Abstract
Although the implication of BCL3 has been disclosed in human chronic lymphocytic leukemia as well as other solid tumors, the diagnostic and prognostic of BCL3 expression in acute myeloid leukemia (AML) remains largely unclear. In this study, we isolated total RNA from bone marrow mononuclear cells collected from 101 de novo AML patients and 27 healthy donors. After reverse transcription, quantitative real-time PCR was performed to detect BCL3 expression level. BCL3 mRNA level was significantly down-regulated in BMMCs of AML patients compared with healthy controls (P = 0.0015). BCL3 was showed a higher level in AML patients with poor-risk karyotypes than that of in patients with favorable/intermediate-risk karyotypes (P = 0.014). ROC analysis demonstrated that BCL3 could effectively differentiate AML patients from normal controls. Among the French-American-British (FAB) subtypes, the frequency of low BCL3 expression in M2 subtypes is significantly higher than that of in the other subtypes M1/M4/M5/M6/M7 (P = 0.006), and mildly lower in myelomonocytic/monocytic subtypes M4/M5 (P = 0.064) than those in M1/M2/M6/M7 subtypes. Chromosome analysis revealed that BCL3low patients had a remarkably higher frequency of t (8;21) abnormality (P = 0.0047) and lower frequency of normal karyotype (P = 0.0059) than BCL3high patients. BCL3high patients showed a significantly higher frequency of FLT3-ITD mutation (P = 0.028) and lower frequency of C-Kit mutation (P = 0.0232) than BCL3low patients. Although there were no significant differences in complete remission and overall survival between BCL3low and BCL3high groups, patients with high BCL3 expression markedly shorter overall survival (OS, P = 0.049), relapse-free survival (RFS, P = 0.027) and disease-free survival (DFS, P = 0.042) in M2 AML than low BCL3 expression patients. Additionally, in AMLs of M2 subtype, high BCL3 expression patients had markedly lower complete remission (CR) rate (P = 0.0317) after the second induction treatment than patients with BCL3 low expression. Thus, these findings indicated that BCL3 appeared as a promising molecular biomarker of pediatric acute myeloid leukemia with unfavorable prognosis.
Keywords: BCL3, Expression, Prognosis, Acute myeloid leukemia
Introduction
Acute myeloid leukemia (AML) is a cancer of the myeloid line of blood cells, which comprises approximately 15–20% of pediatric leukemia[1]. Although the cure rate has improved, treatments are associated with notable morbidity and mortality. About 60–70% of patients achieve complete remission after the induction chemotherapy, only 20–30% of patients achieve long-term disease-free survival (DFS) [2–4]. Each AML patients can be separated into distinct risk subgroup. The diagnostic classification of pediatric AML depends on the combination of morphology, cytochemistry, immunophenotyping and molecular genetics. Moreover, aberrant expression of cancer-related genes has been associated with the prognosis and treatment outcome of AML.
BCL3 gene was initially identified in B cell chronic lymphocytic leukemia (B-CLL) with chromosomal translation of t(14;19)(q32;q13), which encodes a nuclear protein that belongs to the IkB family of inhibitors of NFkB [5]. Nevertheless, latter research has shown that BCL3 rearrangement is recurrent in other hematological malignancies, such as T cell lymphoma, Burlitt-like lymphoma, Hodgkin lymphoma [6–8]. Disregulation of BCL3 without chromosomal translocation has been found to participate in progression in a variety of solid tumors, from breast cancer, nasopharyngeal carcinoma and renal-cell carcinoma to lung cancer [9–12]. Moreover, abnormal expression of BCL3 was associated with prognosis in patients with CLL, breast cancer, clear-cell renal-cell carcinoma and non-small-cell lung cancer [12–15]. Due to its overexpression in numerous types of tumors, as well as its extensive roles in promoting transformation in hematologic malignancies, BCL3 has been recognized as an oncogenic gene. However, up to now, little studies investigated BCL3 expression or its role in myeloid malignancies. The current study was aimed to investigate BCL3 expression and its clinical significance in pediatric patients with de novo AML.
Materials and Methods
Patients and Control Group
This original research study included 100 patients who had previously been diagnosed with AML at the Children’s Hospital Affiliated to Suzhou University as well as 27 normal controls. None of the patients had received any treatment before bone marrow (BM) sample collection. The diagnosis and classification of AML were based on the French-American-British (FAB) and World Health Organization (WHO) criteria. Complete remission (CR) was defined as that the bone marrow contained less than 5% blasts. Overall survival was defined as the period of time after patients were diagnosed with disease. Disease-free survival (DFS) was the length of time after treatment during which no disease is found. Details of clinical characteristics of all AML patients were summary in Table 1.
