Abstract
The entomopathogenic fungus Beauveria bassiana can adopt an endophytic lifestyle by colonising a wide array of plant species. Beauveria-colonised plants can show enhanced resistance against insects and plant pathogens alike. However, little is known about the molecular and physiological mechanisms that govern such interactions. Here, we assessed the effects of two B. bassiana strains (BG11, FRh2) on the growth of Arabidopsis thaliana and its resistance against two herbivore species and a phytopathogen. Plant responses were studied on the transcriptomic and metabolic level using microarrays and by measuring changes in defence-related phytohormones and glucosinolates (GLSs). Root inoculation with B. bassiana BG11 significantly increased plant growth, while FRh2 had no such effect. Both Beauveria strains decreased leaf lesion area caused by the phytopathogen Sclerotinia sclerotiorum but did not affect population growth of the aphid Myzus persicae or the growth of Plutella xylostella caterpillars. Microarray analyses of leaves from endophyte-inoculated A. thaliana provided evidence for transcriptional reprogramming of plant defence pathways, with strain-specific changes in the expression of genes related to pathogenesis, phytoalexin, jasmonic (JA), and salicylic acid (SA) signalling pathways. However, B. bassiana colonisation did not result in higher concentrations of JA and SA or major changes in leaf GLS profiles. We conclude that the endophyte B. bassiana induces plant defence responses and hypothesise that these contribute to enhanced resistance against S. sclerotiorum.
Keywords: endophytes, glucosinolates, induced resistance, phytohormones, plant–microbe interaction, Plutella xylostella, Myzus persicae, Sclerotinia sclerotiorum
Introduction
Entomopathogenic fungi are primarily known for their ability to penetrate the cuticle of insects and to cause disease. Once inside the host, the fungus proliferates and eventually grows out again through the cuticle to produce spores, which often persist in the soil until a new host can be infected (Hajek, 2004). Due to their insect-killing capabilities and the fact that some species are easily mass-produced, entomopathogenic fungi have been widely used as biopesticides (Glare et al., 2012). However, more recently an increasing number of studies have shown that entomopathogenic fungi can also adopt a lifestyle as endophytes by colonising plant tissues without causing disease symptoms (Schulz and Boyle, 2005; Roy et al., 2006; Card et al., 2016). This change in paradigm has opened new avenues of research into the ecology of insect-killing fungi and has raised interest in the question whether endophytic entomopathogens could be useful tools for sustainable crop protection (Lacey et al., 2015; Lugtenberg et al., 2016). Several studies have shown that symbioses between entomopathogens and plants can have positive impacts on plant growth and resistance (Vega et al., 2009; Vidal and Jaber, 2015; Jaber and Ownley, 2018; Vega, 2018).
The fungus Beauveria bassiana (Balsamo-Crivelli) Vuillemin was the first entomopathogen to be reported as a naturally occurring endophyte in maize (Bing and Lewis, 1991) and has since been isolated from several other plant species including coffee (Vega et al., 2010), white pine (Ganley and Newcombe, 2006) and common bean (Ramos et al., 2017). The fungus has also been successfully established as an endophyte in many crop species, with seed or root inoculation leading to systemic colonisation of aboveground plant parts (Vidal and Jaber, 2015; McKinnon et al., 2017). The potential benefits of endophytic B. bassiana on plant health such as improving plant growth and suppressing insect pests and plant pathogens have been demonstrated in several studies (McKinnon et al., 2017; Jaber and Ownley, 2018).
Despite accumulating reports on entomopathogens helping plants to fend off antagonists, very few authors have addressed the mechanisms underlying this phenomenon. Non-mutually exclusive hypotheses for explaining B. bassiana-mediated resistance against attacking organisms include the presence of fungal toxins and mycoparasitism (Vidal and Jaber, 2015; Sword et al., 2017; Jaber and Ownley, 2018). However, such mechanisms have either not been demonstrated or only for endophytic fungi other than B. bassiana. For example, toxic destruxins produced by the entomopathogen Metarhizium robertsii were detected in cowpea plants after root inoculation but whether concentrations would be high enough to affect herbivores remains unclear (Golo et al., 2014). Mycosis through direct infection of herbivores seems another plausible mechanism but has rarely been observed in plants containing endophytic entomopathogens (Vidal and Jaber, 2015). Finally, it has been suggested that fungus-induced plant responses could contribute to resistance against insects and pathogens (Ownley et al., 2008; Jaber and Ownley, 2018). Indeed, limited support for this notion is provided by a proteomic analysis of leaves of Phoenix dactylifera (Arecaceae), demonstrating that B. bassiana colonisation can lead to the induction of proteins related to stress and plant defences (Gomez-Vidal et al., 2009). So far, most of our knowledge on plant resistance mediated by beneficial endophytic fungi is derived from studies involving the root-colonising genus Trichoderma, which can parasitize other fungi but does not infect insects (Steyaert et al., 2003; Mukherjee et al., 2013; Pieterse et al., 2014; Vos et al., 2015).
Here, we assessed the effects of endophytic B. bassiana in promoting plant growth and resistance against three economically important pests and pathogens of Brassicaceae: the fungus Sclerotinia sclerotiorum (Ascomycota: Sclerotiniaceae) and the two herbivore species, Myzus persicae (Homoptera: Aphididae) and Plutella xylostella (Lepidoptera: Plutellidae). We conducted transcriptomic analyses of plant responses to two B. bassiana strains in the model plant Arabidopsis thaliana (Brassicaceae). This was complemented with measurements of defence hormones and glucosinolates (GLSs), the main secondary metabolites characteristic for members of the order Brassicales.
Materials and Methods
Plants and Entomopathogens
Surface sterilised and stratified A. thaliana seeds (ecotype Colombia Col-0) were grown on Murashige and Skoog basal medium (M5519 – Sigma-Aldrich, Castle Hill, NSW, Australia) in a growth chamber at 20–22°C, 60–70% RH under a 12L:12D day/night cycle. Two B. bassiana strains, FRh2 and BG11, were used in this study. B. bassiana strain BG11 was recovered from Bellis perennis (Asteraceae) (Bio-Protection Research Centre, Lincoln University), whereas B. bassiana strain FRh2 was recovered from Hylastes ater (Coleoptera: Curculionidae) (Reay et al., 2010).
Endophyte Colonisation
For growth and plant resistance experiments, roots of 1-week-old A. thaliana seedlings, growing in a gnotobiotic environment, were dipped in a 10 ml of B. bassiana conidia suspension at a final concentration of 1 × 108 conidia/ml for 1 min. Control plants were mock-inoculated with 0.05% Tween 80 in sterile water. Inoculated and control seedlings were grown in gamma-irradiated potting mix for 4 weeks in separate propagation boxes (Mini propagator, Hortlink New Zealand Ltd.).
For microarray, RT-qPCR, phytohormone and GLS analyses, roots of 5-week-old A. thaliana plants grown in a gnotobiotic environment were dipped in B. bassiana conidia suspension as described above. Inoculated and control plants were transplanted into twice autoclaved vermiculite and kept in separate autoclaved 4.2 l cereal containers for another 15 days.
All experiments consisted of three treatments: FRh2 inoculation (F), BG11 inoculation (B) and mock-inoculation (C) and were conducted using a completely randomised design in a growth chamber at 20–22°C, 60–70% RH under a 12L:12D day/night cycle expect for plant growth experiment where a cycle of 16L:8D day/night was used.
