Abstract
The application of bioinformatics in lipase research has the potential to discover robust members from different genomic/metagenomic databses. In this study, we explored the diversity and distribution of alkaliphilic lipases in archaea domain and metagenome data sets through phylogenetic survey. Reconstructed ancestral sequence of alkaphilic lipase was used to search the homologous alkaliphilic lipases among the archaea and metagenome public databases. Our investigation revealed a total 21 unique sequences of new alkaliphilic lipases in the archaeal and environmental metagenomic protein databases that shared significant sequence similarity to the bacterial alkaliphilic lipases. Most of the identified new members of alkaliphilic lipases belong to class Haloarchaea. The searched list of homologs also comprised of one characterized lipase from alkalohyperthermophilic Archaeoglobus fulgidus. All the newly identified alkaliphilic lipase members showed conserved pentapeptide [X-His-Ser-X-Gly] motif, a key feature of lipase family. Furthermore, detailed analysis of all these new sequences showed homology either with thermostable or alkalophilic lipases. The reconstructed ancestral sequence-based searches increased the sensitivity and efficacies to detect remotely homologous sequences. We hypothesize that this study can enrich our current knowledge on lipases in designing more potential thermo-alkaliphilic lipases for industrial applications.
Keywords: Alkaliphilic lipase, Ancestral sequence reconstruction, Archaea, Phylogenetic, Thermostable lipases, Microbial lipase
Introduction
Lipases (Triacylglycerol acyl hydrolase, EC 3.1.1.3) belong to serine hydrolases with immense potential to carry out a variety of industrial processes based on their versatility in catalyzing a variety of reaction including hydrolysis, interesterification, transesterification, alcoholysis, acidolysis and aminolysis (Haki and Rakshit 2003). Besides, these enzymes also exhibit greater selectivities viz. chemo, region- and enantioselectivity (Saxena et al. 2005). Additionally, thermophilic lipases show extreme stability at high temperature, extreme pH (acidic or alkaline pH) and extraordinary salinity (Hasan et al. 2006; Sharma et al. 2011, 2015). Since lipases catalyze a variety of reactions with high specificity, they are widely used in different industries, e.g., oleochemical, food, dairy, detergent, cosmetic, leather processing, pulp and paper, biodiesel, biopolymer, and pharmaceutical (Saxena et al. 2005; Meghwanshi and Vashishtha 2018). Due to global industrial demand of lipases (more than 1000 ton each year which makes lipases third largest group of enzymes based on total sales volume after protease and carbohydrase), a number of researchers are interested to identify, isolate, and introduce new lipases produced by microorganisms (Hasan et al. 2006; Shu et al. 2010; Treichel et al. 2010). Structurally, lipase families share a common α/β hydrolase fold (Ollis et al. 1992) which is also reported in other hydrolases that include the haloalkane dehalogenase, dienelactone hydrolase, acetylcholinesterase, and serine carboxypeptidase. The active site of lipases comprises serine, aspartic acid or glutamic acid and histidine residues. The nucleophilic serine residue is located in a conserved Gly-X-Ser-X-Gly motif. The prokaryote lipases have been classified into eight families based on their sequence similarity and biological properties.
Most of the industrial processes are carried out under harsh conditions and most of them are performed at elevated temperatures and acidic or alkaline pHs. Therefore, the demand for thermostable and alkaliphilic lipases is rapidly increasing in various industries. Exploring thermostable lipases in biotechnological industries/processes has several advantages over thermolabile enzymes. High temperatures decrease the risk of microbial contamination in industrial production processes and enhances the solubility of organic compounds as well as increase the reaction rates (Demirjian et al. 2001; Abol Fotouh et al. 2016). Thermotolerant lipases can also be utilized as prototypes for designing thermostable variants of other proteins through protein engineering (Haki and Rakshit 2003). It is known that such enzymes can be obtained from archaea strains as they are natural inhabitants of such harsh environments. Enzymes obtained from thermophilic archaea generally are more stable to high temperature, presence of organic solvents, and show resistance to proteolysis which are ideal features for industrial applications (Littlechild 2015). However, it is important to mention here that very few archaea have been explored for the production of lipase like enzymes, i.e., the carboxyesterases which acts on short chain fatty acid esters. So, exploring their genomic sequences for finding out true lipases from the archaea kingdom is an important filed of research.
Ancestral sequence reconstruction (ASR) is the approximation of the most likely ancient protein sequences from the existing protein sequences which have resulted due to evolutionary changes, incorporated in the organisms during the millions of years. It involves the construction of phylogenetic evolutionary trees from the extant nucleotide or protein sequences and predicting the most probable putative common ancestral sequence. To date, parsimony and maximum likelihood methods are widely used (Elias and Tuller 2007; Koshi and Goldstein 1996; Cunningham et al. 1998) for reconstruction of ancestor sequences. The most promising use of ASR method is to detect the remotely homologous sequences (Collins et al. 2003). In this study, we explored the diversity and distribution of thermo-alkaliphilic lipases in archaea and metagenomics “dark matter” using ASR approach. The “dark matter” here refers to the nucleotide sequences which are unclassified or poorly understood. The reconstructed putative ancestral sequence-based searches enhanced the probability of detecting the similarities between the distantly related and unknown lipases from different biological databases. Our results indicated that reconstruction of ancestral sequences of lipases is a promising approach to identify phylogenetically diverse and novel lipases.
