Abstract
Background
Non‐small cell lung cancer (NSCLC) accounts for a significant proportion of cancer‐related deaths and lacks an effective treatment strategy. NSCLC tissues are generally found in a low oxygen environment. The NDUFA4L2 protein, located in the mitochondria, is encoded by the nucleus genome and is considered a crucial mediator that regulates cell survival. A better understanding of the mechanism of NDUFA4L2 in NSCLC survival in hypoxic environments is essential to design new therapeutic methods.
Methods
Twenty NSCLC and corresponding paired non‐tumorous lung tissue samples were collected. NSCLC cell lines were cultured in hypoxic conditions to investigate the mechanism of NDUFA4L2 in NSCLC. The role of NDUFA4L2 was confirmed by using Western blotting, reactive oxygen species measurement, flow cytometry, immunofluorescence analysis, and wound healing and colony formation assays.
Results
The expression of HIF‐1α and mitochondrial NDUFA4L2 increased in NSCLC cell lines cultured in hypoxic conditions (1% O2). NDUFA4L2 was drastically overexpressed in human NSCLC tissues and cell lines cultured in hypoxic conditions. HIF‐1α regulated the expression of NDUFA4L2. Knockdown of NDUFA4L2 notably increased mitochondrial reactive oxygen species production, which suppressed the viability of NSCLC.
Conclusion
In conclusion, overexpression of NDUFA4L2 is a key factor for maintaining NSCLC growth, suggesting that mitochondrial NDUFA4L2 may be a potential target for the treatment of lung cancer.
Keywords: HIF‐1α, hypoxia, NDUFA4L2, non‐small cell lung cancer, ROS
Introduction
Non‐small cell lung cancer (NSCLC) is one of the most malignant tumors with the fastest growth in morbidity and mortality rates, and poses the greatest threat to population health and life.1 NSCLC includes large cell carcinoma, adenocarcinoma, and squamous cell carcinoma (SCC).2 The five‐year survival rate of NSCLC patients is dramatically reduced according to the clinical stages of the disease.3, 4, 5 Most cases of NSCLC are diagnosed as a result of tumor metastasis to other organ tissues, which predicts poor treatment outcomes. Surgical resection is mainly used to treat early‐stage NSCLC and its effect is notable; however, its effect is limited in locally advanced cancer patients with high distant metastasis and recurrence rates. Approximately 85% of all lung cancers are considered NSCLC, and most patients are diagnosed with inoperable, locally advanced, or metastatic disease. Chemotherapy, radiation, and immunotherapy are used to treat lung cancer; however, NSCLC is becoming more resistant to treatment. Variability analysis of different types of cancers has revealed that resistance is related to mutated genes. Therefore, mechanisms of NSCLC occurrence and development are necessary to find effective therapies to improve the survival rate of NSCLC.6
The oxygen tension in normal tissue is approximately 1–4%, while the oxygen tension in solid tumors is < 1%. Lung cancer cells generally survive in a hypoxic environment, which is caused mainly by the rapid proliferation of tumor cells.7 Tumor cells exposed to hypoxia initiate hypoxic gene pathways and promote tumor malignancies and chemoresistance, which results in a poor prognosis.8 Cancer cells regulate hypoxic genes to adapt to hypoxia through the HIF‐1 protein. HIF‐1 protein consists of an hypoxia‐regulated α subunit and a non‐hypoxia‐regulated β subunit.9 In normoxia, HIF‐1α is hydroxylated by prolyl hydroxylases, and hydroxylated HIF‐1α is degraded by the proteasome. Activation of prolyl hydroxylases is repressed when cancer cells survive in a hypoxic environment.10, 11, 12 HIF‐1α and its cofactors combine with hypoxia response elements (HREs) to drive the expression of target genes regulating cellular metabolism.13, 14 However, HIF‐1α has numerous target genes, and how target genes regulate NSCLC survival in hypoxia remains unclear.
As a component of the electron transport chain (ETC) complex I subunit, NDUFA4L2 fine‐tunes complex I activity, thereby mediating mitochondrial activation of oxidative phosphorylation and reactive oxygen species (ROS) production. Currently, little is known about NDUFA4L2, particularly its role in cancer development. Mitochondrial ROS production induced by hypoxia is exacerbated because of the knockout of NDUFA4L2, suggesting that NDUFA4L2 protein restrains ROS production and thus could prevent oxidative stress in cancer cells.15 DNA stress or damage caused by high ROS accumulation is harmful to the survival, proliferation, and metastasis of cancer cells. A recent study reported that NDUFA4L2 promoted the survival of hepatocellular carcinoma in a hypoxic environment. The expression of NDUFA4L2 messenger RNA (mRNA) was greater in 31 lung SCC tissue samples than in the neighboring non‐cancerous tissue samples.16 However, the mechanism of NDUFA4L2 in regulating the survival of NSCLC is unknown. Therefore, we hypothesize that HIF‐1α promotes the vitality of lung cancer cells by NDUFA4L2.
Methods
Patients and samples
Twenty human NSCLC and corresponding paired non‐tumorous lung tissue samples were obtained from patients who had undergone surgery from March 2017 to June 2018 in the Xin Hua Hospital Affiliated with the Shanghai Jiao Tong University School of Medicine (Shanghai, China). All human lung samples were snap‐frozen in liquid nitrogen before RNA and protein were extracted. The study protocol and application were fully reviewed by the Ethics Committee of Xin Hua Hospital Affiliated with the Shanghai Jiao Tong University School of Medicine, and this study did not raise any issues of patient risk. Two pathologists evaluated each specimen. Patients recruited to this study did not receive any preoperative treatments.
