Abstract
This study was aimed to explore if lncRNA MALAT1 would modify chemo-resistance of non-small cell lung cancer (NSCLC) cells by regulating miR-197-3p and p120 catenin (p120-ctn). Within this investigation, we totally recruited 326 lung cancer patients, and purchased 4 NSCLC cell lines of A549, H1299, SPC-A-1 and H460. Moreover, cisplatin, adriamycin, gefitinib and paclitaxel were arranged as chemotherapies, and half maximal inhibitory concentration (IC50) values were calculated to evaluate the chemo-resistance of the cells. Furthermore, mice models of NSCLC were also established to assess the impacts of MALAT1, miR-197-3p and p120-ctn on tumor growth. Our results indicated that MALAT1 and miR-197-3p were both over-expressed within NSCLC tissues and cells, when compared with normal tissues and cells (P < 0.05). The A549, H460, SPC-A-1 and SPC-A-1 displayed maximum resistances to cisplatin (IC50 = 15.70 μg/ml), adriamycin (IC50 = 5.58 μg/ml), gefitinib (96.82 μmol/L) and paclitaxel (141.97 nmol/L). Over-expression of MALAT1 and miR-197-3p, or under-expression of p120-ctn were associated with promoted viability and growth of the cancer cells (P < 0.05), and they could significantly strengthen the chemo-resistance of cancer cells (P < 0.05). MALAT1 Wt or p120-ctn Wt co-transfected with miR-197-3p mimic was observed with significantly reduced luciferase activity within NSCLC cells (P < 0.05). Finally, the NSCLC mice models were observed with larger tumor size and weight under circumstances of over-expressed MALAT1 and miR-197-3p, or under-expressed p120-ctn (P < 0.05). In conclusion, MALAT1 could alter chemo-resistance of NSCLC cells by targeting miR-197-3p and regulating p120-ctn expression, which might assist in improvement of chemo-therapies for NSCLC.
Keywords: Animal model, Cell viability, Chemo-sensitivity, LncRNA MALAT1, MiR-197-3p, non-small cell lung cancer, P120-catenin
INTRODUCTION
Lung cancer is a common respiratory cancer, with a high degree of malignancy (Miller et al., 2016). One of its subdivision, non-small cell lung cancer (NSCLC) (Petrelli et al., 2009), occupied around 85% of all lung cancer cases, with the 5-year survival rate of as low as 15% (Camps et al., 2009). Considering that the clinical manifestations of early-stage NSCLC were mostly hidden and non-specific, the subjects have already reached the advanced stage of NSCLC when being diagnosed. Hence, timely discovery of the NSCLC onset was particularly vital. Besides, since the chemo-resistance of tumor cells could greatly influence the curative effect of NSCLC, a deep clarification of the etiology inherent in NSCLC development and chemo-sensitivity also helped immensely.
The lncRNAs, a type of non-coding RNAs with the length of > 200 bp, have been documented to characterize tumorigenesis through epigenetic, transcriptional and post-transcriptional regulations (Wapinski and Chang, 2011; Zhang et al., 2012). Among them, the metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), also called as nuclear enriched abundant transcript (NEAT2), was over-expressed within deteriorated neoplasms, including lung cancer, melanoma, prostate cancer, breast cancer, colon cancer and liver cancer (Fan et al., 2014; Schmidt et al., 2011; Wang et al., 2014). Taking lung cancer for instance, the MALAT1 expression within peripheral blood could be deemed as a reliable criterion for judging the presence of NSCLC, with features of minimal invasion and high specificity (Weber et al., 2013). It was additionally held that MALAT1 might expedite the epithelial-mesenchymal transition (EMT) process (Wu et al., 2018) and metastasis of lung cancer cell lines (Chen et al., 2017; Shen et al., 2015). The above-mentioned findings provided sound evidences for that MALAT1 might heighten the resistance of neoplastic cells. Nevertheless, quite finite chemotherapies and downstream pathways were investigated relevant to MALAT1 and NSCLC.
As was documented before, lncRNAs could serve as the competing endogenous RNA (ceRNA) to elevate or lower the risk of disorders by modifying the biological function of miRNAs (Cesana et al., 2011). The starbase database (Li et al., 2014) was usually depended on to predict the sponging miRNAs of MALAT1. One of the miRNAs was turned out as miR-197, which was involved with the progression of human neoplasms and metabolic disorders, such as NSCLC, diabetes mellitus type 2 and myocardial infarction (Dai et al., 2014; Fiori et al., 2014; Hamada et al., 2013; Zampetaki et al., 2010; 2012; Zheng et al., 2011; Zhou et al., 2014). For instance, miR-197 was verified to facilitate exacerbation of NSCLC through a p53-dependent approach to inhibit apoptosis of cancer cells (Fiori et al., 2014). Of note, this function of miR-197 was hypothesized, to some extent, as a consequence of its mediating expression of tumor-repressive genes [e.g. p120-catanin (p120-ctn)] (Du et al., 2009; Hamada et al., 2013; Zheng et al., 2011). The p120-ctn herein was capable of interacting with certain critical biomarkers of EMT process (e.g. E-cadherin), and it was also responsible for intensifying the proliferative ability of lung cancer cells (Jiang et al., 2012). Taken together, MALAT1, miR-197 and p120-ctn were hypothesized to be interconnected in modifying development and even chemo-resistance of NSCLC, although few investigations have been performed with respect to this point.
Hence, this investigation was performed to testify the contribution of MALAT1, miR-197-3p and p120-ctn to NSCLC progression. Meanwhile, whether they could modulate chemo-sensitivity of NSCLC cells were also validated, which might be conducive to perfecting the chemo-therapy for NSCLC.
MATERIALS AND METHODS
Collection of NSCLC tissues
From September 2016 to August 2018, we gathered 326 pairs of NSCLC samples and para-carcinoma tissues from the pathological department of the First Affiliated Hospital of Xi’an Jiaotong University. The lung cancer tissues were histologically grouped according to the lung cancer classification criteria issued by World Health Organization (Travis, 2004), and they were classified in line with TNM staging criteria of lung cancer, which was formulated by International Union against Cancer (UICC) (7th edition) (Goldstraw, 2011). No patients have received radiotherapy or chemotherapy pre-operatively, and all the participants were given standard surgery post-operatively. The subjects all have signed informed consents, and this investigation was approved by the First Affiliated Hospital of Xi’an Jiaotong University and the ethics committee of the First Affiliated Hospital of Xi’an Jiao-tong University.
Cell culture
Three NSCLC cell lines (i.e.A549, H1299, H460 and SPC-A-1) and normal bronchial epithelial cell line (i.e. HBE) were purchased from American Type Culture Collection (Manassas, USA), and another NSCLC cell line (i.e. SPC-A-1) was taken from Shanghai Cell Bank of Chinese Academy of Sciences (China). The cells were cultured within high-glucose DMEM complete medium (Hyclone, USA) that included 10% fetal bovine serum (FBS), 100U/ml penicillin and 100 Lg/ml streptomycin. Then we maintained the cells in 5% CO2 and saturated humidity at 37°C. The culture solution was changed daily, and cells were managed with secondary culture at the ratio of 1:2 every 3 days.
Cell transfection
After being digested with 0.25% trypsin, the cells at the logarithmic phase were inoculated into 6-well plates at the density of 5 × 105 per well. Moreover, the primer sequences for MALAT1-siRNAs (Genepharma, China) were specifically designed as: 1) siRNA1: 5′-GAGGUGUAAAGGGAUUUAUTT-3′ (upstream) and 5′-AUAAAUCCCUUUACACCUCTT-3′ (downstream); 2) siRNA2: 5′-GGCAUUUGCAUCUUUAAAUTT-3′ (upstream) and 5′-AUUUAAAGAUGCAAAUGCCTT-3′ (downstream); and 3) siRNA3: 5′-CCCUCUAAAUAAGGAAUAATT-3′ (upstream) and 5′-UUAUUCCUUAUUUAGAGGGTT-3′ (downstream). The pcDNA-MALAT1, si-MALAT1 and NC were provided by GenePharma (Shanghai, China). Besides, miR-197-3p mimic (5′-CGGGUAGAGAGGGCAGUGGGAGG-3′) (Dharmacon, USA) and miR-197-3p inhibitor (5′-AAGUGGUGGAAGAGGUGGGUCG-3′) (Dharmacon, USA) were designed for the mature endogenous miR-197, and miR-NC (5′-CAGUACUUUUGUGUAGUACAA-3′) was also provided by Dharmacon (USA). The transfection process was initiated when cell fusion reached 60%-70%, and was conducted following the specification of Lipofectamine™2000 reagent (Invitrogen, USA).
