Abstract
Background
The classification of large cell neuroendocrine carcinoma (LCNEC) has generated considerable debate and has been revised since its recognition as a separate entity. Although it shares clinical features with small cell lung carcinoma (SCLC) and was classified with SCLC in the 2015 World Health Organization classification system, numerous studies have revealed inferior treatment outcomes of LCNEC when it was treated as SCLC. Because the incidence of LCNEC is rare, its mutational landscape has not been comprehensively interrogated.
Methods
We performed capture‐based ultra‐deep targeted sequencing on tumor samples of LCNEC, large cell carcinoma (LCC), and SCLC to elucidate its biological relationship with these subtypes and to identify potentially targetable molecular alterations.
Results
Our data revealed a molecular signature, consisting of RUNX1, ERBB4, BRCA1, and EPHA3, that is distinctively mutated in LCNEC. A majority (60%) of LCNEC patients harbored copy number variations (CNVs). Interestingly, there were no common CNVs shared among the three subtypes: NFкBIA amplification was shared between LCNEC and LCC, while AKT2 amplification was shared between LCNEC and SCLC. Furthermore, genetic alterations in the PI3K/AKT/mTOR pathway were enriched in all three subtypes.
Conclusion
Despite the histological and/or morphological similarities among LCNEC, LCC, and SCLC, our data revealed a molecular signature, consisting of RUNX1, ERBB4, BRCA1, and EPHA3, that is distinctively mutated in LCNEC, which has the potential to be used as a panel of biomarkers to distinguish LCNEC from a molecular perspective. Furthermore, the molecular distinction among the three subtypes can also be reflected from CNV events.
Keywords: Genomic profiling, large cell carcinoma, large cell neuroendocrine carcinoma, small cell lung carcinoma
Introduction
Large cell neuroendocrine carcinoma (LCNEC) of the lung is a highly malignant and poorly differentiated tumor with a poor prognosis occurring in approximately 3% of patients with lung cancer.1 In 1991, Travis et al. first proposed to classify such tumors into a separate single category called LCNEC because of the presence of larger cells with abundant cytoplasm, a high proliferation rate, and neuroendocrine features.2 Prior to that, these tumors were classified into different categories based on a variety of criteria.3 Subsequently, the World Health Organization (WHO) adopted this classification and listed LCNEC as a separate entity.
The classification of large cell neuroendocrine tumors has always been controversial as they exhibit morphological features of large cell carcinoma (LCC) and immunochemical features of neuroendocrine tumors.2, 4, 5 In 2004, LCNEC was classified as a histological variant of LCC, which was defined as an undifferentiated non‐small cell lung cancer (NSCLC), lacking the architectural and cytological features of small cell lung cancer (SCLC).4 The diverse heterogeneity observed between LCC and LCNEC in terms of cytological features and biological and clinical behavior to treatment have been reported. A study reported that LCNEC patients have significantly inferior overall survival (OS) after surgical resection compared to LCC patients, even in stage I disease.6 In addition, LCNEC exhibits differential cytological, clinical, and biological features than those of classic LCC, thus rendering controversy regarding its classification. This classification system placed all lung cancers with large cell morphology in a group; however, it failed to address the biological behavior of these histological variants.
In 2015, WHO separated LCNEC from LCC and reclassified it under neuroendocrine tumors, along with SCLC.7 Previous studies have shown similarities in clinicopathological characteristics and prognoses between LCNEC and SCLC patients, such as higher incidence rates in men and smokers, a high proliferation rate, and immunohistochemical features.8, 9, 10, 11 However, numerous studies have revealed distinctions between the two subtypes, including but not limited to cell size, morphological features, and growth pattern.12 In addition, they also differ significantly in response to certain treatments. There is a wide range of clinical responses to chemotherapy, resulting in a lack of consensus on the management of LCNEC. A multicenter prospective phase II study regarding combination chemotherapy consisting of irinotecan and cisplatin revealed significantly better OS in patients with SCLC than patients with LCNEC.13 Furthermore, a few studies have suggested that LCNEC is a biologically heterogeneous group, which can be further classified into two major subsets: small cell carcinoma‐like and non‐small cell carcinoma‐like.14 A very small fraction of LCNEC is carcinoid‐like.15 Accurate distinction of the histological subtype is of major clinical relevance because subtype directed diagnosis and treatment are well established. Therefore, there is an urgent need to clarify the biological relationship between LCC, LCNEC, and SCLC.
With advances in sequencing technology, genomic profiling has significantly facilitated the classification of many cancers, including lung cancer. However, because LCNEC is rare, comprehensive genomic profiling is limited and few comparisons of genomic profiles of LCNEC and its related subtypes have been conducted.14 In this study, we examined surgically resected samples from patients with LCNEC and related subtypes, including SCLC and LCC, using capture‐based ultra‐deep targeted sequencing to clarify the biological relationship between them and derive molecular signatures associated with each subtype. Our data reveal a distinct genetic profile associated with LCNEC, thus linking the current lung cancer classification scheme with the genetic landscape of this subtype.
