Abstract
Background
Breast cancer brain metastases (BrMs) are defined by complex adaptations to both adjuvant treatment regimens and the brain microenvironment. Consequences of these alterations remain poorly understood, as does their potential for clinical targeting. We utilized genome-wide molecular profiling to identify therapeutic targets acquired in metastatic disease.
Methods
Gene expression profiling of 21 patient-matched primary breast tumors and their associated brain metastases was performed by TrueSeq RNA-sequencing to determine clinically actionable BrM target genes. Identified targets were functionally validated using small molecule inhibitors in a cohort of resected BrM ex vivo explants (n = 4) and in a patient-derived xenograft (PDX) model of BrM. All statistical tests were two-sided.
Results
Considerable shifts in breast cancer cell-specific gene expression profiles were observed (1314 genes upregulated in BrM; 1702 genes downregulated in BrM; DESeq; fold change > 1.5, Padj < .05). Subsequent bioinformatic analysis for readily druggable targets revealed recurrent gains in RET expression and human epidermal growth factor receptor 2 (HER2) signaling. Small molecule inhibition of RET and HER2 in ex vivo patient BrM models (n = 4) resulted in statistically significantly reduced proliferation (P < .001 in four of four models). Furthermore, RET and HER2 inhibition in a PDX model of BrM led to a statistically significant antitumor response vs control (n = 4, % tumor growth inhibition [mean difference; SD], anti-RET = 86.3% [1176; 258.3], P < .001; anti-HER2 = 91.2% [1114; 257.9], P < .01).
Conclusions
RNA-seq profiling of longitudinally collected specimens uncovered recurrent gene expression acquisitions in metastatic tumors, distinct from matched primary tumors. Critically, we identify aberrations in key oncogenic pathways and provide functional evidence for their suitability as therapeutic targets. Altogether, this study establishes recurrent, acquired vulnerabilities in BrM that warrant immediate clinical investigation and suggests paired specimen expression profiling as a compelling and underutilized strategy to identify targetable dependencies in advanced cancers.
Breast cancer brain metastases (BrMs) occur in 10%–30% of patients with metastatic breast cancer. With the advent of better systemic therapies, BrMs are increasing in incidence and confer a dismal prognosis. Treatment options for BrMs are limited, with radiation therapy and surgical excision being the mainstay (1). Although recently small molecule inhibitors of human epidermal growth factor receptor 2 (HER2) have had encouraging activity in HER2-positive BrMs, to date no targeted therapies have demonstrated efficacy in HER2-negative BrM (2–4).
Ongoing temporal genomic analyses of primary and metastatic cancers are beginning to reveal the extent of divergent tumor evolution (5–8). However, despite pressing clinical need, little is known about BrM due in part to limited sample availability. Previous research using experimental models and primary tumor data sets has proposed gene signatures and mechanisms of disease progression relating to BrM (9–11). More recently, targeted mutational analysis on longitudinal breast and BrM samples illustrated acquired mutations affecting the PI3k/AKT/mTOR pathway (12). A comprehensive characterization of the global transcriptional landscape of BrM and its divergence from primary breast cancer nonetheless remains incomplete.
Here, we comprehensively analyze the transcriptome across 21 cases of patient-matched primary breast tumors and their associated BrMs. We explore the transcriptional reprogramming of breast cancer cells as a critical step upon brain metastases and report on clinically actionable alterations acquired in BrMs that warrant immediate clinical investigation.
Methods
Sample Selection
Eligible breast cancer patients had paired formalin-fixed paraffin-embedded (FFPE) tissue from primary and resected BrMs processed for analysis. Informed consent was received from all patients, and the study was approved by institutional review boards (IRBs) from both participating institutions (University of Pittsburgh IRB#PRO15050502, Royal College of Surgeons in Ireland IRB#13/09/ICORG09/07). Tumor tissues were analyzed by a pathologist for histological and tumor cellularity classifications before sequencing. All specimens had a tumor cellularity equal to or greater than 60% except for BM_Pitt_68 (40%) and BM_Pitt_71 (30%). FFPE sections immediately adjacent to the hematoxylin and eosin–analyzed section underwent dual DNA/RNA extraction using Qiagen’s AllPrep kit according to the manufacturer’s instructions.
RNA-Sequencing
Library preparation was performed using 100 ng of RNA and Illumina’s TruSeq RNA Access Library Preparation protocol. Full details on sequencing and bioinformatics analyses are provided in the Supplementary Methods (available online).
DNA Methylation
Details on sequencing and analysis are available in the Supplementary Methods (available online).
Immunohistochemistry
Immunohistochemistry (IHC) was carried out using the Dako EnVisionTM Kit, as described previously (13). Full details on the protocol and antibodies used can be found in the Supplementary Methods (available online).
In Vitro Studies
Full details on in vitro studies and cell lines used are provided in the Supplementary Methods (available online).
Patient-Derived Brain Metastases Ex Vivo Samples
To establish patient-derived BrM ex vivo models, fresh intact tumor tissue was collected, anonymized, and placed in DMEM/F12 on ice immediately after surgical resection from the brain. Establishment and experimental protocol are provided in the Supplementary Methods (available online).
In Vivo
Mouse experiments were conducted under the Institutional Animal Care and Use Committee approval and in collaboration with Champions Oncology. Five- to eight-week-old immunocompromised female nu/nu nude mice (Harlan Laboratories) were implanted subcutaneously into the left flank with the tumor fragments. Tumor growth was monitored twice weekly using digital calipers, and the tumor volume (TV) was calculated. When the TV reached approximately 150–300 mm3, mice were matched by tumor size and assigned into control or treatment groups (n = 4/group). Researchers were not blinded to the treatment groups. Effects on tumor growth were evaluated by measuring percent tumor growth inhibition (TGI). The study was terminated when the mean tumor volume in the control group reached approximately 1500 mm3. Full experimental details are provided in the Supplementary Methods (available online).
Statistical Analysis
All statistical tests were two-sided, and a P value of less than .05 was considered statistically significant. Differentially expressed genes between patient-matched primary tumors and brain metastases were determined with DESeq2 utilizing a negative binomial distribution to assign differential expression P values. For single-gene queries, paired Wilcoxon signed-ranked tests on log2normCPM values were used. For survival analyses, log-rank tests were used to illustrate statistically significant differences in event probabilities (14). For in vitro/ex vivo and in vivo analyses, P values were obtained using a t test and analysis of variance (ANOVA), followed by Newman-Keuls multiple comparison test, respectively (GraphPad Prism), where indicated. No statistical method was used to predetermine sample size. The investigators were blinded for ex vivo and immunohistochemical analyses.
Data Availability
Gene expression data from patient-matched samples were deposited on https://github.com/npriedig.
Results
BrM Transcriptome Characterization
To identify recurrent alterations that can guide improved BrM treatment, we analyzed a cohort of patient-matched primary breast and paired brain metastases resected during routine clinical care (n = 21) (Table 1; Supplementary Table 1, available online). We performed genome-wide exome-capture RNA-seq. This method, developed specifically for FFPE samples, yields a highly concordant transcriptome when compared with standard FFPE protocols (Supplementary Figure 1, A–D, available online) or matched frozen samples (8).
Table 1.