Colonisation of A. thaliana by each B. bassiana strain was assessed using standard isolation techniques on culture medium (Reay et al., 2010). Inoculated and mock-inoculated plants were divided into two parts, rosette and inflorescence. Plant parts were surface-sterilised for 1 min in 70% ethanol, 2 min in 4.2% NaOCl and then rinsed 3× 3 min in 0.01% Triton X-100 in sterile water. The sterilisation method was tested on aliquots of fungal conidia to confirm the effectiveness of the selected exposure time and NaOCl concentration on the viability of conidia. After sterilisation, imprints of each leaf and inflorescence were made on Beauveria-selective medium (BSM) to verify that plant surfaces were not contaminated (Brownbridge et al., 2012). Subsequently, the same leaf tissue and inflorescence were cut into segments of 0.5 to 1 cm, plated on BSM, and cultivated for up to 3 weeks at 20°C in a dark incubator. Emerging mycelia were isolated and identified on the basis of colony and conidia morphology. In addition, genomic DNA was extracted from A. thaliana rosettes and B. bassiana presence was further verified by PCR using SCAR primers developed by Castrillo et al. (2003). Three strain-specific molecular markers were tested to facilitate the detection of B. bassiana in field samples: SCA14445 (F 5′ TCTGTGCTGGCCCTTATCG 3′, R 5′ TCTGTGCTGGGTACTGACGTG 3′), SCA15441 (F 5′ TTCCGAACCCGGTTAAGAGAC 3′, R 5′ TTCCGAACCCATCATCCTGC 3′) and SCB9677 (F 5′ TGGGGGACTCGC AAA CAG 3′, R 5′ TGGGGGACTCAC TCC ACG 3′). Similar to Biswas et al. (2012), we found that SCA15441 amplified B. bassiana DNA best and was therefore used throughout this study. We tested 38 FRh2-inoculated, 34 BG11-inoculated and 51 control plants grown in gamma-irradiated soil for B. bassiana presence that had been used earlier in the bioassays. Additionally, plants grown in vermiculite under gnotobiotic conditions used for molecular (n = 4) and biochemical experiments (n = 6) were also tested for endophyte colonisation.
Plant Growth
Plants were grown in gamma-irradiated soil and inoculated as described above. The following phenotypic traits were measured 4 weeks after inoculation: rosette diameter, number of leaves, shoot and root biomass (fresh weight) and root/shoot ratio. We also measured time until onset of the reproductive phase (appearance of inflorescence) and inflorescence length after 12 weeks. For each treatment n = 20 replicates were carried out.
Testing of Plant Resistance Against Leaf-Chewing and Phloem-Sucking Herbivores
A colony of the diamondback moth Plutella xylostella was reared on potted cabbage plants (Brassica oleracea var. capitata) in a climate chamber at constant 22°C under a 16L: 8D day/night cycle. A single third-instar caterpillar was transferred to a 5-week-old A. thaliana plant inoculated with B. bassiana or a mock-inoculated control with 24–26 plants per treatment. Separate experiments were carried out for each endophyte strain with its corresponding control. Caterpillar body mass was measured before the experiment and after a feeding period of 48, 72, and 96 h.
Myzus persicae was maintained under the same conditions as described for P. xylostella. Five nymphs of M. persicae, characterised by their wide cauda, were carefully transferred to a 5-week-old A. thaliana plant inoculated with B. bassiana or a mock-inoculated control with 12–14 plants per treatment. Nymphs were caged onto a leaf by using a clip cage of 44 mm diameter. The cage was removed 24 h post infestation and only a single nymph, which had inserted its stylet, was left on the leaf to become reproductive. After 5 days the number of next generation nymphs was recorded daily for 10 days.
Testing of Plant Resistance Against a Leaf Pathogen
Five-week-old A. thaliana plants inoculated with one of the B. bassiana strains and mock-inoculated controls were infected with Sclerotinia sclerotiorum using an agar plug method. The S. sclerotiorum strain SsOSR (Bio-Protection Research Centre, Lincoln University) was cultured on potato dextrose agar in a Petri dish of 9 cm diameter for 3 weeks at 20°C under a 12L:12D day/night cycle. A cork borer was used to punch out a 3 mm agar disc with fungal hyphae from a Petri dish. Discs were punched out 1 cm from the edge of the dish. The agar disc was placed on a single leaf surface. All selected leaves for infection were standardised for a length of ca. 7 cm. The agar discs were covered with plastic foil to maintain high relative humidity and plants were incubated in propagation boxes (Mini propagator, Hortlink New Zealand Ltd.). The experiment was carried out with 19–22 plants per treatment. Lesion areas were measured 5 days post infection according to the method of Rostás et al. (2006). The lesions were scanned and the area of each lesion was calculated using the self-written software Surface (available upon request from authors).
Microarrays
Total RNA was extracted from inoculated and control A. thaliana rosettes of four independent biological replicates per treatment using the RNeasy Plant Mini Kit (QIAGEN). Each biological replicate consisted of a pool of eight plants. One-column DNase digestion treatment using the RNase-Free DNase Set (QIAGEN) was incorporated in the extraction protocol to eliminate any DNA contaminations in downstream experiments. RNA samples were sent to OakLabs (Hennigsdorf, Germany) for analysis using Agilent 8 × 60 K microarrays (Agilent Technologies, Santa Clara, CA, United States) with 32072 target IDs representing 30541 gene loci where annotation is based on A. thaliana Genome, TAIR10. RNA quality was confirmed by electrophoretic analysis via the 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, United States) for determination of the RNA integrity number. The cRNA syntheses and microarray hybridisations were performed using the Low Input QuickAmp Labeling Kit and the Agilent Gene Expression Hybridisation Kit (Agilent Technologies). Microarray data were normalised according to the ranked median quantiles (Bolstad et al., 2003) using DirectArray software (OakLabs). Hierarchical clustering analysis (HCA) and principal component analysis (PCA) were used to test whether samples of the same treatment were homogenous and thus clustered together. DEGs were identified using two-sample t-tests with unequal variance (Welch’s t-test), with a p-value of 5% using DirectArray software. Data were further subjected to false discovery rate correction with a threshold of 5% (Benjamini and Hochberg, 1995). GO and enrichment analysis for each group of up- and downregulated DEGs were performed (Ashburner et al., 2000; Mi et al., 2017; The Gene Ontology Consortium, 2017). Gene expression data were visualised in the context of metabolic pathways using MapMan (Thimm et al., 2004; Usadel et al., 2009).
Quantitative Real Time PCR
For microarray validation, the expression of seven genes was evaluated using RT-qPCR (Primer sequences: Supplementary Table S6). The selected genes were related to different defence responses and had shown differential regulation in the FRh2-inoculated plants transcriptome data. AXR5 (Hardtke et al., 2007), ASC4 (Abel et al., 1995), MYB122 (Frerigmann and Gigolashvili, 2014) and ARR11 (Kieber and Schaller, 2014) are involved in auxin, ET, camalexin and cytokinin pathways, respectively. GLIP1 and chitinase gene are known to be involved in resistance against bacteria and fungi (Oh et al., 2005; Lee et al., 2009; Hermosa et al., 2012). WRKY63 is involved in abiotic stress resistance and mediates plant responses to drought tolerance (Ren et al., 2010; Bakshi and Oelmuller, 2014).
