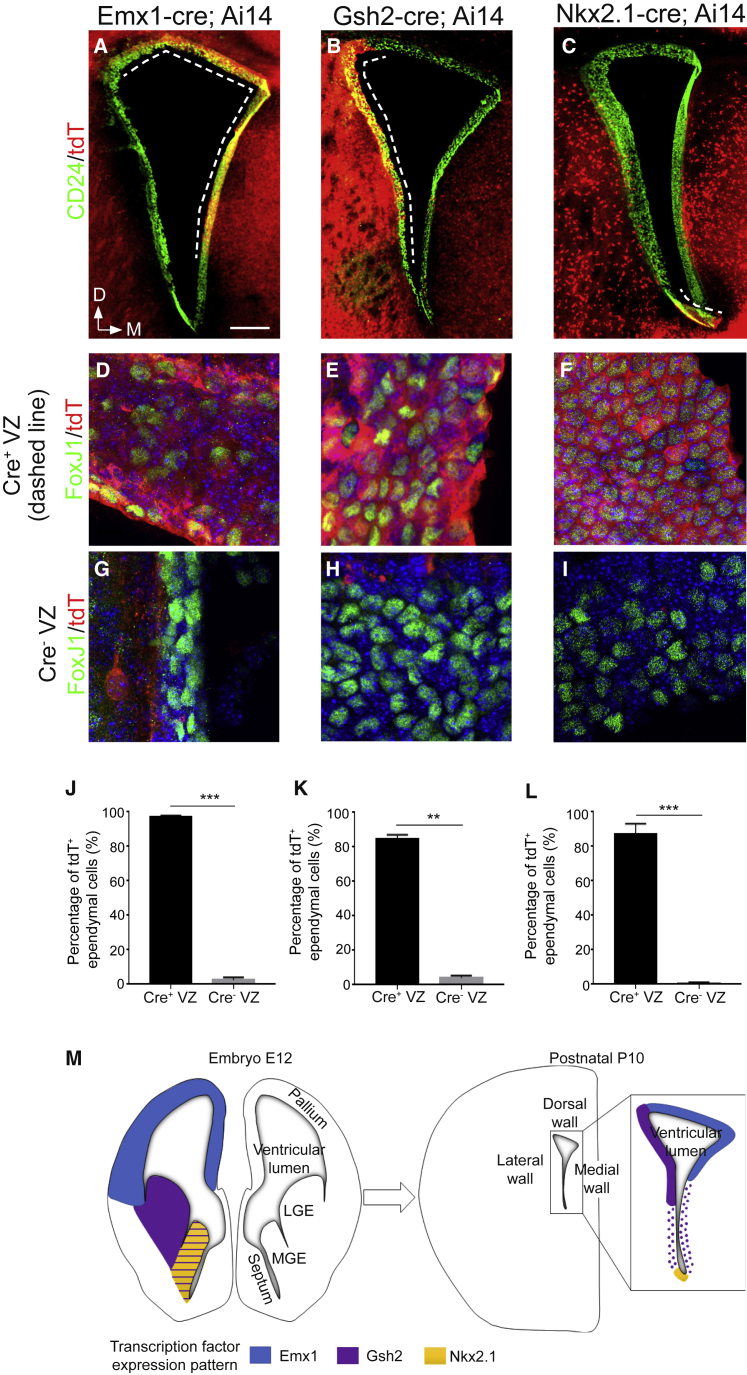
Multiciliated ependymal cells are generated from RGCs around E15 (Spassky et al., 2005). To determine how these cells develop, we performed a single injection of 5-ethynyl-2’-deoxyuridine (EdU) at E15.5 and studied the relative positions of EdU+ ependymal cells on the ventricular walls at post-natal day 15 (P15). EdU+ ependymal cells were often juxtaposed or close to each other (Figures S1A–S1C). To quantitatively assess their spatial distribution, we performed a nearest neighbor distance (NND) analysis on the datasets. The NNDs among EdU+ ependymal cells were significantly shorter than in simulated random datasets, suggesting that ependymal cells born at the same time remain in the same area (Figure S1D). To further test this possibility, we employed a genetic fate-tracing strategy. We crossed the Ai14 transgenic mouse line, which expresses tdTomato after Cre-dependent excision of a “floxed stop” cassette (Madisen et al., 2010), with Emx1-Cre, Gsh2-Cre, or Nkx2.1-Cre transgenic mice, which express Cre recombinase in the dorsal-medial, lateral, and ventral regions of the LVs, respectively (Figures 1A–1C). At P10, almost all ependymal cells were tdT+ in Cre-expressing ventricular walls (Figures 1D–1F), whereas they were tdT− in Cre-negative regions (Figures 1G–1I), showing that ependymal cells do not migrate out of their site of origin during maturation (Figures 1J–1M). We observed similar results in all caudo-rostral regions examined. Together, these results show that ependymal cells are produced locally and do not migrate long distances from their site of origin.
Figure 1.
Ependymal Cell Progenitors Are Produced Locally along the Brain Ventricles
(A–C) Representative images of coronal sections of Emx1-Cre; Ai14 (A), Gsh2-Cre; Ai14 (B), and Nkx2.1-Cre; Ai14 (C) forebrain at P10, immunostained with CD24 (green) and DsRed (tdT, red) antibodies. CD24+tdT+ ependymal cells are only observed in the Cre-expressing domains in each mouse line (indicated by a dashed line).
(D–I) Representative high-magnification images of the Emx1-Cre; Ai14 (D and G), Gsh2-Cre; Ai14 (E and H), and Nkx2.1-Cre; Ai14 (F and I) coronal sections immunostained with FoxJ1 (green) and DsRed (tdT, red) antibodies in the Cre+ domains (D–F) or Cre− domains (G–I), respectively. In the Cre+ domains, almost all ependymal cells are tdT+, whereas very few cells are double-labeled in the Cre− domains in each mouse line.
(J–L) Quantification of the mean percentage of tdT+ ependymal cells in different areas of the ventricular zone from n = 6, n = 4, and n = 5 mice from each of the three transgenic mouse lines: Emx1-Cre; Ai14 (J), Gsh2-Cre; Ai14 (K), and Nkx2.1-Cre; Ai14 (L), respectively. Error bars indicate the SEM. The p values were determined with a Mann-Whitney test; ∗∗p ≤ 0.01 and ∗∗∗p ≤ 0.001.
(M) Schematic of the expression patterns of each transcription factor in the mouse forebrain at E12 and model of the spatial origin of ependymal cells at P10. D, dorsal; M, medial.
The scale bars represent 200 μm (A–C) and 10 μm (D–I).
IUE Labels Ependymal Cells and Type B1 Astrocytes in the V-SVZ
Given that ependymal cells develop locally from RGCs, we labeled their progenitors at E14.5 in the lateral ganglionic eminence (LGE) by in utero electroporation and traced their lineage at later stages. We first verified that cells targeted by in utero electroporation (IUE) are cycling by injecting EdU at E13.5 or E14.5. The next day, 78% ± 2% of electroporated cells were indeed EdU+ (Figure S2), confirming that cycling cells are preferentially transfected by IUE and that progenitor fate can be traced by this technique, as shown previously (Loulier et al., 2014, Stancik et al., 2010).
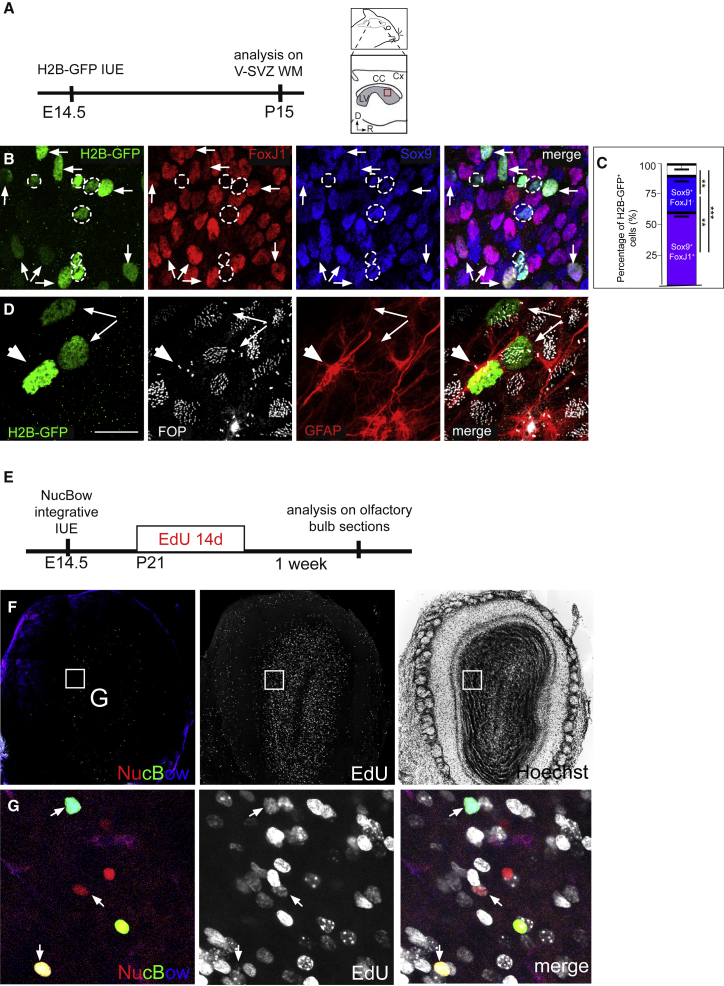
We then characterized the progeny of cells electroporated at E14.5 with the H2B-GFP plasmid by immunostaining the V-SVZ at P10–P15 with FoxJ1 and Sox9 antibodies to distinguish ependymal cells (FoxJ1+Sox9+) from other glial cells (FoxJ1−Sox9+) (Sun et al., 2017; Figures 2A and 2B). We observed that around two-thirds of GFP+ cells were ependymal cells, whereas most of the remaining FoxJ1− cells were Sox9+ astrocytes (Figure 2C). We also performed FGFR1OP (FOP) and glial fibrillary acidic protein (GFAP) staining to distinguish ependymal cells (multiple FOP+ basal bodies and GFAP−) from astrocytes (FOP+ centrosome and GFAP+). Most electroporated cells close to the ventricular surface were either GFAP− ependymal cells containing multiple FOP+ basal bodies or GFAP+ astrocytes with one FOP+ centrosome (Figure 2D). A ventricular contact emitting a primary cilium was also observed on GFP+ astrocytes (Doetsch et al., 1999). The GFP+ astrocytes often had an unusual nuclear morphology with envelope invaginations, as reported recently (Cebrián-Silla et al., 2017). Noteworthy, neuroblasts with their typical migratory morphology were observed deeper in the tissue and at a distance from the electroporated area in the direction of the olfactory bulb (data not shown).
Figure 2.
Radial Glial Cells Generate Ependymal Cells and Adult Neural Stem Cells (Type B1 Astrocytes)
(A) Experimental schematic for (B)–(D). The H2B-GFP-expressing plasmid was electroporated in utero at E14.5 and analyzed on V-SVZ whole-mount (WM) at P15. CC, corpus callosum; Cx, cortex; LV, lateral ventricle; R, rostral; D, dorsal.
(B and D) P15 V-SVZ whole-mounts were double-immunostained with FoxJ1 (red) and Sox9 (blue) antibodies (B) or FOP (white) and GFAP (red) antibodies (D). GFP+FoxJ1+Sox9+ ependymal cells are indicated by arrows, and GFP+FoxJ1−Sox9+ astrocytes are outlined in white (B). GFP+GFAP− ependymal cells with multiple FOP+ dots are indicated by arrows, and a GFP+GFAP+ astrocyte with a FOP+ centrosome is indicated by an arrowhead (D).
(C) Mean percentage of astrocytes (Sox9+FoxJ1−), ependymal cells (Sox9+FoxJ1+), and others (Sox9−FoxJ1−) among H2B-GFP+ electroporated cells. Analyses were done on n = 3 animals; a total of 441 cells were counted. Error bars represent the SEM. The p values were determined with a two-proportion Z test; ∗∗∗p ≤ 0.001, ∗∗p ≤ 0.01.
(E) Experimental schematic for (F) and (G). Nucbow plasmids (PBCAG-Nucbow along with the PiggyBac transposase and the self-excising Cre recombinase) were electroporated in utero at E14.5 and received EdU (through drinking water) for 14 days starting at P21.
