Abstract
Objective
To explore genes potentially co-expressed with cyclin E in gastric cancer and discover possible targets for gastric cancer treatment.
Methods
The Cancer Genome Atlas (TCGA) stomach adenocarcinoma sequencing data were used to predict genes co-expressed with cyclin E. Co-expression genes predicted by cBioPortal online analysis with Pearson correlation coefficient ≥0.4 were analyzed by gene ontology (GO) enrichment annotation using the PANTHER online platform (Ver. 7). Interactions between proteins encoded by these genes were analyzed using the STRING online platform (Ver. 10.5) and Cytoscape software (Ver. 3.5.1). Genes displaying a high degree of connection were analyzed by transcription factor enrichment prediction using FunRich software (Ver. 3). The significant transcription factor and cyclin E expression levels and their impact on gastric cancer progression were analyzed by Western blotting and Kaplan–Meier survival curve analysis.
Results
After filtering the co-expression gene prediction results, 78 predicted genes that included 73 protein coding genes and 5 non-coding genes with Pearson correlation coefficient ≥0.4 were selected. The expressions of the genes were considered to be correlated with cyclin E expression. Among the 78 genes co-expressed with cyclin E, 19 genes at the central of the regulatory network associated with cyclin E were discovered. Nuclear transcription factor Y subunit alpha (NF-YA) was identified as a significant transcription factor associated with cyclin E co-expressing genes. Analysis of specimen donors’ clinical records revealed that high expression of NF-YA tended to be associated with increased cyclin E expression. The expression of both was associated with progression of gastric cancer. Western blotting results showed that compared with normal tissues, NF-YA and cyclin E were highly expressed in tumor tissues (P < 0.001). Survival curve analysis clearly demonstrated relatively poor overall survival of gastric cancer patients with high cyclin E or high NF-YA expression level, compared to patients with low cyclin E or NF-YA expression (P < 0.05).
Conclusions
NF-YA may promote gastric cancer progression by increasing the transcription of cyclin E and other cell cycle regulatory genes. NF-YA might be a potential therapeutically useful prognostic factor for gastric cancer.
Keywords: Cyclin E, Nuclear transcription factor Y subunit alpha, Oncogene, Gastric cancer
Introduction
The incidence of gastric cancer is increasing in developing countries. This imposes a significant financial burden for patients and the healthcare system. Surgical treatment, chemotherapy, and targeted-therapy have been employed in gastric cancer treatment, but the treatment outcome and patients’ post-treatment survival remain disappointing, especially for patients with late stage carcinoma. Cancer cells are characterized by uncontrolled hyper-proliferation. Targeting the cell proliferation machinery or the signal transduction network promoting cell proliferation has been proposed as possible therapeutic options for cancer management. Regulation of the cell cycle is frequently altered in cancer by different genetic and epigenetic causes, and the de-regulated cell cycle progression is crucial in cell proliferation and cancer development, in which cyclins and cyclin dependent kinases are direct promoters.1 Targeting the cell cycle has been accepted in-principle as a potential therapeutic option.2, 3, 4, 5 However, the molecular mechanisms of altered cell cycle machineries that promote gastric cancer progression remain unclear.
Gene amplification and overexpression of cyclin E have been recently linked to gastric cancer development and poor prognosis.6, 7 During the progression of the cell cycle, cyclin E binds to and activates cyclin-dependent kinase 2, which promotes G1/S entry by phosphorylating the appropriate substrates. Cyclin E overexpression has been reported in gastric cancer,8, 9, 10, 11 but the molecular mechanism of the up-regulation remains unclear. In this study, we aimed to discover possible mechanisms that may be involved in regulating cyclin E expression by identifying and investigating genes that are potentially co-expressed with cyclin E in gastric cancer. This was done by querying The Cancer Genome Atlas (TCGA) stomach adenocarcinoma sequencing data with different bioinformatics approaches.
Materials and methods
Bioinformatics analysis pipeline
We used the cBioPortal online platform12, 13 to query the TCGA stomach adenocarcinoma (2017, provisional) sequencing dataset (http://www.cbioportal.org/study?id=stad_tcga#summary). A total of 415 tumor samples (from the TCGA database) with messenger RNA (mRNA) next-generation sequencing data were used. Co-expressed genes predicted by cBioPortal online analysis with Pearson correlation coefficient ≥0.4 were selected for gene ontology (GO) enrichment annotation using the PANTHER online platform (Ver. 7).14, 15 Protein interactions were predicted using the STRING online analysis platform (Ver. 10.5)16 with minimum required interaction score adjusted to 0.15 to obtain the maximal interactions. The acquired protein interaction network was subjected to topological structural analysis using Cytoscape software (Ver. 3.5.1)17 using the default settings. Genes with a high degree of connection were subjected to transcription factor enrichment prediction using FunRich software (Ver. 3)18 using the default settings.
Western blotting
This research was approved by the ethical board of Henan Tumor Hospital (No. 2018132). Twenty-two gastric cancer patients were enrolled. Informed consent was obtained from each patient. Their clinical-pathological records are summarized in Table 1. Gastric cancer biopsies and non-cancerous adjacent biopsies were obtained from these patients. The tissue samples were analyzed by Western blotting to detect cyclin E and nuclear transcription factor Y subunit alpha (NF-YA) protein expression using beta-actin protein as the loading control. Primary antibodies against cyclin E (ab71535), NF-YA (ab23471), beta-actin (ab16039), and correlating secondary antibody (ab205718) were purchased from Abcam Trading Company Ltd. (Cambridge, United Kingdom). Gastric cancer patients were grouped into cyclin E high/low and NF-YA high/low groups based on the gray scale analysis of Western blotting results. Cyclin E or NF-YA expression higher or lower than the average was considered high or low expression, respectively. The NF-YA and cyclin E expression levels in gastric cancer specimens from patients with different TNM stages are presented in Table 2.