Total RNA was extracted from FRh2-inoculated and mock-inoculated Arabidopsis rosette of three additional independent biological replicates using the RNeasy Plant Mini Kit and RNase-Free DNase Set (QIAGEN) as described above. RNA quality check was performed by electrophoresis and photometrical measurement with the Nanodrop 2000 spectrophotometer (Thermo Scientific). A total of 2 μg of DNase-treated RNA was then reverse-transcribed into the first-strand cDNA using SuperScript® III First-Strand Synthesis System (Invitrogen). cDNA synthesis was performed according to the manufacturer’s instructions, using Oligo dB 12-18 primer and including an RNase H digestion as a last step to remove RNA template from the cDNA:RNA hybrid molecule. qPCR was performed in triplicate from three biological replicates with a reaction mixture containing gene specific primers, cDNA template with a dilution value of 1:10, SYBR Green reagent, ROX Reference Dye to normalise the fluorescent reporter signal and the FastStartTM Taq DNA Polymerase, dNTPack (Roche). The thermal cycling conditions were 95°C for 10 min followed by 95°C for 15′′, 60°C for 45′′ and 72°C for 45′′ for 40 cycles, followed by melting curve step at 95°C for 15′′, 60°C for 1′ and 95°C for 15′′ to validate amplicon specificity. Non-template controls were included in each qPCR plate indicating the purity of the reagents. Primers were designed to amplify short cDNA fragments using Primer-BLAST1. The primers were designed to span an exon/exon junction with a product size between 70 and 100 bp using the RefSeq accession database. Three reference genes that were stable and not identified as differentially expressed in the FRh2-inoculated plants microarray data, actin 2 (Act-2); glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and elongation factor (EF1α) genes were used as reference genes to normalise the qPCR data. The relative expression levels were analysed using the 2-ΔΔCT method (Livak and Schmittgen, 2001) and are presented as log2 relative levels of gene expression.
Phytohormone Analysis
Salicylic acid and jasmonic acid levels were quantified in B. bassiana inoculated and mock-inoculated A. thaliana leaves by vapour-phase extraction and subsequent gas chromatography-mass spectrometry (GC-MS) analysis according to Schmelz et al. (2004). Plants were grown and inoculated as described above in section Endophyte colonisation. The experiment was carried out with six independent replicates (plants) per treatment. Each replicate consisted of 150 mg of frozen homogenised leaf tissue. The plant material was extracted in micro-reaction tubes filled with zirconium beads (FastRNA® Pro Green Kit), acidified 1-propanol in water and methylene chloride, using a FastPrep®-24 System (MP Biomedicals, Solon, OH, United States). For quantification, D6-salicylic acid (CDN Isotopes, Pointe-Claire, QC, Canada) and dihydrojasmonic acid (TCI America, United States) were used as internal standards. Trimethylsilyldiazomethane (Sigma-Aldrich, Castle Hill, NSW, Australia) was added to derivatise the two phytohormones into their corresponding methyl esters. Samples were subjected to a vapour-phase extraction procedure consisting of two evaporation steps at 70°C and 200°C using a volatile collection trap packed with 30 mg Super-Q absorbent (Analytical Research Systems, Micanopy, FL, United States). The absorbed methylated compounds were then eluted from the collection trap with methylene chloride and stored at -80°C for subsequent GC-MS analysis according to Maag et al. (2014).
Glucosinolate Analysis
Glucosinolate concentrations were measured in inoculated and mock-inoculated A. thaliana leaves as described above. The experiment was carried out with six independent replicates (plants) per treatment. Freeze dried leaves samples of each replicate were pulverised in 2 ml microcentrifuge tubes containing 2.5 mm zirconium/silica beads (dnature, New Zealand) using a bead mill (TissueLyser II, Qiagene, Hilden, Germany) for 1 min. Leaf samples (10 mg) were then extracted three times in 80% methanol. At the first extraction, 2-propenyl glucosinolate (Phytoplan, Heidelberg, Germany) was added as internal standard. Purified sulfatase (E.C. 3.1.6.1; purification following; Graser et al., 2001) was used to convert glucosinolates to desulfoglucosinolates which were then analysed by high performance liquid chromatography (HPLC) coupled to a diode array detector (HPLC-1200 Series, Agilent Technologies, Inc., Santa Clara, CA, United States) as described by Abdalsamee and Müller (2012). Desulfoglucosinolate separation was performed on a reverse phase Supelcosil LC 18 column (3 μm, 150 × 3 mm, Supelco, Bellefonte, PA, United States) using a gradient of water to methanol, starting at 5% methanol for 6 min, and increasing from 5 to 95% methanol within 13 min with a hold at 95% for 2 min. Metabolites were identified by comparison of retention times and UV spectra to purified standards (Phytoplan, Heidelberg, Germany; Copenhagen, Denmark) or by confirming the identities by ultra-HPLC coupled with a time of flight mass spectrometer (1290 Infinity UHPLC and 6210 TOF-MS Agilent, Technologies, Santa Clara, CA, United States). Desulfoglucosinolates were quantified using the integrated area at 229 nm, applying the response factors as described previously (Brown et al., 2003), and relating the amounts to the sample dry mass.
Statistical Analyses
Differences in presence/absence between B. bassiana strains in A. thaliana tissues was analysed by a Chi-square test. Plant phenotypic traits, concentrations of JA and several GLSs were analysed using one-way ANOVA followed by Fisher’s least significant difference (LSD) post hoc tests. Data not meeting assumptions of normality and homogeneity of variance, such as number of leaves, root/shoot ratio, SA concentration, 4-hydroxyindol-3-ylmethyl GLS and 5-methylsulfinylpentyl GLS levels were analysed using the non-parametric Kruskal–Wallis ANOVA followed by a Student–Newman–Keuls test. A PCA was performed to visualise the effects of B. bassiana strains on glucosinolate concentrations. The biomass of P. xylostella caterpillars and population growth of M. persicae aphids were analysed using Student’s t-tests for independent samples and repeated measures ANOVA, respectively. Differences in mortality between caterpillars reared on endophyte-inoculated and control plants were compared by Chi-square tests. Count data for the population growth of aphids of M. persicae were square root transformed and homogeneity of variance was tested using Cochran’s test. S. sclerotiorum assay was analysed using one-way ANOVA followed by Fisher’s least significant difference (LSD) post hoc tests. S. sclerotiorum lesions area data was log-transformed to meet assumption of normality and homogeneity of variance. Statistical analyses were performed using IBM® SPSS statistics 22 and Statistica 13 software as well as R package version 3.2.02.
Results
Endophytic Colonisation of A. thaliana by the Fungal Entomopathogen B. bassiana
The entomopathogenic fungus was able to endophytically colonise aboveground tissues of the model plant A. thaliana. The root dipping method showed that the fungus can translocate systemically throughout the plant system when roots were dipped in conidia suspension, confirming previous studies (e.g., Muvea et al., 2014; Qayyum et al., 2015).
No statistically significant differences in colonisation rates were found between the strains FRh2 and BG11 (χ2: 2.62, d.f. = 1, P = 0.106). However, the environmental conditions of the inoculated plants had a strong influence on endophyte presence. While 100% of inoculated plants grown in vermiculite and under germ-free conditions were infected with B. bassiana, only 53 ± 6% of plants were tested positive when grown in soil under non-gnotobiotic conditions. No background infection of B. bassiana in A. thaliana seeds and seedlings was found using both molecular detection and the standard isolation techniques on culture medium. The fungus was recovered from rosettes and inflorescences after root inoculation, suggesting systemic colonisation of the entire plant. None of the control plants were colonised by the fungus. Also, tissue imprints and cuts showed no presence of B. bassiana, indicating that the fungus did not colonise the plant surface. Using PCR, we were able to detect B. bassiana in inoculated but not in control plants.
