Abstract
Background
Re‐biopsy is important for exploring resistance mechanisms, especially for non‐small cell lung cancer (NSCLC) patients who develop resistance to EGFR‐tyrosine kinase inhibitors (TKIs). Liquid biopsy using circulating tumor DNA has come into use for this purpose. This retrospective study investigated the status of re‐biopsy and liquid biopsy in NSCLC patients with EGFR mutations and evaluated their effect on clinical strategies and prognosis.
Methods
Five hundred fifty‐five NSCLC patients with resistance to EGFR‐TKIs were included and divided into three groups: re‐biopsy, liquid biopsy, and no re‐biopsy. Amplification refractory mutation system (ARMS) PCR or super ARMS PCR was used to detect EGFR mutations.
Results
Three hundred eight (55.5%) patients underwent re‐biopsy; 45.5% (140/308) were positive for T790M. The most common re‐biopsy procedure was computed tomography‐guided percutaneous core needle biopsy (60.1%), followed by effusion drainage (29.5%) and superficial lymph node biopsy (6.5%). One hundred eighteen (21.3%) patients underwent liquid biopsy; the T790M detection rate was 41.5% (49/118.) Of the 308 patients who underwent re‐biopsy, 69 were examined for EGFR mutations with plasma. The concordance rate of T790M detection between tissue and plasma was 66.7%. A statistical difference in further treatment after EGFR‐TKI failure was observed among all groups (P = 0.014). Patients in the biopsy groups were more likely to receive third‐generation EGFR‐TKIs. Multivariate analysis showed that re‐biopsy had a significant impact on overall survival (P < 0.001).
Conclusion
Re‐biopsy plays a pivotal role in the management of patients with NSCLC and resistance to EGFR‐TKIs. Liquid biopsy may be an alternative if difficulties performing re‐biopsy exist.
Keywords: EGFR‐TKI resistance, liquid biopsy, non‐small cell lung cancer, re‐biopsy
Introduction
In China, lung cancer is the most frequently diagnosed cancer and the leading cause of cancer‐related mortality.1 The five‐year survival rate in patients with lung cancer varies from 3.7–32.9%, depending on stage and regional differences.2 Non‐small cell lung cancer (NSCLC) accounts for 80–85% of lung cancers.3 Personalized therapeutic strategies have dramatically improved the clinical outcome of patients with advanced NSCLC in recent years.4, 5, 6 Activating EGFR mutations are the most common therapeutically tractable driver mutation in lung adenocarcinomas with distinct ethnic differences, occurring at higher frequencies in Asian (40–60%) compared to Caucasian populations (7–10%).7, 8, 9, 10 Many clinical trials have proven that EGFR‐tyrosine kinase inhibitors (TKIs) yield a superior response and more acceptable toxicity than traditional chemotherapy in advanced NSCLC patients with EGFR mutations; however patients inevitably develop resistance after achieving 9–13 months of progression‐free survival (PFS).11, 12, 13, 14 Acquired resistance to EGFR‐TKIs is mainly attributed to secondary EGFR mutations.15, 16, 17 EGFR T790M, a specific point mutation in exon 20, is the most common resistance mutation, and accounts for 33–63%.18, 19, 20, 21, 22, 23, 24 Third‐generation EGFR‐TKIs (3‐TKIs), such as osimertinib, have proven to have promising activity in advanced NSCLC patients with EGFR T790M mutations.25, 26, 27 Based on these facts, repeat biopsy plays an important role in clinical application for exploring resistance mechanisms and determining further therapeutic strategies.28, 29 However, obtaining tissue samples from patients for repeat molecular analysis after EGFR‐TKI failure remains a challenge. In the present study, we evaluated the current status of re‐biopsy and liquid biopsy and their effect on clinical strategies and prognosis in Chinese NSCLC patients with EGFR mutations after EGFR‐TKI failure.
Methods
Patients
We retrospectively reviewed the medical records of NSCLC patients with EGFR sensitive mutations, including 19deletions (19del), L858R, G719X, L861Q, and S768I, treated at the Shanghai Pulmonary Hospital between October 2011 and October 2017. Patients without EGFR mutations, or with EGFR 20 insert, de novo T790M, or known concomitant gene mutations, were excluded. Patients who were lost follow‐up before confirmation of progressive disease (PD) from EGFR‐TKIs were also excluded. Patients were divided into two groups according to the method of mutation testing: the rebiopsy group included patients who underwent tissue or cytologic sampling, while the liquid biopsy group included patients who underwent mutation testing with circulating tumor DNA (Fig 1). A control group of patients who did not undergo re‐biopsy was also included. If mutation had been tested with tissues and plasma, the tissue results were regarded as standard. Progression‐free survival (PFS) was defined as the interval from the initiation of further treatment after EGFR‐TKI failure to first confirmed PD according to Response Evaluation Criteria in Solid Tumors (RECIST) version 1.1. Overall survival (OS) was defined as the interval from diagnosis to death from any cause or to 31 December 2018. The primary objective was to evaluate the status of re‐biopsy in Chinese patients including frequencies, procedure results, and impact on follow‐up treatment strategies. The potential application of plasma‐testing methodologies was also assessed. The second objective was to explore whether patients that underwent re‐biopsy achieved a better clinical outcome. The need for written informed consent was waived because of the retrospective design of the study. The institutional review board of Shanghai Pulmonary Hospital approved the study.
Figure 1.
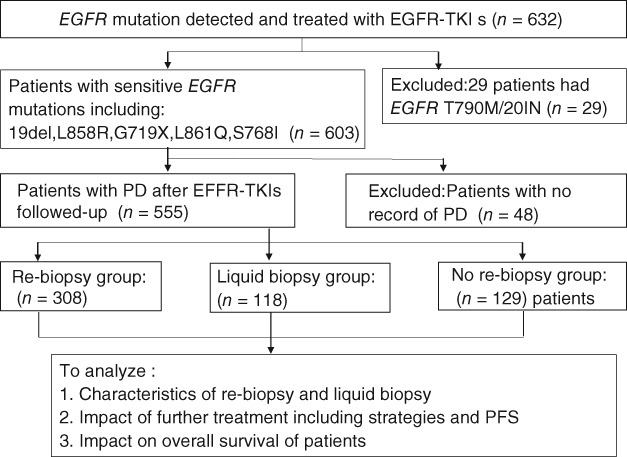
Flow chart of subject enrollment. PFS, progression‐free survival; PD, progressive disease; TKIs, tyrosine kinase inhibitors.