(F and G) Coronal sections of the olfactory bulb (OB) were prepared 1 week after the last day of EdU administration. (G) is a high-magnification image of (F) to show that some Nucbow+ interneurons in the OB are EdU+.
The scale bars represent 40 μm (B), 15 μm (C), 520 μm (F), and 180 μm (G).
To further test whether some of the astrocytes originating from the electroporated RGCs could act as adult neural stem cells (type B1 astrocytes), we permanently labeled RGCs and their progeny by IUE of a transposable Nucbow vector at E14.5 (nuclear MAGIC markers; Loulier et al., 2014) and administered EdU through the animals’ drinking water for 14 days starting at P21 (Figure 2E). One week after the end of EdU administration, EdU+Nucbow+ neurons were observed on each olfactory bulb section, showing that cells derived from electroporated RGCs at E14.5 are adult neural stem cells that give rise to olfactory bulb neurons (Figure 2F and 2G).
These results show that electroporation of RGCs at E14.5 labels multiciliated ependymal cells and adult neural stem cells (type B1 astrocytes) that are retained in the V-SVZ at adult stages.
Lineage Tracing Using MAGIC Markers Shows that Ependymal Cells Derive from Symmetric and Asymmetric Divisions of RGCs
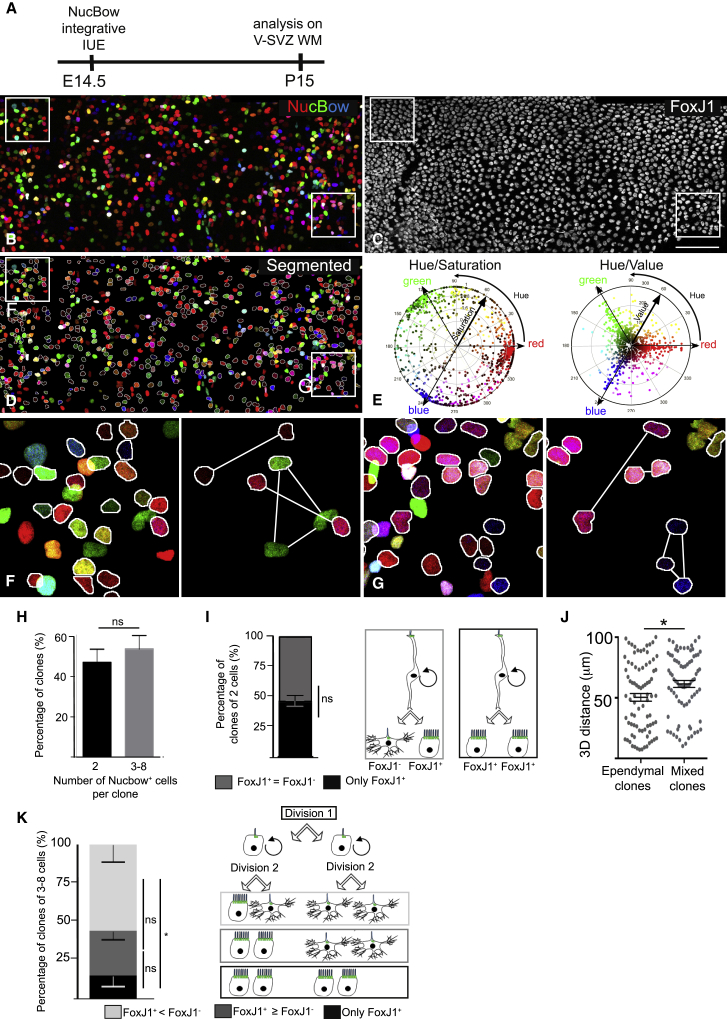
We then took advantage of the large panel of distinct colors produced by the MAGIC markers approach to trace and analyze the lineage of ependymal cells. The V-SVZ of P15–P20 brains electroporated with the Nucbow vector at E14.5 were immunostained with the ependymal marker FoxJ1 in far red, and colors were automatically analyzed to avoid any eye bias (Figures 3A–3C). Briefly, FoxJ1 staining was first used as a reference for the ventricular surface, and 25-μm-thick 3D image stacks of the ventricular whole-mounts were segmented as described previously (Shihavuddin et al., 2017). Nucbow+ cells were then sorted as FoxJ1+ or FoxJ1− (Figure S3; Figure 3D). To define the criteria that identify two cells as sister cells, 2 independent researchers manually picked 49 pairs of cells with similar Nucbow colors (Figure S4A). Both their color content (saturation, value, and hue in the RGB tridimensional space) and their 3D spatial distances were computed (Figure 3E; Figures S4B and S4C). The maximum difference found for each of these parameters was chosen as a threshold for automatic analysis of all Nucbow+ cells in each brain (Figures S4D–S4G). This automatic analysis of all cells from 6 electroporated brains (corresponding to a total of 7,668 Nucbow+ cells and 418 clones of 2 cells or more) showed that more than 80% of clones (with at least one Nucbow+FoxJ1+ cell) contained 8 or less cells, suggesting that most ependymal cells were derived from 3 or less cell divisions (Figures 3F and 3G; Figures S5A–S5F). We excluded the largest clones (9 to 32 cells) because we noted that they were often labeled with the most frequent labels in the dataset (corresponding to the primary colors red, green, and blue), suggesting that merging of juxtaposed clones expressing the same label had occurred (Figure S5G).
Figure 3.
Clonal Analysis of Ependymal Cells with MAGIC Markers Reveals Both Symmetric and Asymmetric Divisions of RGCs
(A) Experimental schematic. Nucbow plasmids were electroporated in utero at E14.5 and analyzed at P15–P20.
(B–D) Representative Z-projected image of an en face view of the V-SVZ (B) immunostained at P15 with anti-FoxJ1 antibody (C). (D) shows a segmented image of (B) and (C), obtained using FoxJ1 staining, as a reference (STAR Methods; Figures S3–S5). FoxJ1+Nucbow+ cells are outlined in white.
(E) Circular hue-saturation and hue-value plots of all Nucbow+ cells from (D).
(F and G) High-magnification images of the insets in (D), showing examples of clones: 2 ependymal doublets and 1 triplet containing 1 ependymal cell and 2 FoxJ1− cells (F) and 3 ependymal doublets, 1 ependymal triplet, and 1 triplet containing 1 ependymal cell and 2 FoxJ1− cells (G).
(H) Mean percentages of clones containing 2 or 3–8 Nucbow+ cells. Error bars represent the SEM of n = 163 clones of 2 cells and n = 186 clones of 3–8 cells; p values were determined by Mann-Whitney test; ns, p > 0.05.
(I) Mean percentages of clones of 2 Nucbow+ cells containing 1 (mixed clones, gray) or 2 (ependymal clones, black) FoxJ1+ cells. Error bars represent the SEM of n = 82 ependymal clones and n = 81 mixed clones from 6 independent experiments; p values were determined by Mann-Whitney test; ns, p > 0.05.
(J) Average 3D distances between the cells composing ependymal or mixed clones of 2 cells. Error bars represent the SEM of n = 82 ependymal clones and n = 81 mixed clones from 6 independent experiments; p values were determined by Mann-Whitney test; ∗p ≤ 0.05.
(K) Mean percentages of clones of 3–8 Nucbow+ cells containing only FoxJ1+ cells (black), more or an equal number of FoxJ1+ compared with FoxJ1− cells (dark gray), or more FoxJ1− cells (light gray) per clone. Error bars represent the SEM of n = 186 clones of 3–8 cells; p values were determined by Mann-Whitney test; ∗∗p ≤ 0.01; ∗p ≤ 0.05; ns, p > 0.05.
The scale bars represent 100 μm (B)–(D) and 22 μm (F) and (G).
Among the 349 clones with 8 or less cells, around half contained only 2 cells, suggesting that, at E14.5, most clones were generated from one terminal cell division of RGCs (n = 6 mice; Figure 3H). These 2-cell clones were composed of 1 or 2 FoxJ1+ cells in a 1:1 ratio, showing that the terminal division could be either symmetric or asymmetric (Figure 3I). Interestingly, the 3D distance between cells was higher in mixed clones (clones composed of ependymal and non-ependymal cells) compared with pure ependymal clones (Figure 3J), showing that FoxJ1− cells were deeper in the SVZ compared with FoxJ1+ cells in the VZ.
Clones containing 3 to 8 cells were generated through 2 or 3 cell divisions, the last of which was either only symmetric (clones containing FoxJ1+ cells only) or both symmetric and asymmetric (clones containing FoxJ1+ and FoxJ1− cells). Interestingly, a majority of these clones contained more FoxJ1− cells than FoxJ1+ cells, suggesting that symmetric divisions giving rise to 2 FoxJ1− cells might have occurred in these clones (Figure 3K).
Lineage tracing experiments of RGCs using the MAGIC markers strategy thus show that ependymal cells originate from either one terminal symmetric division giving rise to 2 ependymal cells or 1 asymmetric division giving rise to 1 ependymal and 1 FoxJ1− cell. Most importantly, this analysis of a large number of clones distributed along the caudo-rostral and ventro-dorsal axis of the lateral wall of the LV of 6 different electroporated brains did not reveal any regional differences. This observation suggests that the ontogeny of the neurogenic niche can be determined by analyzing individual cells along the LV.