Growth Promotion Depends on Entomopathogen Strain
Endophytic colonisation by B. bassiana showed strain-specific effects on several phenotypic traits of A. thaliana. Rosette diameter remained unaffected by the endophyte (one-way ANOVA, d.f. = 2, F = 2.336, P = 0.106; Figure 1A) but plants infected with BG11 produced significantly more leaves than either controls or FRh2-treated A. thaliana (Kruskal–Wallis ANOVA, H = 7.461, d.f. = 2, P = 0.024; BG11 vs. control: P = 0.037, BG11 vs. FRh2: P < 0.001, FRh2 vs. control: P = 0.761; Figure 1B). Likewise, no statistically significant effect was found for shoot biomass (one-way ANOVA, d.f. = 2, F = 2.519, P = 0.089; Figure 1C), while root biomass was increased in BG11 plants (one-way ANOVA, d.f. = 2, F = 10.727, P < 0.001; BG11 vs. control: P < 0.001, BG11 vs. FRh2: P = 0.001, FRh2 vs. control: P = 0.845; Figure 1D). Plants of all three treatments differed in their root/shoot ratios (Kruskal–Wallis ANOVA, d.f. = 2, H = 27.307, P < 0.001; BG11 vs. control: P = 0.002, BG11 vs. FRh2: P < 0.001, FRh2 vs. control: P < 0.001; Figure 1E). The time until plants entered the reproductive phase was independent of endophyte colonisation (Kruskal–Wallis ANOVA, d.f. = 2, H = 2.293, P = 0.318) and reached on average 29 ± 1.0 days (control), 29 ± 1.5 days (BG11) and 29 ± 0.3 days (FRh2). However, BG11 treated plants produced significantly shorter inflorescences (one-way ANOVA, d.f. = 2, F = 9.198, P < 0.001; BG11 vs. control: P < 0.001, BG11 vs. FRh2: P = 0.007, FRh2 vs. control: P = 0.624; Figure 1F).
Figure 1.
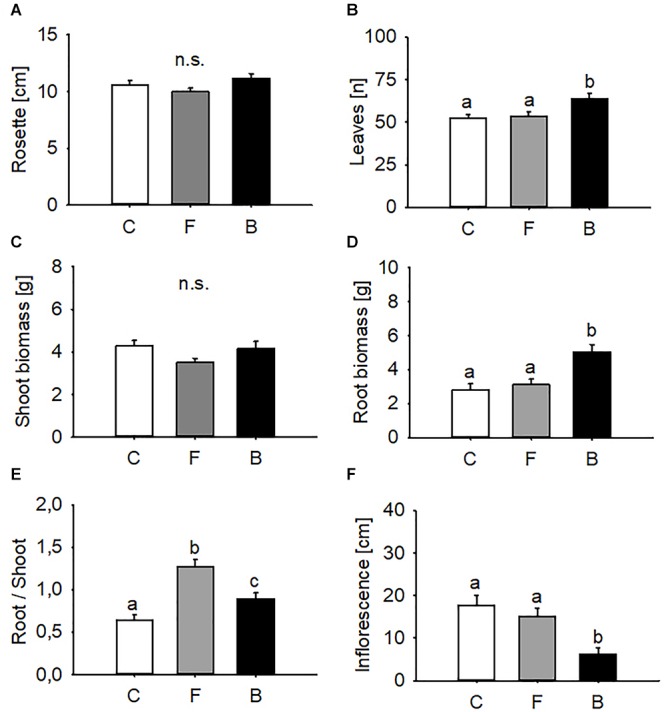
Effects of B. bassiana on A. thaliana growth. Rosette diameter (A), numbers of leaves (B), shoot biomass (C), root biomass (D), root/shoot ratio (E), and inflorescence length (F) in B. bassiana colonised (F, FRh2 and B, BG11) and control (C) plants. Error bars represent the standard error of the mean (N = 20), bars with different letters differ significantly (P < 0.05).
Entomopathogen Colonisation Increases Plant Resistance to Leaf Pathogen but Not to Insect Herbivores
Endophyte colonisation had no effect on the mortality rate of P. xylostella caterpillars, which was 31% and 38% in FRh2- and BG11-treated plants, respectively. Mortality in the corresponding controls was 29% in both experiments (Chi-square tests, FRh2 vs. control: χ2 = 0.02, d.f. = 1, P = 0.902; BG11 vs. control: χ2 = 0.38, d.f. = 1, P = 0.540). No significant differences in P. xylostella caterpillar body mass were observed after 48, 72, and 96 h of feeding on B. bassiana-inoculated plants when compared to insects feeding on control plants (t-test for FRh2 at 0 h; t = 0.39, P = 0.969, n = 24–26; 48 h; t = 0.502, P = 0.618, n = 22–24; 72 h; t = -1.228, P = 0.227, n = 20–19; 96 h; t = -1.959, P = 0.059, n = 17–18; t-test for BG11 at 0 h; t = -0.607, P = 0.547, n = 23–24; 48 h; t = 0.459, P = 0.649, n = 20–22; 72 h; t = 0.377, P = 0.709, n = 20; and 96 h; t = -0.345, P = 0.732, n = 15–17, Figure 2A,B).
Figure 2.
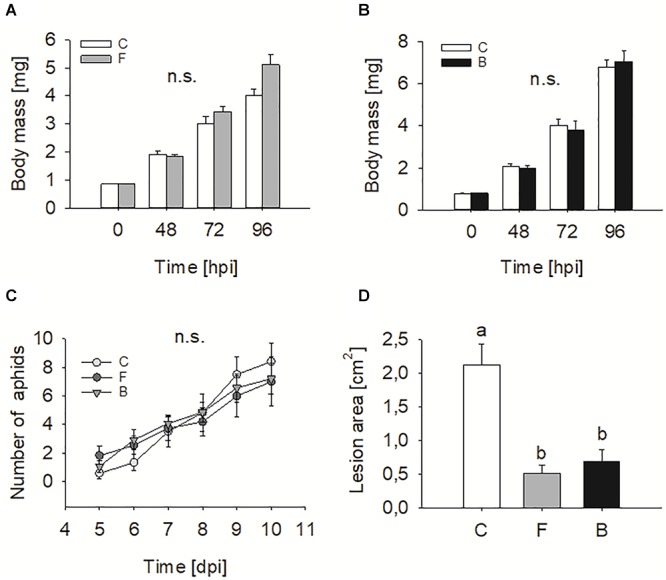
Effects of endophytic B. bassiana on two insect herbivores and a leaf pathogen. P. xylostella body mass (means ± SE) after 48, 72, and 96 h of feeding on B. bassiana FRh2 (A) and BG11 (B) colonised Arabidopsis thaliana (N = 24–26). M. persicae population growth when feeding on control and B. bassiana colonised A. thaliana (C). Symbols represent mean number of M. persicae (N = 12–14). S. sclerotiorum lesion area on A. thaliana leaf measured 5 days post infection (D). Disease intensity was calculated as average lesion area (N = 19–22). Different letters above bars indicate significant differences (P < 0.05). C, control plants; F, FRh2 inoculated plants; B, BG11 inoculated plants. Error bars represent the standard error of the mean. n.s., not significant.
Equally, the population growth of aphids of M. persicae was not significantly affected by B. bassiana presence when fixed on plants within a 10-day period (repeated measures ANOVA, treatment effect: d.f. = 2, F = 0.37, P = 0.692, Figure 2C).
In contrast to insect bioassays, inoculation with B. bassiana resulted in a significant reduction in S. sclerotiorum lesion area on leaves compared to those on mock-inoculated plants (one-way ANOVA, d.f. = 2, F = 22.062, P < 0.001, Figure 2D). The effect was significant for both B. bassiana strains compared to the control plants.
Entomopathogen Colonisation Results in Strain-Specific Remodelling of the A. thaliana Transcriptome
The comparative transcriptome analysis of B. bassiana-inoculated (FRh2, BG11) and mock-inoculated plants revealed 1,166 differentially expressed genes (DEGs) for FRh2-inoculated plants and 552 DEGs for BG11-inoculated plants, indicating a strain-specific effect on the plant’s gene expression. The ratio of upregulated to downregulated A. thaliana transcripts was 58:42% in FRh2-inoculated plants and 52:48% in BG11-inoculated plants (mean of four biological replicates). Fifty-eight DEGs were shared between the FRh2-inoculated plants and BG11-inoculated plants. Thirty-eight of the 58 shared DEGs had a similar expression pattern in the presence of FRh2 or BG11 while the remaining showed diverging expression patterns (Figure 3 and Supplementary Table S5).
Figure 3.

Venn diagram comparing the expression patterns of the shared differentially expressed genes (DEGs) between FRh2- and BG11-inoculated plants.