Table 1.
Correlation between BCL3 expression and clinical parameters of 100 AML patients
| Patients’ parameters | BCL3 expression | ||
|---|---|---|---|
| Low (n = 50) | High (n = 50) | P value | |
| Sex, male/female | 33/16 | 25/26 | 0.063 |
| Median age, month, (range) | 91.67 (9–149) | 78 (7–166) | 0.215 |
| Median WBC, ×109/L (range) | 15.5 (0.42–275.9) | 16.9 (1.05–300.55) | 0.811 |
| Median HGB, g/L (range) | 75 (20–111) | 77.67 (1.96–108) | 0.947 |
| Median PLT,×109/L, (range) | 41 (10–179) | 66.33 (10–586) | 0.017 |
| BM blasts, % (range), | 63 (15–95) | 72.5 (12–98) | 0.699 |
| PB blasts, % (range) | 56.8 (2–91) | 41.33 (2–94) | 0.201 |
| FAB classification | n = 47 | n = 50 | |
| M1 | 1 (1%) | 3 (3.1%) | 0.338 |
| M2 | 30 (30.9%) | 18 (18.6%) | 0.006 |
| M4 | 7 (7.2%) | 13 (13.4%) | 0.177 |
| M5 | 9 (9.3%) | 13 (13.4%) | 0.420 |
| M4 + M5 | 16 | 26 | 0.064 |
| M6 | 0 (0%) | 1 (1%) | 0.330 |
| M7 | 0 (0%) | 2 (2.1%) | 0.166 |
| Karyotypes | n = 50 | n = 50 | |
| Normal | 4 (4%) | 15 (15%) | 0.0059 |
| t(8;21) | 23 (23%) | 10 (10%) | 0.0047 |
| Inv(16) | 5 (5%) | 8 (8%) | 0.3936 |
| + 8 | 0 (0%) | 1 (1%) | 0.3197 |
| -5/5q | 1 (1%) | 0 (0%) | 0.3101 |
| -7/7q | 0 (0%) | 1 (1%) | 0.3197 |
| MLL rearrangement | 4 (4%) | 7 (7%) | 0.3558 |
| Complex | 4 (4%) | 2 (2%) | 0.3860 |
| Other | 5 (5%) | 4 (4%) | 0.7037 |
| No data | 4 (4%) | 3 (2%) | 0.6752 |
| Karyotype classification | n = 46 | n = 51 | 0.278 |
| Favorable | 18 | 17 | |
| Intermediate | 24 | 22 | |
| Unfavorable | 7/ | 12 | |
| Gene mutation | n = 49 | n = 51 | |
| No mutation (± ) | 14 | 8 | 0.1339 |
| C-Kit (± ) | 9 | 2 | 0.0232 |
| FLT3-ITD (± ) | 1 | 5 | 0.0486 |
| CEBPA | 0 | 2 | 0.1573 |
| NPM1 | 1 | 0 | 0.3101 |
| No data | 24 | 34 | 0.0893 |
| CR | |||
| CR1 (CR/PR/NR) | 30/12/7 | 22/21/8 | 0.156 |
| CR2 (CR/PR/NR) | 36/5/8 | 36/9/6 | 0.499 |
AML, acute myeloid leukemia; WB, white blood cells; HB, hemoglobin; PLT, Platelet; BM, bone marrow; PB, peripheral blood; FAB, French-American-British; CR, complete remission; PR, partial remission; NR, no remission
This research was approved by the institutional Ethics Committee of the Children’s Hospital Affiliated to Suzhou University. A written informed consent was also signed from each participant of the study.
RNA Isolation and Reverse Transcription
BM mononuclear cells (BMMCs) were isolated by Lymphocyte Separation reagent (Dakewei, Shenzhen, China) mediated density gradient centrifugation according to the protocol. Total RNA was extracted using Trizol reagent (Invitrogen, Carlsbad, CA, USA), the concentration and purity of total RNA was assessed by using NanoDrop 2000 Spectrophotometer. Reverse transcription was performed to synthesize cDNA using the PrimeScript RT reagent kit (Takara, Dalian, China). The system of Reverse transcription was incubated for 60 min 42 °C, followed by 5 min at 95 °C.