Clinical information for the brain metastases cohort*
| Primary breast tumor |
Pre–brain metastasis treatment |
Recurrence before BrM | Brain metastases |
Post–brain metastasis treatment |
Progression, mo |
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case | Histo | Dx.Age | ER | PR | HER2 | Endo | Rx.HER2 | Radio | Chemo | ER | PR | HER2 | Endo | Rx.HER2 | Radio | Chemo | Status | DFS | BMSF | SPBM | OS | |
| 1_RCS | IDC | 49 | Neg | Neg | Pos | No | Yes | No | Yes | No | Neg | Neg | Pos | No | Yes | Yes | Yes | Dead | 20 | 20 | 11 | 32 |
| 2_RCS | IDC | 58 | Neg | Neg | Pos | No | Yes | Yes | Yes | Yes | Neg | Neg | Pos | No | Yes | Yes | No | Dead | 61 | 67 | 48 | 108 |
| 3_RCS | IDC | 61 | Pos | Neg | Pos | No | Yes | No | Yes | No | Pos | Neg | Pos | Yes | No | Yes | No | Alive | 37 | 37 | 67 | 76 |
| 4_RCS | IDC | 53 | Pos | Neg | Neg | Yes | No | Yes | Yes | No | Pos | Neg | Pos | No | No | Yes | No | Dead | 66 | 66 | 24 | 90 |
| 5_RCS | IDC | 38 | Neg | Neg | Neg | No | No | No | Yes | No | Neg | Neg | Neg | No | No | Yes | Yes | Dead | 23 | 23 | 17 | 40 |
| 6_RCS | IDC | 45 | Pos | Neg | Neg | Yes | No | No | No | Yes | Pos | Neg | Pos | No | No | Yes | No | Dead | 53 | 53 | 21 | 74 |
| 6_Pitt | IDC | 66 | Neg | Neg | Neg | No | No | Yes | Yes | Yes | Neg | Neg | Neg | No | No | Yes | Yes | Dead | 23 | 25 | 12 | 37 |
| 7_Pitt | IDC | 40 | Pos | Neg | Pos | Yes | Yes | Yes | Yes | – | Pos | NA | Pos | NA | Yes | Yes | Yes | Dead | 0 | 5 | 13 | 18 |
| 12_Pitt | IDC | 38 | Neg | Neg | Neg | No | No | Yes | Yes | – | Neg | Neg | Neg | No | No | Yes | Yes | Dead | 0 | 31 | 14 | 46 |
| 17_Pitt | MDC | 36 | Pos | Neg | Pos | No | Yes | Yes | Yes | No | Pos | Pos | Pos | Yes | Yes | Yes | No | Dead | 12 | 12 | 30 | 42 |
| 19_2_Pitt | IDC | 57 | Neg | Neg | Pos | No | No | Yes | Yes | No | Neg | NA | Pos | NA | NA | Yes | Yes | Dead | 17 | 17 | 16 | 33 |
| 25_Pitt | IDC | 66 | Neg | Neg | Neg | No | No | Yes | Yes | Yes | Neg | NA | Neg | No | No | No | Yes | Dead | 24 | 40 | 3 | 43 |
| 29_Pitt | IDC | 52 | Neg | Neg | Neg | No | No | Yes | Yes | No | Neg | NA | Neg | NA | NA | NA | NA | Dead | 22 | 22 | 5 | 28 |
| 47_Pitt | IDC/ILC | 53 | Pos | Pos | Pos | Yes | No | Yes | Yes | Yes | Pos | Neg | Pos | Yes | Yes | Yes | Yes | Dead | 83 | 151 | 74 | 225 |
| 51_Pitt | IDC | 60 | Pos | Neg | Neg | Yes | No | Yes | Yes | Yes | Pos | Pos | Neg | No | No | Yes | Yes | Dead | 18 | 57 | 9 | 66 |
| 52_Pitt | IDC | 62 | Neg | Pos | Pos | No | No | Yes | Yes | Yes | Neg | Pos | Pos | No | Yes | Yes | Yes | Dead | 36 | 55 | 7 | 63 |
| 62_Pitt | IDC | 63 | Pos | Pos | Neg | Yes | No | No | Yes | Yes | Pos | NA | Pos | No | Yes | Yes | Yes | Dead | 39 | 53 | 6 | 60 |
| 64_Pitt | IDC | 39 | Neg | Neg | Neg | Yes | No | Yes | Yes | Yes | Neg | Neg | Neg | No | No | Yes | Yes | Dead | 75 | 89 | 5 | 94 |
| 68_Pitt | IDC | 51 | Neg | Neg | Neg | Yes | No | Yes | Yes | No | Neg | NA | Neg | Yes | No | Yes | No | Dead | 20 | 20 | 114 | 135 |
| 71_Pitt | IDC | 26 | Neg | Neg | Neg | No | No | Yes | Yes | No | Neg | Neg | Neg | No | No | Yes | Yes | Alive | 25 | 25 | 147 | 173 |
| 72_Pitt | ILC | 55 | Pos | Pos | Neg | Yes | No | Yes | Yes | – | Pos | Pos | Neg | NA | No | Yes | Yes | Dead | 0 | 31 | 5 | 37 |
- = not determined; BMFS = brain metastasis–free survival, time from primary diagnosis to death or last follow-up; Chemo = chemotherapy; Dx.Age = age at primary breast diagnosis; BrM = brain metastasis; DFS = disease-free survival, time from primary diagnosis to first recurrence; Endo = endocrine treatment; ER = estrogen receptor; HER2 = human epidermal growth factor receptor 2; IDC = invasive ductal carcinoma; ILC = invasive lobular carcinoma; MDC = mucinous ductal carcinoma; NA = not available; neg = negative; OS = overall survival, time from primary diagnosis to death or last follow-up; pos = positive; PR = progesterone receptor; Radio = radiotherapy; Rx.HER2 = targeted HER2 therapy; SPBM = survival post–brain metastasis, time from brain metastasis to death or last follow-up.
Differential gene expression analyses revealed a catalog of recurrently altered genes in BrM (1314 genes upregulated in BrM; 1702 genes downregulated in BrM; DESeq; fold change > 1.5, Padj < .05) (Supplementary Table 2, available online). Correspondence analysis demonstrated that despite gene expression divergence from primary to BrM, samples grouped based on molecular subtype (Figure 1A). Indeed, unsupervised hierarchical clustering revealed three major clusters: estrogen receptor (ER)–positive, HER2-positive, and ER-negative disease; 38.1% (8/21) of the patient-matched primary and metastatic tumor samples clustered as related pairs in the dendrogram (Figure 1B). Furthermore, 90.5% (19/21) brain metastases retained the intrinsic molecular subtype (PAM50) of the matched primary tumor (Figure 1C), consistent with the previous observations using targeted nanoString analysis (15). Despite this broad conservation, 47.6% (10/21) brain metastases showed deviations (>10%) of PAM50 subtype probabilities from their patient-matched primaries, with the most common shifts being gains in HER2 and LumB profiles (Figure 1C; Supplementary Table 3, available online), in line with recent PAM50 analyses in metastatic tumors (16).