Methods
Patients and samples
Surgically resected LCNEC, SCLC, and LCC samples taken from August 2009 to April 2015 were reviewed by two independent pathologists according to the 2015 (4th) Edition of the WHO Classification of Tumors of the Lung, Pleura, Thymus and Heart. Only histologically pure LCNEC, SCLC, and LCC were included. Specimens containing a minimum of 10% tumor cells were used for analysis.
The Ethics Committee of Shanghai Chest Hospital approved this study. All procedures in studies involving human participants were conducted in accordance with the ethical standards of the Medical Ethics Committee of Shanghai Chest Hospital. Informed consent was obtained from all participants included in the study.
Tissue DNA extraction
DNA was extracted using a QIAamp DNA FFPE tissue kit (Qiagen, Carlsbad, CA, USA) according to the manufacturer's instructions. DNA concentration was measured using Qubit dsDNA assay (Thermo Fisher Scientific, Waltham, MA, USA).
Next generation sequencing library preparation
DNA fragmentation was performed using a Covaris M220 Focused‐ultrasonicator (Woburn, MA, USA), followed by end repair, phosphorylation, and adaptor ligation. Fragments of 200–400 bp were selected using AMPure beads (Agencourt AMPure XP Kit, Beckman Coulter, CA, USA), followed by hybridization with capture probe baits, hybrid selection with magnetic beads, and PCR amplification. Subsequently, high‐sensitivity DNA assay was performed to assess the quality and size of all fragments.
Capture‐based targeted DNA sequencing
Genetic profiles of all tissue samples were assessed by performing capture‐based targeted deep sequencing using the OncoScreen panel (Burning Rock Biotech Ltd., Guangzhou, China), covering 2.02 MB of human genomic regions, including all exons and critical introns of 295 genes. DNA quality and size were assessed by high sensitivity DNA assay using a bioanalyzer. All indexed samples were sequenced on a NextSeq 500 (Illumina, Inc., Madison, WI, USA) with pair‐end reads.
Sequencing data analysis
The sequencing data in the FASTQ format were mapped to the human genome (hg19) using Burrows–Wheeler Aligner 0.7.10. Local alignment optimization, variant calling, and annotation were performed using GATK 3.2, MuTect, and VarScan, respectively. DNA translocation analysis was performed using both Tophat2 and Factera 1.4.3. Gene‐level copy number variation (CNV) was assessed using a statistic after normalizing read depth at each region by total read number and region size, and correcting GC‐bias using a LOESS algorithm.
Tumor mutational burden
The tumor mutational burden (TMB) was defined as the number of somatic, coding, base substation, and indels per megabase of genome examined. Fusions, CNVs, and non‐coding mutations were not counted. Synonymous mutations were counted in order to reduce sampling noise. White blood cells were used to filter germline mutations.
Results
Patient characteristics
This study consisted of a cohort of 14 LCNEC, 10 SCLC, and 5 LCC patients at a median age of 69 (range: 48–76) years. All patients were treatment‐naïve, male, and 83% (24/29) were either current or ex‐smokers.
Representative hematoxylin and eosin (H&E) staining of each subtype is shown in Figure 1a–c. LCC showed a major component of polygonal‐shaped cells with abundant cytoplasm and without definitive cytological and histological features of squamous cell carcinoma, adenocarcinoma, or neuroendocrine tumor (Fig 1a). LCNEC was defined by light microscopy as a tumor with epithelioid cells and neuroendocrine morphology including organoid nesting, rosette‐like structures, trabecular growth, and peripheral palisading patterns. In addition, cells were large with an irregular shape, abundant eosinophilic cytoplasm, and hyperchromatic and prominent nucleoli. Necrosis was frequent and often extensive (Fig 1b). HE staining of SCLC showed malignant epithelial tumor features consisting of small cells with a round‐to‐fusiform shape, scant cytoplasm, fine granular chromatin, and no or inconspicuous nucleoli. The nuclear molding was prominent (Fig 1c). We then examined the immunohistochemical expression of selected markers, including CK, TTF‐1, CD56, P40, and Ki‐67. Immunohistochemistry staining for CK showed a dot‐like cytoplastic staining pattern in SCLC samples, and a mainly diffuse cytoplasmic staining pattern in LCC and LCNEC (Fig 1d–f). LCNEC and SCLC exhibited focal reactivity for TTF‐1, whereas none of the LCC tumors expressed this marker (Fig 1h–i). Both SCLC and LCNEC tumors showed immunoreactivity to CD56 antibody (Fig 1k,l). LCC lacked TTF‐1 and p40 expression (Fig 1g,j). Notably, all tumors displayed high proliferative activity, as revealed by the strong expression of Ki‐67 (Fig 1m–o).
Figure 1.
