Abstract
From an epidemiological standpoint, disruptions to circadian rhythms have been shown to contribute to the development of various disease pathologies, including breast cancer. However, it is unclear how altered circadian rhythms are related to malignant transformations at the molecular level. In this article, a series of isogenic breast cancer cells representing disease progression was used to investigate the expression patterns of core circadian clock proteins BMAL1 and PER2. Our model is indicative of 4 stages of breast cancer and includes the following cells: MCF10A (non-malignant), MCF10AT.Cl2 (pre-malignant), MCF10Ca1h (well-differentiated, malignant), and MCF10Ca1a (poorly differentiated, malignant). While studies of circadian rhythms in cancer typically use low-resolution reverse transcription polymerase chain reaction assays, we also employed luciferase reporters BMAL1:Luc and PER2:Luc in real-time luminometry experiments. We found that across all 4 cancer stages, PER2 showed relatively stable oscillations compared with BMAL1. Period estimation using both wavelet-based and damped-sine-fitting methods showed that the periods are distributed over a wide circadian range and there is no clear progression in mean period as cancer severity progresses. Additionally, we used the K-nearest neighbors algorithm to classify the recordings according to cancer line, and found that cancer stages were largely differentiated from one another. Taken together, our data support that there are circadian discrepancies between normal and malignant cells, but it is difficult and insufficient to singularly use period evaluations to differentiate them. Future studies should employ other progressive disease models to determine whether these findings are representative across cancer types or are specific to this series.
Keywords: breast cancer, isogenic cell lines, disrupted circadian rhythms, luciferase reporters, machine learning, period-estimation
Introduction
In 2017, breast cancer became the most common cancer type (30% incidence) among women in the United States; a woman living in the United States has a 12.4% risk of being diagnosed with the disease over the course of her lifetime.1 There are many risk factors linked to the development of breast cancer, including advancing age and familial history.2,3 Among these, accumulating evidence has shown that circadian alterations elicit pathological changes in breast tissues, resulting in the formation of invasive breast tumors.4,5 Disruption of circadian rhythms can be induced through clock gene mutations or environmental contributors.6 While relevant genetic factors are yet to be identified, it is widely believed that particular activities will trigger circadian disruptions, including night and other shift-work, chronic exposure to light at night, chronic jet lag, exposure to low-frequency electromagnetic waves, and diet disorders.7-10 The International Agency for Research on Cancer states that “shift work that includes circadian disruption is probably carcinogenic to humans (Group 2A).”11
While circadian disruption has been associated with breast cancer development, the molecular mechanisms underlying this relationship are still unclear. To reveal the systematic interactions between circadian rhythms and malignant transformations at the cellular level, several studies have manipulated the expression of core clock proteins in cell culture models of breast cancer, and show that circadian rhythms may have opposing roles. BMAL1 and PER2 have been strongly implicated as tumor suppressors. When BMAL1 and PER2 levels are reduced via siRNA knockdown, breast cancer cells exhibit faster growth and migration rates while overexpression of BMAL1 and PER2 has been shown to decrease cancer cell invasiveness and inhibit the formation of tumors.12,13 PER2 expression has been found to be variable in cancerous tissues in comparison with near-by noncancerous tissue samples,14 whereas BMAL1 has shown no significant changes in comparison with normal cells in patients.15 Conversely, CLOCK and REV-ERBβ have been suggested to be tumor drivers, as they are found to be upregulated in breast cancer cells and downregulated in healthy breast tissues.16 While these studies provide valuable insights toward understanding the roles of circadian rhythms in breast cancer development, in most cases, the dynamic nature of circadian oscillations was not taken into account.
Circadian oscillations are dynamic processes that cycle with a ~24-hour period. The rhythms of cancer cells have been proposed to be altered through effects on period, amplitude, phase, and/or damping rate, which may result in promotion of proliferation and metastasis.17-20 The expression profiles of endogenous mRNA have been widely used to assess changes,21-24 but these experiments inherently suffer from short tracking time, infrequent sampling, and low data resolution, resulting in poor mathematical estimations. To overcome these limitations, real-time bioluminescence monitoring of in vitro cell or ex vivo organ cultures has been utilized to reveal intrinsic circadian oscillations in a time-dependent manner. For example, using a luciferase reporter system in cultured peripheral tissues of PER2::Luc mice, Yoo et al25 were the first to show robust and persistent circadian oscillations for >20 cycles. Their results have led to the employment of luciferase reporter systems to analyze circadian changes in various cell models to study dynamic processes, including signal transduction and tissue development.26,27 However, for circadian studies in the context of cancer, luciferase reporters have largely been used to study how rhythms respond to changes in a single cell line, either via genetic alteration (eg, knock-down or overexpression) or chemical modulation (eg, small molecule antagonist/agonist). Reporter systems have been rarely used with serial and isogenic cancer cell models to track cancer-induced circadian alterations; the only previous instance known assessed a serial model of skin cancer.17
In this study, we use real-time luminometry to uncover changes in core clock gene expression (specifically, BMAL1 and PER2) following malignant transformations in breast cancer. To mimic these genetic alterations we used the MCF10 series of breast cancer cells representing disease progression, which spans from nontumorigenic epithelial to highly malignant metastatic cancer.28 Cell lines within this series are isogenic and originally derived from an immortalized breast epithelial cell line, MCF10A (nontumorigenic). Following H-Ras transformation of MCF10A, the daughter cell line MCF10AT.Cl2 (premalignant) was produced. In turn, these cells were also modified to give rise to MCF10Ca1h (malignant, well-differentiated), and MCF10Ca1a (malignant, poorly differentiated) cells. According to a whole genome, exome, and RNA sequencing study, the driving mutations of TP53 and PIK3CA are acquired during the malignant transformation in this series of cells.29 While MCF10A cells are incapable of forming tumors in vivo, MCF10AT.Cl2 cells yield tumors 25% of the time, and MCF10Ca1h and MCF10Ca1a result in tumors almost 100%.30
We initially obtained lower-resolution results of BMAL1 and PER2 expression patterns using reverse transcription polymerase chain reaction (RT-PCR). We also generated stable BMAL1:Luc and PER2:Luc versions of each cell line in the series, and used real-time luminometry to follow their oscillations for 5 to 6 cycles. We estimated periods using continuous wavelet transformation (CWT) and damped-sine (DS) fitting methods. Overall, we found that both BMAL1 and PER2 were rhythmic, but BMAL1 tended to have more unstable oscillations than PER2, concomitant with cancer progression. Additionally, the durations of periods slightly increased with time of recording/cycles recorded, but no clear trends in period distributions were observed. However, when we used the K-nearest neighbors algorithm to classify each time-series according to its cancer line, we found it was moderately successful. Altogether, our data indicate that while subtle, differences in circadian rhythms indeed exist as cells transform from normal to malignant, and more detailed analyses than period estimations are required.
