Abstract
Thioredoxins (TRX) are traditionally considered as enzymes catalyzing redox reactions. However, redox-independent functions of thioredoxins have been described in different organisms, although the underlying molecular mechanisms are yet unknown. We report here the characterization of the first generated endogenous redox-inactive thioredoxin in an animal model, the TRX-1 in the nematode Caenorhabditis elegans. We find that TRX-1 dually regulates the formation of an endurance larval stage (dauer) by interacting with the insulin pathway in a redox-independent manner and the cGMP pathway in a redox-dependent manner. Moreover, the requirement of TRX-1 for the extended longevity of worms with compromised insulin signalling or under calorie restriction relies on TRX-1 redox activity. In contrast, the nuclear translocation of the SKN-1 transcription factor and increased LIPS-6 protein levels in the intestine upon trx-1 deficiency are strictly redox-independent. Finally, we identify a novel function of C. elegans TRX-1 in male food-leaving behaviour that is redox-dependent. Taken together, our results position C. elegans as an ideal model to gain mechanistic insight into the redox-independent functions of metazoan thioredoxins, overcoming the limitations imposed by the embryonic lethal phenotypes of thioredoxin mutants in higher organisms.
Keywords: Caenorhabditis elegans, Dauer, Food-leaving, Lips-6, Longevity, Male, Redox, Skn-1, Thioredoxin
Highlights
-
•
C. elegans expressing endogenous “redox-dead” TRX-1 are viable.
-
•
The extended lifespan extension of worm daf-2 and eat-2 mutants and the food-leaving behaviour of C. elegans males requires a redox-active TRX-1.
-
•
The SKN-1 nuclear translocation and increased lips-6 expression upon TRX-1 deficiency is redox-independent.
-
•
TRX-1 regulates dauer formation by both redox-dependent and redox-independent mechanisms.
-
•
C. elegans is an ideal model to interrogate on the molecular mechanisms underlying the redox-independent functions of metazoan thioredoxins.
1. Introduction
Thioredoxins are a class of ubiquitous, small redox proteins that function as general disulphide reductases by virtue of the reversible oxidation of the two cysteine residues at their conserved active site Cys-Gly-Pro-Cys (CGPC) [1,2]. Based on this sequence, two different redox inactive thioredoxins can be generated, in which either both or one of the two cysteine residues are substituted by redox-inactive amino acids such as serine (S) or alanine (A). The variants lacking both cysteines cannot bind their redox substrates nor can perform any redox reaction while the variants lacking one of the two cysteine residues can bind their substrates by a disulphide bond but are unable to reduce them. Even though the vast majority of cellular functions assigned to thioredoxins are dependent on their redox capabilities [1,2], there are some exceptions where the role of thioredoxins is independent of the redox cycling of their active site. For instance, in bacteria, E. coli TRX-1 is required for filamentous phage assembly [3] and for supporting phage T7 growth [4,5] and both functions are maintained in E. coli mutants expressing redox-inactive TRX-1 in which one or both cysteine residues are mutated [6,7]. E. coli TRX-1 can also work as a chaperone protein both in vitro and in vivo and this function is independent of a functional redox active site [8,9]. Interestingly, the role of thioredoxins as chaperone appear to be conserved in eukaryotes [10], although the redox-independency of this function has only been additionally demonstrated in plants [11,12]. Yet, this may also apply to mammals as redox-inactive TRX-1 is a functional constituent of the “early pregnancy factor”, a secreted chaperone complex mainly composed of chaperonin cpn10, required for establishment of pregnancy and embryo implantation [13,14]. Other redox-independent functions of thioredoxins in eukaryotes have been reported in yeast, where cytoplasmic TRX-1, complexed with the proteinase inhibitor IB2, is needed for vacuole inheritance [15]. In mammals, TRX-1 works as a monocyte chemoattractant in a redox-dependent manner while it causes monocyte desensitization by a yet unknown redox-independent mechanism [16]. In addition, TRX-1 variants in which either, but not both, of the cysteine residues at the active site are mutated to serine residues can bind to apoptosis signal-regulating kinase ASK1 and promote its ubiquitination-dependent degradation [17].
The identification of novel redox-independent functions of thioredoxins in the context of a complete organism is hampered by the embryonic lethal phenotype of knock-out mice lacking Trx1 or Trx2 genes [18,19]. This phenotype is more likely redox-dependent as mice lacking cytoplasmic or mitochondrial thioredoxin reductase genes, Trxr1 and Trxr2, also die during embryogenesis [20,21]. Similarly, attempts to generate viable trx-2 mutants in zebrafish have also failed [22]. The nematode model Caenorhabditis elegans provides an ideal alternative choice to approach the study of redox-independent functions of thioredoxins as, in this organism, mutants in all thioredoxin genes are viable and have no deleterious phenotype under normal growth conditions [[23], [24], [25], [26]]. Indeed, it has been recently shown that trx-1 null mutant worms fail to avoid the pathogen Pseudomonas aeruginosa while worms expressing a TRX-1(SGPC) variant avoid this pathogen as efficiently as wild type worms [27].
In this study, we report the generation and characterization of a C. elegans trx-1 mutant that expresses an endogenous redox-inactive TRX-1(SGPS) protein instead of wild type TRX-1(CGPC). This has allowed us to confirm the existence of both TRX-1 redox-dependent and redox-independent functions in this model organism.