MADM of V-SVZ Gliogenesis Reveals that Ependymal and B1 Cells Share a Common Lineage
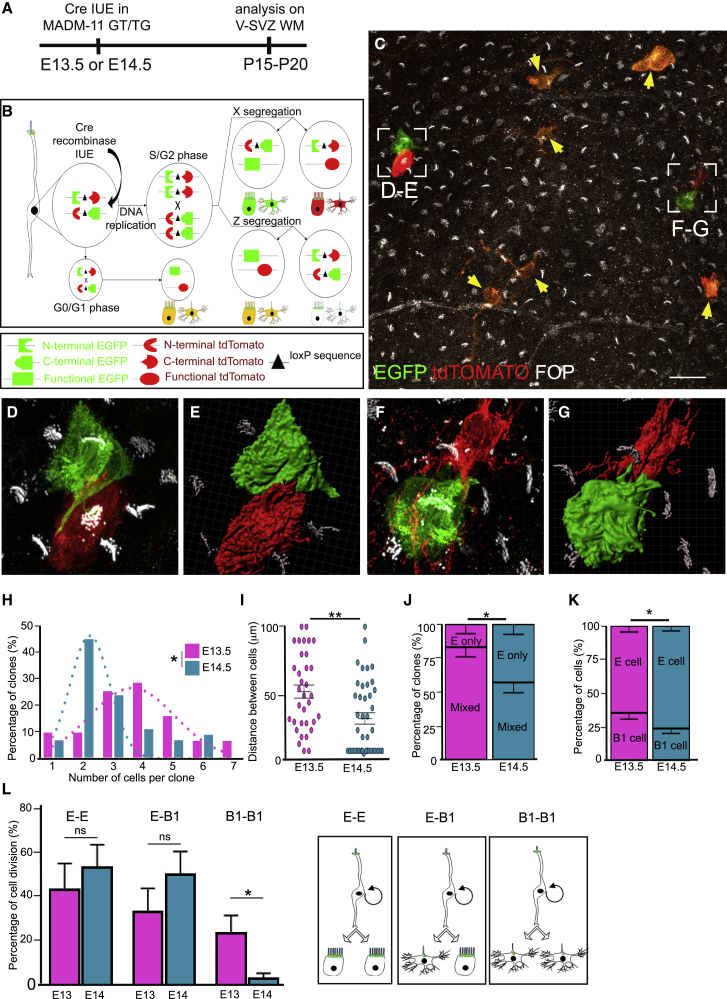
To obtain more insight into the cellular mechanisms and the sequence of symmetric versus asymmetric divisions producing each clone, we used the MADM system coupled with IUE of Cre recombinase at E13.5 or E14.5 (Figure 4A; Gao et al., 2014). In electroporated cells, Cre recombinase mediates interchromosomal recombination, which reconstitutes cytoplasmic enhanced GFP (EGFP, green) or tandem dimer Tomato (tdTomato, red). If recombination occurs in G2 phase of the cell cycle, and each red or green chromosome segregates in separate daughter cells (X segregation), then the two descendent lineages will be permanently labeled green or red by MADM events (Figure 4B). Analysis of cell number and identity will thus allow direct assessment of the division pattern (symmetric versus asymmetric) and cell fate decision of the original dividing progenitors. Otherwise, if recombination occurs in G0/G1, or if both red and green chromosomes segregate in the same cell (Z segregation), then recombined cells appear yellow and will be excluded from the analysis (Figure 4B). We thus induced Cre activity through IUE in MADM pregnant mothers at E13.5 or E14.5 and analyzed the V-SVZ at P15–P20 after immunolabeling of centrioles combined with MADM cytoplasmic staining to identify the cell types composing each clone (Figure 4C). This approach allowed a clonal study of green-red clones because the efficiency of recombination leading to green-red clones was low in these mice (mean number of clones per animal, 5), and most recombined cells were double-labeled (yellow) (Figure 4C). Cells were considered a clone when their spatial distance was less than 100 μm, as defined previously by the Nucbow lineage-tracing experiments. Red or green cells located in the electroporated region of the V-SVZ were either multiciliated ependymal (E) cells, characterized by a few short processes and multiple FOP+ basal bodies in their cytoplasm associated with long cilia, or astrocytes (type B1), whose cell body and multiple long processes were deeper in the SVZ. These astrocytes contained 2 centrosomal centrioles that occasionally contacted the ventricular surface and extended a primary cilium. These cells were thus easily discriminated from multiciliated ependymal cells (identified by multiple centrioles and long cilia) or even neuroblasts, which displayed typical migrating morphologies in the direction of the olfactory bulb and were located deeper in the tissue at larger distances from the clone. When the cells of a clone were in close proximity, their cell body or processes often contacted each other, suggesting that they might maintain communication at the adult stage (Figures 4C–4G; Videos S1 and S2). We observed very few red or green cells alone (clone of 1 cell in Figure 4H) or larger monochrome clones, if any, in the V-SVZ, suggesting that asymmetric divisions giving rise to one ventricular and one non-ventricular cell were rare in these experiments. In contrast, we found that, among the 44 clones of 2–6 cells, 48% of them contained 2 cells (21 clones), and 52% of them contained 3–6 cells (23 clones), which is in line with our findings above showing that half of the RGCs at E14.5 divided once to produce glial cells in the V-SVZ. At E13.5, RGCs also produced V-SVZ cells, but the majority divided twice or more because 90% of clones contained 3 or more cells (Figure 4H). The distance between cells in a clone was higher at E13.5 compared with E14.5, showing that cells disperse as cell divisions proceed (Figure 4I). Both the proportion of mixed clones (containing both ependymal and B1 cells; Figure 4J) and the number of type B1 astrocytes (Figure 4K) in the clones decreased at E14.5 compared with E13.5, suggesting that fewer type B1 astrocytes are produced compared with ependymal cells. Alternatively, type B1 astrocytes might be produced at earlier stages compared with ependymal cells. Noteworthy is that the distribution of astrocytes (B1) and ependymal cells in each clone revealed that astrocytes were produced at a lower rate than ependymal cells and that symmetric divisions producing 2 astrocytes (B1-B1) occurred more frequently at E13.5 than at E14.5 (Figure 4L; Table S1).
Figure 4.
MADM Reveals the Presence of Ependymal-Ependymal and Ependymal-Astrocyte Divisions at E13.5 and E14.5
(A) Experimental schematic. The Cre plasmid was electroporated in utero in MADM-11TG/GT at E13.5 or E14.5, and V-SVZ WMs were analyzed at P15–P20.
(B) Schematic representation of Cre-mediated MADM clone induction in dividing RGCs. A G2-X event results in clones of red- and green-labeled cells, and a G2-Z event generates double-labeled (yellow) and unlabeled clones of cells. Recombination occurring in G0/G1 phases of the cell cycle leads to double-labeled (yellow) cells.
(C–G) Airyscan confocal image of a P15 MADM-labeled V-SVZ whole-mount electroporated with a CRE-expressing plasmid at E14.5. The ventricular wall was stained with EGFP (green), tdTomato (red), and FOP (white) antibodies. (C) Double-labeled yellow cells issued from a G2-Z recombination event are indicated by yellow arrows. Ependymal-ependymal (D and E) and ependymal-astrocyte (F and G) clones of two sister cells are shown at high magnification (D and F) and in a 3D view (E and G). See also Videos S1 and S2.
(H) Mean percentages of all clones generated from in utero electroporation with Cre at E13.5 or E14.5 according to the number of cells per clone (n = 6 and 16 animals at E13.5 and E14.5, respectively) are represented in a histogram. Also shown are dotted curves fitting both the E13.5 and E14.5 distributions; p values were determined with the χ2 test for trend; ∗p ≤ 0.05.
(I) Average distance between cells composing the clones. Error bars represent the SEM of 29 and 44 clones at E13.5 and E14.5, respectively; p values were determined with a Mann-Whitney test; ∗∗p ≤ 0.01.
(J) Mean percentage of all clones generated from E13.5 or E14.5 containing ependymal cells only or a mixed population of ependymal cells and astrocytes (B1). Error bars represent the SEM of 29 and 44 clones at E13.5 and E14.5, respectively; p values were determined with a two-proportion Z test; ∗p ≤ 0.05.
(K) Mean percentage of ependymal and B1 cells in all clones generated from E13.5 or E14.5. Error bars represent the SEM of 117 and 134 cells at E13.5 and E14.5, respectively; p values were determined with a two-proportion Z test; ∗p ≤ 0.05.
(L) Mean percentage of E-E, E-B1, and B1-B1 cell divisions in all clones generated from E13.5 or E14.5. Error bars represent the SEM of 24 and 54 cell divisions at E13.5 and E14.5, respectively; p values were determined with a Mann-Whitney test; ∗p ≤ 0.05.
The scale bars represent 30 μm (C) and 8 μm (D–G).
Together, these results show that ependymal cells and astrocytes are sister cells produced through symmetric (B1-B1 or E-E) and asymmetric (E-B1) divisions of RGCs at mid-gestation in the mouse forebrain.
To gain more insight into the molecular regulation of RGC differentiation into type B1 astrocytes or ependymal cells, we perturbed these divisions with members of the Geminin superfamily, initially described as regulators of DNA replication (Balestrini et al., 2010, Pefani et al., 2011). Two members of this family (Mcidas and GemC1) were recently identified as master regulators of multiciliated ependymal cell fate (Kyrousi et al., 2015), whereas the other member, Geminin, was found to regulate neural cell fate and to be highly expressed in cycling type B1 cells in the adult SVZ (Khatri et al., 2014, Sankar et al., 2016). We also confirmed that GemC1 and Geminin genes are expressed along the LV at E14.5, in the choroid plexus and the ventricular zone, respectively (Figure S6). Moreover, ependymal cell differentiation was totally absent in cultured cells from the GemC1 full mutant, whereas it was slightly (although not significantly) increased in cultured cells from the Geminin conditional mutant (Figures S7B–S7E).
GemC1 Expression Induces Premature Ependymal Cell Differentiation at the Expense of Type B1 Cells
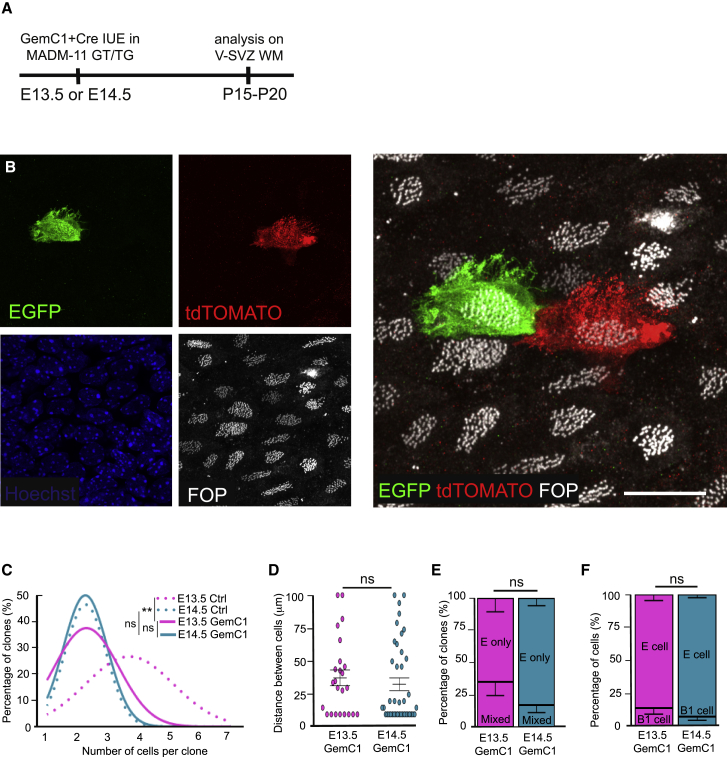
Overexpression of GemC1 through IUE at E13.5 or E14.5 dramatically increased ependymal cell differentiation at the expense of SVZ cells, as shown previously (Figures S7G–S7I; Kyrousi et al., 2015). Interestingly, because B1 cells were absent, pinwheels were not observed in densely GemC1-electroporated regions (Figures S7J and S7K) compared with neighboring areas in which GemC1 electroporation was sparse (Figure S7L). Overexpression of GemC1 together with induction of Cre activity through IUE in MADM embryos at E14.5 did not change the size of the clones compared with controls, suggesting that most RGCs were already undergoing their last division at that stage (Figure 5C). In contrast, when IUE was performed at E13.5, the clones were smaller compared with controls, suggesting that GemC1 induced premature exit from the cell cycle at that stage (Figure 5C). Consistently, the average distance between cells in the GemC1 clones at E13.5 was smaller than in controls (compare Figures 4I and 5D; Mann-Whitney test, ∗∗p ≤ 0.01) and similar to E14.5 GemC1 (Figure 5D). Furthermore, overexpression of GemC1 at E13.5 or E14.5 promoted the ependymal fate because the numbers of both pure ependymal clones and ependymal cells in the clones were dramatically increased compared with controls (two-proportion Z test between controls and GemC1: ∗∗∗p ≤ 0.001; compare Figures 4J and 4K and 5E and 5F). Notably, although astrocytes were occasionally produced through symmetric divisions in controls, they were exclusively generated through asymmetric divisions with ependymal cells after GemC1 overexpression at E13.5 or E14.5. Indeed, no pairs of astrocytes were detected after GemC1 overexpression (Figure 6F; Table S2).
Figure 5.
GemC1 Favors the Formation of Pure Ependymal Clones at Both E13.5 and E14.5
(A) Experimental schematic. GemC1 and Cre plasmids were co-electroporated in utero in MADM-11TG/GT at E13.5 or E14.5, and V-SVZ WMs were analyzed at P15–P20.
(B) Airyscan confocal image of a P15 MADM-labeled V-SVZ whole-mount immunostained with EGFP (green), tdTomato (red), and FOP (white) antibodies showing a clone of 2 ependymal cells.
(C) Fitting curves of the distribution of clone size according to the number of cells per clone, issued from electroporation of Cre at E13.5 or E14.5 (dotted curves; n = 6 and 16 animals at E13.5 and E14.5, respectively) or co-electroporation of Cre with GemC1 at E13.5 or E14.5 (solid curves; n = 4 and 9 animals at E13.5GemC1 and E14.5GemC1, respectively); p values were determined with a χ2 test for trend; ns, p > 0.05, ∗∗p ≤ 0.01.
(D) Average distance between cells composing the clones generated from co-electroporation of Cre and GemC1 at E13.5 or E14.5. Error bars represent the SEM of 20 and 41 clones at E13.5 and E14.5, respectively; p values were determined with a Mann-Whitney test; ns, p > 0.05.
(E) Mean percentage of all clones generated from co-electroporation of Cre and GemC1 at E13.5 or E14.5 containing ependymal cells only or a mixed population of ependymal cells and B1 cells. Error bars represent the SEM of 20 and 41 clones, respectively; p values were determined with a two-proportion Z test; ns, p > 0.05.
(F) Mean percentage of ependymal and B1 cells in all clones generated from co-electroporation of Cre and GemC1 at E13.5 or E14.5. Error bars represent the SEM of 57 and 110 cells at E13.5 and E14.5, respectively; p values were determined with a two-proportion Z test; ns, p > 0.05.
