Abstract
Alcohol use in persons living with HIV (PLWH) worsens the severity of bacterial pneumonia. However, the exact mechanism(s) by which this occurs remain ill-defined. We hypothesized that alcohol in the setting of HIV infection decreases S. pneumoniae clearance from the lung through mechanisms mediated by the gut microbiota. Humanized BLT (bone marrow, liver, thymus) mice were infected with 1×104 TCID50 of HIV (BAL and JRCSF strains) via IP injection. One week post HIV infection animals were switched to a Lieber-DeCarli 5% ethanol diet or an isocaloric control diet for 10 days, alcohol-fed animals were also given 2 binges of 2 g kg−1 ethanol on days 5 and 10. Feces were also collected, banked, and the community structures were analyzed. Mice were then infected with 1×105 CFU of S. pneumoniae and sacrificed 48 hours later. HIV-infected mice had viral loads of ~2×104 copies/mL of blood one week post infection, and exhibited an ~57% decrease in the number of circulating CD4+ T-cells at the time of sacrifice. Fecal microbial community structure was significantly different in each of the feeding groups, as well as with HIV infection. Alcohol-fed mice had a significantly higher burden of S. pneumoniae 48 hours post infection, regardless of HIV status. In follow-up experiments, female C57BL/6 mice were treated with a cocktail of antibiotics daily for two weeks and recolonized by gavage with intestinal microbiota from HIV+ ethanol-fed, HIV+ pair-fed, HIV- ethanol-fed, or HIV- pair-fed mice. Recolonized mice were then infected with S. pneumoniae and sacrificed 48 hours later. The intestinal microbiota from alcohol-fed mice (regardless of HIV status) significantly impaired clearance of S. pneumoniae. Collectively, these data indicate that alcohol-feeding, as well as alcohol-associated intestinal dysbiosis compromise pulmonary host defense against pneumococcal pneumonia. Whether HIV infection acts synergistically with alcohol-use in impairing pulmonary host defense will require additional study.
Keywords: Alcohol, Pulmonary, host-defense, microbiota, S. pnuemoniae
Introduction:
Alcohol use disorders (AUD) and human immunodeficiency virus (HIV) infection are major public health problems in the United States and throughout the world.(“Alcohol Facts and Statistics | National Institute on Alcohol Abuse and Alcoholism (NIAAA),” 2016; “Alcohol Use Disorder | National Institute on Alcohol Abuse and Alcoholism (NIAAA),” 2016; Chander et al., 2008; Fitzpatrick, Brooks, & Kaplan, 2016) The prevalence of alcohol misuse among HIV-infected individuals is greater than the population as a whole, with 33% of HIV-infected subjects reporting any alcohol use and 18.6% meeting criteria for alcohol misuse.(Chander et al., 2008; Wandera et al., 2015) In spite of the high prevalence of alcohol use among HIV-infected patients, the consequence of AUD on HIV progression is ill defined. The most widely accepted consequence of AUD on HIV progression is a significant disruption of the HIV care continuum.(Vagenas et al., 2015) Specifically, diagnosis of HIV infection, engagement and retention in medical care, adherence with antiretroviral therapy, and achieving viral suppression are all disrupted by AUD.(Vagenas et al., 2015)
Further, opportunistic infections of the lung are a frequent complication of HIV infection.(Boyton, 2005; Cilloniz et al., 2017) In fact, in the United States despite the availability of combination antiretroviral therapy (cART), bacterial pneumonia is now the leading cause of community-acquired pneumonia in HIV-infected patients, as Pneumocystis jirovecii has been supplanted by Streptococcus pneumoniae as the leading etiologic agent.(Boyton, 2005; Cilloniz et al., 2017; Fitzpatrick et al., 2016; Hirschtick et al., 1995; Huang & Crothers, 2009; Kohli et al., 2006) Alcohol consumption is a well-known risk factor for pneumococcal pneumonia in the non-HIV population and is a known risk factor for invasive disease and death in HIV-infected patients with pneumococcal bacteremia.(Gamble, Mason, & Nelson, 2006; Happel & Nelson, 2005; Nelson & Kolls, 2002) Our group recently published epidemiological evidence supporting a detrimental impact of alcohol use in HIV-associated pneumonia.(Jolley, Alkhafaf, Hough, & Welsh, 2016) Harmful alcohol use was common, with 57% of patients meeting criteria for binge drinking. There was a direct correlation between AUD risk, as assessed by the Alcohol Use Disorders Identification Test (AUDIT), and pneumonia severity.(Jolley et al., 2016)
However, experimental evidence examining the interactions of alcohol use and HIV infection on pulmonary host defense and development of pneumonia are limited. Alcohol consumption in rhesus macaques increases the plasma viral load set point and progression to end-stage acquired immune deficiency syndrome (AIDS),(Bagby, Zhang, Purcell, Didier, & Nelson, 2006; Nelson et al., 2013; Poonia et al., 2006) and leads to an accelerated decline in peripheral CD4+ T cells, and CD4+ T cell subsets.(Poonia et al., 2006) Chronic EtOH consumption was also associated with enhanced SIV replication in the lung during pneumococcal pneumonia, which persisted after the resolution of bacterial infection.(Nelson et al., 2013)
It is now widely accepted that the composition of the intestinal microbial communities contributes to pulmonary host defense against bacterial pneumonia.(L. W. Chen, Chen, & Hsu, 2011; M. M. Chen et al., 2014; Dickson et al., 2016; Fagundes et al., 2012; Fox et al., 2012; Gauguet et al., 2015; Ichinohe et al., 2011; McAleer et al., 2016; D. R. Samuelson et al., 2017; D.R. Samuelson, Welsh, & Shellito, 2015) HIV and alcohol use have been associated with changes to the resident microbial community structure in the gastrointestinal tract.(Beck et al., 2015; Casafont Morencos et al., 1996; Dillon et al., 2014; Dubourg, Surenaud, Levy, Hue, & Raoult, 2016; Engen, Green, Voigt, Forsyth, & Keshavarzian, 2015; Hartmann, Seebauer, & Schnabl, 2015; Leclercq et al., 2014) In general, ethanol reduces Lactobacillus spp., while Enterococcus, Akkermansia, Corynebacterium, and Alcaligenes spp. increase after alcohol administration.(Bode, Bode, Heidelbach, Durr, & Martini, 1984; Casafont Morencos et al., 1996; P. Chen, Starkel, Turner, Ho, & Schnabl, 2015; Engen et al., 2015; Hartmann et al., 2015; Leclercq et al., 2014; Yan et al., 2011) Several studies suggest that Prevotella species are increased in abundance with a reciprocal decrease in Bacteroides during HIV infection.(Dillon, Frank, & Wilson, 2016; Dillon et al., 2014; Dubourg et al., 2016; Williams, Landay, & Presti, 2016; Williams, Mirmonsef, et al., 2016) However, recent work has shown these changes are associated with sexual behavior rather than HIV infection.(Noguera-Julian et al., 2016) Despite the lack of a complete understanding of the microbial composition of HIV-associated dysbiosis, one finding has remained consistent, that is HIV-1 infection is associated with a reduction in bacterial richness.(Noguera-Julian et al., 2016) Interestingly, cART and immune reconstitution do not restore the normal microbial ecosystem.(Williams, Landay, et al., 2016) Our recently published work demonstrated that alcohol-associated intestinal dysbiosis increases susceptibility to Klebsiella pneumoniae pneumonia in alcohol-fed animals.(D. R. Samuelson et al., 2017) Alcohol-naïve animals recolonized with a microbiota isolated from alcohol-fed mice had an increased K. pneumoniae burden compared to mice recolonized with a control microbiota.(D. R. Samuelson et al., 2017) However, the role of HIV/alcohol-associated dysbiosis on host defense is not known.
Thus, the motivation for this project originates from human epidemiologic observations, as well as from pre-clinical basic science. Using a humanized mouse model, data presented here support the hypothesis that chronic plus binge alcohol impair S. pneumoniae clearance from the lung in both HIV-infected and uninfected mice, and this host defense defect is linked to intestinal dysbiosis.
Methods:
Ethics Statement:
All experiments were approved by the Louisiana State University Health Sciences Center (LSUHSC) Institutional Animal Care and Use Committee, protocol number 3300. LSUHSC utilizes the Public Health Service Policy on Humane Care and Use of Laboratory Animals (PHS) and uses the Guide for the Care and Use of Laboratory Animals as a basis for establishing and maintaining an institutional program for activities involving animals. LSUHSC animal care policies comply with all applicable provisions of the Animal Welfare Act (AWA), guidance from the Office of Laboratory Animal Welfare (OLAW), the American Veterinary Medical Association Guidelines on Euthanasia, and all state and local regulations.