Cell culture
NSCLC cell lines A549 and H1299 were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). Cells were cultured in 5% CO2/95% air in a 37% humidified incubator under normoxic (20%) or hypoxic (1%) conditions. Cell lines were planted in high‐glucose Dulbecco's modified Eagle medium (Gibco, Grand Island, NY, USA). Cells were cultured under low oxygen conditions (1% O2; 4% CO2; 95% N2) as described above in the subsequent experiments. Prior to hypoxia, 20 μmol/L 2‐methoxyestradiol (2‐ME, HY‐12033) and 0.2 mmol/L dimethyloxalylglycine (DMOG, MCE, HY‐15893; MedChem Express, Princeton, NJ, USA) were added to A549 and H1299 cells for two hours at 37°C. Subsequently, cells were cultured in a hypoxic environment in Dulbecco's modified Eagle medium.
Quantitative real‐time PCR
Total RNA was isolated from NSCLC tissue samples and cell lines with TRIzol (D9108, Takara, Dalian, China) according to manufacturer protocol. Reverse transcription (RR037A, Takara) and quantitative PCR reactions were conducted according to the manufacturer's instructions. The 2−ΔΔCt method was used to assess the relative mRNA expression change. The PCR primers used for NDUFA4L2 were: 5′‐TTCTACCGGCAGATCAAAAGACA‐3′ (forward) and 5′‐GGGCGACTCGCAGCAA‐3′ (reverse).
RNA interference and transfection
Three NDUFA4L2‐small interfering RNAs (siRNAs) and one negative control (NC) siRNA were purchased from Sangon Biotech (Shanghai, China). The siRNA sequences were: 5′‐UCAUCCCGAUGAUCGGCUUTT‐3′(sense) and 5′‐ AAGCCGAUCAUCGGGAUGATT‐3′ (anti‐sense) for si‐NDUFA4L2–1, 5′‐ GCUGGGACAGAAAGAACAATT‐3′ (sense) and 5′‐ UUGUUCUUUCUGUCCCAGCTT‐3′ (anti‐sense) for si‐NDUFA4L2–2, and 5′‐GCAGUUUCCACUGACUAUATT‐3′ (sense) and 5′‐UAUAGUCAGUGGAAACUGCTT‐3′ (anti‐sense) for si‐NDUFA4L2–3. Cells were transfected with siRNAs using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to manufacturer protocol. Cells were used in subsequent experiments after 48 hours.
Western blotting
Western blotting was performed according methods previously described.17 Briefly, total protein was isolated using radioimmunoprecipitation assay lysis buffer supplemented with protease inhibitors (Beyotime Biotechnology, Shanghai, China). The primary antibodies used were: anti‐HIF‐1α (#36169, 1:1000), anti‐caspase‐3 (#9662, 1:1000), and anti‐GAPDH (#5174, 1:1000, Cell Signaling Technology, Danvers, MA, USA); and anti‐NDUFA4L2 (A14288, 1:1000, ABclonal, Woburn, MA, USA). Horseradish peroxidase‐conjugated goat anti‐rabbit immunoglobulin G (1:1000, Beyotime Biotechnology) was used as a secondary antibody. All experiments were repeated at least three times.
Reactive oxygen species (ROS) measurement
Mitochondrial ROS production was determined by using the oxidative conversion of cell permeable 2′,7′‐dichlorofluorescein diacetate (DCFH‐DA) to fluorescent dichlorofluorescein (DCF, Beyotime Biotechnology) using a fluorescence microscope (Olympus Fluoview, Tokyo, Japan). A549 and H1299 cells transfected with si‐NDUFA4L2 and si‐NC were planted in six‐well plates under hypoxic conditions (1% O2; 4% CO2; 95% N2). Cell lines were washed with phosphate buffered saline (PBS) and incubated with serum‐free medium containing DCFH‐DA at 37°C for 20 minutes. The DCFH‐DA was then removed and the cells were washed three times with serum‐free medium. DCF fluorescence distribution of cells was detected using a fluorescence microscope. Positive cells emitted green fluorescence.
Flow cytometry
Apoptosis was measured using the Annexin V‐FITC Apoptosis Detection Kit I (Beyotime Biotechnology) according to the manufacturer's instructions. A549 and H1299 cells were digested with 0.25% trypsin and washed twice with cold PBS. Cells were resuspended in 1X binding buffer and 10 μL propidium iodide (PI, 1 mg/mL), and 5 μL annexin V‐FITC solution was then added at 37°C for 30 minutes in the dark. Cell samples were detected by flow cytometry within 30 minutes. Annexin V+/PI‐ cells plus Annexin V+/PI+ cells represented the percentage of the total apoptotic rate in NSCLC cell lines.