Materials and methods
Assessment of alkaliphilic lipase sequences in database
Initially, the well-studied Bacillus subtilis alkaliphilic lipase sequence (Uniport IDs: P37957) was used as an input query to retrieve orthologs (lipase gene of common descent, resulting through the process of speciation). Orthologous protein searches were carried out using 0.00001 e value threshold, sequence identity cutoff 50% and maximums hits 250 were applied against UniPortKB target database using basic local alignment search tool (BLAST) in the UniPort database (https://www.uniprot.org/). Further, we manually crosschecked the retrieved sequence and removed all fragmented and truncated sequences. Unique sequences were assigned at 95% sequences identity cutoff value using the CDHIT (Ying et al. 2010) program (http://weizhongli-lab.org/cd-hit/). All those microbial alkaliphilic orthologs were further used for ancestral sequence reconstruction (ASR).
Ancestral sequence reconstruction
Unique orthologous sequences were aligned using the multiple sequence alignment (MSA) program Muscle (Edgar et al. 2004) in MEGA-X (Kumar et al. 2018). The evolutionary history was inferred using the maximum likelihood (ML) method based on amino acid substitution JTT matrix-based model (Jones et al. 1992). Initial tree(s) for the heuristic search were obtained automatically using Neighbor-Join (NJ) method to a matrix of pairwise distances estimated using a JTT model followed by selecting the topology with superior log likelihood value. MEGA analysis was carried out which predicts ancestral sequences at each node of a given tree. The midpoint node of rooted tree was selected as the most probable putative ancestral sequence.
Identification of potential alkaliphilic lipases from archaea and metagenome
To identify new alkaliphilic lipase enzymes, BLASTp algorithm with default parameters was used, wherein the constructed ancestral sequence was used as a query sequence and was used to search the related sequences in NCBI non-redundant (nr) databases, selecting “archaea (taxid:2157)” under the organism option (Altschul et al. 1990). The BLASTp search output having expected threshold values ≥ 0.01, sequence identity ≥ 25% and query coverage ≥ 50% were considered as potential homologs. In addition, environmental metagenome (env_nr) database was also considered at same parameters. The identified homologs of Archaea lipase gene sequences were merged with the retrieved orthologs that were used for ASR to explore the phylogenetic diversity of the lipase enzyme. The merged data set was used for further multiple sequence alignment with Muscle algorithm. The phylogenetic tree was generated using Maximum Likelihood method based on the JTT matrix-based model in MEGA-X program. Further, we performed NCBI BLASTp search for identification of archaea lipase sequences against the bacterial domain in public database to validate and confirm those putative lipase sequences.
Results and discussion
Phylogenetic tree reconstruction
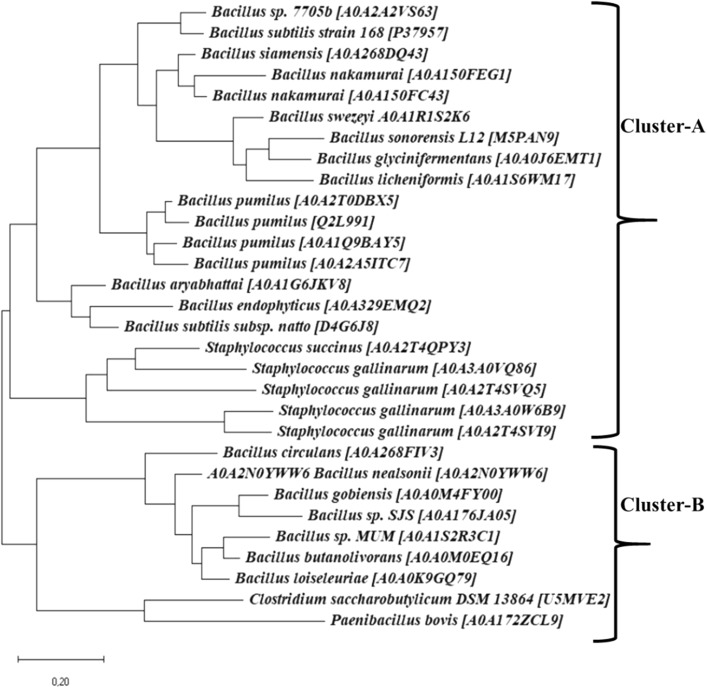
The Bacillus subtilis lipase is remarkably stable at alkaline pH, showing maximum stability at pH 12 and activity at pH 10. (Lesuisse et al. 1993). The lipase sequence of Bacillus subtilis used to find homologous sequences from public database resulted in a total of 250 extant lipase sequences from Uniport database. Nearly 30 lipase sequences showing 45–90% sequence identity were selected; all these retrieved sequences were clustered in two main clusters. All homologous sequences shared a common minimal alpha/beta hydrolase fold and conserved pentapeptide [Gly/Ala-His-Ser-X-Gly] motif and catalytic resides. The larger cluster-A had 21 lipase sequences which were similar to the Bacillus species, subspecies, Staphylococcus succinus and Staphlyococcus gallinarum. Whereas, the smaller cluster-B comprised of five lipase sequences from Bacillus species, subspecies, Clostridium saccharobutylicum and Paenibacillusbovis as shown in Fig. 1. The most probable putative ancestral sequence was generated from midpoint node of rooted tree. The most probable inferred ancestral sequence was used to detect remotely homologous sequences from the NCBI non-redundant (nr) databases within Archaeal, Bacterial and environmental Metagenome domains.