Mice:
Female 24 to 25 week old BLT (bone marrow, liver, thymus) mice were obtained from the Marasco lab at the Dana-Farber Cancer Institute and maintained in a temperature (72oF) controlled room for two days prior to experimental manipulation. The BLT mice are a modification of the SCID-Hu model. Specifically, NSG mice (6 weeks of age purchased from Jackson Labs) are cotransplanted with second trimester human fetal liver and thymus tissues from Advanced Bioscience Resources (Alameda, CA) along with autologous CD34+ hematopoietic stem cells after irradiation (323 Rads). Development of a robust human hemato-lymphoid system is observed between 12–16 weeks post engraftment with human cells and tissues.(Karpel, Boutwell, & Allen, 2015) This model is also being used extensively in HIV research and studying of virus-specific human immune responses.(Biswas et al., 2011; Denton et al., 2012; Deruaz & Luster, 2013; Karpel et al., 2015) Animals were housed in filter-topped cages and were provided autoclaved water and chow ad libitum. The mice were kept in the animal care facility at LSUHSC throughout the experiment. Animals were handled under a laminar flow hood to maintain SPF conditions throughout the course of the experiment.
HIV Infection:
BLT mice were inoculated intraperitoneally with a 1:1 mix of the R5-tropic HIV strain JRCSF and BaL (HIVJRCSF and HIVBaL) at 10,000 50% tissue culture infectious doses [TCID50]. Following inoculation, mice were bled weekly to obtain plasma for HIV viral load measurements and T-cell counts. BLT mice were switched to binge-on-chronic alcohol feeding one week post HIV inoculation.
Viral Quantification:
HIV viral load measurements were performed by using the Cobas Amplicor reverse transcription-PCR (RT-PCR) assay (Roche Diagnostics), as described by the manufacturer. Viral levels were normalized to plasma volume and values log-transformed for comparisons.
Alcohol levels:
Plasma alcohol levels were determined via AM1 analyzer obtained from Analox Instruments USA (Lunenburg, MA), as per manufacturer’s instructions.
CD4 Counts:
The number of peripheral blood human T-cells was determined using the BD Multitest™ for CD3/CD8/CD45/CD4 (Becton Dickinson), as per manufacturer’s instructions. Counts were acquired on a LSRII (Becton Dickinson) and analyzed using manufacturer’s gating strategies.
Binge-on-chronic Alcohol Feeding:
We have adapted and modified the NIAAA chronic-binge alcohol model.(Bertola, Mathews, Ki, Wang, & Gao, 2013) Briefly, female BLT mice were acclimated to liquid diet for 5 days using Lieber-DeCarli ‘82 Shake and Pour control liquid diet (Bioserv, Flemington, NJ). Ethanol feeding began with 1% EtOH vol./vol. on day 1 and ended with 5% EtOH vol./vol. on day 5. Groups of mice (n = 5 per cage) were randomized into ethanol fed (5% vol/vol ethanol liquid diet) or pair-fed groups (control-liquid diet). Pair-fed mice were maintained on control-liquid diet adjusted daily according to the consumption of ethanol-fed mice. Mice were then administered 2 g kg−1 (24.03% vol/vol) ethanol by gavage (binge) following 5 days of chronic-ethanol consumption. Pair-fed control mice were gavaged with 9 g kg−1 (45% wt/vol) maltose dextrin. Mice were maintained on the 5% ethanol diet for an additional 5 day period and on day 10, mice received a second and final ethanol binge (2 g kg−1). Mice were then infected with 1 × 105 CFU of S. pneumoniae via i.n. inoculation and sacrificed 48 hrs. post infection.
Streptococcus pneumoniae Infection:
Streptococcus pneumoniae infections were performed as previously described.(Boe, Nelson, Zhang, Quinton, & Bagby, 2003; Siggins et al., 2011) Briefly, S. pneumoniae (ATCC® BAA-334™, strain TIGR4 [JNR.7/87], Capsular serotype 4; American Type Culture Collection, Manassas, VA) were grown in 100 mL Todd Hewitt Broth (Becton Dickinson, Franklin Lakes, NJ) in a CO2 incubator (5% CO2) at 37oC for 6 hours. Bacteria were then pelleted by centrifugation (2,000 x g for 15 minutes at 4 oC), washed twice with phosphate-buffered saline (PBS), and resuspended in PBS at a concentration of 1 × 105 colony-forming units (CFU)/mL. The actual number of viable bacteria were determined by serial dilutions onto BD BBL™ Trypticase™ Soy Agar (TSA II™) with Sheep Blood and performing standard colony counts. Twenty-four hours after the final ethanol binge, animals were anesthetized with isoflurane and given 1 × 105 CFU bacteria in 50 μL PBS via intranasal (i.n.) administration. Animals were allowed to recover from anesthesia and returned to their cages. Mice were sacrificed 48 hrs. post infection. S. pneumoniae burden was then determined by serial dilutions onto BD BBL™ Trypticase™ Soy Agar (TSA II™) with Sheep Blood and performing standard colony counts.
Microbiota adoptive transfer:
Fecal samples used for community structure and adoptive transfer were collected post binge number two and prior to respiratory infection. Generation of intestinal microbial communities for adoptive transfer experiments were performed as described in our recent publication.(D. R. Samuelson et al., 2017)
DNA sequencing of the 16S rRNA gene:
16S rRNA gene sequencing was performed by the Louisiana State University School of Medicine Microbial Genomics Resource Group (http://metagenomics.lsuhsc.edu/mgrg), as described previously.(D. R. Samuelson et al., 2018; D. R. Samuelson et al., 2016; D. R. Samuelson et al., 2017) Briefly, fecal genomic DNA extraction was performed using the QIAamp DNA Stool Mini Kit (Qiagen Valencia, CA) modified to include bead-beating. 16S ribosomal DNA hypervariable region V4 was amplified using gene-specific sequences, Illumina adaptors, and molecular barcodes primers. Samples were sequenced on an Illumina MiSeq (Illumina, San Diego, CA) using the 2 × 300 bp V4 sequencing kit. In addition, internal sequencing controls (negative water control and a positive mock community control) were included to test for contamination of the sequencing runs.
Sequence curation and analysis:
All curation and analyses were performed using the R platform and the following R packages: DADA2 v1.1.5, Phyloseq v1.16.2, DESeq2 v1.20.0, and vegan v2.3–5.(Anders & Huber, 2010; Callahan et al., 2016; McMurdie & Holmes, 2012, 2014; Oksanen, 2007) Raw sequences were truncated to 250 base pairs, denoised, chimera-filtered, and clustered into sequence variants using DADA2. Median sequencing depth for all sequence variants was 24,740 reads with a min of 2,409 reads and a maximum of 35,241 reads. For taxonomically informed analyses, operational taxonomic units (OTU) were generated in DADA2 by taxonomic classification of sequence variants using the SILVA reference database v132 and a minimum bootstrap confidence level of 80. Relative abundance of each DADA2 16S OTU was examined at phylum, class, order, family, genus, and species levels. Beta (between communities) and alpha (within a community) diversities, as well as taxonomic community assessments were analyzed via the R package Phyloseq. Sequencing depth was included as a technical confounder in all analyses. Number of unique sequence variants in a sample (alpha diversity), which we termed “Observed sample richness”, was calculated using the estimate_richness function in Phyloseq. Bray–Curtis dissimilarity was calculated using the vegdist function in vegan using raw OTU counts to assess beta-diversity.
Inferred functional capacity of the intestinal microbial community:
Inferred metagenomes from 16S rDNA sequencing data were obtained through the Piphillin pipeline, as described previously.(Iwai et al., 2016)
Data Repository:
16S rDNA sequencing data is deposited in the NCBI (National Center for Biotechnology Information) of the National Institutes of Health and the U.S. National Library of Medicine GeneBank (SRA) with accession number (SRP158523).
Statistical Analysis:
Results are presented as mean ± S.E.M. Statistical analyses were performed using GraphPad Prism 5 (La Jolla, CA. USA) and statistical significance was measured at p ≤ 0.05. Specific statistical analyses are indicated in the figure legends. Microbiome statistical analysis was performed as follows. Observed sample richness was modeled using generalized linear models (GLM) available within the R platform. Distance-based redundancy analysis (dbRDA) was performed on sample-wise Bray-Curtis dissimilarity distances to assess beta-diversity. Significant marginal effects of independent model covariates on dbRDA clustering was inferred via permutational multivariate analysis of variance within R.
All pair-wise comparisons were performed with corrections for multiple comparison via False Discovery Rate (FDR).(Benjamini, 1995) DESeq2, which employs sequencing depth normalization and a negative-binomial GLM to account for over-dispersion in sequenced-read count data was used to detect differentially abundant OTUs between HIV+ ethanol-fed, HIV+ pair-fed, HIV- ethanol-fed, or HIV- pair-fed mice. Model variables included feeding, HIV infection, Streptococcus pulmonary burden, inferred functional capacity, and sequencing depth. A p-value < 0.05 and an FDR q-value < 0.1 were considered statistically significant where indicated in the text.