2. Results
2.1. C. elegans TRX-1 lacking the two active-site cysteine residues is redox-dead
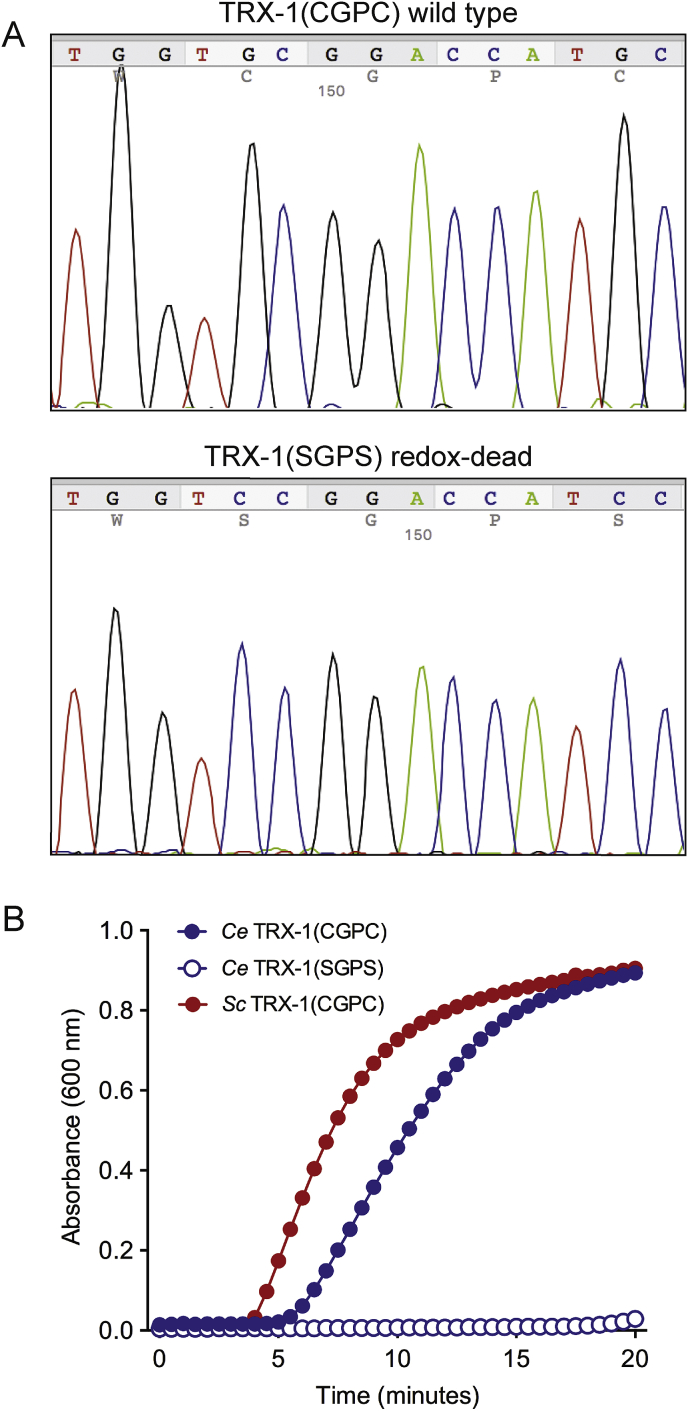
Prior attempting the identification of putative redox-independent roles of C. elegans TRX-1, we first aimed to demonstrate that a TRX-1(SGPS) variant (Fig. 1A), like other metazoan thioredoxins, is indeed redox inactive. For this purpose, we performed the insulin enzymatic assay, which is based on the ability of thioredoxins to use DTT electrons to reduce the disulphide bonds that link together the two insulin polypeptide chains. Once reduced, insulin chains precipitate and the increase in optical density is quantified [28]. We expressed recombinant TRX-1 as GST fusion protein in E. coli and show that, as expected, TRX-1(CGPC) is functional in the insulin assay while TRX-1(SGPS) is unable to reduce insulin chains (Fig. 1B).
Fig. 1.
C. elegans TRX-1(SGPS) is redox dead. A) Chromatogram of plasmids pGEX-4T-1/CeTRX-1(CGPC) (upper panel) and pGEX-4T-1/CeTRX-1(SGPS) (lower panel) used to generate the respective recombinant proteins TRX-1(CGPC) and TRX-1(SGPS). Identical chromatograms were obtained when sequencing the genomic DNA of trx-1(null) and trx-1(sgps) nematodes. B) Enzymatic activity of C. elegans TRX-1(CGPC) and TRX-1(SGPS) recombinant proteins measured by their capacity to reduce insulin using DTT as electron donor. Saccharomyces cerevisiae TRX-1 is used as control.
Next, by CRISPR-Cas9 technology, we generated a C. elegans strain in which the endogenous trx-1 gene is mutated in the same nucleotides as those described for the generation of TRX-1(SGPS) recombinant protein (Fig. 1A). This redox inactive trx-1(syb206) mutant strain [hereafter designated as trx-1(sgps), see Supplementary Table 1 for strain genotype description] only expresses a TRX-1(SGPS) protein, that we have demonstrated to be redox inactive in biochemical activity assays (Fig. 1B) and can then be used to explore which TRX-1 functions are redox-independent.
2.2. C. elegans TRX-1 can regulate dauer formation by redox-dependent and redox-independent mechanisms
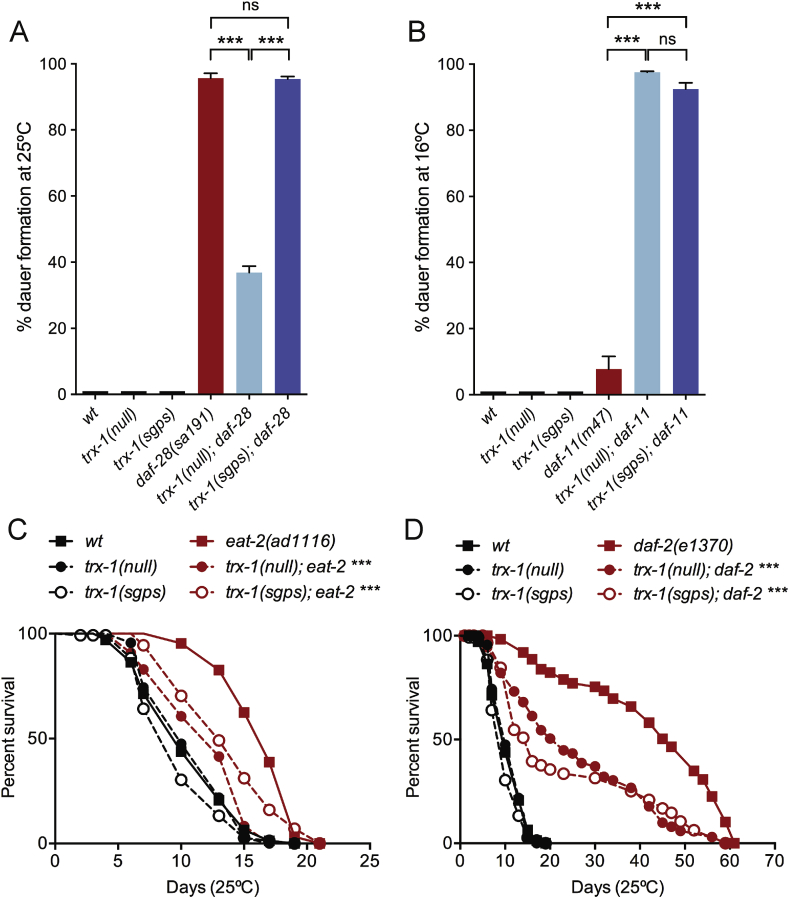
Under harsh environmental conditions like scarce nutrients, high population density and high temperature, C. elegans undergoes a developmental arrest in an endurance larval stage named “dauer”, whose formation is mainly regulated by the cGMP, TGFβ and insulin signalling pathways [29]. Mutations in specific genes of these three pathways result in a dauer-constitutive phenotype as they form dauers under favourable conditions [29]. While mammalian TRX-1 is ubiquitously expressed, C. elegans TRX-1 expression is restricted to worm ASJ sensory neurons [23], which have been shown to regulate dauer entry and exit [30]. Although trx-1(ok1449) deletion mutants [hereafter designated as trx-1(null)] do not show defects on dauer formation under favourable conditions, trx-1(null) worms with additional mutations in genes that function in any of the three dauer pathways mentioned above display a strong synthetic dauer phenotypes in opposite directions [31]. Thus, the trx-1(null) mutation strongly suppresses the dauer-constitutive phenotype of daf-28 insulin mutants while robustly increases the dauer-constitutive phenotype of worms harbouring mutations in the daf-11 gene, encoding a guanylate cyclase [31] (Fig. 2A and B). To test whether the trx-1 synthetic dauer phenotype depends on TRX-1 redox function, we generated animals carrying the trx-1(sgps) mutation in daf-28(sa191) and daf-11(m47) backgrounds, respectively. Interestingly, we found that the TRX-1 dependent dauer formation in daf-28 mutants remains unchanged in redox-dead trx-1(sgps) mutants, showing that this process is redox-independent (Fig. 2A). In contrast, the enhancement of daf-11(m47) dauers upon trx-1 deficiency was the same in trx-1(null) and trx-1(sgps) backgrounds, indicating that this synthetic dauer phenotype is redox-dependent (Fig. 2B). Hence, TRX-1 regulates dauer formation by redox-dependent and redox-independent mechanisms.