Fig. 1.
Molecular phylogenetic tree inferred via maximum likelihood method. The ancestral sequence reconstruction analysis involved 30 amino acid sequences. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site
Identification of potential alkaline lipases from archaea
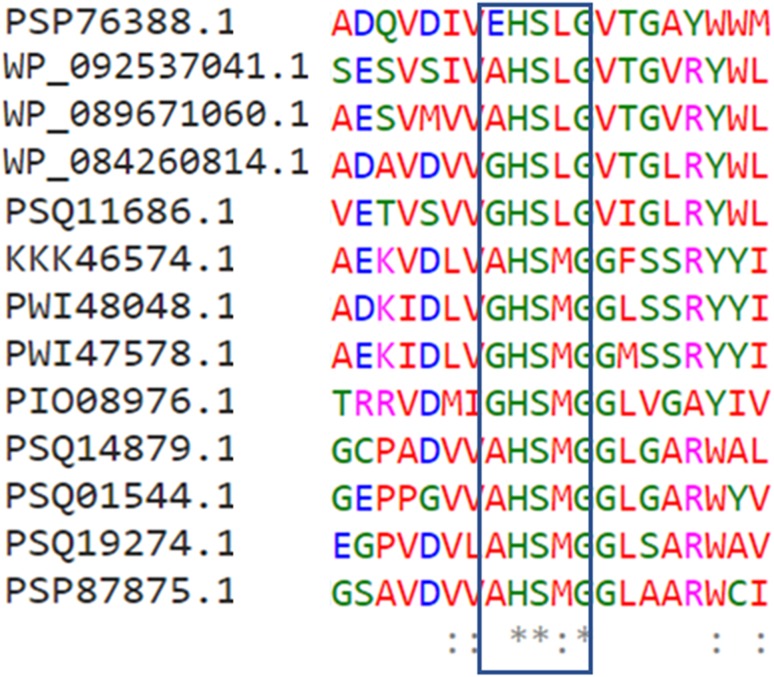
We identified 12 unique hypothetical and 1 putative lipase like protein sequences, which showed significant sequence identity (≥ 25%) and query coverage (≥ 50%) with ancestal query sequence. Interestingly, well characterized alkalohyperthermophilic Archaeoglobus fulgidus lipase (Chen et al. 2009) was also found in our search list which showed ~ 23% sequence identity with ancestoral sequence. These identified archaeal lipase sequences were further taken for phylogenetic analysis including 30 extant bacterial lipase sequences. The optimum MSA analysis showed that the active site catalytic residues that comprises of serine, aspartic acid and histidine were highly conserved in all the identified hypothetical archaeal lipase sequences. Interestingly, conserved pentapeptide sequence [Ala/Gly-His-Ser-Leu/Met-Gly] pattern was observed among the identified archaeal homologous sequences. The first glycine residue of the pentapeptide was replaced by glutamic acid in one identified archaeal sequence as depicted in Fig. 2. This conserved pentapeptide sequence motif [Gly-X-Ser-X-Gly], located in the hydrophobic pocket of the active site of lipase is a key feature of lipase family (Brenner 1988; Derewenda and Derewenda 1991; Verma et al. 2018). In the conserved Ala-X-Ser-X-Gly motif, the role of Ala (Alanine) in thermostability has been reported earlier for Bacillus thermoalkalophilic lipases (Jeong et al. 2002).
Fig. 2.