The scale bar represents 50 μm.
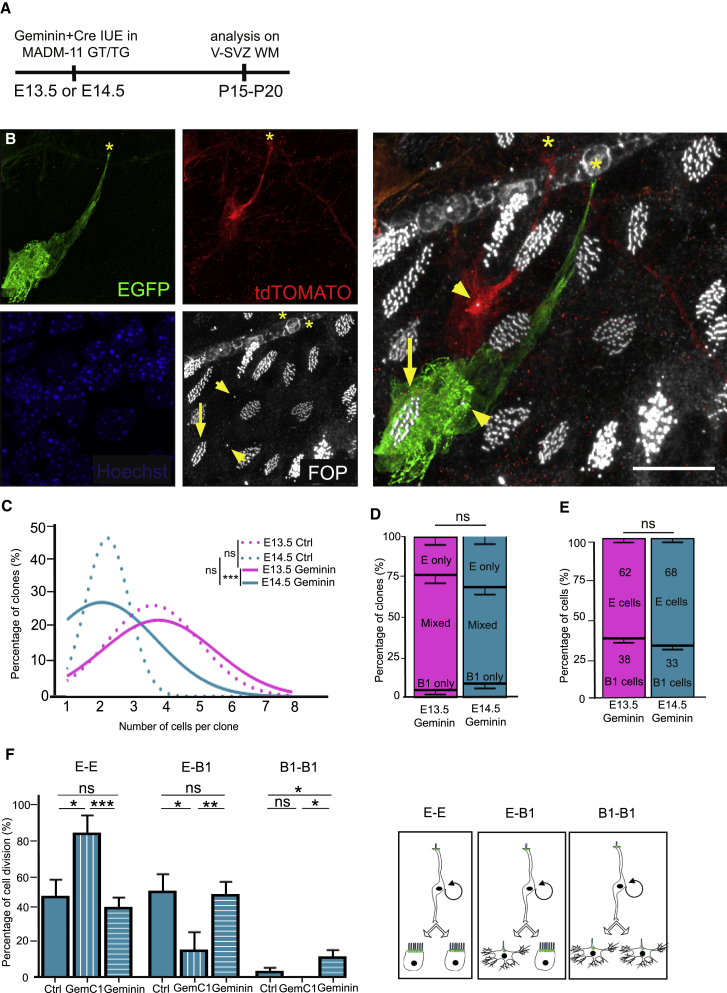
Figure 6.
Geminin Favors the Formation of B1 Cell-Containing Clones at E14.5
(A) Experimental schematic. The Geminin and Cre plasmids were co-electroporated in utero in MADM-11TG/GT at E13.5 and E14.5, and V-SVZ WMs were analyzed at P15–P20.
(B) Airyscan confocal image of a P15 MADM-labeled V-SVZ whole-mount immunostained with EGFP (green), tdTomato (red), and FOP (white) antibodies showing a clone containing 1 GFP+ ependymal cell (arrow) and two B1 cells (one GFP+ and one tdT+, arrowheads). Note that both B1 cells contain a centrosome at the ventricular surface (arrowheads), and they extend a process toward a blood vessel (yellow asterisks). See also Video S3.
(C) Fitting curves of the distribution of clone size according to the number of cells per clone, issued from electroporation of Cre at E13.5 or E14.5 (dotted curves; n = 6 and 16 animals at E13.5 and E14.5, respectively) or co-electroporation of Cre with Geminin at E13.5 or E14.5 (solid curves; n = 8 and 13 animals at E13.5 Geminin and E14.5 Geminin, respectively); p values were determined with a χ2 test for trend; ns, p > 0.05, ∗∗∗p ≤ 0.001.
(D) Mean percentage of all clones generated from co-electroporation of Cre and Geminin at E13.5 and E14.5 and containing either B1 cells only, ependymal cells only, or a mixed population of ependymal cells and B1 cells. Error bars represent the SEM of 73 or 107 clones; the p value was determined with a two-proportion Z test; ns, p > 0.05.
(E) Mean percentage of ependymal and B1 cells in all clones generated from co-electroporation of Cre and Geminin at E13.5 and E14.5. Error bars represent the SEM of 317 or 335 cells, respectively; the p value was determined with a two-proportion Z test; ns, p > 0.05.
(F) Mean percentage of E-E, E-B1, and B1-B1 cell division in clones generated from E14.5 in controls or after overexpression of GemC1 or Geminin. Error bars represent the SEM of 16, 9, and 13 independent animals electroporated with Cre, Cre+GemC1, or Cre+Geminin, respectively; p values were determined with a Mann-Whitney test; ∗∗∗p ≤ 0.001; ∗∗p ≤ 0.01; ∗p ≤ 0.05; ns, p > 0.05.
The scale bar represents 50 μm.
Geminin Expression Favors the Generation of Type B1 Cells
Geminin physically interacts with GemC1 and Mcidas (Caillat et al., 2013, Caillat et al., 2015), but its role during ependymal cell generation is still unknown. We thus tested the influence of Geminin overexpression on the fate of RGCs through IUE with Cre in MADM pregnant mothers at E13.5 or E14.5 (Figures 6A and 6B). Notably, a majority of the clones contained type B1 astrocytes, characterized by an apical contact with a primary cilium and cytoplasmic extensions contacting blood vessels (Figure 6B; Video S3; Table S3). The size of the clones was slightly increased both in E13.5 and E14.5 Geminin-overexpressing clones but similar to that of controls, suggesting that Geminin does not act on the rate of cell division in RGCs (Figure 6C). Interestingly, the proportion of mixed clones and the number of type B1 astrocytes were not significantly increased after Geminin overexpression (two-proportion Z test between controls and Geminin: not significant, p > 0.05; compare Figures 4J and 4K and 6D and 6E; Figure 6F). However, Geminin overexpression led to significant formation of clones containing only B1 cells, which was never observed in controls (two-proportion Z test between E14.5 control and E14.5 Geminin for the B1-only population: ∗p ≤ 0.05; compare Figures 4 and 6D). Consistently, the number of symmetric divisions producing 2 astrocytes (B1-B1) increased significantly after Geminin overexpression at E14.5 compared with controls (Figure 6F; Table S3).
Altogether, these results show that ependymal cells and astrocytes are sister cells produced through symmetric or asymmetric divisions, the balance of which is modulated by the level of expression of Geminin family genes.
Discussion
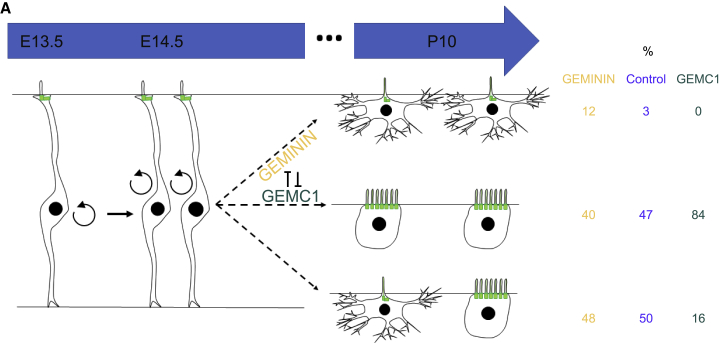
Using a Cre-lox fate mapping technique and complementary MAGIC markers- and MADM-based clonal analysis, our study revealed how glial cells are produced in the V-SVZ during development. First, our results proved that ependymal cells are derived from RGCs all along the embryonic neuroepithelium (pallium and lateral and medial ganglionic eminences) and differentiate locally; ependymal progenitors born in a specific area of the VZ do not migrate long distances to colonize other areas of the neuroepithelium (Figure 1). We then showed that ependymal cells and B1-type astrocytes appear at the end of neurogenesis, mainly through E-B1 asymmetric or E-E symmetric divisions of RGCs. B1-B1 symmetric divisions were less frequent and always combined with E-E or E-B1 divisions (Figure 7). These glial cells have a low migratory capacity and often contact each other, even at the adult stage. Our study thus demonstrates that multiciliated ependymal cells and adult neural stem cells, ultimately forming the adult neurogenic niche, are sister cells that share a common origin. We also provide evidence that these cells are sequentially produced, with the bulk of B1 astrocytes being produced just before the bulk of ependymal cells. Interestingly, their respective numbers are precisely regulated by the Geminin family members. Overexpression of Geminin, a gene expressed more in cycling compared with quiescent neural stem cells (Khatri et al., 2014) and in the ventricular zone at E14.5 (Figure S6), favors B1-B1 symmetric divisions (Figure 6F). On the contrary, overexpression of its antagonist, GemC1, at E14.5 induces premature terminal E-E divisions and leads to a sharp decrease in the final number of B1 cells (Figures 5E and 5F). Given that GemC1 expression is only detected in the choroid plexus at that stage (Figure S6; Arbi et al., 2016), one can hypothesize that it is expressed at very low and/or undetectable levels in these progenitors. Alternatively, GemC1 might be expressed at later stages of development because ependymal differentiation starts postnatally in controls (Spassky et al., 2005). Both possibilities should be tested further, but they might already explain why GemC1 expression at high levels and/or before its normal expression in progenitors leads to premature ependymal differentiation. The sequential expression of Geminin family members could thus be responsible for the temporal differences in glia production. The 2-fold presence of ependymal cells with respect to B1 cells (Mirzadeh et al., 2008) could result from the balance between the levels of expression of these genes. These findings raise the question of the fate decision mechanisms driving RGCs toward symmetric or asymmetric cell division. An analogous question was addressed by others concerning neuronal versus glial cell generation. Interestingly, it has been shown that the number of neurons produced by RG is predictable and that around one in 6 RGCs perform a gliogenic division only when they have exhausted their capacity to proliferate (Gao et al., 2014). At early stages of corticogenesis, RGCs would thus divide asymmetrically to produce neurons and glial progenitors, which would then generate type B1 astrocytes and ependymal cells. Similarly, we found that RGCs generate more mixed clones and more astrocytes when they are electroporated at E13.5 than at E14.5 (Figures 4J and 4K). This suggests that astrocytes are produced earlier than ependymal cells. One might hypothesize that RGCs first give rise to astrocytes until they exhaust their proliferative capacity and yield two ependymal cells through symmetric cell division at later developmental stages. Further lineage studies would be required to identify whether/which neuronal subtypes are lineally related to V-SVZ glial cells (type B1 astrocytes and ependymal cells). Importantly, although ependymal cells become post-mitotic (Spassky et al., 2005), most V-SVZ astrocytes can be reactivated in the adult (Obernier et al., 2018). Altogether, this suggests that RGCs first produce quiescent daughter cells with the potential to enter the cell cycle again (type B1 astrocytes) and then post-mitotic ependymal cells. Interestingly, the description of distinct pathways of glial production via symmetric or asymmetric division unveils the existence of two separate fate decision mechanisms that occur subsequent to the last division of RGCs. This indicates that ependymal versus astrocyte specification might be dependent on the correct segregation of organelles (i.e., centrioles or mitochondria), which have been shown to influence neural stem cell self-renewal and fate decisions (Khacho et al., 2016, Wang et al., 2009). Noteworthy is that Geminin superfamily members were initially described as regulators of DNA replication. It would thus be of interest to determine whether fate decisions in RGCs are driven by DNA replication events following re-entry into the cell cycle.
Figure 7.