Real-Time Quantitative PCR
BCL3 expression was examined by real-time quantitative PCR (Q-PCR) performed on a 7500 thermo cycler (Applied Biosystems, CA, USA). SYBR Premix Ex Taq II kit (Takara, Dalian, China) was used for Q-PCR reactions with the following primers, forward 5’ CGTGAACGCGCAAATGTACT 3′ and reverse GATGTCGATGACCCTGCGG. The Q-PCR reaction was carried out at 95 °C for 5 min, followed by 40 cycles at 95 °C for 10 s, 60 °C for 30 s, 72 °C for 30 s and 70 °C for 30 s to collect fluorescence. Relative BCL3 expression levels were calculated by 2-△△CT method against GAPDH reference gene.
Statistical Analysis
Statistical analysis was performed using the SPSS 19.0 software package. Non-parametric Mann-Whitney U test was used to compare the distributions of BCL3 expression value in AML patients and healthy controls. Person Chi-square analysis or Fisher exact test were carried out to compare the difference of categorical variables. We also assessed the diagnostic potential of BCL3 expression by receiver operating characteristic (ROC) analysis. Thus, a ROC curve was built by plotting sensitivity versus 1-sepecicity, and the respective area under the curve (AUC) was analyzed by the Hanley and McNeil method. Kaplan-Meier overall survival (OS) analysis was performed, and differences between OS curves were evaluated using the Mantel-Cox (log-rank) test.
Results
BCL3 Expression in Controls and AML Patients
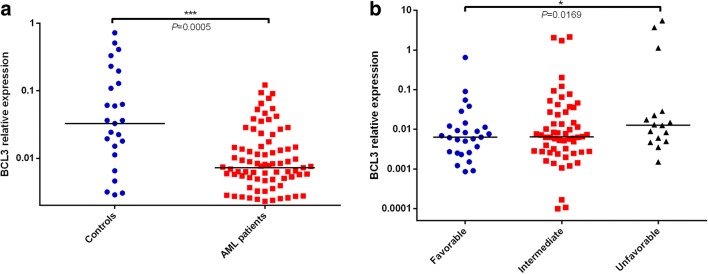
Expression of BCL3 in AML patients and healthy donors was detected by qRT-PCR. In contrast to controls (median 0.0032, range 1.25E5–0.0072), BCL3 (median 0.0071, range 0.00032–5.5277) BCL3 mRNA levels were significantly decreased in AML patients (p < 0.001, Fig. 1a). However, AML patients in unfavorable-risk cytogenetics group had much higher BCL3 expression than that in intermediate-risk or favorable-risk cytogenetics group (p < 0.05, Fig. 1b).
Fig. 1.
Relative expression level of BCL3 in AML patients and controls. a The BCL3 levels in AML patients were lower than those in controls (Mann-Whitney-U test). b The BCL3 levels in AML patients with favorable-risk cytogenetics group (One-Way anova analysis). The distributions of the BCL3 expression were presented with scatter plots. The median level of BCL3 expression in each group was shown with horizontal line
Discriminative Capacity of BCL3 Expression in AML
The discriminative capacity of BCL3 was revealed by ROC curve analysis, and the results indicated that BCL3 with and AUC value of 0.721 (95% CI: 0.56–0.84) might serve as a potential biomarker for distinguishing between AML and controls (P < 0.001, Fig. 2).
Fig. 2.
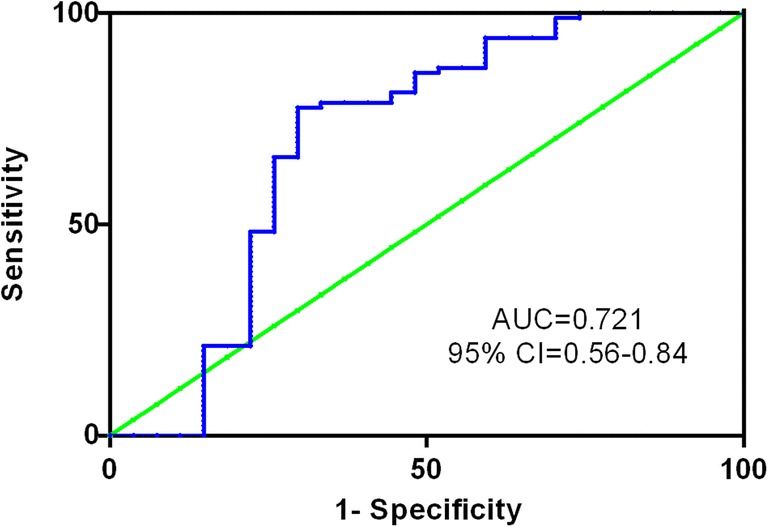