The conserved pentapeptide [X-His-Ser-X-Gly] motif in the identified archaea lipase. Asterisk symbol indicates fully conserved residues. HP hypothetical proteins
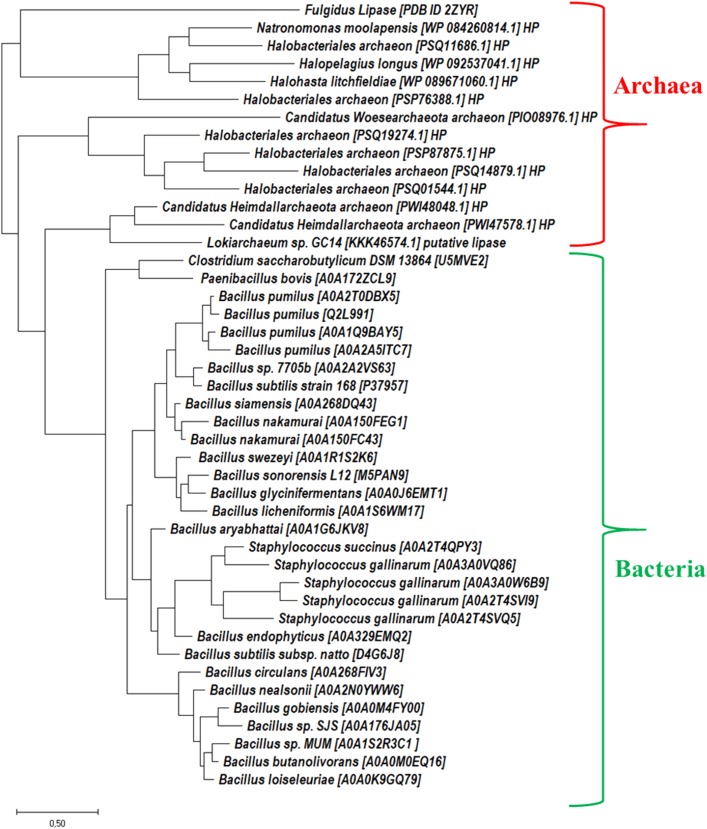
The phylogenetic analysis of identified archaeal lipases showed novel clade; however, some sequences showed more closeness with extant bacterial lipase sequences. This study revealed that the archaeal domain possesses a new and broad reservoir of unseen/unreported extremophilic lipase genes. Analysis of the phylogeny tree of merged extant bacterial and archaeal sequences revealed that they can be clustered mainly into two clades as shown in Fig. 3. We noticed that twelve novel members which segregated from extant lipase enzymes share key features like conserved pentapeptide motif [Gly-X-Ser-X-Gly] and active site residues. Some sequences of new clade were closer to Clostridium saccharobutylicum and Paenibacillus bovis species. These results indicated the presence of a different class of extremophile archaeal lipases in microorganisms inhabiting hot springs, salt lakes, soil, fresh water, marine water, and marshlands.
Fig. 3.
Phylogenetic analysis by maximum likelihood method. The analysis involved a total 44 (30 extant lipase sequences plus 14 sequences searched from archaea). The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. HP hypothetical proteins
The identified hypothetical archaeal lipase sequences were further confirmed and predicted using similarity searches with known bacterial lipases. We performed NCBI BLASTp searches for each identified putative archaeal lipase sequence using publicly available bacterial genomic database. This approach is better than direct bacterial lipase search method, as it can also be used to find extremophilic bacterial lipases using the information in archaeal sequence databases. The BLASTp search results further support that the identified hypothetical proteins are lipases. All those hypothetical lipase sequences showed significant sequences identity ≥ 31% and query coverage ≥ 64% with the bacterial lipase sequence. More detailed information about sequence identity, query coverage, and bacterial homologs is provided in Table 1. Based on the sequence similarity, all these hypothetical archaeal lipase sequences can be considered accurate bacterial homologs. These results further allowed to identify and discover the remotely related homologous sequences.
Table 1.