Ependymal Cells and B1 Astrocytes Form One Common Lineage Regulated by Geminin Family Members
(A) Model of adult neural stem cells (NSCs) and multiciliated ependymal cell generation. RGCs give rise to type B1 cells through symmetric divisions (rare event, 3%) or asymmetric divisions (frequent event, 50%) and to multiciliated ependymal cells through symmetric divisions (frequent event, 47%). The antagonistic Geminin family members Geminin and GemC1 can modulate the cell fate decision. Geminin overexpression favors symmetric divisions giving rise to type B1 astrocytes. On the contrary, GemC1 overexpression triggers symmetric divisions giving rise to ependymal cells. The percentages of E-E, E-B1, and B1-B1 divisions are indicated for IUE at E14.5 in a control situation and upon GemC1 or Geminin overexpression, respectively.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rat Monoclonal Anti-Mouse CD24 | BD Biosciences | Cat#557436; Clone: M1/69; RRID: AB_396700 |
| Mouse IgG1 Monoclonal Anti FoxJ1 | Thermo Fisher Scientific | Cat#14-9965-82; Clone: 2A5; RRID: AB_1548835 |
| Chicken Polyclonal Anti GFP | Aves Labs | Cat#GFP-1020; RRID: AB_10000240 |
| Rabbit Polyclonal Anti DsRed | Clontech Laboratories | Cat#632496; RRID: AB_10013483 |
| Rabbit Polyclonal Anti Sox9 | Millipore | Cat#AB5535; RRID:AB_2239761 |
| Mouse IgG2b Monoclonal Anti FOP | Abnova Corporation | Cat#H00011116-M01; Clone: 2B1 RRID: AB_463883 |
| Mouse IgG1 Monoclonal Anti GFAP | Millipore | Cat#MAB3402; Clone: GA5 RRID: AB_94844 |
| Rabbit Polyclonal Anti ZO1 | Thermo Fischer Scientific | Cat#40-2200; RRID: AB_2533456 |
| Mouse IgG1 Monoclonal Anti Gamma-tubulin | Sigma-Aldrich | Cat#T6557; Clone: GTU88 RRID: AB_477584 |
| Mouse IgG2b Monoclonal Anti Acetylated Tubulin | Sigma-Aldrich | Cat#T6793; Clone: 6-11B-1 RRID: AB_477585 |
| Donkey Polyclonal anti-Chicken IgY (IgG) (H+L) AffiniPure, Alexa Fluor 488 | Jackson ImmunoResearch Labs | Cat#703-545-155; RRID:AB_2340375 |
| Donkey Polyclonal anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 594 | Thermo Fischer Scientific | Cat#A-21207; RRID: AB_141637 |
| Donkey Polyclonal anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 647 | Thermo Fischer Scientific | Cat#A-31573; RRID: AB_2536183 |
| Goat anti-Mouse IgG1 Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Thermo Fischer Scientific | Cat#A-21121; RRID:AB_2535764 |
| Goat anti-Mouse IgG1 Cross-Adsorbed Secondary Antibody, Alexa Fluor 594 | Thermo Fischer Scientific | Cat#A-21125; RRID:AB_2535767 |
| Goat anti-Mouse IgG1 Cross-Adsorbed Secondary Antibody, Alexa Fluor 647 | Thermo Fischer Scientific | Cat#A-21240; RRID:AB_2535809 |
| Goat anti-Mouse IgG2b Cross-Adsorbed Secondary Antibody, Alexa Fluor 647 | Thermo Fischer Scientific | Cat#A-21242; RRID:AB_2535811 |
| Anti-Digoxigenin-AP, Fab fragments | Sigma-Aldrich | Cat#11093274910; RRID: AB_2734716 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| EdU (5-ethynyl-2-deoxyuridine) | Thermo Fisher Scientific | Cat#11590926, CAS: 61135-33-9 |
| Hoechst (bisBenzimide H 33342 trihydrochloride) | Sigma-Aldrich | Cat# B2261 CAS: 23491-52-3 |
| T7 RNA Polymerase | Sigma-Aldrich | Cat#RPOLT7-RO |
| T3 RNA Polymerase | Sigma-Aldrich | Cat#RPOLT3-RO |
| DIG RNA Labeling Mix | Sigma-Aldrich | Cat#11277073910 |
| RNasin Ribonuclease Inhibitors | Promega | Cat#N2511 |
| RQ1 RNase-Free DNase | Promega | Cat#M6101 |
| BCIP (5-bromo-4-chloro-3-indolyl-phosphate) | Sigma-Aldrich | Cat#BCIP-RO, CAS: 6578-06-9 |
| NBT (4-Nitro blue tetrazolium chloride) | Sigma-Aldrich | Cat#11585029001, CAS: 298-83-9 |
| Critical Commercial Assays | ||
| Click-iT EdU Alexa Fluor 488 Imaging Kit | Thermo Fisher Scientific | Cat#C10337 |
| Click-iT EdU Alexa Fluor 594 Imaging Kit | Thermo Fisher Scientific | Cat#C10339 |
| Click-iT EdU Alexa Fluor 647 Imaging Kit | Thermo Fisher Scientific | Cat#C10340 |
| Experimental Models: Organisms/Strains | ||
| B6.129S2-Emx1tm1(cre)Krj/J | The Jackson Laboratory (Gorski et al., 2002) | Cat#JAX:005628, RRID: IMSR_JAX:005628 |
| B6;CBA-Tg(Gsx2-icre)1Kess/J | Gift from the laboratory of N. Kessaris (Kessaris et al., 2006) | Cat#JAX025806 RRID: IMSR_JAX:025806 |
| C57BL/6J-Tg(Nkx2-1-cre)2Sand/J | The Jackson Laboratory (Xu et al., 2008) | Cat#JAX:008661, RRID:IMSR_JAX:008661 |
| B6;129S6-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J | The Jackson Laboratory (Madisen et al., 2010) | Cat#JAX:007908, RRID:IMSR_JAX:007908 |
| MADM-11GT | Hippenmeyer et al., 2010 | Cat#JAX:013749 RRID:IMSR_JAX:013749 |
| MADM-11TG | Hippenmeyer et al., 2010 | Cat#JAX:013751 RRID:IMSR_JAX:013751 |
| RjORL:SWISS | Janvier Labs | N/A |
| GemC1KO/KO | Arbi et al., 2016 | N/A |
| Gemininflox/flox | Spella et al., 2011 | N/A |
| NestinCre ± | Zimmerman et al., 1994 | N/A |
| Recombinant DNA | ||
| PBCAG-Nucbow Plasmid | Loulier et al., 2014 | N/A |
| CAG-hypBase Plasmid | Loulier et al., 2014 | N/A |
| CAG-seCre Plasmid | Loulier et al., 2014 | N/A |
| CAG-H2B-GFP Plasmid | Gift from the laboratory of X. Morin (Hadjantonakis and Papaioannou, 2004) | N/A |
| pCAGGS-Cre Plasmid | Gift from the laboratory of X. Morin (Morin et al., 2007) | N/A |
| pCAGGS-GemC1 Plasmid | Kyrousi et al., 2015 | N/A |
| pCAGGS-Geminin Plasmid | Spella et al., 2011 | N/A |
| pBluesCriptKS-GemC1 Plasmid | This paper | N/A |
| pBluesCriptKS-Geminin Plasmid | Spella et al., 2007 | N/A |
| Software and Algorithms | ||
| Fiji | Schindelin et al., 2012 | https://imagej.nih.gov/ij/download.html; RRID: SCR_003070 |
| MATLAB | MATLAB and Statistics Toolbox Release 2012b, The MathWorks, Natick, Massachusetts, United States | https://fr.mathworks.com/products/matlab.html; RRID: SCR_001622 |
| GraphPad Prism | GraphPad Prism version 7.00 for Windows, GraphPad Software, La Jolla California USA | https://www.graphpad.com/; RRID: SCR_002798 |
| Other | ||
| Glass capillaries (for IUE) | Harvard Apparatus | Cat#30-0019 |
| CUY21EDIT Square Wave Electroporator | Nepagene | N/A |
| ProbeQuant G-50 Micro Columns | Sigma-Aldrich | Cat#GE28-9034-08 |
Contact for Reagent and Resources Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Nathalie Spassky (spassky@biologie.ens.fr).
Experimental Model and Subject Details
Mice were bred and the experiments were performed in conformity with French and European Union regulations and the recommendations of the local ethics committee (Comité d’éthique en experimentation animale n°005). The date of the vaginal plug was recorded as embryonic day (E) 0.5 and the date of birth as postnatal day (P) 0. Healthy, immunocompetent animals were kept in a 12 h light /12 h dark cycle at 22°C and fed ad libitum. All the individuals used in our study were not previously subject to any unrelated experimental procedures. Pregnant females were used for IUE (see below), but their littermates and any other mice of both sexes were randomly used for all experiments in this study. Emx1-Cre+/− (B6.129S2-Emx1tm1(cre)Krj/J, JAX stock #005628, Gorski et al., 2002), Gsh2-Cre+/− (B6;CBA-Tg(Gsx2-icre)1Kess/J, a gift from the laboratory of N. Kessaris, Kessaris et al., 2006) and Nkx2.1-Cre+/− (C57BL/6J-Tg(Nkx2-1-cre)2Sand/J, JAX stock #008661, Xu et al., 2008) transgenic animals were crossed with R26:tdTomatomT/mT homozygous animals, also called Ai14 (B6;129S6-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J, Madisen et al., 2010). The presence of the Cre transgene was assessed at birth by observing the neonatal brain (when the fluorescence is still visible through the skin with no fur) under the fluorescent stereo microscope. MADMGT/+ and MADMTG/+ transgenic animals were a gift from the laboratory of S. Hippenmeyer (Hippenmeyer et al., 2010). Heterozygous mice were crossed to obtain homozygous MADMGT/GT and MADMTG/TG animals. These homozygous mice were then mated to obtain MADMGT/TG embryos. Expression of the Cre Recombinase in MADMGT/TG embryos was achieved by IUE of pcX-Cre plasmid (1μg/μl, Morin et al., 2007) at E13.5 or E14.5. All transgenic mice lines were kept as B6D2F1/J or C57/Bl6 background. For all other experiments involving IUE, RjORL:SWISS pregnant females were used due to their fertility and their maternal instinct. RjORL:SWISS embryos were also used for the in situ hybridization experiment at E14.5 and cell culture. GemC1KO/KO homozygous animals were incrossed to obtain GemC1-deficient cell cultures. Gemininflox/KO mice and NestinCre ± were crossed in order to have Geminin-deficient glial progenitors in our culture system. All animals analyzed in this study were sacrificed at P15-P20, except for the adult mice sacrificed at P42 to assess the neurogenic potential of SVZ astrocytes, the embryos (and consequently, the mother, at E14.5-E15.5) used for the in situ hybridization studies or EdU-mediated assessment of cell cycle stage of electroporated cells, and newborn pups (P0-P2) used for the cell culture.
Method Details
In utero electroporation
In utero electroporation of mouse embryos was performed at E13.5 or E14.5. Pregnant females were injected subcutaneously with buprenorphine (0.1 mg/kg) 15 minutes prior to surgery. They were subsequently anaesthetized by isoflurane inhalation, the abdominal cavity opened and the uterine horns exposed. With a thin glass capillary (Harvard Apparatus), 1μl of plasmid in filtered PBS was injected together with FastGreen (0.025%, Sigma) into the LVs of the embryo. The final concentrations of plasmids were 1 μg/μl PBCAG-Nucbow, 0.5 μg/μl CAG-hypBase, 0.1 μg/μl CAG-seCre (Loulier et al., 2014), 1 μg/μl CAG-H2B-GFP (a gift from the laboratory of X. Morin, Hadjantonakis and Papaioannou, 2004), 1μg/μl pCAGGS-Cre (a gift from the laboratory of X. Morin, Morin et al., 2007), or 1μg/μl pCAGGS-GemC1 or pCAGGS-Geminin (gifts from the laboratory of S. Taraviras, Kyrousi et al., 2015).
Immediately after injection, four pulses of 50 ms and 35 V were applied to the embryos’ telencephalon at 950 ms intervals with an electroporator (CUY21 EDIT, Nepagene). Finally, the embryos were carefully placed back into the abdominal cavity and left to develop before sacrifice.
EdU administration and detection
To determine the spatial disposition of newborn ependymal cells and the cell cycle stage at the time of electroporation, 50 mg/kg body weight (8 mg/ml stock, dissolved in filtered PBS) of EdU (Thermo Fisher Scientific) was administered to pregnant mice by intraperitoneal injection. In the first case, a single injection was administered at E15.5. In the second one two injections were performed; the first one 2 hours before and the second one 2 hours after IUE. To assess the neurogenic potential of SVZ astrocytes, EdU was administered for 14 days through the drinking water (1 mg/ml) of P21 electroporated litters. EdU incorporation was detected using the Click-iT EdU Alexa Fluor imaging kit (Thermo Fisher Scientific for Alexa Fluor 488, 594 or 647 staining), according to manufacturer’s protocol. Briefly, V-SVZ wholemounts or fixed coronal sections of the forebrain or olfactory bulbs were permeabilized in blocking solution with 0.1% Triton X-100 and 10% fetal bovine serum in PBS for 1h. After washing with PBS, sections were incubated for 1 hour with the Click-iT reaction cocktail, protected from light. The sections were washed again and incubated overnight at 4°C with the primary antibodies. After incubation with the secondary antibody for 2 hours and Hoechst staining, slices were mounted with Fluoromount-G (Southern Biotech, 0100-01) mounting medium.