Similarities between FRh2- and BG11-inoculated plants were observed in the upregulation of defence-related processes. Only BG11-inoculated plants showed responses associated to wounding (Table 1), however, both fungal strains induced genes involved in oxidative stress processes. FRh2-inoculated plants furthermore showed transcriptional changes in many additional abiotic stress-related processes that included responses to heat and temperature, while BG11-inoculated plants showed downregulation of genes related to radiation and light processes (Table 1 and Supplementary Tables S1–S4). An apparent difference in DEGs between FRh2- and BG11-inoculated plants was observed in the biological processes relating to plant hormones: DEGs associated with responses to JA, ethylene (ET) and SA were upregulated in BG11-inoculated plants, whereas DEGs associated with gibberellin and auxin stimulus were downregulated in FRh2-inoculated plants compared to control plants (Table 1 and Supplementary Tables S1–S4).
Table 1.
Gene ontology terms and enrichment analysis for FRh2- and BG11-inoculated plants.
 |
Red and blue boxes show up- and downregulated biological process enriched at p-value < 0.05.
Gene ontology (GO) and enrichment analysis were complemented by mapping the transcriptomic data with MapMan. An overview of the plant’s metabolic pathways that were affected by B. bassiana colonisation was generated. MapMan mapped 1,163 and 548 DEGs into the 35 major bins for FRh2- and BG11-inoculated tissues, respectively. Bins 7, 12, 14 (oxidative pentose phosphate, N-metabolism, S-assimilation, polyamine metabolism) and bins 8, 9, 18 (tricarboxylic acid cycle/organic acid transformation, mitochondrial electron transport/ATP synthesis, co-factor and vitamin metabolism) were the only non-represented bins for FRh2- and BG11-inoculated plants, respectively (Figure 4). A total of 367 DEGs (142 upregulated and 225 downregulated) for FRh2-inoculated plants and 159 DEGs (61 upregulated and 98 downregulated) for BG11-inoculated plants were unassigned (bin 35) DEGs (Figure 4).
Figure 4.

Numbers of differentially expressed genes (DEGs) assigned into MapMan bins (bin number in brackets) from FRh2- and BG11-inoculated plants. Downregulated DEGs are negative. Unassigned DEGs were 142, –225, and 61, –98 for FRh2- and BG11-inoculated plants, respectively. OPP, oxidative pentose phosphate; TCA/org transformation, tricarboxylic acid cycle/organic acid transformation.
The biotic stress overview pathway generated by MapMan highlighted the involvement of both FRh2- and BG11-inoculated plants in defence-related responses and gave a visual assessment on how B. bassiana reprograms major parts of the plant immune response (Figure 5, 6). There were changes to expressions of stress-related transcripts associated to biotic and abiotic stress. MapMan identified 374 and 175 DEGs associated with the biotic stress overview pathway for FRh2- and BG11-inoculated plants, respectively. Among these, only 38 DEGs for FRh2-inoculated plants and 13 DEGs for BG11-inoculated plants were related to abiotic stress (bin 20.2) whereas the remaining were related directly or indirectly to biotic stress. The majority of the 38 FRh2-inoculated plants transcripts related to abiotic stress were identified as heat shock protein coding genes. A subset of 23 DEGs for FRh2-inoculated plants and 13 DEGs for BG11-inoculated plants were related directly to biotic stress (bin 20.1) and mapped as pathogenesis-related proteins coding genes (bin 20.1.7). The remaining of FRh2- and BG11-inoculated plants DEGs were indirectly related to biotic stress responses and mapped as signalling (bin 30), transcription factors (bin 27), oxidative stress (bin 21 and bin 26), hormones (bin 17), secondary metabolism-related genes (bin 16) and cell-wall modification genes (bin 10). For both FRh2- and BG11-inoculated plants DEGs, regulation of transcription showed diverging expression patterns in DEGs identified as WRKY factors, MYB domain and ET response factors AP2/ERF coding genes. Transcripts of FRh2- and BG11-inoculated plants associated with oxidative stress were identified as glutaredoxins, peroxidases and glutathione S transferases (GST) coding genes. DEGs associated with the secondary metabolism were related to camalexin and flavonoid metabolism and those associated with hormones metabolism were identified as auxin-responsive protein, ET signal transduction coding genes and JA and SA metabolism-associated genes.
Figure 5.
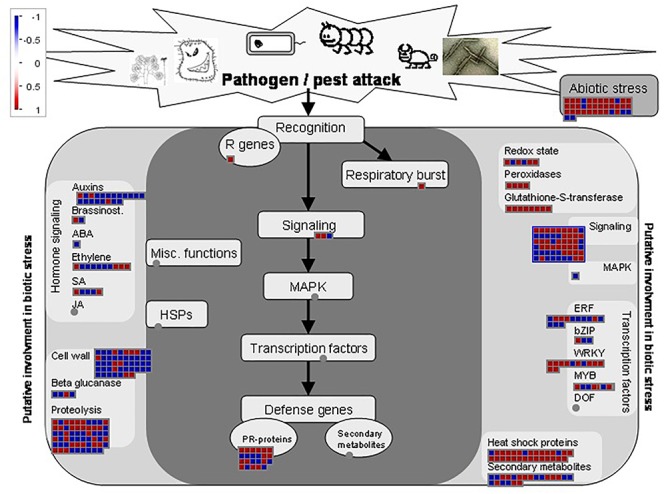
Biotic stress overview pathway for FRh2-inoculated plants DEGs as generated by MapMan software. Red and blue boxes represent up and downregulated genes, respectively. Colour intensity represents the degree of expression given as log2 fold change >1 and <–1.
Figure 6.

Biotic stress overview pathway for BG11-inoculated plants DEGs as generated by MapMan software. Red and blue boxes represent up and downregulated genes, respectively. Colour intensity represents the degree of expression given as log2 fold change >1 and <–1.
The expression levels of the selected genes followed the same pattern of expression as in the FRh2-inoculated plants microarray. Averaging the expression level of each gene over the three biological replicates and over the three reference genes showed that AXR5, ACS4, and ARR1 expression were downregulated, whereas GLIP1, chitinase gene, MYB122 and WRKY63 expression were upregulated (Supplementary Figure S1).
Entomopathogen Colonisation Affects Secondary Metabolite Profiles but Not Defence Hormones
Colonisation with B. bassiana did not result in increased concentrations of JA and SA in A. thaliana leaves 15 days after inoculation. For the three treatments, no significant differences (P > 0.05) were found in SA and JA concentrations (one-way ANOVA for JA d.f. = 2, F = 0.908, P = 0.426, Figure 7; Kruskal–Wallis Test for SA, d.f. = 2, χ2 = 0.1.333, P = 0.513, Figure 7).
Figure 7.
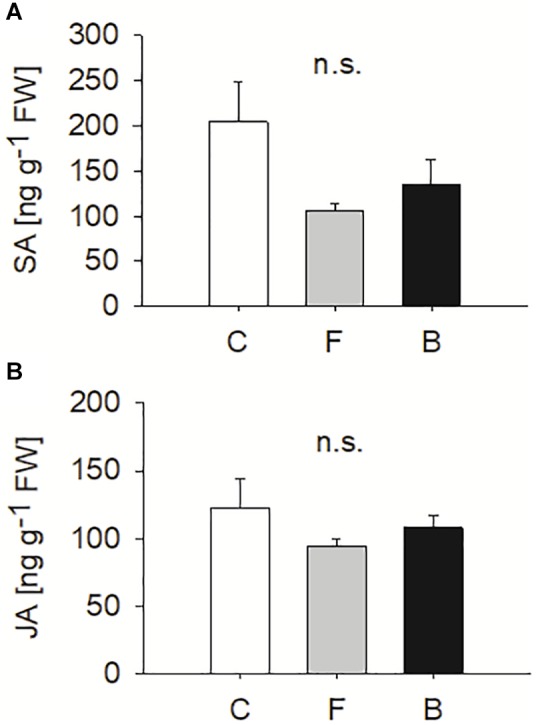
Levels of salicylic acid (A) and jasmonic acid (B) (means ± SE) measured in B. bassiana colonised (F, FRh2 and B, BG11) and control (C) A. thaliana. Error bars represent the standard error of the mean (N = 6), bars with different letters differ significantly (P < 0.05). n.s., not significant.