Table 1.
Clinical and pathological information of 22 patients with gastric cancer.
| Clinical-pathological characteristic | n (%) |
|---|---|
| Age, years | |
| <65 | 12 (54.5) |
| ≥65 | 10 (45.5) |
| Gender | |
| Male | 13 (59.1) |
| Female | 9 (40.9) |
| Tumor size, cm | |
| <5 | 11 (50.0) |
| ≥5 | 11 (50.0) |
| Histopathological grading | |
| Highly differentiated | 5 (22.7) |
| Moderately differentiated | 10 (45.5) |
| Poorly differentiated | 7 (31.8) |
| TNM stage | |
| I | 3 (13.6) |
| II | 14 (63.6) |
| III | 3 (13.6) |
| IV | 2 (9.1) |
| Lymph node metastasis | |
| No | 7 (31.8) |
| Yes | 15 (68.2) |
TNM: Tumor Node Metastasis.
Table 2.
NF-YA and cyclin E expression in gastric cancer specimens from patients with different TNM stages (n = 22).
| Classification | NF-YA |
Cyclin E |
||
|---|---|---|---|---|
| Low | High | Low | High | |
| TNM stage | ||||
| I | 2 | 1 | 2 | 1 |
| IIA | 3 | 2 | 2 | 3 |
| IIB | 4 | 5 | 3 | 6 |
| III | 1 | 2 | 0 | 3 |
| IV | 0 | 2 | 1 | 1 |
| Lymph node metastasis | ||||
| No | 5 | 2 | 6 | 1 |
| Yes | 3 | 12 | 3 | 12 |
NF-YA: nuclear transcription factor Y subunit alpha; TNM: Tumor Node Metastasis.
Statistical analyses
Statistical analyses were performed using Graphpad Prism (Ver. 7). Patients’ survival was compared by Kaplan–Meier curve analysis (log-rank test) using SPSS software ver. 19.0 (IBM, New York, NY, USA). Student’s t-test was adopted for statistical analysis of Western blotting results. A P-value < 0.05 was considered statistically significant.
Results
Analysis of cyclin E co-expression genes in gastric cancer
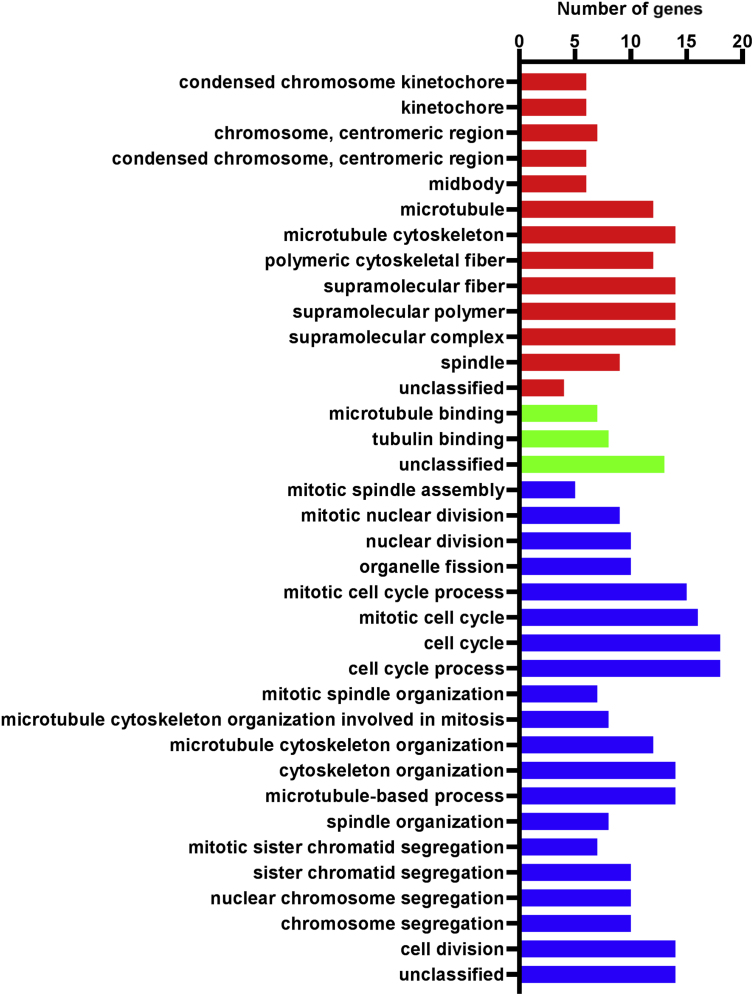
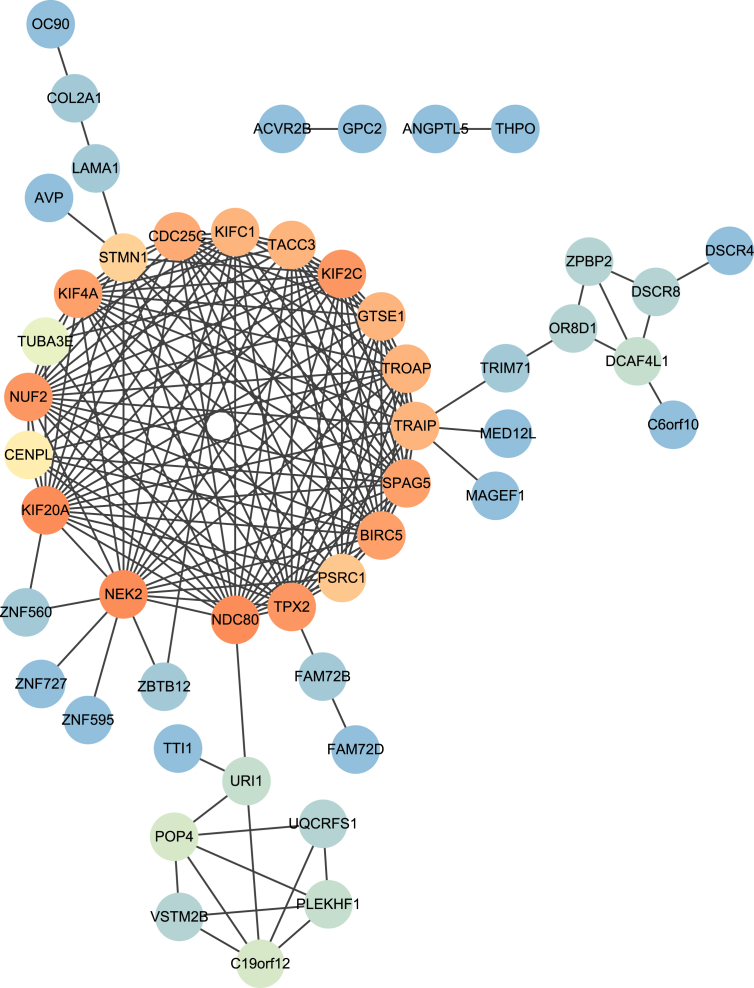
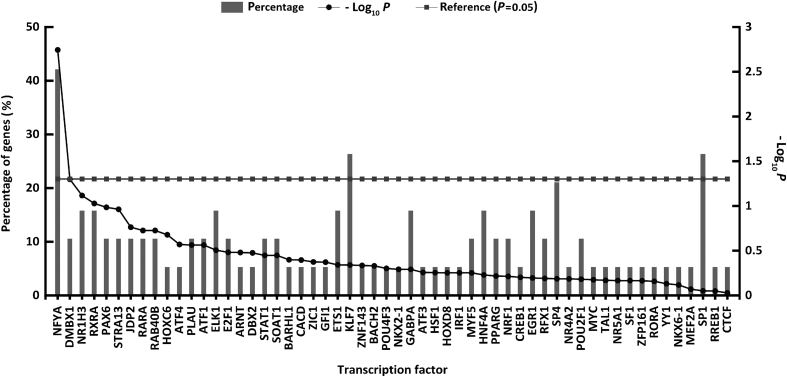