Receiver operating characteristic (ROC) curve analysis of BCL3 expression for discriminating AML patients from controls. ROC plots of BCL3 expression demonstrated the area under the curve (AUC) of 0.721 (95%CI:0.5922–0.85; P < 0.001) with 97.6% sensitivity and 55.6% specificity
Association of BCL3 Expression with Clinical Characteristics in AML
The whole-cohort of AML patients were divided into two groups (BCL3high and BCL3low) by median value. The correlation between BCL3 expression and clinical characteristics was present in Table 1. There were no significant differences in sex, age, white blood cells, hemoglobin, bone marrow blasts, peripheral blood blasts, karyotypes and karyotypic classifications between two groups (BCL3high vs BCL3low, all P > 0.05). AML patients in the BCL3high group had an increased number of platelet (median 66.33, range 10–586) compared that in the BCL3low group (median 41, range 10–179) (P = 0.017). Significant differences were observed in the distribution of FAB subtypes between BCL3high and BCL3low groups. Among the FAB subtypes, the incidence of low BCL3 expression in M2 subtype was significantly higher than in the other subtypes M1/M2/M4/M5/M6/M7 (30/17, P = 0.006), whereas the frequency of high BCL3 expression in myelomonocytic/monocytic leukemia subtypes is mildly higher than leukemia with other cell types (26/50, P = 0.064). Among WHO subtypes, the frequency of low BCL3 expression in AML patients with t(8;21) was the highest (23/27, P = 0.0047), and in AML patients without chromosomal abnormalities was the lowest (4/46, P = 0.006). Moreover, patients with low BCL3 expression harbored a higher frequency of C-kit mutations (9/41, 81.8%, P = 0.0232) and lower frequency of FLT3-ITD mutations (1/49, 16.7%, P = 0.0486).
Prognostic Value of BCL3 Expression in AML
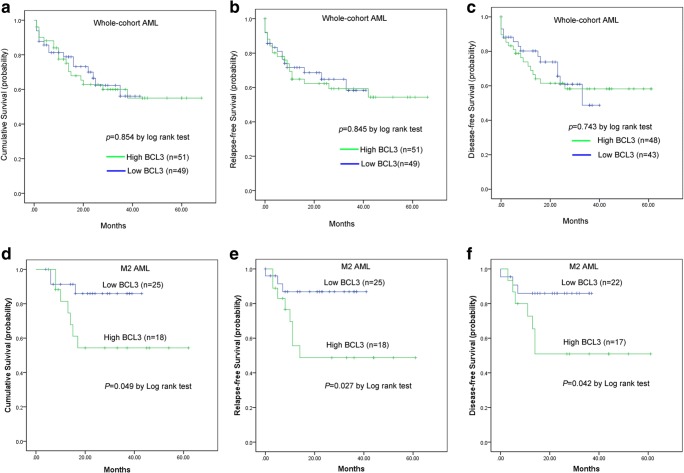
Follow-up data was obtained in 100 AML patients. Due to independent disease entity, acute promyelocytic (M3) leukemia was excluded from the analysis. Among whole-corhort AML, no significant differences were observed in complete remission rate after the induction treatment (CR, P = 0.499) between two groups.
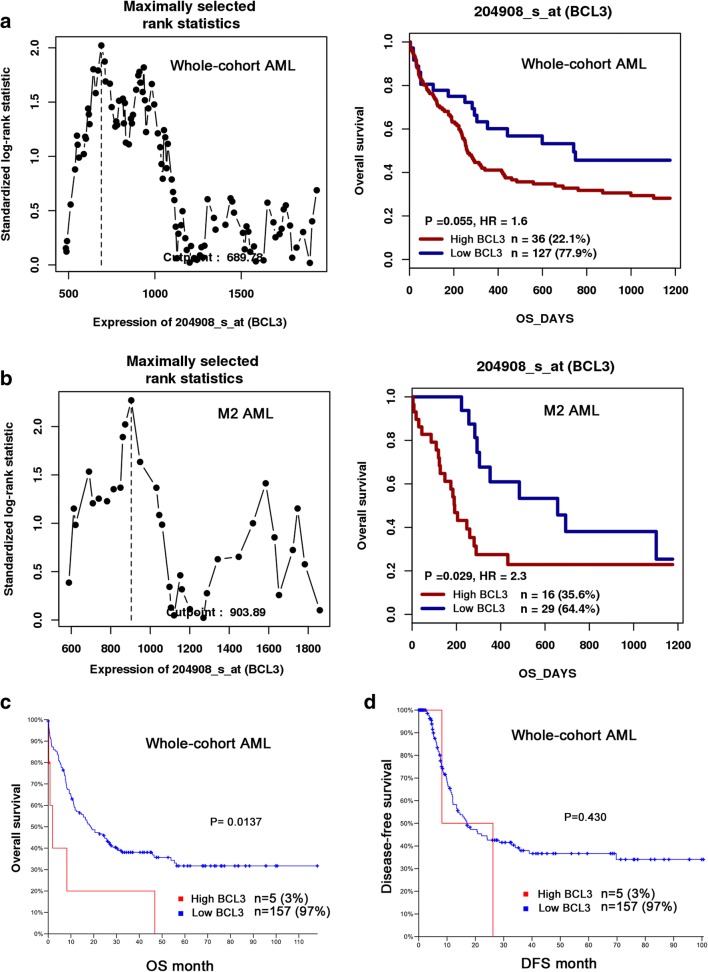
In parallel, Survival analysis was performed in 100 non-APL patients with follow-up data ranged from 1 to 68 months (median 19.5 months). Kaplan-Meier analysis revealed that there was no significant difference in overall survival (OS), relapse-free survival (RFS) as well as disease-free survival (DFS) between BCL3low and BCL3high groups in whole-corhort AML patients (P = 0.854, 0.845 and 0.743, (Figure 3a-c). The distribution frequencies of BCL3low and BCL3high in M2 subtype of AML are statistically different, which suggested an association of BCL3 expression with the type of AML. Among patients with M2, high BCL3 expression presented that