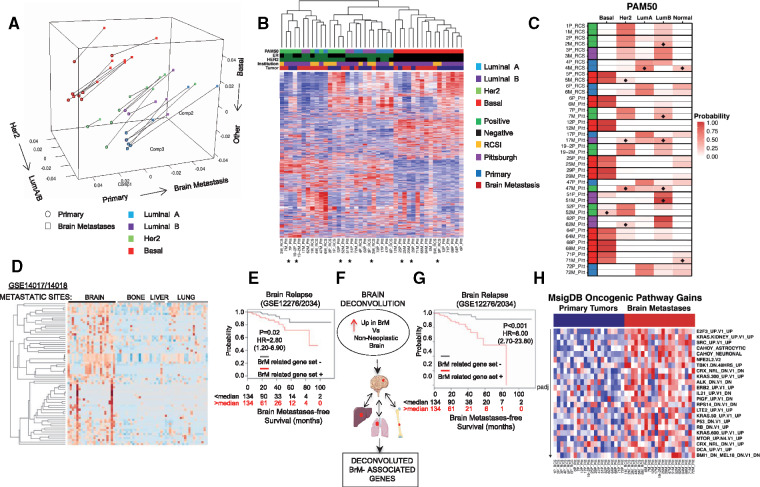
Figure 1.
Transcriptome evolution in breast cancer brain metastasis. A) Correspondence analysis showing overall trends in paired samples (n = 21) using the gene expression of all protein coding genes. The matched primary (circles) and metastasis samples (squares) are paired via a connecting line. The first component (“Comp1”) represents the strongest trend and splits the samples from the primary to the metastasis; the other two components split the samples by intrinsic subtype. B) Unsupervised hierarchical clustering heatmap. Patient-matched primary and metastatic tumor samples (n = 21) that clustered as related pairs in the dendogram are indicated with an asterisk. C) PAM50 intrinsic molecular subtype calls in patient-matched samples (n = 21). Probability for each subtype is the mean of all 20-fold test probabilities; tile plot denotes this probability for each subtype. Diamonds indicate brain metastases with greater than 10% probability gain in PAM50 subtypes. Legend denotes PAM50 subtype (blue = luminal A, purple = luminal B, green = HER2, red = basal), hormone status (green = positive, black = negative), tissue source (yellow = Royal College of Surgeons, Ireland; purple = University of Pittsburgh, United States), and tumor site (blue = primary, red = metastasis). D) Recurrent differentially upregulated genes (n = 1314) were screened in two merged public metastatic cohorts (GSE14017/18). Heatmap displays 62 genes whose expression was upregulated in brain metastases (BrMs) but not in metastases to lung, liver, or bone (BrM-related gene set). E) Kaplan-Meier curves for brain metastasis–free survival of BrM-related gene set status in two cohorts (n = 268; GSE12276/2034). P value based on two-sided log-rank test. F) Schematic of the workflow for uncovering decontaminated brain metastasis–related genes. G) Kaplan-Meier curves for brain metastasis–free survival on the basis of decontaminated BrM-related gene set (n = 11) status in two cohorts (n = 268; GSE12276/2034). P value based on two-sided log-rank test. H) Gene set variation analysis utilizing MsigDB Oncogenic Pathway (MsigDB). Heatmap illustrates brain metastasis–enriched pathways (false discovery rate–adjusted Wilcoxon signed-ranked P < .05) in brain metastases vs primaries. BrM = brain metastasis; ER = estrogen receptor; HER2 = human epidermal growth factor receptor 2; RCSI = Royal College of Surgeons in Ireland.
To identify determinants of brain metastasis proficiency, we interrogated the overexpressed BrM genes in an expression data set with multiple metastatic sites (17). Of the 1314 upregulated in BrM genes, we focused on those expressed in BrM cohorts at a higher level (>1.5-fold) than in metastases from other sites; 7.9% of the genes satisfied these criteria (Figure 1D;Supplementary Figure 2, A and B, available online). Notably, in established cohorts of primary breast cancer tumors with extended follow-up (9,18), expression of this BrM-related gene set statistically significantly was associated with brain (hazard ratio [HR] = 2.80, 95% confidence interval [CI] = 1.20 to 6.90, P = .02) and lung relapse (HR = 2.90, 95% CI = 1.70 to 4.90, P < .001) but not relapse to either bone or the liver (Figure 1E;Supplementary Figure 2B, available online). To further define brain tumor–associated genes, we developed a brain deconvolution approach to remove potentially contaminating non-neoplastic brain genes (Supplementary Figure 2, C–G, Supplementary Table 2, available online). A deconvoluted BrM gene set had a highly statistically significant association with brain relapse (HR = 8.00, 95% CI = 2.70 to 23.80, P < .001) (Figure 1, F and G; Supplementary Figure 3, available online).
Beyond identifying alterations in genes important in the brain metastatic process, including enrichment in genes implicated in vascular co-option (L1CAM) (10) and metastatic outgrowth (SOX2) (19), using gene set variation analysis (GSVA) (20), we further delineated expression changes in BrM from matched primaries by identifying several oncogenic pathway gains in BrM (21). These included gene sets associated with cell cycle dysregulation (E2F3, RB), proto-oncogenes (KRAS, ALK), and kinase-driven pathways (SRC, mTOR, HER2) (Figure 1H).
Recurrent Expression Gains of Clinically Actionable Kinase Pathways in BrM
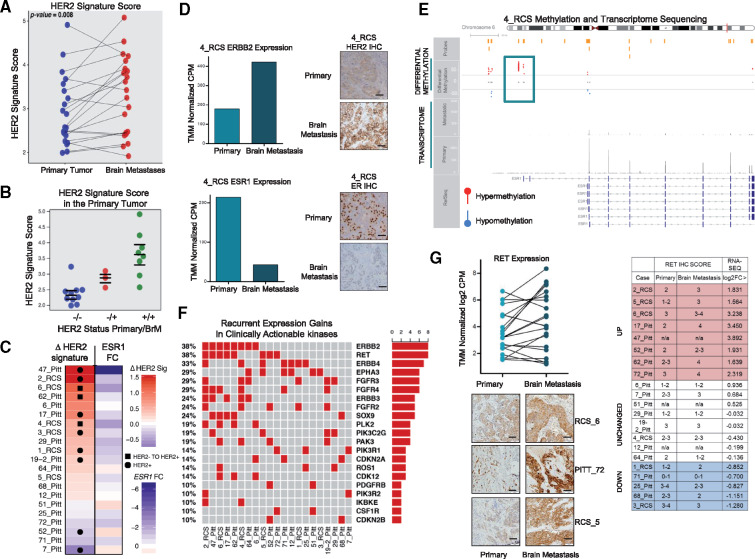
We explored HER2 pathway activation in BrMs using an established HER2 signature (22), given HER2 expression increase in up to 35% of BrMs relative to matched primaries (15) and a statistically significant HER2 pathway enrichment from GSVA (P = .006) (Figure 1H). Here, we show that 71.4% (15/21, P = .008) of pairs harbored elevated HER2 signature scores in BrMs relative to the matched primary (Figure 2A;Supplementary Figure 4A, available online). Indeed, tumors that switched from HER2-negative to HER2-positive in the BrM had intermediate HER2 signature scores in the primary tumor (Figure 2B;Supplementary Figure 4B, available online). Interestingly, 6_RCS and 62_PITT had an ERBB2 amplification gain in BrM compared with the primary (Supplementary Table 1, available online). Loss of ESR1 gene expression, a known mediator of hormone therapy resistance (13,23), correlated with increases in HER2 signature (Figure 2C;Supplementary Figure 4, C and D, available online). In case 4_RCS, loss of ESR1 was accompanied by enhanced ESR1 hypermethylation acquired in BrM compared with the primary tumor (Figure 2, D and E).
Figure 2.