To identify genes co-existing with cyclin E and to identify potential oncogenes in gastric cancer, we used cBioPortal online platform to predict cyclin E co-expression genes among all 415 samples within the stomach adenocarcinoma (TCGA provisional) sample set. After filtering the co-expression gene prediction results, 78 predicted genes, including 73 protein coding genes and 5 non-coding genes with Pearson correlation coefficient ≥0.4 were selected. Their expressions were considered to be correlated with cyclin E expression. The top 15 protein coding genes with their gene symbols, chromosome locations, Pearson correlation, and Spearman correlation coefficients are summarized and listed in Table 3. To better understand the involvement of these predicted genes in cellular composition and function, we annotated them in the GO database using the PANTHER online classification system (Fig. 1). The annotation results suggested that these genes that were presumably co-expressed with cyclin E participated mostly in mitosis and regulation of the cell cycle, consistent with the role of cyclin E in proliferation. We next surveyed the interaction (including predicted association) network among the 73 protein coding genes using the STRING analysis platform and further analyzed the topological structure of the predicted interaction network using Cytoscape software (Ver. 3.5.1). Nineteen genes at the central of the predicted interaction network were identified (Fig. 2 and Table 4). The majority have been previously suggested to promote the progression of gastric cancer or have been associated with poor prognosis. Considering the potential co-expression of these 19 genes with cyclin E and their involvement in gastric cancer progression, we performed a transcription factor enrichment prediction using FunRich software (Fig. 3). The enrichment prediction revealed transcription factor NF-YA as the only one significant result (P < 0.05). It was the most highly related to the 19 queried genes.
Table 3.