significantly shorter OS (median 16 versus 22 months, respectively, P = 0.049, Fig. 3d), RFS (median 11 versus 21 months, respectively, P = 0.027, Fig. 3e) and DFS (median 14 versus 21.5 months, respectively, P = 0.042, Fig. 3f) time compared with low BCL3 expression. Moreover, multivariate analyses were further conducted to determine the prognostic value of BCL3 expression on OS among M2 AML patients. To further validate the prognostic value of BCL3 expression in M2 AML, we focused on a cohort of M2-AML patients from the GEO data (Accession number GSE12417). Using the online web tool Genomicscape, BCL3low patients showed a longer OS than BCL3high among AML (HR = 1.6, P = 0.055) and M2 subtype AML patients (HR = 2.3, P = 0.029, Fig. 4a, b). Survival analysis was also conducted using the online software cBioportal, patients with BCL3 upregulation presented significantly shorter OS time (median 1.9 month) than those without BCL3 upregulation (median18.5 month) in whole-cohort AML (P = 0.0137, Fig. 4c, d).
Fig. 3.
The impact of BCL3 expression on survival of AML patients. Survival analysis was performed by Kaplan-Meier methods. a Overall survival, OS. b Relapse-free survival, RFS. c Disease-free survival, DFS
Fig. 4.
The effect of BCL3 expression on overall survival in AML patients by bioinformatics analysis. a and b survival analysis was performed by using the online web tool GenomicScape (http://genomicscape.com/microarray/survival.php). A cohort of 163 AML patients including 45 M2 subtype was obtained from Gene Expression Omnibus data (http://www.ncbi.nlm.nih.gov/geo/; accession number GSE12417). c and d overall survival and disease-free survival were conducted by the website cBioPortal (http://www.cbioportal.org/index.do)
Discussion
In the present study, we for the first time identified that BCL3 was differentially expressed in de novo pediatric AML patients compared to healthy controls. BCL3 was statistically significant lower expression levels in bone marrow mononuclear cells of pediatric AML patients, and confirmed using ROC curve analysis revealing that BCL3 was a promising candidate biomarker for pediatric AML at prognosis. This is consistent with findings of previously studies reporting dysregulation of BCL3 expression in T/B cell leukemia [16–18], Hodgkin lymphoma [19, 20], T/B cell lymphoma [21, 22]. Apart from hematology malignancies, BCL3 has been shown to be participated in progression of diverse solid tumors, such as breast cancer [15], renal-cell carcinoma [13], non-small-cell lung cancer [12], cervical cancer [11], colorectal cancer [23]. Although dysregulation and oncogenetic roles of BCL3 in hematology malignancies are well established, the diagnostic and prognostic values of this gene are still to be evaluated.