Immunofluorescence analysis
The NSCLC cell line A549 was plated on sheet glasses in a 6 mm plate. Cells on sheet glasses were fixed with 4% paraformaldehyde for 20 minutes and then washed three times with PBS. Cells were permeabilized with PBS containing 0.1% Triton X‐100 for 20 minutes and blocked in 5% bovine serum albumen for one hour. Sheet glasses were incubated with NDUFA4L2 and cytochrome C primary antibodies overnight at 4°C using the following dilutions: cytochrome C (mouse mAb, #12963, 1:200, Cell Signaling Technology) and NDUFA4L2 (rabbit polyclonal antibody, 1:25, Proteintech, Chicago, IL, USA). Sheet glasses were washed with tris‐buffered saline plus tween 20 three times and incubated with Cy3‐AffiniPure Goat Anti‐Rabbit IgG (H + L) and Alexa Fluor 488‐AffiniPure Goat Anti‐Mouse IgG (H + L) (Jackson ImmunoResearch, West Grove, PA, USA). Immunofluorescence was analyzed by fluorescence microscopy (Olympus BX51).
Immunohistochemical staining
Immunohistochemical staining was performed as previously described.18
Wound healing assay
Approximately 2× 106 cells were plated into 6 mm plates and cultured in a 37% humidified incubator until cells reached at least 90% confluence. Wounds were formed by a 1 mL plastic pipette tip and then cultured in medium with 1% fetal calf serum for 24 hours under hypoxic conditions (1% O2; 4% CO2; 95% N2). Cell migration was evaluated by calculating the mean number of migrating cells per field.
Colony formation assay
Approximately 1000 A549 or H1299 cells were plated into each well of six‐well plates and cultured in media containing 10% fetal bovine serum. Cells were incubated with methanol and stained with 0.1% crystal violet for 10 minutes. The proliferative capacity was represented by the number of visible colonies.
Statistical analysis
All data presented as the mean ± standard deviation were analyzed using SPSS version 20.0 (IBM Corp., Armonk, NY, USA). Differences in NDUFA4L2 expression between NSCLC and neighboring non‐cancerous tissues were analyzed by paired sample t‐test. One‐way analysis of variance was used to assess statistical significance, and Tukey's test was subsequently used for comparisons between control and treatment groups. The differences between NSCLC and corresponding paired non‐tumorous lung tissue samples were evaluated using the independent‐samples t‐test. All experimental data were replicated at least three times. Differences were determined statistically significant according to *P < 0.05, **P < 0.01, and ***P < 0.001.
Results
Mitochondrial NDUFA4L2 protein was involved in non‐small cell lung cancer (NSCLC) survival and migration
A recent study reported that NDUFA4L2 was overexpressed in lung SCC tissues16 therefore, we obtained 20 human NSCLC (adenocarcinoma) and corresponding paired non‐tumorous lung tissue samples to detect NDUFA4L2 mRNA. The results confirmed that NDUFA4L2 mRNA was overexpressed in NSCLC (Fig 1a). Western blotting also showed that NDUFA4L2 protein levels were overexpressed in human NSCLC tissue (Fig 1b,c). Immunohistochemical staining suggested that mitochondrial NDUFA4L2 increased in human NSCLC tissues (Fig 1d). The results revealed that NDUFA4L2 was overexpressed in human NSCLC tissues; however, the mechanism of NDUFA4L2 in human NSCLC is not clear.
Figure 1.
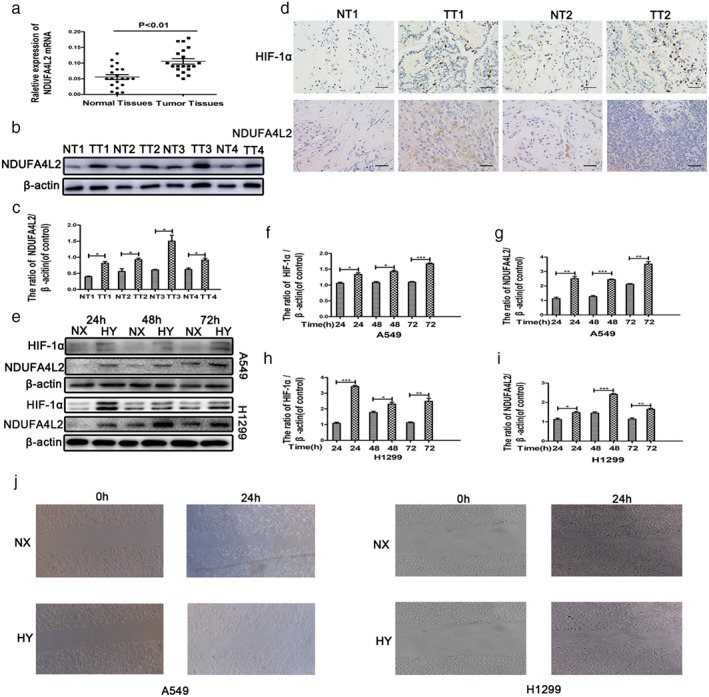
NDUFA4L2 is involved in the survival and migration of non‐small cell lung cancer (NSCLC) in hypoxia. (a) NDUFA4L2 messenger RNA (mRNA) of human NSCLC tissue samples was measured by quantitative PCR. (b) Four pairs of tissue samples were randomly extracted and measured by Western blotting. (c) The results of quantitative analysis of NDUFA4L2 were significant. (d) The expression of NDUFA4L2 and HIF‐1α was measured by immunohistochemical staining. A high percentage of tumor cells and high staining intensity indicate high expression. Pneumocytes were the normal cells. (e) HIF‐1α and NDUFA4L2 protein levels in NSCLC cell lines were measured by Western blotting assays. (f–i) The results of quantitative analysis of NDUFA4L2 and HIF‐1α were significant. ( ) Normoxia (NX), and (
) Normoxia (NX), and ( ) Hypoxia (HY). (j) Wound healing assays in NSCLC cell lines cultured in normoxia and hypoxia are shown. All data were analyzed as the mean ± standard deviation from at least three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001 versus control. H, hours; NT, normal tissues; TT, tumor tissues.