Results:
Alcohol-fed HIV-infected humanized mice:
We adapted the NIAAA chronic-binge alcohol model(Bertola et al., 2013) to assess the effects of chronic-binge alcohol feeding on host defense against S. pneumoniae. The experimental design used in this study is shown in Supplemental Figure 1. Blood alcohol levels were determined 3 hours post binge number 2 (Figure 1A) and at the time of sacrifice (Figure 1B) via AM1 analyzer. Daily food intake was similar in alcohol- and pair-fed mice (Figure 1C). However, some weight loss was seen in the alcohol-fed mice at around 15 days post HIV inoculation and 8 days post alcohol-diet initiation (Figure 1D). HIV-infected mice had a mean viral load 1.4 × 104 copies/mL of blood one week post infection (prior to ethanol-feeding) (Figure 2A), and exhibited an ~57% decrease in the number of circulating CD4+ T-cells at the time of sacrifice (Figure 2B). No significant changes were seen in circulating CD8+ T-cells at the time of sacrifice, in either HIV+ or alcohol-fed mice (Figure 2C).
Figure 1: Binge-on-chronic alcohol feeding in HIV-infected humanized BLT mice.
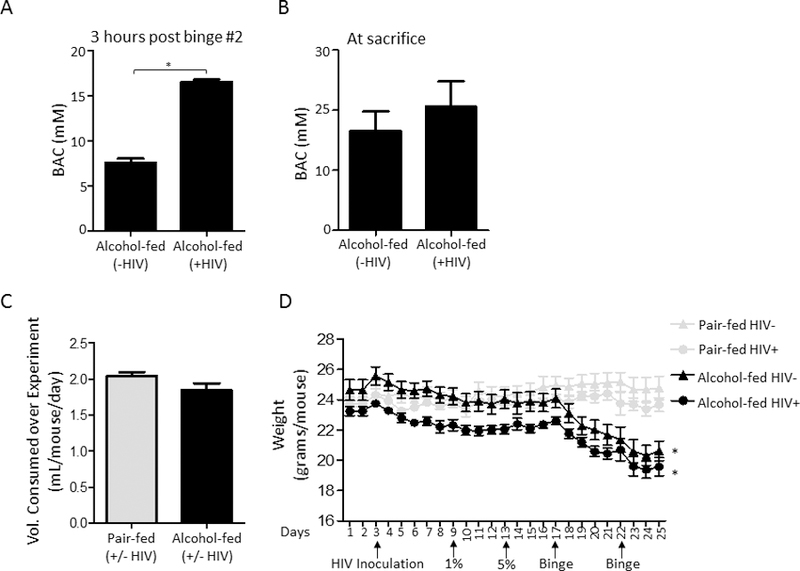
(A) Blood alcohol levels (mM) of chronic alcohol fed mice following 10 days of diet and 3 hrs following the second binge alcohol administration. (B) Blood alcohol levels (mM) of chronic alcohol fed mice at the time of sacrifice. (C) Volume of liquid diet consumed by alcohol-fed and pair-fed mice per day per mouse. (D) Body weights of pair-fed and alcohol-fed mice at baseline and post binge-on-chronic alcohol feeding (10 days chronic + 2x binge). Bars are the mean ± SEM, *indicates P<0.05, by Mann-Whitney U. N=5/group.
Figure 2: HIV infection decreases circulating CD4+ T-cells in humanized BLT mice.
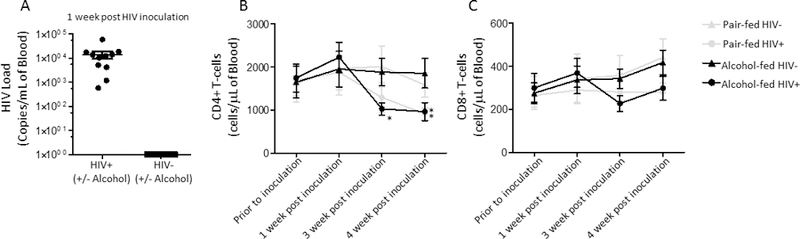
(A) HIV viral load (copies/ml) at 1-week post infection in pair-fed and binge-on-chronic alcohol treated mice. (B) Difference in absolute number of circulating CD4+ T-cells in HIV-infected humanized BLT mice. (C) Difference in absolute number of circulating CD8+ T-cells in HIV-infected humanized BLT mice. Bars represent the mean of the cell counts post infection minus the cell counts prior to infection ± SEM. * indicates P < 0.05, by Mann-Whitney U or by repeated measures 2-way ANOVA with Bonferroni correction. N=5/group. Viral loads in HIV-negative samples are set to one for visualization on the log scale.
Alcohol feeding and HIV infection alters the fecal microbial community structure:
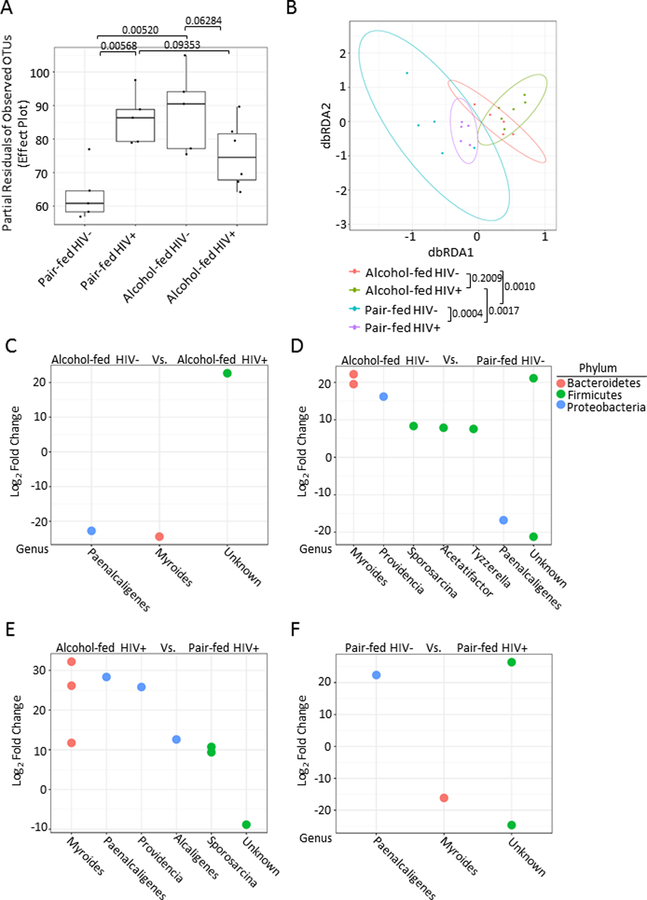
Microbial community structure was then analyzed one day post second binge and 18 days post HIV infection, prior to S. pneumoniae infection. Binge-on-chronic alcohol feeding in HIV-infected mice resulted in changes to the microbial alpha (α)-diversity characterized by a reduction (q = 0.0953) in the number of observed species in HIV-infected alcohol-fed mice compared to HIV-infected pair-fed mice (Figure 3A). Further, microbial α-diversity was significantly higher (q = 0.00568) in HIV-infected pair-fed mice compared to HIV-negative pair-fed mice (Figure 3A). Additionally, α-diversity was significantly higher (q = 0.00520) in alcohol-fed HIV-infected mice compared to HIV-negative pair-fed mice (Figure 3A). Finally, a reduction (q = 0.06284) in the number of observed species in HIV-infected alcohol-fed mice compared to HIV-negative alcohol-fed mice (Figure 3A). Beta (β)-diversity of the microbial communities from alcohol-fed, pair-fed mice, HIV infection alcohol-fed mice, and HIV-infected pair-fed mice was also assessed. Significant differences in the β-diversity of HIV-infected alcohol-fed mice compared to HIV-infected pair-fed mice (q = 0.0017), HIV-negative alcohol-fed mice compared to HIV-negative pair-fed mice (q = 0.0010), as well as HIV-infected pair-fed mice compared to HIV-negative pair-fed mice (q = 0.0004) were observed as determined by the distance-based redundancy analysis (dbRDA) and permutational multivariate analysis of variance analysis via vegan (Figure 3B). However, no significant difference (q = 0.2009) was observed between HIV-infected alcohol-fed mice compared to HIV-negative alcohol-fed mice (Figure 3B). Comparisons between alcohol-fed, pair-fed mice, HIV infection alcohol-fed mice, and HIV-infected pair-fed mice microbial communities demonstrated significant changes in the relative abundance of specific operational taxonomic units (OTUs), via negative binomial mixture model using DESeq2 (Figure 3C-F).