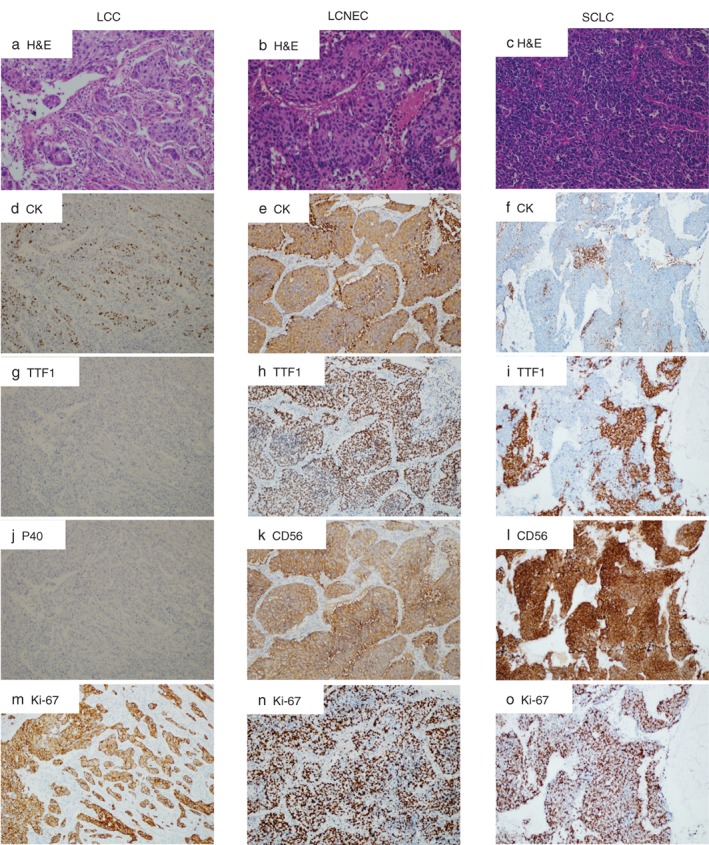
Morphological and immunohistochemical features of (a) large cell carcinoma (LCC), (b) large cell neuroendocrine carcinoma (LCNEC), and (c) small cell lung cancer (SCLC). Representative examples from each subtype were illustrated. (a) LCC consists of sheets or nests of large polygonal cells. The tumor cell has vesicular nuclei, prominent nucleoli, and moderate amounts of cytoplasm (hematoxylin and eosin [H&E]). Immunohistochemistry (IHC) illustrates dot‐like, cytoplastic staining of (d) CK in tumor cells, (g) negative TTF‐1 and (j) P40, and (m) a high Ki‐67 index. (b) Photomicrograph of LCNEC showing solid nests with multiple rosette‐like structures, with generally large tumor cells with moderate to abundant cytoplasm. Nucleoli are frequent, often prominent (H&E). IHC illustrates (e) a diffuse cytoplasmic pattern of CK expression, (h) positive TTF‐1 with diffuse nuclei staining, (k) positive CD56 in a diffuse membranous staining pattern, and (n) a high Ki‐67 rate. (c) SCLC consists of dense sheets of small cells with scant cytoplasm, finely granular nuclear chromatin, and absent or inconspicuous nucleoli. IHC illustrates (f) a dot‐like, cytoplastic expression pattern of CK, (i) focal TTF‐1 expression, (l) positive CD56 in a membranous staining pattern, and (o) a high Ki‐67 index.
Mutation spectrums of LCNEC, LCC, and SCLC
The classification of LCNEC of the lung has generated considerable debate and has been revised since its recognition as a separate entity. LCNEC of the lung is traditionally classified as a histological variant of large cell carcinoma (LCC). However, it exhibits differential cytological, morphological, clinical, and biological features than those of classic LCC, thus it was re‐classified under neuroendocrine tumors in 2015. Because of its rarity, its genomic profile has not been comprehensively interrogated and compared against its counterparts, including LCC and SCLC. To interrogate the molecular landscapes of the three subtypes, we performed capture‐based ultra‐deep targeted sequencing using Burning Rock Biotech's OncoScreen Panel, which consists of all exons and critical introns of 295 cancer‐related genes. This panel can be used to detect multiple classes of somatic mutations, including single nucleotide variation (SNV), rearrangements, CNVs, and insertions and deletions (INDELs) and can be used to detect genomic alterations, both qualitatively and quantitatively. Among the 29 patients, 14 of were diagnosed with LCNEC, 5 with LCC, and 10 with SCLC.