) Hypoxia (HY). (j) Wound healing assays in NSCLC cell lines cultured in normoxia and hypoxia are shown. All data were analyzed as the mean ± standard deviation from at least three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001 versus control. H, hours; NT, normal tissues; TT, tumor tissues.
A recent study reported that NDUFA4L2 regulates the viability of cells in hypoxia.19, 20 Cancer cells can survive, proliferate, and migrate in a hypoxic environment.21, 22 To confirm this classic phenomenon, two lung cancer cell lines, A549 and H1299, were cultured in normoxic (20% O2) and hypoxic (1% O2) conditions. The survival and migration of A549 and H1299 cells were slightly inhibited in hypoxia compared to normoxia (Fig 1j). Subsequently, we focused on mitochondrial NDUFA4L2. Western blotting showed that the protein expression of NDUFA4L2 was increased in A549 and H1299 cells cultured in hypoxic conditions for prolonged times (Fig 1e). The NSCLC cell lines were cultured in hypoxic conditions for 48 hours because the protein expression of NDUFA4L2 was markedly increased after 48 hours. As the definite exposure time was 48 hours in hypoxia, the cells were exposed to hypoxia in all subsequent experiments. Simultaneously, we also determined the protein expression of HIF‐1α in A549 and H1299 cells. Figure 1e shows that HIF‐1α expression increased in hypoxic conditions. Quantitative analysis by Western blotting showed significant protein expression of HIF‐1α and NDUFA4L2 (Fig 1f–i). A wound healing assay (Fig 1j) showed that NSCLC migration was repressed in hypoxia. These results suggest that mitochondrial NDUFA4L2 might play an important role in lung cancer cells.
HIF‐1α mediated the survival and proliferation of NSCLC cell lines through mitochondrial NDUFA4L2.
HIF‐1α is considered to regulate the survival and metabolism of cells in a hypoxic environment. To confirm the relationship between HIF‐1α and NDUFA4L2 in lung cancer cells cultured in hypoxia, we used an HIF‐1α inhibitor (2‐ME, 20 μM)23 and an agonist (DMOG, 1 mM)24 to pretreat A549 and H1299 cells. DMOG repressed and 2‐ME activated the activation of prolyl hydroxylases and were used to investigate the role of HIF‐1α.25, 26, 27 First, we confirmed that NDUFA4L2 was located in mitochondria (Fig 2a). Cytochrome C was marked with green, and NDUFA4L2 was marked with red. Two colors were merged as yellow (Fig 2a). As shown in Figure 2b,c, the protein expression of HIF‐1α was inhibited by 2‐ME and increased by DMOG. Interestingly, the protein expression of NDUFA4L2 was also decreased by 2‐ME and increased by DMOG. The results of quantitative analysis by Western blotting were significant (Fig 2d–i) and revealed that HIF‐1α could regulate NDUFA4L2 protein expression.
Figure 2.
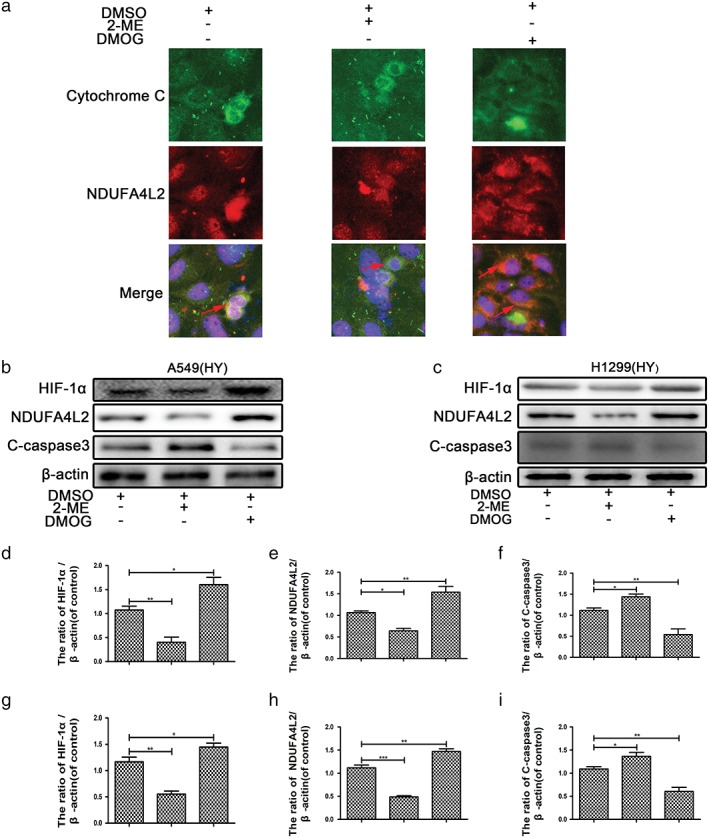
Mitochondrial NDUFA4L2 was activated by HIF‐1α. Non‐small cell lung cancer (NSCLC) cell lines were cultured in hypoxic conditions. (a) DMOG promoted and 2‐ME inhibited NDUFA4L2 expression. (b,c) Western blotting assays showed that apoptosis of NSCLC cell lines was increased by 2‐ME and inhibited by DMOG. (d–i) The results of quantitative analysis of NDUFA4L2, HIF‐1α, and cleaved caspase‐3 (C‐caspase3) were significant. All data were analyzed as the mean ± standard deviation from at least three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001 versus control. DMSO, dimethyl sulfoxide.