Recurrent expression gains of clinically actionable kinase pathways in breast cancer brain metastases. A) Paired ladder plot of established genes represented in the human epidermal growth factor receptor 2 (HER2) signature depicts the expression change in patient-matched cases (P = .008; two-sided Wilcoxon signed-rank test; primaries vs brain metastases). Blue dots represent primary tumor signature scores, and red dots represent metastatic tumor signature scores. B) Scatter plot of HER2 signature score in primary tumors. Blue dots (-/-) represent patient-matched cases that are HER2 negative in both the primary and metastatic tumors, red dots (-/+) represent patient-matched cases that switched from HER2 negative to positive, whereas green dots (+/+) represent HER2-positive tumors that have further activation in the HER2 pathway. C) Tile plot indicates gain of HER2 signature or loss of ESR1 expression. Squares represent patient-matched cases that switched from HER2 negative to positive, whereas circles represent HER2-positive tumors that have further activation in HER2 pathways. D) Primary and metastatic log2normCPM values of ESR1/ERBB2 from case 4_RCS, along with immunohistochemistry protein analysis. Images shown are 20×; scale bars correspond to 50 μm. E) ESR1 gene differentially methylated regions (DMRs) identified with methyl capture sequencing are illustrated and were identified by comparing 4_RCS case primary and brain metastasis. Plot shows regions of hypermethylation and hypomethylation found in the ESR1 gene. F) OncoPrint of clinically actionable kinases (DGIdb) with discrete expression gains in brain metastases. G) Paired ladder plot of RET expression in patient-matched cases. Light green dots represent primary tumor expression values, and dark green dots represent metastatic tumor expression values (log2normCPM). Representative primary and metastatic immunohistochemistry (IHC) staining of RET protein from case 6_RCS, 72_PITT and 5_RCS; along with a graphic displaying RET IHC scores alongside corresponding log2FC RET mRNA scores for the sequenced paired samples. Images shown are 20×; scale bars correspond to 50 μm. BrM = brain metastasis; HER2 = human epidermal growth factor receptor 2; IHC = immunohistochemistry.
Given BrM-acquired gains in multiple kinase-driven signaling pathways, we examined clinically actionable kinases for recurrent expression gains (DGIdb 2.0). The most recurrent expression gains in BrM were RET and ERBB2 (both gained in 38% of BrMs) (Figure 2F). Alterations observed in RET mRNA in BrMs were confirmed in patient tumors at the protein level by IHC (Figure 2G;Supplementary Figure 4E, available online).
Inhibition of RET and HER2 in Breast Cancer Brain Metastases Ex Vivo and In Vivo
We next evaluated the effect of RET and HER2 inhibition in BrM models using the RET inhibitor cabozantinib and pan-HER pathway inhibitor afatinib. In vitro, we observed that treatment with either cabozantinib or afatinib had a statistically significant effect on the cellular viability (P < .001, P < .001) and migratory capacity (P < .001, P = .002) of TNBC MDA-231-BrM2 and ER-positive LY2 (P = .01, P = .002) brain-colonizing cell lines, along with T347-2c primary cells derived from patient BrM tumor (P < .001, P < .001) (Supplementary Figure 5, A–C, available online). Moreover, combination of cabozantinib with afatinib did not provide additional benefit, suggesting that RET inhibition may have sufficient efficacy as a single agent to treat BrM.
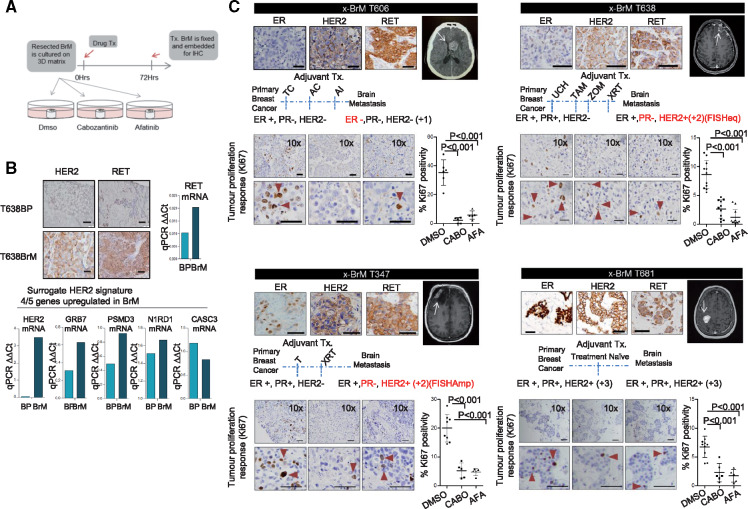
For preclinical assessment of the efficacy of cabozantinib and afatinib on BrM, we developed an ex vivo culture of BrM samples obtained from patients undergoing BrM resection (Figure 3A). The pathology of these metastatic tumors recapitulates the key receptor subtype alterations relevant to our sequencing study. Ex vivo Patient 1 (x-BrM-T606) had endocrine-resistant disease, with loss of ER expression resulting in a triple-negative brain metastatic tumor, whereas Patient 2 (x-BrM-T347) and Patient 3 (x-BrM-T638) lost PR and gained HER2. Ex vivo Patient 4 (x-BrM-T681) was treatment naïve. Where matched primary and metastatic tissue was available (T638), gains in transcript and protein expression of the receptors RET and HER2 were observed, along with elevations in the HER2 signature (Figure 3B). We observed tumor-specific RET expression in all ex vivo models (Figure 3C). Clinically, two BrM explant models harbored ERBB2 amplifications (T347 and T681), and two were non-ERBB2-amplified (T606 and T638) (Supplementary Table 1, available online). HER2 was highly expressed in x-BrM T347, T681, and T638, whereas x-BrM T606 harbored weak expression and was clinically graded as +1. Whole-exome sequencing (WES) of these tumors revealed multiple mutations in HER family members (Supplementary Table 4, available online). However, there was a notable absence of mutations relating to the ERBB2/ERBB3 kinase domain (Supplementary Table 4, available online) or other mutations previously identified as activating (24,25).
Figure 3.
Inhibition of RET and human epidermal growth factor receptor 2 (HER2) in breast cancer brain metastases ex vivo. A) Schematic of the ex vivo experimental set up. B) Immunohistochemistry (IHC) protein analysis of HER2/RET from case T638P (primary breast) and patient-matched T638 brain metastasis (BrM). Images shown are 20×; scale bars correspond to 50 μm. Also shown is mRNA expression levels of RET and key modules of HER2 signature (HER2, PSMD3, CASC3, GRB7, and N1RD1) analyzed by Taqman polymerase chain reaction. The bar chart displays ΔΔCt values for each gene. C) Brain metastatic tissue (x-BrMT606, T347, T638, and T681) was treated with vehicle (0.1% DMSO), 10 nM cabozantinib, and 25 nM afatinib and processed as described. IHC was carried out to profile ER, HER2, and RET of the ex vivo sample. Magnetic resonance/computed tomography images of the brain metastases resected are shown. Estrogen receptor, progesterone receptor, and HER2 status in primary and brain metastases are indicated alongside adjuvant treatment received before resection. Representative images of IHC analyses of Ki67 tumors treated for 72 hours with indicated treatments (positive cells indicated with red triangles). All analyses of variance, followed by Dunnett’s test. All statistical tests were two-sided. AC = cyclophosphamide/doxorubicin; AFA = afatinib; AI = aromatase inhibitor; BrM = brain metastasis; CABO = cabozantinib; ER = estrogen receptor; HER2 = human epidermal growth factor receptor 2; PR = progesterone receptor; qPCR = quantitative polymerase chain reaction; TAM = tamoxifen; T = taxol; TC = taxol/carboplatin; UCH = unknown chemotherapy; xBrM = brain metastases explant; XRT = radiotherapy; ZOM = zometa.