Figure 3: Binge-on-chronic alcohol feeding in HIV-infected humanized BLT mice alters the intestinal microbial community structure.
(A) Alpha-diversity of the fecal microbial communities following binge-on-chronic alcohol feeding and HIV-infected. Significant differences in α-diversity of the microbial communities were seen between; HIV-infected alcohol-fed mice compared to HIV-infected pair-fed mice, HIV-infected pair-fed mice compared to HIV-negative pair-fed mice, alcohol-fed HIV-infected mice compared to HIV-negative pair-fed mice, and HIV-infected alcohol-fed mice compared to HIV-negative alcohol-fed mice. (B) Beta-diversity of the fecal microbial communities following binge-on-chronic alcohol feeding and HIV-infected. Significant differences in the β-diversity were seen between; HIV-infected alcohol-fed mice compared to HIV-infected pair-fed mice, HIV-negative alcohol-fed mice compared to HIV-negative pair-fed mice, as well as HIV-infected pair-fed mice compared to HIV-negative pair-fed mice. No significant difference was observed between HIV-infected alcohol-fed mice compared to HIV-negative alcohol-fed mice. (C) HIV infection alters the realative abundance of specific OTUs compared to HIV-negative pair-fed mice. Postive changes indicate the genus is more abundant in the HIV-infected animals, while negative values indicate that the genus is more abundant in the HIV-negative animals. (D) Alcohol-feeding alters the realative abundance of of specific OTUs compared to pair-feeding in HIV-infected mice. Postive changes indicate the genus is more abundant in the pair-fed animals, while negative values indicate that the genus is more abundant in the alcohol-fed animals (E) HIV infection alters the realative abundance of specific OTUs compared to HIV-negative alcohol-fed mice. Postive changes indicate the genus is more abundant in the HIV-infected animals, while negative values indicate that the genus is more abundant in the HIV-negative animals. (F) Alcohol-feeding alters the realative abundance of specific OTUs compared to pair-feeding in HIV-negative mice. Postive changes indicate the genus is more abundant in the pair-fed animals, while negative values indicate that the genus is more abundant in the alcohol-fed animals.
Alcohol-fed HIV-infected mice have less inferred functional capacity for catalase and alcohol-dehydrogenase:
We also assessed the inferred metagenomes from 16S rRNA sequencing data to determine the functional capacity of the intestinal microbial communities for ethanol metabolism using the publically available Piphillin pipeline. Analysis of the inferred functional capacity of the microbiota for catalase and alcohol-dehydrogenase function demonstrated that catalase (Figure 4A, p = 0.0032) and alcohol-dehydrogenase relative abundances (Figure 4B, p = 0.0601) were decreased in alcohol-fed HIV-infected mice compared to alcohol-fed HIV-negative mice, as determined by multivariate linear mixed-effects modeling. Model variables included feeding, HIV infection, inferred functional capacity, and sequencing depth.
Figure 4: Inferred functional capacity for catalase and alcohol-dehydrogenase is decreased in alcohol-fed HIV-infected humanized BLT mice:
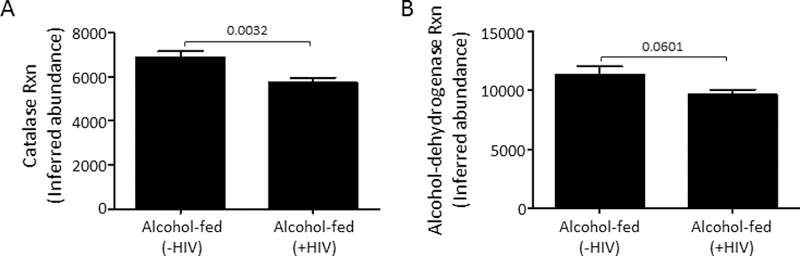
(A) HIV-infected alcohol-fed mice have significantly lower inferred capacity for catalase function compared to HIV-negative alcohol-fed mice. (B) HIV-infected alcohol-fed mice have significantly lower inferred capacity for alcohol-dehydrogenase function compared to HIV-negative alcohol-fed mice. P values are indicated, as determined by Student’s T-test. N=5/group.
Alcohol feeding increases S. pneumoniae burden in HIV-infected humanized mice:
We evaluated the effects of binge-on-chronic alcohol consumption on host susceptibility to S. pneumoniae. Mice receiving binge-on-chronic alcohol feeding had an increased lung burden of S. pneumoniae 48 hours post infection compared to pair-fed control mice (Figure 5). Bacterial burdens based on HIV infection were not statistically different. We also examined the relationship between the composition of the intestinal microbial communities and S. pneumoniae pulmonary burden, however no significant association was observed between either α-diversity or β-diversity and S. pneumoniae pulmonary burden (data not shown).
Figure 5: Binge-on-chronic alcohol use increases host susceptibility to Streptococcus pneumoniae in HIV-infected humanized BLT mice.
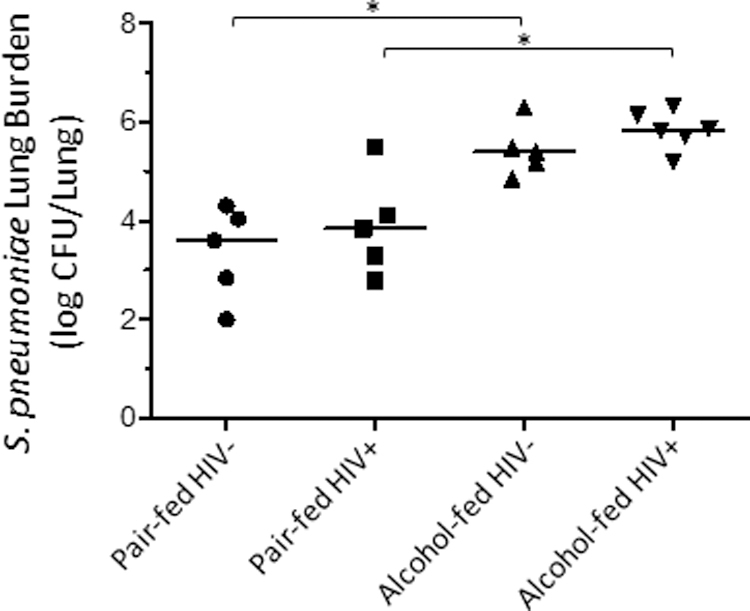
Streptococcus lung burden (Log10 CFU/ml) at 48 hrs. post infection in pair-fed and binge-on-chronic alcohol-treated mice. Bars represent the median log10 CFU/lung of S. pneumoniae. * indicates P < 0.05, by Mann-Whitney U or by ANOVA with Dunn’s correction. N=10/group.
S. pneumoniae correlates with pulmonary viral replication:
We then evaluated the relationship between S. pneumoniae infection and pulmonary HIV viral replication. No difference in pulmonary HIV RNA levels was observed between alcohol-fed and pair-fed mice (p = 0.102) (Figure 6A). However, S. pneumoniae pulmonary burden positively correlated with pulmonary HIV RNA levels (Figure 6B). We also evaluated the correlation between S. pneumoniae pulmonary burden and pulmonary HIV RNA levels in alcohol- and pair-fed mice. The correlation between S. pneumoniae pulmonary burden and pulmonary HIV RNA levels was most pronounced in alcohol-fed mice (p = 0.0544, r2 = 0.6448), however, the correlation was not significantly different from the correlation between S. pneumoniae pulmonary burden and pulmonary HIV RNA levels in pair-fed mice (p = 0.8157, r2 = 0.02110) (Figure 6C).
Figure 6: Streptococcus pneumoniae infection increases pulmonary HIV viral load.
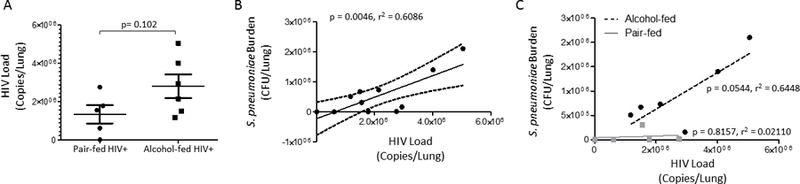
(A) HIV RNA levels in the lungs of alcohol-fed and pair-fed mice. (B) Streptococcus lung burden (Log10 CFU/ml) correlates with pulmonary HIV RNA levels (P = 0.0046 r2 = 0.6086). (C) Streptococcus lung burden (Log10 CFU/ml) correlates with pulmonary HIV RNA levels in alcohol-fed mice but not in pair-fed mice (P = 0.0544 r2 = 0.6448 and P = 0.8157 r2 = 0.0211).