Primary Ependymal Cell culture
Primary culture of ependymal cells was done like previously described (Delgehyr et al., 2015, Al Jord et al., 2014). Briefly, newborn mice (P0–P2) were sacrificed by decapitation. Their brains were dissected in Hank’s solution (10% HBSS, 5% HEPES, 5% sodium bicarbonate, 1% penicillin/streptomycin (P/S) in pure water) and the extracted ventricular walls were cut manually into small pieces, followed by enzymatic digestion (DMEM glutamax, 33% papain (Worthington 3126), 17% DNase at 10 mg/ml, 42% cysteine at 12 mg/ml, using 1 mL of the enzymatic digestion solution per brain) for 45 min at 37°C in a humidified 5% CO2 incubator. Digestion was stopped by addition of a solution of trypsin inhibitors (Leibovitz Medium L15, 10% ovomucoid at 1 mg/ml, 2% DNase at 10 mg/ml, using 1 mL of enzyme inhibiting solution per brain). The cells were then washed in L15 and resuspended in 1 mL per brain dissected of DMEM glutamax supplemented with 10% fetal bovine serum (FBS) and 1% P/S. Cells were then seeded in a Poly-L-lysine (PLL)-coated flask (1 mL per 75 cm2 flask), containing 5 mL of the same medium in which cells were resuspended. Ependymal progenitors proliferated for 5 days until confluence followed by shaking (250rpm) overnight at room temperature. Pure confluent astroglial monolayers were replated at a density of 7 × 104 cells per cm2 in DMEM glutamax, 10% FBS, 1% P/S on PLL-coated coverslides for immunocytochemistry experiments and maintained overnight. The medium was then replaced by serum-free DMEM glutamax 1% P/S, to trigger ependymal differentiation gradually in vitro (DIV 0).
Tissue and cell culture preparation
When the immunostaining was performed on coronal sections of postnatal animals, these were previously anesthetized with a mixture of 100 mg of ketamine and 10 mg of xylazine per kg of body weight, and then were perfused transcardially with 4% PFA. Adult animals used for EdU-retaining olfactory bulb neuron analyses were not perfused, since no immunohistochemistry procedure was performed on them. After overnight fixation of the dissected brain in 4% PFA at 4°C, of either perfused or non-perfused postnatal mice or embryos, 80 μm-thick floating sections were cut on a vibratome. Wholemounts of the lateral walls of the LVs were dissected (Mirzadeh et al., 2010) from animals sacrificed by cervical dislocation and fixed for 15 minutes in 4% PFA at room temperature. Primary cultures of ependymal cells were fixed for 10 minutes in 4% PFA at room temperature.
For in situ hybridization, an E14.5 pregnant female was sacrificed by cervical dislocation, the embryos were retrieved and their whole brains fixed for 3 days in 4% PFA at 4°C. The sectioning of the tissue was done like described above.
Immunostaining
Tissue samples and primary ependymal cell cultures were incubated for 1h in blocking solution (1X PBS with 0.1%–0.2% Triton X-100 and 10% fetal bovine serum) at room temperature. All these were incubated overnight at 4°C in the primary antibodies diluted in blocking solution. The primary antibodies used targeted CD24 (1:200, BD PharMingen), FoxJ1 (1:500, Thermo Fischer Scientific), GFP (1:1600, Aves Labs), Dsred (1:400, Clontech Laboratories), Sox9 (1:1200, Millipore), FOP (1:600, Abnova Corporation), GFAP (1:400, Millipore) ZO1 (1:100, Thermo Fischer Scientific), Gamma-tubulin (1:500, Sigma-Aldrich) and Acetylated-tubulin (1:400, Sigma-Aldrich). The following day, they were stained with species-specific AlexaFluor fluorophore-conjugated secondary antibodies (1:400, Thermo Fischer Scientific or Jackson ImmunoResearch Labs, see Key Resources Table). Nuclei were counterstained with a 1:1500 Hoechst solution (from a 20 mg/ml stock, Sigma-Aldrich), containing the secondary antibodies for 2h at room temperature. Finally, the wholemounts were redissected to keep only the thin lateral walls of the LV (Mirzadeh et al., 2010) which were mounted with Vectashield mounting medium (Sigma, H-1000), for Nucbow samples, or Fluoromount-G mounting medium (Southern Biotech, 0100-01), for other stainings. Fluoromount-mounted slides were stored at 4°C, whereas Vectashield-mounted wholemounts were stored at −20°C to avoid color fading. Cell culture coverslides were mounted with Fluoromount-G.
In situ hybridization
The GemC1 cDNA sequence was subcloned into a pBlueScriptKS plasmid by removing the former from the same pCAGGS-GemC1 plasmid used for GemC1 overexpression (see In Utero Electroporation, Kyrousi et al., 2015). Both plasmids were doubly digested with XbaI and XhoI. Then the DNA fragment corresponding to the GemC1 cDNA size was isolated from an agarose gel and ligated to the pBlueScriptKS backbone, upstream of the T3 promoter sequence. The pBlueScriptKS-Geminin plasmid was a gift from the laboratory of S. Taraviras (Spella et al., 2007). Briefly, in this study, the open reading frame of Geminin was cloned between the EcoRI/BamHI sites of the pBlueScriptKS plasmid, upstream of the T7 promoter sequence.
pBlueScriptKS-GemC1 and pBlueScriptKS-Geminin were linearized with XbaI and EcoRI restriction enzymes. Using the T3 and T7 RNA polymerases (Sigma-Aldrich), respectively, DIG-labeled ribonucleotide mix (Sigma-Aldrich) and a Ribonuclease inhibitor (Promega), a DIG-labeled gene-specific RNA probe was generated, according to manufacturer’s instructions. RNA probes were subsequently treated with a Deoxyribonuclease (Promega) for 20 min at 37°C. Once synthesized, the RNA probes were purified in a ProbeQuant G-50 Micro Column (Sigma-Aldrich).
The in situ hybridization was performed as previously described (de Frutos et al., 2016). Unless stated otherwise, washing steps were performed thrice for 5 min. Floating sections of E14.5 embryos were incubated for 1 hour at room temperature (RT) in the dark in 2% H2O2 in PBS-0.1% Tween-20 (PBT). After washing in PBT, sections were treated with Proteinase K (10 μg/ml in PBT) for 3-4 min at RT and then the reaction was stopped in a 2mg/ml glycine solution in PBT for 5 min at RT. After washes in PBT, samples were postfixed in 0.2% glutaraldehyde in 4% PFA for 30 min at RT. The tissue was washed again in PBT and then incubated for 1 hour at 60°C in hybridization buffer (50% formamide, 5X SSC, 1% SDS, 50 μg/ml heparin and 50 μg/ml yeast tRNA, in water). RNA probes were diluted at 5-10 μg/ml in hybridization buffer and incubated with the samples at 60°C overnight. The next day, sections were washed twice in a 50% formamide, 5X SSC, 1% SDS solution, for 30 min at 60°C. They were washed again twice in a 50% formamide, 2X SSC, 0,5% SDS solution, for 30 min at 60°C. Washing at RT in TBST (0.08% NaCl, 0.002%KCl, 2.5mM Tris, from a 1M Tris pH = 7.5 stock, 0.01% Tween-20) followed and blocking in 10% FBS in TBST for 1h 30 min at RT. An anti-DIG antibody (Sigma-Aldrich) was diluted in blocking solution (1:2000) and incubated with the samples overnight at 4°C. The next day, at least 8 washes in TBST and 3 in NTMT (100 mM NaCl, 100 mM Tris, from a 1M Tris pH = 9.5 stock, 50 mM MgCl2, 0.01% Tween-20) for 10 min were done. Finally, color developing was performed in a 0.35% vol/vol BCIP (Sigma-Aldrich) and 0.34% vol/vol NBT (Sigma-Aldrich) solution, from a 50 and 100 mg/ml stock dilution in dimethylformamide, respectivelty, in NTMT.
Imaging
Fixed slices or LV wholemounts were examined with an upright Zeiss Axio Observer.ZI epifluorescence microscope, using an apochromat 63 × 1.4 NA objective and a Zeiss Apotome with an H/D grid.
Confocal image stacks were collected with a 40 × 1.3 NA water objective on Olympus FV1000 and FV1200 microscopes, or with a 40 × 1.4 NA oil objective or a 63 × 1.4 NA oil objective on an inverted LSM 880 Airyscan Zeiss microscope with 440, 515 and 560 laser lines to excite, independently, Cerulean, mEYFP and mCherry, or Alexa 488, 594 and 633/Cy5.
Finally, images of the in situ hybridization sections were taken with a Leica MZ16 F Fluorescence Stereo Microscope (Leica Microsystems), equipped with a plan-apochromatic objective 1.0x (Leica, 10447157) and a Nikon DS-Ri1 High Resolution Color Camera (Nikon), with the assistance of the NIS-Element F Ver5.502 Imaging Software (Nikon).
Automatic image analysis of MAGIC Markers
For clarity, mCherry, EYFP and Cerulean Nucbow signals are represented as red, green and blue (RGB) values. 1) Local apical layer extraction: to maintain consistency among datasets, only cells within 25 μm of the apical surface were considered using the SME projection tool on the FoxJ1-stained cell nuclei (Shihavuddin et al., 2017). 2) Segmentation of ependymal cells stained with FoxJ1: the 3D volume occupied by each cell nucleus was delineated using FoxJ1 far red staining. RGB information was extracted from the segmented mask using the following steps implemented as a Fiji macro: Noise was reduced in a preprocessing step using 3D Gaussian filtering, where the sigma values of the Gaussian kernel was set to 1/3rd of the estimated mean nuclear radius in 3D. This was followed by Log3D filtering (Sage et al., 2005) to select objects corresponding to nuclear size; the local 3D maximum was then detected to determine the center of each cell nucleus. 3D-seeded watershed segmentation was performed from these maxima to accurately detect the nuclear border in 3D. This 3D segmentation mask was used to compute the volume and the mean color of each nucleus. 3) Segmentation of non-ependymal cells: After elimination of FoxJ1-positive ependymal cells, only FoxJ1-negative non-ependymal cells remained in the 25μm apical layer. Since there is no specific marker for these cells, they were characterized by their color information as follows: Projection: projection of the Nucbow color channels was maximized to obtain a 2D representation of all labeled non-ependymal cells. Color gradient extraction: In order to accentuate nuclear borders, the image gradient was computed from the sum of the intensities of the three RGB channels. The gradient image was further filtered with adaptive Gaussian filtering to improve the signal to noise ratio. The adaptive filter augments smoothing where the image gradient is weak and decreases smoothing where the gradient is high, in order to preserve nuclear edges. Watershed segmentation: Local maxima were extracted from the inverted smoothed gradient response to retrieve one maximum per nucleus. The seeded watershed transform was then used (Ollion et al., 2013) to detect cells in 2D. 4) Color normalization: RGB channels were rescaled linearly from 0 and the 99th percentile of their intensity distribution to ensure alignment of their relative intensity (1% of the most saturated cells were therefore excluded from the analysis of each sample). 5) Determination of clonal lineage: To identify the cell lineage, each cell was characterized by the median R G B values and their spatial location in 3D X, Y, Z. RGB values were converted to their equivalent in the HSV (Hue, Saturation, Value) color space as described in Loulier et al. (2014). This conversion was performed in MATLAB with the HEXCONE model proposed by Smith (1978).
Quantification and Statistical Analyses
Quantification, image and statistical analyses were performed with Fiji (Schindelin et al., 2012), MATLAB (Mathworks, USA), Excel, and GraphPad Prism software. Quantifications throughout the study are represented as the mean value, with the exception of the clone size representation, which indicates the clone size frequency distribution (only Figure 4H), as well as the Gaussian non-linear regression curve fitting the frequency of clones of variable sizes (from 1 to 8 cells per clone, Figure 4H, 5C, 6C). Error bars indicate the Standard Error of the Mean (SEM), except for Figure S1C, in which the Standard Deviation (SD) is depicted. P values in this manuscript present the following star code: ns: p > 0.05 (non-significant), ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001.