EGFR mutation analysis
Amplification refractory mutation system (ARMS) PCR was used to detect EGFR mutations. Our hospital adopted the ARMS‐PCR method on 1 May 2017, thus mutation analysis of plasma was tested by ARMS‐PCR before 1 May 2017 and by super ARMS‐PCR thereafter. Tissue or cytologic specimens were frozen within 30 minutes of sampling and stored at −80°C. Frozen samples were thawed at 37°C, refrozen rapidly to disrupt the cells, and divided into equal aliquots before DNA was extracted using standard procedures according to EGFR mutation test kit instructions.
Statistical analysis
Statistical analyses were performed using SPSS version 19.0. Intergroup comparisons were performed using the Mann–Whitney U test for continuous variables, and Pearson's χ2 or Fisher's exact tests for categorical variables. Multivariate analysis was performed to determine the feasibility of re‐biopsy using multinomial logistic regression. The Kaplan–Meier method was used to estimate OS and PFS. Univariate differences in survival were assessed using the stratified log‐rank method. Hazard ratios (HRs) for PFS and OS and multivariate differences in survival were estimated using a stratified Cox model. P < 0.05 was considered significant.
Results
Patient characteristics
Baseline individual characteristics were retrospectively reviewed in 555 patients with NSCLC after EGFR‐TKI failure (Table 1). The median age was 60 years (range: 53–67), with women comprising 59.3% of the total. Adenocarcinoma (89.4%) was the most common histology. Of the total patients, 292 (52.6%) had 19del, 245 (44.1%) had L858R, 11 (2.0%) had uncommon mutations, and 7 (1.3%) had double sensitive mutations. The EGFR‐TKIs used were gefitinib (62.0%), erlotinib (18.6%), icotinib (17.5%), and afatinib (2.0%); 73.9% patients were administered EGFR‐TKIs at first‐line.
Table 1.
Baseline characteristics of patients
| Characteristics | All (n = 588) | Univariate | Multivariate | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Re‐biopsy | Liquid biopsy | ||||||||
| Re‐biopsy | Liquid biopsy | No biopsy | P | HR | P | HR | P | ||
| Age (years) | 1.046 | 0.847 | 0.660 | 0.220 | |||||
| ≤ 65 | 379 (68.3) | 208 (67.5) | 87 (73.7) | 84 (65.1) | 0.318 | ||||
| > 65 | 176 (31.7) | 100 (32.5) | 31 (26.3) | 45 (34.9) | |||||
| Gender | 0.889 | 0.674 | 1.557 | 0.135 | |||||
| Male | 226 (40.7) | 126 (40.9) | 50 (42.4) | 50 (38.8) | 0.842 | ||||
| Female | 329 (59.3) | 182 (59.1) | 68 (57.6) | 79 (61.2) | |||||
| Smoking | 0.893 | 0.731 | 1.066 | 0.875 | |||||
| Non‐smoker | 438 (78.9) | 242 (78.6) | 94 (79.7) | 102 (79.1) | 0.969 | ||||
| Current or former smoker | 117 (21.1) | 66 (21.4) | 24 (20.3) | 27 (20.9) | |||||
| ECOG | 0.728 | 0.365 | 0.907 | 0.824 | |||||
| 0–1 | 494 (89.0) | 268 (87.0) | 111 (94.1) | 115 (89.1) | 0.114 | ||||
| 2–3 | 61 (11.0) | 40 (13.0) | 7 (5.9) | 14 (10.9) | |||||
| Histology | 0.986 | 0.969 | 1.854 | 0.219 | |||||
| Adenocarcinoma | 496 (89.4) | 276 (89.6) | 105 (89.0) | 115 (89.1) | 0.978 | ||||
| Non‐adenocarcinoma | 59 (10.6) | 32 (10.4) | 13 (11.0) | 14 (10.9) | |||||
| Gene mutation type | |||||||||
| 19del | 292 (52.6) | 178 (57.8) | 54 (45.8) | 60 (46.5) | 0.096 | 4.306 | 0.130 | 0.955 | 0.964 |
| L858R | 245 (44.1) | 121 (39.3) | 59 (50.0) | 65 (50.4) | 3.090 | 0.241 | 1.304 | 0.795 | |
| Other | 11 (2.0) | 7 (2.3) | 3 (2.5) | 1 (0.8) | 11.029 | 0.095 | 4.394 | 0.336 | |
| Double mutation | 7 (1.3) | 2 (0.6) | 2 (1.7) | 3 (2.3) | |||||
| EGFR‐TKIs | |||||||||
| Gefitinib | 344 (62.0) | 198 (64.3) | 72 (61.0) | 74 (57.4) | 0.63 | 1.729 | 0.476 | 1.025 | 0.977 |
| Erlotinib | 103 (18.6) | 58 (18.8) | 18 (15.3) | 27 (20.9) | 1.234 | 0.789 | 0.740 | 0.738 | |
| Icotinib | 97 (17.5) | 47 (15.3) | 25 (21.2) | 25 (19.4) | 1.188 | 0.828 | 1.093 | 0.921 | |
| 2‐TKIs | 11 (2.0) | 5 (1.6) | 3 (2.5) | 3 (2.3) | |||||
| Line of EGFR‐TKIs | 0.752 | 0.279 | 1.208 | 0.570 | |||||
| 1 | 410 (73.9) | 221 (71.8) | 93 (78.8) | 96 (74.4) | 0.328 | ||||
| ≥ 2 | 145 (26.1) | 87 (28.2) | 25 (21.2) | 33 (25.6) | |||||
| Efficacy of EGFR‐TKIs | |||||||||
| PR | 277 (49.9) | 163 (52.9) | 70 (59.3) | 44 (34.1) | <0.001 | 1.032 | 0.953 | 1.652 | 0.452 |
| SD | 214 (38.6) | 112 (36.4) | 42 (35.6) | 60 (46.5) | 0.507 | 0.205 | 0.698 | 0.592 | |
| PD | 36 (6.5) | 15 (4.9) | 1 (0.8) | 20 (15.5) | 0.211 | 0.013 | 0.044 | 0.010 | |
| Not clear | 28 (5.0) | 18 (5.8) | 5 (4.2) | 5 (3.9) | |||||
Non‐adenocarcinoma included: 26 non‐small cell lung cancer (NSCLC), 15 adenosquamous carcinoma, 12 squamous carcinoma, 4 poorly differentiated adenocarcinoma, and 2 large cell lung cancer. Other included 5 G719X, 3 L861Q, and 3 S768I. Double mutation included five 19del and L858R, one 19 del and L861Q, and one S768I and G719X. ECOG, Eastern Cooperative Oncology Group; PD, progressive disease; PR, partial response; SD, stable disease; TKI, tyrosine kinase inhibitor.