Fig. 2.
Effect of TRX-1 redox activity on C. elegans dauer formation and longevity. Percentage of dauer formation of A) daf-28(sa191) and B) daf-11(m47) mutants in trx-1(null) and trx-1(sgps) mutant backgrounds. Bars represent the mean ± S.E.M. of 3 independent experiments, n > 100 worms per assay and strain. ***p < 0.001 by unpaired t-test with Welch's correction. ns, no statistically significant difference. Note that wt, trx-1(null) and trx-1(sgps) worms do not form dauers at any temperature. Lifespan at 25 °C of C) eat-2(ad1116) and D) daf-2(e1370) mutants in trx-1(null) and trx-1(sgps) backgrounds. Graphs represent Kaplan-Meier survival plots of one representative experiment out of three with similar results (Supplementary Table 2, Assay 1). ***p < 0.001 by Log-rank (Mantel-Cox) test compared to eat-2(ad1116) or daf-2(e1370) single mutants, respectively. No statistically significant difference is found when comparing trx-1(null) and trx-1(sgps) mutants in eat-2 or daf-2 backgrounds.
2.3. C. elegans TRX-1 role in longevity is redox-dependent
In addition of being a dauer-inducing condition, reduced nutrient availability also impacts organismal longevity mainly by regulating the insulin/IGF-1 and mTOR pathways [32]. Consistently, besides their role in dauer formation, C. elegans ASJ neurons have also been implicated in the modulation of worm longevity [33]. In this context, we have previously shown that trx-1(null) mutants are slightly short-lived on their own [23] but profoundly reduce the extended lifespan of long-lived daf-2(e1370) insulin receptor mutants or dietary restricted eat-2(ad1116) mutants [34]. As shown in Fig. 2C and D and Supplementary Table 2, the suppressive effect of trx-1 deficiency on daf-2(e1370) and eat-2(ad1116) mutants longevity is redox dependent as the lifespan of trx-1(spgs); daf-2(e1370) and trx-1(sgps); eat-2(ad1116) double mutants is not significantly different from those of trx-1(null); daf-2(e1370) and trx-1(null); eat-2(ad1116) worms.
2.4. C. elegans TRX-1 controls SKN-1 subcellular localization and lips-6 expression in a redox-independent manner
SKN-1 is the C. elegans orthologue of the Nrf2 transcription factor, which is well known to function in stress responses [35]. During larval and adult stages, SKN-1 is predominantly cytoplasmic but strongly accumulates in nuclei upon stress [36]. We have previously shown that the trx-1(null) mutation induces the nuclear translocation of the SKN-1 transcription factor under non-stress conditions [37]. This SKN-1 nuclear localization can be rescued by a transgene expressing a TRX-1(SGPS) redox-dead protein, suggesting that the effect of trx-1 on SKN-1 subcellular localization is redox-independent [37]. To confirm this finding with the endogenous gene, we generated a worm strain expressing the SKN-1::GFP fluorescent reporter in a trx-1(sgps) background which, in contrast to trx-1(null) mutants, was unable to target SKN-1::GFP into the nucleus of intestinal cells (Fig. 3A and B), hence confirming that the regulation of SKN-1 subcellular localization by TRX-1 is redox-independent.
Fig. 3.