To confirm whether HIF‐α regulated NSCLC cell line proliferation, a colony formation assay was carried out. DMOG promoted and 2‐ME inhibited NSCLC cell line proliferation (Fig 3a). To determine whether HIF‐1α regulated NSCLC apoptosis, A549 cells were pretreated with 2‐ME and DMOG. Western blotting showed that the mitochondrial apoptosis marker (cleaved caspase‐3) was enhanced by 2‐ME and weakened by DMOG (Fig 2b,c). The results of flow cytometric analysis suggested that early and late stage apoptosis were enhanced strongly by 2‐ME and reduced by DMOG (Fig 3b). The results of quantitative analysis of flow cytometry were significant (Fig 3c) and suggest that HIF‐1α might mediate the survival and proliferation of NSCLC cell lines through mitochondrial NDUFA4L2.
Figure 3.
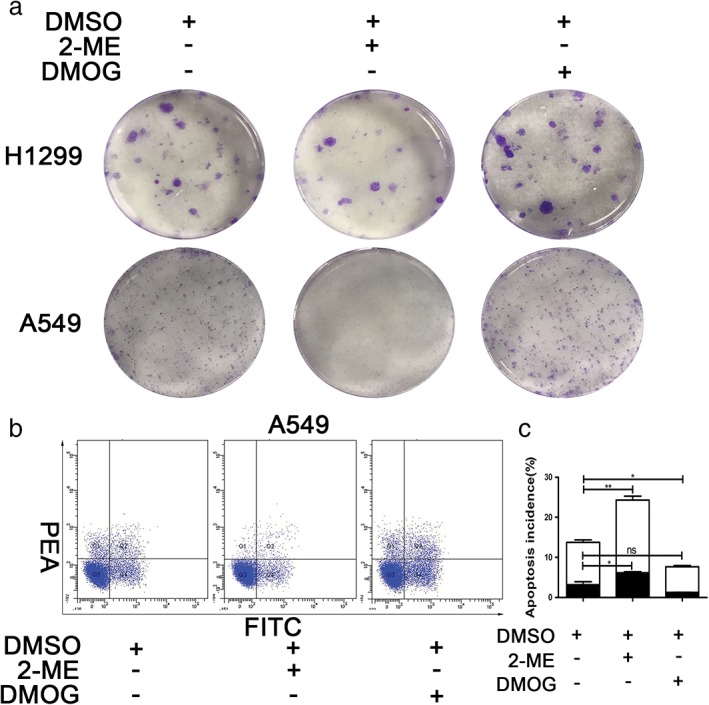
HIF‐1α regulated the survival and proliferation of non‐small cell lung cancer (NSCLC) cell lines in hypoxia. NSCLC cell lines were cultured in hypoxia. (a) Colony formation assay showed that the number of colonies formed was inhibited by 2‐ME and facilitated by DMOG. (b) Flow cytometry showed that the percentage of early (Annexin V+/propidium iodide [PI]‐) and late (Annexin V+/PI+) apoptotic cells was increased by 2‐ME and decreased by DMOG. (c) The results of quantitative analysis of the percentage of early (Annexin V+/PI‐) and late (Annexin V+/PI+) apoptotic cells were significant. All data were analyzed as the mean ± standard deviation from at least three independent experiments ( ) Annexin V (+) PI (+), and (
) Annexin V (+) PI (+), and ( ) Annexin V (+) PI (−). *P < 0.05, **P < 0.01, ***P < 0.001 vs. control. DMSO, dimethyl sulfoxide.
) Annexin V (+) PI (−). *P < 0.05, **P < 0.01, ***P < 0.001 vs. control. DMSO, dimethyl sulfoxide.
Induction of NDUFA4L2 by HIF‐1α regulated the survival and epithelial‐to‐mesenchymal transition of NSCLC cell lines by repressing ROS
A recent study reported that NDUFA4L2 regulates cell apoptosis and proliferation in a hypoxic environment.15, 16 To investigate whether induction of NDUFA4L2 by HIF‐1α regulates phenotypes of NSCLC cell lines, we designed three sequences of siRNA to silence NDUFA4L2. The protein levels of NDUFA4L2 were silenced by si‐NDUFA4L2‐2 and si‐NDUFA4L2‐3. The results showed that silencing NDUFA4L2 did not influence HIF‐1α protein expression (Fig 4a), which was upstream of NDUFA4L2. Therefore, si‐NDUFA4L2–3 was selected for the following experiments. NDUFA4L2 was only slightly expressed in A549 cells cultured in normoxia, thus the effect of si‐NDUFA4L2 was not confirmed; however, it was obvious that NDUFA4L2 was silenced by si‐NDUFA4L2‐3 in a hypoxic environment (Fig 4b).15 Knockdown of NDUFA4L2 increased apoptosis in A549 cells (Fig 4b). Quantitative analysis of Western blotting showed that the protein levels of cleaved caspase‐3, Bcl‐2, Bax, and NDUFA4L2 were significant (Fig 4c–h). Some studies have suggested that NDUFA4L2 represses the production of ROS in mitochondria.15, 16, 20 As shown in Figure 4i,j, silencing of NDUFA4L2 promoted the production of ROS in A549 cells cultured in hypoxia but not in normoxia. In addition, we also found that silencing NDUFA4L2 regulated migration and epithelial‐to‐mesenchymal transition (EMT) in A549 cells (Fig 4k–n). The mechanism of this phenomenon requires further study. These results suggest that induction of NDUFA4L2 by HIF‐1α regulated the survival, proliferation, and EMT progression of NSCLC cell lines by fine‐tuning ROS.
