Abstract
Microaerobic cultivation conditions have been shown experimentally and theoretically to improve the performance of a number of bioproduction systems. However, under these conditions, the production of l-valine by Escherichia coli is decreased mainly because of a redox cofactor imbalance and a decreased l-glutamate supply. The synthesis of one mole of l-valine from one mole of glucose generates two moles of NADH via glycolysis but consumes a total of two moles of NADPH, one in the ketol-acid reductoisomerase (KARI) reaction and the other in the regeneration of l-glutamate as an amino group donor for the branched-chain amino acid aminotransferase (BCAT) reaction. The improvement of l-valine synthesis under oxygen deprivation may be due to solving these problems. Increased l-valine synthesis under oxygen deprivation conditions was previously shown in Corynebacterium glutamicum (Hasegawa et al., 2012). In this study, we have proposed the use of NADH-dependent leucine dehydrogenase (LeuDH; EC 1.4.1.9) Bcd from B. subtilis instead of the native NADPH-dependent pathway including aminotransferase encoded by ilvE to improve l-valine production in E. coli under microaerobic conditions. We have created l-valine-producing strains on the base of the aminotransferase B-deficient strain V1 (B-7 ΔilvBN ΔilvIH ΔilvGME::PL-ilvBNN17KDA) by introducing one chromosomal copy of the bcd gene or the ilvE gene. Evaluation of the l-valine production by the obtained strains under microaerobic and aerobic conditions revealed that leucine dehydrogenase Bcd had a higher potential for l-valine production under microaerobic conditions. The Bcd-possessing strain exhibited 2.2-fold higher l-valine accumulation (up to 9.1 g/L) and 2.0-fold higher yield (up to 35.3%) under microaerobic conditions than the IlvE-possessing strain. The obtained results could be interpreted as follows: an altering of redox cofactor balance in the l-valine biosynthesis pathway increased the production and yield by E. coli cells under microaerobic conditions. Thus, the effective synthesis of l-valine by means of “valine fermentation” was shown in E. coli. This methodology has the advantages of being an economical and environmentally friendly process.
Keywords: Biochemistry, Microbiology, Genetics
1. Introduction
l-Valine (hereinafter, valine) is a branched-chain amino acid (BCAA) that is widely used in dietary products, pharmaceuticals, and cosmetics, as an animal feed additive and as a precursor in the chemical synthesis of antibiotics and herbicides (Park and Lee, 2010). In addition, an immediate precursor of valine, 2-ketoisovalerate (Fig. 1), is an initial compound in the biosynthesis of isobutanol, a promising biofuel (Atsumi et al., 2008; Savrasova et al., 2011). To date, valine has been produced by microbial synthesis, mainly by using engineered E. coli and Corynebacterium glutamicum. General strategies to develop efficient valine-producing strains have been reported (Blombach et al., 2008; Park et al., 2007, 2011; Park and Lee, 2010; Wang et al., 2018).
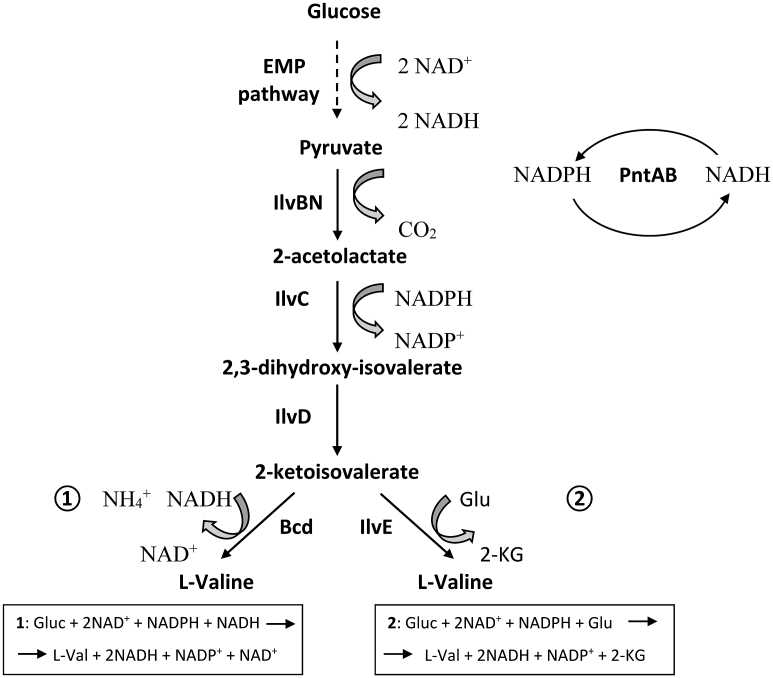
Fig. 1.
Pathway of l-valine biosynthesis. EMP pathway, Embden-Meyerhof-Parnas pathway; IivBN, acetolactate synthase I; IlvC, ketol-acid reductoisomerase; IlvD, dihydroxyacid dehydratase; IlvE, branched-chain amino acid aminotransferase; Bcd, leucine dehydrogenase; PntAB, pyridine nucleotide transhydrogenase.
Usually, amino acids are produced from sugars by microbes during aerobic cultivation. Particularly for E. coli cells, aerobic conditions are preferable from the viewpoint of cell energetic and growth rate. Oxygen is an effective electronic acceptor and can provide a significantly higher ATP/glucose yield (more than 30 ATP per glucose under aerobic conditions vs only 2 ATP from the glycolysis pathway under anaerobic conditions). However, in some cases, anaerobic cultivation may increase the product yield, as E. coli is a metabolically versatile bacterium able to respond to changes in oxygen availability. This approach exploits a flexible biochemistry in which aerobic respiration is preferred to anaerobic respiration, which in turn is preferred to fermentation (Partridge et al., 2007). E. coli cells can be intentionally adapted to microaerobic conditions, e.g., by laboratory adaptive evolution (Partridge et al., 2007).
Microaerobic conditions have been shown experimentally and theoretically to improve the performance of a number of bioproduction systems. However, under oxygen deprivation conditions, NADH is oxidized in mixed fermentation pathways, resulting in ethanol, acetate, lactate, and succinate production (Böck and Sawers, 1996); otherwise, excess NADH inhibits glycolysis, particularly its NAD+-dependent glyceraldehyde-3-phosphate dehydrogenase reaction. In this respect, the potential use of NADH formed under microaerobic conditions as a driving force for synthesis of a target compound is of particular interest. The microbial production of valine seems to be one of the most appropriate models. Previously, the production of different compounds (ethanol, lactate, succinate, organic acid, l-alanine, l-valine) by C. glutamicum was shown under oxygen deprivation conditions (Hasegawa et al., 2012, 2013; Inui et al., 2004a, 2004b, 2007; Jojima et al., 2010). In addition, improved isobutanol synthesis in E. coli under anaerobic conditions via the NADH-dependent pathway was shown (Bastian et al., 2011).
In some cases, E. coli is preferable over C. glutamicum as a host for the microbial production of useful compounds because of faster cell growth and better developed genetic tools. In this work, we demonstrated the effective synthesis of valine by means of so-called “valine fermentation” under microaerobic conditions (Fig. 1).
The valine biosynthetic pathway in E. coli consists of four reactions catalyzed by enzymes (Fig. 2): acetohydroxy acid synthase, which catalyzes the first common step in BCAA biosynthesis (isoenzymes AHAS I, II, III, encoded by ilvBN, ilvGM, and ilvIH, respectively); ketol-acid reductoisomerase (KARI), encoded by ilvC; dihydroxy acid dehydratase (DHAD), encoded by ilvD; and BCAA aminotransferase (BCAT, hereinafter AT), encoded by ilvE. The pathway is also responsible for the biosynthesis of other BCAAs (l-leucine and l-isoleucine) and D-pantothenate (Park et al., 2007; Park and Lee, 2010). The key enzyme among the four is AHAS because it is subject to different regulation (Umbarger, 1996). Expression of the ilvGMEDA operon is controlled by transcriptional attenuation mediated by all three BCAAs (Lawther and Hatfield, 1980; Lawther et al., 1987), whereas the ilvBN operon is controlled by attenuation mediated only by l-valine and l-leucine (Umbarger, 1996; Wek et al., 1985). In this work, we have created E. coli valine-producing strains containing feedback-resistant AHAS I encoded by the ilvBNN17K genes as a part of the artificial operon PL-ilvBNN17KDA in the chromosome (Sycheva et al., 2009).
