Abstract
The discovery of long noncoding RNAs (lncRNA) has provided a new perspective on gene regulation in diverse biological contexts. lncRNAs are remarkably versatile molecules that interact with RNA, DNA or proteins to promote or restrain expression of protein-coding genes. Activation of immune cells is associated with dynamic changes in gene expression, the products of which combat infectious microorganisms, initiate repair and resolve inflammatory responses in cells and tissues. Recent evidence indicates that lncRNAs play important roles in directing the development of diverse immune cells and controlling the dynamic transcriptional programs that are a hallmark of immune cell activation. The importance of these molecules is underscored by their newly recognized roles in inflammatory diseases. In this review, we discuss the contribution of lncRNAs to immune cell lineage development, immune cell activation and immune related diseases. We also discuss the challenges faced in identifying biological functions for this large and complex class of genes.
INTRODUCTION
The innate and adaptive immune systems consist of diverse immune cells that collaborate to protect the host from pathogenic microorganisms. The innate immune system relies on a surveillance system of neutrophils, monocytes, macrophages, and dendritic cells which recognize and restrict pathogens and instruct the adaptive immune system. Adaptive immune cells (e.g. T and B cells) undergo somatic hypermutation, which allows them to detect specific antigens and ultimately eliminate pathogens and pathogen-infected cells. The timing of these events is carefully coordinated and involves the differentiation and activation of immune cells in response to distinct external stimuli (microbial products, cytokines or endogenous mediators)(1). Each type of immune cell expresses a specific repertoire of receptors (pattern recognition receptors, antigen receptors, cytokine receptors) that detect these stimuli, and activate downstream signaling pathways, chromatin modifying complexes, and transcription factors. These events lead to rapid and dynamic changes in gene expression that are a hallmark of activated immune cells. Recently, long noncoding RNAs (lncRNAs) have been discovered which form regulatory complexes that coordinate the development of immune cell lineages and control the gene expression programs that are unleashed in these cells. Here we review this exciting area, which emphasizes the importance of these regulatory RNAs in the immune system.
Identification and classification of lncRNAs
Noncoding RNAs (ncRNA) are non-protein coding transcripts that function as RNA molecules. lncRNAs are arbitrarily defined as non-coding RNAs that are at least 200 nucleotides, a cut-off that distinguishes lncRNAs from smaller noncoding RNAs such as tRNA, miRNA and piRNA (Piwi-interacting RNAs). Similar to mRNAs, most lncRNAs are capped, polyadenylated, and spliced (2, 3). Genome-wide transcriptome studies (RNA-seq, microarray and tilling arrays) have led to the discovery of thousands of noncoding RNAs in animals. The current estimates, as per the latest GENCODE release (version 21) (http://www.gencodegenes.org), indicate that ~ 2% of the mammalian genome is comprised of protein-coding genes, and 75–90% of the genome is transcribed as noncoding RNAs (4, 5). Most annotated lncRNAs are expressed in specific cell types and are often expressed at lower levels than protein-coding genes. lncRNAs are often classified as long intergenic ncRNAs (lincRNAs), natural antisense transcripts, (NATs), transcripts of uncertain coding potential (TUCP), enhancer RNAs (eRNAs), and pseudogene-derived lncRNAs. With the exception of lincRNAs, which are located in the intergenic region between two protein-coding genes, most other lncRNAs are located near a protein-coding gene. For example, intronic lncRNAs are transcribed from the introns of protein-coding genes and NATs are often transcribed from the opposite complementary strand of a protein-coding gene. Antisense (AS) lncRNAs are particularly common, and it is thought that up to ~ 72% of genomic loci in mice show evidence of divergent transcription leading to the generation of antisense lncRNAs (6). Figure 1 describes the genomic locations and abundance of lncRNA genes.
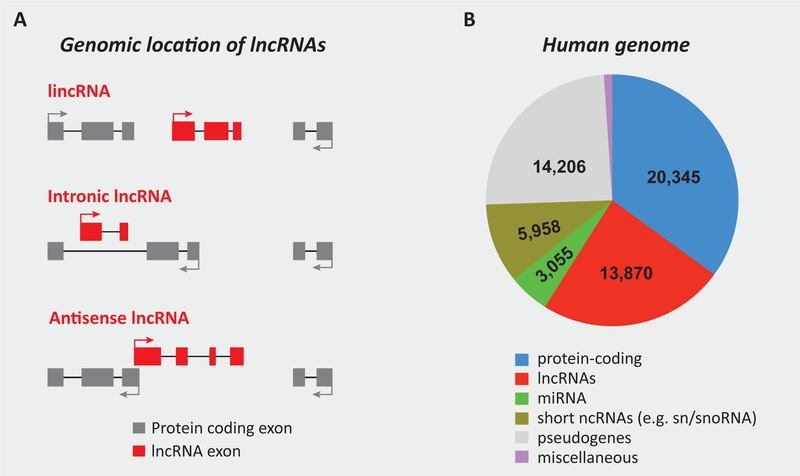
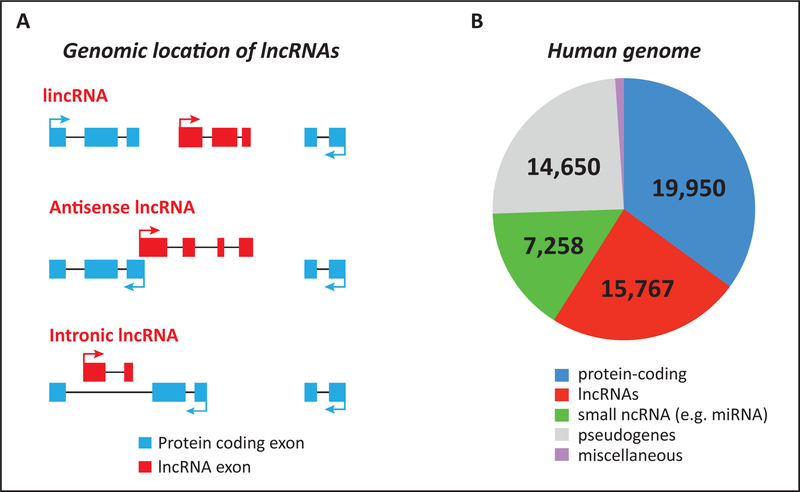
Figure 1. Classification and abundance of lncRNA genes.
(A) The classification of lncRNAs based on their genomic location with respect to nearby protein-coding genes. An intergenic lncRNA (lincRNA) is located between two protein-coding genes. All other sub-types of lncRNAs exhibit some degree of overlap with another gene located either on the same or opposite strand. Such lncRNAs may contains region(s) of complementary sequences with the mature, spliced mRNA of the overlapping protein-coding gene (antisense lncRNA), or are transcribed within the intron of a protein-coding gene, and therefore do not contain sequences complementary to the mature, spliced mRNA of the protein-coding gene. (B) The abundance of protein- and noncoding genes in the human genome. The data represents the latest GENCODE estimates (version 21) (http://www.gencodegenes.org). miRNA: micro-RNA; snRNA: small nuclear RNA; snoRNA: small nucleolar RNA.
Assessing the coding potential of lncRNAs
There are several computational methods that predict coding potential of a given lncRNA transcript using a variety of features that include homology with known proteins and phylogenetic models of codon/triplet evolution (7, 8). Purely computational methods provide a tentative assessment of coding potential that should be followed up with more robust experimental methods. The Ribosome release score (RSS) (9), uses ribosome-profiling data to measure ribosome release at stop codons. The RSS is based on the rationale that ribosome binding to an ORF will decrease abruptly at putative stop codons. Individual lncRNAs can be compared to known mRNAs using polysome profiling. Here, cells are treated with cyclohexamide, which traps ribosomes along their RNA strands and the RNA is detected in the heavier fractions of a sucrose density gradient. In parallel, cells are treated with harringtonine prior to cyclohexamide, to inhibit translation and polysome formation by causing ribosomes to accumulate at their initiation site. The latter treatment causes mRNAs to shift to lighter fractions of a sucrose gradient, whereas lncRNAs are retained in the same fraction. In vitro translation assays also represent another potential tool to assess protein or peptide coding capacity, however transcripts can produce peptides in these assays that may not be translated in a cellular environment. Certain genomic loci express a bifunctional RNA that exert biological activity as both an RNA molecule and a small peptide (10). One such bifunctional RNA is a 227-nt long RNA called SgrS (sugar-transport related small RNA) that plays an important role responding to metabolic stress in E. coli (11, 12). A clear demonstration that the biological activity of a lncRNA is driven by the RNA molecule itself should be the gold standard for the characterization of bona fide functional lncRNAs (discussed in greater detail below).
Evolution of lncRNAs
It is difficult to detect homologous lncRNAs among species that diverged more than forty million years ago (4, 13–15). Mice, the most commonly used model organism in immunology, shared a common ancestor with humans approximately 91 million years ago (16). Detecting homologous relationships between human and mouse lncRNAs is therefore challenging. It is possible that many annotated lncRNAs will not perform critical functions and consequently their sequences are not constrained during evolution (due to the absence of purifying selection). However, lncRNAs with defined biological functions are also difficult to align to homologous regions of another species genome and there are several potential reasons for this: First, the sequence constraints imposed by RNA folding are not as strict as those for protein folding. In theory, a multitude of sequences can produce the same RNA fold as long as base-complementarity is conserved. Thus, it is possible that homologous lncRNAs have undergone extensive correlated mutations at nucleotide positions that base-pair with each other. Second, it is possible that many lncRNAs are species-specific genes that have originated from scratch. These putative de novo genes might account for species-specific differences between the human and mouse immune systems. Third, repetitive elements are a major component of most lncRNAs (17, 18) and these repetitive sequences are difficult to align to the genome. Furthermore, rodents lack the primate-specific Alu element that is derived from 7SL RNA. The Alu element is the most abundant mobile element in the human genome and is found in many human lncRNAs (19). Rodent genomes contain B1 elements that are also derived from 7SL RNA and B2 elements that are derived from tRNAs. In summary, the evolutionary relationships between human and mouse lncRNAs are poorly understood and require further investigation.