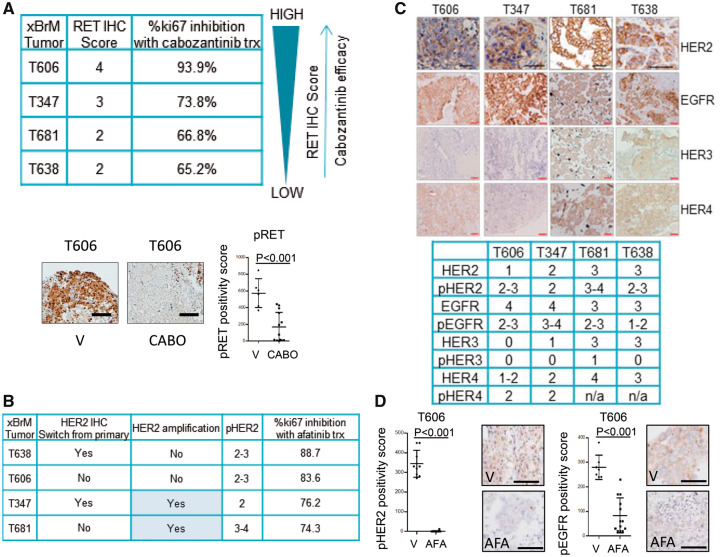
A statistically significant tumor response to afatinib inhibition in each of the x-BrM models was observed (mean difference [SD], T606 = 33.27 [6.93], T347 = 14.76 [2.34], T638 = 5.96 [0.87], T681 = 4.51 [0.9], all P < .001) (Figure 3C). Similarly, cabozantinib demonstrated substantial antitumor efficacy in each of the x-BrM models, as demonstrated by a statistically significant decrease in proliferating cells (ki67+) compared with vehicle-treated tumors (mean difference [SD], T606 = 29.62 [5.98], T347 = 15.26 [2.22], T638 = 7.42 [0.84], T681 = 5.01 [0.9], all P < .001) (Figure 3C). Sensitivity to cabozantinib treatment was found to be directly related to RET expression of the BrM tumor (Figure 4A).
Figure 4.
Ex vivo effects of RET and human epidermal growth factor receptor 2 (HER2) inhibition on downstream signaling pathways. A) Analysis of cabozantinib antitumor efficacy in xBrM explants. xBrM explants were designated an RET immunohistochemistry (IHC) positivity score. The cabozantinib efficacy with respect to RET IHC score was calculated based on %ki67 inhibition where (100%- (mean ki67% cabozantinib-treated samples/mean ki67% DMSO-treated)*100 = %ki67 inhibition with cabozantinib treatment. xBrM T606 was analyzed for pRET(Y1062) IHC expression in vehicle (DMSO; V)- and cabozantinib (CABO)-treated samples. Representative images of IHC analyses of the tumors treated for 72 hours with indicated treatments. All scale bars = 50 μm. Error bars represent mean (SD) (n = 5–10 images per group), two-tailed paired t test. B) Afatinib efficacy with respect to ERBB2 amplification status and HER2 and pHER2 IHC score. C) Representative IHC staining of HER family members alongside key phosphorylated proteins pEGFR (Y1068), pHER2 (Y1221), pHER3 (Y1289), and pHER4 (Y1284), along with a graphic displaying IHC scores for each of the brain metastatic tissues utilized in the study. Images shown are 20×; scale bars correspond to 50 μm. D) T606 was analyzed for pHER2 (Y1221) and pEGFR (Y1068) IHC expression in vehicle (V)- and afatinib (AFA)-treated samples. Representative images of IHC analyses of the tumors treated for 72 hours with indicated treatments. All scale bars = 50 μm. Error bars represent mean (SD) (n = 5–10 images per group), two-tailed paired t test. AFA = afatinib; BrM = brain metastasis; CABO = cabozantinib; EGFR = epidermal growth factor receptor; HER2 = human epidermal growth factor receptor 2; IHC = immunohistochemistry; V = vehicle.
Of note, afatinib had an antiproliferative effect independent of HER2 amplification, as evidenced in x-BrM-T606 (and T638) (Figure 4B), though loss of both phospho-HER2 and phospho-EGFR on treatment was observed in T606 (Figure 4D). To understand the mechanism underlying the antiproliferative response to cabozantinib and afatinib in the triple-negative xBrM-T606, we explored downstream effectors and key signaling nodes. Cabozantinib treatment successfully abrogated AKT/mTOR and SRC pathways, downstream of RET (indicated by reductions in pAKT, p70S6K, and pSRC) (Supplementary Figure 5D, available online). These pathways were not statistically significantly inhibited by afatinib (Supplementary Figure 5D, available online). Notably, we do see a reduction in pRAF and pERK signaling with afatinib treatment, indicating that antitumor effects of afatinib may be in part due to inhibition of EGFR, in addition to phospho-HER2 (Supplementary Figure 5E, available online).
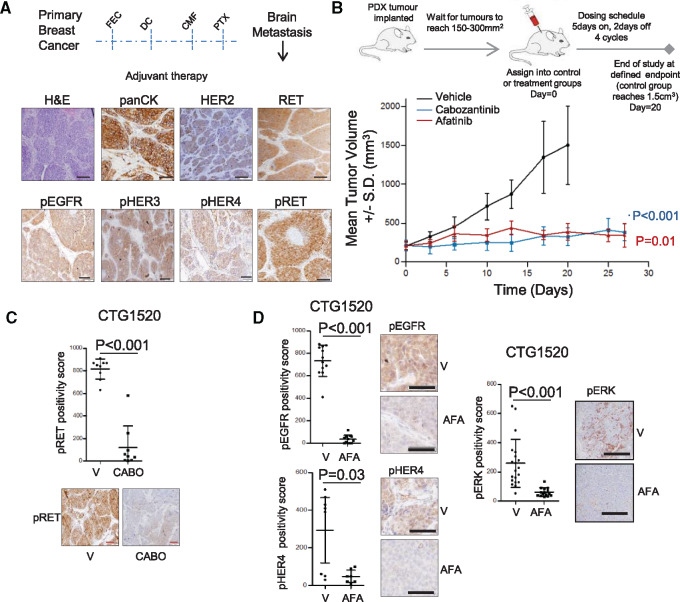
We next evaluated the effect of cabozantinib and afatinib in a BrM patient–derived xenograft (CTG-1520) established from a triple-negative tumor (Supplementary Table 1, available online). The metastatic tumor, though clinically HER2 negative (IHC +1; non-ERBB2 amplified), expressed high levels of phospho-RET, phospho-EGFR, phospho-HER3, and phospho-HER4 (Figure 5A). Cabozantinib and afatinib showed similar and statistically significant antitumor activity leading to stable disease (no progression and no regression of treated tumors) compared with vehicle treatment in the BrM PDX model (n = 4, % tumor growth inhibition [mean difference; SD], cabozantinib = 86.3% [1176; 258.3], P < .001; afatinib = 91.2% [1114; 257.9], P < .01) (Figure 5B). Treatment with cabozantinib statistically significantly reduced phospho-RET expression (P < .001) (Figure 5C), whereas afatinib inhibited phospho-EGFR, phospho-HER4, and phospho-ERK (Figure 5D) and induced a loss in key HER2-related genes (Supplementary Figure 5F, available online).