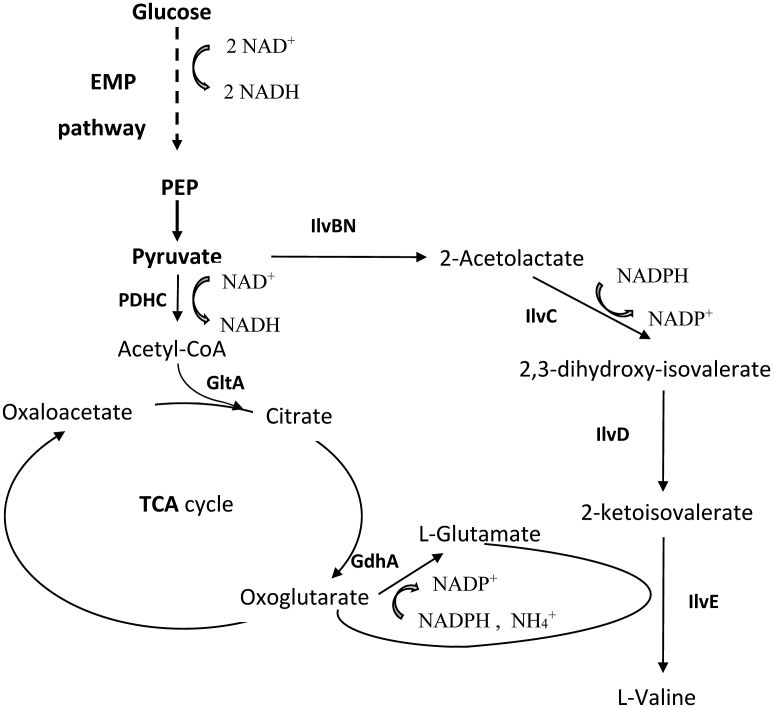
Fig. 2.
Metabolic pathway of E. coli and l-valine biosynthetic pathway. EMP pathway, Embden–Meyerhof–Parnas pathway; PDHC, pyruvate dehydrogenase complex; GltA, citrate synthase; IivBN, acetolactate synthase I; IlvC, ketol-acid reductoisomerase; IlvD, dihydroxyacid dehydratase; IlvE, branched-chain amino acid aminotransferase; GdhA, glutamate dehydrogenase.
The biosynthesis of one mole of valine requires two moles of NADPH, one of which is consumed in the glutamate dehydrogenase (GDH; EC 1.4.1.4) (Sakamoto et al., 1975; Helling, 1994) reaction, which yields l-glutamate as a universal amino group donor for aminotransferase reactions, including the BCAA aminotransferase (BCAT; EC 2.6.1.42)-mediated (Inoue et al., 1988) synthesis of valine from its immediate keto precursor (Fig. 2). As an alternative, valine synthesis can proceed via the NADH-dependent BCAA dehydrogenase reaction, thereby providing the NADH oxidation necessary for the function of the Embden-Meyerhof-Parnas (EMP) pathway yielding pyruvate, a starting compound for valine synthesis. A range of microorganisms other than E. coli possess such dehydrogenases, and NADH-specific BCAA dehydrogenase (leucine dehydrogenase; LeuDH; EC 1.4.1.9) enables the reversible reductive amination of BCAA keto precursors using ammonia directly as a substrate (Livesey and Lund, 1988; Nagata et al., 1988; Ohshima et al., 1994; Debarbouille et al., 1999).
In the case of valine synthesis by E. coli under oxygen deprivation conditions via the native metabolic pathway, including the aminotransferase reaction, the redox cofactor imbalance - two moles of NADH are synthesized via the EMP pathway, and two moles of NADPH are consumed in valine synthesis - should be overcome by the native enzyme systems of a cell (Fig. 1). There are various strategies to solve this problem. One of the methods is changing the cofactor requirement of valine biosynthetic reactions from NADPH to NADH to improve the redox status of a cell. Alternatively, a heterologous BCAA dehydrogenase can be used to provide NADH-dependent branched-chain keto acid amination instead of ordinary NADPH-dependent metabolic pathway, including the BCAA aminotransferase reaction (Fig. 1). In this case, glycolysis is simultaneously promoted by NADH oxidation due to so-called “valine fermentation” by analogy with traditional fermentation types, such as ethanol fermentation, lactate fermentation, etc. Additionally, the change in KARI cofactor specificity from NADPH to NADH is also useful, as was realized earlier for C. glutamicum (Hasegawa et al., 2012, 2013). However, for E. coli, this approach is affected by the presence of a membrane-bound pyridine nucleotide transhydrogenase PntAB, which catalyzes the energy-dependent transfer of reducing power from NADH to NADP+ (Sauer et al., 2004). In contrast, C. glutamicum does not possess a chromosomally encoded nicotinamide nucleotide transhydrogenase to catalyze the reversible interconversion between NADH and NADPH (Kabus et al., 2007), and NADPH formation from NADH via malic enzyme would play only a minor role (Bartek et al., 2010; Georgi et al., 2005).
In this work, we showed an increase in valine production in E. coli under microaerobic conditions due to the improvement of the redox cofactor balance (NADH synthesis/NADPH consumption) by the introduction of NAD-specific LeuDH from B. subtilis encoded by the bcd gene instead of AT encoded by ilvE. Under these conditions, we achieved a 2.2-fold increase in valine production and a 2.0-fold increase in yield (m/m) compared to the use of the traditional metabolic pathway including the BCAA aminotransferase reaction.
2. Materials and methods
2.1. Strains and media
All bacterial strains and plasmids used in this study are listed in Table 1. The XL1-Blue E. coli strain was used for cloning. The CC118 (λpir+) E. coli strain was used for the maintenance of pir-dependent recombinant plasmids. The following media were used for bacteria culture: lysogenic broth (LB), SOB, SOC and M9 medium (Sambrook and Russell, 2001). Glucose (0.4%) was added to minimal media as a carbon source. Ampicillin (Ap, 100 mg/L), chloramphenicol (Cm, 20 mg/L), tetracycline (Tc, 20 mg/L) and kanamycin (Km, 40 mg/L), were used for selection as necessary. Cultivation of valine-producing strains was carried out in fermentation medium (FM) at pH 7.0 containing the following (g/L): (NH4)2SO4, 15; KH2PO4, 1.5; MgSO4, 1; CaCO3, 20; B1, 0.01; glucose, 60.
Table 1.
Bacterial strains and plasmids used in present study.
| Strain or plasmid | Relevant genotype | Source or reference |
|---|---|---|
| Strains | ||
| XL-Blue | E. coli (recA1 endA1 gyrA96 (NalR) thi-1 hsdR17 (rk_ mk_)glnV44 relA1 lac [F’::Tn10(TetR)'proAB lacIq Δ(lacZ)M15]) | Stratagene |
| CC118 (λpir+) | E. coli Δ(ara-leu) araD ΔlacX74 galE galK phoA20 thi-1 rpsE rpoB argE(Am) recAl, lysogenized with λpir phage | Herrero et al. (1990) |
| MG1655 | E. coli K12 wild-type | VKPM B-6195 |
| MG1655 (Δφ80-attBnative)φ80-attB trs5-7 | E. coli K-12 MG1655 with deleted native φ80-attB site and φ80-attB in trs5-7 locus | Minaeva et al. (2008) |
| MG1655 4Δ | MG1655 ΔilvE ΔtyrB ΔavtA::KmR ΔaspC::TetR | Laboratory collection |
| BW25113 ΔilvE::KmR | E. coli K12 Δ(araD-araB)567 Δ(rhaD-rhaB)568 ΔlacZ4787 (::rrnB-3) hsdR514 rph-1 lacI+ ΔilvE::FRT-kan-FRT | Keio Collection |
| Baba et al. (2006),Grenier et al. (2014) | ||
| MG1655 trs5-7::bcd-TetR | MG1655 (Δφ80-attBnative) trs5-7::bcd-λattL-TetR- λattR | Present study |
| MG1655 trs5-7::bcd | MG1655 (Δφ80-attBnative) trs5-7::bcd -TetS | Present study |
| MG1655 trs5-7::bcd ΔilvE::KmR | MG1655 (Δφ80-attBnative) trs5-7::bcd -TetS ΔilvE:: FRT-kan-FRT | Present study |
| MG1655 cat-PL-bcd5.7 | MG1655 (Δφ80-attBnative) trs5-7:: λattL-CmR- λattR-PL-bcd | Present study |
| MG1655 cat-PL-ilvE5.7 | MG1655 (Δφ80-attBnative) trs5-7:: λattL-CmR- λattR- PL-ilvE ΔilvE:: FRT-kan-FRT | Present study |
| B-7 ΔilvBN ΔilvGM ΔilvIH | E. coli K12 ΔilvBN ΔilvGM ΔilvIH | Sycheva et al. (2009) |
| B-7 ΔilvIH ΔilvGM cat-PL-ilvBNN17K | E. coli K12 ΔilvIH ΔilvGM λattL-CmR- λattR-PL- ilvBNN17K | Sycheva et al. (2009) |
| MG1655 cat-PL-ilvBNN17KDA | MG1655ΔilvGME::λattL-CmR- λattR-PL- ilvBNN17KDA | Laboratory collection, Serebrianyi |
| B-7 ΔilvBN ΔilvGM ΔilvIH cat-PL-ilvBNN17KDA | E. coli K12 ΔilvIH ΔilvBN ΔilvGME:: λattL-CmR- λattR-PL-ilvBNN17KDA | Present study |
| V1 | E. coli K12 ΔilvIH ΔilvBN ΔilvGME::PL-ilvBNN17KDA-CmS | Present study |
| V1 cat-PL-bcd5.7 | E. coli K12 ΔilvIH ΔilvBN | Present study |
| ΔilvGME::PL-ilvBNN17KDA trs5-7:: λattL-CmR- λattR-PL-bcd | ||
| V1 cat-PL-ilvE5.7 | E. coli K12 ΔilvIH ΔilvBN ΔilvGME::PL-ilvBNN17KDA trs5-7:: λattL-CmR- λattR- PL-ilvE | Present study |
| Plasmids | ||
| pUC57-bcd-Bsub | pMB1 ori; AmpR; bcd | Present study |
| pMW118 | pSC101 ori; AmpR; MCS | GenBank accession number AB005475 |
| pMW118-bcd | pSC101 ori; AmpR; Plac-bcd | Present study |
| pAH162- λattL-TetR- λattR -2Ter | oriR; λattL-TetR- λattR; attP φ80; MCS | Minaeva et al. (2008) |
| pAH162- λattL-TetR- λattR-2Ter-bcd | oriR; λattL-TetR- λattR; attP φ80; bcd | Present study |
| pAH123 | oriR101, repA101ts, λcIts857, λPR→φ80-int, AmpR | Haldimann and Wanner (2001), GenBank accession number AY048726 |
| pKD46 | oriR101, repA101ts, araC, ParaB-[γ, β, exo of phage λ], AmpR | Datsenko and Wanner (2000), GenBank accession number AY048746 |
| pMW-Int-Xis | oriR101, repA101ts, λcIts857, λPR→λxis-int, AmpR | Minaeva et al. (2008) |
2.2. Cultivation conditions
Cells were preseeded in test tubes containing 3 ml of LB medium and incubated at 32 °C for 3 h on a rotary shaker (250 rpm). The preseeded cultures were then diluted 1:20 into 2 ml of FM medium in 20 × 200-mm test tubes. Strains V1 cat-PL-bcd5.7 and V1 cat-PL-ilvE5.7 were cultivated at 32 °C for 68 h on a rotary shaker (250 rpm). To provide microaerobic conditions, rubber stoppers were used instead of cotton stoppers.