Materials and Methods
Cell Culture
The MCF10 series of cells was obtained from the Barbara Ann Karmanos Cancer Institute, Detroit, MI. MCF10A, MCF10AT.Cl2, MCF10Ca1h, and MCF10Ca1a breast cancer cells were maintained in DMEM/F12 culture medium (Gibco), supplemented with 1% HEPES (Gibco), 5% fetal bovine serum (Gibco), 1% penicillin-streptomycin (Gibco), 1% L-glutamine (Gibco), 15 µg/mL gentamicin (Fisher Scientific), 10 µg/mL insulin (Sigma), 20 ng/mL human EGF (Gibco), 0.1 µg/mL cholera enterotoxin (Sigma), and 0.5 µg/mL hydrocortisone (Sigma). All cells were incubated at 37°C with 5% CO2 atmosphere. For all experiments conducted, no cell line was passaged more than 5 times.
Synchronization of Cells by Serum Shock
Cells were seeded in 35 mm culture dishes at a density of 1 × 105 cells/mL and incubated for approximately 24 hours to reach 100% confluence. Culture media was removed and cells were washed with phosphate-buffered saline (Gibco). Cells were then starved in DMEM/F12 medium without any growth supplements for 12 hours (for both RT-PCR and real-time luminometry experiments). After starvation, cells were serum shocked using growth medium containing 50% fetal bovine serum (batch bought) for 2 hours, followed by replacement with standard growth or recording media (described below).
RNA Extraction and cDNA Synthesis
Following synchronization of cells by serum shock as described above, cells were washed with phosphate-buffered saline and returned to starvation conditions. Cells were harvested with the first time point (T = 0) taken prior to serum shock, and every 4 hours thereafter for 48 or 60 hours. Cells were harvested from separate dishes at each time point to avoid longitudinal effects/trends. Total RNA was extracted via TRIzol Reagent (Gibco) according to the manufacturer’s instructions. Briefly, 1 mL TRIzol Reagent was added to lyse the cells. Cell lysates were incubated at room temperature for 5 minutes to allow complete dissociation of nucleoprotein complexes. After addition of 200 µL chloroform per 1 mL TRIzol, samples were shaken vigorously by hand for 15 seconds and incubated at room temperature for 3 minutes. Samples were then centrifuged at 12 000 × g for 15 minutes at 4°C to separate the RNA-containing, upper aqueous phase, from the lower chloroform phase. The RNA samples were further purified via PureLink RNA kit (Ambion) according to the manufacturer’s instructions. Total RNA concentration was determined via Nanodrop UV/Vis (Thermo Fisher Scientific). Overall, 1 µg of total RNA was reverse-transcribed to cDNA using 50 µM random hexamers, 40 U/µL RNaseOut, 10 mM dNTPs, and 200 U/µL SuperScript IV Reverse Transcriptase (Thermo Fisher Scientific).
Quantitative RT-PCR
RT-PCR was performed in 96-well plates. The reaction (20 µL per well) consisted of 100 ng cDNA, 10 µL iTaq universal SYBR Green Supermix (Biorad), 4 µM of forward and reverse primers each, and RNAse free water to a final volume of 20 µL. All DNA primers were purchased from Integrated DNA Technologies (Coralville, IA). The following sequences were used: GAPDH Forward (5′-CTT CTT TTG CGT CGC CAG CC-3′), Reverse (5′-ATT CCG TTG ACT CCG ACC TTC-3′); BMAL1 Forward (5′- CTA CGC TAG AGG GCT TCC TG-3′), Reverse (5′-CTT TTC AGG CGG TCA GCT TC-3′); and PER2 Forward (5′-TGT CCC AGG TGG AGA GTG GT-3′), Reverse (5′-TGT CAC CGC AGT TCA AAC GAG-3′). After brief centrifugation, samples were analyzed via CFX Connect real-time system (Biorad) programmed with an initial activation at 95°C for 3 minutes, followed by 40 cycles of 95°C denaturation for 10 seconds, and 60°C annealing/extension for 30 seconds. Relative BMAL1 and PER2 expression were determined by comparing the Ct values of BMAL1 and PER2 with GAPDH control via the 2∧ΔΔCt method.31 Three biological replicates and three technical replicates per biological replicate were analyzed for each condition. RAIN was used to classify RT-PCR recordings as rhythmic (P < .05) or arrhythmic with a circadian period (24 hours).32
Plasmid and Recombinant DNA
The BMAL1-luciferase reporter construct (pABpuro-BluF, herein referred to as BMAL1:Luc) was obtained from Addgene (plasmid #46824, deposited by Dr Steven Brown). For generation of PER2-luciferase (PER2:Luc) plasmid, a 159-bp EcoRI/NotI fragment was isolated from a pGL3 basic PER2 construct, obtained from Addgene (plasmid #48747, deposited by Dr Joseph Takahashi). The PER2 promoter-containing fragment was subcloned into the lentiviral construct pMA3160 (Addgene plasmid #35043, deposited by Dr Mikhail Alexeyev) to generate a PER2-luciferase reporter construct using the following primers: Forward primer containing EcoRI restriction site (5′-CCG GAA TTC AGC GTA GCT CTC AGG TTC CG-3′) and Reverse primer containing NotI restriction site (5′-ATA AGA ATG CGG CCG CGG AGC CGC TAG TCC CAG TAG-3′). Lentiviral packaging (psPAX2) and envelope (pMD2.G) plasmids were obtained from Prof. D. Joseph Jerry (UMass Amherst).
Lentiviral Transductions
Overall, 3 × 106 HEK293T cells were seeded in 60 mm culture dishes and transiently transfected with 3 µg psPAX2 packaging plasmid, 2 µg pMD2.G envelope plasmid, and 3 µg BMAL1:Luc or PER2:Luc reporter constructs using Lipofectamine3000 (Thermo Fisher Scientific) according to the manufacturer’s instructions. After 48 hours of incubation, lentiviral particles were harvested from the supernatant and passed through a 45 µm filter. 9 mL of lentivirus-containing supernatant was combined with 9 mL of DMEM/F12 culture medium containing 10 µg/mL polybrene (Sigma). All MCF10 series cell lines were seeded in T25 culture flasks at a density of 2 × 105 cells/mL and incubated under standard cell culture conditions until 70% to 80% confluence was reached. Culture medium was removed and 6 mL of lentiviral-containing media was added to each flask. After 2 days of infection, the medium was replaced with selection medium (DMEM/F12 with all growth supplements plus 4 µg/mL puromycin). At the beginning of selection (within the first ~3 days), approximately 10% of cell population survived, which contained BMAL1:Luc and PER2:Luc in their genomes. The selection medium was changed every 3 days for 4 weeks. Following transfection/selection, cell lines were frozen and stored in liquid nitrogen at passage 10. All cells were incubated at 37°C under 5% CO2 atmosphere.