Fecal microbial community structure following xenotransplantation:
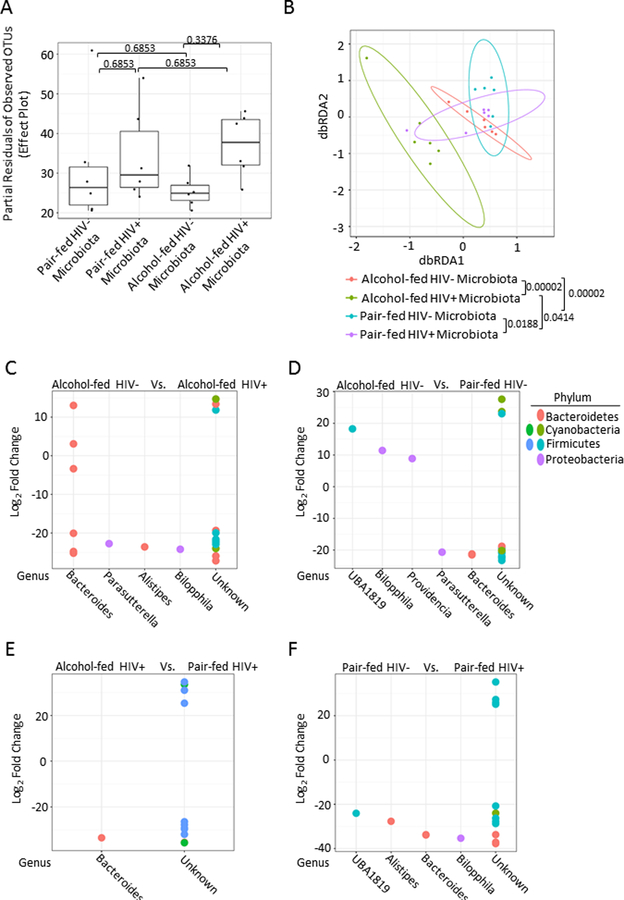
Microbial community structure was analyzed 1 week following xenotransplantation into alcohol and HIV-naïve mice, prior to S. pneumoniae infection. Following adoptive transfer no significant changes to the microbial α-diversity where observed between any group (Figure 7A). β-diversity of the xenotransplanted microbial communities from alcohol-fed, pair-fed mice, HIV infection alcohol-fed mice, and HIV-infected pair-fed mice was also assessed. Significant differences in the β-diversity of the xenotransplanted microbial communities was seen between; (1) HIV-infected alcohol-fed mice compared to HIV-infected pair-fed mice (q = 0.0414), (2) HIV-negative alcohol-fed mice compared to HIV-negative pair-fed mice (q = 0.00002), (3) HIV-infected pair-fed mice compared to HIV-negative pair-fed mice (q = 0.0188), and (4) HIV-infected alcohol-fed mice compared to HIV-negative alcohol-fed mice (q = 0.00002) were observed as determined by dbRDA and ADONIS analysis via vegan (Figure 7B). Comparisons between the xenotransplanted microbial communities from alcohol-fed, pair-fed mice, HIV infection alcohol-fed mice, and HIV-infected pair-fed mice demonstrated significant changes in the relative abundance of specific operational taxonomic units (OTUs), via negative binomial mixture model using DESeq2 (Figure 7C-F).
Figure 7: Fecal microbial community structure following xenotransplantation.
Microbial community structure was analyzed 1 week following xenotransplantation into alcohol and HIV-naïve mice, prior to S. pneumoniae infection. (A) No significant changes to the microbial α-diversity where observed between any group. (B) β-diversity of the xenotransplanted microbial communities from alcohol-fed, pair-fed mice, HIV infection alcohol-fed mice, and HIV-infected pair-fed mice. Significant differences in the β-diversity of the xenotransplanted microbial communities were seen between; HIV-infected alcohol-fed mice compared to HIV-infected pair-fed mice, HIV-negative alcohol-fed mice compared to HIV-negative pair-fed mice, HIV-infected pair-fed mice compared to HIV-negative pair-fed mice, and HIV-infected alcohol-fed mice compared to HIV-negative alcohol-fed mice via vegan. (C) Xenotransplanted microbiota from HIV-infected mice alters the realative abundance of specific OTUs compared to xenotransplanted microbiota from HIV-negative pair-fed mice. Postive changes indicate the genus is more abundant in the HIV-infected xenotransplanted microbiota, while negative values indicate that the genus is more abundant in the HIV-negative xenotransplanted microbiota. (D) Xenotransplanted microbiota from alcohol-fed mice alters the realative abundance of of specific OTUs compared to xenotransplanted microbiota from pair-fed HIV-infected mice. Postive changes indicate the genus is more abundant in the pair-fed xenotransplanted microbiota, while negative values indicate that the genus is more abundant in the alcohol-fed xenotransplanted microbiota (E) Xenotransplanted microbiota from HIV-infected mice alters the realative abundance of specific OTUs compared to xenotransplanted microbiota from HIV-negative alcohol-fed mice. Postive changes indicate the genus is more abundant in the HIV-infected xenotransplanted microbiota, while negative values indicate that the genus is more abundant in the HIV-negative xenotransplanted microbiota. (F) Xenotransplanted microbiota from alcohol-fed mice alters the realative abundance of specific OTUs compared to xenotransplanted microbiota from HIV-negative pair-fed mice. Postive changes indicate the genus is more abundant in the pair-fed xenotransplanted microbiota, while negative values indicate that the genus is more abundant in the alcohol-fed xenotransplanted microbiota.
Alcohol-dysbiosis increases S. pneumoniae burden in mice:
We evaluated the effects of alcohol and HIV-associated intestinal dysbiosis on host susceptibility to S. pneumoniae. Normal mice recolonized with the intestinal microbiota from alcohol-fed mice had an increased lung burden of S. pneumoniae 48 hours post infection compared to mice recolonized with microbiota from pair-fed control mice (Figure 8). HIV infection did not cause a statistical difference within the feeding groups. An additional group of mice was recolonized with microbiota treated with amphotericin B to assess the impacts of fungal communities. We found no statistical differences between mice recolonized with the full microbial community compared to mice recolonized with microbiota deficient fungal organism (Supplemental Figure 2). We also examined the relationship between the composition of the intestinal microbial communities and S. pneumoniae pulmonary burden. We found that S. pneumoniae pulmonary burden was significantly association with both α-diversity (Figure 9A, p = 0.0283) and β-diversity (Figure 9B, p = 0.0415), as determined by multiple general linear regression and permutational multivariate analysis of variance within R, respectively. Model covariables included xenotransplanted microbial community origin, S. pneumoniae pulmonary burden, and sequencing depth.
Figure 8: Alcohol-associated intestinal dysbiosis increases host susceptibility to Streptococcus pneumoniae in mice, independent of alcohol or HIV infection.
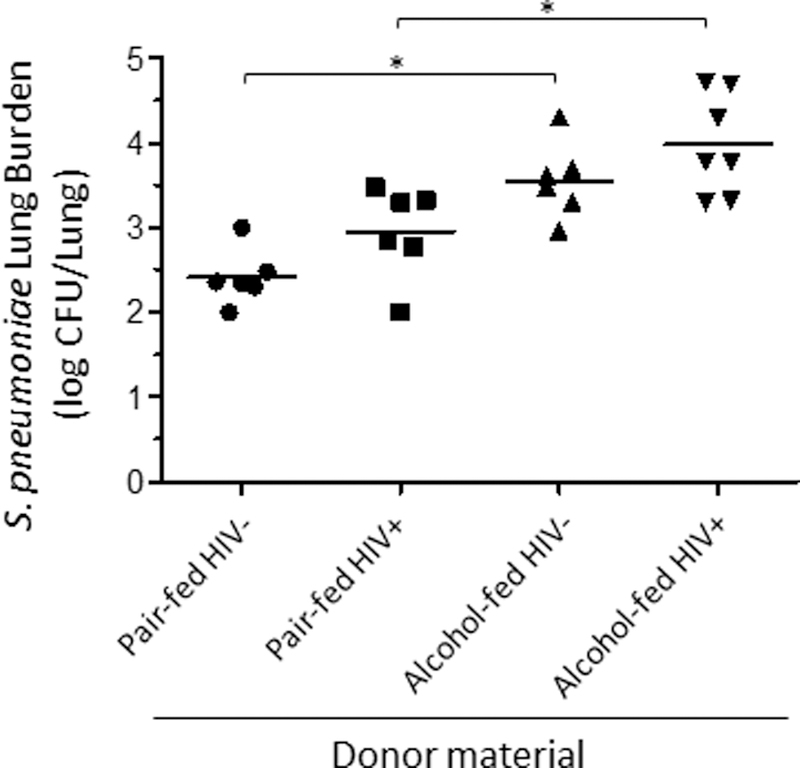
Streptococcus lung burden (Log10 CFU/ml) at 48 hrs. post infection in animals recolonized with microbiota from HIV+ ethanol-fed, HIV+ pair-fed, HIV- ethanol-fed, or HIV- pair-fed. Bars represent the median log10 CFU/lung of S. pneumoniae. * indicates P < 0.05, by Mann-Whitney U or by ANOVA with Dunn’s correction. N=10/group.