Figure 4.
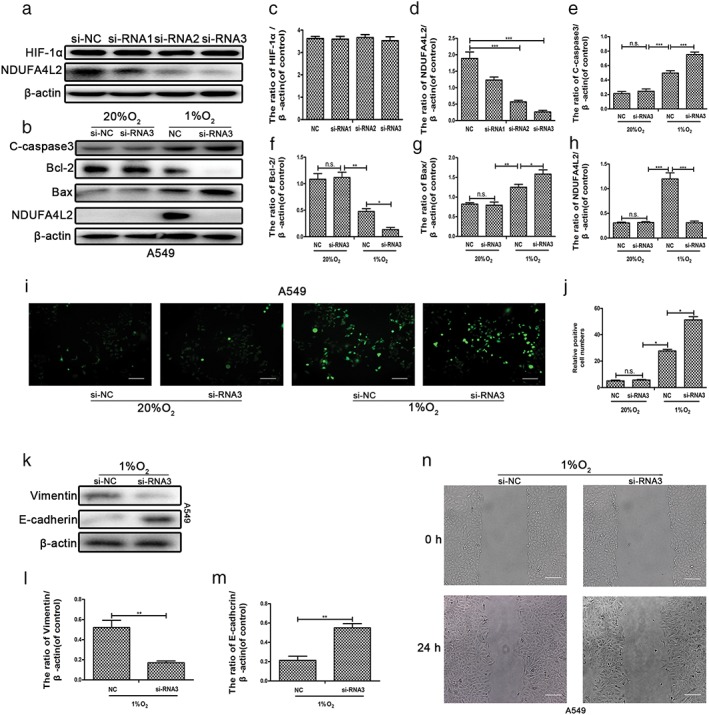
Induction of NDUFA4L2 by HIF‐1α maintained the survival, epithelial‐to‐mesenchymal transition, and migration of non‐small cell lung cancer (NSCLC) through fine‐tuning the production of reactive oxygen species (ROS). A549 cells transfected with small interfering negative control (si‐NC) and si‐NDUFA4L2‐1, si‐NDUFA4L2‐2, and si‐NDUFA4L2 were cultured in hypoxic conditions. (a) Western blotting showed that NDUFA4L2 was knocked down by si‐NDUFA4L2‐2 and si‐NDUFA4L2‐3, and there were no changes in HIF‐1α expression. (b) Knockdown of NDUFA4L2 by si‐NDUFA4L2‐2 and si‐NDUFA4L2‐3 was significant. (c) The expression of cleaved caspase‐3 (C‐caspase3), Bcl‐2, Bax, and NDUFA4L2 was determined by Western blotting assays. (c–h) Quantitative analysis of the expression of cleaved caspase‐3, Bcl‐2, Bax, HIF‐1α, and NDUFA4L2. (i) Mitochondrial production of ROS was detected by a Reactive Oxygen Detection Kit. (j) Quantitative analysis of the mitochondrial production of ROS by ImageJ. (k) The expression of vimentin and E‐cadherin in A549 cells transfected with si‐NC or si‐NDUFA4L2‐3 was determined by Western blotting assays. (l,o) Quantitative analysis of the expression of vimentin and E‐cadherin. (m) Wound healing assays in A549 cells cultured in hypoxia are shown. (n) Wound healing assays show the migration of NSCLC cell lines transfected with si‐NC or si‐NDUFA4L2 ‐3 and cultured in hypoxia. All data were analyzed as the mean ± standard deviation from at least three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001 versus control. si‐RNA1: si‐NDUFA4L2‐1; si‐RNA2: si‐NDUFA4L2; si‐RNA3: si‐NDUFA4L2‐3. N.s., not significant.
Discussion
It is well known that tumors resist the hypoxic environment and even resist chemotherapy drug toxicity with the help of hypoxia.28, 29 In cancer cells, oxygen metabolism is converted to oxygen‐free anaerobic metabolism, which is known as the Warburg effect. Many genes depending on HIF‐1α reprogram the Warburg effect. For example, the PHD oxygen‐sensing pathway upregulates PDH kinase isoforms PDK1, PDK3, and PDK4 to reduce the conversion of pyruvate into acetyl‐CoA, which represses the TCA cycle and, ultimately, the source of NADH. Being downstream of the TCA cycle, ETC plays an important role in the Warburg effect. NDUFA4L2, a component of the ETC complex I subunit, is thought to cooperate with PDKs to repress mitochondrial complex I activity in a hypoxic microenvironment. NDUFA4L2 genes, including HRE location, are also regulated by HIF‐1α.