2.3. Determination of amino acid and glucose concentrations
Accumulated valine was measured by thin-layer chromatography (TLC). TLC plates (10 х 15) cm were coated with 0.11-mm layers of Sorbfil silica gel containing no fluorescent indicator (Stock Company Sorbpolymer, Krasnodar, Russia). The Sorbfil plates were developed with a mobile phase consisting of isopropanol-ethylacetate-25% aqueous ammonia-water (16:16:5:10 v/v). A solution of ninhydrin (2% w/v) in acetone was used as the visualizing reagent. Residual glucose was measured by a Biosen glucose and lactate analyzer (EKF Diagnostics, UK).
2.4. DNA handling procedures
Protocols for the genetic manipulation of E. coli and techniques for isolating and manipulating nucleic acids were described previously (Sambrook and Russell, 2001). Restriction enzymes, T4 DNA ligase, High Fidelity PCR Enzyme Mix and 1-kb DNA Ladder were purchased from Thermo Scientific Inc (USA). Plasmids and genomic DNA were isolated using QIAGEN Plasmid Mini Kits (QIAGEN GmbH, Germany) and Bacterial Genomic DNA Kits (Sigma, USA), respectively. QIAquick Gel Extraction Kits (QIAGEN GmbH, Germany) were used to isolate DNA from agarose gels. Oligonucleotides were purchased from Sintol (Russia). The sequences of oligonucleotide primers are presented in Table 2. Synthesis of the bcd gene was performed by Sloning BioTechnology GmbH (Germany).
Table 2.
Oligonucleotides used in this study.
| Primers | Sequence (5′- 3′) | Purpose |
|---|---|---|
| P1 | AAAGGATGAACTACGAGGAAGGGAACAACATTC-ATACGCTCAAGTTAGTATAAAAAAGCTGAAC | creation of cat-PL-ilvBNN17K |
| P2 | ATGGGACGGTGCGTGCCGTCCCATTTTTTGTATTTACTGAAAAAACACCGCGATCTTGTTAAAC | creation of cat-PL-ilvBNN17K |
| P3 | GTAAAGCGCTTACGCGTCGA | verification of cat-PL-ilvBNN17K integration |
| P4 | TGCAAGTGAAGTTGAGTTGTTC | verification of cat-PL-ilvBNN17K integration |
| P5 | GAATGATATCCATATCCTCGAC | verification of bcd integration |
| P6 | GTCTTCTCACGGGAACGGTT | verification of bcd integration |
| P7 | CGAAAGTGATTGCGCCTACCCGGATATTATCGTGAGCGCTCAAGTTAGTATAAAAAAGCTGAAC | creation of cat-PL upstream bcd and ilvE |
| P8 | TATATTTAAAAAGTTCCATACATAGATCTCCTTCTTCGGCCAATGCTTCGTTTCGTATCACACA | creation of cat-PL upstream bcd |
| P9 | CCATATTACGACCATGAGTGG | verification of ilvE deletion |
| P10 | CCAGTAATTCAGAAATGTTGG | verification of ilvE deletion |
| P11 | AGATAGATCTCCTTCTTCGGCCAATGCTTC | creation of cat-PL upstream of ilvE |
| P12 | ATTGGCCGAAGAAGGAGATCTATCTATGACCACGAAGAAAGCTGATTACATTT | amplification of ilvE |
| P13 | AAGCTTGCATGCCTGCAGGTCGACTCTAGAGGATCCTTATTGATTAACTTGATCTAACCAGCCC | amplification of ilvE |
| P14 | GTTCGTTGCAACAAATTGATAAG | sequencing of PL-ilvE |
| P15 | CAGGGAAGAGAGGTAGTTACC | sequencing of PL-ilvE |
| P16 | GATCGATGCGATGGTTTCCTC | sequencing of PL-ilvE |
2.5. Plasmid construction
All plasmids used or constructed in this study are listed in Table 1.
2.5.1. Construction of pUC57-bcd-Bsub
To express leucine dehydrogenase Bcd from B. subtilis in E. coli, the rare codon-free variant of the bcd gene was synthesized. To clone bcd, a bcd gene (GenBank accession number BSU24080) from B. subtilis with a modified nucleotide sequence codon-optimized for E. coli and with SacI and BamHI restriction sites (5′-GAGCTCAAGAAGGAGATCTATGT-3′, 5′-GGATCC-3′) was synthesized by Sloning BioTechnology. In the modified bcd sequence, the following 29 codons were optimized: 8 Arg positions, 43, 59, 62, 108, 155, 341, 349 and 362 (codons AGA (7) and CGG (1) were replaced with CGC); 9 Gly positions, 23, 41, 78, 104, 143, 156, 166, 172 and 195 (codon GGA was replaced with GGC); 4 Pro positions, 137, 147, 222 and 329 (codons CCA (1) and CCT (3) were replaced with CCG); and 8 Thr positions, 33, 46, 80, 117, 129, 133, 149 and 266 (codon ACA was replaced with ACC). The resulting DNA fragment containing the bcd gene was digested with SacI and BamHI and cloned into the pUC57 vector (GenBank accession number Y14837) cut with the same enzymes, yielding the plasmid pUC57-bcd-Bsub. The bcd gene was cloned in the opposite orientation relative to the Lac promoter to reduce the potential toxicity of the gene expression.
2.5.2. Construction of pMW118-bcd
To clone bcd, pUC57-bcd-Bsub was digested with SacI and BamHI. The DNA fragment containing the bcd gene was cloned into the pMW118 vector (under control of the Plac promoter) and cut with the same enzymes, creating pMW118-bcd. The resulting plasmid was shown to complement the Val and Ile auxotrophy of the aminotransferase-deficient strain MG1655 4Δ under IPTG induction during growth on M9 minimal medium supplemented with appropriate amino acids.
2.5.3. Construction of pAH162-λattL-TetR-λattR-2Ter-bcd
The construction of the integrative vector pAH162 λattL-TetR-λattR-2Ter was previously described (Minaeva et al., 2008). To clone bcd, pUC57-bcd-Bsub was digested with SacI and BamHI. The DNA fragment containing the promoter-less bcd gene was cloned into the integrative vector pAH162-λattL-TetR-λattR-2Ter cut with the same enzymes, yielding pAH162-λattL-TetR-λattR-2Ter-bcd.