Bioluminescence Recording and Analysis
All bioluminescence experiments were conducted at passage number 15 or less. Cells were seeded in 35 mm culture dishes at a density of 1 × 105 cells/mL and incubated to reach 100% confluence; cells were starved and serum shocked as described above. After 2 hours of synchronization, cells were incubated in recording medium (phenol-red free DMEM/F12 [Gibco] containing 20% of normal growth supplement concentrations, 6.5 mM sodium bicarbonate, 10 mM HEPES, 50 units/mL penicillin/streptomycin, and 0.5 mM luciferin; Thermo Fisher Scientific). Dishes were sealed with 40 mm sterile cover glass using silicon vacuum grease and subjected to continuous monitoring using a LumiCycle 32 (Actimetrics) at 36.5°C for 5 to 7 days.
Bioluminescence recordings were preprocessed by discarding the first 24 hours, removing spikes (a point that is more than one third the range of the data greater than the previous point), and filling in missing points (using the mean of the previous and subsequent points). To remove both low-frequency trends and high-frequency noise, the maximum overlap discrete wavelet transform (12-tap symmlet) with reflective boundary condition isolated the signal in the frequency band that contains a period of 24 hours (periods of 21 to 42 hours; wavelet methods33 implemented by Charles Cornish in the WMTSA MATLAB package). Using the same discrete wavelet analysis, time-series were determined to be rhythmic if the energy in the 24-hour band was greater than the sum of the energies in all higher frequency bands.34 If a time-series failed this initial test, it was visually inspected to determine if the initial transient had lasted beyond hour 24 (ie, that there was an initial peak that appeared to be biasing the wavelet results). If so, we repeated the analysis, using data after hour 36.
A bioluminescence recording was labeled as an outlier if its first trough or peak was out of phase with the majority (for BMAL1:Luc outliers first trough after t = 48 hours, for PER2:Luc outliers peak before t = 35 hours). Periods were estimated using 2 methods: (1) continuous wavelet analysis and (2) DS curve fitting. Continuous wavelet analysis was performed with the WAVOS package,35 using the Morlet wavelet, excluding edge data, with a tuning parameter η of 5, seeking a period within the range of 6 to 60 hours. DS curve fitting was performed by minimizing the least squares distance between a sine curve with exponential damping (Ae-λtsin(2π/τ + φ) + c) and the preprocessed, de-trended time series, using scipy.optimize.differential_evolution (Scipy version 0.19.0) to determine a rough estimate, and then scipy.optimize.minimize to find the precise estimate (using SLSQP method with 10 000 maximum iterations). Each parameter was constrained to a given range: the period τ between 10 and 50 hours, the phase offset φ between −π and π radians, the amplitude offset c between the maximum of the time-series and its negative, the amplitude A between half and twice the range of the time series, and the damping rate λ between 0 and 0.04 1/h.
K-nearest neighbors (KNN) classifiers were trained to classify preprocessed bioluminescence recordings by cancer lines.36 Because the numbers of recordings were small (N = 84 for BMAL1:Luc and N = 80 for PER2:Luc across all cell lines), K-fold cross-validation was used to assess their effectiveness. All code was written in Python 3.6 using Scikit-learn.36 For each of the BMAL1:Luc and PER2:Luc recordings, we used stratified K-fold cross-validation (K = 8 folds) with a KNN classifier (with K = 3 nearest neighbors) to compute the accuracy (number of correctly classified recordings divided by the total number of recordings). To determine whether this accuracy could be achieved by random chance, we repeated the cross-validation for 1000 permutations of the cancer line labels. If the largest accuracy from randomly permuted labels was smaller than the accuracy from the correct labels, then we deemed the classifier acceptable.
Results
BMAL1 and PER2 Display Different Circadian Phenotypes Across the MCF10 Series of Cells
The advantage of using the MCF10 series of cells is that it can recapitulate genetic changes that may occur during breast cancer progression. This isogenic cell model reflects characteristics of in vivo human breast lesions during progressive breast tumorigenesis. We have assessed the phenotypic characteristics of the component cell lines via colony formation (Figure S1; available online), proliferation (Figure S2), and wound-healing (Figure S3) assays. We found that the premalignant MCF10AT.Cl2 cells have the fastest proliferation and migration rates. As expected, the nontumorigenic MCF10A cells display the slowest proliferation and migration rates.
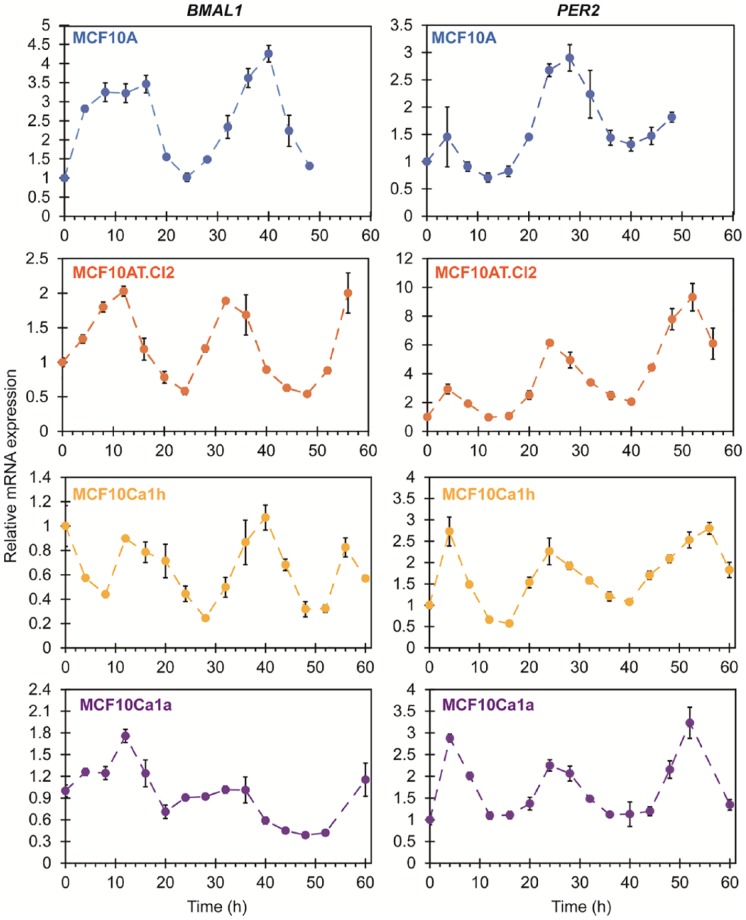
As it is the most common strategy for assessing circadian rhythms in breast and other cancer cell models, we first determined the endogenous mRNA expression patterns for BMAL1 and PER2 in cells across the MCF10 series via RT-PCR (Figure 1). While it is a relatively low-resolution method, it can be used to reveal relative changes in gene expression. Following synchronization, the mammary epithelial, nontumorigenic MCF10A cells showed BMAL1 (ARNTL; positive clock oscillator) peaks at ~8 to 12 hours and ~40 hours, while PER2 (negative clock oscillator) showed peaks at ~0 to 4 hours and ~24 to 28 hours. Therefore, the phase difference between BMAL1 and PER2 was approximately 12 hours, and the two had expression profiles in opposition to one another, as expected.23 Similar mRNA expression patterns for BMAL1 and PER2 were observed in the premalignant MCF10AT.Cl2 cells and malignant MCF10Ca1h and MCF10Ca1a cells. It is noteworthy that the fold changes in amplitude of BMAL1 were significantly higher (~4-fold) in MCF10A cells versus the cancer cells (<2-fold in MCF10AT.Cl2, MCF10Ca1h, and MCF10Ca1a). However, the relative changes in expression amplitudes of PER2 were similar (~3-fold) across the cells, with the exception of MCF10AT.Cl2. Most expression patterns were found to be rhythmic with a period of 24 hours (P < .05) using RAIN.32 Only BMAL1 expression in MCF10Ca1a was not clearly rhythmic (P = .055). However, the coarse resolution (time points every 4 hours) of the data obtained via RT-PCR cannot facilitate more accurate analysis of circadian oscillations, including a precise period estimation. In addition, the short duration (≤60 hours) means that the number of cycles that can be observed via RT-PCR is typically low (1-2 cycles), increasing uncertainty for assessments.