Overall, we identified 335 mutations spanning 157 genes, including 248 SNVs, 29 INDELs, and 22 copy‐number amplifications (CNAs). The mutation spectrum of each subtype is shown in Figure 2a, with TP53 being the most frequently mutated gene in all subtypes, occurring in 60%, 86%, and 70% of LCC, LCNEC, and SCLC, respectively. RB1 mutation, a hallmark of SCLC, was also detected in 36% of LCNEC and 20% of LCC patients. Interestingly, four LCNEC patients exhibited concurrent TP53 and RB1 mutations, a hallmark of SCLC. Mutations in LRP1B, a member of the low‐density lipoprotein receptor family and a putative tumor suppressor, were also frequently observed in all three subtypes: 40%, 36%, and 40% of LCC, LCNEC, and SCLSC, respectively. Other mutations shared by all three subtypes included but were not limited to FAT3, SMARCA4, NOTCH3, PIK3CG, PIK3CA, and KMT2D. The most frequently mutated genes are shown in Figure 2b. It is important to point out that no classic NSCLC driver mutations were identified in this cohort. In addition to the above‐mentioned common features, our data revealed a unique molecular signature, consisting of four genes, RUNX1, ERBB4, BRCA1, and EPHA3, that were only mutated in LCNEC (Fig 2a). Such a signature can potentially be used to distinguish LCNEC from its counterparts.
Figure 2.
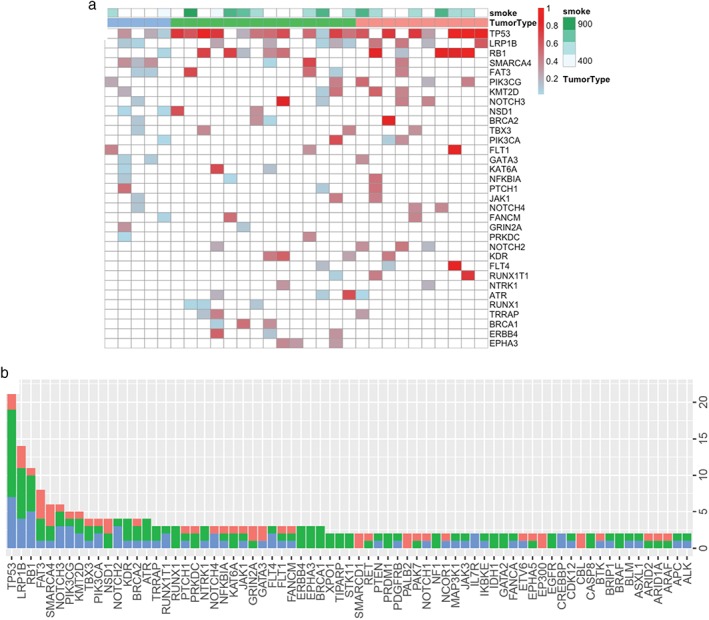
Mutation spectrum of large cell carcinoma (LCC), large cell neuroendocrine carcinoma (LCNEC), and small cell lung cancer (SCLC). (a) Mutation profiles of each subtype. Smoking history and pathological subtype are denoted on top of the oncoprint. Each column represents a patient and each row represents a gene. The color gradient represents allelic fraction. (b) Bar graph summarizes the mutation frequency of the most frequently mutated genes in this cohort. 
 , LCC;
, LCC; 
 , LCNEC;
, LCNEC; 
 , SCLC.
, SCLC.
Copy number variation analysis
CNVs have the potential to underlie diseases by altering the diploid status of DNA. We performed CNV analysis to compare and contrast the genomic variability generated by deletions and amplifications. Our data revealed a higher prevalence of CNA in LCNEC, with 60% of cases harboring such alteration, including SOX2, KRAS, EGFR, KIT, PDGFRA, NFKBIA, and NKX2‐1. Among them, amplification of KRAS, EGFR, KIT, PDGFRA, and NKX2‐1 only occurred in LCNEC. Interestingly, no common CNVs are shared among the three subtypes. NFкBIA amplification is the only CNV found in both LCNEC and LCC, and AKT2 amplification is only CNV shared by LCNEC and SCLC. Collectively, our data demonstrate that most CNVs are subtype‐specific.
Pathway enrichment analysis
We next performed pathway enrichment analysis based on all of the mutated genes identified from each subtype to reveal functionally aberrant pathways. Collectively, 16 genes for SCLC, 23 genes for LCNEC, and 9 genes for LCC were used for the analysis. Enrichment analysis was performed using the Database for Annotation, Visualization and Integrated Discovery based on all metabolic and non‐metabolic pathways in the Kyoto Encyclopedia of Genes and Genomes. The cut‐off criteria chosen as the default included an EASE score (a modified Fisher's exact P value proposed by the software) of 0.1 and a minimum of two genes belonging to a pathway. Our data revealed 53 enriched pathways for at least one subtype. The most enriched pathways are shown in Figure 3b. All three subtypes have mutations in genes participating in the following pathways: PI3K‐Akt, small cell lung cancer, MAPK signaling, and apoptosis (Fig 3). The PI3K‐Akt pathway was the most enriched pathway. Interestingly, we also observed a number of pathways that were only shared by LCNEC and SCLC, including but not limited to ERBB signaling, focal adhesion, and Jak‐STAT signaling. Among the 11 most enriched pathways listed, only one pathway, the p53 signaling pathway, was shared between LCNEC and LCC. Collectively, the pathway analyses show that LCNEC and SCLC have more functionally aberrant pathways in common, suggesting their mutational commonality.