2.6. Strain construction
The primers and strains used and constructed in this study are listed in Tables 1 and 2. Chromosomal gene deletions and insertions in the chromosome of the E. coli strain MG1655 K-12 were prepared via the method developed by Datsenko and Wanner called “λRed-mediated recombination” (Datsenko and Wanner, 2000) combined with the phage λ Int/Xis-mediated marker excision. The plasmid pKD46 carrying the arabinose-inducible λ-Red genes was used to provide λ-Red recombination. φ80-Mediated integration was carried out according to (Haldimann and Wanner, 2001; Minaeva et al., 2008). The CRIM helper plasmid pAH123 containing the thermoinducible φ80-Int gene was used to provide φ80-mediated integration (Haldimann and Wanner, 2001). Specifically designed cassettes with the CmRex marker were transferred into the E. coli strains by P1 phage-mediated transduction (Miller, 1972). The DNA fragments CmRex and TetRex flanked by λattL/R were eliminated from the E. coli chromosome using a λ-Int/Xis site-specific recombination system with the pMWts-λInt/Xis helper plasmid (Minaeva et al., 2008).
2.6.1. Construction of the E. coli strain MG1655 ΔilvGME::λattL-CmR-λattR-PL-ilvBNN17KDA
The artificial ilv operon cat-PL-ilvBNN17KDA was created by introducing the PCR fragment cat-PL-ilvBNN17K to replace the ilvGME genes of the ilvGMEDA operon in the E. coli strain MG1655 via λ Red-mediated recombination (Datsenko and Wanner, 2000). The PCR fragment cat-PL-ilvBNN17K (3.97 kbp) was created with primers P1 and P2 and the chromosome of strain B-7ΔilvGMΔilvIH λattL-CmR-λattR-PL-ilvBNN17K as the template (Sycheva et al., 2009). The obtaining DNA fragment was introduced by electroporation into strain MG1655/pKD46, resulting in the strain MG1655 ΔilvGME::λattL-CmR-λattR-PL-ilvBNN17KDA (MG1655 cat-PL-ilvBNN17KDA). Integration was verified using primers P3 and P4.
2.6.2. Construction of E. coli strain MG1655 trs5-7::λattL-CmR-λattR-PL-bcd
The plasmid pAH162 λattL-TcR-λattR-2Ter-bcd was integrated by the φ80-Int system into MG1655 φ80-attB trs5-7 Δφ80-attBnative using the TcR marker for selection to obtain the strain MG1655 trs5-7::λattL-TcR-λattR-bcd. Integration was verified by using primers P5 and P6. The vector part of the integrated plasmid containing the TcR marker was excised by means of the pMWts-λInt/Xis helper plasmid, and the strain MG1655 trs5-7::bcd was obtained. The modification was verified by using primers P5 and P6. After that, the λ phage PL promoter marked with Cmex upstream of the bcd gene was introduced by λ Red-mediated recombination (Datsenko and Wanner, 2000). The PCR fragment 1.97 kb containing the modification λattL-CmR-λattR-PL was created by using primers P7 and P8 with the chromosome of MG1655 λattL-CmR-λattR-PL-leuABCD as a template. The obtained DNA fragment was introduced by electroporation into MG1655 trs5-7::bcd/pKD46, resulting in the strain MG1655 trs5-7::λattL-CmR-λattR-PL-bcd (MG1655 trs5-7::cat-PL-bcd). The modification was checked by using primers P5 and P6.
2.6.3. Construction of E. coli strain MG1655 trs5-7::λattL-CmR-λattR-PL-ilvE ΔilvE::FRT-kan-FRT
To create an aminotransferase-overexpressing strain, we used the strain MG1655 trs5-7::bcd Δφ80-attBnative mentioned above. First, the introduction of ilvE deletion (Keio collection) marked with KmRex by P1 transduction was performed. The presence of this deletion was verified with primers P9 and P10. Then, the PCR fragment λattL-CmR-λattR-PL-ilvE was introduced into this strain instead of the bcd gene by λ Red-mediated recombination (Datsenko and Wanner, 2000). The PCR fragment λattL-CmR-λattR-PL-ilvE5.7 was created by overlap extension PCR with primers P7 and P13 by using the PCR fragments λattL-CmR-λattR-PL (created with primers P7 and P11) and ilvE (created with primers P12 and P13) as templates. The resulting 2.9 kb DNA fragment λattL-CmR-λattR-PL-ilvE5.7 was introduced by electroporation into the strain MG1655 trs5-7::bcd ΔilvE::FRT-kan-FRT/pKD46. The selection of integrants was performed on M9 minimal medium with 0.4% glucose to obtain the strain MG1655 trs5-7::λattL-CmR-λattR-PL-ilvE ΔilvE::FRT-kan-FRT (MG1655 trs5-7::cat-PL-ilvE) containing the PL promoter marked with CmRex upstream of ilvE. The modification was checked by using primers P6 and P13. The obtained structure λattL-CmR-λattR-PL-ilvE5.7 was verified by sequencing with primers P14, P15, and P16.
2.7. Preparation of cell extracts
Strains MG1655 cat-PL-bcd5.7 and MG1655 cat-PL-ilvE5.7 ΔilvE::KmR were cultured overnight, and 0.1 ml was used to inoculate fresh medium (10 ml of M9 medium supplemented with 1/10 v/v LB). Inoculated cultures were grown for 4.5 h until an optical density at 540 nm of 0.8 was reached. Cells were harvested by centrifugation and washed twice in 1 M sodium chloride and then in 0.1 M potassium–phosphate buffer (pH 7.0). Cells were suspended in 0.1 M potassium–phosphate buffer (pH 7.0) and disrupted by sonication. The supernatant after centrifugation at 12,000 rpm for 30 min was used as the cell extract. All steps were performed at temperatures <4 °C. Protein concentrations were determined by the Bradford assay (Bio-Rad protein assay, GmbH).
2.8. Branched-chain amino acid aminotransferase assay
Cell extracts were incubated for 15 min at 37 °C in 2-ml vials at pH 7.5 [0.5 M Tris hydrochloride (HCl) buffer + 1 mM dithiothreitol (DTT)] with 50 mM l-Phe and 50 mM 2-ketoisovalerate as the substrate and with 0.5 mM pyridoxal phosphate (PLP) as a cofactor in a total volume of 200 μl. Then, to stop the reaction, the samples were placed on ice, and 0.8 ml of 1.25 N NaOH was added. The formation of phenylpyruvate was analyzed at 320 nm against a control to which NaOH had been added at 0 min. The OD was read immediately after the addition of NaOH. A molar extinction coefficient of 17,500 M-1 cm-1 was used (Collier and Kohlhaw, 1972). Specific activity is defined as nanomoles of phenylpyruvate formed per minute per milligram of protein.
2.9. Leucine dehydrogenase assay: oxidative deamination and reductive amination
The enzyme activity was measured by spectrophotometrically monitoring the production of NADH in oxidative deamination or the consumption of NADH in reductive amination at 340 nm (ɛ = 6,220 M−1 cm−1). The activity was defined as the number of nanomoles of NADH produced (or consumed) in 1 min by 1 mg of enzyme (nmol min−1 mg−1).
The assay mixture for the deamination reaction contained 100 mM Tris buffer pH 9.0, 3.5 mM NAD+, 10 mM l-Leu (or l-Ile, l-Val) and enzyme solution in a final volume of 1 ml. The assay mixture for the reductive amination reaction contained 100 mM Tris buffer pH 7.5, 600 mM NH4Cl, 1 mM DTT, 0.125 mM NADH, 5 mM KIV (KMV, KIC) and enzyme solution in a final volume of 1 ml (Livesey and Lund, 1988).
2.10. Determination of volumetric mass transfer coefficient of oxygen (kLa) in test tubes cultivation conditions
The sulfite method (Cooper et al., 1944) was used to determine the volumetric mass transfer coefficient of oxygen (kLa) for test tubes cultivation according to Garcia-Ochoa and Gomez (2009); Cu2+ ions (1 mM CuSO4) were applied as catalyst; 1 ml samples were taken, mixed with an excess of standard iodine reagent (5 ml of 0.2N I2 solution) and finally titrated with tiosulfate solution (0.1N Na2S2O3). The measurements were performed at the range 60–120 min of incubation. The experiments were performed in triplicate.