New lipase sequences identified from the archaeal domain using the putative ancestor query sequence
| S. no | Organism name | IDs | Protein name | Query cover/ SEq. Ident. with bacteria (%) | Bacterial homologs (protein/organism name) |
|---|---|---|---|---|---|
| 1 | Candidatus Heimdallarchaeota archaeon | PWI48048.1 | Hypothetical | 80/39 | Triacylglycerol lipase [Bacillus sp. 17,376] |
| 2 | Candidatus Heimdallarchaeota archaeon | PWI47578.1 | Hypothetical | 77/38 | Triacylglycerol lipase [Bacillus jeotgali] |
| 3 | Halopelagiuslongus | WP_092537041.1 | Hypothetical | 74/32 | Triacylglycerol esterase/lipase [Bradyrhizobium erythrophlei] |
| 4 | Candidatus Woesearchaeota archaeon CG08 | PIO08976.1 | Hypothetical | 79/32 | Alpha/beta fold hydrolase [Blastococcus sp. TF02-8] |
| 5 | Natronomonas moolapensis | WP_084260814.1 | Hypothetical | 69/36 | Triacylglycerol lipase [Streptomyces ipomoeae] |
| 6 | Lokiarchaeum sp. GC14 | KKK46574.1 | Putative lipase | 87/34 | Triacylglycerol lipase [Bacillus jeotgali] |
| 7 | Halobacteriales archaeon QS_5_70_15 | PSQ11686.1 | Hypothetical | 64/34 | Triacylglycerol lipase [Salinispora pacifica] |
| 8 | Halobacteriales archaeon QS_8_69_26 | PSQ19274.1 | Hypothetical | 93/37 | Triacylglycerol lipase [Bacillus selenatarsenatis] |
| 9 | Halobacteriales archaeon QS_4_69_34 | PSP87875.1 | Hypothetical | 80/33 | Triacylglycerol lipase [Bacillus selenatarsenatis] |
| 10 | Halobacteriales archaeon QS_1_68_20 | PSP76388.1 | Hypothetical | 71/31 | Hypothetical protein [Sinobacteraceae bacterium] |
| 11 | Halohasta litchfieldiae | WP_089671060.1 | Hypothetical | 70/32 | Triacylglycerol lipase [Nocardia pneumoniae] |
| 12 | Halobacteriales archaeon QS_5_70_17 | PSQ01544.1 | Hypothetical | 93/39 | Alpha/beta fold hydrolase [Deltaproteobacteria bacterium] |
| 13 | Halobacteriales archaeon QS_7_69_60 | PSQ14879.1 | Hypothetical | 74/31 | Triacylglycerol lipase [Bacillus jeotgali] |
| 14 | Archaeoglobus fulgidus | PDB ID: 2ZYR | Lipase | 94/37 | Alpha/beta fold hydrolase [Actinomadura amylolytica] |
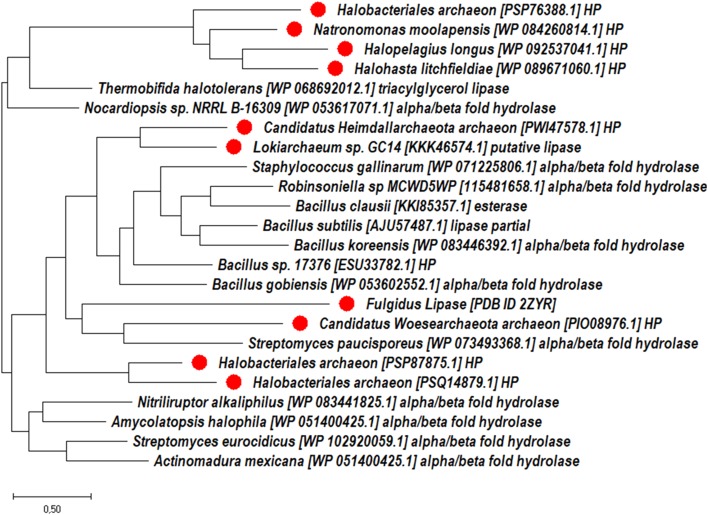
In addition, we searched distantly related homologous lipase gene sequences in bacterial domain using ancestor query sequence in BLASTp program. Top 500 hits were taken and clustered at 50% sequence identity to select the representative set. To investigate the diversity and distribution of archaeal alkaliphilic lipase with respect to the bacterial domain, we performed separate phylogenomic analysis including both the sets of identified lipase protein sequences as shown in Fig. 4. We observed that identified archaeal lipase sequences clustered with some bacterial lipase, and most of them belongs to class Haloarchaea. The archaeal enzymes have the unique stability at high temperature, salt concentrations and high pH, hence, the enzymes of archaeal origin are expected to be super biocatalysts and can catalyze hydrolytic and synthetic reactions under harsh conditions suitable in industrial processes. Mainly, the halophilic archaea are most likely source of extremophilic lipolytic enzymes due to salt tolerance and thermophilic properties. The diversity in Gram-positive halophilic bacteria producing extracellular hydrolytic enzymes has been reported by Sanchez-Porro et al. 2003. The halophilic archaeal microorganisms that can live in saline habitats are expected to showcase multitude of potential applications in various fields of biotechnology. Enzymes of halophilic archaea can offer distinct rewards over their conventional complements in the development of new bioconversion methods, potentially overring resistance to high salt and high temperatures conditions (Margesin and Schinner 2001; Oren 2002). The Ozcan et al. 2009 reported that the lipolytic enzymes from halophilic isolates were active at high temperature (60–65 °C) and pH conditions (pH 8–8.5). In addition, the isolates were found to have higher esterase activity compared to lipase activity on the basis of kinetic parameters (Ozcan et al. 2009).
Fig. 4.