Figure 3.
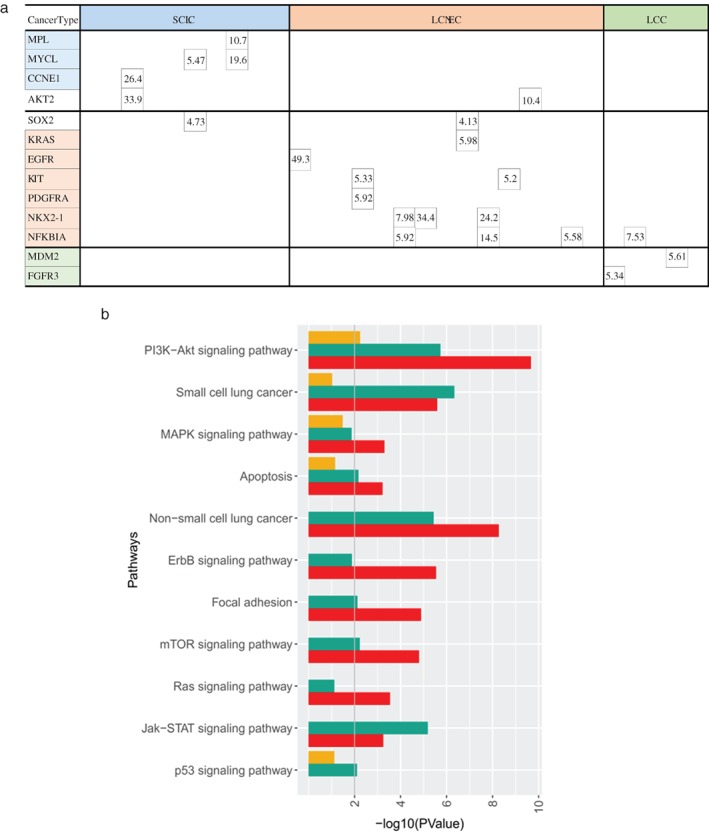
Copy number variation analysis. (a) Copy number amplification identified in each patient. Each column represents one sample, and each row represents one gene. Only genes in which a copy number variation was detected in one or more samples are depicted. (b) Pathway enrichment analysis based on all mutated genes identified from each subtype. Pathways were ranked according to P values. Different color bars represent different pathological subtypes.  , SCLC;
, SCLC;  , LCNEC;
, LCNEC;  , LCC.
, LCC.
Tumor mutational burden analysis
The association between high TMB and the efficacy of immune checkpoint inhibitors has been established in an array of cancer types, including lung cancer. TMB has been shown as a better biomarker for predicting PD‐1 and PD‐L1 blockade immunotherapy than PD‐1 or PD‐L1 expression. We investigated the landscape of TMB associated with each subtype. A previous study reported that TMB can be accurately assessed using targeted comprehensive genomic profiling. Across the entire data, the median TMB was 8.75 mutations/Mb (range: 2.5–32.5 mutations/Mb). Our data is comparable to the median TMB reported for lung cancer, which is 7.2 mutations/Mb. The median TMBs for LCNEC, SCLC, and LCC are 10, 5, and 10 mutations/Mb, respectively. TMB is comparable between LCC and SCLC, as well as between LCC and LCNEC. LCNEC has a higher TMB than SCLC (P = 0.021) (Fig 4). There is a very substantial range of TMB in LCC, ranging from 5 to 32.5 mutations/Mb, suggesting great heterogeneity within this subtype. Larger cohort studies are needed to accurately assess the range. The TMB of each patient is included in the supplemental material (Table S1). In summary, we examined TMB for each subtype and revealed a higher TMB in LCNEC.
Figure 4.