3. Results and discussion
3.1. Construction of aminotransferase-deficient E. coli strain harboring feedback-resistant AHAS I
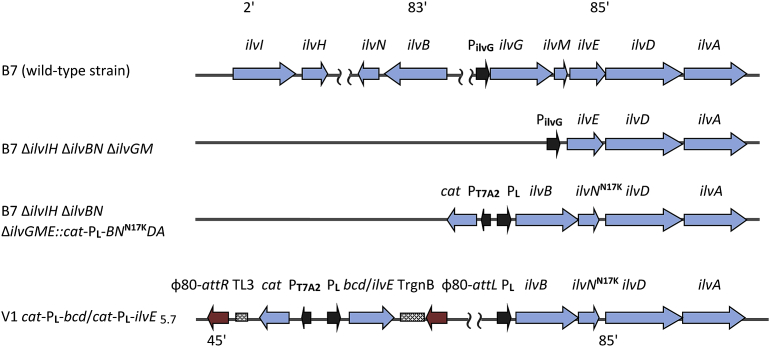
At the first step, the aminotransferase-deficient E. coli strain harboring feedback-resistant AHAS I was designed as a platform for the further construction of valine-producing strains overexpressing aminotransferase or, alternatively, leucine dehydrogenase genes. The AHAS-deficient E. coli strain K-12 B-7ΔilvBN ΔilvGM ΔilvIH with deletions of the genes encoding acetolactate synthase types I, II and III was used as a starting point (Sycheva et al., 2009) (Fig. 3). In the native locus of the chromosome, harboring the isoleucine-valine operon (ilv operon, PilvG-ilvGMEDA), this strain contains a deletion of AHAS II-encoding genes (PilvG-EDA). To provide valine oversynthesis and, simultaneously, to inactivate the aminotransferase-encoding gene in the native locus, the operon PilvG-EDA of this strain was replaced with the artificial construct cat-PL-ilvBNN17KDA harboring the feedback-resistant AHAS I-encoding genes ilvBNN17K under control of the “strong” λ phage PL promoter (Sycheva et al., 2009); the ilvE gene encoding aminotransferase was simultaneously deleted (for the construct design, see Materials and Methods) (Fig. 3). After elimination of the CmR marker by λ-Int/Xis site-specific recombination, the aminotransferase IlvE-deficient strain V1 (B-7ΔilvBNΔilvIHΔilvGME::PL-ilvBNN17KDA) was obtained. The strain V1 is a BCAA auxotroph due to aminotransferase B (IlvE) deficiency. This strain was then used to construct the valine-producing strains carrying AT or, alternatively, LeuDH.
Fig. 3.
Modifications in E. coli chromosome. The structural parts of the genes are represented by blue arrows, the promoters are designated by black arrows, the φ80-att sites are represented by red arrows, the terminators are marked by shaded rectangles.
3.2. Construction of valine-producing E. coli strains harboring overexpressed aminotransferase or heterologous leucine dehydrogenase genes
For valine synthesis in E. coli under oxygen deprivation, the redox cofactor imbalance (two moles of NADH is synthesized via the EMP pathway/two moles of NADPH is consumed in valine synthesis) and the l-glutamate supply for the AT reaction are the main problems (Fig. 2). To avoid these difficulties, a replacement of the Glu-dependent aminotransferase reaction at the final step of valine biosynthesis with the NADH-dependent NH4+ assimilating dehydrogenase reaction was proposed. A range of microorganisms possess NADH-specific dehydrogenases (leucine dehydrogenase; LeuDH; EC 1.4.1.9) that carry out the reversible reductive amination of BCAA keto precursors by directly utilizing ammonia as a substrate (Livesey and Lund, 1988; Nagata et al., 1988; Ohshima et al., 1994).
Evidently, a major physiological function of these enzymes during aerobic growth on glucose is BCAA degradation (Nagata et al., 1988); however, LeuDH could be supposed to enable the amination of BCAA keto precursors and the formation of the corresponding amino acids, particularly valine, under microaerobic conditions. Considering that, in contrast to NADPH-dependent GDH, LeuDH is an NADH-dependent enzyme, the application of LeuDH seems to be preferable to the AT-exploiting metabolic pathway for the microaerobic production of valine from the viewpoint of redox cofactor balance.
3.2.1. Expression of LeuDH from B. subtilis in E. coli cells
For the above purpose, in this work, LeuDH encoded by the bcd gene from B. subtilis was applied. The native nucleotide sequence of bcd contains a number of codons that are rare in E. coli; therefore, to express this LeuDH in E. coli, the codon-optimized variant of the bcd gene was chemically synthesized (see Materials and Methods) and subcloned into the integrative vector pAH162-λattL-TetR-λattR-2Ter (Minaeva et al., 2008) for the φ80-Int-mediated insertion of the bcd gene into the chromosome of the wild-type strain MG1655 (see Materials and Methods). As a result, the MG1655-derived strain containing the bcd gene integrated into the artificial φ80-attB site inside the trs5-7 locus was designed by means of the φ80-Int system. After the elimination of the TcR-marker-harboring part of the integrative vector by means of λ-Int/Xis site-specific recombination, the bcd gene was placed under control of the λ phage PL promoter marked with CmRex by λRed-mediated recombination. As a result, the strain MG1655 cat-PL-bcd 5.7 harboring the chromosomal copy of the overexpressed bcd gene from B. subtilis encoding NADH-dependent leucine dehydrogenase was obtained.
Analysis of the Bcd activity for the strain MG1655 cat-PL-bcd5.7 in the reaction of oxidative deamination revealed practically the same high level of enzymatic activity towards both substrates Val and Leu; this activity was approximately 1.8-fold higher than that using Ile as a substrate. Measurement of Bcd activity in the direction of reductive amination showed that the level of enzymatic activity was practically the same towards all the tested substrates, KIC, KIV and KMV (Tables 3 and 4).
Table 3.
Specific leucine dehydrogenase activity in the direction of oxidative deamination in strain MG1655 cat-PL-bcd5.7.a
| Strain | leucine dehydrogenase activity (nmol min−1 mg−1) |
||
|---|---|---|---|
| Substrate | |||
| l-Val | l-Leu | l-Ile | |
| MG1655 cat-PL-bcd5.7 | 8270 ± 410 | 7900 ± 350 | 4410 ± 230 |
| MG1655 | 11 ± 2 | 8 ± 2 | 6 ± 1 |
Data represent the means of three separate experiments.
Table 4.
Specific leucine dehydrogenase activity in the direction of reductive amination in strain MG1655 cat-PL-bcd5.7.a
| Strain | leucine dehydrogenase activity (nmol min−1 mg−1) |
||
|---|---|---|---|
| Substrateb | |||
| KIV | KIC | KMV | |
| MG1655 cat-PL-bcd5.7 | 650 ± 35 | 620 ± 25 | 560 ± 30 |
| MG1655 | 6 ± 2 | 5 ± 1 | 4 ± 1 |
Data represent the means of three separate experiments.
KIV, 2-ketoisovalerate; KIC, 2-ketoisocaproate; KMV, 2-keto-3-methylvalerate.
To fulfill a similar task, valine production under oxygen deprivation in C. glutamicum cells, LeuDH from Bacillus sphaericus was previously applied (Hasegawa et al., 2012). This enzyme shares 79% amino acid sequence identity and 90% amino acid sequence positives with the LeuDH from B. subtilis studied in the present work. The majority of differences (23 residues) are located in domain II (residues 141–348), which resembles the classical nucleotide-binding domain of lactate dehydrogenase (Baker et al., 1995). The substrate specificity of LeuDH from B. sphaericus for both reductive amination and oxidative deamination was studied previously (Li et al., 2009). The substrate affinity profile for LeuDH from B. subtilis used in this work is similar to that reported for LeuDH from B. sphaericus (Li et al., 2009).
3.2.2. Overexpression of ilvE gene encoding BCAA aminotransferase in E. coli chromosome
To provide a level of ilvE gene expression similar to that of the aforementioned bcd gene from B. subtilis in E. coli chromosome, the ilvE gene was inserted into the same trs5-7 locus and placed under control of the same “strong” promoter. To this end, the ilvE gene deletion (ΔilvE::KmRex) was first introduced into the chromosome of the strain MG1655 trs5-7::bcd by P1 transduction, yielding the BCAA auxotrophic ilvE-deficient strain MG1655 ΔilvE::KmRex trs5-7::bcd. Notably, in this case, the promoter-less bcd gene did not provide growth on minimal medium in the absence of BCAAs. Then, the PCR fragment λattL-CmR-λattR-PL-ilvE, containing the ilvE gene under the control of the λ phage PL promoter, was directly inserted into the chromosome of this strain in place of the bcd gene by λRed-mediated recombination. Integrants were selected on M9 minimal medium with 0.4% glucose to obtain the strain MG1655 cat-PL-ilvE5.7 ΔilvE::KmR harboring the aminotransferase gene under the control of the “strong” λPL promoter.
The obtained chromosomal construct cat-PL-ilvE5.7 was shown to provide essentially higher (more than 100-fold) BCAA TA activity than the native copy of the same gene (Table 5). KIV, an immediate precursor of valine, was used as a substrate in these experiments.
Table 5.
Specific BCAA aminotransferase activity in strain MG1655 cat-PL-ilvE5.7 ΔilvE::KmR.
| Strain | BCAA aminotransferase activity (nmol min−1 mg−1) |
|---|---|
| KIVa | |
| MG1655 cat-PL-ilvE5.7 | 1060 ± 45 |
| MG1655 | 4 ± 0.5 |
KIV- 2-ketoisovalerate as a substrate; data represent the means of three separate experiments.