Figure 1.
Endogenous mRNA expression of BMAL1 and PER2 in synchronized cell lines belonging to the MCF10 series. Relative mRNA expression levels were determined via RT-PCR, normalized to GAPDH, using 3 biological replicates with 3 technical replicates each. Error bars represent relative error.
To overcome these issues, we generated luciferase reporter systems composed of these cell lines in order to track BMAL1 and PER2 expression in real time. This method also enables the recording of circadian oscillations over higher cycle numbers (ie, 5-7 days). Firefly luciferase has been widely used in many organisms and cells for circadian rhythms analysis.37-39 Unlike fluorescent reporter systems (ie, GFP), which require external excitation and may have problems associated with photo-bleaching and signal-to-noise ratios,40 luciferase rapidly generates bioluminescence in the presence of its substrate luciferin, and is highly sensitive. In the present study, all cells in the MCF10 series were separately lentivirally transduced with BMAL1- and PER2-driven luciferase reporters. Luciferase expression was validated via luciferase assay (Figure S4). All transfected cells showed significant levels of luciferase compared with nontransfected cells (P < .001), indicating successful insertion of BMAL1:Luc and PER2:Luc into host genomes.
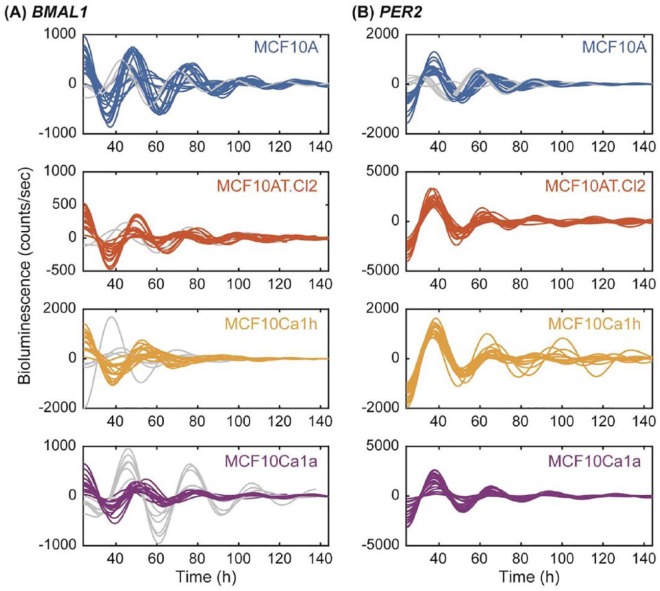
The expression of luciferase protein driven by BMAL1- (Figure 2A and Figure S5A) and PER2-promoters (Figure 2B and Figure S5B) were tracked for 6 days via LumiCycle 32 (Actimetrics). In all cell lines, BMAL1 and PER2 exhibited antiphase oscillations. Using a discrete wavelet transform, we determined that all recordings were rhythmic, with all but 3 passing a test for rhythmicity that took into consideration the entire time-series and the remaining 3 passing when only data after t = 36 hours was considered (see Methods and Leise and Harrington34). Of the 84 total BMAL1:Luc recordings (21 per cancer line), 16 troughed too late (t > 48 hours) and were classified as outliers (Figure 3A; 1 from MCF10A, 2 from MCF10Cl2, 4 from MCF10Ca1h, and 6 from MCF10Ca1a). Of the 80 total PER2:Luc recordings (20 per cancer line), 5 (all from nontumorigenic MCF10A cells) peaked too early (t < 35 hours) in the first cycle and were classified as outliers (Figure 3B). This may be the result of inherent heterogeneity in the MCF10A cell line.41 Our data show that PER2 displays relatively stable oscillations at the transcriptional level across all 4 cancer stages, while BMAL1 appears to be more unstable. The consistency of PER2 expression was corroborated by western blot (Figure S6). We found that PER2 protein levels were relatively similar in all cell lines, while BMAL1 was significantly increased in premalignant MCF10AT.Cl2 cells. Our results lead to an inference that BMAL1 may be disrupted to a greater extent than PER2 when certain malignant transformations occur, or in particular cancer subtypes. Our data also raise an interesting conjecture, that in some cases, BMAL1 may play a more dominant role in suppressing tumors than PER2, which is universally thought as a tumor suppressor in breast cancer.14,42,43
Figure 2.
Circadian oscillations of (A) BMAL1:Luc and (B) PER2:Luc in the MCF10 series of cells. Shown for each cell line are the 21 time series for BMAL1 and 20 for PER2 obtained, de-trended and de-noised using a discrete wavelet transform. Both reporters show stable oscillations across cellular transformations from nonmalignant MCF10A (blue) to premalignant MCF10AT.Cl2 (orange) to malignant MCF10Ca1h (yellow) and MCF10Ca1a cells (purple). However, BMAL1 has more time-series out of phase with the majority (gray; N = 4 for MCF10A, N = 2 for MCF10A.Cl2, N = 4 for MCF10Ca1h, and N = 6 for MCF10Ca1a) versus PER2 (gray; N = 5 for MCF10A).