Phylogenetic analysis by maximum likelihood method. The analysis involved a total 24 searched representative set at 50% sequence identity (14 from bacterial and 10 from archaea). The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The red colored branches represent archaeal members
Identification of potential alkaliphilic lipases from metagenomic database
A total 9 unique lipase like protein sequences were searched from NCBI environmental metagenome (env_nr) database, which showed significant sequence identity ≥ 30% with ancestoral query sequence. All obtained proteins sequences are either of marine or sediment metagenome. Interestingly, all these sequences have highly conserved pentapeptide sequence [Ala/Gly-His-Ser-X-Gly] motif. The first glycine/alanine residue of the pentapeptide is replaced by cysteine amino acid in metagenome sequence Ids: ECA41166.1 and ECZ28584. First time, at this position cysteine residue was observed in pentapeptide motif. More conserved pentapeptide sequence information is depicted in Fig. 5. All those obtained hypothetical predicted protein sequences were annotated based on the sequence homology. We observed that all these metagenomic protein sequences showed significant identity and more than 75% query coverages with alpha/beta hydrolase fold. The six-protein sequences out of nine showed > 97% sequence identity with NCBI non-redundant protein sequence database. All these sequences encode proteins similar to alkaliphilic lipases.
Fig. 5.
The conserved pentapeptide [X-His-Ser-X-Gly] motif in the lipase identified through metagenomic sequences. Asterisk symbol indicates fully conserved residues. HP hypothetical proteins
Recently, alkane hydroxylase, lipase and esterase were reported in bacteria Alcanivorax borkumensis (Kadri et al. 2018). Lipase activity has been reported from a marine proteobacterium Alteromonas species isolated from Bay of Marseille, France (Duflos et al. 2009). Extracellular lipase activity detected on tributyrin agar has been identified from Aeromonas hydrophila (Ingham and Pemberton 1995). All those reported lipase activities of highly homologous sequences from metagenomics searches validate our hypothesis. The lipase sequence from archaeum Lokiarchaeum sp. GC14 is found common in metagenomic and archaea searches. Three sequences showing sequence identity in the range between 33–66% and were considered as new sequences. The sequence Id: GAG97039.1 showed sequence homology with archaeal origin while the sequences id EBE45432.1 showed sequence similarity with marine cyanobacteria Prochlorococcus marinus. More details about sequence identity and homology is shown in Table 2. The present study illustrates large diversity of lipases in different biological genomic databases.
Table 2.
Homologs sequences identified from the environmental non-redundant metagenomics proteins database
| S. no | Metagenome Seq ID |
Protein name/metagenome | Query cover/ SEq. ident. (%) | Homologs (protein/organism name) |
|---|---|---|---|---|
| 1 | ECZ28584.1 | HP, partial [marine] | 100/100 | Alpha/beta hydrolase fold [Alcanivoraxborkumensis] |
| 2 | KKM83358.1 | HP [marine sediment] | 100/100 | Putative esterase/lipase with alpha/beta hydrolase [Lokiarchaeum sp. GC14_75] |
| 3 | ECZ99212.1 | HP [marine metagenome] | 100/99 | Triacylglycerol lipase [Alteromonas macleodii] |
| 4 | EBS88226.1 | HP, partial [marine] | 83/97 | Triacylglycerol lipase [Alteromonas] |
| 5 | EDA82741.1 | HP, partial [marine] | 100/100 | Triacylglycerol lipase [Aeromonas hydrophila] |
| 6 | GAG97039.1 | HP, partial [marine sediment] | 96/68 | Putative esterase/lipase with alpha/beta hydrolase [Lokiarchaeum sp. GC14_75] |
| 7 | ECA41166.1 | HP, partial [marine] | 81/33 | Triacylglycerol lipase [Alcanivorax sp. KX64203] |
| 8 | EDA34272.1 | HP, partial [marine] | 93/99 | Alpha/beta hydrolase [Paraburkholderia fungorum] |
| 9 | EBE45432.1 | HP, partial [marine] | 75/65 | Putative esterase/lipase protein [Prochlorococcus marinus] |
The occurrence of alkaliphilic lipase homologs in the archaea and metagenomic database points at unexplored new lipolytic proteins with enhanced thermostability and alkalinity. This work was carried out with the help of most probable ancestral sequence derived from the extant alkaliphilic lipase sequences, which could detect remotely homologous lipase sequences with higher efficiency. The metagenomic database is a valuable resource to discover novel protein families that greatly assist the annotation and identification of remotely homologous sequences (Lobb et al. 2015). The identified numbers of hypothetical unannotated lipase like protein sequences, showed significant sequence similarity with ancestral sequence. In upcoming years, the more powerful sequencing technologies in combination with advanced bioinformatics algorithms will allow a high-throughput data analysis and will endure to revolutionize the exploration of microbial dark matter (Saw et al. 2015). Nevertheless, the validation of the data obtained from reconstruction of ancestral sequences can be carried out by heterologous expression of some of these sequences and evaluating the expressed enzymes for their activities and other properties. Mainly lipase research is concentrated on identifying highly thermostable lipases from different thermophilic microbial sources. However, the studies related to thermoalkaliphilic lipases from hyperthermoalkaliphilic organisms are inadequate. The ancestral sequence reconstruction-based approach can be more systematic and useful than the preliminary high throughput screening of distinct homologs sequences.