Figure 5.
Inhibition of RET and human epidermal growth factor receptor 2 (HER2) in breast cancer brain metastases in vivo. A) Schematic indicates clinical information pertaining to brain metastasis (BrM) patient-derived xenograft (PDX) CTG-1520 and the experimental design of the in vivo experiment. Representative immunohistochemistry (IHC) images of hematoxylin and eosin, pan cytokeratin, HER2, RET, pRET(Y1062), pEGFR (Y1068), pHER3 (Y1289), and pHER4 (Y1284) are shown. Scale bars = 50 μm. B) Schematic of the in vivo experimental setup. Treatment schedule was four cycles (QD×5 on/2 off) via oral gavage of vehicle (black line), 30 mg/kg cabozantinib (red line), and 20 mg/kg afatinib (blue line). Effects on tumor growth were evaluated with % tumor growth inhibition (%TGI). The tumor growth curve shows mean tumor volume +/-SD (n = 4 per treatment group). The analysis of variance test was followed by Newman-Keuls multiple comparison test. All statistical tests were two-sided. C) CTG1520 PDX was analyzed for pRET(Y1062) IHC expression in vehicle (V)- and cabozantinib (CABO)-treated samples. Representative images of IHC analyses of the tumors treated analyzed at the conclusion of the experiment. All scale bars = 100 μm. Error bars represent mean ±SD (n = 5–10 images per group), two-sided paired t test. D) CTG1520 PDX was analyzed for pEGFR (Y1068), pHER4 (Y1284), and pERK (T202/Y204) IHC expression in vehicle (V)- and afatinib (AFA)-treated samples. Representative images of IHC analyses of the tumors analyzed at the conclusion of the experiment. All scale bars = 50 μm. Error bars represent mean (SD) (n = 5–10 images per group), two-tailed paired t test. AFA = afatinib; BrM = brain metastasis; CABO = cabozantinib; CMF = cyclophosphamide, methotrexate, 5-fluorouracil; DC = docetaxel/carboplatin; EGFR = epidermal growth factor receptor; FEC = fluorouracil (5FU), epirubicin, cyclophosphamide; H&E = hematoxylin and eosin; HER2 = human epidermal growth factor receptor 2; PTX = paclitaxel; V = vehicle.
Discussion
Brain relapse can occur rapidly or many years after primary diagnosis, a facet of BrM latency reflected in our clinical cohort. Genomically, analyses of BrM suggest that cancer cells continue to evolve upon colonization of the brain parenchyma, with mutations that are both common and distinct to originating tumors (12). The observations presented here expand upon these findings and establish recurrent, longitudinal transcriptional remodeling events in breast cancer cells following brain colonization, shedding new light on the biology of BrM and potential therapeutic targets. Previous gene expression profiling approaches utilizing targeted gene panels and specific pathways highlighted key features of BrM biology but failed to yield direct actionable targets (26–29).
Our studies revealed a comprehensive list of genes enriched in BrM, including genes previously implicated in experimental models in the early events of vascular co-option (11) and those found to be essential for early survival and brain metastatic outgrowth (19). Our work also points to many novel candidate BrM genes, whose exact role in BrM is open to further analyses but that appear specific to cancer cells in the brain parenchyma. Indeed, a number of the clinically actionable targets investigated here were not found to be recurrently enriched in similar transcriptome analysis of cases that included primaries and extracranial metastatic sites (8). This BrM-related gene set statistically significantly associated with brain relapse in primary tumors. Given the overlap with lung relapse and the limited available data sets, these observations are not interpreted as a gene signature capable of predicting brain relapse with high selectivity. More complete analyses can be undertaken as further relevant cohorts become available. Nevertheless, these collective shifts in gene expression signify a molecularly dynamic tumor adapting to its new microenvironment that have a large degree of metastatic selectivity and clinical relevance.
Metastatic colonization and BrM outgrowth merges key adaptive pathways and alterations, and we demonstrate recurrent enrichment in druggable kinase-driven signaling. We show conclusive activation of the HER2 pathway in BrM, especially important given the increasing case reports of HER2-negative to HER2-positive switching (30,31) and the acquired HER2 mutational burden verified in BrM (12,15). Similarly, in a pan-cancer expression analysis of unmatched BrM, Saunus et al. reported that breast cancer BrMs have higher ERBB2 expression than BrMs from other sites (32). Additionally, the preclinical data presented here indicate that BrM tumors could potentially benefit from pan-HER inhibition, even in the absence of ERBB2 amplifications, a finding that is not surprising considering reports that HER pathway activation may occur independently of receptor amplification in BrM (32–35). Future trials therefore may need to evaluate the status of other HER family members in addition to HER2 in patients before the use of pan-HER inhibitors, in a similar manner to the SUMMIT trial (36), to better understand the potential role of these drugs in BrMs.
Notably, our transcriptional approach revealed no loss in PTEN expression, which has been proposed as a potential driver of PI3K/AKT activation in BrM (37,38). This concordance in PTEN expression in patient-matched samples has previously been reported (39) and does not rule out its potential biological significance in BrM, particularly in PTEN-mutated BrM. Perhaps more importantly, ESR1, a key clinically actionable gene, demonstrated consistent depletion in BrM compared with primary tumor. This loss of ESR1 gene expression, a known feature of hormone therapy–resistant disease, correlated with increases in HER2 signature. We further show that ER loss in brain metastases can be epigenetically driven, suggesting that further mechanistic studies into this process may be informative. The exact point at which these ESR1/ERBB2 alterations are acquired in the multistep metastatic process is unclear and could be addressed in longitudinal liquid biopsies or circulating tumor cell studies of patients with BrM going forward. Overall, these observations reinforce the dynamic regulatory interactions between ESR1 and HER2 (40) and expand their importance to the clinical setting of brain metastases.
Lastly, we define recurrent RET enrichment as a novel target for breast cancer BrMs. Expression and activation of RET contribute to disease progression in multiple tumor types and have been implicated in therapy resistance in breast cancer models (41–43), but RET mutations are rare in advanced breast cancer (44) and we detected no RET fusions within our own sequencing cohort. Cabozantinib, a multikinase RET inhibitor, has shown efficacy against various RET-driven tumors including extracranial advanced breast cancer (45–48). Here, we demonstrate statistically significant antitumor efficacy of RET targeting in vivo, leading to disease stabilization.
Our study is not without limitations. Given the relatively short duration of the in vivo intervention experiment due to ethical considerations, it remains to be determined whether BrM patients on RET inhibitor therapy would experience prolonged stabilization despite the reported concordances between PDX studies and clinical responses (49–52).
While a larger therapeutic data set that includes matching primary tumors with low expression of RET would have been desirable, the BrM ex vivo models utilized in this study demonstrated an antiproliferative response to RET targeting and they represent an important modeling tool as they recapitulate the cellular and molecular components observed in our comprehensive characterization of BrM. Though the response to cabozantinib could likely be augmented by the inhibition of other receptor tyrosine kinases and/or downstream pathways such as mTOR (37,43,53), the results reported here suggest no additional benefit of combined HER inhibitor treatment. The data presented here raise the possibility of anti-RET treatment as a single agent for the treatment of BrM. In future studies, the impact of the tumor cell–brain parenchyma interaction could be further assessed in intracranial PDX models that may provide additional translational findings on reactive astrocytes/neuro-inflammatory responses to this therapeutic intervention and the enduring effects of RET inhibition in aging neuron function (54,55).