Status of re‐biopsy and liquid biopsy
A total of 308 (55.5%) patients underwent re‐biopsy procedures, including: 185 (60.1%) computed tomography (CT)‐guided percutaneous core needle biopsy (PCNB); 91 (29.5%) effusion drainage (including 81 pleural effusion, 5 pericardial effusion, and 5 peritoneal effusion); 20 (6.5%) superficial lymph node biopsy; and 7 (2.3%) transbronchial needle aspiration (TBNA) or endobronchial ultrasonography (EBUS)‐guided TBNA (Fig 2a). Of the 308 patients, 134 (43.5%) were positive for EGFR T790M, 134 (43.5%) patients only harbored their initial mutation, and 25 (8.1%) patients were wild type (Fig 2b). A total of 118 (21.3%) patients underwent liquid biopsy: 49 (41.5%) patients were positive for T790M, 35 (29.7%) patients only harbored their initial mutation, and 32 (27.1%) patients were wild type (Fig 2c). Of the 308 patients who underwent re‐biopsy, 69 patients were examined for EGFR mutation using plasma. The specificity of liquid biopsy for T790M detection was 84.4% and the sensitivity was 33.3%. The concordance of T790M detection between tissue and plasma was 66.7%. The prevalence of acquired T790M mutation was statistically different between patients with uncommon and common mutations (19del/L858R: 44.8% vs. 11.1%; P = 0.04), but further studies are required to verify these results, as our samples of patients with uncommon mutations were few. In addition, univariate and multivariate analysis showed that patients in the re‐biopsy and liquid biopsy groups exhibited a better previous response to EGFR‐TKIs compared to the control (P < 0.05) (Table 1).
Figure 2.
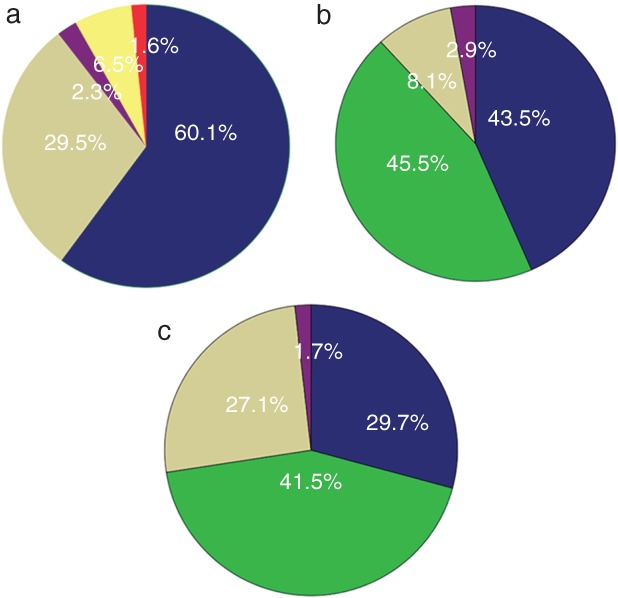
(a) The proportions of different sampling procedures in patients who underwent re‐biopsy, ( ) CT‐guided PCNB, (
) CT‐guided PCNB, ( ) Effusion drainage, (
) Effusion drainage, ( ) TBNA/EBUS‐TBNA, (
) TBNA/EBUS‐TBNA, ( ) SLNB and (
) SLNB and ( ) other metastasis biopsy. The distribution of results in patients who were tested for EGFR mutation via (b) tissue (c) or plasma. (
) other metastasis biopsy. The distribution of results in patients who were tested for EGFR mutation via (b) tissue (c) or plasma. ( ) Consistent with baseline, (
) Consistent with baseline, ( ) Mut plus T790M/T790M, (
) Mut plus T790M/T790M, ( ) Wild type, and (
) Wild type, and ( ) others. “Others” in (b) include three patients with small‐cell lung cancer, two with baseline mutations and c‐MET, one with an exon 20 insert, one with translation from 19del to L858R, one with KRAS and one with ALK. “Others” in (c) include one patient with an exon 20 insert and one with baseline mutations (L858R), S768I, and T790M. CT, computed tomography; EBUS, endobronchial ultrasonography; mut plus T790M, patients harbored a baseline mutation and a T790M mutation when re‐tested; PCNB, percutaneous core needle biopsy; SLNB, superficial lymph node biopsy; TBNA, transbronchial needle aspiration; T790M, patients only harbored a T790M mutation and the baseline sensitive mutation disappeared.