Fate mapping of the spatial origin of ependymal cells
In order to characterize the spatial origin of ependymal cells and the presence or absence of ependymal progenitor cell migration, we considered two areas along the ventricular wall in the Cre-expressing animals; a Cre-positive area, or the anatomical part of the ventricle directly derived from the embryonic Cre-expressing area (Dorsal and Dorsal Medial Walls in the Emx1Cre mice, the Lateral Wall in the Gsh2Cre mice and the ventral-most region of the wall in the Nkx2.1Cre animals), and a Cre-negative area, or the anatomical part of the ventricle that is not derived from the embryonic Cre-expressing area, according to the literature. In the Emx1-cre; Ai14 group, 14 images from n = 6 animals were analyzed, with 1615 counted cells in the Cre-positive area and 1723 cells in the Cre-negative area. For the Gsh2-cre; Ai14 individuals, 15 images from n = 4 animals were used for quantification, with 383 and 895 cells counted on the Cre-positive and Cre-negative anatomical areas, respectively. Finally, 16 images from n = 5 Nkx2.1-cre; Ai14 animals were used, with a total of 496 and 2367 cells analyzed in the Cre-positive and negative areas, respectively.
To determine whether the differences between the Cre-positive and negative areas were significant, we performed a Mann-Whitney test.
Characterization of the cell types in the electroporated V-SVZ
19 images containing 441 V-SVZ electroporated (H2B-GFP positive) cells were analyzed in n = 3 different animals. The differences between cell types (astrocytes or FoxJ1-Sox9+, ependymal or FoxJ1+Sox9+, and unknown cell type or FoxJ1-Sox9-) were determined in pairs via the Mann-Whitney test.
Automatic image analysis of MAGIC markers
We assessed n = 6 V-SVZ wholemounts electroporated with the MAGIC markers (Nucbow). The automated analysis of such samples yielded 7668 Nucbow+ cells, which could be regrouped in clones of cells, i.e., cells with a common progenitor, based on their color characteristics (see Method Details). 1142 Nucbow+ cells that belonged to 163 clones with 2 cells (326 cells) or 186 clones with 3 to 8 cells (816 cells) and that contained at least one FoxJ1+ cell were taken into account. These 349 clones represented the 83% of all 418 clones found by the automated analysis with at least one FoxJ1+ cell. The 17% remaining clones had 9 to 32 cells and they were excluded from the analysis. Clones of 2 cells (163 of the 349 total clones) were categorized in clones formed by 2 ependymal cells (only FoxJ1+) or 1 ependymal and 1 non-ependymal cells (FoxJ1+ = FoxJ1-). Clones of 3 to 8 cells (186 of the 349 total clones) were subdivided in clones formed by exclusively ependymal cells (only FoxJ1+), clones with as many or more ependymal cells as non-ependymal cells (FoxJ1+ ≥ FoxJ1-), or clones with less ependymal than non-ependymal cells (FoxJ1+ < FoxJ1-). The difference between the frequency of clones with 2 or 3 to 8 cells, as well as the difference between the percentage of types of clones (only FoxJ1+, FoxJ1+ = FoxJ1-, FoxJ1+ ≥ FoxJ1-, FoxJ1+ < FoxJ1-), were determined in two-by-two comparisons with the Mann-Whitney test.
The 3D-distance between cells in pure ependymal clones and between cells in mixed clones (with at least one FoxJ1- cell) was calculated automatically and the p value was assessed using the Mann-Whitney test, as well.
MADM transgenic image analysis
In all, 314 clones of 2 or more cells were analyzed (29 E13Ctrl, 20 E13GemC1, 73 E13Geminin, 44 E14Ctrl, 41 E14GemC1 and 107 E14 Geminin), which counted for 1069 cells (117 E13Ctrl, 56 E13GemC1, 317 E13Geminin, 134 E14Ctrl, 110 E14GemC1 and 335 E14Geminin), obtained from 52 electroporated embryos (6 E13Ctrl, 4 E13GemC1, 8 E13Geminin, 16 E14Ctrl, 9 E14GemC1 and 13 E14Geminin). To assess the percentage of types of clones (Ependymal only versus Mixed and versus B1 astrocytic only), and cells (Ependymal versus B1 astrocytes), all clones were grouped, independently of animals, since the efficiency of the IUE technique and the Cre recombination in MADM mice are highly variable. This resulted in the problem of having animals with a very small number of clones (one or two) and animals with a very large number (up to 26) and, hence, not having the same weight in the statistical analysis. In order to study the difference of clone types and cell proportion among the different categories, a two-proportion Z-test was performed in each case.
The differences in cell division type (Ependymal symmetric, E-E, B1 astrocytic symmetric, B1-B1, or asymmetric, E-B1) were assessed with the Mann-Whitney test.
Finally, the clone size distribution (number of cells per clone) for each category was represented as a Gaussian non-linear regression curve, fitting the frequency of clones with several sizes (from 1 to 8 cells per clone). The differences in the clone size distribution were determined via a Chi2 test for trend.
The distance between cells in a clone was determined by assessing the mean distance between pairs of cells in a clone, when they possessed more than 2 cells, or the only distance between the unique pair of cells in clones with 2 cells. The significance of the difference in such distance was calculated using the Mann-Whitney test.
EdU incorporation in the V-SVZ analysis
17 and 12 coronal sections of electroporated brains with stained EdU were analyzed for the E13.5 and E14.5 brains (n = 3 for each category). In these, the percentage of EdU retaining cells was assessed. The p value was calculated using the Mann-Whitney test.
Characterization of the differentiation status in the V-SVZ with or without GemC1
41 and 15 coronal sections were analyzed for the H2B-GFP and GemC1/H2B-GFP-electroporated brains, respectively. A total of 4434 and 1953 H2B-GFP+ ependymal (multi-FOP stained) and non-ependymal cells (two-dot FOP stained) were counted in the V-SVZ of 3 control and 3 GemC1 brains. Even though the number of animals was the same, the difference in analyzed sections and counted cells is due to the variability of the electroporation, which causes that some brains are electroporated over a wide area, whereas others are targeted by the electroporation in a restrained zone. The difference between the percentage of electroporated ependymal cells (over the total electroporated cells) in both categories was determined with a Mann-Whitney test.
Assessment of the differentiation status of GemC1 KO and Geminin cKO primary cultures
n = 4 cultures for WT, n = 2 for GemC1KO/KO and n = 4 for GemininFL/KO;NestinCre ± were quantified. In all 1015, 637 and 1638 cells were counted for each one of the genotypes, respectively. The percentage of differentiation in each condition was normalized to the WT (control). The differences between genotypes were determined in pairs using the Mann-Whitney test.
Data and Software Availability
Several macros were created using the MATLAB software to use for the automatic analysis of MAGIC markers. They will be available upon request to the corresponding author.
Acknowledgments
We thank all members of the Spassky laboratory as well as the Jean-François Brunet, Sonia Garel, and Xavier Morin laboratories for comments and discussions. We thank X. Morin for the pCAAGS-H2B-GFP and pCAGGS-Cre plasmids. We thank A.-K. Konate and R. Nagalingum for administrative support and the IBENS Animal Facility for animal care. The team received support from Agence Nationale de la Recherche (ANR) Investissements d’Avenir (ANR-10-LABX-54 MEMO LIFE and ANR-11-IDEX-0001-02 PSL∗ Research University). The Spassky laboratory is supported by INSERM, CNRS, École Normale Supérieure (ENS), ANR (ANR-17-CE12-0021-03), FRM (Equipe FRM grant 20140329547), European Research Council (ERC Consolidator grant 647466), and Cancéropôle Île-de-France (2014-1-PL BIO-11-INSERM 121). The Livet and Hippenmeyer laboratories are supported by the European Research Council (ERC Consolidator grants 649117 and 725780, respectively). G.O.-A. and M.D. received fellowships from Labex MEMOLIFE and the French Ministry of Higher Education and Research, respectively. S.C. received fellowships from the Région Ile-de-France and Association pour la Recherche sur le Cancer (ARC).
Author Contributions
N.S. designed, funded, and supervised the research. G.O.-A., M.D., P.L., A.F., M.F., S.C., J.L., and N.S. designed the experiments and performed the research. G.O.-A., M.D., P.L., A.F., A.S., A.M., A.G., and N.S. analyzed the data. S.T. and S.H. provided resources. G.O.-A., M.D., and N.S. wrote the manuscript with input from all authors.
Declaration of Interests
The authors declare no competing interests.
Published: February 26, 2019
Footnotes
Supplemental Information can be found with this article online at https://doi.org/10.1016/j.neuron.2019.01.051.