Top 15 genes with highest Pearson correlation coefficient in 78 genes predicted to co-express with cyclin E.
| Gene symbol | Cytoband | Pearson Co. | Spearman Co. |
|---|---|---|---|
| C19orf12 | 19q12 | 0.82 | 0.24 |
| URI1 | 19q12 | 0.8 | 0.5 |
| POP4 | 19q12 | 0.69 | 0.44 |
| UQCRFS1 | 19q12 | 0.68 | 0.48 |
| PLEKHF1 | 19q12 | 0.6 | 0.12 |
| DSCR8 | 21q22.13 | 0.53 | 0.31 |
| NUF2 | 1q23.3 | 0.52 | 0.61 |
| C21orf58 | 21q22.3 | 0.52 | 0.33 |
| AMIGO3 | 3p21.31 | 0.52 | 0.17 |
| OR8A1 | 11q24.2 | 0.52 | 0.27 |
| C6orf10 | 6p21.32 | 0.51 | 0.06 |
| PSRC1 | 1p13.3 | 0.51 | 0.5 |
| COL2A1 | 12q13.11 | 0.5 | 0.23 |
| NEK2 | 1q32.3 | 0.49 | 0.64 |
| VSTM2B | 19q12 | 0.48 | 0.18 |
Pearson Co.: Pearson correlation coefficient; Spearman Co.: Spearman correlation coefficient.
Fig. 1.
Gene Ontology (GO) enrichment analysis of 78 predicted cyclin E co-expressing genes. GO terms of cellular components (red), molecular functions (green), and biological processes (blue) are plotted in different colors.
Fig. 2.
Predicted protein–protein interaction network within the 78 predicted cyclin E co-expressing genes.
Table 4.
Top 19 genes with highest connectivity in predicted interaction network.
| Gene Symbol | Connectivity (Degree) | Pearson Co. | Spearman Co. |
|---|---|---|---|
| NDC80 | 19 | 0.44 | 0.55 |
| KIF20A | 19 | 0.42 | 0.61 |
| NEK2 | 19 | 0.49 | 0.64 |
| NUF2 | 18 | 0.52 | 0.61 |
| KIF2C | 18 | 0.45 | 0.61 |
| TPX2 | 18 | 0.46 | 0.6 |
| BIRC5 | 17 | 0.43 | 0.59 |
| KIF4A | 17 | 0.41 | 0.62 |
| SPAG5 | 17 | 0.47 | 0.66 |
| CDC25C | 16 | 0.42 | 0.57 |
| KIFC1 | 15 | 0.42 | 0.56 |
| TRAIP | 15 | 0.43 | 0.6 |
| TROAP | 15 | 0.4 | 0.59 |
| GTSE1 | 15 | 0.45 | 0.56 |
| TACC3 | 15 | 0.45 | 0.48 |
| PSRC1 | 13 | 0.51 | 0.5 |
| STMN1 | 12 | 0.4 | 0.43 |
| CENPL | 9 | 0.44 | 0.6 |
| TUBA3E | 6 | 0.41 | 0.01 |
Pearson Co.: Pearson correlation coefficient; Spearman Co.: Spearman correlation coefficient.
Fig. 3.
Transcription factor enrichment analysis of 19 predicted cyclin E co-expressing genes. Percentage of genes enriched for each transcription factor is plotted on the left axis and -log10P on the right axis. The dotted horizontal line represents the P = 0.05 threshold.
High expression of cyclin E and NF-YA is associated with gastric cancer progression and poor prognosis
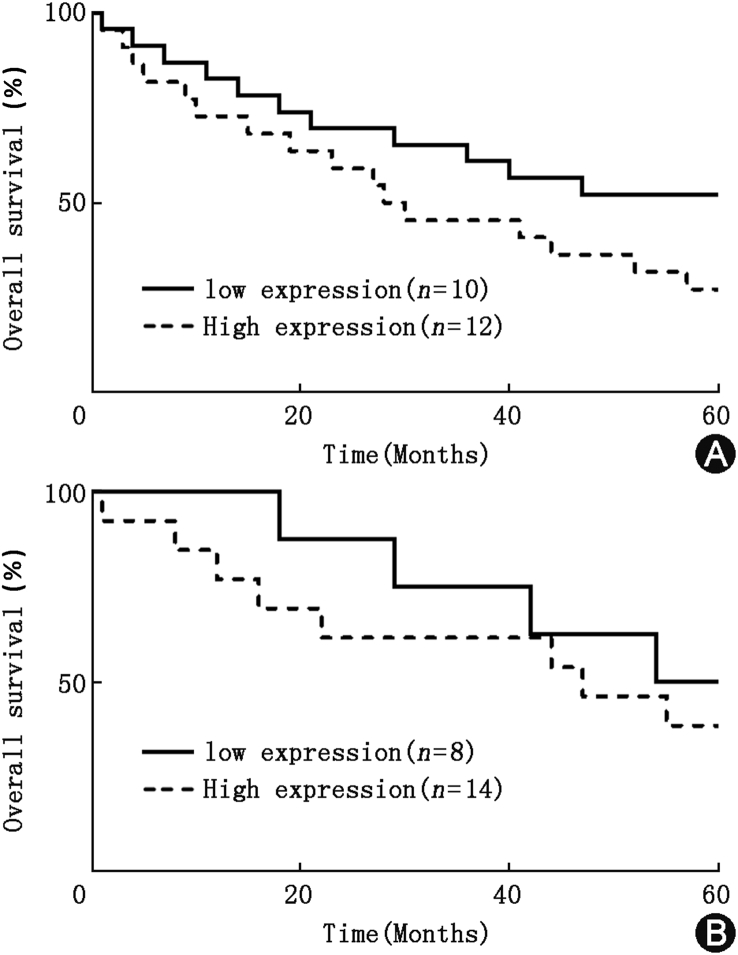
Based on the previous results, we hypothesized that cyclin E and NF-YA may be related to the promotion of the progression of gastric cancer. As a proof-of-concept, we examined cyclin E and NF-YA protein expression levels in 22 pairs of gastric cancer specimens and non-cancerous counterparts. Significant up-regulation and co-expression (P < 0.001) of cyclin E and NF-YA in gastric cancer were revealed by Western blotting (Fig. 4). The results of Fig. 4A and B showed that cyclin E and NF-YA were highly expressed in gastric cancer tissues compared with adjacent tissues. Fig. 4C showed that high expression of NF-YA tended to be associated with increased cyclin E expression. Analysis of specimen donors' clinical records also revealed that the high expression of NF-YA tended to be associated with increased cyclin E expression, both of which were associated with the progression of gastric cancer (Table 2). We further compared the survival curves of patients with different cyclin E and NF-YA expression levels based on the Western blotting analysis results by Kaplan–Meier curve analysis (Fig. 5). Gastric cancer patients with high cyclin E or high NF-YA expression level clearly showed relatively low overall survival compared to patients with low cyclin E or NF-YA expression (P < 0.05).