To measure the efficiency in E. coli of the native valine biosynthetic pathway, including NADPH-dependent aminotransferase-mediated valine formation and the artificial one, including NADH-dependent leucine dehydrogenase-mediated NH4+ assimilation at the final step, we designed valine-producing strains containing one chromosomal copy of the bcd gene or the ilvE gene. The aminotransferase IlvE-deficient strain V1 (B-7 ΔilvBN ΔilvIH ΔilvGME::PL-ilvBNN17KDA) was used as the recipient for construction, as it can produce Val upon restoration its ability to perform the last step of Val synthesis, 2-ketoisovalerate (KIV) amination. To this end, the expression units cat-PL-bcd5.7 and cat-PL-ilvE5.7 were separately introduced into the chromosome of V1 by P1 transduction, resulting in the strains V1 cat-PL-bcd5.7 and V1 cat-PL-ilvE5.7 (Fig. 3). Both of these strains demonstrated similar growth on M9 plates with 0.4% glucose in the absence of Val, Ile and Leu addition, which indicated the ability of the heterologous leucine dehydrogenase Bcd from B. subtilis to enable the in vivo formation of valine by E. coli cells.
3.3. Valine accumulation by valine-producing E. coli strains under different cultivation conditions
LeuDH effectively catalyzes the reversible reductive amination of BCAA keto precursors using NADH as a cofactor and ammonia as a substrate (Fig. 1). Therefore, the introduction of LeuDH in place of native AT should improve the intracellular redox balance by reoxidizing NADH and, in the case of a sufficient NH4+ supply, increase the valine production under deprivation conditions. We evaluated the obtained strains V1 cat-PL-bcd5.7 and V1 cat-PL-ilvE5.7 under different cultivation conditions, aerobic and microaerobic, for 68 h at 32 °C (Table 6). As shown in Table 6, the expression of leucine dehydrogenase Bcd or aminotransferase IlvE in strain V-1 (B-7 ΔilvBN ΔilvIH ΔilvGME::PL-ilvBNN17KDA) in both cases resulted in the production of valine.
Table 6.
Valine accumulation by Bcd- or IlvE- harboring E. coli strains under different aeration conditions.a
| Strain | O2 conditionsb | Val, g/L | OD540 | Glc residual, g/L | Y m/m, % |
|---|---|---|---|---|---|
| V1 cat-PL-bcd5.7 | microaerobic | 9.1 ± 0.3 | 19.5 ± 0.3 | 20.3 ± 0.4 | 35.3 ± 0.8 |
| aerobic | 6.9 ± 0.1 | 34.5 ± 0.8 | 0 | 17.7 ± 0.3 | |
| V1 cat-PL-ilvE5.7 | microaerobic | 4.1 ± 0.1 | 14.5 ± 0.3 | 24.6 ± 0.3 | 17.8 ± 0.3 |
| aerobic | 9.8 ± 0.4 | 31.3 ± 1.8 | 0 | 25.1 ± 1.1 |
Cultivation time was 48 h; data represent the means of three separate experiments; kLa was determined by sulfite method as described in p2.10 “Materials and methods”.
Microaerobic conditions (kLa = 5.4 ± 0.2 × 10−4, mmole O2 ml−1 min−1); aerobic conditions (kLa = 14.2 ± 0.4 × 10−4, mmole O2 ml−1 min−1).
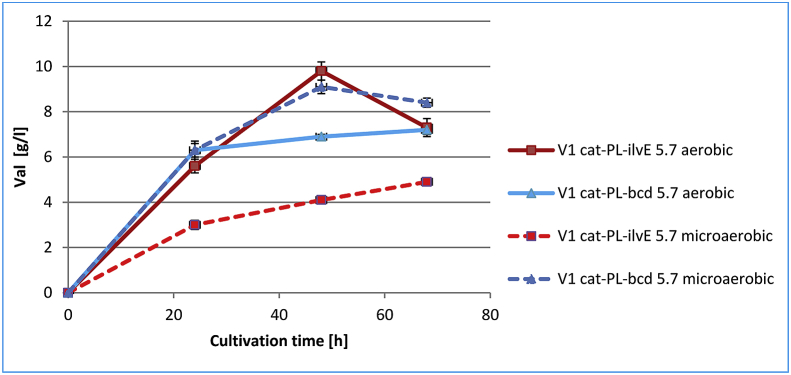
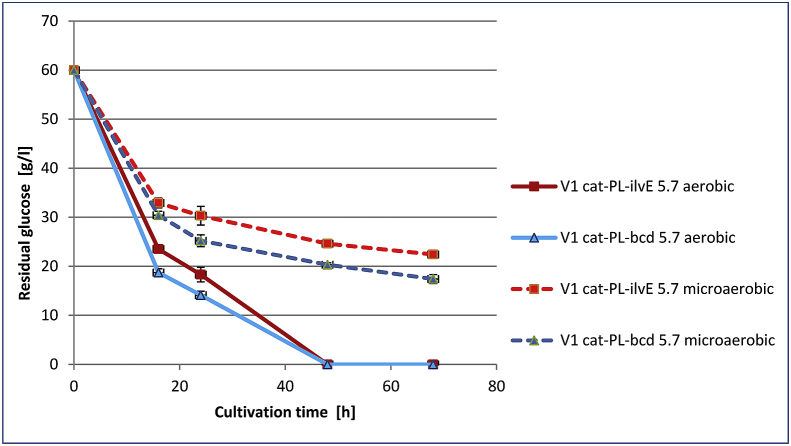
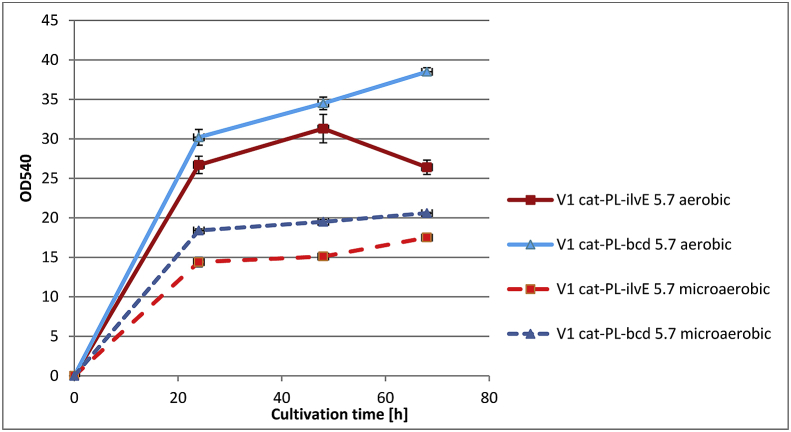
Time-course profiles of Val accumulation, glucose consumption and cell growth were analyzed for the strains V1 cat-PL-bcd5.7 and V1 cat-PL-ilvE5.7 under microaerobic and aerobic conditions (Figs. 4, 5, and 6). For both the tested strains glucose was completely consumed at 48h under aerobic conditions in contrast to that under microaerobic conditions. The maximum level of Val accumulation was observed for tested strains V1 cat-PL-bcd5.7 and V1 cat-PL-ilvE5.7 at 48h under microaerobic and aerobic conditions, respectively (Fig. 4).
Fig. 4.
Time-course profiles of valine accumulation by Bcd- or IlvE- harboring E. coli strains under different aeration conditions. Three independent cultivations were performed.
Fig. 5.
Time-course profiles of glucose consumption by Bcd- or IlvE- harboring E. coli strains under different aeration conditions. Three independent cultivations were performed.
Fig. 6.
Time-course profiles of cell growth for Bcd- or IlvE- harboring E. coli strains under different aeration conditions. Three independent cultivations were performed.
Under microaerobic cultivation conditions, the Bcd-possessing strain V1 cat-PL-bcd5.7 accumulated 2.2-fold more valine than the IlvE-possessing strain V1 cat-PL-ilvE5.7. Additionally, decreasing the oxygen supply resulted in a 32% increase in Val accumulation by the Bcd-possessing strain V1 cat-PL-bcd5.7 compared with that in aerobic conditions. Thus, the Bcd-possessing strain was capable of the efficient synthesis of valine under both aeration conditions. In contrast, Val production by the IlvE-possessing strain V1 cat-PL-ilvE5.7 was decreased by 2.4 times under microaerobic cultivation conditions, which clearly indicated a drawback of the AT-mediated step limited by an insufficient supply of l-glutamate. A decrease in optical density (∼2-fold) and a significant amount of residual glucose were observed under microaerobic cultivation conditions for both the tested strains. Under these conditions, the Bcd-containing strain accumulated Val with a 2.0-fold increased yield (Y, m/m) over that of V1 cat-PL-ilvE5.7. Additionally, the Bcd-possessing strain accumulated Val with a 1.4-fold increased yield under microaerobic conditions compared to that of V1 сat-PL-ilvE5.7 under aerobic conditions, but Val accumulation was decreased by 7.1%. LeuDH catalyzed the amination of KIV much more efficiently than AT under microaerobic conditions. The IlvE-possessing strain showed higher Val accumulation (9.8 g/L) under aerobic conditions, which can be explained by the level of NADPH in the cells.
The application of LeuDH instead of AT at the final step of valine biosynthesis altered the amino group donor and cofactor requirement for valine synthesis from l-glutamate and NADPH to NH4+ and NADH, respectively (Fig. 2), which increased both valine accumulation and yield under microaerobic conditions. In this case, a sufficient NH4+ supply and sufficient intracellularly accumulated NADH are the main requirements for efficient valine synthesis by LeuDH.