Slight changes in leaf GLS concentrations were observed due to entomopathogen colonisation after 15 days of inoculation. No significant differences (P ≤ 0.05) in total indole glucosinolate (IGS) levels (sum of four individual indole GLS: 4MOI3M, 4-methoxyindol-3-ylmethyl GLS; 4OHI3M, 4-hydroxyindol-3-ylmethyl GLS; I3M, indol-3-ylmethyl GLS; 1MOI3M, 1-methoxyindol-3-ylmethyl GLS) were observed between B. bassiana-treated and control plants. However, total aliphatic glucosinolate (AGS) levels (sum of seven individual aliphatic GLS: 3MSOP, 3-methylsulfinylpropyl GLS; 4MSOB, 4-methylsulfinylbutyl GLS; 5MSOP, 5-methylsulfinylpentyl GLS; 6MSOH, 6-methylsulfinyl-heptyl GLS; 7MSOH, 7-methylsulfinylheptyl GLS; 4MTB, 4-methylthiobutyl GLS; 8MSOO, 8-methylsulfinyl-octyl GLS) were affected by the fungus (one-way ANOVA, IGS: d.f. = 2, F = 2.886, P = 0.087; AGS: d.f. = 2, F = 7.958, P = 0.004, Figure 8). FRh2-colonised plants showed lower levels of AGS compared to controls with a significantly low level of 4MSOB (one-way ANOVA, d.f. = 2, F = 6.676, P = 0.008, Table 2). Despite the absence of a significant change in the total amount of AGS when compared to controls, BG11-colonised plants showed a significant increase in the level of 7MSOH, 4MTB, 8MSOO) when compared to FRh2-colonised plants (one-way ANOVA, d.f. = 2, 7MSOH: F = 14.146, P < 0.001; 4MTB: F = 7.843, P = 0.005, 8MSOO: F = 8.619, P = 0.003, Table 2). PCA resulted in distinct clusters of FRh2- and BG11-treated plants, which overlapped with the cluster of mock-inoculated controls (Figure 9).
Figure 8.
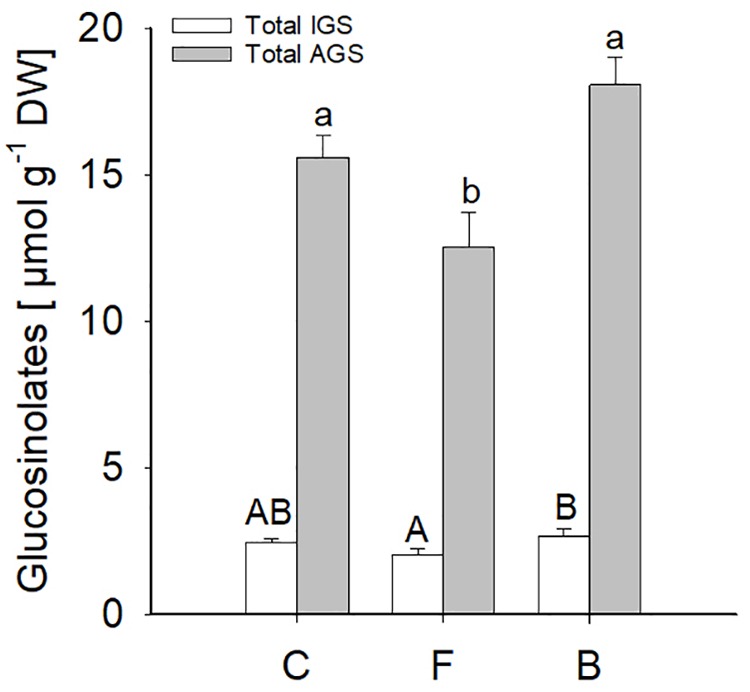
Indole (white bars) and aliphatic (grey bars) glucosinolate levels (means ± SE) measured in B. bassiana colonised (F, FRh2 and B, BG11) and control (C) A. thaliana leaves. Error bars represent the standard error of the mean (N = 6), bars with different letters differ significantly, (P < 0.05).
Table 2.
Mean (±SE) concentration of glucosinolates (GLS) (μmol g-1 dry weight) in Arabidopsis thaliana leaves of Beauveria bassiana-colonised plants (F, FRh2; B, BG11; C, control).
| Glucosinolatesa | C | F | B |
|---|---|---|---|
| Total AG | 15.60 ± 0.75 | 12.54 ± 1.18* | 18.07 ± 0.95 |
| Total IG | 2.44 ± 0.11 | 2.01 ± 0.21 | 2.67 ± 0.23 |
| 3MSOP | 1.27 ± 0.05 | 0.99 ± 0.12 | 1.47 ± 0.12 |
| 4MSOB | 12.32 ± 0.56 | 9.91 ± 0.91* | 13.79 ± 0.76 |
| 5MSOP | 1.07 ± 0.03 | 0.90 ± 0.10 | 1.17 ± 0.06 |
| 6MSOH | 0.08 ± 0.02 | 0.06 ± 0.01 | 0.09 ± 0.01 |
| 7MSOH | 0.19 ± 0.03 | 0.13 ± 0.02 | 0.30 ± 0.02* |
| 4MTB | 0.15 ± 0.04 | 0.11 ± 0.02 | 0.38 ± 0.08* |
| 8MSOO | 0.52 ± 0.09 | 0.43 ± 0.08 | 0.88 ± 0.07* |
| 4MOI3M | 0.47 ± 0.04 | 0.48 ± 0.06 | 0.62 ± 0.08 |
| 1MOI3M | 0.29 ± 0.08 | 0.15 ± 0.03 | 0.31 ± 0.09 |
| 4OHI3M | 0.02 ± 0.01 | 0.04 ± 0.01 | 0.00 ± 0.00 |
| I3M | 1.66 ± 0.10 | 1.34 ± 0.20 | 1.74 ± 0.021 |
Asterisk indicates statistically significant difference compared to control plant. One-way ANOVA followed by Fisher’s least significant difference (P ≤ 0.05). aAG, aliphatic glucosinolates; IG, indole glucosinolates; 3MSOP, 3-methylsulfinylpropyl GLS; 4MSOB, 4-methylsulfinylbutyl GLS; 5MSOP, 5-methylsulfinylpentyl GLS; 6MSOH, 6-methylsulfinyl-heptyl GLS; 7MSOH, 7-methylsulfinylheptyl GLS; 4MTB, 4-methylthiobutyl-GLS; 8MSOO, 8-methylsulfinyl-octyl GLS; 4MOI3M, 4-methoxyindol-3-ylmethyl GLS; 1MOI3M, 1-methoxyindol-3-ylmethyl GLS; 4OHI3M, 4-hydroxyindol-3-ylmethyl GLS; I3M, indol-3-ylmethyl GLS.
Figure 9.

Bi-plot of principal component analysis (PCA) of glucosinolates in A. thaliana leaves. Plants were mock-inoculated (control) or colonised by B. bassiana strains FRh2 and BG11, respectively. Glucosinolates (GLS): 3MSOP, 3-methylsulfinylpropyl GLS; 4MSOB, 4-methylsulfinylbutyl GLS; 5MSOP, 5-methylsulfinylpentyl GLS; 6MSOH, 6-methylsulfinyl-heptyl GLS; 7MSOH, 7-methylsulfinylheptyl GLS; 4MTB, 4-methylthiobutyl GLS; 8MSOO, 8-methylsulfinyl-octyl GLS; 4MOI3M, 4-methoxyindol-3-ylmethyl GLS; 1MOI3M, 1-methoxyindol-3-ylmethyl GLS; 4OHI3M, 4-hydroxyindol-3-ylmethyl GLS; I3M, indol-3-ylmethyl GLS. PC1 = 49%, PC2 = 17%.