Fig. 4.
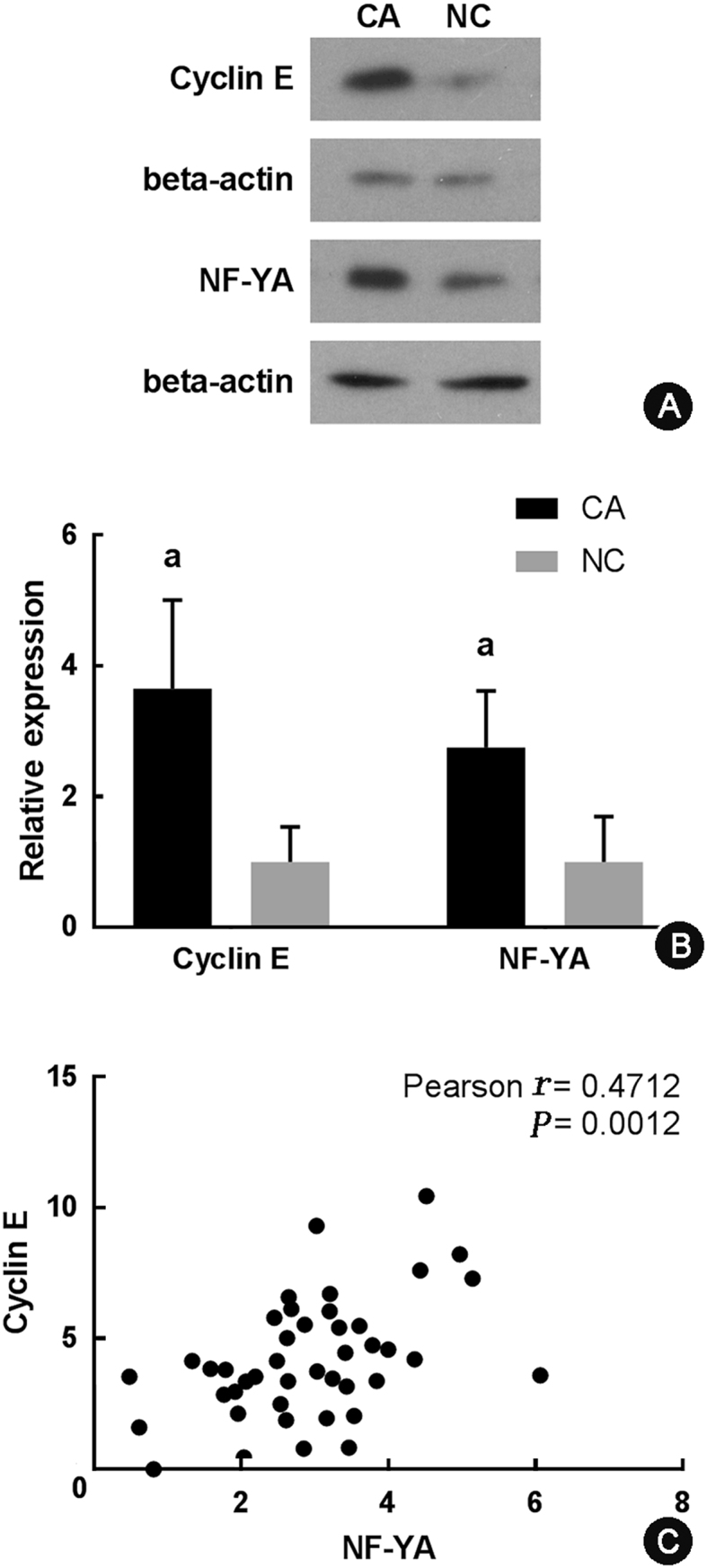
Western blotting analysis of cyclin E and NF-YA protein expression levels in 22 pairs of gastric cancer specimens (CA) and their non-cancerous counterparts (NC). (A) Representative results of cyclin E and NF-YA protein expression levels in CA and NC samples from one patient. (B) Statistical analysis of cyclin E and NF-YA expression in 22 pairs of CA and NC samples. The gray scale analysis of each band was performed using ImageJ software, and gray scale of each band was normalized to the mean value of that in NC group. P value was calculated automatically. aCompared with NC, P < 0.001. (C) Correlation analysis of cyclin E and NF-YA protein expression based on Western blotting results.