As for the construction of p120-ctn siRNA, the DNA oligos that respectively contained siRNA sequences for p120-ctn were annealed. The primers for synthesizing siRNAs of p120-ctn incorporated: A) 5′-GGATCACAGTCACCTTCTA-3′ (upstream) and 5′-TAGAAGGTGACTGTGATCC-3′ (downstream); B) 5′-GCACTTGTATTACAGACAA-3′ (upstream) and 5′-TTGTCTGTAATACAAGTGC-3′ (downstream); and C) 5′-GGATAACAAGATTGCCATA-3′ (upstream) and 5′-TATGGCAATCTTGTTATCC-3′ (downstream). Subsequently, the pGCsi vectors that were digested by BamHI and HindIII enzymes (GeneChem, China) were connected to them. The vectors were transduced into host bacteria (i.e. DH5α), and were sequenced by virtue of TakaRa (Japan). After being transduced into lung cancer cell lines with Lipofectamine™2000 reagent (Invitrogen, USA), G148 (Invitrogen, USA) was applied to screen positive cells for cloning. Western blot and RT-PCR were then conducted to identify the transfection results, and the mono-clones that were successfully transfected were continuously cultivated within RPMI 1640 medium.
Conduction of quantitative real time-polymerase chain reaction (qRT-PCR)
We extracted total RNA from cultured cells and transplanted tumor tissues using Trizol reagent (Invitrogen, USA). The RNAs were reversely transcribed to cDNAs in accordance with the manual of the reverse transcription kit provided by TaKaRa (Japan). Subsequently, PCR reaction was carried out following the guidance of SYBR Green PCR kit (Toyobo, Japan), and was run on the PCR iCycler device (Bio-Rad, USA). The particularized PCR conditions were: 1) pre-degeneration at 95°C for 10 min; 2) 40 cycles of degeneration at 95°C for 50s, annealing at 60°C for 50s and extension at 72°C for 60s; and 3) final extension at 72°C for 5 min. Moreover, the relative expression of target genes were signified through 2−ΔΔCt method and the primer sequences were listed in Table 1. GAPDH was set as the internal control for MALAT1 and p120-ctn, whereas U6 was arranged the reference for miR-197-3p.
Table 1.
Primer sequences of for quantitative real-time PCR
| Gene | Sequence |
|---|---|
| MALAT1 | F:5′-GCGCTATTATCCTAAGGTCA-3′ |
| R:5′-CTTGGCCAAGTCTGTTATGT-3′ | |
| GAPDH | F:5′-GGAGTCCACTGGCGTCTTCACCACC-3′ |
| R:5′-GCAGGTCAGGTCCACCACTGACACG-3′ | |
| miR-197-3p | F: 5′-AGTTGTTCACCACCTTCTCCAC-3′ |
| R: 5′-TATCGTTGTACTCCAGTCCAAGTC-3′ | |
| U6 | F: 5′-GTGCTCGCTTCGGCAGCACATATAC-3′ |
| R: 5′-AAAAATATGGAACGCTCACGAATTTG-3′ |
Implementation of western blotting
Total protein was extracted from the cultivated cells and transplanted tumor tissues with application of RIPA lysis buffer (Beyotime, China). After determining the concentration of proteins with BCA protein assay kit, the proteins were supplemented with 5×SDS loading buffer (Beyotime, China) that has been boiled for 10 min before being stored at −80°C. Subsequently, 30 μg protein under each treatment was electrophoretically separated on the sodium dodecyl sulfate-polyacrylamide gel (SDS-PAGE) (Beyotime, China). The resultants were then transferred onto the polyvinylidene fluoride (PVDF) membrane following the semi-dry process. After 2-h blockage at room temperature with 5% skim milk, the mouse-anti-human primary antibodies against E-cadherin (1: 1000, Abcam, USA), N-cadherin (1: 1 000, Abcam, USA), vimentin (1:1000, Abcam, USA), p120-ctn (1:1000, BD Bioscience, USA) and GAPDH (1:1 000, Abcam, USA) were added. After being left at -4°C for overnight, the membrane was washed with TBST for 3 times, and then the secondary antibodies marked by goat-anti-mouse horseradish peroxidase (HRP) (1: 2000, Abcam, USA) were added for another 2-h incubation at 37°C. In order to develop images in the dark room, the chemi-luminescence reagent was supplemented. The images of bands were collected utilizing Chem-iDocXRS imaging system (Bio-Rad, USA), and were analyzed using Quantity One 4.6.2 software.
Evaluation of chemo-resistance of NSCLC cells by MTT assay
Cells at the density of 1 × 104/ml (100 μl per well) were seeded into the 96-well plates for overnight. The nutrient solution with distinct concentrations of cisplatin, adriamycin, gefitinib, paclitaxel were added into the plate (200 μl/cells) for 24-h cultivation. Subsequently, 20 μl MTT (5 mg/ml) that was prepared with pH-7.5 PBS was added into each well for 4-h incubation. Then the liquid waste was carefully removed, and 150 μl DMSO was added into each well for 10-min shaking at 60 r/min. After that, the optical values of each well at the wavelength of 490 nm were measured by means of enzyme-linked immune-metric meter. The inhibitory rate of cells was calculated based on the formula of [1-treatment groupOD490/control groupOD490] × 100%, and the half inhibition concentrations (IC50s) of the cells were also calculated when different drugs were treated.
CCK8 assay
Cells at the logarithmic growth phase were inoculated into 96-well plates at the density of 1 × 103/well, and then they were cultivated in 5%CO2 at 37°C for overnight. Next, 10 μl CCK8 solution (Takara, USA) was added to each well for 4-h culture, and the absorbance value of each well was determined at the wavelength of 450 nm.
Colony formation assay
The cells after 24-h transfection were inoculated into 6-well plates at the density of 500 per well. After 12-d culture in 5% CO2 and saturated humidity at 37°C, we terminated the cultivation and discarded the medium. After washing the cells with PBS for twice, 4% paraformaldehyde solution was added for 15-min fixation at room temperature. Then the stationary liquid was removed, and crystal violet solution was added for 20-min staining. Finally, the 6-well plate was put upside down, and a transparent film with grids was superimposed to count cell colonies by naked eye.
Cell apoptosis
Cells at the logarithmic phase were inoculated into 6-well plates at the density of 5 × 105/well. After 24-h cultivation, cells were washed with pre-cooled phosphate buffer solution (PBS) for 2–3 times, and were re-suspended after addition of 100 μl buffer solution. According to the guidance of Annexin V-fluorescein isothiocyanate (FITC) apoptosis detection kit (Beyotime, China), 10 μl FITC (50 mg/L) and 5 μl propidium iodide (PI) (50 mg/L) were added to culture cells at room temperature for 15 min. Afterwards, 200 μl binding buffer was supplemented, and the solution was loaded into the flow cytometry (model: FACSCalibur, BD Biosciences, Australia) after being set aside for 5 min.
Dual-luciferase reporter gene assay
The MALAT1 Wt was obtained by amplifying the MALAT1 fragments that included binding sites with miR-197-3p via PCR, and MALAT1 Mut was constructed similarly other than mutating the binding sites. Then cells at the logarithmic phase were classified under treatments of pmirGLO-MALAT1 Wt+miR-197-3p mimic, pmirGLO-MALAT1 Wt+miR-NC, pmirGLO-MALAT1 Mut+miR-197-3p mimic and pmirGLO-MALAT1 Mut+miR-NC. In addition, p120-ctn Wt was gained by amplifying the p120-ctn fragments that included binding sites with miR-197-3p via PCR. The p120-ctn Mut was constructed analogous, except that the binding sites were mutated. This series of cells were categorized into pmirGLO-p120-ctn Wt+miR-197-3p mimic group, pmirGLO-p120-ctn Wt+miR-NC group, pmirGLO-p120-ctn Mut+miR-197-3p mimic group and pmirGLO-p120-ctn Mut+miR-NC group. After 24-h transfection, we measured the fluorescence intensity of each group in accordance with the procedures of dual luciferase detection kit (Promega, USA).