Conclusion
The result of this work has provided valuable information in the form of phylogenetic tree. The putative ancestral sequence of extant alkaliphilic lipase provides an intuitive way to inspect homologous sequences in microbial communities and have discovered new members of thermo-alkaliphilic lipase sequences from published genomics content available in the public databases. The identified new protein sequences are evolutionarily distant from known alkaliphilic lipases, but based on the sequence similarity, conserved active amino acid residues, and conserved pentapeptide [Gly/Ala-His-Ser-X-Gly] motif, that is key feature of lipase family, we assume that their catalytic activity will be identical or very similar to the reported lipases with additional advantage of the higher activity and stability in thermoalkaline environments. The study would enrich the current knowledge on lipases with respect to designing more potential thermo-alkaliphilic lipases for industrial applications.
Acknowledgements
We are thankful to Dr. Athar Alam from Umeå University, Sweden, for his help to improve the language of this manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare no conflicts of interest for publishing this work.
Contributor Information
Swati Verma, Email: swati.shush@gmail.com.
Rajender Kumar, Email: rajbioinformatics@gmail.com.
Gautam Kumar Meghwanshi, Email: gotumm@gmail.com.
References
- Abol Fotouh DM, Bayoumi RA, Mohamed Hassan A. Production of thermoalkaliphilic lipase from Geobacillus thermoleovorans DA2 and application in leather industry. Enzym Res. 2016;2016:1–9. doi: 10.1155/2016/9034364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Brenner S. The molecular evolution of genes and proteins: a tale of two serines. Nature. 1988;334:528–530. doi: 10.1038/334528a0. [DOI] [PubMed] [Google Scholar]
- Chen CK, Lee GC, Ko TP, Guo RT, Huang LM, Liu HJ, Ho YF, Shaw JF, Wang AH. Structure of the alkalohyperthermophilic Archaeoglobusfulgidus lipase contains a unique C-terminal domain essential for long-chain substrate binding. J Mol Biol. 2009;390:672–685. doi: 10.1016/j.jmb.2009.05.017. [DOI] [PubMed] [Google Scholar]
- Collins LJ, Poole AM, Penny D. Using ancestral sequences to uncover potential gene homologues. Appl Bioinform. 2003;2:S85–S95. [PubMed] [Google Scholar]
- Cunningham CW, Omland KE, Oakley TH. Reconstructing ancestral character states: a critical reappraisal. Trends Ecol Evol. 1998;13:361–366. doi: 10.1016/S0169-5347(98)01382-2. [DOI] [PubMed] [Google Scholar]
- Demirjian DC, Moris F, Cassidy CS. Enzymes from extremophiles. Curr Opin Chem Biol. 2001;5:144–151. doi: 10.1016/S1367-5931(00)00183-6. [DOI] [PubMed] [Google Scholar]
- Derewenda Z, Derewenda U. Relationships among serine hydrolases: evidence for a common structural motif in triacylglyceride lipases and esterases. Biochem Cell Biol. 1991;69:842–851. doi: 10.1139/o91-125. [DOI] [PubMed] [Google Scholar]
- Duflos M, Goutx M, Van Wambeke F. Determination of lipid degradation by marine lipase-producing bacteria: critical evaluation of lipase activity assays. Lipids. 2009;44:1113–1124. doi: 10.1007/s11745-009-3358-7. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias I, Tuller T. Reconstruction of ancestral genomic sequences using likelihood. J Comput Biol. 2007;14:216–237. doi: 10.1089/cmb.2006.0101. [DOI] [PubMed] [Google Scholar]
- Haki GD, Rakshit SK. Developments in industrially important thermostable enzymes: a review. Bioresour Technol. 2003;89:17–34. doi: 10.1016/S0960-8524(03)00033-6. [DOI] [PubMed] [Google Scholar]
- Hasan F, Shah AA, Hameed A. Industrial applications of microbial lipases. Enzym Microb Technol. 2006;39:235–251. doi: 10.1016/j.enzmictec.2005.10.016. [DOI] [Google Scholar]
- Huang Y, Niu B, Gao Y, Fu L, Li W. CD-HIT Suite: a web server for clustering and comparing biological sequences. Bioinformatics. 2010;26:680–682. doi: 10.1093/bioinformatics/btq003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham AB, Pemberton JM. A lipase of Aeromonas hydrophila showing nonhemolytic phospholipase C activity. Curr Microbiol. 1995;31:28–33. doi: 10.1007/BF00294630. [DOI] [PubMed] [Google Scholar]