TRX-1 regulates SKN-1 nuclear translocation and LIPS-6 induction in a redox-independent manner. A) Fluorescence images of wt, trx-1(null) and trx-1(sgps) L4 worms expressing a SKN-1::GFP reporter. The double dots with very bright GFP expression in worms head denote the constitutive expression of the SKN-1::GFP reporter in ASI neurons [36] and the brownish labelling corresponds to the intestinal autofluorescence. Scale bar 200 μm. B) Percentage of SKN-1::GFP nuclear localization categorically scored and quantified as described in Materials and Methods. Percentages are the average of three biological replicates with n ≥ 50 worms per assay and strain. ***p < 0.001 by Fisher's exact test. ns, no statistically significant difference. C) Differential interference contrast (upper panel) and fluorescence (lower panel) images of wt, trx-1(null) and trx-1(sgps) L4 worms expressing a LIPS-6::GFP reporter. Scale bar 100 μm. D) Quantification of LIPS-6::GFP fluorescence intensity (A.B.U., arbitrary brightness units) in whole animals. Dots represent individuals pooled from two independent experiments, n = 25 per assay and strain. ***p < 0.001 by unpaired t-test with Welch's correction.
Interestingly, despite having SKN-1 in the nucleus, the transcriptional profile of trx-1(null) animals is substantially different from the typical SKN-1 stress-dependent transcriptional program. Specifically, transcriptomics analysis of trx-1(null) mutants revealed a significant downregulation of genes encoding cuticle components and proteins involved in lipid localization and transport while only three genes, encoding the lipase-related proteins LIPS-6, LIPS-11 and LBP-8, are upregulated [37]. lips-6 is the most upregulated gene upon trx-1 deficiency and this induction is skn-1 dependent. We asked whether lips-6 induction in a trx-1 mutant background is also redox-independent, similarly to SKN-1 subcellular localization. For this purpose, we generated a worm strain expressing GFP under the control of the lips-6 promoter from the integrated transgene sybIs6 [Plips-6::gfp; Pmyo-2::mCherry]. Using this transgenic strain, we found that lips-6 is strongly expressed only in few cells in the vulva and tail, while the rest of the worm tissues are devoid of signal or express lips-6 very faintly (Fig. 3C), consistent with previous results using a non-integrated lips-6 GFP reporter [38]. However, in a trx-1(null) mutant worm, we found a strong induction of the GFP labelling in intestinal cells at the L4 stage which did not occur in a trx-1(sgps) mutant background (Fig. 3C and D), indicating that lips-6 induction upon trx-1 deficiency, like SKN-1 subcellular localization, is redox-independent.
2.5. The modulation of C. elegans male food-leaving behaviour by TRX-1 is redox-dependent
We have found that C. elegans males have 4-fold increased basal levels of TRX-1 expression in ASJ neurons as compared with their hermaphrodite siblings (Fig. 4A–D). C. elegans males display an active mate-searching behaviour, which is inhibited in the presence of mates [39]. This behaviour can be quantified using the food-leaving assay, a method that measures the time one isolated adult male takes to abandon a restricted patch of food in search of hermaphrodite mates (Fig. 4E) [40]. The circuit underlaying mate-searching has been only partly elucidated [[41], [42], [43], [44]]. Interestingly, ASJ neurons have been implicated in male mate-searching behaviour by a daf-7 dependent mechanism [45]. Thus, given the TRX-1 sexual dimorphism in ASJ neurons, we asked whether trx-1 regulates mate-searching behaviour and found that the probability of trx-1(null) mutants males to leave a patch of food depleted of mates was significantly reduced (Fig. 4F). This behaviour appears to be redox-dependent as the time taken by trx-1(sgps) males to abandon the food patch does not significantly differ from that of trx-1(null) males (Fig. 4F).
Fig. 4.
C. elegans male food-leaving behaviour requires TRX-1 redox activity. A) Differential interference contrast and fluorescence overlapping image of first day adult hermaphrodite and male worms expressing a TRX-1::GFP reporter. Scale bar 100 μm. Magnification images of B) hermaphrodites and C) males heads to highlight the labelled ASJ neurons (asterisks). D) Quantification of TRX-1::GFP fluorescence intensity (A.B.U., arbitrary brightness units) in ASJ neurons. Dots represent the maximum fluorescence values in ASJ neurons pooled from two independent experiments, n = 20. ***p < 0.001 by unpaired t-test with Welch's correction. E) Graphical scheme of C. elegans male food-leaving behaviour. F) Probability of leaving (PL) of wt, trx-1(null) and trx-1(sgps) first day adult males. Bars represent the mean ± S.E.M. of 6 independent experiments, n = 20 worms per assay for wt and trx-1(null) males and 4 independent experiments, n = 20 per assay for trx-1(sgps) males. Maximum likelihood statistical analysis was used to compare PL values. **p < 0.01; ***p < 0.001; ns, no statistically significant difference.
3. Discussion
Since their discovery in E. coli [46], thioredoxins have been found in virtually all organisms investigated. By far, most thioredoxin functions have been found to rely on their redox capabilities, although redox-independent functions have also been reported. The identification of redox-independent functions of thioredoxins may have been greatly underestimated as many thioredoxin functions have not yet been tested using redox-inactive variants. Furthermore, the embryonic lethal phenotypes of thioredoxin knock-out mice and zebrafish [18,19,22] present an additional hurdle to the use of vertebrate models in the identification of novel redox-independent thioredoxin functions. In this work, we validate C. elegans as a suitable model to address this question, taking advantage that a null mutation in worm trx-1 gene does not cause any deleterious phenotype [23].