Establishment of mice models
The BALB/C (nu-nu) nude mice aged at 4–5 weeks old and weighing 15–19 g were purchased from the experimental animal center of the First Affiliated Hospital of Xi’an Jiaotong University. They were tested and reared within specific-pathogen-free (SPF) circumstances that were featured by constant temperature and humidity. The rule of 12-h circadian rhythm was abided by, and these nude mice were allowed to eat and drink free. For another, we cultured NSCLC cells through the conventional approach mentioned above, and the concentration of viable cells was adjusted to 5 × 106/ml. Subsequently, the cell suspension (1 × 106 cells) was inoculated into the subcutaneous area of right anterior axilla within nude mice. Then the formation of the transplanted tumors was observed, and tumors with diameter ≥ 0.5 cm were regarded as neoplasia. At last, tumor tissues were cut off, and their long diameter (a, mm), short diameter (b, mm) and weight were measured. The volume (mm3) of the transplantation tumor was calculated as per the formula of πab2/6.
Statistical analyses
We adopted the SPSS 17.0 software to conduct data analyses throughout the manuscript. The homoscedasticity test was performed among groups, and all quantitative data were expressed in the form of mean ± standard deviation (SD). Comparisons between two groups were assessed utilizing SNK-q test, and analysis of variance (ANOVA) was performed to evaluate the differences among ≥ 3 groups. Moreover, the relationship between genetic expressions and clinically pathological features was appraised by Chi-square test. Also the Kaplan-Meier curve was established to determine the association of genetic expressions with overall survival post-operatively. It was considered statistically significant when P < 0.05.
RESULTS
Association of MALAT1 and miR-197-3p expressions with baseline characteristics of NSCLC patients
Among the NSCLC patients enrolled, the expressions of MALAT1 and miR-197-3p were apparently higher within their NSCLC tissues than within corresponding normal tissues (P < 0.05) (Fig. 1A). Simultaneously, both MALAT1 and miR-197-3p were expressed higher within A549, H1299, H460 and SPC-A-1 cell lines than within HBE cell line (Fig. 1B). According to the expressional quantity of MALAT1 and miR-197-3p, we divided these 326 NSCLC patients into highly-expressed MALAT1 group (> median MALAT1 expression, n = 232) and lowly-expressed MALAT1 group (≤ median MALAT1 expression, n = 94). The same crowd was also categorized into highly-expressed miR-197-3p group (> median miR-197-3p expression, n = 205) and lowly-expressed miR-197-3p group (≤ median miR-197-3p expression, n = 121). It was derived that highly-expressed MALAT1 and miR-197-3p were both positively correlated with larger tumor size (> 3 cm), poor differentiation and advanced TNM stage (III–IV) of NSCLC patients (P < 0.05), whereas hardly any association was found between the genetic expressions and age, gender and histological type (P > 0.05) (Table 2). Through application of Kaplan-Meier analysis, we found a positive correlation between highly-expressed MALAT1 or miR-197-3p and shorter overall survival of NSCLC patients, with lowly-expressed MALAT1 or miR-197-3p, respectively, as the reference (P <0.05) (Fig. 1C). Ultimately, abnormally higher expression of MALAT1 or miR-197-3p, smoking larger tumor size (> 3 cm) and poor differentiation could be regarded as potent candidates for predicting poor prognosis of lung cancer patients (all P < 0.05) (Table 3).
Fig. 1. Expressions of lncRNA MALAT1 and miR-197-3p within lung cancer tissues.
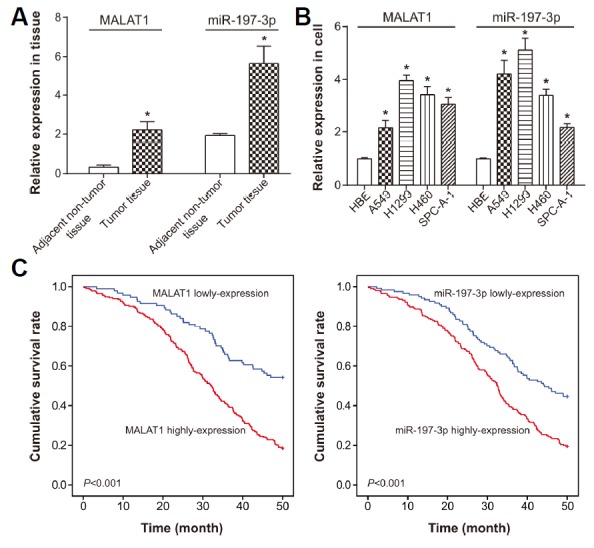
(A) MALAT1 and miR-197-3p expressions were compared between lung cancer tissues and adjacent normal tissues. *P < 0.05 when compared with adjacent normal tissues. (B) MALAT1 and miR-197-3p expressions were compared between lung cancer cell lines (i.e. A549, H1299, H460 and SPC-A-1) and HBE. *P < 0.05 when compared with HBE. (C) Highly-expressed MALAT1 and miR-197-3p were correlated with poorer prognosis of lung cancer patients, when compared with lowly-expressed MALAT1 and miR-197-3p, respectively.
Table 2.
Linkage of lncRNA MALAT1 and miR-197-3p expression with clinical characteristics of non-small cell lung cancer patients.
| Characteristics N = 326 |
LncRNA MALAT1 expression | miR-197-3p expression | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| Low | High | P value | Low | High | P value | |
| Age (years) | ||||||
| >60 | 54 | 149 | 0.253 | 81 | 122 | 0.181 |
| ≤60 | 40 | 83 | 40 | 83 | ||
| Gender | ||||||
| Male | 66 | 161 | 0.885 | 79 | 148 | 0.19 |
| Female | 28 | 71 | 42 | 57 | ||
| Smoking | ||||||
| Yes | 66 | 124 | 0.005 | 59 | 131 | 0.007 |
| No | 28 | 108 | 62 | 74 | ||
| Tumor size | ||||||
| >3 cm | 60 | 197 | <0.001 | 85 | 172 | 0.004 |
| ≤3 cm | 34 | 35 | 36 | 33 | ||
| Histology grade | ||||||
| Poorly and unknown | 14 | 63 | 0.018 | 17 | 60 | 0.002 |
| Well and moderate | 80 | 169 | 104 | 145 | ||
| Histological type | ||||||
| Adenocarcinoma | 48 | 134 | 0.270 | 60 | 122 | 0.217 |
| Squamous cell carcinoma | 30 | 73 | 44 | 59 | ||
| Others | 16 | 25 | 17 | 24 | ||
| TNM stage | ||||||
| III–IV | 53 | 166 | 0.008 | 49 | 170 | <0.001 |
| I–II | 41 | 66 | 72 | 35 | ||
| Lymph node metastasis | ||||||
| Yes | 44 | 84 | 0.076 | 35 | 93 | 0.003 |
| No | 50 | 148 | 86 | 112 | ||
Table 3.