- Jeong ST, Kim HK, Kim SJ, Chi SW, Pan JG, Oh TK, Ryu SE. Novel zinc-binding center and a temperature switch in the Bacillus stearothermophilus L1 lipase. J Biol Chem. 2002;277:17041–17047. doi: 10.1074/jbc.M200640200. [DOI] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- Kadri T, Rouissi T, Magdouli S, Brar SK, Hegde K, Khiari Z, Daghrir R, Lauzon JM. Production and characterization of novel hydrocarbon degrading enzymes from Alcanivorax borkumensis. Int J Biol Macromol. 2018;112:230–240. doi: 10.1016/j.ijbiomac.2018.01.177. [DOI] [PubMed] [Google Scholar]
- Koshi JM, Goldstein RA. Probabilistic reconstruction of ancestral protein sequences. J Mol Evol. 1996;42:313–320. doi: 10.1007/BF02198858. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesuisse E, Schanck K, Colson C. Purification and preliminary characterization of the extracellular lipase of Bacillus subtilis 168, an extremely basic pH-tolerant enzyme. Eur J Biochem. 1993;216:155–160. doi: 10.1111/j.1432-1033.1993.tb18127.x. [DOI] [PubMed] [Google Scholar]
- Littlechild JA. Archaeal enzymes and applications in industrial biocatalysts. Archaea. 2015;2015:10. doi: 10.1155/2015/147671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobb B, Kurtz D, Moreno-Hagelsieb G, Doxey AC. Remote homology and the functions of metagenomic dark matter. Front Genet. 2015;6:234. doi: 10.3389/fgene.2015.00234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margesin R, Schinner F. Potential of halotolerant and halophilic microorganisms for biotechnology. Extremophiles. 2001;5:73–83. doi: 10.1007/s007920100184. [DOI] [PubMed] [Google Scholar]
- Meghwanshi GK, Vashishtha A. Fungi and their role in sustainable development: current perspectives. Singapore: Springer Nature; 2018. Biotechnology of fungal lipases; pp. 383–411. [Google Scholar]
- Ollis DL, Cheah E, Cygler M, Dijkstra B, Frolow F, Franken SM. The alpha/beta hydrolase fold. Protein Eng. 1992;5:197–211. doi: 10.1093/protein/5.3.197. [DOI] [PubMed] [Google Scholar]
- Oren A. Diversity of halophilic microorganisms: environments, phylogeny, physiology, and applications. J Ind Microbiol Biotechnol. 2002;28:56–63. doi: 10.1038/sj/jim/7000176. [DOI] [PubMed] [Google Scholar]
- Ozcan B, Ozyilmaz G, Cokmus C, Caliskan M. Characterization of extracellular esterase and lipase activities from five halophilic archaeal strains. J Ind Microbiol Biotechno. 2009;36:105–110. doi: 10.1007/s10295-008-0477-8. [DOI] [PubMed] [Google Scholar]
- Sanchez-Porro C, Martin S, Mellado E, Ventosa A. Diversity of moderately halophilic bacteria producing extracellular hydrolytic enzymes. J Appl Microbio. 2003;l94:295–300. doi: 10.1046/j.1365-2672.2003.01834.x. [DOI] [PubMed] [Google Scholar]
- Saw JH, Spang A, Zaremba-Niedzwiedzka K, Juzokaite L, Dodsworth JA, Murugapiran SK, Colman DR, Takacs-Vesbach C, Hedlund BP, Guy L, Ettema TJ. Exploring microbial dark matter to resolve the deep archaeal ancestry of eukaryotes. Philos Trans R Soc Lond B Biol Sci. 2015;2:370. doi: 10.1098/rstb.2014.0328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena RK, Agarwal L, Meghwanshi GK (2005) Diversity of fungal and yeast lipases: present and future scenario for the 21st century. In: Microbial diversity: current perspectives and potential applications. I.K. International Pvt. Ltd 791–814. ISBN 9788188237432
- Sharma PK, Kumar R, Kumar R, Mohammad O, Singh R, Kaur J. Engineering of a metagenome derived lipase towards thermal tolerance: effect of aspargine to lysine mutation on the protein surface. Gene. 2011;491:264–271. doi: 10.1016/j.gene.2011.09.028. [DOI] [PubMed] [Google Scholar]
- Sharma PK, Kumar R, Garg P, Kaur J. Insights into controlling role of substitution mutation, E315G on thermostability of a lipase cloned from metagenome of hot spring soil. 3Biotech. 2015;4:189–196. doi: 10.1007/s13205-013-0142-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu ZY, Jiang H, Lin RF, Jiang YM, Lin L, Huang JZ. Technical methods to improve yield, activity and stability in the development of microbial lipases. J Mol Catal B: Enzym. 2010;62:1–8. doi: 10.1016/j.molcatb.2009.09.003. [DOI] [Google Scholar]
- Treichel H, de Oliveira D, Mazutti MA, Di Luccio M, Oliveira JV. A review on microbial lipases production. Food Bioprocess Tech. 2010;3:182–196. doi: 10.1007/s11947-009-0202-2. [DOI] [Google Scholar]
- Verma S, Meghwanshi GK, Kumar R. Structural homogeneity in microbial lipases. Microbiol Curr Res. 2018;2:12–13. [Google Scholar]