Role of lncRNAs in the innate immune system
It is well established that lncRNAs coordinate diverse aspects of cell and tissue development (20). lncRNAs regulate the development of cardiomyocytes (21), stem cells (22), epithelial cells (23), erythrocytes (24, 25), and adipocytes (26). Perhaps not surprisingly, they also regulate the development and differentiation of several different immune cell lineages. Unlike miRNAs, which primarily rely on base complementarity to interact with their target RNAs, lncRNAs use a variety of mechanisms to regulate these processes. Thus, the biological function of any lncRNA needs to be interrogated on a case-by-case basis.
Myeloid and dendritic cell development
Hematopoietic stem cells (HSC) residing in the bone marrow act as the precursor to the development of most immune cells at steady state as well as during inflammatory responses. The maturation of HSCs into specific immune cell populations is directed by the stepwise and combinatorial actions of lineage-determining transcription factors like PU.1. The role of noncoding RNAs in these processes are beginning to be understood. HOTAIRM1 (HOXA transcript, antisense RNA myeloid-specific 1), an antisense lncRNA expressed in the HOXA gene locus, was the earliest lncRNA implicated in myeloid cell development. HOTAIRM1 regulates the expression of the developmentally important HOX genes (27). HOTAIRM1 is selectively expressed in myeloid cells, and its expression is upregulated during retinoic acid (RA)-induced differentiation of promyelocytic NB4 leukemia cells into granulocytes. Gene perturbation studies revealed that HOTAIRM1 controls the expression of neighboring genes HOXA1 and HOXA2, as well CD11b and CD18, which are crucial for myeloid cell differentiation. Based on these observations, and the fact that the HOXA cluster genes are involved in the regulation of normal as well as malignant hematopoiesis (28–30), HOTAIRM1 is thought to regulate the development of myeloid cells.
Another lncRNA, Morrbid (Myeloid RNA regulator of Bim-induced death), regulates the lifespan of short-lived myeloid cells (e.g. neutrophils, eosinophils and classical monocytes) in response to prosurvival cytokines (31). Morrbid mediates allele-specific transcriptional repression of its neighboring gene Bcl2l11 (Bim). Morrbid mediates these function by interacting with the polycomb repressive complex 2 (PRC2) to promote H3K27me3 modification at the Bcl2l11 promoter. These actions maintain Bcl2l11 gene in a poised state. In humans, MORRBID levels are dysregulated in patients with hypereosinophilic syndrome, a disease characterized by persistently elevated numbers of eosinophils.
A lncRNA has also been implicated in the differentiation of dendritic cells in humans (32). Classical DC (cDC; myeloid DC) act as professional antigen presenting cells (APC) to control adaptive immunity (33); whilst plasmacytoid DCs (pDCs), produce high levels of type I IFN important during viral and bacterial infections (34). lnc-DC was identified using RNA-sequencing as a non-coding RNA transcript expressed more than 100 fold higher in cDCs than monocytes (32). Expression of lnc-DC was driven by PU.1, a critical driver of DC differentiation (35). lnc-DC also contributes to the activation status of DCs by regulating STAT3 and controlling the STAT3 transcriptional program in DCs (described in more detail below). Interestingly, lnc-DC is transcribed from the Wdnm1-like pseudogene that is unique to humans (36). All other mammals appear to have an orthologous protein-coding gene that encodes a Wdnm1-like protein (36). Together, these few examples underscore the importance of lncRNAs in processes controlling the differentiation of immune cells. It is likely that additional lncRNAs coordinate the differentiation of other innate cells such as NK cells and innate lymphoid cells (ILC). A schematic detailing the role of lncRNAs in controlling the development of innate cells is shown in figure 2A.
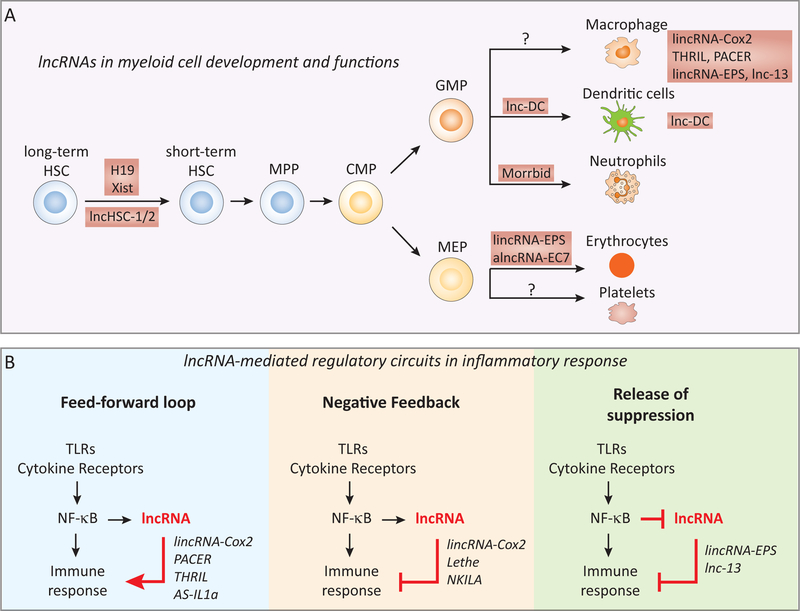
Figure 2. lncRNAs in the development and functions of myeloid cells.
(A) lncRNAs (e.g. H19, Xist, and lnc-HSC1 and lnc-HSC2) regulate the quiescence and the self-renewal of hematopoietic stem cells (HSC), and their differentiation into specific myeloid cell lineages, including DCs (lnc-DC), short-lived myeloid cells such as neutrophils, eosinophils and monocytes (Morrbid), and erythrocytes (lincRNA-EPS and alncRNA-EC7). Examples of lncRNAs that regulate the functions of specific myeloid cell lineages such as macrophages (e.g. lincRNA-Cox2, PACER, THRIL and lincRNA-EPS) are also illustrated. (B) lncRNAs represent a novel layer of the regulatory circuit that controls the inflammatory response of innate immune cells. The activation of intracellular signaling pathways (e.g. NF-κB) upon TLR or cytokine receptor engagement in innate immune cells (e.g. macrophage, DCs or epithelial cells) elaborates immune responses through a highly coordinated program of gene expression. These pathways can also induce the expression of lncRNAs in specific cell-type(s) to turn on a lncRNA-directed regulatory circuit, which acts in a “feed-forward” (e.g. PACER, THRIL and AS-IL1α), “negative feedback” (e.g. Lethe and NKILA), or “release of suppression” (e.g. lincRNA-EPS and lnc-13) manner. HSC, hematopoietic stem cell; CMP, common myeloid progenitor; GMP, granulocyte-monocyte progenitor; MEP, megakaryocyte-erythrocyte progenitor.
Regulation of myeloid and DC functions
lncRNAs have been studied most extensively in the context of TLR responses. Genome-wide transcriptome studies (RNA-seq and microarrays) have identified a large number of lncRNAs in macrophages and DCs exposed to microbial products, cytokines and other immune stimuli. lncRNAs are typically co-expressed with neighboring protein-coding genes. There is growing evidence that lncRNAs contribute in both positive and negative ways to the regulation of immune gene expression induced in cells ligated through TLRs and cytokine receptors
lincRNA-Cox2, THRIL and PACER in the induction of immune genes
One of the first examples of a lncRNA involved in the immune system came from studies in DCs and later macrophages exposed to lipopolysaccharide and other TLR ligands. TLR ligands are potent inducers of the Ptgs2 gene, which encodes cyclooxygenase-2 (COX2). Fifty kilobases upstream of Ptgs2, lincRNA-Cox2 expression is co-regulated with this protein-coding gene. Perturbations in expression of lincRNA-Cox2 using shRNA altered the expression of both basal and TLR-induced genes macrophages. lincRNA-Cox2 interacts with two heterogeneous ribonucleoprotein family members, hnRNPA/B and hnRNPA2/B1 to mediate some of these effects. More recently lincRNA-Cox2 was found to associate with the switch/sucrose nonfermentable (SWI/SNF) chromatin-remodeling complex, to control the expression of late-primary NFκB regulated genes (37). In epithelial cells, lincRNA-Cox2 engages the Mi-2/nucleosome remodeling and deacetylase (Mi2/NuRD) complex, which it helps recruit to the IL12β promoter to block IL12β expression (38). Collectively, these studies indicate that lincRNA-Cox2 helps turn on and off distinct classes of immune genes using different protein partners.
In human macrophages, the nuclear lncRNA PACER (p50-associated COX2 extragenic RNA) promotes the transcription of PTGS2 (COX2) by acting as a decoy molecule in the NF-κB signaling pathway (39). PACER is a 793 bp, single-exon lncRNA that is transcribed upstream of the PTGS2 transcription start site (TSS) in the opposite orientation. PACER is expressed in human mammary epithelial cells and PMA-activated monocytes. Upon induction by LPS and other inflammatory ligands, PACER interacts with the NF-κB repressor subunit p50, which sequester p50 from the PTGS2 promoter. This enables the active NF-κB complex to bind and induce transcription of PTGS2. This mechanism is analogous to that of the lncRNA JPX, which turns on expression of Xist during the initiation of X chromosome inactivation (XCI) in females (40). JPX interacts with the transcriptional repressor and chromatin insulator protein CTCF (CCCTC-binding factor), and titrates it away from the Xist promoter, allowing the transcriptional induction of Xist during XCI. The precise mechanisms involved in selectively recruiting PACER to the PTGS2 promoter remain to be fully elucidated. In mice, Ptgs2 divergent (Ptgs2 opposite strand; Ptgs2os) is thought to be the murine homologue of PACER based on it’s genomic location and expression profile (41). Ptgs2os is induced in mouse embryonic fibroblasts following activation of NF-κB by TLR signaling. It is not known whether Ptgs2os works in a manner similar to PACER in humans is currently unknown.