Fig. 5.
Kaplan–Meier survival estimates. (A) Kaplan–Meier curve analysis of overall survival of patients with different cyclin E expression levels, P = 0.0417. (B) Kaplan–Meier curve analysis of overall survival of patients with different NF-YA expression levels, P = 0.0325.
Discussion
This study sought to identify genes potentially co-expressed with the cyclin E oncogene in gastric cancer and to clarify the probable regulatory mechanisms. By querying the public TCGA stomach adenocarcinoma sequencing data, 78 genes were implicated possibly being co-expressed with cyclin E. Pearson correlation coefficient evaluation of the correlation between the co-expression of these genes and cyclin E used a coefficient threshold set at 0.4.19, 20 The 78 genes included 5 non-coding genes (LOC642852, PHF2P1, BAIAP2-AS1, CSNK1A1P1, and MIMT1). These non-coding genes and their transcripts (i.e. non-coding RNAs) are important for regulating gene transcription or translation, and their changes in expression level often have a strong influence on phenotype. However, due to their relatively low coefficients (<0.4) and weak correlations with cyclin E, the focus shifted from the non-coding genes to the protein-coding genes with the highest correlation. The top 15 of these genes comprise those implicated in the development of gastric cancer. Jun et al21 reported the association of UQCRFS1 amplification with gastric cancer progression and unfavorable prognosis. Yu et al22 subsequently demonstrated that zinc finger protein 331 may suppress gastric carcinogenesis by down-regulating UQCRFS1 as well as other genes involved in cell cycle promotion. Kaneko et al23 reported that NUF2 (also known as CDCA1) is frequently upregulated in gastric cancer, and targeting this gene by small interfering RNA may induce cell cycle arrest and apoptosis in gastric cancer cells in vitro. Similar results have been reported for PSRC1 and NEK2.24, 25, 26 GO annotation is a powerful method to help understand cellular participation and function of a set of genes with potential allocations.27, 28 GO annotation of the 73 protein coding genes potentially co-expressed with cyclin E showed that these genes were mostly enriched in the promotion of the cell cycle and in mitosis. These results strongly suggest that genes co-expressed with cyclin E may have a similar function in promoting cell proliferation and gastric cancer progression.
We further hypothesized that these genes playing similar roles and with potential correlation might be involved or regulated by the same signal regulatory network, and their transcription might be regulated by some shared transcriptional factors. To reveal the interaction network of proteins coded by the 73 discovered cyclin E co-expressing protein coding genes, we employed the STRING online analysis platform to examine the interactions that were experimentally verified or predicted by an algorithm. We further analyzed the topological structure of the summarized interaction network using Cytoscape software.29, 30, 31, 32 We discovered 19 genes at the central of the interaction network of the 73 genes based on the topological structure. Genes with a degree of connection exceeding 15, except for SPAG5, have been previously linked to the development of gastric cancer.33, 34, 35, 36, 37 The result demonstrates the robustness of our bioinformatics analysis methods and reveals a gastric cancer-promoting interaction network associated with cyclin E.
A transcription factor enrichment prediction performed using the FunRich software identified a novel transcription factor, NF-YA, that might be the most significant transcription factor associated with genes in this interaction network that we have discovered. The association of NF-YA with cancer progression has been preliminarily reported in other cancer models.38, 39, 40, 41 The role of NF-YA seems to mainly involve facilitating the transcription of genes in the cell cycle and cell proliferation. Previously, a role of NF-YA in gastric cancer has not been defined.42, 43, 44 NF-YA is a subunit of the NF-Y heterotrimer, which has been suggested to aid tumor development by binding to promoter or enhancer regions of related genes. Its impact on gastric cancer development has not been described. Based on the bioinformatics analysis results, we examined the status of NF-YA and cyclin E expression in paired samples of gastric cancer biopsies and non-cancerous counterparts acquired from 22 gastric cancer patients. Our results strongly suggest that NF-YA is an independent prognostic factor for gastric cancer patients. These results suggest that NF-YA might be involved in increasing the expression of cyclin E and in promoting gastric cancer development by increasing the expression of cell proliferation related genes.
The collective findings support the potential relationship between cyclin E overexpression and the transcription factor NF-YA, both of which have strong prognostic value. NF-YA may promote the progression of gastric cancer by increasing the transcription of cyclin E and other cell cycle promoting genes. Targeting NF-YA may be a feasible therapeutic strategy in treating gastric cancer and further studies are warranted.
Edited by Pei-Fang Wei
Footnotes
Peer review under responsibility of Chinese Medical Association.
Contributor Information
Xiao-Bing Chen, Email: 2290773710@qq.com.
Su-Xia Luo, Email: luosxrm@163.com.