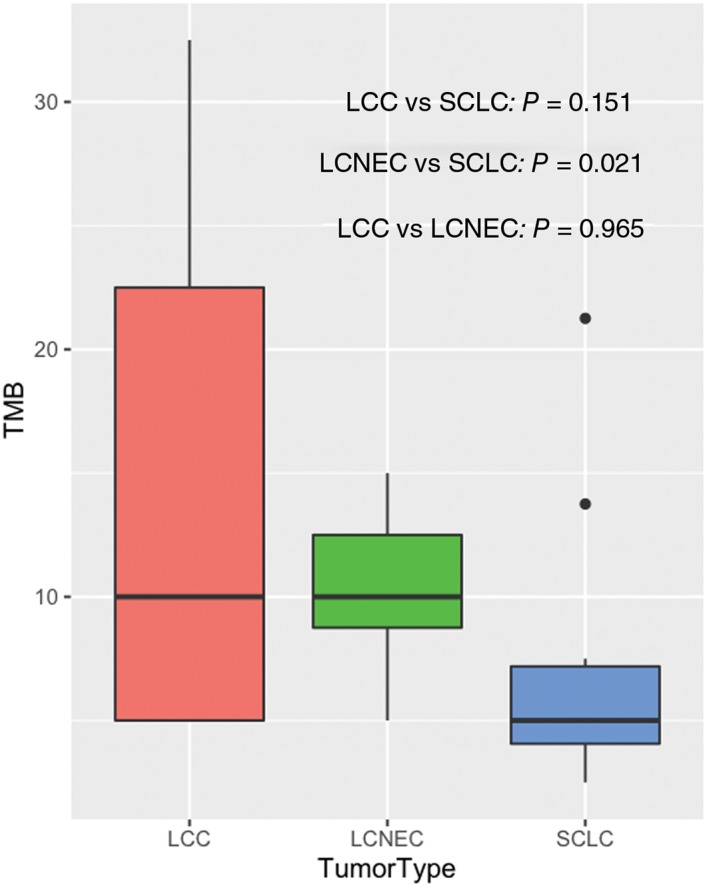
Tumor mutational burden (TMB) analysis. The black lines in each box denote the median. The bottom and top of each box denote the first and third quartile, respectively. Different colors represent different pathological subtypes. P values were calculated using Wilcoxon rank sum test.  , large cell carcinoma (LCC);
, large cell carcinoma (LCC);  , large cell neuroendocrine carcinoma (LCNEC);
, large cell neuroendocrine carcinoma (LCNEC);  , small cell lung cancer (SCLC).
, small cell lung cancer (SCLC).
Discussion
In this study, we performed capture‐based ultra‐deep targeted sequencing to compare the molecular profiles of histologically classified primary LCNEC, LCC, and SCLC. To the best of our knowledge, this is the first study to compare and contrast these three subtypes. Our data reveal a distinct molecular profile associated with LCNEC, evident by a high prevalence of CNA and a set of four genes: RUNX1, ERBB4, BRCA1, and EPHA3. In addition, our analysis also revealed similarity among the three subtypes. For example, the high frequency of inactivating mutations in RB1 was observed in all three tumor types. The mutation frequency of RB1 identified in our study was consistent with the findings of previous studies, which reported a mutation frequency of 39–69.5% in SCLC16, 17, 18, 19, 20 and 38% in LCNEC.21 We revealed 50% and 36% prevalence in SCLC and LCNEC patients, respectively. At present, once diagnosed, LCNEC is often treated with SCLC chemotherapy regimens. Some studies have suggested that LCNEC can be further classified into two major subgroups: small‐cell like carcinomas and non‐small cell like carcinomas, with distinct chemotherapy treatment outcomes based on RB1 status.22 In our LCNEC cohort, four patients had concurrent TP53 and RB1 mutations, a hallmark of SCLC. Compared to the literature, our cohorts exhibited both similarities and distinctions. We did not observe KRAS mutation in our LCC patients, which is reported as the most frequently mutated gene.23 This disparity can potentially be explained by ethnicity differences and the limited cohort size. Unfortunately, we only included five LCC patients. Larger cohort studies are necessary to confirm our findings.
Furthermore, our pathway analyses suggested more commonalty between LCNEC and SCLC than LCNEC and LCC. Among the 11 most enriched pathways listed, four were shared by all three subtypes, six were shared by LCNEC and SCLC, and only one pathway was shared by LCC and LCNEC, suggesting more commonality between LCNEC and SCLC. The most frequently mutated pathway was the PI3K‐AKT signaling pathway, suggesting the important role this pathway plays in the development of all three subtypes. Genomic alterations in this pathway have been reported as a promising therapeutic target in SCLC.16, 24, 25, 26 Therefore, inhibitors targeting this pathway may potentially also offer therapeutic efficacy to LCNEC and LCC patients.
It is becoming evident that a subset of LCNEC tumors shares mutational patterns with SCLC and LCC, whereas others carry mutations typically altered in non‐neuroendocrine tumors.14, 21, 27, 28 Currently, the standard of care for advanced or refractory LCNEC patients is combination chemotherapy. It is interesting to note that a large percentage (64.29%) of LCNEC patients harbor mutations in genes either with Food and Drug Administration approved drugs or with drugs in advanced trials, such as BRCA1 and ERBB4 mutations, and EGFR, KIT, and PDGFRA amplifications, respectively.29, 30, 31, 32, 33, 34 Our analysis highlighted the potential of using targeted therapy in patients with LCNEC.