) others. “Others” in (b) include three patients with small‐cell lung cancer, two with baseline mutations and c‐MET, one with an exon 20 insert, one with translation from 19del to L858R, one with KRAS and one with ALK. “Others” in (c) include one patient with an exon 20 insert and one with baseline mutations (L858R), S768I, and T790M. CT, computed tomography; EBUS, endobronchial ultrasonography; mut plus T790M, patients harbored a baseline mutation and a T790M mutation when re‐tested; PCNB, percutaneous core needle biopsy; SLNB, superficial lymph node biopsy; TBNA, transbronchial needle aspiration; T790M, patients only harbored a T790M mutation and the baseline sensitive mutation disappeared.
Strategies and progression‐free survival of further treatment
A total of 414 (74.6%) patients were followed‐up after further treatment subsequent to EGFR‐TKI failure. Pearson's χ2 test revealed a statistical difference between the three groups (P = 0.014) (Table 2). Further analysis indicated that patients in the biopsy groups were more likely to receive third‐generation TKIs (re‐biopsy 26.4% vs. liquid biopsy 31.4% vs. no re‐biopsy 6.1%): 87.5% (56/64) of patients administered 3‐TKIs were positive for T790M in the re‐biopsy group and 85.7% in the liquid biopsy group (Table 2). However, overall only 52.3% (56/107) of patients positive for T790M received 3‐TKIs in the re‐biopsy and 75.0% (24/32) in the liquid biopsy group (Table 3).
Table 2.
Further treatment after EGFR‐TKI failure
| Further treatment | Re‐biopsy (n = 242) | Liquid biopsy (n = 90) | No re‐biopsy (n = 82) | Total | P |
|---|---|---|---|---|---|
| 3‐TKIs | 64 (26.4) | 28 (31.1) | 5 (6.1) | 97 (23.4) | 0.014 |
| TKI + chemotherapy/radiotherapy | 24 (9.9) | 10 (11.1) | 10 (12.2) | 44 (10.6) | |
| Other TKIs | 7 (2.9) | 2 (2.4) | 2 (2.4) | 12 (2.9) | |
| Former TKIs | 11 (4.5) | 7 (7.8) | 8 (9.8) | 26 (6.3) | |
| Chemotherapy/radiotherapy | 123 (50.8) | 40 (44.4) | 53 (64.6) | 216 (52.2) | |
| Other | 13 (5.4) | 2 (2.2) | 4 (4.9) | 19 (4.6) |
Other tyrosine kinase inhibitors (TKIs) refers to other first or second‐generation EGFR‐TKIs; Other includes best supportive treatment, traditional Chinese medicine, crizotinib, etc. 3‐TKIs, third‐generation TKIs.
Table 3.
Further treatment after EGFR‐TKI failure according to biopsy results
| Result of re‐biopsy/liquid biopsy | ||||||
|---|---|---|---|---|---|---|
| Further treatment | Consistent with baseline | Mut plus T790M/T790M | Wild type | Others | Total | P |
| 3‐TKIs | 6 (5.5)/2 (6.5) | 56 (52.3)/24 (75.0) | 1 (5.9)/1 (4.0) | 1 (12.5)/1 (50.0) | 64 (26.4)/28 (31.1) | <0.001/<0.001 |
| TKI + chemotherapy/radiotherapy | 16 (14.5)/6 (19.4) | 5 (4.7)/1 (3.1) | 2 (11.8)/3 (12.0) | 1 (12.5)/0 (0.0) | 24 (9.9)/10 (11.1) | |
| Other TKIs | 4 (3.6)/2 (6.5) | 1 (0.9)/0 (0.0) | 0 (0.0)/1 (4.0) | 2 (25.0)/0 (0.0) | 7 (2.9)/3 (3.3) | |
| Former TKIs | 6 (5.5)/3 (9.7) | 4 (3.7)/1 (3.1) | 4 (9.5)/3 (12.0) | 0 (0.0)/0 (0.0) | 11 (4.5)/7 (7.8) | |
| Chemotherapy/radiotherapy | 73 (66.4)/17 (54.8) | 35 (32.7)/5 (15.6) | 12 (70.6)/17 (68.0) | 3 (37.5)/0 (0.0) | 123 (50.8)/40 (44.4) | |
| Others | 5 (4.5)/1 (3.2) | 6 (5.6)/1 (3.1) | 1 (5.9)/0 (0.0) | 1 (12.5)/0 (0.0) | 13 (5.4)/2 (2.2) | |
Other tyrosine kinase inhibitors (TKIs) refers to other first or second‐generation EGFR‐TKIs; Other includes best supportive treatment, traditional Chinese medicine, crizotinib, etc. 3‐TKIs, third‐generation TKIs.
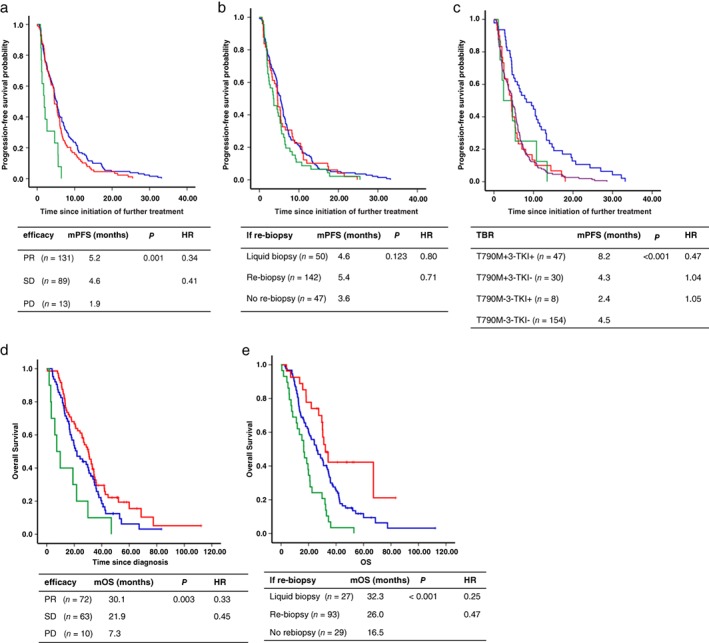
Re‐biopsy and liquid biopsy could impact further treatment strategies, as indicated above. To explore the specific impact, we analyzed PFS after further treatment. Univariate analysis proved that the previous efficacy of EGFR‐TKIs had a significant impact on PFS (P = 0.001) (Table 4, Fig 3a). However, the difference in PFS among the three groups was not statistically significant (P = 0.123) (Table 4, Fig 3b). Considering that the deviation resulted from patients who were positive for T790M but did not receive 3‐TKIs, we introduced a new variable – treatment‐based result (TBR) – and further divided the patients into four groups according to whether T790M was positive and 3‐TKIs were administered. Patients in the T790M + 3‐TKI+ group achieved significantly longer PFS than in other groups (P < 0.001) (Table 4, Fig 3c). Multivariate analysis confirmed that the previous efficacy of EGFR‐TKIs and TBR were independent significant factors of PFS after further treatment (Table 4).