In the correlation analysis of BCL3 expression with clinic-pathologic features, the M2 subtype of acute myeloblastic leukemia with maturation had a significantly higher frequency of low BCL3 expression than other subtypes. Furthermore, a mildly increased incidence of high BCL3 expression was observed in M4/M5 subtypes of acute myelomonocytic/monocytic leukemia. Consistent with our findings, two groups reported that BCL3 deficient myeloid progenitors demonstrated an enhanced capacity to differentiate into granulocytes following G-CFS stimulation, indicating that BCL3 played an important role in myelopoiesis/granulopoiesis [24, 25]. Coincidentally, Strauss L et al. stated that BCL3 was involved in granulo-and monocytopoiesis in tumor microenvironment [26]. In addition, our results demonstrated that patients with low BCL3 expression had a tendency of higher proportion of peripheral blood blasts. Taken together, these findings suggested a crucial role of BCL3 in regulation of myeloid differentiation. Andrew S et al. showed that BCL3 could affect the count and function of human platelet via mTOR-dependent pathway [27]. Consistently, our observation also revealed a relationship between BCL3 expression and platelet count, which is associated with overall survival, might explain its relation to disease prognosis.
In addition to the diagnostic value of BCL3 in cancers, prognostic value of BCL3 remains controversial in human cancers. Elevated expression of BCL3 acted as unfavorable prognostic indicator has been reported in chronic lymphocytic leukemia, non-Hodgkin’s lymphoma, colorectal cancer, breast cancer [14, 20, 23, 15, 12]. Additionally, subcellualar location of BCL3 also is related to prognosis. Karunakar et al. stated there was a difference in protein level and subcellular location of BCL3 between neoplastic tissues and adjacent normal tissue from the same colon cancer patients, and pointed out that analysis of the subcellular localization of BCL3 could be a potential-early diagnostic marker in colon cancer [23]. Consistently, our study also observed an impact of BCL3 expression on OS among AML subtype of FAB M2. BCL3 is a well established factor for maintaining cancer cell proliferation, ant-apoptosis and survival, might explain its relation to disease prognosis.
In summary, our findings suggest that BCL3 is overexpressed and presents a poor prognosis, could be also used a potential biomarker monitoring disease surveillance of Chinese pediatric AML patients.
Acknowledgements
This work was supported by National Natural Science Foundation of China (Grant No. 81701554), and Natural Science Foundation of Henan Province (Grant No. 162300410213).
Funding
This work was supported by National Natural Science Foundation of China (Grant No. 81701554 and 81471559), and Natural Science Foundation of Henan Province (Grant No. 162300410213).
Compliance with Ethical Standards
Conflict of Interests
These authors disclose no conflicts of interests.
Contributor Information
Yuna Niu, Email: niuyuna@126.com.
Xue Yang, Email: xueyangxxmu@163.com.
Yifei Chen, Email: irischen916@163.com.
Linbo Zhang, Email: 924173342@qq.com.
Xinyue Jin, Email: xinyuejinxxmu@163.com.
Youjing Tang, Email: 942810542@qq.com.
Li Li, Email: lilyleevvcx@163.com.
Lu Yu, Email: yulu19940130@163.com.
Yilin Guo, Email: gouyilin_xxmu@qq.com.
Hui Wang, Phone: +86-373-3029977, Email: huiwang65@yeah.net, Email: wanghui@xxmu.edu.cn.
References
- 1.de Rooij JD, Zwaan CM, van den Heuvel-Eibrink M. Pediatric AML: From Biology to Clinical Management. Journal of clinical medicine. 2015;4(1):127–149. doi: 10.3390/jcm4010127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rasche M, Zimmermann M, Borschel L, Bourquin JP, Dworzak M, Klingebiel T, Lehrnbecher T, Creutzig U, Klusmann JH, Reinhardt D (2018) Successes and challenges in the treatment of pediatric acute myeloid leukemia: a retrospective analysis of the AML-BFM trials from 1987 to 2012. Leukemia. 10.1038/s41375-018-0071-7 [DOI] [PMC free article] [PubMed]