TNFα and hnRNPL related immunoregulatory lncRNA (THRIL) controls basal and TLR2-inducible expression of TNFα in human monocytes (42). THRIL interacts with hnRNPL at the promoter of TNFα to drive transcription following TLR2 activation. Knockdown of THRIL alters expression of a number of immune genes. Interestingly, the levels of THRIL are decreased in patients with Kawasaki disease, a condition that causes inflammation in the arteries supplying blood to the heart.
Another lncRNA that regulates the cytokine response in human cells is nuclear enriched abundant transcript 1 (NEAT1) (43). NEAT1 is an abundant lncRNA involved in the assembly of nuclear structures known as ‘paraspeckles’ associated with heterochromatic regions of the genome (44). NEAT1 also has immune-related functions in humans (43, 45). NEAT1 coordinates expression of IL-8 in cells infected with herpes simplex virus (HSV-1) and influenza A virus (IAV), and in response to dsRNA (43). NEAT1 promotes IL-8 transcription by relocating the transcriptional repressor SFPQ (splicing factor proline glutamine-rich) from the IL-8 promoter to the nuclear paraspeckle (43).
Antisense and enhancer lncRNAs in the regulation of cytokine genes
Antisense lncRNAs are thought to have a more restricted impact on gene expression regulating only their complementary protein-coding transcripts. This concept is clearly highlighted by the discovery of the noncoding RNAs, AS-IL1α and IL1β-RBT46, which coordinate the expression of IL-1α and IL-1β, respectively. In contrast to lincRNA-Cox2, which is believed to regulate the expression of a large number of immune genes, located on different genomic loci, AS-IL-1α appears to regulate the transcription of a single gene (IL-1α). The molecular basis for this feed-forward loop is not entirely clear. These transcripts could be part of a general regulatory mechanism that fine-tunes immune responses by titrating the expression of overlapping protein coding immune genes.
NKILA and Lethe act as negative feedback regulators of inflammation:
NKILA (NF-κB interacting lncRNA) acts in the cytosol at the post-translational level to attenuate NF-κB dependent gene expression in both resting and activated human breast cancer cells (46). NKILA expression is induced in MCF7 and MD-MB-221 cells in response to TNFα and IL-1β. NKILA then binds to the NF-κB/IκBα complex to inhibit NF-κB activation. This interaction masks residues in IκBα that are normally phosphorylated by the IκBα kinase complex (IKK) and in turn control the ubiquitin-mediated degradation of IκBα. Degradation of IκBα liberates NF-κB heterodimers to translocate to the nucleus and turn on κB target genes. Consistently, knockdown and overexpression studies indicated that NKILA restrains NF-κB driven gene expression and inflammation in breast cancer cells. Therefore, NKILA acts as a feedback negative regulator of NF-κB signaling and contributes to cancer-associated inflammation. Further, low NKILA expression is associated with increased breast cancer metastasis and poor patient prognosis. Whether NKILA restrains NF-κB signaling using similar strategies in immune cells is still unexplored.
The lncRNA Lethe is transcribed from the Rps15a-ps4 pseudogene and was found to function as a feedback negative regulator of NF-κB dependent gene expression in mouse embryonic fibroblasts (MEF) (41). Lethe is a 697 bp, unspliced RNA that is localized in the nucleus. The expression of Lethe is driven by NFκB signaling downstream of TNFα and IL-1β or by dexamethasone a glucocorticoid receptor agonist. Knockdown of Lethe by antisense oligonucleotides (ASO) leads to enhanced expression of the NF-κB dependent genes, Nfkbia and Nfkb2, in cells exposed to TNFα. Conversely, ectopic expression of Lethe interferes with NF-κB driven luciferase reporter gene activity and blocks the recruitment of NF-κB p65 to the promoters of Sod2, IL-6 and IL-8. Lethe physically associates with the NFkB p65 (RelA) subunit in the nucleus. Lethe therefore also acts as a feedback negative regulator of the NF-κB signaling pathway to restrain inflammation. It is not known whether Lethe or other pseudogene-derived lncRNAs mediate similar functions in other immune cells..
lnc-DC and regulation of STAT3 activity:
As described earlier, lnc-DC regulates the differentiation of DCs in humans (32). In addition, lnc-DC also controls the expression of genes associated with DC activation. Depletion of lnc-DC expression by shRNA impaired the uptake and processing of antigens by DCs resulting in decreased DC proliferation, activation and reduced cytokine production of allogenic CD4+ T cells in co-culture experiments. lnc-DC exerts this effect on DCs by binding to STAT3 (signal transducer and activator of transcription 3) in the cytoplasm. STAT3 drives the differentiation and activation of mDCs. To be an active signal transducer STAT-3 must be phosphorylated, which is normally regulated by the tyrosine phosphatase SHP1. lnc-DC blocks STAT3 dephosphorylation resulting in the maintenance of phospho-STAT3 and transcription of STAT3-dependent DC gene expression programs.
lincRNA-EPS and lnc-13 in the transcriptional repression of inflammation:
lincRNA-EPS (erythroid prosurvival), originally identified as a regulator of erythrocyte differentiation (24), was recently shown to function as a transcriptional brake to restrain immune gene expression in macrophages and in mice (47). lincRNA-EPS is localized in the nucleus in chromatin rich fractions. Upon TLR activation, the expression of lincRNA-EPS is reduced concomitant with the inducible transcription of immune response genes. In macrophages derived from lincRNA-EPS-deficient mice, immune response genes were found to be expressed at higher levels than those found in wild type cells. Consistently, lincRNA-EPS deficient macrophages displayed elevated levels of H3K4me3 (a mark associated with transcriptionally active/poised promoters) at the promoters of IRGs. Detailed biochemical and molecular studies demonstrated that lincRNA-EPS interacted with hnRNPL to exert these regulatory effects. Using ATAC-seq (assay for transposase accessible chromatin), which measures chromatin accessibility (48, 49), it was shown that cells lacking lincRNA-EPS had altered nucleosome positioning at the promoters of these IRG targets. lincRNA-EPS localizes at regulatory regions of IRGs to silence their expression in the absence of activating triggers. Consistent with its cellular functions in vitro, lincRNA-EPS deficient mice had more robust inflammatory responses in vivo. Together, these findings indicated lincRNA-EPS restrains the expression of IRGs in unstimulated myeloid cells by controlling chromatin accessibility.
A second lncRNA, lnc-13 (50), functions in a manner similar to lincRNA-EPS. lnc-13 was found expressed in human macrophages and is rapidly degraded upon TLR4 activation by the RNA decapping enzyme DCP2. In resting cells, lnc-13 interacts with hnRNPD and HDAC1 to suppress the transcription of immune genes. Interestingly, patients with Celiac Disease have a SNP in lnc-13 that disrupts its interaction with HDAC1. This inability to engage HDAC1 results in enhanced expression of immune genes that are a feature of Celiac Disease pathogenesis. Although lincRNA-EPS and lnc-13 control expression of different classes of immune genes, they both function by forming ribonucleoprotein complexes with hnRNP family members to subdue expression of immune genes. Several key examples highlighting the contribution of lncRNAs to the regulation of innate immune cell function are shown in Figure 2B.
Regulation of adaptive immune cells by lncRNAs
Numerous studies over the last few years have revealed that lymphocytes also express several lncRNAs that coordinate the development, differentiation and activation state of both T and B cells. Here, we review functions of lncRNAs in these cells, touching upon a few key examples to emphasize the importance of lncRNAs in T and B lymphocytes.
lncRNA transcriptomes in T cells:
The cytokine milieu coordinates the inducible expression and activation of lineage-defining transcription factor(s) involved in the differentiation of naïve T helper cells into specialized effector populations of effector cells including TH1, TH2, TH17 and regulatory T cells (Treg) (51). Numerous profiling studies have characterized the lncRNA transcriptome in T cells. Distinct T cell subsets express unique profiles of lncRNAs at different stages of their development.
lincRNA-MAF-4 in TH1 cell development:
lincRNA-MAF-4 controls the development of TH1 cells in humans through epigenetic silencing of the transcription factor c-MAF4 (52). lincRNA-MAF-4 is located ~140 kb upstream of the gene encoding c-MAF, a transcription factor involved in the differentiation of naïve CD4+ T cells to TH2 (53) and TH17 cells (54). The expression of lincRNA-MAF-4 is restricted to TH1 cells, and is not expressed in naïve T cells or other lymphoid cells. The kinetics of lincRNA-MAF-4 and MAF expression are inversely correlated in TH1 and TH2 cells in vitro, suggesting potential functional roles in the development of these helper T cells. Depletion of lincRNA-MAF-4 in naïve CD4+ T cells skews their differentiation towards TH2 cells by enhancing the expression of MAF. Chromosome conformation assays (3C), a technique that measures inter-chromosomal interactions in cross-linked cells (55), indicate that lincRNA-MAF-4 acts in cis to control long-range chromatin looping between lincRNA-MAF-4 and MAF genomic loci. Further, lincRNA-MAF-4 interacts with the transcriptional repressors LSD1 and EZH2 to deposit H3K27me3 marks at the promoter of MAF to silence its expression in TH1 cells. It is not known whether lincRNA-MAF-4 regulates other genes or the immune response in vivo.
NeST in TH1 cell function and host defense against pathogens:
Cytokines control the differentiation of naïve CD4+ T cells into T helper lineages. The lincRNA NeST (56), (originally identified as Tmevpg1 (57)) has been shown to regulate TH1 cell differentiation. NeST was identified as a gene that controls the persistence of Theiler’s virus in the central nervous system of mice. Positional cloning studies in susceptible SJL/J mice which express NeST, develop persistent Theiler’s virus infection while the resistant B10.S strain was found to lack NeST and clear Theiler’s virus infection. The generation of B10.S mice expressing a NeST transgene provided compelling genetic evidence that NeST was the host factor responsible for the persistence of Theiler’s virus (56). Tmevpg1/NeST is encoded in the IFN-γ gene locus and like IFNγ is expressed in CD4+ TH1, CD8+ T cells and NK cells (56–58). NeST expression is dependent on the transcription factors T-bet and STAT4 both of which control the differentiation of TH1 cells (58). NeST binds WDR5 (WD repeat-containing protein 5), a histone methyltransferase that deposits H3K4me3 marks (reflective of transcriptionally active promoters) at the IFN-γ promoter, to turn on transcription in CD8+ T cells (56). NeST is regarded as an enhancer RNA because of its close proximity to the IFN-γ locus, and its ability to promote IFN-γ expression in cis. However, NeST expression alone is not sufficient to drive IFN-γ expression since it requires the TH1-specific transcription factor T-bet for its function (58). The critical role of NeST in determining host susceptibility to an infectious disease was amongst the first to shed light on the importance of lncRNA genes in the immune system (Figure 3B).