Table 4.
Univariate and multivariate analyses of clinical parameters of PFS of further treatment
| Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|
| Factors | HR | 95% CI | P | HR | 95% CI | P |
| Gender (female/male) | 0.881 | 0.773–1.004 | 0.057 | 0.882 | 0.641–1.214 | 0.441 |
| Age (< 65/≥ 65) | 1.308 | 0.989–1.730 | 0.060 | 1.130 | 0.847–1.507 | 0.407 |
| Smoking (never/ever) | 0.700 | 0.512–0.957 | 0.025 | 0.849 | 0.574–1.256 | 0.413 |
| ECOG (0–1/2–3) | 0.785 | 0.496–1.244 | 0.319 | |||
| Histology (adenocarcinoma/non‐adenocarcinoma) | 0.771 | 0.492–1.208 | 0.256 | |||
| EGFR mutation | 0.511 | |||||
| 19del/double mutation | 0.593 | 0.188–1.872 | 0.373 | |||
| L858R/double mutation | 0.611 | 0.193–1.933 | 0.402 | |||
| Other/double mutation | 1.129 | 0.252–5.055 | 0.874 | |||
| EGFR‐TKIs | 0.224 | |||||
| Gefitinib/afatinib | 0.552 | 0.203–1.502 | 0.245 | |||
| Erlotinib/afatinib | 0.455 | 0.162–1.280 | 0.136 | |||
| Icotinib/afatinib | 0.672 | 0.239–1.889 | 0.451 | |||
| Treatment line of EGFR‐TKIs (1/≥ 2) | 0.927 | 0.696–1.235 | 0.605 | |||
| Efficacy of EGFR‐TKIs | 0.001 | 0.005 | ||||
| PR/PD | 0.336 | 0.188–0.603 | 0.000 | 0.392 | 0.216–0.712 | 0.002 |
| SD/PD | 0.410 | 0.227–0.740 | 0.003 | 0.500 | 0.273–0.916 | 0.025 |
| If re‐biopsied | 0.123 | |||||
| Re‐biopsy/no re‐biopsy | 0.709 | 0.508–0.989 | 0.043 | |||
| Liquid biopsy/no re‐biopsy | 0.798 | 0.535–1.190 | 0.268 | |||
| TBR | 0.000 | 0.002 | ||||
| T790M+ 3‐TKI+/T790M‐3‐TKI‐ | 0.467 | 0.331–0.659 | 0.000 | 0.520 | 0.364–0.745 | 0.000 |
| T790M + 3‐TKI−/T790M‐3‐TKI‐ | 1.041 | 0.704–1.542 | 0.839 | 1.108 | 0.741–1.657 | 0.618 |
| T790M‐3‐TKI+/T790M‐3‐TKI‐ | 1.053 | 0.516–2.146 | 0.888 | 1.212 | 0.590–2.490 | 0.600 |
ECOG, Eastern Cooperative Oncology Group; PD, progressive disease; PFS, progression‐free survival; PR, partial response; SD, stable disease; TBR treatment‐based result; TKI, tyrosine kinase inhibitor.
Figure 3.
Kaplan–Meier estimates of the duration of progression‐free survival (PFS) of further treatment in (a) patients who exhibited an objective response to EGFR‐tyrosine kinase inhibitors (TKIs), efficacy ( ) PR, (
) PR, ( ) SD and (
) SD and ( ) PD, (b) patients who underwent re‐biopsy or not, if re‐biopsy (
) PD, (b) patients who underwent re‐biopsy or not, if re‐biopsy ( ) re‐biopsy, (
) re‐biopsy, ( ) liquid biopsy and (
) liquid biopsy and ( ) no biopsy and (c) patients divided into different groups according to whether T790M was positive and third‐generation TKIs (3‐TKIs) were administered, TBR (
) no biopsy and (c) patients divided into different groups according to whether T790M was positive and third‐generation TKIs (3‐TKIs) were administered, TBR ( ) T790M +3‐TKI+, (
) T790M +3‐TKI+, ( ) T790M +3‐TKI−, (
) T790M +3‐TKI−, ( ) T790M −3‐TKI+ and (
) T790M −3‐TKI+ and ( ) T790M −3‐TKI−. Kaplan–Meier estimates of the duration of overall survival (OS) of (d) patients who exhibited an objective response to EGFR‐TKIs, efficacy (
) T790M −3‐TKI−. Kaplan–Meier estimates of the duration of overall survival (OS) of (d) patients who exhibited an objective response to EGFR‐TKIs, efficacy ( ) PR, (
) PR, ( ) SD, (
) SD, ( ) PD, (
) PD, ( ) PR‐censored, (
) PR‐censored, ( ) SD‐censored and (
) SD‐censored and ( ) PD‐censored and (e) patients who underwent re‐biopsy or not, if re‐biopsy (
) PD‐censored and (e) patients who underwent re‐biopsy or not, if re‐biopsy ( ) re‐biopsy, (
) re‐biopsy, ( ) liquid biopsy, (
) liquid biopsy, ( ) no rebiopsy, (
) no rebiopsy, ( ) re‐biopsy‐censored, (
) re‐biopsy‐censored, ( ) liquid biopsy‐censored and (
) liquid biopsy‐censored and ( ) no rebiopsy‐censored. HR, hazard ratio; mOS, median OS; mPFS, median PFS; mut plus T790M, patients harbored a baseline mutation and a T790M mutation when re‐tested; PD, progressive disease; PR, partial response; SD, stable disease; T790M, patients only harbored a T790M mutation and the baseline sensitive mutation disappeared.