- 3.Yu MG, Zheng HY ((2017)) Acute Myeloid Leukemia: Advancements in Diagnosis and Treatment. Chinese medical journal 130(2):211–218. 10.4103/0366-6999.198004 [DOI] [PMC free article] [PubMed]
- 4.Dombret H, Gardin C. An update of current treatments for adult acute myeloid leukemia. Blood. 2016;127(1):53–61. doi: 10.1182/blood-2015-08-604520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ohno H, Doi S, Yabumoto K, Fukuhara S, McKeithan TW. Molecular characterization of the t(14;19)(q32;q13) translocation in chronic lymphocytic leukemia. Leukemia. 1993;7(12):2057–2063. [PubMed] [Google Scholar]
- 6.Martin-Subero JI, Wlodarska I, Bastard C, Picquenot JM, Hoppner J, Giefing M, Klapper W, Siebert R. Chromosomal rearrangements involving the BCL3 locus are recurrent in classical Hodgkin and peripheral T cell lymphoma. Blood. 2006;108(1):401–402. doi: 10.1182/blood-2005-09-3843. [DOI] [PubMed] [Google Scholar]
- 7.Mottok A, Steidl C. Biology of classical Hodgkin lymphoma: implications for prognosis and novel therapies. Blood. 2018;131(15):1654–1665. doi: 10.1182/blood-2017-09-772632. [DOI] [PubMed] [Google Scholar]
- 8.Au WY, Horsman DE, Ohno H, Klasa RJ, Gascoyne RD. Bcl-3/IgH translocation (14;19)(q32;q13) in non-Hodgkin’s lymphomas. Leukemia & lymphoma. 2002;43(4):813–816. doi: 10.1080/10428190290016935. [DOI] [PubMed] [Google Scholar]
- 9.Wakefield A, Soukupova J, Montagne A, Ranger J, French R, Muller WJ, Clarkson RW. Bcl3 selectively promotes metastasis of ERBB2-driven mammary tumors. Cancer research. 2013;73(2):745–755. doi: 10.1158/0008-5472.CAN-12-1321. [DOI] [PubMed] [Google Scholar]
- 10.Chung GT, Lou WP, Chow C, To KF. Choy KW, Leung AW, Tong CY, Yuen JW, Ko CW, Yip TT, Busson P, Lo KW. Constitutive activation of distinct NF-kappaB signals in EBV-associated nasopharyngeal carcinoma. The Journal of pathology. 2013;231(3):311–322. doi: 10.1002/path.4239. [DOI] [PubMed] [Google Scholar]
- 11.Zhao H, Wang W, Zhao Q, Hu G, Deng K, Liu Y. BCL3 exerts an oncogenic function by regulating STAT3 in human cervical cancer. OncoTargets and therapy. 2016;9:6619–6629. doi: 10.2147/OTT.S118184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dimitrakopoulos FI, Antonacopoulou AG, Kottorou A, Marousi S, Koukourikou I, Kalofonou M, Panagopoulos N, Scopa C, Dougenis D, Papadaki H, Papavassiliou AG, Kalofonos HP. Variant of BCL3 gene is strongly associated with five-year survival of non-small-cell lung cancer patients. Lung cancer. 2015;89(3):311–319. doi: 10.1016/j.lungcan.2015.06.006. [DOI] [PubMed] [Google Scholar]
- 13.Dai J, Lu Y, Wang J, Yang L, Han Y, Wang Y, Yan D, Ruan Q, Wang S (2016) A four-gene signature predicts survival in clear-cell renal-cell carcinoma. Oncotarget 7 (50):82712–82,726. 10.18632/oncotarget.12631 [DOI] [PMC free article] [PubMed]
- 14.Nguyen-Khac F, Chapiro E, Lesty C, Grelier A, Luquet I, Radford-Weiss I, Lefebvre C, Fert-Ferrer S, Callet-Bauchu E, Lippert E, Raggueneau V, Michaux L, Barin C, Collonge-Rame MA, Mugneret F, Eclache V, Taviaux S, Dastugue N, Richebourg S, Struski S, Talmant P, Baranger L, Gachard N, Gervais C, Quilichini B, Settegrana C, Maloum K, Davi F, Merle-Beral H. Specific chromosomal IG translocations have different prognoses in chronic lymphocytic leukemia. American journal of blood research. 2011;1(1):13–21. [PMC free article] [PubMed] [Google Scholar]
- 15.Schulten HJ, Bangash M, Karim S, Dallol A, Hussein D, Merdad A, Al-Thoubaity FK, Al-Maghrabi J, Jamal A, Al-Ghamdi F, Choudhry H, Baeesa SS, Chaudhary AG, Al-Qahtani MH. Comprehensive molecular biomarker identification in breast cancer brain metastases. Journal of translational medicine. 2017;15(1):269. doi: 10.1186/s12967-017-1370-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nishida Y, Takeuchi K, Tsuda K, Ugai T, Sugihara H, Yamakura M, Takeuchi M, Matsue K. Acquisition of t(11;14) in a patient with chronic lymphocytic leukemia carrying both t(14;19)(q32;q13.1) and + 12. European journal of haematology. 2013;91(2):179–182. doi: 10.1111/ejh.12119. [DOI] [PubMed] [Google Scholar]