A transcriptomic analysis of trx-1(null) mutants revealed that trx-1 deficiency causes upregulation of genes involved in lipid mobilization and downregulation of genes required for lipid localization and transport [37], consistent with a perceived status of starvation. TRX-1 is expressed in ASJ neurons where it regulates dauer formation and worm longevity, traits that are modulated by nutrient availability. Indeed, the trx-1 gene is responsive to nutrient availability as TRX-1 levels are robustly increased upon starvation at any developmental stage, including dauer (Supplementary Fig. 1). Interestingly, we have found that the function of TRX-1 in these nutrient availability related traits is dual: a) TRX-1 redox activity is needed for daf-2 and eat-2 mutants extended lifespan, the enhancement of dauer formation in a daf-11 mutant background and male food-leaving behaviour whereas b) the decrease of dauer formation in daf-28 insulin mutants and the SKN-1 dependent induction of lips-6 expression upon trx-1 deficiency is redox-independent. Although insulins are well known thioredoxin substrates [28], the redox-independent synthetic phenotype of trx-1 and daf-28 mutants suggests that TRX-1 may act in regulating daf-28 expression levels, as previously reported [31], or secretion. On the other hand, impairment of DAF-28 secretion has been associated with increased SKN-1 activity [47]. Given that lips-6 is the most upregulated gene in trx-1(null) mutants in a SKN-1 dependent manner, it is tempting to speculate that these two redox-independent functions of TRX-1 may be mechanistically linked. Indeed, bidirectional signalling between ASJ neurons and the intestine has been previously reported in other nutrient-dependent traits like cold-acclimation, L1 and dauer diapause or foraging behaviour [[48], [49], [50]]. Further support for using C. elegans as model to identify novel redox-independent functions of TRX-1 comes from the recent finding that TRX-1 mediates worm avoidance to Pseudomonas aeuroginosa by responding to nitric oxide produced by this pathogen [27]. While trx-1(null) mutants fail to avoid Pseudomonas, a worm strain producing an endogenous redox-dead TRX-1(CGPS) avoids the pathogen as efficiently as wild type animals. TRX-3 is another C. elegans thioredoxin expressed exclusively in the intestine which is strongly induced upon exposure to different worm pathogens and, consistently, increased levels of TRX-3 provide partial protection to infection [25]. Null trx-3 mutants are viable, thus allowing to test whether TRX-3 role in pathogen resistance, like TRX-1, is also independent of its redox activity.
In summary, our results provide a proof of concept for the use of C. elegans as a model of choice not only to identify novel redox-independent functions of metazoan thioredoxins but, given the genetic amenability of this model, to delve deeper into their molecular mechanisms. Importantly, in addition to the trx-1 and trx-3 genes discussed above, three more thioredoxin-encoding genes (trx-2, trx-4 and trx-5) are present in C. elegans and, interestingly, putative null mutants of these three genes are also viable under normal growth conditions [24,26] thus expanding the potential of the model in this unexplored scenario of redox-independent functions of thioredoxins.
4. Materials and methods
4.1. Recombinant protein purification and enzymatic activity assay
The plasmid pGEX-4T-1/CeTRX-1(SGPS) was obtained by site-directed mutagenesis of trx-1 active site using as bait a pGEX-4T-1/CeTRX-1(CGPC) construct [23] and the mutagenic primers 5′-TCTATGCAACTTGGTCCGGACCATCCAAAGCAATTGCACC-3´ (forward) and 5′-GGTGCAATTGCTTTGGATGGTCCGGACCAAGTTGCATAGA-3′(reverse), following manufacturer's instructions (QuickChange II Site-Directed Mutagenesis Kit, Agilent). Mutagenesis was confirmed by sequencing of the pGEX-4T-1/CeTRX-1(SGPS) plasmid in both directions (Fig. 1A).
For recombinant protein purification, E. coli BL21(DE3) transformed with the corresponding pGEX-4T-1/CeTRX1 construct were inoculated in LB medium containing 0.1 mg ampicillin/ml and grown at 37 °C until Abs600 = 0.5. Then the recombinant protein was induced by the addition 0.5 mM isopropyl-1-thio-β-d-galatopyranoside, and growth was continued at 37 °C for 3.5 h. Cells were collected by centrifugation and resuspended in phosphate buffer saline (PBS) containing 0.5 mg lysozyme/ml and 0.1 μg DNase I/ml (Lysozyme and DNase I from Sigma). Then cells were lysed by sonication and the homogenate was clarified by centrifugation at 10,000×g for 30 min. The cell free extract was filtered and loaded onto a Glutathione Sepharose 4B™ column (Amersham Biosciences) equilibrated with PBS. The column was washed with PBS and the recombinant protein was eluted with PBS containing 10 mM GSH. To separate the GST-tag from the TRX-1 protein, 1 unit of thrombin/ml was added to the purified recombinant protein, incubated for 2 h at room temperature and dialyzed against PBS. Finally, the sample was passed through a Glutathione Sepharose 4B™ column buffered with PBS and the flow-through containing TRX-1 protein was collected. Protein concentration was determined according with the molar extinction coefficient at 280 nm for TRX1(CGPC) (10,220 M−1 cm−1) and for TRX1(SXXS) (10,095 M−1 cm−1). The insulin reduction assay for redox activity of thioredoxin was monitored in an Ultrospect 4000 UV/Visible Spectrophotometer (Pharmacia Biotech) by the increase of light scattering at 600 nm. Assay conditions were 20 mM TrisHCl pH 7.5, 1 mM EDTA, 1 mg porcine insulin/ml (Sigma), 5 μM TRX-1 and the reaction was initiated by addition of 1 mM DTT.
4.2. C. elegans strains
The standard methods used for culturing and maintenance of C. elegans were as described [51]. A list of all strains used and generated in this study is provided in Supplementary Table 1. The strain PHX206, trx-1(syb206) II was generated at SunyBiotech (http://www.sunybiotech.com) by CRISPR-Cas9 using the sgRNAs sg1-GATTTCTATGCAACTTGGTGCGG and sg2-CCATGCAAAGCAATTGCACCATT and the editing was confirmed by sequencing the trx-1 gene in both directions. All VZ strains are 6x backcrossed with N2 wild type. Unless otherwise noted, all experiments were performed on synchronized worms generated by allowing 10 to 15 gravid hermaphrodites to lay eggs during two to three hours on seeded plates at 20 °C.
4.3. Analysis of dauer formation
For dauer formation, worms were incubated at the respective assay temperatures after synchronized egg-lay. Dauers and non-dauers (L3, L4 and adult animals) on the agar and side of the plate were counted after 50 h at 25 °C for the daf-28 dauer assay and after 96 h at 16 °C for the daf-11 dauer assay. In all cases, dauers were discriminated from non-dauers based on the absence of pharyngeal pumping and radial shrinkage of the body [52]. More than 100 animals were counted in three independent experiments per genotype.
4.4. Lifespan determinations
Strains were synchronized by hypochlorite treatment of gravid adults and the progeny was grown during two generations at 16 °C. F2 L4 animals were shifted to 25 °C (t = 0). During the first week, animals were transferred to a fresh plate every two days. Afterwards, worms were transferred at least once per week and scored every two days until death. Animals lost or dead by non-physiological causes were censored. GraphPad Prism 7 (Version 7.00) was used for statistical analysis and graphs. Survival curves were generated using the product-limit method of Kaplan and Meier. The log-rank (Mantel-Cox) test was used to evaluate differences in survival and p values lower than 0.05 were considered as significant.