) no rebiopsy‐censored. HR, hazard ratio; mOS, median OS; mPFS, median PFS; mut plus T790M, patients harbored a baseline mutation and a T790M mutation when re‐tested; PD, progressive disease; PR, partial response; SD, stable disease; T790M, patients only harbored a T790M mutation and the baseline sensitive mutation disappeared.
Overall survival of patients
Overall survival was analyzed in 149 patients and was longer in patients with a previous response to EGFR‐TKIs than those with PD (partial response [PR] vs. stable disease [SD] vs. PD: 30.1 vs. 20.9 vs. 7.3 months, respectively; HR 0.33 for PR vs. PD, HR 0.45 for SD vs. PD; P = 0.003) (Fig 3d). The OS of patients in the liquid and re‐biopsy groups was superior to patients in the no re‐biopsy group (32.3 vs. 26.0 vs. 16.5 months, respectively; HR 0.25 for liquid biopsy vs. no re‐biopsy, HR 0.47 for re‐biopsy vs. no re‐biopsy; P < 0.001) (Fig 3e). Multivariate analysis revealed that previous efficacy to EGFR‐TKIs and re‐biopsy had a significant impact on OS (Table 5). Multivariate analysis showed that patients with good performance status and those administered 2‐TKIs achieved longer survival (Table 5).
Table 5.
Univariate and multivariate analyses of clinical parameters of OS
| Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|
| Factors | HR | 95% CI | P | HR | 95% CI | P |
| Gender (female/male) | 1.002 | 0.702–1.430 | 0.991 | |||
| Age (< 65/≥ 65) | 1.027 | 0.697–1.514 | 0.892 | |||
| Smoking (never/ever) | 0.872 | 0.580–1.310 | 0.509 | |||
| ECOG (0–1/2–3) | 0.604 | 0.374–0.977 | 0.040 | 0.598 | 0.364–0.980 | 0.042 |
| Histology (adenocarcinoma/non‐adenocarcinoma) | 0.962 | 0.568–1.629 | 0.884 | |||
| EGFR mutation | 0.278 | |||||
| 19del/double mutation | 1.273 | 0.460–3.523 | 0.642 | |||
| L858R/double mutation | 1.655 | 0.597–4.591 | 0.333 | |||
| Other/double mutation | 2.827 | 0.624–12.813 | 0.178 | |||
| EGFR‐TKIs | 0.097 | 0.029 | ||||
| Gefitinib/2‐TKIs | 7.595 | 1.045–55.222 | 0.045 | 6.179 | 0.833–45.820 | 0.075 |
| Erlotinib/2‐TKIs | 6.225 | 0.849–45.635 | 0.072 | 4.024 | 0.538–30.118 | 0.175 |
| Icotinib/2‐TKIs | 4.221 | 0.566–36.114 | 0.155 | 2.882 | 0.345–24.111 | 0.329 |
| Treatment line of EGFR‐TKIs (1/≥ 2) | 1.185 | 0.824–1.704 | 0.359 | |||
| Efficacy of EGFR‐TKIs | 0.005 | 0.002 | ||||
| PR/PD | 0.334 | 0.170–0.658 | 0.002 | 0.276 | 0.134–0.569 | 0.000 |
| SD/PD | 0.454 | 0.231–0.894 | 0.022 | 0.319 | 0.153–0.664 | 0.002 |
| If re‐biopsied | <0.001 | 0.000 | ||||
| Re‐biopsy/no re‐biopsy | 0.466 | 0.300–0.725 | 0.001 | 0.358 | 0.220–0.582 | 0.000 |
| Liquid biopsy/no re‐biopsy | 0.276 | 0.146–0.520 | <0.001 | 0.279 | 0.145–0.535 | 0.000 |
| TBR | 0.450 | |||||
| T790M+ 3‐TKI+/T790M‐3‐TKI‐ | 0.707 | 0.407–1.229 | 0.219 | |||
| T790M+ 3‐TKI‐/T790M‐3‐TKI‐ | 0.769 | 0.483–1.225 | 0.269 | |||
| T790M‐ 3‐TKI+/T790M‐3‐TKI‐ | 0.673 | 0.246–1.842 | 0.441 | |||
Variables of P < 0.1 in univariate analysis were included into multivariate analysis. Treatment‐based result (TBR): patients were divided into four groups according to whether T790M was positive and third‐generation tyrosine kinase inhibitors (3‐TKIs) were used. 2‐TKIs, second‐generation TKIs; ECOG, Eastern Cooperative Oncology Group; OS, overall survival; PD, progressive disease; PR, partial response; SD, stable disease.
Discussion
Re‐biopsy has a pivotal role for exploring resistance mechanisms, particularly for patients with resistance to EGFR‐TKIs. However, re‐biopsy is not always feasible for either patients or physicians. Kawamura et al. reported that 120 patients (63%) in their sample had undergone re‐biopsy,30 while Ichihara et al. reported that 55 (53.9%) patients underwent re‐biopsy.21 We analyzed re‐biopsy status in 555 patients and the rate (55.5%) was similar to the reported data. CT‐guided PCNB (60.1%) was the most common re‐biopsy procedure used in our study sample, likely because of its safety and operability. The frequency of T790M mutations detected in patients who underwent re‐biopsy was 45.5% in the clinical setting at our institution, which is also consistent with previous studies (range: 33–63%).18, 19, 20, 21, 22, 23, 24 The remaining patients did not undergo re‐biopsy because of inaccessible tumors (n = 27), deterioration in performance status (n = 21), patient refusal (n = 124), or unknown reasons (n = 75).