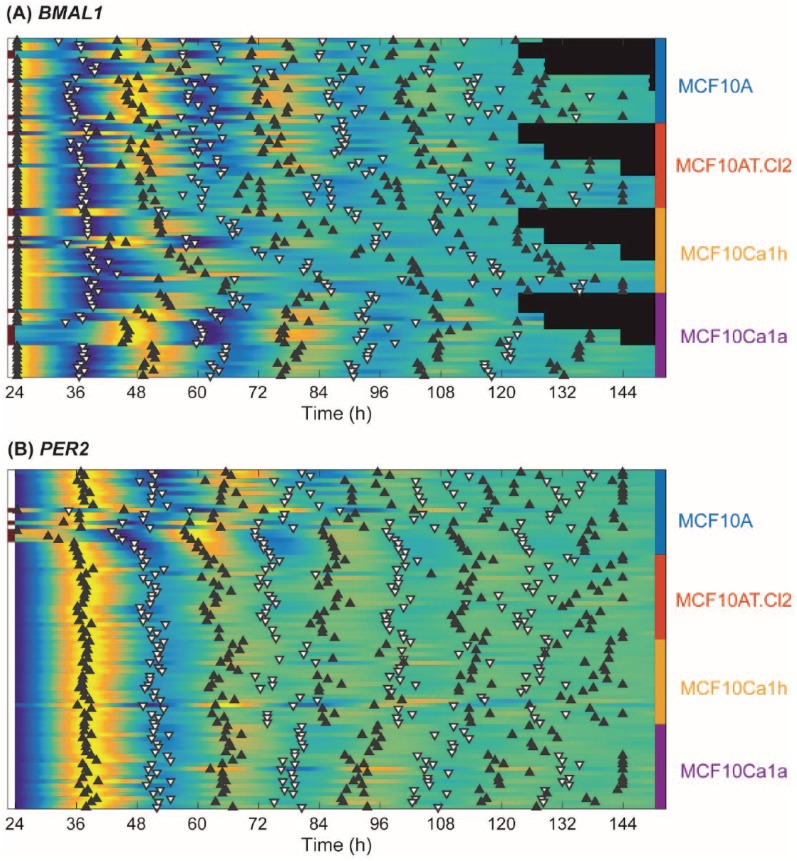
Figure 3.
Peaks and troughs of normalized (A) BMAL1:Luc and (B) PER2:Luc show that the first cycles of most oscillations are in phase and that PER2 and BMAL1 are antiphase. Heat maps are included, representing low (blue) to high (yellow) bioluminescence with one row per time series, organized by cell line (color bar on right indicating blue for MCF10A, orange for MCF10AT.Cl2, yellow for MCF10Ca1h, and purple for MCF10Ca1a cells). Black bars indicate recordings where no data were recorded at that time. Peaks (black triangles) and troughs (white triangles with black outline) are indicated for each recording and are used to determine outliers (for BMAL1:Luc, outliers trough after t = 48 hours, for PER2:Luc, outliers peak before t = 35 hours). Outliers are indicated by a gray entry in the first column, versus white for non-outliers.
To provide a clear view of how the oscillations of BMAL1 and PER2 change in each cancer cell line with time/cycle, we generated heat maps of the peaks and troughs in each recording trace (after removing the trend and noise with a discrete wavelet transform). As shown in Figure 3, the first 3 cycles of BMAL1 in nontumorigenic MCF10A cells and poorly differentiated malignant MCF10Ca1a cells showed stronger relative amplitudes than premalignant MCF10AT.Cl2 and well-differentiated malignant MCF10Ca1h cells. While in the case of the MCF10AT.Cl2 cells, the relative change in amplitude may be attributed to lower transfection efficiency, MCF10Ca1a cells resulted in the highest average levels of luciferase produced across all cell lines (Figure S4). In all cell lines evaluated, the peaks and troughs in BMAL1 traces grew quickly out of phase. For PER2, only MCF10A cells showed 2 strong cycles, while the other 3 cell lines showed only one high-amplitude cycle before losing amplitude (ANOVA [analysis of variance] permutation test comparing ratio of heights of second and third peaks with 100 000 trials, P < .05). Of note, most of the peaks and troughs in PER2 were in phase with each other. This result indicates again that BMAL1 is a more unstable oscillator than PER2 and is highly disrupted as breast epithelial cells (eg, MCF10A) transform into more aggressive and malignant stages (eg, MCF10Ca1a).
Cells in the MCF10 Series Exhibit Periodic Oscillations in a Wide Circadian Range
To further understand how circadian oscillations change across cancer progression, we analyzed changes in the periods of BMAL1 and PER2 when all data were included (from day 1 to day 6) using 2 different methods. The first is the wavelet-based method, CWT (Figure 4), which provides reliable estimations when the speed of rhythms change over time (eg, nonstationary oscillations). However, if the rhythms are dramatically unstable (eg, exhibit amplitude variations), the wavelet-based method may fail to estimate a period. The second approach used is the DS (Figure S7) fitting method, which can always estimate a period from an oscillation curve (eg, stationary oscillation), but the goodness of fit decreases when rhythms change over time. DS fitting is widely used, including in commercially available software, for example, LumiCycle Analysis (Actimetrics). The CWT-based method showed that the periods of both BMAL1 and PER2 changed from t = 0 hours to t = 144 hours, but not dramatically, across all cell lines (Figure 4). Similarly, the DS fitting period estimates for both BMAL1 and PER2 were largely consistent regardless of how many cycles were fit (Figure S7). All CWT period estimates and 88% of DS fitting estimates were within the circadian range of 16 to 32 hours even though neither method was constrained to that range (DS fitting was constrained to 10 to 50 hours and the CWT method was implicitly constrained to 21 to 42 hours by discrete wavelet de-trending). This supports the notion that the circadian oscillations are relatively stable. Taken together, our data indicate that the cells still express periodic oscillations during cancer progression, but the cycling time subtly changes as recording time increases.
Figure 4.
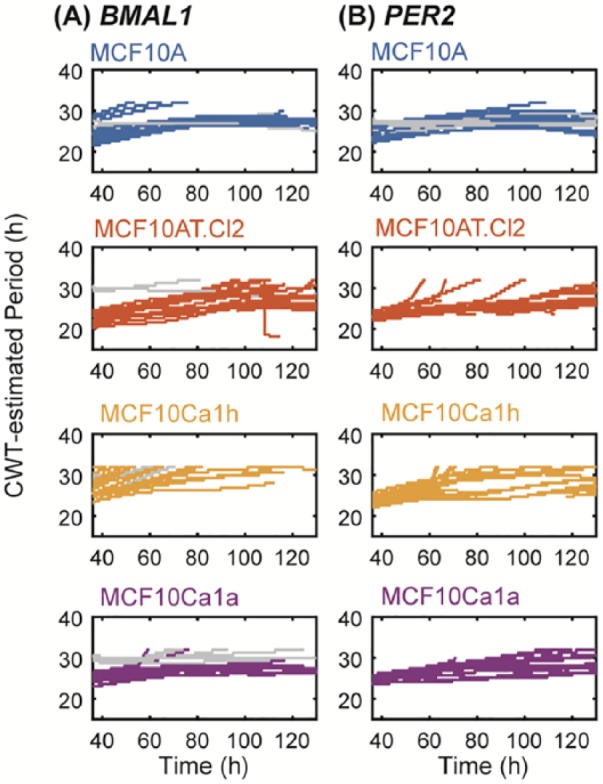
Continuous wavelet transforms (CWT) estimate circadian periods for (A) BMAL1 and (B) PER2 over time. Shown are Morlet-wavelet estimates of the period of oscillation for each cell line and reporter (blue for MCF10A, orange for MCF10AT.Cl2, yellow for MCF10Ca1h, and purple for MCF10Ca1a). The periods of outlier time series are shown in gray.