Figure 9: Streptococcus pneumoniae burden is associated with microbial community structure following xenotransplantation.
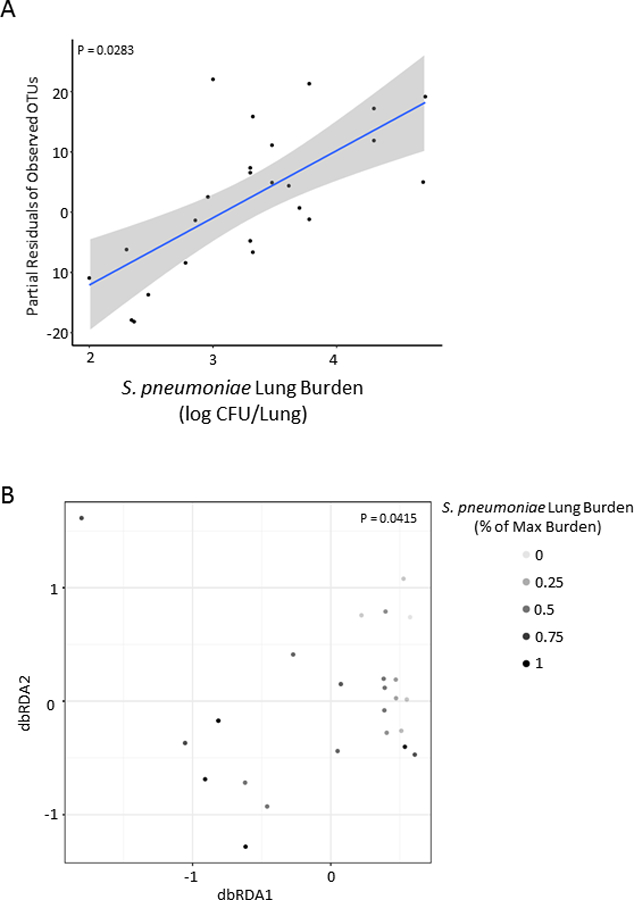
S. pneumoniae pulmonary burden was significantly association with both (A) α-diversity and (B) β-diversity, as determined by multiple general linear regression and permutational multivariate analysis of variance within R, respectively. Model covariables included xenotransplanted microbial community origin, S. pneumoniae pulmonary burden, and sequencing depth.
Conclusions:
This study examines the gut-lung axis and S. pneumoniae clearance from the lung in an alcohol-fed HIV-infected humanized mouse model. We found that the fecal microbial community structure was significantly different in each of the feeding groups, as well as with HIV infection. Alcohol-fed mice had a significantly higher burden of S. pneumoniae 48 hours post infection, regardless of HIV status. Additionally, mice recolonized with intestinal microbiota from alcohol-fed mice also had a significantly higher burden of S. pneumoniae in both HIV-infected and uninfected animals.
Considerable work has detailed the deleterious effects of alcohol-consumption on elements of pulmonary host defenses.(Burnham et al., 2011; Elliott, Sisson, & Wyatt, 2007; Gamble et al., 2006; Happel & Nelson, 2005; Sisson, 2007) As alcohol use disorders are prevalent in HIV-infected persons, it is critical to understand the consequences of alcohol abuse on respiratory tract infections in these individuals. Previous work, has shown that chronic alcohol feeding in the rhesus macaques SIV model increases plasma viral load set point and accelerates the progression to end-stage AIDS.(Bagby et al., 2006; Nelson et al., 2013; Poonia et al., 2006) Chronic alcohol consumption also enhances SIV replication in the lung during pneumococcal pneumonia, which persists after the resolution of bacterial infection.(Nelson et al., 2013) Interestingly, SIV-infected macaques given acute binge alcohol, exhibit prolonged blood ethanol levels and a diminished rate of alcohol elimination.(Simon et al., 2018) Our current work supports these previous studies. First, we observed an accelerated decrease in CD4 T-cells in alcohol-fed mice, as alcohol-fed mice had a significant decreased CD4 T-cell count 3 weeks post infection compared to 4 weeks in pair-fed animals. Second, while we did not observe an alcohol-dependent increase in viral replication in the lung during pneumococcal pneumonia, we did observe a correlation between viral load in the lung and pneumococcal burden, which may be partially driven by alcohol-feeding, although additional studies are need to fully address this possibility. Third, we also observed a prolonged duration of elevated blood ethanol levels in HIV-infected mice following binge alcohol administration, suggesting that HIV infection or HIV associated intestinal dysbiosis may interfere with alcohol clearance rates following binge alcohol consumption. Consistent with these results, we observed a decrease in the inferred functional capacity for catalase and alcohol-dehydrogenase in the microbiota from alcohol-fed HIV-infected mice compared to alcohol-fed HIV-negative mice. Previous work has shown that intestinal aerobes and facultative anaerobes play an important role in the oxidation of ethanol by microbial alcohol dehydrogenases and/or catalases in the colon and rectum.(Tsuruya et al., 2016) Taken together, these results suggest that decreased alcohol clearance rates seen in HIV-infected mice may be due, in part, to a corresponding decrease in the functional capacity for catalase and/or alcohol-dehydrogenase within the intestinal microbial community. While we did observe changes to fecal microbial diversity, which were associated with HIV, as well as alcohol feeding these changes differ from those observed in human studies or other animal model systems.(Dubourg et al., 2016; Engen et al., 2015) For example, both HIV and alcohol use typically lead to a decrease in bacterial richness, however in our cohort of animals we observed an increase in bacterial richness, which may be a unique aspect of this particular model. As there is a clear difference between the human and mouse microbiota, the clinical relevance needs to be investigated with additional models and studies. However, this model represents a unique opportunity to investigate the relationship between HIV, alcohol-use, pneumonia, and the microbial communities while controlling for many of the social and environmental confounders that make human studies difficult. Additionally, the data and methods from this study can be applied to other xenotransplation experiments (i.e., human donors into mice). We did observe a shift in the microbial community structure following xenotransplantation. Yet, this is to be expected given that the microbial communities from an HIV infected humanized mouse consuming alcohol experience vastly different microbial selective pressures than seen in alcohol and HIV-naïve wild-type mice. This is also consistent with other studies that have used xenotransplantation.(Ellekilde et al., 2014) Importantly we still see a difference in the microbial diversity between the four comparison groups, which allows us to investigate the role of these microbial communities on host defense. Finally, our current work adds to the current literature by demonstrating that alcohol/HIV-associated intestinal dysbiosis contributes to the impaired pulmonary host defense seen in HIV-infected individuals with AUD. In addition, our study provides a bases and a new model system to study the effects of HIV and alcohol use in a murine model.
To our knowledge, this is the only study that has investigated the effect of alcohol use and HIV infection on pulmonary infection in a humanized murine model. Additionally, to our knowledge, this is the only study that has sought to understand the role of alcohol/HIV-associated microbiota on pulmonary infection. However, we acknowledge several limitations to the current study. First, alcohol-fed humanized mice began to lose weight after the first EtOH binge administration regardless of HIV infection, when compared to control HIV positive and HIV-negative animals. We are unsure of the direct mechanism by which the animals lost weight. However, animals still appeared robust and did not exhibit any signs of morbidity and no mortality was observed. Second, we were unable to assess pulmonary immune responses, as an insufficient number of immune cells for flow cytometry were harvested at sacrifice. Third, our HIV infection model represents an acute HIV infection not a chronic infection and may not adequately represent bacterial pneumonia, or the effects of alcohol consumption in chronic HIV disease. Finally, while there was an increase in the mean HIV RNA levels in the lungs of alcohol-fed S. pneumoniae infected mice compared to pair-fed mice; this study was not sufficiently powered to investigate this relationship. We also believe limited power affected our ability to distinguish an interaction effect between alcohol feeding and HIV infection on bacterial clearance, as alcohol-fed HIV-infected mice had the highest median pulmonary bacterial burden. BLT mice have a limited availability and high cost, which prevented the acquisition of a sufficient number of animals to adequately power the study. However, our data support additional studies to dissect the mechanisms that regulate host defense against pneumonia, especially in the context of HIV-infection and alcohol-use.
In summary, our results show that binge-on-chronic alcohol feeding results in greater pulmonary S. pneumoniae burden in both HIV-infected and HIV-negative mice. Our results also show that the intestinal microbial communities from alcohol-fed HIV-infected and HIV-negative mice contribute to S. pneumoniae burden. Given AUD is common in HIV-infected persons, further study of the mechanisms by which alcohol modulates host–bacterial-viral responses during disease progression are clearly needed.