More broadly, though limited overall intrinsic clinical subtype switching was observed, our study demonstrates that BrMs undergo a biologically significant transcriptome shift upon colonization. Enhanced cancer cell dependency on aberrant kinase pathways facilitates survival and outgrowth advantages, thereby presenting therapeutic opportunities for BrMs that are distinct from their matched primary tumors. These translational preclinical results deliver compelling proof of principle for exploiting longitudinal transcriptional changes in advanced cancer, which is especially important given the field’s current focus on DNA-level changes in tumor profiling.
Funding
We kindly acknowledge funding support from Irish Cancer Society Collaborative Cancer Research Centre grant CCRC13GAL (LY), Science Foundation Ireland Investigator Award 12/IA/1294 (LY), the Breast Cancer Ireland Programme Grant (LY), the Breast Cancer Research Foundation (AVL, SO), the National Cancer Institute (NCI) of the National Institutes of Health (NIH; P30CA047904), Fashion Footwear Association of New York (FFANY), the Nicole Meloche Memorial Breast Cancer Research Foundation, the Shear Family Foundation, and the Magee-Womens Research Institute and Foundation. AVL and SO are recipients of Scientific Advisory Council awards from Susan G. Komen for the Cure, and AVL is a Hillman Foundation Fellow. NP was supported by a training grant from the NIH/National Institute of General Medical Science (2T32GM008424-21) and an individual fellowship from the NIH/NCI (5F30CA203095).
Notes
Affiliations of authors: Endocrine Oncology Research Group, Department of Surgery, Royal College of Surgeons in Ireland, Dublin, Ireland (DV, AF, SP, NC, EW, SC, LSY); Department of Neurosurgery, National Neurosurgical Center, Beaumont Hospital, Dublin, Ireland (PJO); Departments of Pathology (PCL, RLH), Medicine (SLP, AMB, AM, JPL), Pharmacology and Chemical Biology (NP, RH, AB, SO, AVL), Human Genetics (AVL), Women’s Cancer Research Center, Magee-Women’s Research Institute (NP, CAC, SO, AVL), and Biostatistics (LZ, GCT), University of Pittsburgh Cancer Institute, University of Pittsburgh, PA; Departments of Neuropathology (PGB, MF), Molecular and Cellular Therapeutics (SD, DPO), and Surgical Research (LH, ADKH), Royal College of Surgeons in Ireland, Dublin, Ireland; Discipline of Surgery, School of Medicine, Lambe Institute for Translational Research, National University of Ireland, Galway, Ireland (RD).
The funders had no role in the design of the study; the collection, analysis, or interpretation of the data; the writing of the manuscript; or the decision to submit the manuscript for publication.
We are thankful to the patients who generously provided tumor tissue for our studies and to our surgical, pathology, and tissue bank colleagues for their substantial assistance and support. This project used the University of Pittsburgh HSCRF Genomics Research Core with the support of William Horne and Jay Kolls and the UPCI Tissue and Research Pathology Services, which is supported in part by award P30CA047904. We thank the UPMC Cancer Registry for clinical abstraction.
The authors declare that there are no related competing financial interests. RH is an employee of and has ownership interest in Foundation Medicine. SLP has consulted for AbbVie, MedImmune, Celldex, Puma, Pfizer, AstraZeneca, and Esai and received funding from AbbVie, Pfizer, Lilly, Novartis, Incyte, Covance-Bayer, AstraZeneca, Genentech, and Medivation.
Author contributions: study concept and design (DV, NP, SO, AVL, LSY); acquisition, analysis, or interpretation of data (DV, NP, AF, SP, NC, EW, SC, RH, SO, AVL, LSY), bioinformatic analyses (NP, AF, NC, LZ, GCT); provision of administrative, technical, or material support (JPL, PCL, RLM, PO, SC, LH, RD, SD, DPO, PB, MF, ADKH); drafting of the manuscript (DV, NP, AVL, LSY), critical revision of the manuscript (all authors); study supervision (AVL, LSY).
Supplementary Material
References
- 1. Lin NU, Bellon JR, Winer EP.. CNS metastases in breast cancer. J Clin Oncol. 2004;2217:3608–3617. [DOI] [PubMed] [Google Scholar]
- 2. Bachelot T, Romieu G, Campone M, et al. Lapatinib plus capecitabine in patients with previously untreated brain metastases from HER2-positive metastatic breast cancer (LANDSCAPE): A single-group phase 2 study. Lancet Oncol. 2013;141:64–71. [DOI] [PubMed] [Google Scholar]
- 3. Partridge AH, Rumble RB, Carey LA, et al. Chemotherapy and targeted therapy for women with human epidermal growth factor receptor 2-negative (or unknown) advanced breast cancer: American Society of Clinical Oncology Clinical Practice Guideline. J Clin Oncol. 2014;3229:3307–3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pivot X, Manikhas A, Zurawski B, et al. CEREBEL (EGF111438): A phase III, randomized, open-label study of lapatinib plus capecitabine versus trastuzumab plus capecitabine in patients with human epidermal growth factor receptor 2-positive metastatic breast cancer. J Clin Oncol. 2015;3314:1564–1573. [DOI] [PubMed] [Google Scholar]
- 5. Gundem G, Van Loo P, Kremeyer B, et al. The evolutionary history of lethal metastatic prostate cancer. Nature. 2015;5207547:353–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. McCreery MQ, Halliwill KD, Chin D, et al. Evolution of metastasis revealed by mutational landscapes of chemically induced skin cancers. Nat Med. 2015;2112:1514–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Patch AM, Christie EL, Etemadmoghadam D, et al. Whole-genome characterization of chemoresistant ovarian cancer. Nature. 2015;5217553:489–494. [DOI] [PubMed] [Google Scholar]
- 8. Priedigkeit N, Watters RJ, Lucas PC, et al. Exome-capture RNA sequencing of decade-old breast cancers and matched decalcified bone metastases. JCI Insight. 2017;217: pii: 95703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bos PD, Zhang XH, Nadal C, et al. Genes that mediate breast cancer metastasis to the brain. Nature. 2009;4597249:1005–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen Q, Boire A, Jin X, et al. Carcinoma-astrocyte gap junctions promote brain metastasis by cGAMP transfer. Nature. 2016;5337604:493–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Valiente M, Obenauf AC, Jin X, et al. Serpins promote cancer cell survival and vascular co-option in brain metastasis. Cell. 2014;1565:1002–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brastianos PK, Carter SL, Santagata S, et al. Genomic characterization of brain metastases reveals branched evolution and potential therapeutic targets. Cancer Discov. 2015;511:1164–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Vareslija D, McBryan J, Fagan A, et al. Adaptation to AI therapy in breast cancer can induce dynamic alterations in ER activity resulting in estrogen-independent metastatic tumors. Clin Cancer Res. 2016;2211:2765–2777. [DOI] [PubMed] [Google Scholar]
- 14. Bland JM, Altman DG.. The logrank test. BMJ. 2004;3287447:1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Priedigkeit N, Hartmaier RJ, Chen Y, et al. Intrinsic subtype switching and acquired ERBB2/HER2 amplifications and mutations in breast cancer brain metastases. JAMA Oncol. 2017;35:666–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cejalvo JM, Martinez de Duenas E, Galvan P, et al. Intrinsic subtypes and gene expression profiles in primary and metastatic breast cancer. Cancer Res. 2017;779:2213–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang XH, Wang Q, Gerald W, et al. Latent bone metastasis in breast cancer tied to Src-dependent survival signals. Cancer Cell. 2009;161:67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang Y, Klijn JG, Zhang Y, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;3659460:671–679. [DOI] [PubMed] [Google Scholar]