The predictive value of TMB of anti‐PD1 therapies has been confirmed in an array of cancers, including lung cancer. In this study we characterized the TMB for each subtype, revealing that LCNEC harbors a higher TMB than the other two subtypes, consistent with the results of a previous study, suggesting that such patients may benefit from immune checkpoint blockade.19
In conclusion, our study revealed a distinct genomic profile for LCNEC compared to LCC and SCLC. LCNEC harbors more CNVs and contains a panel of genes, including RUNX1, ERBB4, BRCA1, and EPHA3, which are distinctively mutated. Such a signature can potentially be used to distinguish LCNEC. Large studies are needed to validate the signature.
Disclosure
No authors report any conflict of interest.
Supporting information
Table S1. Tumor mutation burden in patients with lung cancer of different subtypes.
Acknowledgments
This work was supported by the National Key R&D Program of China (2016YFC1303300, 2016YFC1303800).
We thank Dr. Wanglong Deng for constructive discussion and comments.
References
- 1. Fernandez FG, Battafarano RJ. Large‐cell neuroendocrine carcinoma of the lung: An aggressive neuroendocrine lung cancer. Semin Thorac Cardiovasc Surg 2006; 18: 206–10. [DOI] [PubMed] [Google Scholar]
- 2. Travis WD, Linnoila RI, Tsokos MG et al Neuroendocrine tumors of the lung with proposed criteria for large‐cell neuroendocrine carcinoma. An ultrastructural, immunohistochemical, and flow cytometric study of 35 cases. Am J Surg Pathol 1991; 15: 529–53. [DOI] [PubMed] [Google Scholar]
- 3. McDowell EM, Wilson TS, Trump BF. Atypical endocrine tumors of the lung. Arch Pathol Lab Med 1981; 105: 20–8. [PubMed] [Google Scholar]
- 4. Varlotto JM, Medford‐Davis LN, Recht A et al Should large cell neuroendocrine lung carcinoma be classified and treated as a small cell lung cancer or with other large cell carcinomas? J Thorac Oncol 2011; 6: 1050–8. [DOI] [PubMed] [Google Scholar]
- 5. Rekhtman N. Neuroendocrine tumors of the lung: An update. Arch Pathol Lab Med 2010; 134: 1628–38. [DOI] [PubMed] [Google Scholar]
- 6. Sun YH, Lin SW, Hsieh CC, Yeh YC, Tu CC, Chen KJ. Treatment outcomes of patients with different subtypes of large cell carcinoma of the lung. Annals of Thoracic Surgery 2014; 98: 1013–9. [DOI] [PubMed] [Google Scholar]
- 7. Travis WDBE, Burke AP, Marx A, Nicholson AG. WHO Classification of Tumours of the Lung, Pleura, Thymus and Heart. IARC, Lyon: 2015. [DOI] [PubMed] [Google Scholar]
- 8. Fasano M, Della Corte CM, Papaccio F, Ciardiello F, Morgillo F. Pulmonary large‐cell neuroendocrine carcinoma: From epidemiology to therapy. J Thorac Oncol 2015; 10: 1133–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Travis WD, Rush W, Flieder DB et al Survival analysis of 200 pulmonary neuroendocrine tumors with clarification of criteria for atypical carcinoid and its separation from typical carcinoid. Am J Surg Pathol 1998; 22: 934–44. [DOI] [PubMed] [Google Scholar]
- 10. Rossi G, Cavazza A, Marchioni A et al Role of chemotherapy and the receptor tyrosine kinases KIT, PDGFRalpha, PDGFRbeta, and Met in large‐cell neuroendocrine carcinoma of the lung. J Clin Oncol 2005; 23: 8774–85. [DOI] [PubMed] [Google Scholar]
- 11. Asamura H, Kameya T, Matsuno Y et al Neuroendocrine neoplasms of the lung: A prognostic spectrum. J Clin Oncol 2006; 24: 70–6. [DOI] [PubMed] [Google Scholar]
- 12. Battafarano RJ, Fernandez FG, Ritter J et al Large cell neuroendocrine carcinoma: An aggressive form of non‐small cell lung cancer. J Thorac Cardiovasc Surg 2005; 130: 166–72. [DOI] [PubMed] [Google Scholar]
- 13. Niho S, Kenmotsu H, Sekine I et al Combination chemotherapy with irinotecan and cisplatin for large‐cell neuroendocrine carcinoma of the lung: A multicenter phase II study. J Thorac Oncol 2013; 8: 980–4. [DOI] [PubMed] [Google Scholar]