Supplementary Material
Humanized BLT mice were administered binge-on-chronic alcohol (10 days chronic and 2x binges). Following alcohol feeding groups of mice were sacrificed 24 hrs following the final binge or infected intranasal with S. pneumoniae (~1 × 105 CFU in 50 μl of PBS) and sacrificed at 48 hours post infection. 5% EtOH diet was maintained continuously throughout the experiment.
Streptococcus lung burden (Log10 CFU/ml) at 48 hrs. post infection in animals recolonized with both full microbial or fungal-depleted microbiota from HIV+ ethanol-fed, HIV+ pair-fed, HIV- ethanol-fed, or HIV- pair-fed. Bars represent the mean of the cell counts post infection minus the cell counts prior to infection ± SEM. Closed symbols = full community, open symbols = communities without fungal organisms.
Acknowledgments
Funding: This work was supported by The National Institute of General Medical Sciences of the National Institutes of Health, which funds the Louisiana Clinical and Translational Science Center grant #U54-GM104940, The National Institute on Alcohol Abuse and Alcoholism grants #P60-AA009803, #UH2-AA026226, and #K99-AA026336, by the National Heart Lung and Blood Institute grant #P01-HL076100, and by a LSU LIFT Grant #15A4. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
List Abbreviations:
- PLWH
Persons living with HIV
- BLT mice
Bone marrow, liver, thymus
- AUD
Alcohol use disorders
- HIV
Human immunodeficiency virus
- cART
Combination antiretroviral therapy
- AUDIT
Alcohol Use Disorders Identification Test
- AIDS
Acquired immune deficiency syndrome
- SIV
Simian immunodeficiency virus
- RT-PCR
Reverse transcription-polymerase chain reaction
- TSA
Trypticase Soy Agar
- i.n.
Intranasal
- dbRDA
Distance based Redundancy Analysis
- TCID50
50% tissue culture infectious doses
Footnotes
Competing interests: The authors have declared that no competing interests exist.
Works Cited:
- Alcohol Facts and Statistics | National Institute on Alcohol Abuse and Alcoholism (NIAAA) (2016). from https://www.niaaa.nih.gov/alcohol-health/overview-alcohol-consumption/alcohol-facts-and-statistics
- Alcohol Use Disorder | National Institute on Alcohol Abuse and Alcoholism (NIAAA) (2016). from http://www.ncbi.nlm.nih.gov/pubmed/
- Anders S, & Huber W (2010). Differential expression analysis for sequence count data. Genome Biol, 11(10), R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagby GJ, Zhang P, Purcell JE, Didier PJ, & Nelson S (2006). Chronic binge ethanol consumption accelerates progression of simian immunodeficiency virus disease. Alcohol Clin Exp Res, 30(10), 1781–1790. [DOI] [PubMed] [Google Scholar]
- Beck JM, Schloss PD, Venkataraman A, Twigg H 3rd, Jablonski KA, Bushman FD, et al. (2015). Multicenter Comparison of Lung and Oral Microbiomes of HIV-infected and HIV-uninfected Individuals. Am J Respir Crit Care Med, 192(11), 1335–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Yosef. (1995). Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing on JSTOR. Journal of the Royal Statistical Society. Series B, 57(1), 289–300. [Google Scholar]
- Bertola A, Mathews S, Ki SH, Wang H, & Gao B (2013). Mouse model of chronic and binge ethanol feeding (the NIAAA model). Nat Protoc, 8(3), 627–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas S, Chang H, Sarkis PT, Fikrig E, Zhu Q, & Marasco WA (2011). Humoral immune responses in humanized BLT mice immunized with West Nile virus and HIV-1 envelope proteins are largely mediated via human CD5+ B cells. Immunology, 134(4), 419–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode JC, Bode C, Heidelbach R, Durr HK, & Martini GA (1984). Jejunal microflora in patients with chronic alcohol abuse. Hepatogastroenterology, 31(1), 30–34. [PubMed] [Google Scholar]
- Boe DM, Nelson S, Zhang P, Quinton L, & Bagby GJ (2003). Alcohol-induced suppression of lung chemokine production and the host defense response to Streptococcus pneumoniae. Alcohol Clin Exp Res, 27(11), 1838–1845. [DOI] [PubMed] [Google Scholar]
- Boyton RJ (2005). Infectious lung complications in patients with HIV/AIDS. Curr Opin Pulm Med, 11(3), 203–207. [DOI] [PubMed] [Google Scholar]
- Burnham EL, Phang TL, House R, Vandivier RW, Moss M, & Gaydos J (2011). Alveolar macrophage gene expression is altered in the setting of alcohol use disorders. Alcohol Clin Exp Res, 35(2), 284–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, & Holmes SP (2016). DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods, 13(7), 581–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casafont Morencos F, de las Heras Castano G, Martin Ramos L, Lopez Arias MJ, Ledesma F, & Pons Romero F (1996). Small bowel bacterial overgrowth in patients with alcoholic cirrhosis. Dig Dis Sci, 41(3), 552–556. [DOI] [PubMed] [Google Scholar]
- Chander G, Josephs J, Fleishman JA, Korthuis PT, Gaist P, Hellinger J, et al. (2008). Alcohol use among HIV-infected persons in care: results of a multi-site survey. HIV Med, 9(4), 196–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LW, Chen PH, & Hsu CM (2011). Commensal microflora contribute to host defense against Escherichia coli pneumonia through Toll-like receptors. Shock, 36(1), 67–75. [DOI] [PubMed] [Google Scholar]
- Chen MM, Zahs A, Brown MM, Ramirez L, Turner JR, Choudhry MA, et al. (2014). An alteration of the gut-liver axis drives pulmonary inflammation after intoxication and burn injury in mice. Am J Physiol Gastrointest Liver Physiol, 307(7), G711–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, Starkel P, Turner JR, Ho SB, & Schnabl B (2015). Dysbiosis-induced intestinal inflammation activates tumor necrosis factor receptor I and mediates alcoholic liver disease in mice. Hepatology, 61(3), 883–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cilloniz C, Torres A, Manzardo C, Gabarrus A, Ambrosioni J, Salazar A, et al. (2017). Community-Acquired Pneumococcal Pneumonia in Virologically Suppressed HIV-Infected Adult Patients: A Matched Case-Control Study. Chest, 152(2), 295–303. [DOI] [PubMed] [Google Scholar]
- Denton PW, Olesen R, Choudhary SK, Archin NM, Wahl A, Swanson MD, et al. (2012). Generation of HIV latency in humanized BLT mice. J Virol, 86(1), 630–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deruaz M, & Luster AD (2013). BLT humanized mice as model to study HIV vaginal transmission. J Infect Dis, 208 Suppl 2, S131–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson RP, Singer BH, Newstead MW, Falkowski NR, Erb-Downward JR, Standiford TJ, et al. (2016). Enrichment of the lung microbiome with gut bacteria in sepsis and the acute respiratory distress syndrome. Nat Microbiol, 1(10), 16113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon SM, Frank DN, & Wilson CC (2016). The gut microbiome and HIV-1 pathogenesis: a two way street. Aids [DOI] [PMC free article] [PubMed]
- Dillon SM, Lee EJ, Kotter CV, Austin GL, Dong Z, Hecht DK, et al. (2014). An altered intestinal mucosal microbiome in HIV-1 infection is associated with mucosal and systemic immune activation and endotoxemia. Mucosal Immunol, 7(4), 983–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubourg G, Surenaud M, Levy Y, Hue S, & Raoult D (2016). Microbiome of HIV-infected people. Microb Pathog [DOI] [PubMed]
- Ellekilde M, Selfjord E, Larsen CS, Jakesevic M, Rune I, Tranberg B, et al. (2014). Transfer of gut microbiota from lean and obese mice to antibiotic-treated mice. Sci Rep, 4, 5922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott MK, Sisson JH, & Wyatt TA (2007). Effects of cigarette smoke and alcohol on ciliated tracheal epithelium and inflammatory cell recruitment. Am J Respir Cell Mol Biol, 36(4), 452–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engen PA, Green SJ, Voigt RM, Forsyth CB, & Keshavarzian A (2015). The Gastrointestinal Microbiome: Alcohol Effects on the Composition of Intestinal Microbiota. Alcohol Res, 37(2), 223–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagundes CT, Amaral FA, Vieira AT, Soares AC, Pinho V, Nicoli JR, et al. (2012). Transient TLR activation restores inflammatory response and ability to control pulmonary bacterial infection in germfree mice. J Immunol, 188(3), 1411–1420. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick M, Brooks JT, & Kaplan JE (2016). Epidemiology of HIV-Associated Lung Disease in the United States. Semin Respir Crit Care Med, 37(2), 181–198. [DOI] [PubMed] [Google Scholar]