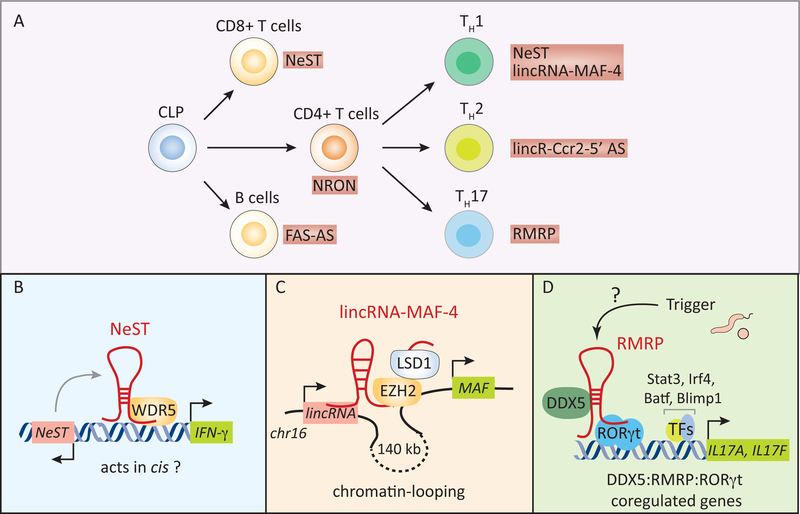
Figure 3. lncRNAs in the development and functions of lymphoid cells.
(A) lncRNAs regulate the development of specific lymphoid cells (e.g. lincRNA-MAF-4 in CD4+ TH1 cells) as well their effector functions. Examples of functional lncRNAs in B cells and T cells are shown. (B-D) The mechanism of actions of lncRNAs in CD4+ T helper cells. NeST interacts with the histone methyltransferase WDR5 to regulate the transcription of the neighboring IFN-γ gene in TH1 cells (B). lincRNA-MAF-4 silences the expression of the transcription factor c-MAF (associated with TH2 polarization) by promoting chromatin-looping between the genomic loci containing lincRNA-MAF-4 and MAF genes spaced ~140 kb apart to recruiting the PRC2 (polycomb repressive complex 2) component EZH2, and the histone demethylase LSD1 at the MAF gene locus (B). The lincRNA-MAF-4:EZH2:LSD1 complex leads to the epigenetic silencing of MAF4 expression in cis to direct the differentiation of naïve CD4+ T cells into TH1 cells. The lncRNA, RMRP, acts as a molecular scaffold to recruit DDX5 and RORγt to genes associated with the effector functions of TH17 cells (C). The RMRP:DDX5:RORγt axis regulates the transcriptional induction of a subset (e.g. IL17A and IL17F) of TH17 genes. CLP, common lymphoid progenitors; TFs, transcription factors.
lincR-Ccr2–5’ AS in TH2 cell function:
lincR-Ccr2–5’ AS is transcribed in an antisense direction immediately upstream of the TSS of the Ccr2 gene, and is selectively expressed in TH2 cells. Knockdown of this lncRNA does not impact the development of TH2 cells in vitro but leads to impaired expression of chemokine genes Ccr1, Ccr2, Ccr3 and Ccr5 located in the same genomic loci that also contains lincR-Ccr2–5’ AS. The role of lncRNAs in directing lymphocyte development and differentiation is shown in Figure 3A.
DDX5-RMRP axis in TH17 effector functions:
TH17 cells mediate pathogen-specific immunity at mucosal surfaces and contribute to the pathogenesis of multiple autoimmune diseases in humans (59). The differentiation and activation state of TH17 cells is driven by the cytokines IL-6 and TGFβ, and is dependent on the induced expression of the nuclear hormone receptor RORγt (Retinoic acid receptor-related orphan nuclear receptor gamma t). RORγt in turn controls the expression of IL-17A, IL-17F and IL-22, which play important roles in the development and function of TH17 cells (60, 61). A DEAD box RNA helicase DDX5 (p68) is an important activator of RORγt-dependent transcriptional responses in TH17 cells (62). DDX5 is a member of a large family of evolutionarily conserved RNA helicases that utilize energy generated from the hydrolysis of ATP to unwind RNAs (63). DDX5 interacts with RORγt in an RNA-dependent manner, and this complex is enriched at the promoters of RORγt-regulated genes to turn on their transcription in TH17 cells. The interaction of DDX5 with RORγt requires a lncRNA called RMRP (RNA component of the mitochondrial RNase complex) (Figure 3D). RMRP is a 274 bp unspliced transcript that is highly conserved, abundant and ubiquitously expressed. RMRP is predominantly localized in the mitochondria and nucleolus and was previously shown to function as the RNA component of the RNase complex involved in mitochondrial RNA processing, and also the processing of 5.8S pre-rRNA into the mature 5S rRNA in the nucleolus. A large number of genetic mutations in RMRP are associated with an early onset autosomal-recessive genetic disease, cartilage-hair hypoplasia (CHH). Patients with CHH display skeletal malformations and have some evidence of immune dysfunction (64). Notably, mice expressing a mutant version of RMRP (270 G > T), corresponding to a SNP in patients with CHH, had defective DDX5:RORγt-dependent effector functions in TH17 cells. The purification of endogenous RMRP in TH17 polarized cells in vitro followed by DNA-sequencing revealed that RMRP was localized to DDX5 and RORγT occupied genomic sites (e.g. IL-17A and IL-17F). Therefore, the DDX5:RMRP complex is essential for RORγt-mediated effector functions of TH17 cells. The signals driving the assembly of DDX5:RMRP complexes in these cells remains to be further elucidated. The identification of DDX5 and RMRP raise the intriguing possibility that additional RNA helicase-lncRNA complexes might function similarly to control gene expression in other immune cells.
NRON regulation of NFAT signaling in resting T cells:
NRON (noncoding RNA repressor of NFAT) is a nuclear lncRNA that restrains T cell activation by keeping the transcription factor NFAT (nuclear factor of activated T cells) in an inactive state under homeostatic conditions (65). NRON was originally identified in an in vitro shRNA screen aimed at examining the functions of 512 lncRNAs in T cell activation. NFAT is a calcium-dependent transcription factor that controls IL-2 production in activated T cells. In resting cells, NFAT is kept in an inactive, phosphorylated state. Upon T cell activation, NFAT is dephosphorylated by the Ca2+-dependent phosphatase calcineurin allowing its nuclear translocation and transcription of NFAT-response genes. NRON was shown to interfere with the nuclear translocation of NFAT by interacting with the importin-beta superfamily protein, KPNB1, which is involved in the nucleocytoplasmic shuttling of a wide-range of cargos including NFAT (66). Subsequent studies however indicate that NRON sequesters inactive NFAT in the cytoplasm via the assembly of a large cytosolic RNA-protein complex containing NRON, NFAT, IQGAP (IQ motif containing GTPase activating protein) and a number of protein kinases including LRRK2 (leucine-rich repeat kinase 2) (67). Consistent with its role in modulating T cell responses, NRON is highly expressed in peripheral lymphoid tissues (65). The precise mechanism by which NRON regulates NFAT activity during T cell activation is still open to debate. The molecular function of NRON in T cells is similar to the function of lnc-DC in DCs, as both lncRNAs help sequester inactive proteins in the cytoplasm (32). These studies highlight the ability of lncRNAs to restrain the activity of key signaling proteins in the absence of activating triggers.
The intergenic lncRNA NTT (noncoding transcript in CD4+ T cells) is another lncRNA expressed in human CD4+ T cells (68). NTT is a 17-kb, unspliced, polyadenylated RNA localized in the nucleus. Although the physiological functions of NTT are still unknown, it is co-expressed with its neighboring protein-coding gene IFN-γR (Interferon-γ receptor) in activated CD4+ T cells.
B lymphocytes.
B cells mediate long-lasting adaptive immunity to pathogens by secreting highly specific antibodies. miRNAs (e.g. miR-155 and miR-150) mediate crucial functions in the development and functions of B cells (69); however, our understanding of lncRNAs in B cells is still in its infancy. Similar to T cells, there have been several transcriptome profiling studies which have explored the expression profiles of lncRNAs in B cells in mice (70) and humans (71, 72). The transcription factor PAX5, which is crucial for B cell development, acts as an important driver of lncRNA expression in pro-B and mature B cells in mice (70). Hundreds of lncRNAs are expressed at different stages of B cell development, further underscoring the cell type specificity of lncRNA expression. One lncRNA with known functions in B cells is the antisense lncRNA, FAS-AS1, which regulates FAS receptor (CD95; TNFRSF6) signaling in B cell lymphomas (73). The engagement of FAS receptor by soluble FAS (sFAS) ligand leads to cell death via apoptosis. Not surprisingly, the development of FAS resistance in B cells is associated with uncontrolled cellular proliferation and lympho-proliferative diseases (74). FAS-AS1 binds RBM5 to block the alternative splicing of FAS pre-mRNA, which is required for the production of sFAS. Therefore, FAS-AS1 expression is inversely correlated with sFAS-induced apoptosis in B cell lymphomas. Since serum sFas levels are associated with poor prognosis in non-Hodgkin’s lymphoma (75), the Fas-AS1 lncRNA could represent a potential therapeutic target in this setting.
lncRNA transcription is also associated with the development and activation status of B cells (76, 77). B cells acquire immunoglobulin receptor during their development in the bone marrow through a process known as V(D)J recombination. In peripheral lymphoid organs, activation of B cells during inflammation or infection is mediated through the enzyme AID (activation-induced deaminase), which controls two crucial processes, namely somatic hypermutation and class switch recombination (78). These processes are crucial for the generation of a vast repertoire of highly specific antibodies by B cells that regulate the humoral arm of adaptive immunity. lncRNA transcription (76, 77) as well as the exosome-dependent regulation of lncRNA abundance (79) are thought to regulate AID-mediated somatic hypermutation and class switch recombination.
lncRNAs in Host Defense against Pathogens.