- 17.Alpatov R, Carstens B, Harding K, Jarrett C, Balakhani S, Lincoln J, Brzeskiewicz P, Guo Y, Ohene-Mobley A, LeRoux J, McDaniel V, Meltesen L, Minka D, Patel M, Manavi C, Swisshelm K. Rare double-hit with two translocations involving IGH both, with BCL2 and BCL3, in a monoclonal B cell lymphoma/leukemia. Molecular cytogenetics. 2015;8:101. doi: 10.1186/s13039-015-0203-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin-Subero JI, Ibbotson R, Klapper W, Michaux L, Callet-Bauchu E, Berger F, Calasanz MJ, De Wolf-Peeters C, Dyer MJ, Felman P, Gardiner A, Gascoyne RD, Gesk S, Harder L, Horsman DE, Kneba M, Kuppers R, Majid A, Parry-Jones N, Ritgen M, Salido M, Sole F, Thiel G, Wacker HH, Oscier D, Wlodarska I, Siebert R. A comprehensive genetic and histopathologic analysis identifies two subgroups of B cell malignancies carrying a t(14;19)(q32;q13) or variant BCL3-translocation. Leukemia. 2007;21(7):1532–1544. doi: 10.1038/sj.leu.2404695. [DOI] [PubMed] [Google Scholar]
- 19.Nishikori M, Maesako Y, Ueda C, Kurata M, Uchiyama T, Ohno H. High-level expression of BCL3 differentiates t(2;5)(p23;q35)-positive anaplastic large cell lymphoma from Hodgkin disease. Blood. 2003;101(7):2789–2796. doi: 10.1182/blood-2002-08-2464. [DOI] [PubMed] [Google Scholar]
- 20.Slovak ML, Bedell V, Hsu YH, Estrine DB, Nowak NJ, Delioukina ML, Weiss LM, Smith DD, Forman SJ. Molecular karyotypes of Hodgkin and Reed-Sternberg cells at disease onset reveal distinct copy number alterations in chemosensitive versus refractory Hodgkin lymphoma. Clinical cancer research: an official journal of the American Association for Cancer Research. 2011;17(10):3443–3454. doi: 10.1158/1078-0432.CCR-10-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang TP, Vancurova I. Bcl3 regulates pro-survival and pro-inflammatory gene expression in cutaneous T cell lymphoma. Biochimica et biophysica acta. 2014;1843(11):2620–2630. doi: 10.1016/j.bbamcr.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ibrahim HA, Amen F, Reid AG, Naresh KN. BCL3 rearrangement, amplification and expression in diffuse large B cell lymphoma. European journal of haematology. 2011;87(6):480–485. doi: 10.1111/j.1600-0609.2011.01684.x. [DOI] [PubMed] [Google Scholar]
- 23.Saamarthy K, Bjorner S, Johansson M, Landberg G, Massoumi R, Jirstrom K, Masoumi KC. Early diagnostic value of Bcl-3 localization in colorectal cancer. BMC cancer. 2015;15:341. doi: 10.1186/s12885-015-1342-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kreisel D, Sugimoto S, Tietjens J, Zhu J, Yamamoto S, Krupnick AS, Carmody RJ, Gelman AE. Bcl3 prevents acute inflammatory lung injury in mice by restraining emergency granulopoiesis. The Journal of clinical investigation. 2011;121(1):265–276. doi: 10.1172/JCI42596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Croker BA, Metcalf D, Robb L, Wei W, Mifsud S, DiRago L, Cluse LA, Sutherland KD, Hartley L, Williams E, Zhang JG, Hilton DJ, Nicola NA, Alexander WS, Roberts AW. SOCS3 is a critical physiological negative regulator of G-CSF signaling and emergency granulopoiesis. Immunity. 2004;20(2):153–165. doi: 10.1016/S1074-7613(04)00022-6. [DOI] [PubMed] [Google Scholar]
- 26.Strauss L, Sangaletti S, Consonni FM, Szebeni G, Morlacchi S, Totaro MG, Porta C, Anselmo A, Tartari S, Doni A, Zitelli F, Tripodo C, Colombo MP, Sica A. RORC1 Regulates Tumor-Promoting “Emergency” Granulo-Monocytopoiesis. Cancer cell. 2015;28(2):253–269. doi: 10.1016/j.ccell.2015.07.006. [DOI] [PubMed] [Google Scholar]
- 27.Weyrich AS, Denis MM, Schwertz H, Tolley ND, Foulks J, Spencer E, Kraiss LW, Albertine KH, McIntyre TM, Zimmerman GA. mTOR-dependent synthesis of Bcl-3 controls the retraction of fibrin clots by activated human platelets. Blood. 2007;109(5):1975–1983. doi: 10.1182/blood-2006-08-042192. [DOI] [PMC free article] [PubMed] [Google Scholar]