Some studies have reported that NDUFA4L2 is overexpressed in many tumors and plays a crucial role in the resistance of hypoxia.15, 16, 19, 30, 31 Lai et al. showed that NDUFA4L2 mRNA is overexpressed in lung cancer16 however, the mechanism by which NDUFA4L2 regulates lung cancer cells is unclear. As a component of the ETC complex I subunit, NDUFA4L2 mediates complex I activity in hypoxia. Complex I is the first step of the respiratory ETC, which transfers electrons from NADH to a non‐covalently bound flavin mononucleotide. There is substantial ROS production in the ubiquinone reduction site of complex I and the outer quinone‐binding site of the Q cycle in complex III.32, 33, 34 ROS production is a major source of redox homeostasis that plays an important role in the survival of other phenotypes in cancer cells. Our results show that mitochondrial NDUFA4L2 represses the production of ROS in NSCLC cells. Meanwhile, a generation of antioxidants, including glutathione, NADPH, and thioredoxin, prevent NSCLC cells from redox damage.
In this study, we showed that mitochondrial NDUFA4L2, a component of the ETC complex I subunit, was overexpressed in NSCLC tumor tissues. NDUFA4L2 was also overexpressed in NSCLC cell lines cultured in hypoxic conditions. We further confirmed that HIF‐1α facilitates the survival, proliferation, and migration of NSCLC cell lines in a hypoxic environment that is ubiquitous in tumor tissues. Moreover, we found that HIF‐1α induced by hypoxia regulates the phenotype of NSCLC cell lines under hypoxic conditions through mitochondrial NDUFA4L2. DMOG promotes and 2‐ME inhibits the expression of NDUFA4L2. Interestingly, we also showed that NDUFA4L2 fine‐tunes ROS produced by the oxidation respiratory chain in mitochondria. Silencing of NDUFA4L2 promoted ROS production, apoptosis, and EMT progression in NSCLC cell lines. Notably, NDUFA4L2 induced by HIF‐1α provides a better understanding of the occurrence and development of tumors in hypoxia.
Acknowledgments
We wish to thank our lab colleagues. This study was supported by the Natural Science Foundation of China (No. 8177100384) and Science and Technology Commission of Shanghai Municipality (No. 16411973100).
References
- 1. Ma L, Huang Y, Zhu W et al An integrated analysis of miRNA and mRNA expressions in non‐small cell lung cancers. PLoS One 2011; 6: e26502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mountain CF, Lukeman JM, Hammar SP et al Lung cancer classification: The relationship of disease extent and cell type to survival in a clinical trials population. J Surg Oncol 1987; 35: 147–56. [DOI] [PubMed] [Google Scholar]
- 3. Martini N, Bains MS, Burt ME et al Incidence of local recurrence and second primary tumors in resected stage I lung cancer. J Thorac Cardiovasc Surg 1995; 109: 120–9. [DOI] [PubMed] [Google Scholar]
- 4. Flehinger BJ, Kimmel M, Melamed MR. The effect of surgical treatment on survival from early lung cancer. Implications for screening. Chest 1992; 101: 1013–8. [DOI] [PubMed] [Google Scholar]
- 5. Mountain CF. Revisions in the international system for staging lung cancer. Chest 1997; 111: 1710–7. [DOI] [PubMed] [Google Scholar]
- 6. Kan Z, Jaiswal BS, Stinson J et al Diverse somatic mutation patterns and pathway alterations in human cancers. Nature 2010; 466: 869–73. [DOI] [PubMed] [Google Scholar]
- 7. Ebbesen P, Eckardt KU, Ciampor F, Pettersen EO. Linking measured intercellular oxygen concentration to human cell functions. Acta Oncologica (Stockholm, Sweden) 2004; 43: 598–600. [DOI] [PubMed] [Google Scholar]
- 8. Keith B, Simon MC. Hypoxia‐inducible factors, stem cells, and cancer. Cell 2007; 129: 465–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Goda N, Ryan HE, Khadivi B, McNulty W, Rickert RC, Johnson RS. Hypoxia‐inducible factor 1alpha is essential for cell cycle arrest during hypoxia. Mol Cell Biol 2003; 23: 359–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Huang LE, Gu J, Schau M, Bunn HF. Regulation of hypoxia‐inducible factor 1alpha is mediated by an O2‐dependent degradation domain via the ubiquitin‐proteasome pathway. Proc Natl Acad Sci U S a 1998; 95: 7987–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Maxwell PH, Wiesener MS, Chang GW et al The tumour suppressor protein VHL targets hypoxia‐inducible factors for oxygen‐dependent proteolysis. Nature 1999; 399: 271–5. [DOI] [PubMed] [Google Scholar]
- 12. Jaakkola P, Mole DR, Tian YM et al Targeting of HIF‐alpha to the von Hippel‐Lindau ubiquitylation complex by O2‐regulated prolyl hydroxylation. Science (New York, NY) 2001; 292: 468–72. [DOI] [PubMed] [Google Scholar]