NEAT1 has been shown to enhance HIV1 replication by transporting newly synthesized HIV-1 mRNA from the nucleus to the cytoplasm (45). Furthermore, the antisense lncRNA NRAV (negative regulator of antiviral response; also known as DYNLL1-AS1) is involved in antiviral immunity (80). NRAV was identified as a lncRNA expressed in human alveolar epithelial A549 cells infected with IAV. A series of functional screening assays examined the potential role of 9 candidate lncRNA genes as regulators of IAV replication in human cells. In humans NRAV is a 1,177 bp long, spliced, polyadenylated and nuclear-localized RNA. The NRAV gene overlaps with the first intron of the protein-coding gene DYNLL1 (dynein light chain 1), and is transcribed in the antisense orientation. NRAV expression in human cells is downregulated by several viruses including IAV. Ectopic expression of NRAV in human cells or mice enhanced IAV replication and virulence suggesting that the downregulation of NRAV is an important component of the host antiviral immune system to efficiently control viral replication. NRAV mediates these functions by repressing the transcription of several ISGs including IFITM3 and MxA by modulating histone modifications at the promoters of these genes. NRAV interacts with ZONAB (ZO-1-associated Y-box factor), a known transcriptional repressor associated with proliferation of epithelial cells. Knockdown of ZONAB leads to elevated levels of ISG expression in IAV-infected A549 cells indicating the functional importance of the NRAV:ZONAB interaction. However, is not known how these complexes regulate gene transcription. Although hundreds of additional lncRNAs are differentially expressed in response to infection with respiratory viruses such as coronavirus and IAV in vivo (81), their functions in anti-viral immunity are presently unknown.
Microbe encoded lncRNAs
There is a constant evolutionary arms race between pathogens and their hosts. The host is equipped with the ability to detect and halt the replication and growth of a pathogen by mounting rapid inflammatory responses. Likewise a successful pathogen must evade, subvert or redirect these responses if they are to establish an infection and ensure their survival by keeping the host alive. Accordingly successful pathogens have evolved strategies to disrupt host immunity. It is not too surprising that several microbial species express lncRNAs, which in some cases subvert host immunity (82). For example, Kaposi’s sarcoma-associated herpesvirus (KSHV) expresses polyadenylated nuclear (PAN) RNA that is functionally linked with the conversion of latent to lytic (active) viral infection. Although the underlying molecular basis of these observations is unclear, it is thought that PAN RNA acts by either facilitating the dissociation of LANA (latency associated nuclear antigen) from the KSHV genome, or by recruiting the demethylase proteins, JMJD3 and UTX, to epigenetically repressed regions of the KSHV genome to facilitate the expression of the KSHV genome. The KSHV PAN lncRNA has also been shown to suppress the expression of host antiviral genes (e.g. IFN-α, IFN-γ and RNaseL) through its interaction with PRC2 leading to the subversion of host immune responses, and enhanced viral replication in infected cells. Herpesviruses encode HSUR1 and HSUR2 (herpesvirus saimiri U-rich ncRNAs) that target miR-27 and miR-16 respectively to alter their cellular functions (83). An antisense lncRNA transcribed from the 5’ long terminal repeat (LTR) promoter of HIV1 suppresses the transcription of viral genes by recruiting Dnmt3a, HDAC1 and EZH2 to form a transcriptional repressor complex (84). Plasmodium falciparum, the causative agent of malaria also encodes approximately 60 lncRNAs (85). These include 22 telomere-associated lncRNAs that are specifically enriched in repetitive sequences. The functions of these lncRNAs during parasite infection are yet to be determined.
Common mechanisms of action - Defined Themes
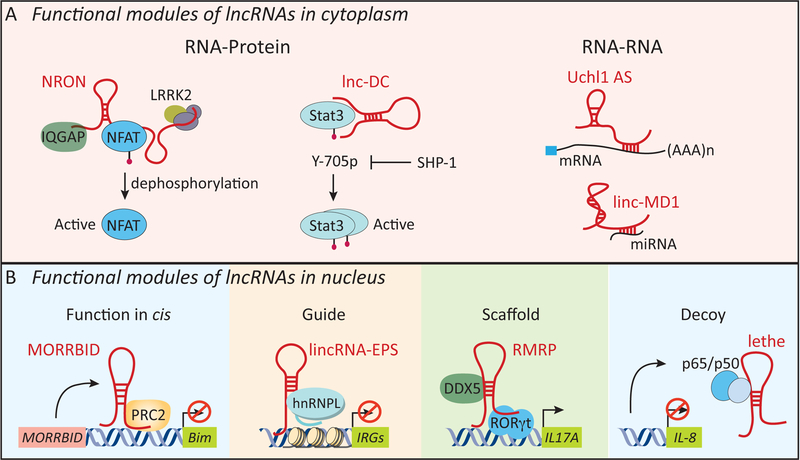
lncRNAs are tremendously versatile molecules and work through RNA-protein, RNA-RNA or RNA-DNA interactions (Figure 4). Based on the cellular location of lncRNAs, such modes of action are restricted to either the cytosol or the nucleus. lncRNAs can target all levels of gene regulation including transcription, mRNA stability and translation. In the cytosol, lncRNAs are known to interact with RNAs or proteins to carry out their molecular functions. Some lncRNAs base pair with mRNAs and this interaction can lead to altered levels of these mRNAs. For example, the lincRNA linc-MD1 expressed in skeletal muscle acts as a competing endogenous lncRNA (ceRNA) to sponge miR-133 and regulate muscle cell differentiation (86). The antisense lncRNA Uchl1-AS promotes the translation of Uchl1 mRNA by enhancing it’s association with polysomes (87). In contrast, certain lncRNAs like lincRNA-p21 base pair with target mRNAs to repress their translation (88). The more common mode of lncRNA action however involves interactions with specific protein(s). In the immune system, several lncRNAs interact with transcription factors. These include lnc-DC which binds STAT3 in DCs (32), NRON which binds NFAT1 in T cells (65) and Lethe and NKILA which complex with NF-κB (46) keeping the activities of these factors in-check in the absence of activating triggers. In the nucleus, lncRNAs regulate gene expression through a multitude of mechanisms. Here, we discuss examples of nuclear localized lncRNAs and their impact on immune gene expression.
Figure 4. Mechanism of action for lncRNAs.
lncRNAs mediate their molecular functions through a multitude of mechanisms in cytoplasm (A) or the nucleus (B). In the cytoplasm, lncRNAs act through RNA-protein (e.g. NRON and lnc-DC) or RNA-RNA (e.g. Uchl1 AS and linc-MD1) interactions. NRON and lnc-DC act as molecular scaffolds for the transcription factors, NFAT and STAT3. Uchl1 AS interacts with target mRNAs through base-paring to enhance their translation. In the nucleus, lncRNAs can act in cis or trans (B). Morrbid interacts with PRC2 to repress the transcription of the neighboring gene, Bim (Bcl2l11), in cis in short-lived myeloid cells such as neutrophils and monocytes. lncRNAs can interact with their protein partners as guide (e.g. lincRNA-EPS: hnRNPL), scaffold (e.g. RMRP interaction with DDX5 and RORγt) or a decoy molecule (e.g. Lethe: NF-κB p65) to mediate their molecular functions. Uchl1, ubiquitin carboxy-terminal hydrolase L1; lMD1: muscle differentiation 1.
lncRNAs as modular guides and scaffolds for proteins:
lncRNAs can recruit proteins or RNAs and these complexes in turn assemble higher order protein/RNA complexes in cells. For example, HOTAIR employs two protein-interacting modules located on its 5’- and 3’ ends to recruit PRC2 and LSD1 (89)- these proteins cooperate with each other to deposit repressive histone marks to silence gene expression at target gene loci. RMRP exhibits a similar mode of action wherein it interacts with DDX5 and RORγt to control the transcriptional program in TH17 cells (62). Because lncRNAs guide protein(s) to specific genomic loci or act as molecular scaffolds to stabilize protein complexes, it is possible that lncRNAs also contribute to the functional diversity of DNA-binding proteins. Specific lncRNAs could base-pair with either DNA or nascent RNA to facilitate recruitment and/or activity of DNA-binding proteins.
lncRNA as signaling molecules.
lncRNAs can also modulate signaling pathways to alter the expression of immune genes. NRON provides an excellent example of such a mechanism (67). In CD8+ T cells, NRON supports a molecular scaffold to sequester NFAT in an inactive state in the cytosol. Similar to NRON, two other lncRNAs, lnc-DC (32) and NKILA (46), have been shown to scaffold STAT3 and NF-κB respectively to retain these factors in an inactive state in the cytosol. Whether lncRNAs represent a general theme by which signaling pathways are regulated remains to be explored further. Exosomes package a variety of RNA species, including lncRNAs, which have the capacity to mediate effects on cells distal to the site of their synthesis. For example, one scenario could be that the packaged lncRNA molecules in RNA exosomes are delivered to bystander or even distantly located cells to modulate signaling pathways, transcription factors or gene expression.
Defining RNA functions – Genetic tools and caveats
Determining how a particular lncRNA influences gene expression remains a major challenge since there are many potential ways these RNAs can function. Although many lncRNAs are reported to function in the immune system, only a handful of lncRNAs have detailed biochemical and genetic evidence supporting their functions. Convincingly demonstrating that a lncRNA functions as an RNA molecule is essential. Genetic tools including the insertion of poly-A termination sites to disrupt the transcription of the lncRNA gene, CRISPR-Cas9 system to overexpress a lncRNA from its endogenous locus or to generate knockout cells, and rescue of lncRNA functions in genetic knockout cells through ectopic expression are some of the powerful approaches that can be used to address these questions. lncRNAs can act in cis to control local allele-specific functions, or act in trans at one or more genomic loci to regulate gene expression. In general, antisense and enhancer lncRNAs act in cis to control expression of neighboring gene loci. Some lncRNAs, such as HOTAIR (90) and lincRNA-EPS (47) however clearly alter distant gene loci in trans.