Discussion
In this study, we assessed the ability of B. bassiana to promote plant growth and we tested whether systemic colonisation of A. thaliana by the insect-pathogenic fungus B. bassiana induces resistance against insect and fungal plant antagonists. Furthermore, we investigated whether the resistance can be explained by correlational changes in gene expression, signalling molecules and secondary metabolites related to plant defences.
Our analysis of different plant parameters showed that growth promotion is strain-dependent as only BG11 but not FRh2 colonisation resulted in an increased number of leaves, shorter inflorescences and higher root biomass. Growth promotion by entomopathogens also varied in other studies and in some systems depended on the species or strain used as well as on soil nutrient availability (Jaber and Enkerli, 2017; Tall and Meyling, 2018). Gomez-Vidal et al. (2009) showed that B. bassiana can induce proteins related to photosynthesis and energy metabolism which could enhance plant growth. However, in the present study subsequent transcriptomic analyses did not indicate enhanced gene expression related to primary metabolism or photosynthesis and further research will be necessary to elucidate the underlying mechanisms of plant growth promotion by BG11.
Regarding the question whether the endophyte confers resistance against different plant antagonists, we observed that inoculation of A. thaliana roots with FRh2 and BG11, respectively, did not result in any negative effects on insect herbivores. Neither B. bassiana strain affected P. xylostella development or M. persicae population growth in this study and suggests that herbivore resistance is species-specific. This lack of effect could have been due to the low presence of B. bassiana after 4 weeks of endophytic growth, which was around 50% in the tested plant population. It should be noted that mortality of P. xylostella larvae in the controls was relatively high (29%) due to first instar losses. High baseline mortality could have been due to switching from B. oleracea to A. thaliana as a host plant at the start of the experiment. It is also conceivable that handling the small delicate insects with a soft paintbrush, despite taking great care, might have had some adverse effects. Baseline mortality of P. xylostella seems to vary considerably between 10 and 43% (Haas et al., 2018; Sun et al., 2018). Nevertheless, both endophyte strains caused a significant decrease in the area of necrotic leaf lesions by S. sclerotiorum. We suggest that, to some extent, this may be due to the induction of local and/or systemic plant resistance. The genome-wide expression analysis of A. thaliana provided evidence for the transcriptional reprogramming of plant defence pathways following B. bassiana colonisation, which may explain the observed effects against the pathogen. Interestingly, gene responses to both strains differed in several aspects, despite the fact that pathogen resistance was conferred by both B. bassiana fungi. We also found that, overall, the transcriptional responses to B. bassiana colonisation seemed to resemble those of other endophytic, non-entomopathogenic fungi, such as Trichoderma spp. and Piriformospora indica which are known inducers of systemic resistance against plant pathogens (Molitor et al., 2011; Mathys et al., 2012; Nongbri et al., 2012; Brotman et al., 2013; Gill et al., 2016).
Gene ontology and enrichment analyses suggested that both B. bassiana strains initiate microbial-associated molecular pattern triggered immunity (MTI) in A. thaliana as indicated by differential gene expression in the GO categories “response to innate immune response”, and “defence response” (Table 1 and Supplementary Tables S1–S4), suggesting that B. bassiana was recognised as an invading fungus. Both fungal strains induced a number of genes for receptor-like kinases, including a LysM domain-containing protein, which is necessary for chitin signalling and fungal resistance in A. thaliana (Wan et al., 2008). The induction of these genes may indicate that Beauveria-derived chitin is recognised by the plant as microbial-associated molecular pattern (MAMP) and could have contributed to enhanced resistance against S. sclerotiorum. Mathys et al. (2012) and Brotman et al. (2013) reported responses to chitin as one of the significantly upregulated biological processes in endophytic Trichoderma spp.–A. thaliana interactions. In addition, the upregulation of different pattern recognition receptors (PRRs) indicative for MTI was observed (Yang et al., 2012; Tang et al., 2017), such as the receptor-like kinase FRK1 (in FRh2-inoculated plants) that is involved in the early defence response against the 22-amino-acid peptide Flg22 (Asai et al., 2002), and AtRLP23 (in BG11-inoculated plants), which confers Nep1-like proteins (NLPs) recognition. These proteins are widely distributed among bacteria, fungi and oomycetes (Albert et al., 2015). WRKY transcription factors act as important regulators of complex defence networks and are central players of many aspects of MTI (Rushton et al., 2010; Pieterse et al., 2012). WRKY29, which was induced in FRh2-inoculated plants, is linked to enhanced resistance to bacterial and fungal pathogens (Asai et al., 2002) while WRKY75, induced in FRh2- and BG11-inoculated plants, is associated with defence responses against necrotrophic pathogens (Gechev et al., 2005; Encinas-Villarejo et al., 2009). In particular the induction of WRKY75 could have played a role in the observed resistance against S. sclerotiorum, since Chen et al. (2013) showed that WRKY75 overexpression lines conferred enhanced resistance to the same fungal species. In addition, FRh2- and BG11-inoculated plants were characterised by altered expression patterns of nucleotide binding site-leucine-rich repeat (NBS-LRR) genes, which act as plant immune receptors and are responsible for the initiation of a secondary line of plant defence called effector-triggered immunity (ETI). ETI is mediated by a complex recognition process of pathogen effectors by corresponding resistance proteins in the host plant (Jones and Dangl, 2006).
Downstream of MTI activation, different plant hormones such SA, JA, and ET are key players in the regulation of plant defence responses (Bari and Jones, 2009; Pieterse et al., 2012). GO analysis of the transcriptomic responses to BG11 suggested activation of the SA signalling pathway (“response to SA”), which is involved in systemic acquired resistance against pathogens and sucking herbivores such as aphids. Specifically, we observed the induction of SA marker genes such as PR1, PR2, GRX480, and WRKY6 (Maleck et al., 2000; Robatzek and Somssich, 2002; Ndamukong et al., 2007). In addition, BG11 induced genes associated with the GO categories “response to JA”, “response to ET”, “response to wounding” and “JA-mediated signalling pathway” (Supplementary Tables S1–S4), namely PDF1.2, ORA 59, ERF1, ERF2 and WRKY 40 (Pre et al., 2008). This aligns with many studies that have shown similar effects of Trichoderma spp. colonisation on SA and JA–ET pathway activation (Salas-Marina et al., 2011; Mathys et al., 2012; Yoshioka et al., 2012; Brotman et al., 2013). A different picture emerged in the transcriptome of FRh2-inoculated plants: despite FRh2 inducing downregulation of JA–ET-regulated genes, this did not affect the expression of marker genes such as PDF 1.2. Similarly, FRh2 induced no effect on SA-pathway marker genes. The unaltered expression of SA and JA–ET related genes does not necessarily exclude the involvement of both pathways in the FRh2-triggered resistance against S. sclerotiorum. Using mutant analysis, Mathys et al. (2012) demonstrated the involvement of the JA-pathway in the T. hamatum T382–A. thaliana interaction, despite the fact that none of the JA-pathway marker genes were induced.