4.5. SKN-1::GFP nuclear localization
To visualize SKN-1 expression, synchronized L4 worms expressing a SKN-1B/C::GFP fusion protein (abbreviated as SKN-1::GFP) were washed from plates and anesthetized with 50 mM tetramisole. Anesthetized worms were mounted on 2% agarose pads and visualized and imaged using an Olympus Fluoview fv3000 confocal microscope. For quantification of SKN-1::GFP localization, the percentage of intestinal SKN-1::GFP nuclear localization was categorically scored as follows: score 0: no localization; score 1: marginal posterior or anterior intestinal localization; score 2: expanded posterior or anterior intestinal localization; score 3: posterior and anterior intestinal localization; score 4: localization throughout the entire intestine (modified from Ref. [53]).
4.6. LIPS-6::GFP and TRX-1::GFP expression
Synchronized L4 worms (48 h after egg-lay) expressing a LIPS-6::GFP fusion protein and synchronized first day adult worms (72 h after egg-lay) expressing a TRX-1::GFP fusion protein were blindly picked at the stereoscope and anesthetized with 10 mM levamisole. Anesthesized worms were mounted on 2% agarose pads and visualized using an Olympus BX61 fluorescence microscope equipped with a DP72 digital camera coupled to CellSens Software for image acquisition and analysis. ImageJ Software (NIH) was used to quantify the fluorescence of the worms either in the whole body (for LIPS-6::GFP) or in ASJ neurons (for TRX-1::GFP).
4.7. Male food-leaving behaviour
The assay was performed and scored as previously described [40] with some modifications. A population of 20 L4 males were transferred to new plates the day before the assay (afternoon) and grown overnight at 20 °C. The assay was performed by placing each male individually in a Petri dish (9 cm in diameter and 12 ml of agar) in the center of a 15 μl (9 mm diameter approximately) patch of food (E. coli OP50 seeded the evening before). Plates were kept at 20 °C during the 24 h assay, and the proportion of males that had left the food was scored at four time points (at 2 h, 5 h, 8 h and 24 h). A male was considered a leaver and left censored if at the scoring time point it had reached a 1 cm distance from the edge of the plate.
Author contribution
ASM and AMV designed the study. JRP purified the recombinant proteins and carried out the thioredoxin enzymatic activity assays. ASM generated all worm trx-1(sgps) derivative strains and performed the dauer and LIPS-6::GFP quantifications. JMM and MJM performed the lifespan experiments. OK and DAG quantified SKN-1 nuclear translocation. DGG and RCN imaged and quantified TRX-1::GFP transgenic worms. RCN and AB performed the male food leaving assay. JCFG and PSW performed starvation assays and contributed with strains and reagents. All authors edited and revised the manuscript.
Competing interests
Authors declare no competing interest.
Acknowledgements
We thank the Caenorhabditis Genetics Center (CGC), which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440), for providing worm strains and SunyBiotech Corporation for the generation of the trx-1(sgps) strain. We thank Prof. Stefan Taubert for providing the lips-6::gfp reporter strain. AMV was supported by a grant from the Spanish Ministry of Economy and Competitiveness (BFU2015-64408-P), cofinanced by the Fondo Social Europeo (FEDER). DAG was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award number R01AI076406. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. AMV is a member of the GENIE and EU-ROS Cost Actions of the European Union.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2019.101178.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Lee S., Kim S.M., Lee R.T. Thioredoxin and thioredoxin target proteins: from molecular mechanisms to functional significance. Antioxidants Redox Signal. 2013;18(10):1165–1207. doi: 10.1089/ars.2011.4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Collet J.F., Messens J. Structure, function, and mechanism of thioredoxin proteins. Antioxidants Redox Signal. 2010;13(8):1205–1216. doi: 10.1089/ars.2010.3114. [DOI] [PubMed] [Google Scholar]
- 3.Russel M., Model P. Thioredoxin is required for filamentous phage assembly. Proc. Natl. Acad. Sci. U.S.A. 1985;82(1):29–33. doi: 10.1073/pnas.82.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chamberlin M. Isolation and characterization of prototrophic mutants of Escherichia coli unable to support the intracellular growth of T7. J. Virol. 1974;14(3):509–516. doi: 10.1128/jvi.14.3.509-516.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Modrich P., Richardson C.C. Bacteriophage T7 Deoxyribonucleic acid replication in vitro. A protein of Escherichia coli required for bacteriophage T7 DNA polymerase activity. J. Biol. Chem. 1975;250(14):5508–5514. [PubMed] [Google Scholar]
- 6.Russel M., Model P. The role of thioredoxin in filamentous phage assembly. Construction, isolation, and characterization of mutant thioredoxins. J. Biol. Chem. 1986;261(32):14997–15005. [PubMed] [Google Scholar]
- 7.Huber H.E., Russel M., Model P., Richardson C.C. Interaction of mutant thioredoxins of Escherichia coli with the gene 5 protein of phage T7. The redox capacity of thioredoxin is not required for stimulation of DNA polymerase activity. J. Biol. Chem. 1986;261(32):15006–15012. [PubMed] [Google Scholar]
- 8.Kern R., Malki A., Holmgren A., Richarme G. Chaperone properties of Escherichia coli thioredoxin and thioredoxin reductase. Biochem. J. 2003;371(Pt 3):965–972. doi: 10.1042/BJ20030093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jurado P., de Lorenzo V., Fernandez L.A. Thioredoxin fusions increase folding of single chain Fv antibodies in the cytoplasm of Escherichia coli: evidence that chaperone activity is the prime effect of thioredoxin. J. Mol. Biol. 2006;357(1):49–61. doi: 10.1016/j.jmb.2005.12.058. [DOI] [PubMed] [Google Scholar]