RNA-directed technologies to probe lncRNA function.
Advances in RNA-sequencing efforts have provided insights into lncRNA expression across diverse cell-types and tissues. Additional RNA-centric tools are now needed to elucidate the molecular functions of lncRNAs by interrogating their biochemical partners (e.g. proteins and RNAs), genomic localization and secondary structures. Novel approaches described over the last few years including ChIRP (chromatin isolation by RNA purification) (91, 92), RAP (RNA antisense purification) (93–95) and CHART (capture hybridization analysis of RNA targets) (96, 97) enable purification of endogenous lncRNAs and identification of their interactomes under native conditions. These approaches have defined lncRNA-bound protein(s), RNAs and genome-wide binding sites for some of the most highly expressed lncRNAs. The development of new as well further refinement of existing RNA technologies will be essential to advance our understanding of lncRNA function.
Human Diseases and lncRNA.
Unchecked immune activation is linked to the pathophysiology of human diseases ranging from diabetes to cancer. Genome-wide association studies (GWAS) and exome-sequencing have identified that the majority of disease-associated genetic polymorphisms (single nucleotide polymorphism; SNP) are located in noncoding regions of the genome. In particular, only ~7% of disease-associated SNPs identified-to-date are localized to protein-coding genes (98), whereas the vast majority of these SNPs are localized to the noncoding regions of the genome including genomic loci expressing lncRNAs (99). It should be noted that the majority of identified SNPs fail to explain the variance in their respective studies. Thus, the genetics of most diseases is complex and difficult to determine. Nevertheless, these genetic variations could affect the expression and/or function of lncRNAs that are relevant in human physiology and disease. Recent studies suggest that altered expression of lncRNAs occur in a number of immune-related diseases such as inflammatory bowel disease (IBD), diabetes and multiple sclerosis (100). Moreover, the discovery of lnc13 as a regulator of inflammatory genes and SNPs associated with Celiac disease further underscore the potential roles of lncRNAs in disease.
Aberrant expression and/or function of lncRNAs has been linked to the progression of hematological malignancies. A comprehensive transcriptome analysis of human T cell acute lymphoblastic leukemia (T-ALL) identified more than 1,000 lncRNAs that were selectively expressed in T-ALL leukemia cells but not in untransformed T cells (101). One such lncRNA, LUNAR1 (leukemia-induced noncoding activator RNA), is highly expressed in T-ALL cells downstream of the oncogenic NOTCH1 receptor. LUNAR1 promotes leukemogenesis by regulating the expression of IGFR1 (Insulin growth factor receptor 1), a receptor that is critical for leukemia initiating activity of T-ALL cells (102). Analysis of higher-order chromatin structure by Hi-C demonstrated a close nuclear association between LUNAR1 and the neighboring IGFR1 gene loci. Chromatin-looping of LUNAR1 and IGFR1 gene loci was shown to control IGFR1 expression, which further highlights the ability of lncRNAs in maintaining higher-order nuclear structures containing functionally linked TADs. In addition, dysregulation of Xist mediated XCI is also associated with myeloid cell malignancies. The casual relationship between Xist and cancer was demonstrated by the conditional deletion of Xist in HSC in mice, which showed that deletion of Xist led to a de-repression of gene expression from the X chromosome. Further, expression profiles of lncRNAs might help in disease diagnosis, classification as well as markers for therapeutic intervention.
CONCLUDING REMARKS
Long noncoding RNAs regulate gene expression in many contexts. As detailed in this review, lncRNAs are expressed in many different classes of immune cells with broad functionality in controlling the development, differentiation and effector functions of these cells. While there has been considerable progress made in describing the expression profiles of lncRNAs in specific cell types, elaboration of lncRNA function in immune cells has lagged far behind. Several major issues must be resolved to fully appreciate the spectrum of lncRNA functions in the immune system. There remains a huge gap in our understanding of lncRNA functions and their molecular mechanisms. Development of new as well as the further refinement of existing experimental and computational technologies will be instrumental in advancing our understanding of the biology of lncRNAs. RNA-directed technologies (e.g. ChIRP and RAP) will help uncover lncRNA interactomes including protein partners and genomic binding sites. In addition, lessons learned from different biological contexts will need to be merged to generate unified concepts and paradigms of lncRNA functionality. The mammalian immune system is extremely complex and lncRNAs represent an additional layer of complexity. It is important that we understand the function of lncRNAs in the immune systems of model organisms and humans.
SUMMARY POINTS.
lncRNAs exhibit striking cell-type specific expression profiles.
lncRNAs control the development of specific immune cell-types including DCs (lnc-DC), neutrophils, eosinophils and monocytes (MORRBID), and CD4+ TH1 cells (lincRNA-MAF-4).
lncRNAs regulate transcriptional programs of innate immune cells (lincRNA-EPS, lnc-13, lincRNA-Cox2, THRIL and NKILA).
lncRNAs act locally (e.g. MORRBID and lincRNA-MAF-4) to impact expression of neighboring genes or globally to regulate multiple genes across the genome (e.g. RMRP and lincRNA-EPS).
The majority of disease-associated SNPs identified in GWAS studies are located in noncoding regions of the human genome.
FUTURE ISSUES.
Elucidating the in vivo functions of lncRNAs is a major challenge. Currently, only a few lncRNAs including NeST, lincRNA-EPS and RMRP have demonstrated in vivo functions in the immune system.
Assessment of the functional consequences of disease-associated SNPs in noncoding regions of the genome and lncRNA expression or function is required.
The biochemical and molecular basis of lncRNA function in the immune system required to define unifying principles of lncRNA-dependent regulation of gene expression in immunity.
A comprehensive map of lncRNAs that play essential roles in immune cell development and functions during homeostasis and infections remains to be fully elucidated.
Determinations of RNA functions versus the role of functional DNA elements in studies using lncRNA knockout animals.
Better guidelines for the annotation of lncRNA genes and more stringent analysis for the discovery of functional lncRNAs.
BIBLIOGRAPHY
- 1.Medzhitov R, Horng T. 2009. Transcriptional control of the inflammatory response. Nat Rev Immunol 9: 692–703 [DOI] [PubMed] [Google Scholar]
- 2.Ulitsky I, Bartel DP. 2013. lincRNAs: genomics, evolution, and mechanisms. Cell 154: 26–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rinn JL, Chang HY. 2012. Genome regulation by long noncoding RNAs. Annu Rev Biochem 81: 145–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, Lagarde J, Veeravalli L, Ruan X, Ruan Y, Lassmann T, Carninci P, Brown JB, Lipovich L, Gonzalez JM, Thomas M, Davis CA, Shiekhattar R, Gingeras TR, Hubbard TJ, Notredame C, Harrow J, Guigo R. 2012. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 22: 1775–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, Barnes I, Bignell A, Boychenko V, Hunt T, Kay M, Mukherjee G, Rajan J, Despacio-Reyes G, Saunders G, Steward C, Harte R, Lin M, Howald C, Tanzer A, Derrien T, Chrast J, Walters N, Balasubramanian S, Pei B, Tress M, Rodriguez JM, Ezkurdia I, van Baren J, Brent M, Haussler D, Kellis M, Valencia A, Reymond A, Gerstein M, Guigo R, Hubbard TJ. 2012. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res 22: 1760–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Werner A, Carlile M, Swan D. 2009. What do natural antisense transcripts regulate? RNA Biol 6: 43–8 [DOI] [PubMed] [Google Scholar]
- 7.Lin MF, Jungreis I, Kellis M. 2011. PhyloCSF: a comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics 27: i275–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kong L, Zhang Y, Ye ZQ, Liu XQ, Zhao SQ, Wei L, Gao G. 2007. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res 35: W345–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guttman M, Russell P, Ingolia NT, Weissman JS, Lander ES. 2013. Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell 154: 240–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dinger ME, Pang KC, Mercer TR, Mattick JS. 2008. Differentiating protein-coding and noncoding RNA: challenges and ambiguities. PLoS Comput Biol 4: e1000176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wadler CS, Vanderpool CK. 2007. A dual function for a bacterial small RNA: SgrS performs base pairing-dependent regulation and encodes a functional polypeptide. Proc Natl Acad Sci U S A 104: 20454–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morita T, Aiba H. 2007. Small RNAs making a small protein. Proc Natl Acad Sci U S A 104: 20149–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Washietl S, Kellis M, Garber M. 2014. Evolutionary dynamics and tissue specificity of human long noncoding RNAs in six mammals. Genome Res 24: 616–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wiberg RA, Halligan DL, Ness RW, Necsulea A, Kaessmann H, Keightley PD. 2015. Assessing Recent Selection and Functionality at Long Noncoding RNA Loci in the Mouse Genome. Genome Biol Evol 7: 2432–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Necsulea A, Soumillon M, Warnefors M, Liechti A, Daish T, Zeller U, Baker JC, Grutzner F, Kaessmann H. 2014. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature 505: 635–40 [DOI] [PubMed] [Google Scholar]