Liquid biopsy has shown promising advantages of noninvasiveness and accessibility. Some studies have suggested that a plasma‐based test is a useful method for noninvasive assessment and monitoring of T790M resistance mutations.31, 32, 33 Our results indicate that detection of T790M in plasma using the super ARMS‐PCR method exhibits low sensitivity (33.3%) but high specificity (84.4%), with a satisfactory T790M detection rate (41.5%). The low sensitivity was attributed to the fact that 83 (70.3%) patients were tested by ARMS‐PCR. Moreover, these results suggest that re‐biopsy and liquid biopsy could provide guidance for further treatment strategies. As expected, patients who harbor T790M are more inclined to receive 3‐TKIs.
As the development of new agents has improved the overall survival duration of patients with malignant tumors, management of cancer patients throughout the entire course of disease has become a focus. As an essential part of the process, whether re‐biopsy could improve the clinical outcomes of patients with cancer remains to be determined. To the best of our knowledge, our study is the first and the largest to report the relationship between re‐biopsy and survival, including PFS of further treatment and OS in patients with NSCLC after EGFR‐TKI failure. As mentioned above, the previous efficacy of EGFR‐TKIs is an independent significant factor affecting not only PFS of further treatment but also OS. This may be because tumors with primary resistance to EGFR‐TKIs are genetically complex, with high heterogeneity and drug resistance.34, 35 What is interesting is that our data suggest that re‐biopsy has a significant impact on OS but not on PFS of further treatment, while TBR was a significant factor for PFS of further treatment but not for OS. The lack of a significant difference in PFS after further treatment among the three initial groups was attributed to the low proportion of patients positive for T790M that were administered 3‐TKIs in the re‐biopsy (23.1%) and liquid biopsy (26.7%) groups, but the very value of re‐biopsy is to determine the resistance mechanism, on which basis physicians can choose the appropriate therapy, thus the TBR variable was applied. However, any type of treatment a patient is administered can affect OS, which is why re‐biopsy is important. Not all of the patients identified with T790M were administered 3‐TKIs as further treatment after EGFR‐TKI failure, but they were more likely to receive subsequent treatment. Thus, patients who underwent rebiopsy were administered more appropriate subsequent therapies and achieved longer OS.
This study is limited by its retrospective nature and that our cohort was from a single institution. We used ARMs and super ARMS to test EGFR mutation in tissues and plasma, which has limited sensitivity. Although we used a large sample overall, the sample sizes of some of the subgroups were small. Our results require further verification in prospective studies.
In conclusion, re‐biopsy plays a pivotal role in the management of patients with NSCLC and resistance to EGFR‐TKIs and improves the clinical outcome to some extent. Liquid biopsy may be an alternative if difficulties performing re‐biopsy exist.
Disclosure
No authors report any conflict of interest.
Contributor Information
Xuefei Li, Email: bug_lily2003@163.com.
Chunxia Su, Email: susu_mail@126.com.
References
- 1. Chen W, Zheng R, Baade PD et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016; 66 (2): 115–32. [DOI] [PubMed] [Google Scholar]
- 2. Allemani C, Matsuda T, Di Carlo V et al. Global surveillance of trends in cancer survival 2000–14 (CONCORD‐3): Analysis of individual records for 37 513 025 patients diagnosed with one of 18 cancers from 322 population‐based registries in 71 countries. Lancet 2018; 391 (10125): 1023–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med 2008; 359 (13): 1367–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dolly SO, Collins DC, Sundar R et al. Advances in the development of molecularly targeted agents in non‐small‐cell lung cancer. Drugs 2017; 77 (8): 813–27. [DOI] [PubMed] [Google Scholar]
- 5. Jiang W, Cai G, Hu PC, Wang Y. Personalized medicine in non‐small cell lung cancer: A review from a pharmacogenomics perspective. Acta Pharm Sin B 2018; 8 (4): 530–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bui KT, Cooper WA, Kao S et al. Targeted molecular treatments in non‐small cell lung cancer: A clinical guide for oncologists. J Clin Med. 2018; 7 (8): 192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shigematsu H, Lin L, Takahashi T et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst 2005; 97 (5): 339–46. [DOI] [PubMed] [Google Scholar]
- 8. Clinical lung Cancer Genome Project (CLCGP), Network Genomic Medicine (NGM) . A genomics‐based classification of human lung tumors. Sci Transl Med 2013; 5 (209): 209ra153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jain A, Lim C. Gan EM3, impact of smoking and brain metastasis on outcomes of advanced EGFR mutation lung adenocarcinoma patients treated with first line epidermal growth factor receptor tyrosine kinase inhibitors. PLoS One 2015; 10 (5): e0123587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kris MG, Johnson BE, Berry LD et al. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA 2014; 311 (19): 1998–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mok TS, Wu YL, Thongprasert S et al. Gefitinib or carboplatin‐paclitaxel in pulmonary adenocarcinoma. N Engl J Med 2009; 361 (10): 947–57. [DOI] [PubMed] [Google Scholar]
- 12. Zhou C, Wu YL, Chen G et al. Erlotinib versus chemotherapy as first‐line treatment for patients with advanced EGFR mutation‐positive non‐small‐cell lung cancer (OPTIMAL, CTONG‐0802): A multicentre, open‐label, randomised, phase 3 study. Lancet Oncol 2011; 12 (8): 735–42. [DOI] [PubMed] [Google Scholar]
- 13. Hu X, Han B, Gu A et al. A single‐arm, multicenter, safety‐monitoring, phase IV study of icotinib in treating advanced non‐small cell lung cancer (NSCLC). Lung Cancer 2014; 86 (2): 207–12. [DOI] [PubMed] [Google Scholar]