Impacts of MALAT1 expression, miR-197-3p expression and clinical characteristics on the survival rates of non-small cell lung cancer patients
| Characteristics | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| Hazard Ratio | 95% CI | P value | Hazard Ratio | 95% CI | P value | |
| LncRNA MALAT1 expression | ||||||
| High vs. Low | 5.21 | 3.09–8.77 | <0.001 | 5.56 | 2.94–10.00 | <0.001 |
| miR-197-3p expression | ||||||
| High vs. Low | 3.32 | 2.02–5.46 | <0.001 | 2.63 | 1.35–5.00 | 0.004 |
| Age | ||||||
| >60 vs. ≤60 | 1.10 | 0.67–1.80 | 0.699 | 1.27 | 0.72–2.25 | 0.415 |
| Gender | ||||||
| Female vs. Male | 0.96 | 0.57–1.62 | 0.885 | 0.82 | 0.44–1.54 | 0.533 |
| Smoking | ||||||
| Yes vs. No | 1.71 | 1.05–2.77 | 0.030 | 2.15 | 1.18–3.93 | 0.013 |
| Tumor size | ||||||
| >3 cm vs. ≤3 cm | 2.95 | 1.69–5.12 | <0.001 | 2.02 | 1.05–3.90 | 0.036 |
| Histology grade | ||||||
| Poorly and unknown vs. Well and moderate | 3.92 | 1.86–8.23 | <0.001 | 2.53 | 1.14–5.62 | 0.023 |
| Histological type | ||||||
| Adenocarcinoma vs. Others | 0.82 | 0.48–1.41 | 0.473 | 0.69 | 0.36–1.29 | 0.241 |
| Squamous cell carcinoma vs. Others | 0.96 | 0.45–2.01 | 0.904 | 0.68 | 0.29–1.63 | 0.390 |
| TNM stage | ||||||
| III–IV vs. I–II | 1.95 | 1.18–3.20 | 0.009 | 0.83 | 0.42–1.63 | 0.581 |
| Lymph node metastasis | ||||||
| Yes vs. No | 1.20 | 0.73–1.97 | 0.467 | 1.00 | 0.55–1.81 | 0.989 |
Inhibitory effects of cisplatin, adriamycin, gemcitabine and paclitaxel on the growth of NSCLC cell lines
According to the results of CCK8 technique (Fig. 2A), the IC50 values of A549 (15.70 μg/ml), H1299 (9.17 μg/ml), H460 (3.00 μg/ml) and SPC-A-1 (0.69 μg/ml) implied that SPC-A-1 displayed the highest sensitivity to cisplatin (P < 0.05), with the tolerance of A549 to cisplatin standing on the top (P < 0.05). The IC50 values of cells after treatments with adriamycin were successively enlisted as: H460 (5.58 μg/ml) > H1299 (2.98 μg/ml) > SPC-A-1 (1.71 μg/ml) > A549 (1.09 μg/ml), suggesting H460 and A549, respectively, as most tolerant and sensitive cell lines to adriamycin (Fig. 2B). Furthermore, the tolerance of SPC-A-1 (IC50 = 96.82 μmol/L) to gefitinib was more evident than A549 (IC50 = 8.64 μmol/L), H1299 (IC50 = 35.73 μmol/L) and H460 (IC50 = 7.51 μmol/L) (Fig. 2C). Also SPC-A-1 (IC50 = 141.97 nmol/L) presented a tolerance to paclitaxel that was more pronounced than any other cell line (Fig. 2D). Considering the tolerance of H1299 and SPC-A-1 to 4 chemo-therapies being at the forefront, they were chosen for follow-up experiments.
Fig. 2.
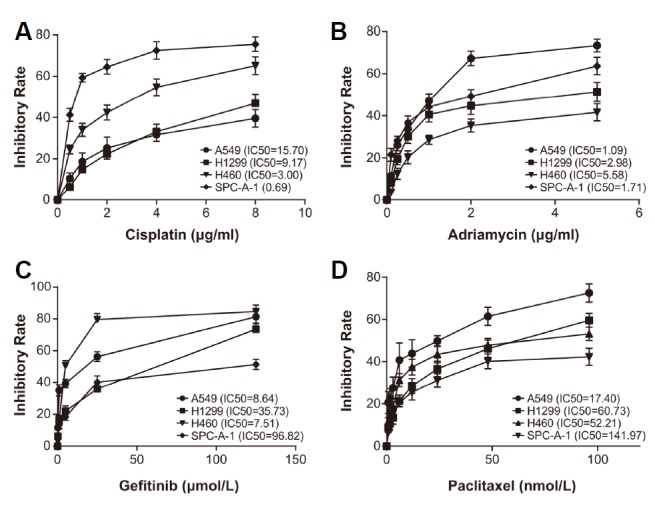
The lung cancer cells were compared regarding their sensitivities to chemotherapies, including cisplatin (A), adriamycin (B), gefitinib (C) and paclitaxel (D).
Effects of MALAT1 and miR-197-3p on the chemo-resistance of lung cancer cells
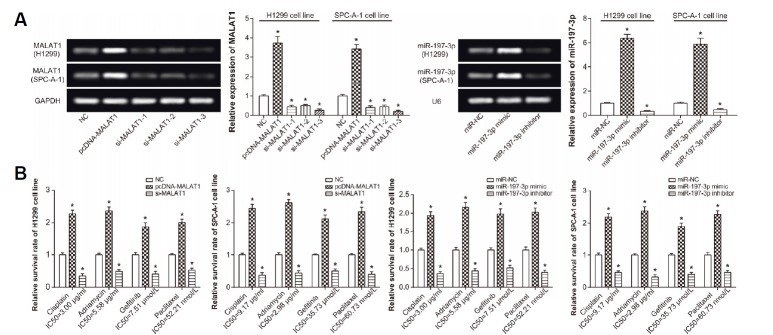
Among the three MALAT1-siRNAs, siRNA3 was indicated to be associated with the highest interfering efficiency of up to 80% (Fig. 3A). Besides, the expressional level of MALAT1 was raised to about 3.8 times of the original condition after transfection of pcDNA-MALAT1 (P < 0.05). Meanwhile, after transfection of miR-197-3p mimic, the expression quantity of miR-197-3p was elevated to 6.1 times of the control group (P < 0.05) (Fig. 3B). Nonetheless, cells transfected with miR-197-3p inhibitor possessed a miR-197-3p expression that was merely a quarter of the control group (P < 0.05).
Fig. 3. Impacts of MALAT1 and miR-197-3p on the sensitivity of lung cancer cells to chemo-therapies.
(A) The MALAT1 expression was assessed after treatments with pcDNA-MALAT1 and si-MALAT1s, and miR-197-3p expression was determined after transfection of miR-197-3p mimic and miR-197-3p inhibitor. *P < 0.05 when compared with NC. (B) The effects of MALAT1 and miR-197-3p on survival rates of lung cancer cells were evaluated after treatments with cisplatin, adriamycin, gefitinib and paclitaxel. *P < 0.05 when compared with NC. NC: negative control.
Furthermore, it was insinuated that transfection of pcDNA-MALAT1 produced higher survival rate of H1299 and SPC-A-1 cell lines than NC group, even after treatments of cisplatin, adriamycin, gefitinib and paclitaxel at their IC50 values (P < 0.05) (Fig. 3C). Conversely, silencing of MALAT1 degraded the viability of cells in comparison to NC group (P < 0.05). What’s more, transfection of miR-197-3p inhibitor, contrary to miR-197-3p mimic, could restrain cell growth to a greater degree than transfection of miR-NC, when chemotherapies were given (P < 0.05).
Effects of MALAT1 and miR-197-3p on proliferation, viability and apoptosis of NSCLC cells, as well as expressions of EMT-specific proteins
In line with Figs. 4A and 4B, the viability and proliferation of cells in the pcDNA-MALAT1 group and miR-197-3p mimic group markedly exceeded that in the NC group (P < 0.05), and suppression of MALAT1 or miR-197-3p expression could suppress viability and proliferation of the cells (P < 0.05). Furthermore, pcDNA-MALAT1 group and miR-197-3p mimic group were accompanied with over-expressed N-cadherin and vimentin, as well as under-expressed E-cadherin (P < 0.05) (Fig. 4C). By contrast, treatments of si-MALAT1 and miR-197-3p inhibitor generated contrary results (P < 0.05). In addition, the apoptotic proportion of cells that underwent transfection of si-MALAT1 or miR-197-3p inhibitor was above that of cells in the NC group (P < 0.05) (Fig. 4D). Following an opposite direction, cells of pcDNA-MALAT1 group and miR-197-3p mimic group were observed with obviously lower apoptosis than NC group (P < 0.05).
Fig. 4.
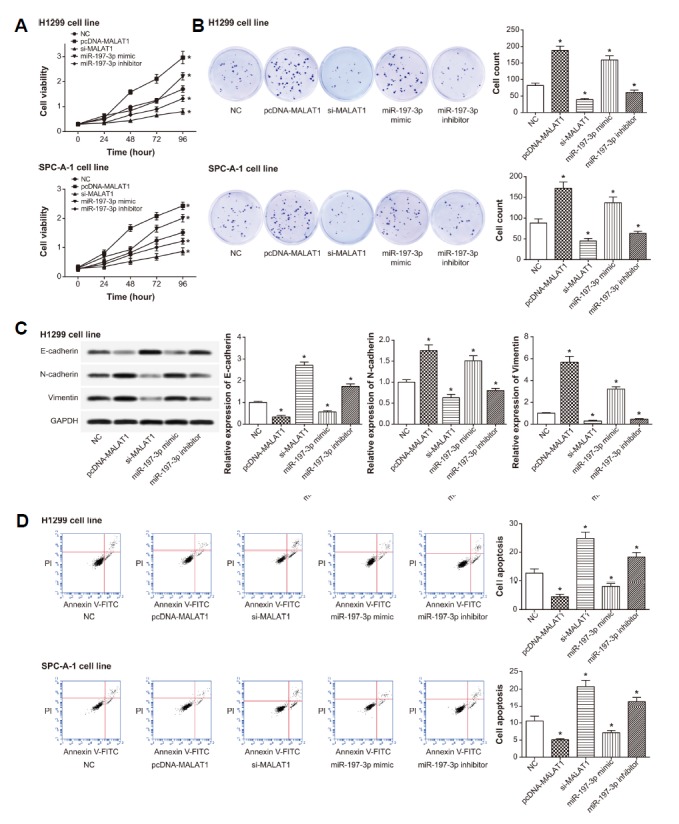
The viability (A), proliferation (B), expressions of EMT-specific proteins (C) and apoptosis (D) of lung cancer cells were examined among pcDNA-MALAT1, si-MALAT1, miR-197-3p mimic, miR-197-3p inhibitor and NC groups. *P < 0.05 when compared with NC. NC: negative control.