- 16.Hedges SB. 2002. The origin and evolution of model organisms. Nat Rev Genet 3: 838–49 [DOI] [PubMed] [Google Scholar]
- 17.Kapusta A, Kronenberg Z, Lynch VJ, Zhuo X, Ramsay L, Bourque G, Yandell M, Feschotte C. 2013. Transposable elements are major contributors to the origin, diversification, and regulation of vertebrate long noncoding RNAs. PLoS Genet 9: e1003470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kelley D, Rinn J. 2012. Transposable elements reveal a stem cell-specific class of long noncoding RNAs. Genome Biol 13: R107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim EZ, Wespiser AR, Caffrey DR. 2016. The domain structure and distribution of Alu elements in long noncoding RNAs and mRNAs. RNA 22: 254–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu W, Alvarez-Dominguez JR, Lodish HF. 2012. Regulation of mammalian cell differentiation by long non-coding RNAs. EMBO Rep 13: 971–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grote P, Wittler L, Hendrix D, Koch F, Wahrisch S, Beisaw A, Macura K, Blass G, Kellis M, Werber M, Herrmann BG. 2013. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell 24: 206–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, Yang X, Amit I, Meissner A, Regev A, Rinn JL, Root DE, Lander ES. 2011. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 477: 295–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kretz M, Siprashvili Z, Chu C, Webster DE, Zehnder A, Qu K, Lee CS, Flockhart RJ, Groff AF, Chow J, Johnston D, Kim GE, Spitale RC, Flynn RA, Zheng GX, Aiyer S, Raj A, Rinn JL, Chang HY, Khavari PA. 2013. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature 493: 231–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu W, Yuan B, Flygare J, Lodish HF. 2011. Long noncoding RNA-mediated anti-apoptotic activity in murine erythroid terminal differentiation. Genes Dev 25: 2573–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alvarez-Dominguez JR, Hu W, Yuan B, Shi J, Park SS, Gromatzky AA, van Oudenaarden A, Lodish HF. 2014. Global discovery of erythroid long noncoding RNAs reveals novel regulators of red cell maturation. Blood 123: 570–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun L, Goff LA, Trapnell C, Alexander R, Lo KA, Hacisuleyman E, Sauvageau M, Tazon-Vega B, Kelley DR, Hendrickson DG, Yuan B, Kellis M, Lodish HF, Rinn JL. 2013. Long noncoding RNAs regulate adipogenesis. Proc Natl Acad Sci U S A 110: 3387–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang X, Lian Z, Padden C, Gerstein MB, Rozowsky J, Snyder M, Gingeras TR, Kapranov P, Weissman SM, Newburger PE. 2009. A myelopoiesis-associated regulatory intergenic noncoding RNA transcript within the human HOXA cluster. Blood 113: 2526–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bei L, Lu Y, Bellis SL, Zhou W, Horvath E, Eklund EA. 2007. Identification of a HoxA10 activation domain necessary for transcription of the gene encoding beta3 integrin during myeloid differentiation. J Biol Chem 282: 16846–59 [DOI] [PubMed] [Google Scholar]
- 29.Eklund EA. 2006. The role of HOX genes in myeloid leukemogenesis. Curr Opin Hematol 13: 67–73 [DOI] [PubMed] [Google Scholar]
- 30.Rice KL, Licht JD. 2007. HOX deregulation in acute myeloid leukemia. J Clin Invest 117: 865–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kotzin JJ, Spencer SP, McCright SJ, Kumar DB, Collet MA, Mowel WK, Elliott EN, Uyar A, Makiya MA, Dunagin MC, Harman CC, Virtue AT, Zhu S, Bailis W, Stein J, Hughes C, Raj A, Wherry EJ, Goff LA, Klion AD, Rinn JL, Williams A, Flavell RA, Henao-Mejia J. 2016. The long non-coding RNA Morrbid regulates Bim and short-lived myeloid cell lifespan. Nature [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang P, Xue Y, Han Y, Lin L, Wu C, Xu S, Jiang Z, Xu J, Liu Q, Cao X. 2014. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science 344: 310–3 [DOI] [PubMed] [Google Scholar]
- 33.Steinman RM, Hemmi H. 2006. Dendritic cells: translating innate to adaptive immunity. Curr Top Microbiol Immunol 311: 17–58 [DOI] [PubMed] [Google Scholar]
- 34.Schmidt SV, Nino-Castro AC, Schultze JL. 2012. Regulatory dendritic cells: there is more than just immune activation. Front Immunol 3: 274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murphy KM. 2013. Transcriptional control of dendritic cell development. Adv Immunol 120: 239–67 [DOI] [PubMed] [Google Scholar]
- 36.Dijkstra JM, Ballingall KT. 2014. Non-human lnc-DC orthologs encode Wdnm1-like protein. F1000Res 3: 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu G, Gong AY, Wang Y, Ma S, Chen X, Chen J, Su CJ, Shibata A, Strauss-Soukup JK, Drescher KM, Chen XM. 2016. LincRNA-Cox2 Promotes Late Inflammatory Gene Transcription in Macrophages through Modulating SWI/SNF-Mediated Chromatin Remodeling. J Immunol 196: 2799–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tong Q, Gong AY, Zhang X, Lin C, Ma S, Chen J, Hu G, Chen XM. 2015. LincRNA-Cox2 modulates TNF-alpha-induced transcription of Il12b gene in intestinal epithelial cells through regulation of Mi-2/NuRD-mediated epigenetic histone modifications. FASEB J [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krawczyk M, Emerson BM. 2014. p50-associated COX-2 extragenic RNA (PACER) activates COX-2 gene expression by occluding repressive NF-kappaB complexes. Elife 3: e01776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun S, Del Rosario BC, Szanto A, Ogawa Y, Jeon Y, Lee JT. 2013. Jpx RNA activates Xist by evicting CTCF. Cell 153: 1537–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rapicavoli NA, Qu K, Zhang J, Mikhail M, Laberge RM, Chang HY. 2013. A mammalian pseudogene lncRNA at the interface of inflammation and anti-inflammatory therapeutics. Elife 2: e00762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Z, Chao TC, Chang KY, Lin N, Patil VS, Shimizu C, Head SR, Burns JC, Rana TM. 2014. The long noncoding RNA THRIL regulates TNFalpha expression through its interaction with hnRNPL. Proc Natl Acad Sci U S A 111: 1002–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hirose T, Virnicchi G, Tanigawa A, Naganuma T, Li R, Kimura H, Yokoi T, Nakagawa S, Benard M, Fox AH, Pierron G. 2014. NEAT1 long noncoding RNA regulates transcription via protein sequestration within subnuclear bodies. Mol Biol Cell 25: 169–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB. 2009. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell 33: 717–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Q, Chen CY, Yedavalli VS, Jeang KT. 2013. NEAT1 long noncoding RNA and paraspeckle bodies modulate HIV-1 posttranscriptional expression. MBio 4: e00596–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu B, Sun L, Liu Q, Gong C, Yao Y, Lv X, Lin L, Yao H, Su F, Li D, Zeng M, Song E. 2015. A cytoplasmic NF-kappaB interacting long noncoding RNA blocks IkappaB phosphorylation and suppresses breast cancer metastasis. Cancer Cell 27: 370–81 [DOI] [PubMed] [Google Scholar]
- 47.Atianand MK, Hu W, Satpathy AT, Shen Y, Ricci EP, Alvarez-Dominguez JR, Bhatta A, Schattgen SA, McGowan JD, Blin J, Braun JE, Gandhi P, Moore MJ, Chang HY, Lodish HF, Caffrey DR, Fitzgerald KA. 2016. A Long Noncoding RNA lincRNA-EPS Acts as a Transcriptional Brake to Restrain Inflammation. Cell 165: 1672–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. 2013. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 10: 1213–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schep AN, Buenrostro JD, Denny SK, Schwartz K, Sherlock G, Greenleaf WJ. 2015. Structured nucleosome fingerprints enable high-resolution mapping of chromatin architecture within regulatory regions. Genome Res 25: 1757–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Castellanos-Rubio A, Fernandez-Jimenez N, Kratchmarov R, Luo X, Bhagat G, Green PH, Schneider R, Kiledjian M, Bilbao JR, Ghosh S. 2016. A long noncoding RNA associated with susceptibility to celiac disease. Science 352: 91–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nakayamada S, Takahashi H, Kanno Y, O’Shea JJ. 2012. Helper T cell diversity and plasticity. Curr Opin Immunol 24: 297–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ranzani V, Rossetti G, Panzeri I, Arrigoni A, Bonnal RJ, Curti S, Gruarin P, Provasi E, Sugliano E, Marconi M, De Francesco R, Geginat J, Bodega B, Abrignani S, Pagani M. 2015. The long intergenic noncoding RNA landscape of human lymphocytes highlights the regulation of T cell differentiation by linc-MAF-4. Nat Immunol 16: 318–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ho IC, Lo D, Glimcher LH. 1998. c-maf promotes T helper cell type 2 (Th2) and attenuates Th1 differentiation by both interleukin 4-dependent and -independent mechanisms. J Exp Med 188: 1859–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sato K, Miyoshi F, Yokota K, Araki Y, Asanuma Y, Akiyama Y, Yoh K, Takahashi S, Aburatani H, Mimura T. 2011. Marked induction of c-Maf protein during Th17 cell differentiation and its implication in memory Th cell development. J Biol Chem 286: 14963–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miele A, Dekker J. 2009. Mapping cis- and trans- chromatin interaction networks using chromosome conformation capture (3C). Methods Mol Biol 464: 105–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gomez JA, Wapinski OL, Yang YW, Bureau JF, Gopinath S, Monack DM, Chang HY, Brahic M, Kirkegaard K. 2013. The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-gamma locus. Cell 152: 743–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vigneau S, Rohrlich PS, Brahic M, Bureau JF. 2003. Tmevpg1, a candidate gene for the control of Theiler’s virus persistence, could be implicated in the regulation of gamma interferon. J Virol 77: 5632–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Collier SP, Collins PL, Williams CL, Boothby MR, Aune TM. 2012. Cutting Edge: Influence of Tmevpg1, a Long Intergenic Noncoding RNA, on the Expression of Ifng by Th1 Cells. J Immunol [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Patel DD, Kuchroo VK. 2015. Th17 Cell Pathway in Human Immunity: Lessons from Genetics and Therapeutic Interventions. Immunity 43: 1040–51 [DOI] [PubMed] [Google Scholar]