- 10.Berndt C., Lillig C.H., Holmgren A. Thioredoxins and glutaredoxins as facilitators of protein folding. Biochim. Biophys. Acta. 2008;1783(4):641–650. doi: 10.1016/j.bbamcr.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 11.Park S.K., Jung Y.J., Lee J.R., Lee Y.M., Jang H.H., Lee S.S. Heat-shock and redox-dependent functional switching of an h-type Arabidopsis thioredoxin from a disulfide reductase to a molecular chaperone. Plant Physiol. 2009;150(2):552–561. doi: 10.1104/pp.109.135426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Du H., Kim S., Hur Y.S., Lee M.S., Lee S.H., Cheon C.I. A cytosolic thioredoxin acts as a molecular chaperone for peroxisome matrix proteins as well as antioxidant in peroxisome. Mol. Cell. 2015;38(2):187–194. doi: 10.14348/molcells.2015.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cavanagh A.C. Identification of early pregnancy factor as chaperonin 10: implications for understanding its role. Rev. Reprod. 1996;1(1):28–32. doi: 10.1530/ror.0.0010028. [DOI] [PubMed] [Google Scholar]
- 14.Tonissen K., Wells J., Cock I., Perkins A., Orozco C., Clarke F. Site-directed mutagenesis of human thioredoxin. Identification of cysteine 74 as critical to its function in the “early pregnancy factor” system. J. Biol. Chem. 1993;268(30):22485–22489. [PubMed] [Google Scholar]
- 15.Xu Z., Mayer A., Muller E., Wickner W. A heterodimer of thioredoxin and I(B)2 cooperates with Sec18p (NSF) to promote yeast vacuole inheritance. J. Cell Biol. 1997;136(2):299–306. doi: 10.1083/jcb.136.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bizzarri C., Holmgren A., Pekkari K., Chang G., Colotta F., Ghezzi P. Requirements for the different cysteines in the chemotactic and desensitizing activity of human thioredoxin. Antioxidants Redox Signal. 2005;7(9–10):1189–1194. doi: 10.1089/ars.2005.7.1189. [DOI] [PubMed] [Google Scholar]
- 17.Liu Y., Min W. Thioredoxin promotes ASK1 ubiquitination and degradation to inhibit ASK1-mediated apoptosis in a redox activity-independent manner. Circ. Res. 2002;90(12):1259–1266. doi: 10.1161/01.res.0000022160.64355.62. [DOI] [PubMed] [Google Scholar]
- 18.Matsui M., Oshima M., Oshima H., Takaku K., Maruyama T., Yodoi J. Early embryonic lethality caused by targeted disruption of the mouse thioredoxin gene. Dev. Biol. 1996;178(1):179–185. doi: 10.1006/dbio.1996.0208. [DOI] [PubMed] [Google Scholar]
- 19.Nonn L., Williams R.R., Erickson R.P., Powis G. The absence of mitochondrial thioredoxin 2 causes massive apoptosis, exencephaly, and early embryonic lethality in homozygous mice. Mol. Cell. Biol. 2003;23(3):916–922. doi: 10.1128/MCB.23.3.916-922.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jakupoglu C., Przemeck G.K., Schneider M., Moreno S.G., Mayr N., Hatzopoulos A.K. Cytoplasmic thioredoxin reductase is essential for embryogenesis but dispensable for cardiac development. Mol. Cell. Biol. 2005;25(5):1980–1988. doi: 10.1128/MCB.25.5.1980-1988.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Conrad M., Jakupoglu C., Moreno S.G., Lippl S., Banjac A., Schneider M. Essential role for mitochondrial thioredoxin reductase in hematopoiesis, heart development, and heart function. Mol. Cell. Biol. 2004;24(21):9414–9423. doi: 10.1128/MCB.24.21.9414-9423.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang J., Cui X., Wang L., Liu F., Jiang T., Li C. The mitochondrial thioredoxin is required for liver development in zebrafish. Curr. Mol. Med. 2014;14(6):772–782. doi: 10.2174/1566524014666140724103927. [DOI] [PubMed] [Google Scholar]
- 23.Miranda-Vizuete A., Fierro Gonzalez J.C., Gahmon G., Burghoorn J., Navas P., Swoboda P. Lifespan decrease in a Caenorhabditis elegans mutant lacking TRX-1, a thioredoxin expressed in ASJ sensory neurons. FEBS Lett. 2006;580(2):484–490. doi: 10.1016/j.febslet.2005.12.046. [DOI] [PubMed] [Google Scholar]
- 24.Cacho-Valadez B., Munoz-Lobato F., Pedrajas J.R., Cabello J., Fierro-Gonzalez J.C., Navas P. The characterization of the Caenorhabditis elegans mitochondrial thioredoxin system uncovers an unexpected protective role of thioredoxin reductase 2 in beta-amyloid peptide toxicity. Antioxidants Redox Signal. 2012;16(12):1384–1400. doi: 10.1089/ars.2011.4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jimenez-Hidalgo M., Kurz C.L., Pedrajas J.R., Naranjo-Galindo F.J., Gonzalez-Barrios M., Cabello J. Functional characterization of thioredoxin 3 (TRX-3), a Caenorhabditis elegans intestine-specific thioredoxin. Free Rad. Biol. Med. 2014;68:205–219. doi: 10.1016/j.freeradbiomed.2013.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arodin L., Miranda-Vizuete A., Swoboda P., Fernandes A.P. Protective effects of the thioredoxin and glutaredoxin systems in dopamine-induced cell death. Free Rad. Biol. Med. 2014;73:328–336. doi: 10.1016/j.freeradbiomed.2014.05.011. [DOI] [PubMed] [Google Scholar]
- 27.Hao Y., Yang W., Ren J., Hall Q., Zhang Y., Kaplan J.M. Thioredoxin shapes the C. elegans sensory response to Pseudomonas produced nitric oxide. eLife. 2018;7 doi: 10.7554/eLife.36833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holmgren A. Thioredoxin catalyzes the reduction of insulin disulfides by dithiothreitol and dihydrolipoamide. J. Biol. Chem. 1979;254(19):9627–9632. [PubMed] [Google Scholar]
- 29.Hu P.J. Dauer. WormBook. 2007:1–19. doi: 10.1895/wormbook.1.144.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bargmann C.I., Horvitz H.R. Control of larval development by chemosensory neurons in Caenorhabditis elegans. Science. 1991;251(4998):1243–1246. doi: 10.1126/science.2006412. [DOI] [PubMed] [Google Scholar]