MALAT1 was complementary to miR-197-3p in certain sites
The fluorescent activity of lung cancer cells was held up after co-transfection of miR-197-3p mimic and MALAT1-Wt, when compared with the miR-NC+MALAT1-Wt group (P < 0.05) (Fig. 5A). Nevertheless, the fluorescence activity was unaffected in the miR-197-3p mimic and MALAT1-Mut group, which suggested a failed binding of MALAT1 to miR-197-3p Wt (P < 0.05). Moreover, miR-197-3p expression was up-regulated and down-regulated after respective transfection of MALAT1-siRNA and pcDNA-MALAT1, when compared with NC group (P < 0.05) (Fig. 5B). Nevertheless, no significant distinctions of MALAT1 expression were present among miR-197-3p mimic group, miR-197-3p inhibitor group and NC group (P > 0.05) (Fig. 5C).
Fig. 5. The correlations among MALAT1, miR-197-3p and p120-catenin within lung cancer cells.
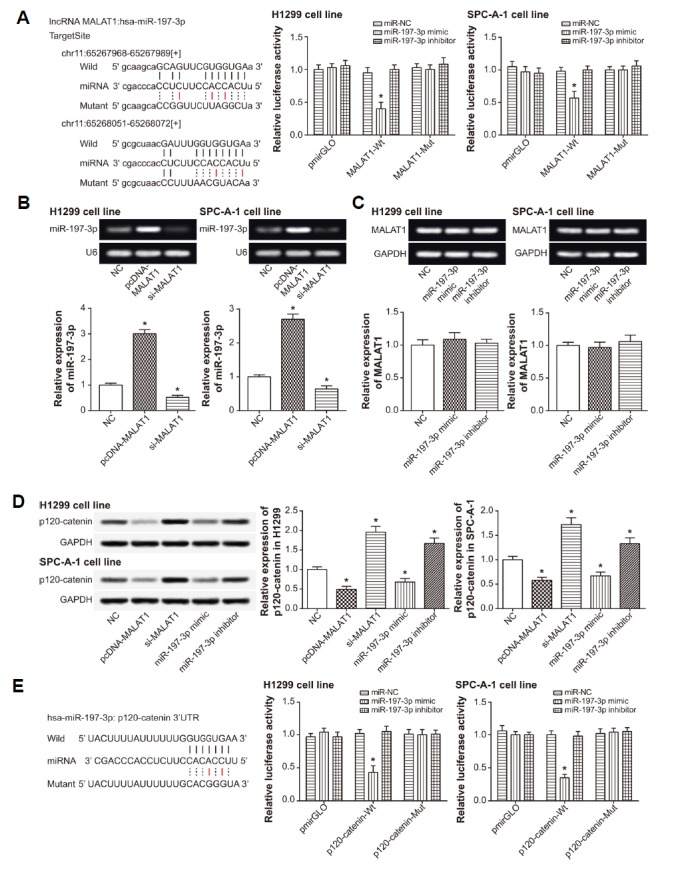
(A) MALAT1 targeted miR-197-3p in a series of sites, and the luciferase activity of lung cancer cells were compared between miR-197-3p mimic+MALAT1 Wt group and miR-197-3p mimic+MALAT1 Mut group. *P < 0.05 when compared with miR-NC+MALAT1 Wt group. (B) The miR-197-3p expression was evaluated after transfection of pcDNA-MALAT1 and si-MALAT1. *P < 0.05 when compared with NC. (C) The MALAT1 expression was assessed after transfection of miR-197-3p mimic and miR-197-3p inhibitor. *P < 0.05 when compared with NC. (D) The expression of p120-catenin was assessed after transfection of NC, pcDNA-MALAT1, si-MALAT1, miR-197-3p mimic and miR-197-3p inhibitor. *P < 0.05 when compared with NC. (E) The p120-catenin was targeted by miR-197-3p in specific sites, and the luciferase activity of lung cancer cells were compared between miR-197-3p mimic+ p120-catenin Wt group and miR-197-3p mimic+ p120-catenin Mut group. *P < 0.05 when compared with miR-NC+p120-catenin Wt group. NC: negative control.
P120-ctn mediated the contribution of MALAT1 and miR-197-3p to the chemo-sensitivity of NSCLC cells
It was implied that over-expression of MALAT1 and miR-197-3p could restrain expression of p120-ctn within NSCLC cell strains (P < 0.05) (Fig. 5D). Moreover, through co-transfection of p120-ctn Wt or p120-ctn Mut together with miR-197-3p mimic into NSCLC cells, we observed that an obviously lower fluorescence of p120-ctn Wt+miR-197-3p mimic group than p120-ctn Mut+miR-197-3p mimic group (P < 0.05) (Fig. 5E). Taken together, p120-ctn might be a downstream molecule targeted by miR-197-3p and its expression was modified by miR-197-3p.
Besides, the proliferation and viability of cells in the miR-NC+si-p120-ctn group was suppressed prominently, when compared with cells of miR-NC group and miR-197-3p mimic group (P < 0.05) (Figs. 6A and 6B). Meanwhile, the miR-NC+si-p120-ctn group was also determined with higher E-cadherin expression, as well as lower N-cadherin and vimentin expression, than miR-NC group (P < 0.05) (Fig. 6C). More than that, the apoptotic rate of miR-NC+si-p120-ctn group was observably higher than that of miR-NC group and miR-197-3p mimic group (P < 0.05) (Fig. 6D).
Fig. 6.
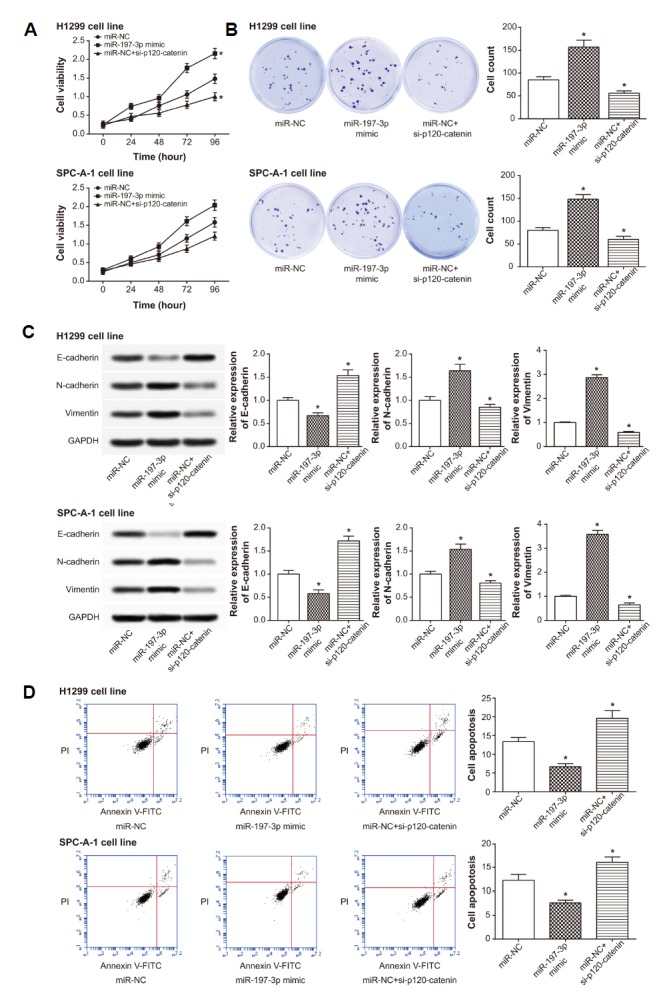
The viability (A), proliferation (B), expressions of EMT-specific proteins (C) and apoptosis (D) of lung cancer cells were examined among miR-NC, miR-197-3p mimic and miR-NC+si-p120-catenin groups. *P < 0.05 when compared with miR-NC. NC: negative control.