The obtained results are in agreement with the data obtained by the application of a similar approach in C. glutamicum (Hasegawa et al., 2012, 2013), where the improvement of the redox cofactor balance in valine synthesis by the replacement of NADPH-dependent reactions (AT and KARI) with NADH-dependent reactions resulted in increased Val accumulation, yield and glucose consumption rate.
The Bcd-containing E. coli strain used in our experiments possessed unmodified pathways for mixed fermentation (lactate, ethanol, etc.); therefore, further steps in the improvement of valine production under microaerobic conditions should include the restriction of these pathways’ functioning to make “valine fermentation” the main route for the required NADH oxidation. Additionally, changes in the KARI reaction cofactor specificity from NADPH to NADH and/or enhancement of PntAB transhydrogenase functioning seem promising for further improvement of the redox cofactor balance in the process of valine production under microaerobic conditions.
4. Conclusions
LeuDH from different species is widely used for the synthesis of a range of compounds by biotransformation, such as l-ABA from threonine (Тао et al., 2014) and l-tert-leucine from trimethylpyruvate (TMP) (Zhu et al., 2016). Additionally, NADH-dependent LeuDH from B. sphaericus was used instead of endogenous NADPH-dependent AT for successful valine synthesis in C. glutamicum under oxygen deprivation (Hasegawa et al., 2012, 2013). l-Alanine synthesis was shown under oxygen deprivation conditions in C. glutamicum by using the alanine dehydrogenase AlaD from B. sphaericus (Jojima et al., 2010). Changing the cofactor requirements from NADPH to NADH in AA biosynthesis resulted in increased accumulation and glucose consumption rates under oxygen deprivation in C. glutamicum (Hasegawa et al., 2012, 2013; Jojima et al., 2010). The efficient isobutanol synthesis in E. coli under anaerobic conditions at theoretical yield by using the NADH-dependent pathway was shown (Bastian et al., 2011).
Microaerobic conditions may be preferable for the production of valine considering that pyruvate, generated by the EMP pathway, is simultaneously a starting compound for the synthesis of this amino acid and a substrate for pyruvate dehydrogenase (PDH), which involves pyruvate in the respiratory process. At the same time, AHASes, which are responsible for the first step of valine synthesis from pyruvate, cannot compete with PDH for this substrate due to their approximately two orders of magnitude lower affinity (Bisswanger, 1981). Thus, limiting PDH activity by cultivation under oxygen deprivation conditions seems promising for valine production.
In this study, we have shown the application of leucine dehydrogenase Bcd from B. subtilis for valine synthesis in E. coli under microaerobic conditions. We have demonstrated the effective synthesis of valine by means of so-called “valine fermentation” (Fig. 1). The Bcd-possessing valine-producing strain containing one chromosomal copy of the artificial operon PL-ilvBNN17KDA was able to accumulate a 2.2-fold higher amount of valine with a 2.0-fold increased yield (m/m) compared with the IlvE-possessing strain under microaerobic cultivation conditions. Additionally, the Bcd-possessing strain accumulated Val with 1.4-fold increased yield (m/m) under microaerobic conditions compared to that of the IlvE-possessing strain under aerobic conditions, although Val accumulation was decreased by 7.1%. Thus, microaerobic fermentation can be favorable as an economical, environmentally friendly process for production at scale with high yield.
Declarations
Author contribution statement
Ekaterina A. Savrasova: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Nataliya V. Stoynova: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Contributor Information
Ekaterina A. Savrasova, Email: ekaterina_savrasova@agri.ru.
Nataliya V. Stoynova, Email: nataliya_stoynova@agri.ru.
References
- Atsumi S., Hanai T., Liao J.C. Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature. 2008;451:86–90. doi: 10.1038/nature06450. [DOI] [PubMed] [Google Scholar]
- Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K.A., Tomita M., Wanner B.L., Mori H. Construction of Escherichia coli K-12 in-frame single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006;2 doi: 10.1038/msb4100050. 2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker P.J., Turnbull A.P., Sedelnikova S.E., Stillman T.J., Rice D.W. A role for quaternary structure in the substrate specificity of leucine dehydrogenase. Structure (Lond.) 1995;3:693–705. doi: 10.1016/s0969-2126(01)00204-0. [DOI] [PubMed] [Google Scholar]
- Bartek T., Blombach B., Zönnchen E., Makus P., Lang S., Eikmanns B.J., Oldiges M. Importance of NADPH supply for improved L-valine formation in Corynebacterium glutamicum. Biotechnol. Prog. 2010;26:361–371. doi: 10.1002/btpr.345. [DOI] [PubMed] [Google Scholar]
- Bastian S., Liu X., Meyerowitz J.T., Snow C.D., Chen M.M., Arnold F.H. Engineered ketol-acid reductoisomerase and alcohol dehydrogenase enable anaerobic 2-methylpropan-1-ol production at theoretical yield in Escherichia coli. Metab. Eng. 2011;13(3):345–352. doi: 10.1016/j.ymben.2011.02.004. [DOI] [PubMed] [Google Scholar]
- Bisswanger H. Substrate specificity of the pyruvate dehydrogenase complex from Escherichia coli. J. Biol. Chem. 1981;256(2):815–822. [PubMed] [Google Scholar]
- Blombach B., Schreiner M.E., Bartek T., Oldiges M., Eikmanns B.J. Corynebacterium glutamicum tailored for high-yield L-valine production. Appl. Microbiol. Biotechnol. 2008;79(3):471–479. doi: 10.1007/s00253-008-1444-z. [DOI] [PubMed] [Google Scholar]
- Böck A., Sawers G. Fermentation. In: Neidhardt F.C., Curtiss R. III., Ingraham J.L., Lin E.C.C., Low K.B., Magasanik B., Reznikoff W.S., Riley M., Schaechter M., Umbarger H.E., editors. Escherichia coli and Salmonella: Cellular and Molecular Biology. second ed. ASM; Washington: 1996. pp. 262–282. [Google Scholar]
- Collier R.H., Kohlhaw G. Nonidentity of the aspartate and the aromatic amino- transferase components of transaminase A in Escherichia coli. J. Bacteriol. 1972;112(1):365–371. doi: 10.1128/jb.112.1.365-371.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper C.M., Fernstorm G.A., Miller S.A. Performance of agitated gas–liquid contactors. Ind. Eng. Chem. 1944;36:504–509. [Google Scholar]
- Datsenko K.A., Wanner B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 2000;97(12):6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debarbouille M., Gardan R., Arnaud M., Rapoport G. Role of BkdR, a trans-criptional activator of the SigL-dependent isoleucine and valine degradation pathway in Bacillus subtilis. J. Bacteriol. 1999;181(7):2059–2066. doi: 10.1128/jb.181.7.2059-2066.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Ochoa F., Gomez E. Bioreactor scale-up and oxygen transfer rate in microbial processes: an overview. Biotechnol. Adv. 2009;27:153–176. doi: 10.1016/j.biotechadv.2008.10.006. [DOI] [PubMed] [Google Scholar]
- Georgi T., Rittmann D., Wendisch V.F. Lysine and glutamate production by Corynebacterium glutamicum on glucose, fructose and sucrose: roles of malic enzyme and fructose-1,6-bisphosphatase. Metab. Eng. 2005;7:291–301. doi: 10.1016/j.ymben.2005.05.001. [DOI] [PubMed] [Google Scholar]
- Grenier F., Matteau D., Baby V., Rodrigue S. Complete genome sequence of Escherichia coli BW25113. Genome Announc. 2014;2(5) doi: 10.1128/genomeA.01038-14. e01038–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldimann A., Wanner B. Conditional-replication, integration, excision, and retrieval plasmid-host systems for gene structure-function studies of bacteria. J. Bacteriol. 2001;183(21):6384–6393. doi: 10.1128/JB.183.21.6384-6393.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa S., Uematsu K., Natsuma Y., Suda M., Hiraga K., Jojima T., Inui M., Yukawa H. Improvement of the redox balance increases L-valine production by Corynebacterium glutamicum under oxygen deprivation conditions. Appl. Environ. Microbiol. 2012;78:865–875. doi: 10.1128/AEM.07056-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa S., Suda M., Uematsu K., Natsuma Y., Hiraga K., Jojima T., Inui M., Yukawa H. Engineering of Corynebacterium glutamicum for high-yield L-valine production under oxygen deprivation conditions. Appl. Environ. Microbiol. 2013;79:1250–1257. doi: 10.1128/AEM.02806-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helling R.B. Why does Escherichia coli have two primary pathways for synthesis of glutamate? J. Bacteriol. 1994;176(15):4664–4668. doi: 10.1128/jb.176.15.4664-4668.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero M., de Lorenzo V., Timmis K.N. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 1990;172(11):6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K., Kuramitsu S., Aki K., Watanabe Y., Takagi T., Nishigai M., Ikai A., Kagamiyama H. Branched-chain amino acid aminotransferase of Escherichia coli: overproduction and properties. J. Biochem. 1988;104:777–784. doi: 10.1093/oxfordjournals.jbchem.a122549. [DOI] [PubMed] [Google Scholar]