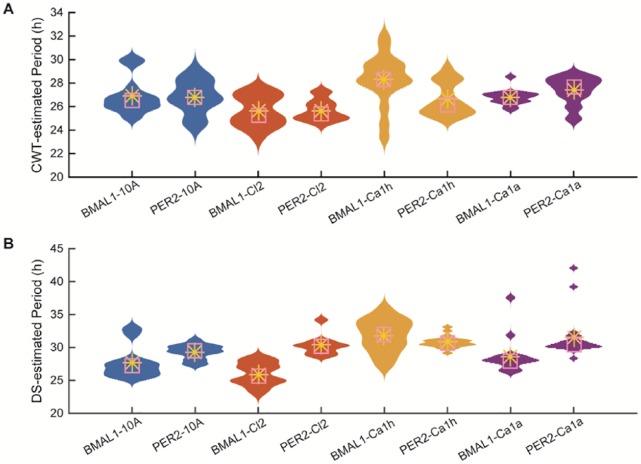
Because both period estimation methods led to a wide range of values, we sought to determine if their distributions differed across cancer lines and reporters (Figure 5). We evaluated both the average CWT estimate over time (Figure 5A) and the DS fitting period estimate (Figure 5B) for the longest time-series (t = 24 hours to t = 144 hours). Visual inspection reveals that the distributions are not consistently Gaussian and that period estimates cover a wide range of values. In general, the wavelet-based estimates fall into a narrower range than the damped sine curve-based estimates, but there are clear outliers in the sine-based estimates of MCF10Ca1a for both reporters. For individual traces, the DS-estimates were longer and, for MCF10A and MCF10Ca1h, weakly correlated (R2 = 0.39 and 0.28, respectively) with CWT estimates. The widest range estimates occur for MCF10Ca1h, which are 23 to 32 hours using CWT (Figure 5A), and 25 to 40 hours using DS (Figure 5B). There is no obvious trend in the distributions of period estimation as cancer progresses.
Figure 5.
Period estimates from 2 methods reveal similar distributions across the MCF10 series. For each cell line, shown are the distributions of period estimates computed using (A) the average CWT-estimated period over hours t = 24 hours to t = 144 hours of recording and (B) the period of best-fit damped sine curve over hours t = 24 hours to t = 144 hours of recording. The distributions are represented as violin plots (the wider the violin, the more frequent the period) with their means (yellow asterisks) and medians (pink boxes). The periods of outlier time series are excluded from these distributions.
We sought to quantify the differences in distributions by testing whether they shared the same means. Because the data did not meet the requirements for a stand-alone ANOVA, we used ANOVA to compute an F-statistic and a permutation test (100 000 permutations) to determine the P value. We first quantified differences between reporters. Both the DS curve fitting and wavelet-based methods computed different means (Bonferroni-corrected P < .05) for MCF10Ca1h and MCF10Ca1a. We then quantified differences across cancer lines with pairwise comparisons between cancer lines for each reporter and each period estimation method. For data collected with the PER2 reporter, there were no statistically significant (Bonferroni-corrected P < .05) differences between period estimates for different cancer lines. For data collected with the BMAL1 reporter, both methods led to different mean estimates when MCF10Ca1a was compared with either MCF10A or MCF10AT.Cl2 (Bonferroni-corrected P < .05).
Oscillations in MCF10 Series of Cells are Characterized by Cell Line
The period distributions from bioluminescence analyses exhibited no clear pattern differentiating cells lines, but were clearly varied within and among cell lines. Given that period-estimation methods provide a summary statistic and therefore may not reflect important characteristics, we sought an alternative approach that would analyze the time-series directly to determine whether they could be characterized by cell line. Thus, we tested whether a classifier could be trained to classify time-series according to their cell lines. We chose a KNN algorithm (see Materials and Methods) because it relies on the data directly rather than on a summarizing statistic, and is straight-forward to train and test.
In separate analyses, we considered the pre-processed (but not de-trended) and the DWT-de-trended and de-noised time-series. In both cases, the time-series were normalized to the range of 0 to 1 so that baseline amplitude would not have an effect. We tested classifiers built to classify time-series of similar amplitude and waveform (using the squared Euclidean distance measure) and to classify time-series of similar phase (using the correlation distance measure). Using K-fold cross-validation, we measured the accuracy (percent of time-series classified correctly) of each classifier and found that the values ranged from 57% to 75% (Table S1). For reference, a perfect classifier would have an accuracy of 100% and a random classifier would have an accuracy of 25%.
We also sought to quantify the probability that we would reach an accuracy as high as that observed by random chance. For each classifier, we used a permutation test to build a distribution of accuracy values for time-series with randomly assigned cancer line labels and found that all accuracies were lower than the accuracy of the data with the true cancer line labels (permutation test, P = 0). This result held regardless of data de-trending, reporter used, inclusion of outliers, and distance measure.
Discussion
Circadian disruptions have been implied to elicit pathological developments, including but not limited to breast cancer. Conversely, malignant transformation(s) in cells have also been proposed to provoke alterations to circadian functions, resulting in primary tumor formation and metastases. While genetic manipulations (eg, knockdown, overexpression, and mutation) of circadian genes/proteins can provide valuable insights into their roles in regulating cancer development, a basic understanding of the kinetic profiles of circadian disruptions occurring in cancer is still lacking. To investigate the dynamic changes of circadian clocks in cancer cells, most studies have characterized mRNA expression patterns using RT-PCR. As shown in Figure 1, we follow this commonly used strategy to observe the expression profiles of BMAL1 and PER2 in 4 isogenic breast cancer cells representing disease progression: MCF10A (non-tumorigenic), MCF10AT.Cl2 (pre-malignant), MCF10Ca1h (well-differentiated malignant), and MCF10Ca1a (poorly differentiated malignant). Our data show that BMAL1 and PER2 are rhythmic with a period of 24 hours (P < .05 using RAIN), and only BMAL1 expression in MCF10Ca1a was not clearly rhythmic (P = .055). Although this analysis method can provide fundamental information on rhythmic expression, the coarse resolution (eg, 4 hours) and short tracking duration (eg, 48-60 hours) raise the uncertainty of period estimation, and make evaluation of other circadian parameters (eg, phase, amplitude, and decay rate) difficult.
To overcome these issues, we used firefly luciferase reporters, whose expressions are driven via BMAL1 or PER2 promoters, to perform real-time analyses of circadian oscillations in the MCF10 series of cells. With this method, we collected nearly 900 data points over the course of 5 to 7 recording days of BMAL1 and PER2 oscillations (Figure 2 and Figure S5), allowing us to use period-estimation methods that take into account damping or nonstationary rhythms. After de-trending the raw data via DWT, BMAL1 and PER2 displayed antiphase oscillation patterns in all cancer lines, which can be clearly observed on a heat map annotated with peaks and troughs of BMAL1 (Figure 3A) and PER2 (Figure 3B). We further examined rhythmicity and found that 16 out of 84 total BMAL1 traces were classified as outliers, while the same held for only 5 out of 80 total PER2 traces (see Methods and Figure 3). This result indicates that BMAL1 is more unstable than PER2 across different cancer stages. In addition to its instability, BMAL1 also exhibits smaller relative amplitude changes than PER2. While in luminometry this may be attributable to lower transfection efficiencies, reduced levels were also observed using RT-PCR (Figure 1) and western blots (Figure S6). Across the series’ progression, our data indicate that as cells transform into premalignant and malignant states, BMAL1 is downregulated at both transcriptional and translational levels, while PER2 remains fairly consistent. However, it is unclear why premalignant MCF10AT.Cl2 cells exhibited significantly higher BMAL1 protein levels than found in the other 3 cell lines (Figure S6B). Future studies will include assessment of the translational profiles of clock proteins in a time-dependent manner.