- 19. Malladi S, Macalinao DG, Jin X, et al. Metastatic latency and immune evasion through autocrine inhibition of WNT. Cell. 2016;1651:45–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hanzelmann S, Castelo R, Guinney J.. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liberzon A, Subramanian A, Pinchback R, et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;2712:1739–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Desmedt C, Haibe-Kains B, Wirapati P, et al. Biological processes associated with breast cancer clinical outcome depend on the molecular subtypes. Clin Cancer Res. 2008;1416:5158–5165. [DOI] [PubMed] [Google Scholar]
- 23. Osborne CK, Schiff R.. Mechanisms of endocrine resistance in breast cancer. Annu Rev Med. 2011;62:233–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bose R, Kavuri SM, Searleman AC, et al. Activating HER2 mutations in HER2 gene amplification negative breast cancer. Cancer Discov. 2013;32:224–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hyman DM, Piha-Paul SA, Won H, et al. HER kinase inhibition in patients with HER2- and HER3-mutant cancers. Nature. 2018;5547691:189–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lee JY, Park K, Lee E, et al. Gene expression profiling of breast cancer brain metastasis. Sci Rep. 2016;6:28623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Palmieri D, Bronder JL, Herring JM, et al. Her-2 overexpression increases the metastatic outgrowth of breast cancer cells in the brain. Cancer Res. 2007;679:4190–4198. [DOI] [PubMed] [Google Scholar]
- 28. Sevenich L, Bowman RL, Mason SD, et al. Analysis of tumour- and stroma-supplied proteolytic networks reveals a brain-metastasis-promoting role for cathepsin S. Nat Cell Biol. 2014;169:876–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Woditschka S, Evans L, Duchnowska R, et al. DNA double-strand break repair genes and oxidative damage in brain metastasis of breast cancer. J Natl Cancer Inst. 2014;1067:dju145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Duchnowska R, Dziadziuszko R, Trojanowski T, et al. Conversion of epidermal growth factor receptor 2 and hormone receptor expression in breast cancer metastases to the brain. Breast Cancer Res. 2012;144:R119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Thomson AH, McGrane J, Mathew J, et al. Changing molecular profile of brain metastases compared with matched breast primary cancers and impact on clinical outcomes. Br J Cancer. 2016;1147:793–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Saunus JM, Quinn MC, Patch AM, et al. Integrated genomic and transcriptomic analysis of human brain metastases identifies alterations of potential clinical significance. J Pathol. 2015;2373:363–378. [DOI] [PubMed] [Google Scholar]
- 33. Da Silva L, Simpson PT, Smart CE, et al. HER3 and downstream pathways are involved in colonization of brain metastases from breast cancer. Breast Cancer Res. 2010;124:R46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kodack DP, Askoxylakis V, Ferraro GB, et al. The brain microenvironment mediates resistance in luminal breast cancer to PI3K inhibition through HER3 activation. Science Translat Med. 2017;9391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sun M, Behrens C, Feng L, et al. HER family receptor abnormalities in lung cancer brain metastases and corresponding primary tumors. Clin Cancer Res. 2009;1515:4829–4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hyman DM, Piha-Paul SA, Won H, et al. HER kinase inhibition in patients with HER2- and HER3-mutant cancers. Nature. 5547691:189–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ni J, Ramkissoon SH, Xie S, et al. Combination inhibition of PI3K and mTORC1 yields durable remissions in mice bearing orthotopic patient-derived xenografts of HER2-positive breast cancer brain metastases. Nat Med. 2016;227:723–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang L, Zhang S, Yao J, et al. Microenvironment-induced PTEN loss by exosomal microRNA primes brain metastasis outgrowth. Nature. 2015;5277576:100–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Adamo B, Deal AM, Burrows E, et al. Phosphatidylinositol 3-kinase pathway activation in breast cancer brain metastases. Breast Cancer Res. 2011;136:R125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hurtado A, Holmes KA, Geistlinger TR, et al. Regulation of ERBB2 by oestrogen receptor-PAX2 determines response to tamoxifen. Nature. 2008;4567222:663–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mulligan LM. RET revisited: Expanding the oncogenic portfolio. Nat Rev Cancer. 2014;143:173–186. [DOI] [PubMed] [Google Scholar]
- 42. Morandi A, Martin LA, Gao Q, et al. GDNF-RET signaling in ER-positive breast cancers is a key determinant of response and resistance to aromatase inhibitors. Cancer Res. 2013;7312:3783–3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Plaza-Menacho I, Morandi A, Robertson D, et al. Targeting the receptor tyrosine kinase RET sensitizes breast cancer cells to tamoxifen treatment and reveals a role for RET in endocrine resistance. Oncogene. 2010;2933:4648–4657. [DOI] [PubMed] [Google Scholar]
- 44. Zehir A, Benayed R, Shah RH, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;236:703–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Drilon A, Wang L, Hasanovic A, et al. Response to Cabozantinib in patients with RET fusion-positive lung adenocarcinomas. Cancer Discov. 2013;36:630–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lipson D, Capelletti M, Yelensky R, et al. Identification of new ALK and RET gene fusions from colorectal and lung cancer biopsies. Nat Med. 2012;183:382–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Takeuchi K, Soda M, Togashi Y, et al. RET, ROS1 and ALK fusions in lung cancer. Nat Med. 2012;183:378–381. [DOI] [PubMed] [Google Scholar]
- 48. Tolaney SM, Nechushtan H, Ron IG, et al. Cabozantinib for metastatic breast carcinoma: Results of a phase II placebo-controlled randomized discontinuation study. Breast Cancer Res Treat. 2016;1602:305–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gao H, Korn JM, Ferretti S, et al. High-throughput screening using patient-derived tumor xenografts to predict clinical trial drug response. Nat Med. 2015;2111:1318–1325. [DOI] [PubMed] [Google Scholar]
- 50. Murphy B, Yin H, Maris JM, et al. Evaluation of alternative in vivo drug screening methodology: A single mouse analysis. Cancer Res. 2016;7619:5798–5809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Townsend EC, Murakami MA, Christodoulou A, et al. The public repository of xenografts enables discovery and randomized phase II-like trials in mice. Cancer Cell. 2016;294:574–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Williams JA. Patient-derived xenografts as cancer models for preclinical drug screening In: Wang Y, Lin D, Gout PW, eds. Patient-Derived Xenograft Models of Human Cancer. Cham, Switzerland: Springer International Publishing; 2017:141–154. [Google Scholar]
- 53. Gild ML, Landa I, Ryder M, et al. Targeting mTOR in RET mutant medullary and differentiated thyroid cancer cells. Endocr Relat Cancer. 2013;205:659–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jain S, Golden JP, Wozniak D, et al. RET is dispensable for maintenance of midbrain dopaminergic neurons in adult mice. J Neurosc.i 2006;2643:11230–11238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kramer ER, Aron L, Ramakers GM, et al. Absence of Ret signaling in mice causes progressive and late degeneration of the nigrostriatal system. PLoS Biol. 2007;53:e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Gene expression data from patient-matched samples were deposited on https://github.com/npriedig.