- 31.Fierro-Gonzalez J.C., Cornils A., Alcedo J., Miranda-Vizuete A., Swoboda P. The thioredoxin TRX-1 modulates the function of the insulin-like neuropeptide DAF-28 during dauer formation in Caenorhabditis elegans. PLoS One. 2011;6(1) doi: 10.1371/journal.pone.0016561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gems D., Partridge L. Genetics of longevity in model organisms: debates and paradigm shifts. Annu. Rev. Physiol. 2013;75:621–644. doi: 10.1146/annurev-physiol-030212-183712. [DOI] [PubMed] [Google Scholar]
- 33.Alcedo J., Kenyon C. Regulation of C. elegans longevity by specific gustatory and olfactory neurons. Neuron. 2004;41(1):45–55. doi: 10.1016/s0896-6273(03)00816-x. [DOI] [PubMed] [Google Scholar]
- 34.Fierro-Gonzalez J.C., Gonzalez-Barrios M., Miranda-Vizuete A., Swoboda P. The thioredoxin TRX-1 regulates adult lifespan extension induced by dietary restriction in Caenorhabditis elegans. Biochem. Biophys. Res. Commun. 2011;406(3):478–482. doi: 10.1016/j.bbrc.2011.02.079. [DOI] [PubMed] [Google Scholar]
- 35.Blackwell T.K., Steinbaugh M.J., Hourihan J.M., Ewald C.Y., Isik M. SKN-1/Nrf, stress responses, and aging in Caenorhabditis elegans. Free Rad. Biol. Med. 2015;88(Pt B):290–301. doi: 10.1016/j.freeradbiomed.2015.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.An J.H., Blackwell T.K. SKN-1 links C. elegans mesendodermal specification to a conserved oxidative stress response. Genes Dev. 2003;17(15):1882–1893. doi: 10.1101/gad.1107803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McCallum K.C., Liu B., Fierro-Gonzalez J.C., Swoboda P., Arur S., Miranda-Vizuete A. TRX-1 regulates SKN-1 nuclear localization cell non-autonomously in Caenorhabditis elegans. Genetics. 2016;203(1):387–402. doi: 10.1534/genetics.115.185272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee K., Goh G.Y., Wong M.A., Klassen T.L., Taubert S. Gain-of-Function alleles in Caenorhabditis elegans nuclear hormone receptor nhr-49 are functionally distinct. PLoS One. 2016;11(9) doi: 10.1371/journal.pone.0162708. e0162708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barrios A. Exploratory decisions of the Caenorhabditis elegans male: a conflict of two drives. Semin. Cell Dev. Biol. 2014;33:10–17. doi: 10.1016/j.semcdb.2014.06.003. [DOI] [PubMed] [Google Scholar]
- 40.Lipton J., Kleemann G., Ghosh R., Lints R., Emmons S.W. Mate searching in Caenorhabditis elegans: a genetic model for sex drive in a simple invertebrate. J. Neurosci. Offic. J. Soc. Neurosci. 2004;24(34):7427–7434. doi: 10.1523/JNEUROSCI.1746-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barrios A., Nurrish S., Emmons S.W. Sensory regulation of C. elegans male mate-searching behavior. Curr. Biol. 2008;18(23):1865–1871. doi: 10.1016/j.cub.2008.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barrios A., Ghosh R., Fang C., Emmons S.W., Barr M.M. PDF-1 neuropeptide signaling modulates a neural circuit for mate-searching behavior in C. elegans. Nat. Neurosci. 2012;15(12):1675–1682. doi: 10.1038/nn.3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ryan D.A., Miller R.M., Lee K., Neal S.J., Fagan K.A., Sengupta P. Sex, age, and hunger regulate behavioral prioritization through dynamic modulation of chemoreceptor expression. Curr. Biol. 2014;24(21):2509–2517. doi: 10.1016/j.cub.2014.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hilbert Z.A., Kim D.H. PDF-1 neuropeptide signaling regulates sexually dimorphic gene expression in shared sensory neurons of C. elegans. eLife. 2018;7 doi: 10.7554/eLife.36547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hilbert Z.A., Kim D.H. Sexually dimorphic control of gene expression in sensory neurons regulates decision-making behavior in C. elegans. eLife. 2017;6 doi: 10.7554/eLife.21166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Laurent T.C., Moore E.C., Reichard P. Enzymatic synthesis of deoxyribonucleotides. Iv. Isolation and characterization of thioredoxin, the hydrogen donor from Escherichia coli B. J. Biol. Chem. 1964;239:3436–3444. [PubMed] [Google Scholar]
- 47.Olahova M., Veal E.A. A peroxiredoxin, PRDX-2, is required for insulin secretion and insulin/IIS-dependent regulation of stress resistance and longevity. Aging Cell. 2015;14(4):558–568. doi: 10.1111/acel.12321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ohta A., Ujisawa T., Sonoda S., Kuhara A. Light and pheromone-sensing neurons regulates cold habituation through insulin signalling in Caenorhabditis elegans. Nat. Commun. 2014;5:4412. doi: 10.1038/ncomms5412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen Y., Baugh L.R. Ins-4 and daf-28 function redundantly to regulate C. elegans L1 arrest. Dev. Biol. 2014;394(2):314–326. doi: 10.1016/j.ydbio.2014.08.002. [DOI] [PubMed] [Google Scholar]
- 50.O'Donnell M.P., Chao P.H., Kammenga J.E., Sengupta P. Rictor/TORC2 mediates gut-to-brain signaling in the regulation of phenotypic plasticity in C. elegans. PLoS Genet. 2018;14(2) doi: 10.1371/journal.pgen.1007213. e1007213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stiernagle T. Maintenance of C. elegans. WormBook. 2006:1–11. doi: 10.1895/wormbook.1.101.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cassada R.C., Russell R.L. The dauerlarva, a post-embryonic developmental variant of the nematode Caenorhabditis elegans. Dev. Biol. 1975;46(2):326–342. doi: 10.1016/0012-1606(75)90109-8. [DOI] [PubMed] [Google Scholar]
- 53.Inoue H., Hisamoto N., An J.H., Oliveira R.P., Nishida E., Blackwell T.K. The C. elegans p38 MAPK pathway regulates nuclear localization of the transcription factor SKN-1 in oxidative stress response. Genes Dev. 2005;19(19):2278–2283. doi: 10.1101/gad.1324805. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.