References
- 1.Otto T., Sicinski P. Cell cycle proteins as promising targets in cancer therapy. Nat Rev Cancer. 2017;17:93–115. doi: 10.1038/nrc.2016.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Patel T.N., Roy S., Ravi R. Gastric cancer and related epigenetic alterations. Ecancermedicalscience. 2017;11:714. doi: 10.3332/ecancer.2017.714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Figueiredo C., Camargo M.C., Leite M., Fuentes-Pananá E.M., Rabkin C.S., Machado J.C. Pathogenesis of gastric cancer: genetics and molecular classification. Curr Top Microbiol Immunol. 2017;400:277–304. doi: 10.1007/978-3-319-50520-6_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lazăr D.C., Tăban S., Cornianu M., Faur A., Goldiş A. New advances in targeted gastric cancer treatment. World J Gastroenterol. 2016;22:6776–6799. doi: 10.3748/wjg.v22.i30.6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berger H., Marques M.S., Zietlow R., Meyer T.F., Machado J.C., Figueiredo C. Gastric cancer pathogenesis. Helicobacter. 2016;21(suppl 1):34–38. doi: 10.1111/hel.12338. [DOI] [PubMed] [Google Scholar]
- 6.Ooi A., Oyama T., Nakamura R. Gene amplification of CCNE1, CCND1, and CDK6 in gastric cancers detected by multiplex ligation-dependent probe amplification and fluorescence in situ hybridization. Hum Pathol. 2017;61:58–67. doi: 10.1016/j.humpath.2016.10.025. [DOI] [PubMed] [Google Scholar]
- 7.Alsina M., Landolfi S., Aura C. Cyclin E amplification/overexpression is associated with poor prognosis in gastric cancer. Ann Oncol. 2015;26:438–439. doi: 10.1093/annonc/mdu535. [DOI] [PubMed] [Google Scholar]
- 8.Huang L., Ren F., Tang R., Feng Z., Chen G. Prognostic value of expression of cyclin E in gastrointestinal cancer: a systematic review and meta-analysis. Technol Cancer Res Treat. 2016;15:12–19. doi: 10.1177/1533034614568098. [DOI] [PubMed] [Google Scholar]
- 9.Schraml P., Bucher C., Bissig H. Cyclin E overexpression and amplification in human tumours. J Pathol. 2003;200:375–382. doi: 10.1002/path.1356. [DOI] [PubMed] [Google Scholar]
- 10.Liang B., Wang S., Yang X., Ye Y., Yu Y., Cui Z. Expressions of cyclin E, cyclin dependent kinase 2 and p57(KIP2) in human gastric cancer. Chin Med J (Engl). 2003;116:20–23. [PubMed] [Google Scholar]
- 11.Sutter T., Dansranjavin T., Lubinski J. Overexpression of cyclin E protein is closely related to the mutator phenotype of colorectal carcinoma. Int J Colorectal Dis. 2002;17:374–380. doi: 10.1007/s00384-002-0390-y. [DOI] [PubMed] [Google Scholar]
- 12.Gao J., Aksoy B.A., Dogrusoz U. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cerami E., Gao J., Dogrusoz U. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thomas P.D., Campbell M.J., Kejariwal A. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mi H., Dong Q., Muruganujan A., Gaudet P., Lewis S., Thomas P.D. PANTHER version 7: improved phylogenetic trees, orthologs and collaboration with the Gene Ontology Consortium. Nucleic Acids Res. 2010;38:D204–D210. doi: 10.1093/nar/gkp1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Szklarczyk D., Franceschini A., Wyder S. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Killcoyne S., Carter G.W., Smith J., Boyle J. Cytoscape: a community-based framework for network modeling. Methods Mol Biol. 2009;563:219–239. doi: 10.1007/978-1-60761-175-2_12. [DOI] [PubMed] [Google Scholar]
- 18.Pathan M., Keerthikumar S., Ang C.S. FunRich: an open access standalone functional enrichment and interaction network analysis tool. Proteomics. 2015;15:2597–2601. doi: 10.1002/pmic.201400515. [DOI] [PubMed] [Google Scholar]
- 19.Bo L., Wei B., Li C., Wang Z., Gao Z., Miao Z. Identification of potential key genes associated with glioblastoma based on the gene expression profile. Oncol Lett. 2017;14:2045–2052. doi: 10.3892/ol.2017.6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Couto C.M.V., Comin C.H., Costa L.D.F. Effects of threshold on the topology of gene co-expression networks. Mol Biosyst. 2017;13:2024–2035. doi: 10.1039/c7mb00101k. [DOI] [PubMed] [Google Scholar]
- 21.Yu J., Liang Q.Y., Wang J. Zinc-finger protein 331, a novel putative tumor suppressor, suppresses growth and invasiveness of gastric cancer. Oncogene. 2013;32:307–317. doi: 10.1038/onc.2012.54. [DOI] [PubMed] [Google Scholar]
- 22.Jun K.H., Kim S.Y., Yoon J.H., Song J.H., Park W.S. Amplification of the UQCRFS1 gene in gastric cancers. J Gastric Cancer. 2012;12:73–80. doi: 10.5230/jgc.2012.12.2.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaneko N., Miura K., Gu Z. siRNA-mediated knockdown against CDCA1 and KNTC2, both frequently overexpressed in colorectal and gastric cancers, suppresses cell proliferation and induces apoptosis. Biochem Biophys Res Commun. 2009;390:1235–1240. doi: 10.1016/j.bbrc.2009.10.127. [DOI] [PubMed] [Google Scholar]
- 24.Dun B., Sharma A., Xu H. Transcriptomic changes induced by mycophenolic acid in gastric cancer cells. Am J Transl Res. 2013;6:28–42. [PMC free article] [PubMed] [Google Scholar]