- 60.Ivanov II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, Cua DJ, Littman DR. 2006. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell 126: 1121–33 [DOI] [PubMed] [Google Scholar]
- 61.Korn T, Bettelli E, Oukka M, Kuchroo VK. 2009. IL-17 and Th17 Cells. Annu Rev Immunol 27: 485–517 [DOI] [PubMed] [Google Scholar]
- 62.Huang W, Thomas B, Flynn RA, Gavzy SJ, Wu L, Kim SV, Hall JA, Miraldi ER, Ng CP, Rigo FW, Meadows S, Montoya NR, Herrera NG, Domingos AI, Rastinejad F, Myers RM, Fuller-Pace FV, Bonneau R, Chang HY, Acuto O, Littman DR. 2015. DDX5 and its associated lncRNA Rmrp modulate TH17 cell effector functions. Nature 528: 517–22 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 63.Linder P, Jankowsky E. 2011. From unwinding to clamping - the DEAD box RNA helicase family. Nat Rev Mol Cell Biol 12: 505–16 [DOI] [PubMed] [Google Scholar]
- 64.Martin AN, Li Y. 2007. RNase MRP RNA and human genetic diseases. Cell Res 17: 219–26 [DOI] [PubMed] [Google Scholar]
- 65.Willingham AT, Orth AP, Batalov S, Peters EC, Wen BG, Aza-Blanc P, Hogenesch JB, Schultz PG. 2005. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science 309: 1570–3 [DOI] [PubMed] [Google Scholar]
- 66.Gorlich D, Kutay U. 1999. Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol 15: 607–60 [DOI] [PubMed] [Google Scholar]
- 67.Sharma S, Findlay GM, Bandukwala HS, Oberdoerffer S, Baust B, Li Z, Schmidt V, Hogan PG, Sacks DB, Rao A. 2011. Dephosphorylation of the nuclear factor of activated T cells (NFAT) transcription factor is regulated by an RNA-protein scaffold complex. Proc Natl Acad Sci U S A 108: 11381–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu AY, Torchia BS, Migeon BR, Siliciano RF. 1997. The human NTT gene: identification of a novel 17-kb noncoding nuclear RNA expressed in activated CD4+ T cells. Genomics 39: 171–84 [DOI] [PubMed] [Google Scholar]
- 69.Mehta A, Baltimore D. 2016. MicroRNAs as regulatory elements in immune system logic. Nat Rev Immunol 16: 279–94 [DOI] [PubMed] [Google Scholar]
- 70.Brazao TF, Johnson JS, Muller J, Heger A, Ponting CP, Tybulewicz VL. 2016. Long non-coding RNAs in B cell development and activation. Blood [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tayari MM, Winkle M, Kortman G, Sietzema J, de Jong D, Terpstra M, Mestdagh P, Kroese FG, Visser L, Diepstra A, Kok K, van den Berg A, Kluiver J. 2016. Long Noncoding RNA Expression Profiling in Normal B-Cell Subsets and Hodgkin Lymphoma Reveals Hodgkin and Reed-Sternberg Cell-Specific Long Noncoding RNAs. Am J Pathol 186: 2462–72 [DOI] [PubMed] [Google Scholar]
- 72.Bonnal RJ, Ranzani V, Arrigoni A, Curti S, Panzeri I, Gruarin P, Abrignani S, Rossetti G, Pagani M. 2015. De novo transcriptome profiling of highly purified human lymphocytes primary cells. Sci Data 2: 150051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sehgal L, Mathur R, Braun FK, Wise JF, Berkova Z, Neelapu S, Kwak LW, Samaniego F. 2014. FAS-antisense 1 lncRNA and production of soluble versus membrane Fas in B-cell lymphoma. Leukemia [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rudin CM, Thompson CB. 1997. Apoptosis and disease: regulation and clinical relevance of programmed cell death. Annu Rev Med 48: 267–81 [DOI] [PubMed] [Google Scholar]
- 75.Niitsu N, Sasaki K, Umeda M. 1999. A high serum soluble Fas/APO-1 level is associated with a poor outcome of aggressive non-Hodgkin’s lymphoma. Leukemia 13: 1434–40 [DOI] [PubMed] [Google Scholar]
- 76.Bolland DJ, Wood AL, Johnston CM, Bunting SF, Morgan G, Chakalova L, Fraser PJ, Corcoran AE. 2004. Antisense intergenic transcription in V(D)J recombination. Nat Immunol 5: 630–7 [DOI] [PubMed] [Google Scholar]
- 77.Verma-Gaur J, Torkamani A, Schaffer L, Head SR, Schork NJ, Feeney AJ. 2012. Noncoding transcription within the Igh distal V(H) region at PAIR elements affects the 3D structure of the Igh locus in pro-B cells. Proc Natl Acad Sci U S A 109: 17004–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. 2000. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell 102: 553–63 [DOI] [PubMed] [Google Scholar]
- 79.Pefanis E, Wang J, Rothschild G, Lim J, Kazadi D, Sun J, Federation A, Chao J, Elliott O, Liu ZP, Economides AN, Bradner JE, Rabadan R, Basu U. 2015. RNA exosome-regulated long non-coding RNA transcription controls super-enhancer activity. Cell 161: 774–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ouyang J, Zhu X, Chen Y, Wei H, Chen Q, Chi X, Qi B, Zhang L, Zhao Y, Gao GF, Wang G, Chen JL. 2014. NRAV, a long noncoding RNA, modulates antiviral responses through suppression of interferon-stimulated gene transcription. Cell Host Microbe 16: 616–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Peng X, Gralinski L, Armour CD, Ferris MT, Thomas MJ, Proll S, Bradel-Tretheway BG, Korth MJ, Castle JC, Biery MC, Bouzek HK, Haynor DR, Frieman MB, Heise M, Raymond CK, Baric RS, Katze MG. 2010. Unique signatures of long noncoding RNA expression in response to virus infection and altered innate immune signaling. MBio 1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Scaria V, Pasha A. 2012. Long Non-Coding RNAs in Infection Biology. Front Genet 3: 308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cazalla D, Steitz JA. 2010. Down-regulation of a host microRNA by a viral noncoding RNA. Cold Spring Harb Symp Quant Biol 75: 321–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Saayman S, Ackley A, Turner AM, Famiglietti M, Bosque A, Clemson M, Planelles V, Morris KV. 2014. An HIV-Encoded Antisense Long Noncoding RNA Epigenetically Regulates Viral Transcription. Mol Ther [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Broadbent KM, Park D, Wolf AR, Van Tyne D, Sims JS, Ribacke U, Volkman S, Duraisingh M, Wirth D, Sabeti PC, Rinn JL. 2011. A global transcriptional analysis of Plasmodium falciparum malaria reveals a novel family of telomere-associated lncRNAs. Genome Biol 12: R56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I. 2011. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 147: 358–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Carrieri C, Cimatti L, Biagioli M, Beugnet A, Zucchelli S, Fedele S, Pesce E, Ferrer I, Collavin L, Santoro C, Forrest AR, Carninci P, Biffo S, Stupka E, Gustincich S. 2012. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature 491: 454–7 [DOI] [PubMed] [Google Scholar]
- 88.Yoon JH, Abdelmohsen K, Srikantan S, Yang X, Martindale JL, De S, Huarte M, Zhan M, Becker KG, Gorospe M. 2012. LincRNA-p21 suppresses target mRNA translation. Mol Cell 47: 648–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. 2010. Long noncoding RNA as modular scaffold of histone modification complexes. Science 329: 689–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY. 2010. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464: 1071–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. 2011. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell 44: 667–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chu C, Zhang QC, da Rocha ST, Flynn RA, Bharadwaj M, Calabrese JM, Magnuson T, Heard E, Chang HY. 2015. Systematic discovery of Xist RNA binding proteins. Cell 161: 404–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Engreitz JM, Pandya-Jones A, McDonel P, Shishkin A, Sirokman K, Surka C, Kadri S, Xing J, Goren A, Lander ES, Plath K, Guttman M. 2013. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science 341: 1237973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Engreitz JM, Sirokman K, McDonel P, Shishkin AA, Surka C, Russell P, Grossman SR, Chow AY, Guttman M, Lander ES. 2014. RNA-RNA interactions enable specific targeting of noncoding RNAs to nascent Pre-mRNAs and chromatin sites. Cell 159: 188–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.McHugh CA, Chen CK, Chow A, Surka CF, Tran C, McDonel P, Pandya-Jones A, Blanco M, Burghard C, Moradian A, Sweredoski MJ, Shishkin AA, Su J, Lander ES, Hess S, Plath K, Guttman M. 2015. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature 521: 232–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Simon MD, Wang CI, Kharchenko PV, West JA, Chapman BA, Alekseyenko AA, Borowsky ML, Kuroda MI, Kingston RE. 2011. The genomic binding sites of a noncoding RNA. Proc Natl Acad Sci U S A 108: 20497–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Simon MD, Pinter SF, Fang R, Sarma K, Rutenberg-Schoenberg M, Bowman SK, Kesner BA, Maier VK, Kingston RE, Lee JT. 2013. High-resolution Xist binding maps reveal two-step spreading during X-chromosome inactivation. Nature 504: 465–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Consortium EP, Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ricano-Ponce I, Wijmenga C. 2013. Mapping of immune-mediated disease genes. Annu Rev Genomics Hum Genet 14: 325–53 [DOI] [PubMed] [Google Scholar]
- 100.Kumar V, Westra HJ, Karjalainen J, Zhernakova DV, Esko T, Hrdlickova B, Almeida R, Zhernakova A, Reinmaa E, Vosa U, Hofker MH, Fehrmann RS, Fu J, Withoff S, Metspalu A, Franke L, Wijmenga C. 2013. Human disease-associated genetic variation impacts large intergenic non-coding RNA expression. PLoS Genet 9: e1003201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Trimarchi T, Bilal E, Ntziachristos P, Fabbri G, Dalla-Favera R, Tsirigos A, Aifantis I. 2014. Genome-wide mapping and characterization of Notch-regulated long noncoding RNAs in acute leukemia. Cell 158: 593–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Medyouf H, Gusscott S, Wang H, Tseng JC, Wai C, Nemirovsky O, Trumpp A, Pflumio F, Carboni J, Gottardis M, Pollak M, Kung AL, Aster JC, Holzenberger M, Weng AP. 2011. High-level IGF1R expression is required for leukemia-initiating cell activity in T-ALL and is supported by Notch signaling. J Exp Med 208: 1809–22 [DOI] [PMC free article] [PubMed] [Google Scholar]