- 14. Wu YL, Zhou C, Hu CP et al. Afatinib versus cisplatin plus gemcitabine for first‐line treatment of Asian patients with advanced non‐small‐cell lung cancer harbouring EGFR mutations (LUX‐Lung 6): An open‐label, randomised phase 3 trial. Lancet Oncol 2014; 15 (2): 213–22. [DOI] [PubMed] [Google Scholar]
- 15. Yu HA, Suzawa K, Jordan E et al. Concurrent alterations in EGFR‐mutant lung cancers associated with resistance to EGFR kinase inhibitors and characterization of MTOR as a mediator of resistance. Clin Cancer Res 2018; 24 (13): 3108–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lee CK, Kim S, Lee JS et al. Next‐generation sequencing reveals novel resistance mechanisms and molecular heterogeneity in EGFR‐mutant non‐small cell lung cancer with acquired resistance to EGFR‐TKIs. Lung Cancer 2017; 113: 106–14. [DOI] [PubMed] [Google Scholar]
- 17. Lim SM, Syn NL, Cho BC, Soo RA. Acquired resistance to EGFR targeted therapy in non‐small cell lung cancer: Mechanisms and therapeutic strategies. Cancer Treat Rev 2018; 65: 1–10. [DOI] [PubMed] [Google Scholar]
- 18. Yu HA, Arcila ME, Rekhtman N et al. Analysis of tumor specimens at the time of acquired resistance to EGFR‐TKI therapy in 155 patients with EGFR‐mutant lung cancers. Clin Cancer Res 2013; 19 (8): 2240–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ko R, Kenmotsu H, Serizawa M et al. Frequency of EGFR T790M mutation and multimutational profiles of rebiopsy samples from non‐small cell lung cancer developing acquired resistance to EGFR tyrosine kinase inhibitors in Japanese patients. BMC Cancer 2016; 16 (1): 864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kuiper JL, Heideman DA, Thunnissen E et al. Incidence of T790M mutation in (sequential) rebiopsies in EGFR‐mutated NSCLC‐patients. Lung Cancer 2014; 85 (1): 19–24. [DOI] [PubMed] [Google Scholar]
- 21. Ichihara E, Hotta K, Kubo T et al. Clinical significance of repeat rebiopsy in detecting the EGFR T790M secondary mutation in patients with non‐small cell lung cancer. Oncotarget 2018; 9 (50): 29525–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kawamura T, Kenmotsu H, Omori S et al. Clinical factors predicting detection of T790M mutation in rebiopsy for EGFR‐mutant non‐small‐cell lung cancer. Clin Lung Cancer 2018; 19 (2): e247–52. [DOI] [PubMed] [Google Scholar]
- 23. Babu Koyyala VP, Batra U, Jain P et al. Frequency of T790M mutations after progression on epidermal growth factor receptor tyrosine kinase inhibitor in metastatic non‐small cell lung cancer in Indian patients: Real‐time data from tertiary cancer hospital. Lung India 2018; 35 (5): 390–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Arcila ME, Oxnard GR, Nafa K et al. Rebiopsy of lung cancer patients with acquired resistance to EGFR inhibitors and enhanced detection of the T790M mutation using a locked nucleic acid‐based assay. Clin Cancer Res 2011; 17 (5): 1169–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yang JC, Ahn MJ, Kim DW et al. Osimertinib in pretreated T790M‐positive advanced non‐small‐cell lung cancer: AURA study phase II extension component. J Clin Oncol 2017; 35 (12): 1288–96. [DOI] [PubMed] [Google Scholar]
- 26. Wang S, Cang S, Liu D et al. Third‐generation inhibitors targeting EGFR T790M mutation in advanced non‐small cell lung cancer. J Hematol Oncol 2016; 9: 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wu YL, Ahn MJ, Garassino MC et al. CNS efficacy of osimertinib in patients with T790M‐positive advanced non‐small‐cell lung cancer: Data from a randomized phase III trial (AURA3). J Clin Oncol 2018; 36 (26): 2702–9. [DOI] [PubMed] [Google Scholar]
- 28. Jekunen AP. Role of rebiopsy in relapsed non‐small cell lung cancer for directing oncology treatments. J Oncol 2015; 2015: 809835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. González‐Larriba JL, Lázaro‐Quintela M, Cobo M. Clinical management of epidermal growth factor receptor mutation‐positive non‐small cell lung cancer patients after progression on previous epidermal growth factor receptor tyrosine kinase inhibitors: The necessity of repeated molecular analysis. Transl Lung Cancer Res 2017; 6 (Suppl 1): S21–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kawamura T, Kenmotsu H, Taira T et al. Rebiopsy for patients with non‐small‐cell lung cancer after epidermal growth factor receptor‐tyrosine kinase inhibitor failure. Cancer Sci 2016; 107 (7): 1001–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Karlovich C, Goldman JW, Sun JM et al. Assessment of EGFR mutation status in matched plasma and tumor tissue of NSCLC patients from a phase I study of rociletinib (CO‐1686). Clin Cancer Res 2016; 22: 2386–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li C, Jia R, Liu H, Zhang B, Wang C. EGFR T790M detection and osimertinib treatment response evaluation by liquid biopsy in lung adenocarcinoma patients with acquired resistance to first generation EGFR tyrosine kinase inhibitors. Diagn Pathol 2018; 13 (1): 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhu L, Zhang S, Xun Y et al. Comparison of the amplification refractory mutation system, super amplification refractory mutation system, and droplet digital PCR for T790 M mutation detection in non‐small cell lung cancer after failure of tyrosine kinase inhibitor treatment. (Published erratum appears in Pathol Oncol Res 2018;24:951). Pathol Oncol Res 2018; 24: 843–51. [DOI] [PubMed] [Google Scholar]
- 34. Soucheray M, Capelletti M, Pulido I et al. Intratumoral heterogeneity in EGFR‐mutant NSCLC results in divergent resistance mechanisms in response to EGFR tyrosine kinase inhibition. Cancer Res 2015; 75 (20): 4372–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jin Y, Shi X, Zhao J et al. Mechanisms of primary resistance to EGFR targeted therapy in advanced lung adenocarcinomas. Lung Cancer 2018; 124: 110–6. [DOI] [PubMed] [Google Scholar]