To further characterize the oscillation patterns of BMAL1 and PER2 across the cell lines, we used 2 distinct mathematical methods to estimate periods: CWT and DS fitting methods. Both showed that the periods of BMAL1 and PER2 exhibit minor changes with time. CWT-based estimation shows that the periods increase slightly over time (Figure 4). DS fitting-based estimation indicates even more subtle changes: BMAL1 periods increase slightly across the first few cycles and then plateau, while PER2 appears to remain mostly stable (Figure S7). We hypothesize that this period deviation may be caused by (1) desynchronization between individual cells or (2) loss of regular entraining stimuli in the culture environment. Desynchronization may be the result of changes in the cycling rate of individual oscillators or changes in the relationships among oscillators. Future studies could use alternative synchronization methods (ie, temperature entrainment, heat shock, or chemical stimulations) to synchronize inherent clocks, which can provide additional data for period calculations. The latter hypothesis could be tested via media change every few cycles to re-stimulate cellular clock oscillations. Single-cell analysis may also be used to study desynchronization among rhythmic cells.
Since all estimated periods are within the circadian range, we computed the distributions of periods to investigate differences across cancer lines and reporters. All cell lines with either reporter exhibited non-Gaussian distributions with a wide range (Figure 5), and there is no clear trend in the distributions of period estimation concomitant with cancer progression. Together, our data illustrate the difficulty of estimating the “true” period within each oscillator in these cancer cells. Furthermore, the “true” period may not be fixed. Additionally, the mean period appears to be different between MCF10Ca1a and 2 others (MCF10A and MCF10AT.Cl2) when BMAL1 is used as the reporter. However, if we use the KNN algorithm to classify traces with the entire time-series, we can classify each cancer line with moderate success (Table S1). This is true regardless of what preprocessing methods were applied to the raw data.
Altogether, we show that circadian rhythms are altered as cancer progresses from normal to malignant state. However, the changes to BMAL1 and PER2 oscillations are not limited to periodic ones. Other factors, such as amplitude changes, stochasticity, and loss of synchrony, need to be taken into consideration. Single-cell-level analysis could provide further insights, since cancer is heterogeneous even within the same culture environment.41 Additionally, next-generation sequencing could elucidate the genetic changes throughout the entire cell. Potential driver mutations also need to be considered. As mentioned above, the generation of MCF10 series of cells was initiated by H-Ras mutation, followed by other genetic alterations, including TP53 and PIK3CA.29 Although studies of the influence of mutations across isogenic and serial cell lines are limited, Relógio et al17 reported that Ras mutations can cause changes to the period of BMAL1 in a series of human HaCaT skin keratinocytes. Collectively, the data presented here and in that study only address Ras mutations, albeit in 2 different types of cancer. Future studies should evaluate other mutations and cancer types. By correlating circadian rhythm alterations with cancer severity, we may be able to offer significant insights to this devastating disease, leading to improved means for prevention, and the development of new drugs and targets.
Supplemental Material
Supplemental material, Lin_et_al_Supporting_Information_Revision_1 for Analyses of BMAL1 and PER2 Oscillations in a Model of Breast Cancer Progression Reveal Changes With Malignancy by Hui-Hsien Lin, Maan Qraitem, Yue Lian, Stephanie R. Taylor and Michelle E. Farkas in Integrative Cancer Therapies
Acknowledgments
We gratefully acknowledge Prof Tanya Leise (Mathematics and Statistics, Amherst College) for helpful comments on the manuscript, Prof D. Joseph Jerry (Veterinary and Animal Sciences, UMass Amherst) for advice on stable transfections and use of cell culture models, and Prof Jungwoo Lee (Chemical Engineering, UMass Amherst) and Prof Yubing Sun (Mechanical Engineering, UMass Amherst) for plate reader use.
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: HHL was supported by a University of Massachusetts Amherst Chemistry-Biology Interface (CBI) training fellowship. MQ and YL received funding and SRT received travel support from the Office of the Provost of Colby College.
Supplemental Material: Supplemental material for this article is available online.
ORCID iD: Michelle E. Farkas  https://orcid.org/0000-0001-5824-1243
https://orcid.org/0000-0001-5824-1243
References
- 1. American Cancer Society. Cancer Facts & Figures 2018. Atlanta, GA: American Cancer Society; 2018. [Google Scholar]
- 2. Colditz GA, Willett WC, Hunter DJ, et al. Family history, age, and risk of breast cancer. Prospective data from the Nurses Health Study. JAMA. 1993;270:338-343. [PubMed] [Google Scholar]
- 3. McPherson K, Steel CM, Dixon JM. ABC of breast diseases. Breast cancer—epidemiology, risk factors, and genetics. BMJ. 2000;321:624-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Greene MW. Circadian rhythms and tumor growth. Cancer Lett. 2012;318:115-123. [DOI] [PubMed] [Google Scholar]
- 5. Lin HH, Farkas ME. Altered circadian rhythms and breast cancer: from the human to the molecular level. Front Endocrinol (Lausanne). 2018;9:219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shanmugam V, Wafi A, Al-Taweel N, Büsselberg D. Disruption of circadian rhythm increases the risk of cancer, metabolic syndrome and cardiovascular disease. J Local Glob Health Sci. 2013;3. doi: 10.5339/jlghs.2013.3 [DOI] [Google Scholar]
- 7. James P, Bertrand KA, Hart JE, Schernhammer ES, Tamimi RM, Laden F. Outdoor light at night and breast cancer incidence in the Nurses’ Health Study II. Environ Health Perspect. 2017;125:087010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wright KP, Jr, Bogan RK, Wyatt JK. Shift work and the assessment and management of shift work disorder (SWD). Sleep Med Rev. 2013;17:41-54. [DOI] [PubMed] [Google Scholar]
- 9. Kojo K, Pukkala E, Auvinen A. Breast cancer risk among Finnish cabin attendants: a nested case-control study. Occup Environ Med. 2005;62:488-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Stevens RG, Brainard GC, Blask DE, Lockley SW, Motta ME. Breast cancer and circadian disruption from electric lighting in the modern world. CA Cancer J Clin. 2014;64:207-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Baan R, Grosse Y, Straif K, et al. ; WHO International Agency for Research on Cancer Monograph Working Group. A review of human carcinogens—part F: chemical agents and related occupations. Lancet Oncol. 2009;10:1143-1144. [DOI] [PubMed] [Google Scholar]