- Inui M., Kawaguchi H., Murakami S., Vertès A.A., Yukawa H. Metabolic engineering of Corynebacterium glutamicum for fuel ethanol production under oxygen-deprivation conditions. J. Mol. Microbiol. Biotechnol. 2004;8:243–254. doi: 10.1159/000086705. [DOI] [PubMed] [Google Scholar]
- Inui M., Murakami S., Okino S., Kawaguchi H., Vertès A.A., Yukawa H. Metabolic analysis of Corynebacterium glutamicum during lactate and succinate productions under oxygen deprivation conditions. J. Mol. Microbiol. Biotechnol. 2004;7:182–196. doi: 10.1159/000079827. [DOI] [PubMed] [Google Scholar]
- Inui M., Suda M., Okino S., Nonaka H., Puskás L.G., Vertès A.A., Yukawa H. Transcriptional profiling of Corynebacterium glutamicum metabolism during organic acid production under oxygen deprivation conditions. Microbiology. 2007;153:2491–2504. doi: 10.1099/mic.0.2006/005587-0. [DOI] [PubMed] [Google Scholar]
- Jojima T., Fujii M., Mori E., Inui M., Yukawa H. Engineering of sugar metabolism of Corynebacterium glutamicum for production of amino acid L-alanine under oxygen deprivation. Appl. Microbiol. Biotechnol. 2010;87:159–165. doi: 10.1007/s00253-010-2493-7. [DOI] [PubMed] [Google Scholar]
- Kabus A., Georgi T., Wendisch V.F., Bott M. Expression of the Escherichia coli pntAB genes encoding a membrane-bound transhydrogenase in Corynebacterium glutamicum improves L-lysine formation. Appl. Microbiol. Biotechnol. 2007;75:47–53. doi: 10.1007/s00253-006-0804-9. [DOI] [PubMed] [Google Scholar]
- Lawther R.P., Hatfield G.W. Multivalent translational control of transcription termination at attenuator of ilvGEDA operon of Escherichia coli K-12. Proc. Natl. Acad. Sci. U.S.A. 1980;77(4):1862–1866. doi: 10.1073/pnas.77.4.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawther R.P., Wek R.C., Lopes J.M., Pereira R., Taillon B.E., Hatfield G.W. The complete nucleotide sequence of the ilvGMEDA operon of Escherichia coli K-12. Nucleic Acids Res. 1987;15(5):2137–2155. doi: 10.1093/nar/15.5.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Zhu D., Hyatt B.A., Malik F.M., Biehl E.R., Hua L. Cloning, protein sequence clarification, and substrate specificity of a leucine dehydrogenase from Bacillus sphaericus ATCC4525. Appl. Biochem. Biotechnol. 2009;158:343–351. doi: 10.1007/s12010-008-8304-2. [DOI] [PubMed] [Google Scholar]
- Livesey G., Lund P. Isolation and characterization of leucine dehydrogenase from Bacillus subtilis. Methods Enzymol. 1988;166:282–288. doi: 10.1016/s0076-6879(88)66038-1. [DOI] [PubMed] [Google Scholar]
- Miller J.H. Cold Spring Harbor Laboratory Press; U.S: 1972. Experiments in Molecular Genetics. [Google Scholar]
- Minaeva N.I., Gak E.R., Zimenkov D.V., Skorokhodova A.Y., Biryukova I.V., Mashko S.V. Dual-In/Out strategy for genes integration into bacterial chromosome: a novel approach to step-by-step construction of plasmid-less marker-less recombinant E. coli strains with predesigned genome structure. BMC Biotechnol. 2008;8:63. doi: 10.1186/1472-6750-8-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata S., Tanizawa K., Esaki N., Sakamoto Y., Ohshima T., Tanaka H., Soda K. Gene cloning and sequence determination of leucine dehydrogenase from Bacillus stearo-thermophilus and structural comparison with other NAD(P)+-dependent dehydrogenases. Biochemistry. 1988;27(25):9056–9062. doi: 10.1021/bi00425a026. [DOI] [PubMed] [Google Scholar]
- Ohshima T., Nishida N., Bakthavatsalam S., Kataoka K., Takada H., Yoshimura T., Esaki N., Soda K. The purification, characterization, cloning and sequencing of the gene for a halostable and thermostable leucine dehydrogenase from Thermoactinomyces intermedius. Eur. J. Biochem. 1994;222:305–312. doi: 10.1111/j.1432-1033.1994.tb18869.x. [DOI] [PubMed] [Google Scholar]
- Park J.H., Lee S.Y. Fermentative production of branched chain amino acids: a focus on metabolic engineering. Appl. Microbiol. Biotechnol. 2010;85:491–506. doi: 10.1007/s00253-009-2307-y. [DOI] [PubMed] [Google Scholar]
- Park J.H., Lee K.H., Kim T.Y., Lee S.Y. Metabolic engineering of Escherichia coli for the production of L-valine based on transcriptome analysis and in silico gene knockout simulation. Proc. Natl. Acad. Sci. U.S.A. 2007;104:7797–7802. doi: 10.1073/pnas.0702609104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.H., Kim T.Y., Lee K.H., Lee S.Y. Fed-batch culture of Escherichia coli for L-valine production based on in silico flux response analysis. Biotechnol. Bioeng. 2011;108(4):934–946. doi: 10.1002/bit.22995. [DOI] [PubMed] [Google Scholar]
- Partridge J.D., Sanguinetti G., Dibden D.P., Roberts R.E., Poole R.K., Green J. Transition of Escherichia coli from aerobic to micro-aerobic conditions involves fast and slow reacting regulatory components. J. Biol. Chem. 2007;282(15):11230–11237. doi: 10.1074/jbc.M700728200. [DOI] [PubMed] [Google Scholar]
- Sakamoto N., Kotre A.M., Savageau M.A. Glutamate dehydrogenase from Escherichia coli: purification and properties. J. Bacteriol. 1975;124(2):775–783. doi: 10.1128/jb.124.2.775-783.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Russell D.W. Cold Spring Harbor Laboratory Press; U.S: 2001. Molecular Cloning: a Laboratory Manual. [Google Scholar]
- Sauer U., Canonaco F., Heri S., Perrenoud A., Fischer E. The soluble and membrane-bound transhydrogenases UdhA and PntAB have divergent functions in NADPH metabolism of Escherichia coli. J. Biol. Chem. 2004;279:6613–6619. doi: 10.1074/jbc.M311657200. [DOI] [PubMed] [Google Scholar]
- Savrasova E.A., Kivero A.D., Shakulov R.S., Stoynova N.V. Use of the valine biosynthetic pathway to convert glucose into isobutanol. J. Ind. Microbiol. Biotechnol. 2011;38(9):1287–1294. doi: 10.1007/s10295-010-0907-2. [DOI] [PubMed] [Google Scholar]
- Sycheva E.V., Serebryanyy V.A., Yampolskaya T.A., Preobrazhenskaya E.S., Stoynova N.V. 2009. A Mutant Acetolactate Synthase and a Method for Producing Branched-Chain L-Amino Acids. EP 1942183 B1. [Google Scholar]
- Тао R., Jiang Y., Zhu F., Yang S. А one-pot system for production of L-2-aminobutyric acid from L-threonine by L-threonine deaminase and а NADH-regeneration system based on L-leucine dehydrogenase and formate dehydrogenase. Вiotechnol. Lett. 2014;36(4):835–841. doi: 10.1007/s10529-013-1424-y. [DOI] [PubMed] [Google Scholar]
- Umbarger H.E. Biosynthesis of the branched-chain amino acids. In: Neidhardt F.C., Curtiss R. III., Ingraham J.L., Lin E.C.C., Low K.B., Magasanik B., Reznikoff W.S., Riley M., Schaechter M., Umbarger H.E., editors. Escherichia coli and Salmonella: Cellular and Molecular Biology. second ed. ASM; Washington: 1996. pp. 442–457. [Google Scholar]
- Wang X., Zhang H., Quinn P.J. Production of L-valine from metabolically engineered Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 2018;102(10):4319–4330. doi: 10.1007/s00253-018-8952-2. [DOI] [PubMed] [Google Scholar]
- Wek R.C., Hauser C.A., Hatfield G.W. The nucleotide sequence of the ilvBN operon of Escherichia coli: sequence homologies of the acetohydroxy acid synthase isozymes. Nucleic Acids Res. 1985;13(11):3995–4010. doi: 10.1093/nar/13.11.3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L., Wu Z., Jin J., Tang S. Directed evolution of leucine dehydrogenase for improved efficiency of L-tert-leucine synthesis. Appl. Microbiol. Biotechnol. 2016;100(13):5805–5813. doi: 10.1007/s00253-016-7371-5. [DOI] [PubMed] [Google Scholar]