- Fox AC, McConnell KW, Yoseph BP, Breed E, Liang Z, Clark AT, et al. (2012). The endogenous bacteria alter gut epithelial apoptosis and decrease mortality following Pseudomonas aeruginosa pneumonia. Shock, 38(5), 508–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble L, Mason CM, & Nelson S (2006). The effects of alcohol on immunity and bacterial infection in the lung. Med Mal Infect, 36(2), 72–77. [DOI] [PubMed] [Google Scholar]
- Gauguet S, D’Ortona S, Ahnger-Pier K, Duan B, Surana NK, Lu R, et al. (2015). Intestinal Microbiota of Mice Influences Resistance to Staphylococcus aureus Pneumonia. Infect Immun, 83(10), 4003–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Happel KI, & Nelson S (2005). Alcohol, immunosuppression, and the lung. Proc Am Thorac Soc, 2(5), 428–432. [DOI] [PubMed] [Google Scholar]
- Hartmann P, Seebauer CT, & Schnabl B (2015). Alcoholic liver disease: the gut microbiome and liver cross talk. Alcohol Clin Exp Res, 39(5), 763–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschtick RE, Glassroth J, Jordan MC, Wilcosky TC, Wallace JM, Kvale PA, et al. (1995). Bacterial pneumonia in persons infected with the human immunodeficiency virus. Pulmonary Complications of HIV Infection Study Group. N Engl J Med, 333(13), 845–851. [DOI] [PubMed] [Google Scholar]
- Huang L, & Crothers K (2009). HIV-associated opportunistic pneumonias. Respirology, 14(4), 474–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichinohe T, Pang IK, Kumamoto Y, Peaper DR, Ho JH, Murray TS, et al. (2011). Microbiota regulates immune defense against respiratory tract influenza A virus infection [DOI] [PMC free article] [PubMed]
- Iwai S, Weinmaier T, Schmidt BL, Albertson DG, Poloso NJ, Dabbagh K, et al. (2016). Piphillin: Improved Prediction of Metagenomic Content by Direct Inference from Human Microbiomes. PLoS One, 11(11), e0166104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolley SE, Alkhafaf Q, Hough C, & Welsh DA (2016). Presence of an Alcohol Use Disorder is Associated with Greater Pneumonia Severity in Hospitalized HIV-Infected Patients. Lung, 194(5), 755–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpel ME, Boutwell CL, & Allen TM (2015). BLT humanized mice as a small animal model of HIV infection. Curr Opin Virol, 13, 75–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli R, Lo Y, Homel P, Flanigan TP, Gardner LI, Howard AA, et al. (2006). Bacterial pneumonia, HIV therapy, and disease progression among HIV-infected women in the HIV epidemiologic research (HER) study. Clin Infect Dis, 43(1), 90–98. [DOI] [PubMed] [Google Scholar]
- Leclercq S, Matamoros S, Cani PD, Neyrinck AM, Jamar F, Starkel P, et al. (2014). Intestinal permeability, gut-bacterial dysbiosis, and behavioral markers of alcohol-dependence severity. Proc Natl Acad Sci U S A, 111(42), E4485–4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAleer JP, Nguyen NL, Chen K, Kumar P, Ricks DM, Binnie M, et al. (2016). Pulmonary Th17 Antifungal Immunity Is Regulated by the Gut Microbiome. J Immunol, 197(1), 97–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurdie PJ, & Holmes S (2012). Phyloseq: a bioconductor package for handling and analysis of high-throughput phylogenetic sequence data. Pac Symp Biocomput, 235–246. [PMC free article] [PubMed]
- McMurdie PJ, & Holmes S (2014). Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Comput Biol, 10(4), e1003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson S, Happel KI, Zhang P, Myers L, Dufour JP, & Bagby GJ (2013). Effect of bacterial pneumonia on lung simian immunodeficiency virus (SIV) replication in alcohol consuming SIV-infected rhesus macaques. Alcohol Clin Exp Res, 37(6), 969–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson S, & Kolls JK (2002). Alcohol, host defence and society. Nat Rev Immunol, 2(3), 205–209. [DOI] [PubMed] [Google Scholar]
- Noguera-Julian M, Rocafort M, Guillen Y, Rivera J, Casadella M, Nowak P, et al. (2016). Gut Microbiota Linked to Sexual Preference and HIV Infection. EBioMedicine, 5, 135–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J, Roeland Kindt, Pierre Legendre, Bob O’Hara, Henry M Stevens H (2007). “The Vegan Package.” Community Ecology Package 10: 631–37. [Google Scholar]
- Poonia B, Nelson S, Bagby GJ, Zhang P, Quniton L, & Veazey RS (2006). Chronic alcohol consumption results in higher simian immunodeficiency virus replication in mucosally inoculated rhesus macaques. AIDS Res Hum Retroviruses, 22(6), 589–594. [DOI] [PubMed] [Google Scholar]
- Samuelson DR, Burnham EL, Maffei VJ, Vandivier RW, Blanchard EE, Shellito JE, et al. (2018). The respiratory tract microbial biogeography in alcohol use disorder. Am J Physiol Lung Cell Mol Physiol, 314(1), L107–l117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuelson DR, Charles TP, de la Rua NM, Taylor CM, Blanchard EE, Luo M, et al. (2016). Analysis of the intestinal microbial community and inferred functional capacities during the host response to Pneumocystis pneumonia. Exp Lung Res, 42(8–10), 425–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuelson DR, Shellito JE, Maffei VJ, Tague ED, Campagna SR, Blanchard EE, et al. (2017). Alcohol-associated intestinal dysbiosis impairs pulmonary host defense against Klebsiella pneumoniae. PLoS Pathog, 13(6), e1006426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuelson DR, Welsh DA, & Shellito JE (2015). Regulation of lung immunity and host defense by the intestinal microbiota. [Review]. Frontiers in Microbiology, 6(1085). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siggins RW, Melvan JN, Welsh DA, Bagby GJ, Nelson S, & Zhang P (2011). Alcohol suppresses the granulopoietic response to pulmonary Streptococcus pneumoniae infection with enhancement of STAT3 signaling. J Immunol, 186(7), 4306–4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon L, Siggins R, Winsauer P, Brashear M, Ferguson T, Mercante D, et al. (2018). Simian Immunodeficiency Virus Infection Increases Blood Ethanol Concentration Duration After Both Acute and Chronic Administration. AIDS Res Hum Retroviruses, 34(2), 178–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisson JH (2007). Alcohol and airways function in health and disease. Alcohol, 41(5), 293–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuruya A, Kuwahara A, Saito Y, Yamaguchi H, Tenma N, Inai M, et al. (2016). Major Anaerobic Bacteria Responsible for the Production of Carcinogenic Acetaldehyde from Ethanol in the Colon and Rectum. Alcohol Alcohol, 51(4), 395–401. [DOI] [PubMed] [Google Scholar]
- Vagenas P, Azar MM, Copenhaver MM, Springer SA, Molina PE, & Altice FL (2015). The Impact of Alcohol Use and Related Disorders on the HIV Continuum of Care: a Systematic Review: Alcohol and the HIV Continuum of Care. Curr HIV/AIDS Rep, 12(4), 421–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wandera B, Tumwesigye NM, Nankabirwa JI, Kambugu AD, Parkes-Ratanshi R, Mafigiri DK, et al. (2015). Alcohol Consumption among HIV-Infected Persons in a Large Urban HIV Clinic in Kampala Uganda: A Constellation of Harmful Behaviors. PLoS One, 10(5), e0126236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams B, Landay A, & Presti RM (2016). Microbiome alterations in HIV infection a review. Cell Microbiol, 18(5), 645–651. [DOI] [PubMed] [Google Scholar]
- Williams B, Mirmonsef P, Boucher CA, Bushman F, Carrington-Lawrence S, Collman RG, et al. (2016). A Summary of the First HIV Microbiome Workshop 2015. AIDS Res Hum Retroviruses, 32(10–11), 935–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan AW, Fouts DE, Brandl J, Starkel P, Torralba M, Schott E, et al. (2011). Enteric dysbiosis associated with a mouse model of alcoholic liver disease. Hepatology, 53(1), 96–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Humanized BLT mice were administered binge-on-chronic alcohol (10 days chronic and 2x binges). Following alcohol feeding groups of mice were sacrificed 24 hrs following the final binge or infected intranasal with S. pneumoniae (~1 × 105 CFU in 50 μl of PBS) and sacrificed at 48 hours post infection. 5% EtOH diet was maintained continuously throughout the experiment.
Streptococcus lung burden (Log10 CFU/ml) at 48 hrs. post infection in animals recolonized with both full microbial or fungal-depleted microbiota from HIV+ ethanol-fed, HIV+ pair-fed, HIV- ethanol-fed, or HIV- pair-fed. Bars represent the mean of the cell counts post infection minus the cell counts prior to infection ± SEM. Closed symbols = full community, open symbols = communities without fungal organisms.