- 12. Yang X, Wood PA, Oh EY, Du-Quiton J, Ansell CM, Hrushesky WJ. Down regulation of circadian clock gene Period 2 accelerates breast cancer growth by altering its daily growth rhythm. Breast Cancer Res Treat. 2009;117:423-431. [DOI] [PubMed] [Google Scholar]
- 13. Jung CH, Kim EM, Park JK, et al. Bmal1 suppresses cancer cell invasion by blocking the phosphoinositide 3-kinase-Akt-MMP-2 signaling pathway. Oncol Rep. 2013;29:2109-2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen ST, Choo KB, Hou MF, Yeh KT, Kuo SJ, Chang JG. Deregulated expression of the PER1, PER2 and PER3 genes in breast cancers. Carcinogenesis. 2005;26:1241-1246. [DOI] [PubMed] [Google Scholar]
- 15. Lesicka M, Jabłońska E, Wieczorek E, et al. Altered circadian genes expression in breast cancer tissue according to the clinical characteristics. PLoS One. 2018;13:e0199622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xiao L, Chang AK, Zang MX, et al. Induction of the CLOCK gene by E2-ERα signaling promotes the proliferation of breast cancer cells. PLoS One. 2014;9:e95878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Relógio A, Thomas P, Medina-Pérez P, et al. Ras-mediated deregulation of the circadian clock in cancer. PLoS Genet. 2014;10:e1004338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mormont MC, Lévi F. Circadian-system alterations during cancer processes: a review. Int J Cancer. 1997;70:241-247. [DOI] [PubMed] [Google Scholar]
- 19. Ticher A, Haus E, Ron HG, Sackett-Lundeen L, Ashkenazi IE. The pattern of hormonal circadian time structure (acrophase) as an assessor of breast-cancer risk. Int J Cancer. 1996;65:591-593. [DOI] [PubMed] [Google Scholar]
- 20. Otálora BB, Madrid JA, Alvarez N, Vicente V, Rol MA. Effects of exogenous melatonin and circadian synchronization on tumor progression in melanoma-bearing C57BL6 mice. J Pineal Res. 2008;44:307-315. [DOI] [PubMed] [Google Scholar]
- 21. Rossetti S, Esposito J, Corlazzoli F, Gregorski A, Sacchi N. Entrainment of breast (cancer) epithelial cells detects distinct circadian oscillation patterns for clock and hormone receptor genes. Cell Cycle. 2012;11:350-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cadenas C, van de Sandt L, Edlund K, et al. Loss of circadian clock gene expression is associated with tumor progression in breast cancer. Cell Cycle. 2014;13:3282-3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gutiérrez-Monreal MA, Treviño V, Moreno-Cuevas JE, Scott SP. Identification of circadian-related gene expression profiles in entrained breast cancer cell lines. Chronobiol Int. 2016;33:392-405. [DOI] [PubMed] [Google Scholar]
- 24. Chacolla-Huaringa R, Moreno-Cuevas J, Trevino V, Scott SP. Entrainment of breast cell lines results in rhythmic fluctuations of MicroRNAs. Int J Mol Sci. 2017;18:E1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yoo SH, Yamazaki S, Lowrey PL, et al. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci U S A. 2004;101:5339-5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Imamura K, Yoshitane H, Hattori K, et al. ASK family kinases mediate cellular stress and redox signaling to circadian clock. Proc Natl Acad Sci U S A. 2018;115:3646-3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ono D, Honma S, Nakajima Y, Kuroda S, Enoki R, Honma KI. Dissociation of Per1 and Bmal1 circadian rhythms in the suprachiasmatic nucleus in parallel with behavioral outputs. Proc Natl Acad Sci U S A. 2017;114:E3699-E3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Soule HD, Maloney TM, Wolman SR, et al. Isolation and characterization of a spontaneously immortalized human breast epithelial cell line, MCF-10. Cancer Res. 1990;50:6075-6086. [PubMed] [Google Scholar]
- 29. Maguire SL, Peck B, Wai PT, et al. Three-dimensional modelling identifies novel genetic dependencies associated with breast cancer progression in the isogenic MCF10 model. J Pathol. 2016;240:315-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Santner SJ, Dawson PJ, Tait L, et al. Malignant MCF10CA1 cell lines derived from premalignant human breast epithelial MCF10AT cells. Breast Cancer Res Treat. 2001;65:101-110. [DOI] [PubMed] [Google Scholar]
- 31. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔC(T)) method. Methods. 2001;25:402-408. [DOI] [PubMed] [Google Scholar]
- 32. Thaben PF, Westermark PO. Detecting rhythms in time series with RAIN. J Biol Rhythms. 2014;29:391-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Percival DB, Walden AT. Wavelet Methods for Time Series Analysis. Cambridge, England: Cambridge University Press; 2000. [Google Scholar]
- 34. Leise TL, Harrington ME. Wavelet-based time series analysis of circadian rhythms. J Biol Rhythms. 2011;26:454-463. [DOI] [PubMed] [Google Scholar]
- 35. Harang R, Bonnet G, Petzold LR. WAVOS: a MATLAB toolkit for wavelet analysis and visualization of oscillatory systems. BMC Res Notes. 2012;5:163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pedregosa F, Varoquaux G, Gramfort A, et al. Scikit-learn: machine learning in python. J Mach Learn Res. 2011;12:2825-2830. [Google Scholar]
- 37. Welsh DK, Imaizumi T, Kay SA. Real-time reporting of circadian-regulated gene expression by luciferase imaging in plants and mammalian cells. Methods Enzymol. 2005;393:269-288. [DOI] [PubMed] [Google Scholar]
- 38. Mei L, Fan Y, Lv X, Welsh DK, Zhan C, Zhang EE. Long-term in vivo recording of circadian rhythms in brains of freely moving mice. Proc Natl Acad Sci U S A. 2018;115:4276-4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hamada T, Sutherland K, Ishikawa M, et al. In vivo imaging of clock gene expression in multiple tissues of freely moving mice. Nature Commun. 2016;7:11705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hilšer M, Trylčová J, Bušek P, Šedo A. Limitations of macroscopic fluorescence imaging for the estimation of tumour growth in an orthotopic glioma mouse model. Folia Biol (Praha). 2012;58:128-133. [PubMed] [Google Scholar]
- 41. Qu Y, Han B, Yu Y, et al. Evaluation of MCF10A as a reliable model for normal human mammary epithelial cells. PLoS One. 2015;10:e0131285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fu L, Pelicano H, Liu J, Huang P, Lee CC. The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell. 2002;111:41-50. [DOI] [PubMed] [Google Scholar]
- 43. Hwang-Verslues WW, Chang PH, Jeng YM, et al. Loss of corepressor PER2 under hypoxia up-regulates OCT1-mediated EMT gene expression and enhances tumor malignancy. Proc Natl Acad Sci U S A. 2013;110:12331-12336. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, Lin_et_al_Supporting_Information_Revision_1 for Analyses of BMAL1 and PER2 Oscillations in a Model of Breast Cancer Progression Reveal Changes With Malignancy by Hui-Hsien Lin, Maan Qraitem, Yue Lian, Stephanie R. Taylor and Michelle E. Farkas in Integrative Cancer Therapies