Regulatory effects of MALAT1, miR-197-3p and p120-ctn on tumor growth in nude mice
We subcutaneously injected NSCLC strains featured by stably over-expressed/under-expressed MALAT1, miR-197-3p or p120-ctn into nude mice, so that neoplasms were formed on nude mice. During the treatments, normal drinking and eating were maintained for nude mice, and no death occurred. As was shown in Fig. 7A, nude mice injected with si-MALAT1, miR-197-3p inhibitor or si-p120-ctn grew quite smaller neoplasms than NC group (P < 0.05). The tumor size and tumor weight of nude mice injected with si-MALAT1, miR-197-3p inhibitor and si-p120-ctn were also below that of NC group (P < 0.05) (Figs. 7B and 7C), while pcDNA-MALAT1 and miR-197-3p mimic groups displayed apparently larger tumor size and tumor weight than NC group (P < 0.05).
Fig. 7. Effects of MALAT1, miR-197-3p and p120-catenin on tumor formation within mice models of lung cancer.
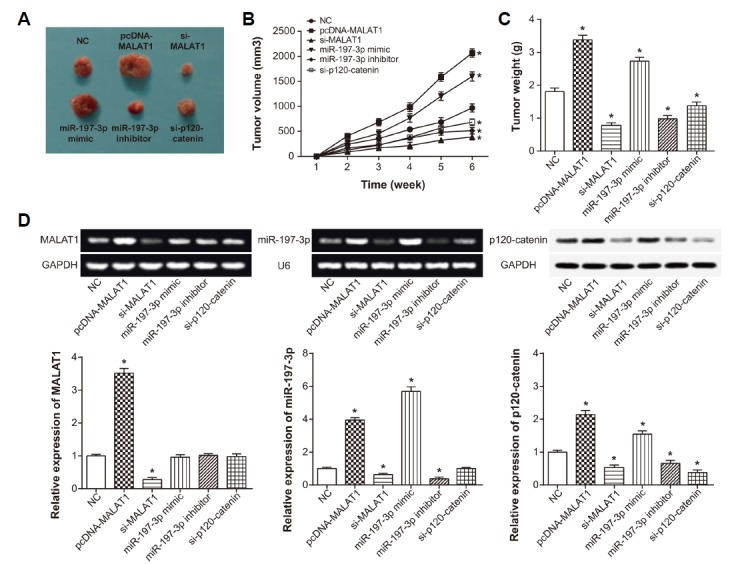
(A) The tumors of pcDNA MALAT1, si-MALAT1, miR-197-3p mimic, miR-197-3p inhibitor, si-p120-catenin and NC groups were illustrated. (B–C) The tumor size and tumor weight of mice models were compared among pcDNA MALAT1, si-MALAT1, miR-197-3p mimic, miR-197-3p inhibitor, si-p120-catenin and NC groups. *P < 0.05 when compared with NC. (D) Expressions of MALAT1, miR-197-3p and p120-catenin were determined within tissues of mice models in the pcDNA MALAT1, si-MALAT1, miR-197-3p mimic, miR-197-3p inhibitor, si-p120-catenin and NC groups. *P < 0.05 when compared with NC. NC: negative control.
Additionally, MALAT1 expression was up-regulated in the pcDNA-MALAT1 group when compared with control group (P < 0.05) (Fig. 7D). Also miR-197-3p and p120-ctn expressions were reduced obviously within tumors in the si-MALAT1 group (P < 0.05), while the pcDNA-MALAT1 group also was discovered with elevated miR-197-3p and p120-ctn expressions (P < 0.05). Furthermore, p120-ctn expression was promoted when miR-197-3p mimic was injected, and miR-197-3p inhibitor group was observed with down-regulated p120-ctn expression (P < 0.05).
DISCUSSION
Currently, the chief control for NSCLC was implemented through early-stage screening and removal of potential hazards. Nonetheless, the complicated and diversified etiologies impeded the prevention and treatment for NSCLC. In consequence, further exploring the generation and progression of NSCLC could be of profound meaning for clinically diagnosing and thereby treating NSCLC.
The MALAT1 investigated here was positioned within cell nucleus, and it lacked full-length open reading frame, which made itself unable to serve as a template for encoding proteins (Hutchinson et al., 2007; Wilusz et al., 2008). As a non-coding RNA, the high expression of MALAT1 could be deemed as a predictor for shortened survival of NSCLC patients in clinical practices (Ji et al., 2003), which was consistent with our investigation (Fig. 1). Moreover, knockdown of MALAT1 was suspected to decelerate the formation of tumors obviously within the established mice models (Liu et al., 2016; Schmidt et al., 2011). The above-mentioned strengthening effects of MALAT1 on tumor progression could be ascribed to its 3′-end functional region of 6918–8441 nt (Xu et al., 2011). Based on this region, MALAT1 could function to improve the viability and proliferation of lung cancer cells (Fig. 4). The particularized etiology might be, to some degree, explained by its up-regulating expression of genes that were relevant to cell movement, including AIM1, LAYN, SLC26A2, CCT4 and so on (Tano et al., 2010). For another, MALAT1 also could interfere with the regular expression of anti-apoptotic proteins (e.g. caspase-3, caspase-8 and Bax) (Guo et al., 2010; Wilusz et al., 2009), and thereby inducing blockage of cell cycle (Lu et al., 2016). The above-mentioned molecular mechanisms appeared well-founded to account for the role of MALAT1 in decreasing the chemo-sensitivity of cancer cells. Virtually, up-regulated expression of MALAT-1 was formerly determined within acute lymphoblastic leukemia cells that were tolerant to vincristine (Akbari Moqadam et al., 2013) and cancer stem cells that displayed resistance to cisplatin (Lopez-Ayllon et al., 2014). Our investigation further elaborated that MALAT-1 could contribute to incremental resistance of lung cancer cells to cisplatin, adriamycin, gefitinib and paclitaxel (Fig. 3), which could be attributed to the accelerating effect of MALAT1 on the viability and proliferation, also possibly the EMT process of NSCLC cells (Shen et al., 2015).
In addition, miR-197-3p was designated as the sponged molecule of MALAT1 within this study (Fig. 5), and it seemed to mediate the impacts exerted by MALAT1 on NSCLC progression (Fig. 4). Actually, miR-197 has been documented as a significant player involved with the pathogenesis of gastric cancer (Li et al., 2011), pancreatic cancer (Hamada et al., 2013) and follicular thyroid cancer (Hu et al., 2011; Kannan and Atreya, 2010; Stokowy et al., 2014). Also its expression was enormously raised within the plasma of lung cancer patients (Zheng et al., 2011), which is similar to the results drawn from our study (Fig. 1). This phenomenon might be ascribed to that miR-197-3p tended to suppress expression of cancer-resistant genes (e.g. FUS1) (Du et al., 2009), which functioned through the mitochondria-dependent approach and Apaf-1-relevant signaling to induce cell apoptosis (Ji and Roth, 2008). Another account could be that miR-197 was able to drive the expression of programmed cell death ligand-1 (PD-L1) by mediating downstream CKS1B and STAT3 (Fujita et al., 2015). It might be because of these vital parameters that miR-197 could boost the chemo-resistance of NSCLC cells (Fig. 3), just as what it did for attenuating the chemo-resistance of colorectal cancer cells (Zhou et al., 2010) and head and neck cancer cells (Dai et al., 2011). Of course, the mechanism behind this was conjectured as the result of MALAT1 targeting miR-197, and the proliferative capacity and viability of NSCLC cells were thus intensified.
More than that, the p120-ctn that was verified as the downstream molecule of MALAT1 and miR-197-3p (Fig. 5) was situated within cyto-membrane, inter-cellular junction and cell nucleus (Cheung et al., 2010), which further explaining its significance in modifying inter-cellular adhesion and thus the EMT process underlying tumorgenesis (Esser et al., 1998; Grossmann et al., 2004). For instance, p120-ctn could directly combine with JMD region of E-cadherin to form a complex that mainly regulated cell adhesion (Davis et al., 2003; Davis and Reynolds, 2006; Ireton et al., 2002). Besides, p120-ctn also could connect to cellular microtube and cytocentrium, whose stability was critical for depressing cell growth (Chartier et al., 2007). Generally speaking, p120-ctn mattered in modulating proliferation of cancer cells by modulating inter-cellular adhesion and cell cycle, and it was reasonable that p120-ctn could mediate the contribution of MALAT1 and miR-197-3p to promoted progression and chemo-resistance of NSCLC cells (Fig. 6). Above all, we certified the underlying role of MALAT1/miR-197-3p/p120-ctn axis in regulating NSCLC etiology, which might introduce a direction for ameliorating the prognosis of NSCLC patients who were treated with chemo-drugs. Of note, the whole study proved the contributions of MALAT1, miR-197-3p and p120-ctn from both sides, specifically explained as up-regulation and down-regulation of the biomarkers. Furthermore, the whole experiments were conducted for ≥ 3 times, which effectively decreased experimental errors and guaranteed data reliability. Nonetheless, the type of drugs used was limited, and radio-sensitivity failed to be explored, so more investigations were in demand.