- 14. Rekhtman N, Pietanza MC, Hellmann MD et al Next‐generation sequencing of pulmonary large cell neuroendocrine carcinoma reveals small cell carcinoma‐like and non‐small cell carcinoma‐like subsets. Clin Cancer Res 2016; 22: 3618–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rekhtman N, Pietanza CM, Sabari J et al Pulmonary large cell neuroendocrine carcinoma with adenocarcinoma‐like features: Napsin a expression and genomic alterations. Mod Pathol 2018; 31: 111–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Umemura S, Mimaki S, Makinoshima H et al Therapeutic priority of the PI3K/AKT/mTOR pathway in small cell lung cancers as revealed by a comprehensive genomic analysis. J Thorac Oncol 2014; 9: 1324–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ross JS, Wang K, Elkadi OR et al Next‐generation sequencing reveals frequent consistent genomic alterations in small cell undifferentiated lung cancer. J Clin Pathol 2014; 67: 772–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bergsland EK, Roy R, Stephens P, et al Genomic Profiling to Distinguish Poorly Differentiated Neuroendocrine Carcinomas Arising in Different Sites. J Clin Oncol 2016; 34: 15_suppl, 4020. [Google Scholar]
- 19. Chae YK, Tamragouri K, Chung J, et al Genomic Alterations (GA) and Tumor Mutational Burden (TMB) in Large Cell Neuroendocrine Carcinoma of Lung (L‐LCNEC) as Compared to Small Cell Lung Carcinoma (SCLC) as Assessed Via Comprehensive Genomic Profiling (CGP) J Clin Oncol 2017; 35: 15_suppl, 8517. [Google Scholar]
- 20. Miyoshi T, Umemura S, Matsumura Y et al Genomic profiling of large‐cell neuroendocrine carcinoma of the lung. Clin Cancer Res 2016; 23: 757–65. [DOI] [PubMed] [Google Scholar]
- 21. Thomas RK, Büttner R, Wolf J et al A genomics‐based classification of human lung tumors. Sci Transl Med 2013; 5: 209ra153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Derks JL, Leblay N, Thunnissen E et al Molecular subtypes of pulmonary large‐cell neuroendocrine carcinoma predict chemotherapy treatment outcome. Clin Cancer Res 2018; 24: 33–42. [DOI] [PubMed] [Google Scholar]
- 23. Karlsson A, Brunnström H, Lindquist KE et al Mutational and gene fusion analyses of primary large cell and large cell neuroendocrine lung cancer. Oncotarget 2015; 6: 22028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wakuda K, Kenmotsu H, Serizawa M et al Molecular profiling of small cell lung cancer in a Japanese cohort. Lung Cancer 2014; 84: 139–44. [DOI] [PubMed] [Google Scholar]
- 25. Cardnell RJ, Feng Y, Mukherjee S et al Activation of the PI3K/mTOR pathway following PARP inhibition in small cell lung cancer. Plos One 2016; 11: e0152584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Umemura S, Tsuchihara K, Goto K. Genomic profiling of small‐cell lung cancer: The era of targeted therapies. Jpn J Clin Oncol 2015; 45: 513. [DOI] [PubMed] [Google Scholar]
- 27. Miyoshi T, Umemura S, Matsumura Y et al Genomic profiling of large‐cell neuroendocrine carcinoma of the lung. Clin Cancer Res 2017; 23: 757–65. [DOI] [PubMed] [Google Scholar]
- 28. Michele S, Andrea M, Katarzyna OS et al Lung neuroendocrine tumours: Deep sequencing of the four World Health Organization histotypes reveals chromatin‐remodelling genes as major players and a prognostic role for TERT, RB1, MEN1 and KMT2D. J Pathol 2017; 241: 488–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tutt A, Robson M, Garber JE et al Oral poly (ADP‐ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: A proof‐of‐concept trial. The Lancet 2010; 376: 235–44. [DOI] [PubMed] [Google Scholar]
- 30. Isakoff SJ, Mayer EL, He L et al TBCRC009: A multicenter phase II clinical trial of platinum monotherapy with biomarker assessment in metastatic triple‐negative breast cancer. J Clin Oncol 2015; 33: 1902–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Engelman JA, Zejnullahu K, Gale C‐M et al PF00299804, an irreversible pan‐ERBB inhibitor, is effective in lung cancer models with EGFR and ERBB2 mutations that are resistant to gefitinib. Cancer Res 2007; 67: 11924–32. [DOI] [PubMed] [Google Scholar]
- 32. Wu Y‐L, Zhou C, Hu C‐P et al Afatinib versus cisplatin plus gemcitabine for first‐line treatment of Asian patients with advanced non‐small‐cell lung cancer harbouring EGFR mutations (LUX‐lung 6): An open‐label, randomised phase 3 trial. Lancet Oncol 2014; 15: 213–22. [DOI] [PubMed] [Google Scholar]
- 33. Yang JC‐H, Wu Y‐L, Schuler M et al Afatinib versus cisplatin‐based chemotherapy for EGFR mutation‐positive lung adenocarcinoma (LUX‐lung 3 and LUX‐lung 6): Analysis of overall survival data from two randomised, phase 3 trials. Lancet Oncol 2015; 16: 141–51. [DOI] [PubMed] [Google Scholar]
- 34. Guo J, Si L, Kong Y et al Phase II, open‐label, single‐arm trial of imatinib mesylate in patients with metastatic melanoma harboring c‐kit mutation or amplification. J Clin Oncol 2011; 29: 2904–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Tumor mutation burden in patients with lung cancer of different subtypes.