Supplemental Information
References
- Al Jord A., Lemaître A.-I., Delgehyr N., Faucourt M., Spassky N., Meunier A. Centriole amplification by mother and daughter centrioles differs in multiciliated cells. Nature. 2014;516:104–107. doi: 10.1038/nature13770. [DOI] [PubMed] [Google Scholar]
- Arbi M., Pefani D.E., Kyrousi C., Lalioti M.E., Kalogeropoulou A., Papanastasiou A.D., Taraviras S., Lygerou Z. GemC1 controls multiciliogenesis in the airway epithelium. EMBO Rep. 2016;17:400–413. doi: 10.15252/embr.201540882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balestrini A., Cosentino C., Errico A., Garner E., Costanzo V. GEMC1 is a TopBP1-interacting protein required for chromosomal DNA replication. Nat. Cell Biol. 2010;12:484–491. doi: 10.1038/ncb2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boon M., Wallmeier J., Ma L., Loges N.T., Jaspers M., Olbrich H., Dougherty G.W., Raidt J., Werner C., Amirav I. MCIDAS mutations result in a mucociliary clearance disorder with reduced generation of multiple motile cilia. Nat. Commun. 2014;5:4418. doi: 10.1038/ncomms5418. [DOI] [PubMed] [Google Scholar]
- Caillat C., Pefani D.-E., Gillespie P.J., Taraviras S., Blow J.J., Lygerou Z., Perrakis A. The Geminin and Idas coiled coils preferentially form a heterodimer that inhibits Geminin function in DNA replication licensing. J. Biol. Chem. 2013;288:31624–31634. doi: 10.1074/jbc.M113.491928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caillat C., Fish A., Pefani D.-E., Taraviras S., Lygerou Z., Perrakis A. The structure of the GemC1 coiled coil and its interaction with the Geminin family of coiled-coil proteins. Acta Crystallogr. D Biol. Crystallogr. 2015;71:2278–2286. doi: 10.1107/S1399004715016892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebrián-Silla A., Alfaro-Cervelló C., Herranz-Pérez V., Kaneko N., Park D.H., Sawamoto K., Alvarez-Buylla A., Lim D.A., García-Verdugo J.M. Unique Organization of the Nuclear Envelope in the Post-natal Quiescent Neural Stem Cells. Stem Cell Reports. 2017;9:203–216. doi: 10.1016/j.stemcr.2017.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Frutos C.A., Bouvier G., Arai Y., Thion M.S., Lokmane L., Keita M., Garcia-Dominguez M., Charnay P., Hirata T., Riethmacher D. Reallocation of Olfactory Cajal-Retzius Cells Shapes Neocortex Architecture. Neuron. 2016;92:435–448. doi: 10.1016/j.neuron.2016.09.020. [DOI] [PubMed] [Google Scholar]
- Delgehyr N., Meunier A., Faucourt M., Bosch Grau M., Strehl L., Janke C., Spassky N. Ependymal cell differentiation, from monociliated to multiciliated cells. Methods Cell Biol. 2015;127:19–35. doi: 10.1016/bs.mcb.2015.01.004. [DOI] [PubMed] [Google Scholar]
- Doetsch F., Caillé I., Lim D.A., García-Verdugo J.M., Alvarez-Buylla A. Subventricular zone astrocytes are neural stem cells in the adult mammalian brain. Cell. 1999;97:703–716. doi: 10.1016/s0092-8674(00)80783-7. [DOI] [PubMed] [Google Scholar]
- Ferri A.L., Cavallaro M., Braida D., Di Cristofano A., Canta A., Vezzani A., Ottolenghi S., Pandolfi P.P., Sala M., DeBiasi S., Nicolis S.K. Sox2 deficiency causes neurodegeneration and impaired neurogenesis in the adult mouse brain. Development. 2004;131:3805–3819. doi: 10.1242/dev.01204. [DOI] [PubMed] [Google Scholar]
- Fuentealba L.C., Rompani S.B., Parraguez J.I., Obernier K., Romero R., Cepko C.L., Alvarez-Buylla A. Embryonic Origin of Postnatal Neural Stem Cells. Cell. 2015;161:1644–1655. doi: 10.1016/j.cell.2015.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furutachi S., Miya H., Watanabe T., Kawai H., Yamasaki N., Harada Y., Imayoshi I., Nelson M., Nakayama K.I., Hirabayashi Y., Gotoh Y. Slowly dividing neural progenitors are an embryonic origin of adult neural stem cells. Nat. Neurosci. 2015;18:657–665. doi: 10.1038/nn.3989. [DOI] [PubMed] [Google Scholar]
- Gao P., Postiglione M.P., Krieger T.G., Hernandez L., Wang C., Han Z., Streicher C., Papusheva E., Insolera R., Chugh K. Deterministic progenitor behavior and unitary production of neurons in the neocortex. Cell. 2014;159:775–788. doi: 10.1016/j.cell.2014.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorski J.A., Talley T., Qiu M., Puelles L., Rubenstein J.L.R., Jones K.R. Cortical excitatory neurons and glia, but not GABAergic neurons, are produced in the Emx1-expressing lineage. J. Neurosci. 2002;22:6309–6314. doi: 10.1523/JNEUROSCI.22-15-06309.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjantonakis A.-K., Papaioannou V.E. Dynamic in vivo imaging and cell tracking using a histone fluorescent protein fusion in mice. BMC Biotechnol. 2004;4:33. doi: 10.1186/1472-6750-4-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hippenmeyer S., Youn Y.H., Moon H.M., Miyamichi K., Zong H., Wynshaw-Boris A., Luo L. Genetic mosaic dissection of Lis1 and Ndel1 in neuronal migration. Neuron. 2010;68:695–709. doi: 10.1016/j.neuron.2010.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessaris N., Fogarty M., Iannarelli P., Grist M., Wegner M., Richardson W.D. Competing waves of oligodendrocytes in the forebrain and postnatal elimination of an embryonic lineage. Nat. Neurosci. 2006;9:173–179. doi: 10.1038/nn1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khacho M., Clark A., Svoboda D.S., Azzi J., MacLaurin J.G., Meghaizel C., Sesaki H., Lagace D.C., Germain M., Harper M.E. Mitochondrial Dynamics Impacts Stem Cell Identity and Fate Decisions by Regulating a Nuclear Transcriptional Program. Cell Stem Cell. 2016;19:232–247. doi: 10.1016/j.stem.2016.04.015. [DOI] [PubMed] [Google Scholar]
- Khatri P., Obernier K., Simeonova I.K., Hellwig A., Hölzl-Wenig G., Mandl C., Scholl C., Wölfl S., Winkler J., Gaspar J.A. Proliferation and cilia dynamics in neural stem cells prospectively isolated from the SEZ. Sci. Rep. 2014;4:3803. doi: 10.1038/srep03803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrousi C., Arbi M., Pilz G.-A., Pefani D.-E., Lalioti M.-E., Ninkovic J., Götz M., Lygerou Z., Taraviras S. Mcidas and GemC1/Lynkeas are key regulators for the generation of multiciliated ependymal cells in the adult neurogenic niche. Development. 2015;142:3661–3674. doi: 10.1242/dev.126342. [DOI] [PubMed] [Google Scholar]
- Loulier K., Barry R., Mahou P., Le Franc Y., Supatto W., Matho K.S., Ieng S., Fouquet S., Dupin E., Benosman R. Multiplex cell and lineage tracking with combinatorial labels. Neuron. 2014;81:505–520. doi: 10.1016/j.neuron.2013.12.016. [DOI] [PubMed] [Google Scholar]
- Ma L., Quigley I., Omran H., Kintner C. Multicilin drives centriole biogenesis via E2f proteins. Genes Dev. 2014;28:1461–1471. doi: 10.1101/gad.243832.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L., Zwingman T.A., Sunkin S.M., Oh S.W., Zariwala H.A., Gu H., Ng L.L., Palmiter R.D., Hawrylycz M.J., Jones A.R. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarry T.J., Kirschner M.W. Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell. 1998;93:1043–1053. doi: 10.1016/s0092-8674(00)81209-x. [DOI] [PubMed] [Google Scholar]
- Mirzadeh Z., Merkle F.T., Soriano-Navarro M., Garcia-Verdugo J.M., Alvarez-Buylla A. Neural stem cells confer unique pinwheel architecture to the ventricular surface in neurogenic regions of the adult brain. Cell Stem Cell. 2008;3:265–278. doi: 10.1016/j.stem.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzadeh Z., Doetsch F., Sawamoto K., Wichterle H., Alvarez-Buylla A. The subventricular zone en-face: wholemount staining and ependymal flow. J. Vis. Exp. 2010;39:1938. doi: 10.3791/1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin X., Jaouen F., Durbec P. Control of planar divisions by the G-protein regulator LGN maintains progenitors in the chick neuroepithelium. Nat. Neurosci. 2007;10:1440–1448. doi: 10.1038/nn1984. [DOI] [PubMed] [Google Scholar]
- Obernier K., Cebrian-Silla A., Thomson M., Parraguez J.I., Anderson R., Guinto C., Rodas Rodriguez J., Garcia-Verdugo J.-M., Alvarez-Buylla A. Adult Neurogenesis Is Sustained by Symmetric Self-Renewal and Differentiation. Cell Stem Cell. 2018;22:221–234.e8. doi: 10.1016/j.stem.2018.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollion J., Cochennec J., Loll F., Escudé C., Boudier T. TANGO: a generic tool for high-throughput 3D image analysis for studying nuclear organization. Bioinformatics. 2013;29:1840–1841. doi: 10.1093/bioinformatics/btt276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pefani D.-E., Dimaki M., Spella M., Karantzelis N., Mitsiki E., Kyrousi C., Symeonidou I.-E., Perrakis A., Taraviras S., Lygerou Z. Idas, a novel phylogenetically conserved geminin-related protein, binds to geminin and is required for cell cycle progression. J. Biol. Chem. 2011;286:23234–23246. doi: 10.1074/jbc.M110.207688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage D., Neumann F.R., Hediger F., Gasser S.M., Unser M. Automatic tracking of individual fluorescence particles: application to the study of chromosome dynamics. IEEE Trans. Image Process. 2005;14:1372–1383. doi: 10.1109/tip.2005.852787. [DOI] [PubMed] [Google Scholar]
- Sankar S., Yellajoshyula D., Zhang B., Teets B., Rockweiler N., Kroll K.L. Gene regulatory networks in neural cell fate acquisition from genome-wide chromatin association of Geminin and Zic1. Sci. Rep. 2016;6:37412. doi: 10.1038/srep37412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah P.T., Stratton J.A., Stykel M.G., Abbasi S., Sharma S., Mayr K.A., Koblinger K., Whelan P.J., Biernaskie J. Single-Cell Transcriptomics and Fate Mapping of Ependymal Cells Reveals an Absence of Neural Stem Cell Function. Cell. 2018;173:1045–1057.e9. doi: 10.1016/j.cell.2018.03.063. [DOI] [PubMed] [Google Scholar]
- Shen Q., Wang Y., Kokovay E., Lin G., Chuang S.-M., Goderie S.K., Roysam B., Temple S. Adult SVZ stem cells lie in a vascular niche: a quantitative analysis of niche cell-cell interactions. Cell Stem Cell. 2008;3:289–300. doi: 10.1016/j.stem.2008.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shihavuddin A., Basu S., Rexhepaj E., Delestro F., Menezes N., Sigoillot S.M., Del Nery E., Selimi F., Spassky N., Genovesio A. Smooth 2D manifold extraction from 3D image stack. Nat. Commun. 2017;8:15554. doi: 10.1038/ncomms15554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A.R. Color gamut transform pairs. ACM SIGGRAPH Comput. Graph. 1978;12:12–19. [Google Scholar]
- Spassky N., Meunier A. The development and functions of multiciliated epithelia. Nat. Rev. Mol. Cell Biol. 2017;18:423–436. doi: 10.1038/nrm.2017.21. [DOI] [PubMed] [Google Scholar]
- Spassky N., Merkle F.T., Flames N., Tramontin A.D., García-Verdugo J.M., Alvarez-Buylla A. Adult ependymal cells are postmitotic and are derived from radial glial cells during embryogenesis. J. Neurosci. 2005;25:10–18. doi: 10.1523/JNEUROSCI.1108-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spella M., Britz O., Kotantaki P., Lygerou Z., Nishitani H., Ramsay R.G., Flordellis C., Guillemot F., Mantamadiotis T., Taraviras S. Licensing regulators Geminin and Cdt1 identify progenitor cells of the mouse CNS in a specific phase of the cell cycle. Neuroscience. 2007;147:373–387. doi: 10.1016/j.neuroscience.2007.03.050. [DOI] [PubMed] [Google Scholar]
- Spella M., Kyrousi C., Kritikou E., Stathopoulou A., Guillemot F., Kioussis D., Pachnis V., Lygerou Z., Taraviras S. Geminin regulates cortical progenitor proliferation and differentiation. Stem Cells. 2011;29:1269–1282. doi: 10.1002/stem.678. [DOI] [PubMed] [Google Scholar]
- Stancik E.K., Navarro-Quiroga I., Sellke R., Haydar T.F. Heterogeneity in ventricular zone neural precursors contributes to neuronal fate diversity in the postnatal neocortex. J. Neurosci. 2010;30:7028–7036. doi: 10.1523/JNEUROSCI.6131-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbs J.L., Vladar E.K., Axelrod J.D., Kintner C. Multicilin promotes centriole assembly and ciliogenesis during multiciliate cell differentiation. Nat. Cell Biol. 2012;14:140–147. doi: 10.1038/ncb2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W., Cornwell A., Li J., Peng S., Osorio M.J., Aalling N., Wang S., Benraiss A., Lou N., Goldman S.A., Nedergaard M. SOX9 Is an Astrocyte-Specific Nuclear Marker in the Adult Brain Outside the Neurogenic Regions. J. Neurosci. 2017;37:4493–4507. doi: 10.1523/JNEUROSCI.3199-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavazoie M., Van der Veken L., Silva-Vargas V., Louissaint M., Colonna L., Zaidi B., Garcia-Verdugo J.M., Doetsch F. A specialized vascular niche for adult neural stem cells. Cell Stem Cell. 2008;3:279–288. doi: 10.1016/j.stem.2008.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terré B., Piergiovanni G., Segura-Bayona S., Gil-Gómez G., Youssef S.A., Attolini C.S., Wilsch-Bräuninger M., Jung C., Rojas A.M., Marjanović M. GEMC1 is a critical regulator of multiciliated cell differentiation. EMBO J. 2016;35:942–960. doi: 10.15252/embj.201592821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Tsai J.-W., Imai J.H., Lian W.-N., Vallee R.B., Shi S.-H. Asymmetric centrosome inheritance maintains neural progenitors in the neocortex. Nature. 2009;461:947–955. doi: 10.1038/nature08435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q., Tam M., Anderson S.A. Fate mapping Nkx2.1-lineage cells in the mouse telencephalon. J. Comp. Neurol. 2008;506:16–29. doi: 10.1002/cne.21529. [DOI] [PubMed] [Google Scholar]
- Zhou F., Narasimhan V., Shboul M., Chong Y.L., Reversade B., Roy S. Gmnc Is a Master Regulator of the Multiciliated Cell Differentiation Program. Curr. Biol. 2015;25:3267–3273. doi: 10.1016/j.cub.2015.10.062. [DOI] [PubMed] [Google Scholar]
- Zimmerman L., Parr B., Lendahl U., Cunningham M., McKay R., Gavin B., Mann J., Vassileva G., McMahon A. Independent regulatory elements in the nestin gene direct transgene expression to neural stem cells or muscle precursors. Neuron. 1994;12:11–24. doi: 10.1016/0896-6273(94)90148-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Several macros were created using the MATLAB software to use for the automatic analysis of MAGIC markers. They will be available upon request to the corresponding author.