REFERENCES
- Akbari Moqadam F., Lange-Turenhout E.A., Aries I.M., Pieters R., den Boer M.L. MiR-125b, miR-100 and miR-99a co-regulate vincristine resistance in childhood acute lymphoblastic leukemia. Leukemia Research. 2013;37:1315–1321. doi: 10.1016/j.leukres.2013.06.027. [DOI] [PubMed] [Google Scholar]
- Camps C., del Pozo N., Blasco A., Blasco P., Sirera R. Importance of quality of life in patients with non-small-cell lung cancer. Clinical Lung Cancer. 2009;10:83–90. doi: 10.3816/CLC.2009.n.010. [DOI] [PubMed] [Google Scholar]
- Cesana M., Cacchiarelli D., Legnini I., Santini T., Sthandier O., Chinappi M., Tramontano A., Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartier N.T., Oddou C.I., Laine M.G., Ducarouge B., Marie C.A., Block M.R., Jacquier-Sarlin M.R. Cyclin-dependent kinase 2/cyclin E complex is involved in p120 catenin (p120ctn)-dependent cell growth control: a new role for p120ctn in cancer. Cancer Research. 2007;67:9781–9790. doi: 10.1158/0008-5472.CAN-07-0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Zhao W., Zhang L., Wang L., Wang J., Wan Z., Hong Y., Yu L. MALAT1-miR-101-SOX9 feedback loop modulates the chemo-resistance of lung cancer cell to DDP via Wnt signaling pathway. Oncotarget. 2017;8:94317–94329. doi: 10.18632/oncotarget.21693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung L.W., Leung P.C., Wong A.S. Cadherin switching and activation of p120 catenin signaling are mediators of gonadotropin-releasing hormone to promote tumor cell migration and invasion in ovarian cancer. Oncogene. 2010;29:2427–2440. doi: 10.1038/onc.2009.523. [DOI] [PubMed] [Google Scholar]
- Dai W., Wang C., Wang F., Wang Y., Shen M., Chen K., Cheng P., Zhang Y., Yang J., Zhu R., et al. Anti-miR-197 inhibits migration in HCC cells by targeting KAI 1/CD82. Biochemical and Biophysical Research Communications. 2014;446:541–548. doi: 10.1016/j.bbrc.2014.03.006. [DOI] [PubMed] [Google Scholar]
- Dai Y., Xie C.H., Neis J.P., Fan C.Y., Vural E., Spring P.M. MicroRNA expression profiles of head and neck squamous cell carcinoma with docetaxel-induced multidrug resistance. Head & Neck. 2011;33:786–791. doi: 10.1002/hed.21540. [DOI] [PubMed] [Google Scholar]
- Davis M.A., Ireton R.C., Reynolds A.B. A core function for p120-catenin in cadherin turnover. The Journal of Cell Biology. 2003;163:525–534. doi: 10.1083/jcb.200307111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M.A., Reynolds A.B. Blocked acinar development, E-cadherin reduction, and intraepithelial neoplasia upon ablation of p120-catenin in the mouse salivary gland. Developmental Cell. 2006;10:21–31. doi: 10.1016/j.devcel.2005.12.004. [DOI] [PubMed] [Google Scholar]
- Du L., Schageman J.J., Subauste M.C., Saber B., Hammond S.M., Prudkin L., Wistuba II, Ji L., Roth J.A., Minna J.D., et al. miR-93, miR-98, and miR-197 regulate expression of tumor suppressor gene FUS1. Mol Cancer Res. 2009;7:1234–1243. doi: 10.1158/1541-7786.MCR-08-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esser S., Lampugnani M.G., Corada M., Dejana E., Risau W. Vascular endothelial growth factor induces VE-cadherin tyrosine phosphorylation in endothelial cells. Journal of Cell Science. 1998;111(Pt 13):1853–1865. doi: 10.1242/jcs.111.13.1853. [DOI] [PubMed] [Google Scholar]
- Fan Y., Shen B., Tan M., Mu X., Qin Y., Zhang F., Liu Y. TGF-beta-induced upregulation of malat1 promotes bladder cancer metastasis by associating with suz12. Clinical cancer research: an official journal of the American Association for Cancer Research. 2014;20:1531–1541. doi: 10.1158/1078-0432.CCR-13-1455. [DOI] [PubMed] [Google Scholar]
- Fiori M.E., Barbini C., Haas T.L., Marroncelli N., Patrizii M., Biffoni M., De Maria R. Antitumor effect of miR-197 targeting in p53 wild-type lung cancer. Cell Death and Differentiation. 2014;21:774–782. doi: 10.1038/cdd.2014.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y., Yagishita, S., Hagiwara K., Yoshioka Y., Kosaka N., Takeshita F., Fujiwara T., Tsuta K., Nokihara H., Tamura T., et al. The clinical relevance of the miR-197/CKS1B/STAT3-mediated PD-L1 network in chemoresistant non-small-cell lung cancer. Molecular Therapy: The Journal of the American Society of Gene Therapy. 2015;23:717–727. doi: 10.1038/mt.2015.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstraw P. Updated staging system for lung cancer. Surgical Oncology Clinics of North America. 2011;20:655–666. doi: 10.1016/j.soc.2011.07.005. [DOI] [PubMed] [Google Scholar]
- Grossmann K.S., Grund C., Huelsken J., Behrend M., Erdmann B., Franke W.W., Birchmeier W. Requirement of plakophilin 2 for heart morphogenesis and cardiac junction formation. The Journal of Cell Biology. 2004;167:149–160. doi: 10.1083/jcb.200402096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F., Li Y., Liu Y., Wang J., Li Y., Li G. Inhibition of metastasis-associated lung adenocarcinoma transcript 1 in CaSki human cervical cancer cells suppresses cell proliferation and invasion. Acta Biochimica et Biophysica Sinica. 2010;42:224–229. doi: 10.1093/abbs/gmq008. [DOI] [PubMed] [Google Scholar]
- Hamada S., Satoh K., Miura S., Hirota M., Kanno A., Masamune A., Kikuta K., Kume K., Unno J., Egawa S., et al. miR-197 induces epithelial-mesenchymal transition in pancreatic cancer cells by targeting p120 catenin. Journal of Cellular Physiology. 2013;228:1255–1263. doi: 10.1002/jcp.24280. [DOI] [PubMed] [Google Scholar]
- Hu Z., Shu Y., Chen Y., Chen J., Dong J., Liu Y., Pan S., Xu L., Xu J., Wang Y., et al. Genetic polymorphisms in the precursor MicroRNA flanking region and non-small cell lung cancer survival. American Journal of Respiratory and Critical Care Medicine. 2011;183:641–648. doi: 10.1164/rccm.201005-0717OC. [DOI] [PubMed] [Google Scholar]
- Hutchinson J.N., Ensminger A.W., Clemson C.M., Lynch C.R., Lawrence J.B., Chess A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007;8:39. doi: 10.1186/1471-2164-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ireton R.C., Davis M.A., van Hengel J., Mariner D.J., Barnes K., Thoreson M.A., Anastasiadis P.Z., Matrisian L., Bundy L.M., Sealy L., et al. A novel role for p120 catenin in E-cadherin function. The Journal of Cell Biology. 2002;159:465–476. doi: 10.1083/jcb.200205115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji L., Roth J.A. Tumor suppressor FUS1 signaling pathway. Journal of thoracic oncology: official publication of the International Association for the Study of Lung Cancer. 2008;3:327–330. doi: 10.1097/JTO.0b013e31816bce65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji P., Diederichs S., Wang W., Boing S., Metzger R., Schneider P.M., Tidow N., Brandt B., Buerger H., Bulk E., et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- Jiang G., Wang Y., Dai S., Liu Y., Stoecker M., Wang E., Wang E. P120-catenin isoforms 1 and 3 regulate proliferation and cell cycle of lung cancer cells via beta-catenin and Kaiso respectively. PloS one. 2012;7:e30303. doi: 10.1371/journal.pone.0030303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan M., Atreya C. Differential profiling of human red blood cells during storage for 52 selected microRNAs. Transfusion. 2010;50:1581–1588. doi: 10.1111/j.1537-2995.2010.02585.x. [DOI] [PubMed] [Google Scholar]