- 13. Semenza GL, Jiang BH, Leung SW et al Hypoxia response elements in the aldolase A, enolase 1, and lactate dehydrogenase a gene promoters contain essential binding sites for hypoxia‐inducible factor 1. J Biol Chem 1996; 271: 32529–37. [DOI] [PubMed] [Google Scholar]
- 14. Wang GL, Jiang BH, Rue EA, Semenza GL. Hypoxia‐inducible factor 1 is a basic‐helix‐loop‐helix‐PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci U S A 1995; 92: 5510–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tello D, Balsa E, Acosta‐Iborra B et al Induction of the mitochondrial NDUFA4L2 protein by HIF‐1α decreases oxygen consumption by inhibiting complex I activity. Cell Metab 2011; 14: 768–79. [DOI] [PubMed] [Google Scholar]
- 16. Lai R, Xu I, Chiu D et al NDUFA4L2 fine‐tunes oxidative stress in hepatocellular carcinoma. Clin Cancer Res 2016; 22: 3105–17. [DOI] [PubMed] [Google Scholar]
- 17. Guo Q, Lan F, Yan X, Xiao Z, Wu Y, Zhang Q. Hypoxia exposure induced cisplatin resistance partially via activating p53 and hypoxia inducible factor‐1alpha in non‐small cell lung cancer A549 cells. Oncol Lett 2018; 16: 801–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang Z, Ma X, Cai Q et al MiR‐199a‐3p promotes gastric cancer progression by targeting ZHX1. FEBS Lett 2014; 588: 4504–12. [DOI] [PubMed] [Google Scholar]
- 19. Piltti J, Bygdell J, Qu C, Lammi M. Effects of long‐term low oxygen tension in human chondrosarcoma cells. J Cell Biochem 2018; 119: 2320–32. [DOI] [PubMed] [Google Scholar]
- 20. Zheng J, Zhang M, Weng H. Induction of the mitochondrial NDUFA4L2 protein by HIF‐1a regulates heart regeneration by promoting the survival of cardiac stem cell. Biochem Biophys Res Commun 2018; 503: 2226–33. [DOI] [PubMed] [Google Scholar]
- 21. Zhu B, Cao X, Zhang W et al MicroRNA‐31‐5p enhances the Warburg effect via targeting FIH. FASEB J 2019; 33: 545–56. [DOI] [PubMed] [Google Scholar]
- 22. Zhu G, Zhou L, Liu H, Shan Y, Zhang X. MicroRNA‐224 promotes pancreatic cancer cell proliferation and migration by targeting the TXNIP‐mediated HIF1α pathway. Cell Physiol Biochem 2018; 48: 1735–46. [DOI] [PubMed] [Google Scholar]
- 23. Chen JW, Ni BB, Li B, Yang YH, SD J, Jiang LS. The responses of autophagy and apoptosis to oxidative stress in nucleus pulposus cells: Implications for disc degeneration. Cell Physiol Biochem 2014; 34: 1175–89. [DOI] [PubMed] [Google Scholar]
- 24. Xie L, Pi X, Mishra A, Fong G, Peng J, Patterson C. PHD3‐dependent hydroxylation of HCLK2 promotes the DNA damage response. J Clin Invest 2012; 122: 2827–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Palazon A, Martinez‐Forero I, Teijeira A et al The HIF‐1alpha hypoxia response in tumor‐infiltrating T lymphocytes induces functional CD137 (4‐1BB) for immunotherapy. Cancer Discov 2012; 2: 608–23. [DOI] [PubMed] [Google Scholar]
- 26. Taniguchi CM, Miao YR, Diep AN et al PHD inhibition mitigates and protects against radiation‐induced gastrointestinal toxicity via HIF2. Sci Transl Med 2014; 6: 236ra264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Aquino‐Galvez A, Gonzalez‐Avila G, Delgado‐Tello J et al Effects of 2‐methoxyestradiol on apoptosis and HIF‐1alpha and HIF‐2alpha expression in lung cancer cells under normoxia and hypoxia. Oncol Rep 2016; 35: 577–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Swinson D, Jones J, Richardson D et al Carbonic anhydrase IX expression, a novel surrogate marker of tumor hypoxia, is associated with a poor prognosis in non‐small‐cell lung cancer. J Clin Oncol 2003; 21: 473–82. [DOI] [PubMed] [Google Scholar]
- 29. Lu Y, Liu Y, Oeck S, Glazer P. Hypoxia promotes resistance to EGFR inhibition in NSCLC cells via the histone demethylases, LSD1 and PLU‐1. Mol Cancer Res 2018; 16: 1458–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang L, Peng Z, Wang K et al NDUFA4L2 is associated with clear cell renal cell carcinoma malignancy and is regulated by ELK1. PeerJ 2017; 5: e4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lv Y, Nie S, Zhou J et al Overexpression of NDUFA4L2 is associated with poor prognosis in patients with colorectal cancer. ANZ J Surg 2017; 87: E251–5. [DOI] [PubMed] [Google Scholar]
- 32. Liu Y, Fiskum G, Schubert D. Generation of reactive oxygen species by the mitochondrial electron transport chain. J Neurochem 2002; 80: 780–7. [DOI] [PubMed] [Google Scholar]
- 33. Raha S, Robinson BH. Mitochondria, oxygen free radicals, disease and ageing. Trends Biochem Sci 2000; 25: 502–8. [DOI] [PubMed] [Google Scholar]
- 34. St‐Pierre J, Buckingham JA, Roebuck SJ, Brand MD. Topology of superoxide production from different sites in the mitochondrial electron transport chain. J Biol Chem 2002; 277: 44784–90. [DOI] [PubMed] [Google Scholar]