- 25.Tong H., Wang J., Chen H., Wang Z., Fan H., Ni Z. Transcriptomic analysis of gene expression profiles of stomach carcinoma reveal abnormal expression of mitotic components. Life Sci. 2017;170:41–49. doi: 10.1016/j.lfs.2016.12.001. [DOI] [PubMed] [Google Scholar]
- 26.Fang Y., Kong Y., Xi J. Preclinical activity of MBM-5 in gastrointestinal cancer by inhibiting NEK2 kinase activity. Oncotarget. 2016;7:79327–79341. doi: 10.18632/oncotarget.12687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arnaud M.B., Costanzo M.C., Shah P., Skrzypek M.S., Sherlock G. Gene Ontology and the annotation of pathogen genomes: the case of Candida albicans. Trends Microbiol. 2009;17:295–303. doi: 10.1016/j.tim.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thomas P.D., Mi H., Lewis S. Ontology annotation: mapping genomic regions to biological function. Curr Opin Chem Biol. 2007;11:4–11. doi: 10.1016/j.cbpa.2006.11.039. [DOI] [PubMed] [Google Scholar]
- 29.Crosara K.T.B., Moffa E.B., Xiao Y., Siqueira W.L. Merging in-silico and in vitro salivary protein complex partners using the STRING database: a tutorial. J Proteomics. 2018;171:87–94. doi: 10.1016/j.jprot.2017.08.002. [DOI] [PubMed] [Google Scholar]
- 30.Lubec G., Afjehi-Sadat L., Yang J.W., John J.P. Searching for hypothetical proteins: theory and practice based upon original data and literature. Prog Neurobiol. 2005;77:90–127. doi: 10.1016/j.pneurobio.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 31.Miryala S.K., Anbarasu A., Ramaiah S. Discerning molecular interactions: a comprehensive review on biomolecular interaction databases and network analysis tools. Gene. 2018;642:84–94. doi: 10.1016/j.gene.2017.11.028. [DOI] [PubMed] [Google Scholar]
- 32.Suderman M., Hallett M. Tools for visually exploring biological networks. Bioinformatics. 2007;23:2651–2659. doi: 10.1093/bioinformatics/btm401. [DOI] [PubMed] [Google Scholar]
- 33.Bai T., Yokobori T., Altan B. High STMN1 level is associated with chemo-resistance and poor prognosis in gastric cancer patients. Br J Cancer. 2017;116:1177–1185. doi: 10.1038/bjc.2017.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Imai T., Oue N., Yamamoto Y. Overexpression of KIFC1 and its association with spheroid formation in esophageal squamous cell carcinoma. Pathol Res Pract. 2017;213:1388–1393. doi: 10.1016/j.prp.2017.09.009. [DOI] [PubMed] [Google Scholar]
- 35.Yun M., Rong J., Lin Z.R. High expression of transforming acidic coiled coil-containing protein 3 strongly correlates with aggressive characteristics and poor prognosis of gastric cancer. Oncol Rep. 2015;34:1397–1405. doi: 10.3892/or.2015.4093. [DOI] [PubMed] [Google Scholar]
- 36.Subhash V.V., Tan S.H., Tan W.L. GTSE1 expression represses apoptotic signaling and confers cisplatin resistance in gastric cancer cells. BMC Cancer. 2015;15:550. doi: 10.1186/s12885-015-1550-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen F., Jin X., Zhao J., Gou S. DN604: a platinum(II) drug candidate with classic SAR can induce apoptosis via suppressing CK2-mediated p-cdc25C subcellular localization in cancer cells. Exp Cell Res. 2018;364:68–83. doi: 10.1016/j.yexcr.2018.01.031. [DOI] [PubMed] [Google Scholar]
- 38.Ma H., Yue X., Gao L. ZHX2 enhances the cytotoxicity of chemotherapeutic drugs in liver tumor cells by repressing MDR1 via interfering with NF-YA. Oncotarget. 2015;6:1049–1063. doi: 10.18632/oncotarget.2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou S.J., Deng Y.L., Liang H.F., Jaoude J.C., Liu F.Y. Hepatitis B virus X protein promotes CREB-mediated activation of miR-3188 and Notch signaling in hepatocellular carcinoma. Cell Death Differ. 2017;24:1577–1587. doi: 10.1038/cdd.2017.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Han L., Zhang E.B., Yin D.D. Low expression of long noncoding RNA PANDAR predicts a poor prognosis of non-small cell lung cancer and affects cell apoptosis by regulating Bcl-2. Cell Death Dis. 2015;6:e1665. doi: 10.1038/cddis.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garipov A., Li H., Bitler B.G., Thapa R.J., Balachandran S., Zhang R. NF-YA underlies EZH2 upregulation and is essential for proliferation of human epithelial ovarian cancer cells. Mol Cancer Res. 2013;11:360–369. doi: 10.1158/1541-7786.MCR-12-0661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maity S.N. NF-Y (CBF) regulation in specific cell types and mouse models. Biochim Biophys Acta. 2017;1860:598–603. doi: 10.1016/j.bbagrm.2016.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gurtner A., Manni I., Piaggio G. NF-Y in cancer: impact on cell transformation of a gene essential for proliferation. Biochim Biophys Acta. 2017;1860:604–616. doi: 10.1016/j.bbagrm.2016.12.005. [DOI] [PubMed] [Google Scholar]
- 44.Dolfini D., Gatta R., Mantovani R. NF-Y and the transcriptional activation of CCAAT promoters. Crit Rev Biochem Mol Biol. 2012;47:29–49. doi: 10.3109/10409238.2011.628970. [DOI] [PubMed] [Google Scholar]