- Li J.H., Liu S., Zhou H., Qu L.H., Yang J.H. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Research. 2014;42:D92–97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Zhang Y., Zhang H., Liu X., Gong T., Li M., Sun L., Ji G., Shi Y., Han Z., et al. miRNA-223 promotes gastric cancer invasion and metastasis by targeting tumor suppressor EPB41L3. Molecular Cancer Research: MCR. 2011;9:824–833. doi: 10.1158/1541-7786.MCR-10-0529. [DOI] [PubMed] [Google Scholar]
- Liu M., Sun W., Liu Y., Dong X. The role of lncRNA MALAT1 in bone metastasis in patients with non-small cell lung cancer. Oncology Reports. 2016;36:1679–1685. doi: 10.3892/or.2016.4909. [DOI] [PubMed] [Google Scholar]
- Lopez-Ayllon B.D., Moncho-Amor V., Abarrategi A., Ibanez de Caceres I., Castro-Carpeno J., Belda-Iniesta C., Perona R., Sastre L. Cancer stem cells and cisplatin-resistant cells isolated from non-small-lung cancer cell lines constitute related cell populations. Cancer Medicine. 2014;3:1099–1111. doi: 10.1002/cam4.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H., He Y., Lin L., Qi Z., Ma L., Li L., Su Y. Long non-coding RNA MALAT1 modulates radiosensitivity of HR-HPV+ cervical cancer via sponging miR-145. Tumour biology: the Journal of the International Society for Oncodevelopmental Biology and Medicine. 2016;37:1683–1691. doi: 10.1007/s13277-015-3946-5. [DOI] [PubMed] [Google Scholar]
- Miller K.D., Siegel R.L., Lin C.C., Mariotto A.B., Kramer J.L., Rowland J.H., Stein K.D., Alteri R., Jemal A. Cancer treatment and survivorship statistics, 2016. CA: A Cancer Journal for Clinicians. 2016;66:271–289. doi: 10.3322/caac.21349. [DOI] [PubMed] [Google Scholar]
- Petrelli N.J., Winer E.P., Brahmer J., Dubey S., Smith S., Thomas C., Vahdat L.T., Obel J., Vogelzang N., Markman M., et al. Clinical Cancer Advances 2009: major research advances in cancer treatment, prevention, and screening--a report from the American Society of Clinical Oncology. Journal of Clinical Oncology: Official Journal of the American Society of Clinical Oncology. 2009;27:6052–6069. doi: 10.1200/JCO.2009.26.6171. [DOI] [PubMed] [Google Scholar]
- Schmidt L.H., Spieker T., Koschmieder S., Schaffers S., Humberg J., Jungen D., Bulk E., Hascher A., Wittmer D., Marra A., et al. The long noncoding MALAT-1 RNA indicates a poor prognosis in non-small cell lung cancer and induces migration and tumor growth. Journal of Thoracic Oncology: Official Publication of the International Association for the Study of Lung Cancer. 2011;6:1984–1992. doi: 10.1097/JTO.0b013e3182307eac. [DOI] [PubMed] [Google Scholar]
- Shen L., Chen L., Wang Y., Jiang X., Xia H., Zhuang Z. Long noncoding RNA MALAT1 promotes brain metastasis by inducing epithelial-mesenchymal transition in lung cancer. Journal of Neuro-Oncology. 2015;121:101–108. doi: 10.1007/s11060-014-1613-0. [DOI] [PubMed] [Google Scholar]
- Stokowy T., Eszlinger M., Swierniak M., Fujarewicz K., Jarzab B., Paschke R., Krohn K. Analysis options for high-throughput sequencing in miRNA expression profiling. BMC Research Notes. 2014;7:144. doi: 10.1186/1756-0500-7-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tano K., Mizuno R., Okada T., Rakwal R., Shibato J., Masuo Y., Ijiri K., Akimitsu N. MALAT-1 enhances cell motility of lung adenocarcinoma cells by influencing the expression of motility-related genes. FEBS Letters. 2010;584:4575–4580. doi: 10.1016/j.febslet.2010.10.008. [DOI] [PubMed] [Google Scholar]
- Travis W.D. Pathology and genetics of tumours of the lung, pleura, thymus and heart. IARC Press; 2004. [Google Scholar]
- Wang J., Wang H., Zhang Y., Zhen N., Zhang L., Qiao Y., Weng W., Liu X., Ma L., Xiao W., et al. Mutual inhibition between YAP and SRSF1 maintains long non-coding RNA, Malat1-induced tumourigenesis in liver cancer. Cellular Signalling. 2014;26:1048–1059. doi: 10.1016/j.cellsig.2014.01.022. [DOI] [PubMed] [Google Scholar]
- Wapinski O., Chang H.Y. Long noncoding RNAs and human disease. Trends in Cell Biology. 2011;21:354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- Weber D.G., Johnen G., Casjens S., Bryk O., Pesch B., Jockel K.H., Kollmeier J., Bruning T. Evaluation of long noncoding RNA MALAT1 as a candidate blood-based biomarker for the diagnosis of non-small cell lung cancer. BMC Research Notes. 2013;6:518. doi: 10.1186/1756-0500-6-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz J.E., Freier S.M., Spector D.L. 3′ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell. 2008;135:919–932. doi: 10.1016/j.cell.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz J.E., Sunwoo H., Spector D.L. Long noncoding RNAs: functional surprises from the RNA world. Genes & Development. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Weng Y., He F., Liang D., Cai L. LncRNA MALAT-1 competitively regulates miR-124 to promote EMT and development of non-small-cell lung cancer. Anti-Cancer Drugs. 2018;29:628–636. doi: 10.1097/CAD.0000000000000626. [DOI] [PubMed] [Google Scholar]
- Xu C., Yang M., Tian J., Wang X., Li Z. MALAT-1: a long non-coding RNA and its important 3′ end functional motif in colorectal cancer metastasis. Int J Oncol. 2011;39:169–175. doi: 10.3892/ijo.2011.1007. [DOI] [PubMed] [Google Scholar]
- Zampetaki A., Kiechl S., Drozdov I., Willeit P., Mayr U., Prokopi M., Mayr A., Weger S., Oberhollenzer F., Bonora E., et al. Plasma microRNA profiling reveals loss of endothelial miR-126 and other microRNAs in type 2 diabetes. Circ Res. 2010;107:810–817. doi: 10.1161/CIRCRESAHA.110.226357. [DOI] [PubMed] [Google Scholar]
- Zampetaki A., Willeit P., Tilling L., Drozdov I., Prokopi M., Renard J.M., Mayr A., Weger S., Schett G., Shah A., et al. Prospective study on circulating MicroRNAs and risk of myocardial infarction. J Am Coll Cardiol. 2012;60:290–299. doi: 10.1016/j.jacc.2012.03.056. [DOI] [PubMed] [Google Scholar]
- Zhang J., Zhang A., Wang Y., Liu N., You Y., Kang C., Pu P. New insights into the roles of ncRNA in the STAT3 pathway. Future Oncol. 2012;8:723–730. doi: 10.2217/fon.12.52. [DOI] [PubMed] [Google Scholar]
- Zheng D., Haddadin S., Wang Y., Gu L.Q., Perry M.C., Freter C.E., Wang M.X. Plasma microRNAs as novel biomarkers for early detection of lung cancer. Int J Clin Exp Pathol. 2011;4:575–586. [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Zheng Q., Xu T., Liao D., Zhang Y., Yang S., Hu J. Associations between physical activity-related miRNAs and metabolic syndrome. Horm Metab Res. 2014;46:201–205. doi: 10.1055/s-0033-1357139. [DOI] [PubMed] [Google Scholar]
- Zhou J., Zhou Y., Yin B., Hao W., Zhao L., Ju W., Bai C. 5-Fluorouracil and oxaliplatin modify the expression profiles of microRNAs in human colon cancer cells in vitro. Oncol Rep. 2010;23:121–128. [PubMed] [Google Scholar]