Following pathogen recognition and hormone signalling, plant immunity relies on the production of antimicrobial compounds, known as phytoalexins, that act against intrusive pathogens (Dodds and Rathjen, 2010). Both B. bassiana strains FRh2 and BG11 induced biosynthesis genes of the phytoalexin camalexin, such as CYP71A12/13 and CYP71B15 (PAD3), respectively. Similarly, Salas-Marina et al. (2011) reported an upregulation in PAD3 expression in both roots and leaves after treatment of A. thaliana with T. atroviride, whereas Contreras-Cornejo et al. (2011) found that A. thaliana seedlings colonised by T. virens and T. atroviride accumulate camalexin in their leaves. It is possible that camalexin-induction or priming by B. bassiana could have resulted in enhanced resistance against S. sclerotiorum. Stotz et al. (2011) showed that A. thaliana genes associated with the formation of camalexin were induced in leaves challenged with S. sclerotiorum, whereas in mutant lines deficient in camalexin biosynthesis was hyper-susceptible to this phytopathogen.
Furthermore, Beauveria-colonised plants showed responses to oxidative stress that are likewise characteristic for Trichoderma–A. thaliana interactions (Salas-Marina et al., 2011; Mathys et al., 2012; Brotman et al., 2013). Multiple scavenger genes such as peroxidase, GST and other enzymes that protect against reactive oxygen species (ROS) were induced. A key regulator of the ascorbate-glutathione pathway for ROS detoxification (Yoon et al., 2004), the mono-dehydro-ascorbate reductase (MHDAR), was also upregulated in BG11-inoculated plants. This reductase is involved in the interaction between A. thaliana roots and the endophyte P. indica (Vadassery et al., 2009). ROS scavengers help plants not only to cope with biotic but also with abiotic stresses (Apel and Hirt, 2004; Daudi et al., 2012; Das and Roychoudhury, 2014). Interestingly, FRh2 colonisation differed from BG11 by triggering a stronger expression of various genes associated with abiotic stress. Changes in the expression of CYP707A3, WRKY63, RAP2.6L, RAV2, HSP101 and MBF1c suggest that FRh2 may protect its host against drought or salinity (Umezawa et al., 2006; Krishnaswamy et al., 2011) and heat (Queitsch et al., 2000; Suzuki et al., 2008; Fu et al., 2014; Park and Seo, 2015). However, this hypothesis awaits further testing. Endophytes such as Trichoderma spp. and P. indica are well-known for their abilities to alleviate osmotic, salt, temperature or other abiotic stresses (Waller et al., 2005; Baltruschat et al., 2008; Brotman et al., 2013; Contreras-Cornejo et al., 2014).
The induction of SA and JA–ET dependent transcriptional responses in A. thaliana following BG11 colonisation did not result in increased levels of SA and JA metabolites in A. thaliana leaves. It confirms that gene expression does not necessarily reflect metabolite levels, which can be regulated at various post-expression stages (Pieterse et al., 2000; Van der Ent et al., 2009; Withers and Dong, 2017). However, this finding is in line with FRh2-inoculated plants, where genes of the defence pathways remained unaffected. Since both strains were able to enhance resistance against S. sclerotiorum, maybe signalling pathways other than those requiring SA and JA were involved. Future work, using SA and JA mutants of A. thaliana, will be needed to elucidate this question. At this point it is unclear whether two different resistance mechanisms have resulted in the same outcome or whether the plant responded similarly to both fungal strains with regard to pathogen defence. In any case, there is a certain overlap in the induction of SAR related genes, including, for instance ALD1, which plays a key role in local and systemic defence in A. thaliana and is involved in SA-independent signalling upstream of SA biosynthesis (Song et al., 2004; Návarová et al., 2012). This could point to a similar resistance mechanism in both cases.
As expected from the observed transcriptomic responses, B. bassiana colonisation did not result in major changes in the plant’s GLS profiles. Nevertheless, three AGS were attenuated in plants colonised by FRh2. Glucosinolates play an important role in the resistance of Brassicales against pathogens and insects (Hopkins et al., 2009; Wittstock and Burow, 2010; Buxdorf et al., 2013) and the fact that glucosinolates were not enhanced could explain to some extent why B. bassiana did not affect the performance of the generalist M. persicae, which has been shown to be susceptible to these compounds (Pfalz et al., 2009). Less likely GLS effects, on the other hand, would be expected in the case of P. xylostella, as this herbivore produces a sulfatase which prevents the formation of toxic GLS hydrolysis products (Ratzka et al., 2002). Regarding S. sclerotiorum, glucosinolates have been found to be induced by this fungus and also restricted pathogenesis (Stotz et al., 2011). Inoculation with B. bassiana may have either primed the plants to respond more strongly after S. sclerotiorum infection or other factors were important in this interaction. In a different endophyte system, Maag et al. (2014) likewise found no changes in GLS levels in Brassica napus after T. atroviride inoculation, and subsequent insect feeding did not lead to priming.
While this study provides correlational evidence for induced resistance against S. sclerotiorum based on the upregulation of pathogenesis-related (PR) proteins, ROS scavengers, camalexin, phytohormones and other defence-related genes, direct mechanisms that were not explored here cannot be ruled out entirely. Mycoparasitism within the plant tissue, competition for nutrients and toxic effects caused by B. bassiana-produced secondary metabolites could have exerted an inhibitory effect on the pathogen (Jaber and Ownley, 2018). However, evidence that such effects occur in planta is still missing.
In summary, our results demonstrate that B. bassiana protects A. thaliana against fungal disease but not against two herbivorous insect species. Endophytic colonisation resulted in profound changes in plant defence gene expression but in only few modifications in defence hormone and GLS levels. The presented study is one of the first to report plant responses to endophytic entomopathogens and contributes to a better understanding of B. bassiana ecology. The findings are also important for assessing the potential of endophytic entomopathogens as biological control agents.
Data Availability
The datasets generated for this study can be found in Lincoln University Research Archive, https://researcharchive.lincoln.ac.nz/handle/10182/8097.
Author Contributions
MRa, MRo, and TG conceived the study. MRa, HB, and CM carried out the experimental work. MRa, MRo, and HB analysed the data. MRa and MRo wrote the manuscript with contributions from all authors.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Dr. Artemio Mendoza Mendoza and Dr. Andrew Holyoake (Bio-Protection Research Centre, New Zealand) for advice concerning quantitative real time PCR. Furthermore, we thank Jason Breitmeyer (Lincoln University, New Zealand) for assistance with GC-MS analysis of phytohormones and Jenny Brookes (Bio-Protection Research Centre, New Zealand) for insect rearing.
Funding. This research was funded by the Tertiary Education Commission, New Zealand, through the Centres of Research Excellence programme to the Bio-Protection Research Centre.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00615/full#supplementary-material
References
- Abdalsamee M. K., Müller C. (2012). Effects of indole glucosinolates on performance and sequestration by the sawfly Athalia rosae and consequences of feeding on the plant defense system. J. Chem. Ecol. 38 1366–1375. 10.1007/s10886-012-0197-4 [DOI] [PubMed] [Google Scholar]
- Abel S., Nguyen M. D., Chow W., Theologis A. (1995). ASC4, a primary indoleacetic acid-responsive gene encoding 1-aminocyclopropane-1-carboxylate synthase in Arabidopsis thaliana. J. Biol. Chem. 270 19093–19099. 10.1074/jbc.270.32.19093 [DOI] [PubMed] [Google Scholar]
- Albert I., Böhm H., Albert M., Feiler C. E., Imkampe J., Wallmeroth N., et al. (2015). An RLP23–SOBIR1–BAK1 complex mediates NLP-triggered immunity. Nat. Plants 1:15140. 10.1038/nplants.2015.140 [DOI] [PubMed] [Google Scholar]
- Apel K., Hirt H. (2004). Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55 373–399. 10.1146/annurev.arplant.55.031903.141701 [DOI] [PubMed] [Google Scholar]
- Asai T., Tena G., Plotnikova J., Willmann M. R., Chiu W. L., Gomez-Gomez L., et al. (2002). MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415 977–983. 10.1038/415977a [DOI] [PubMed] [Google Scholar]
- Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., et al. (2000). Gene ontology: tool for the unification of biology. Nat. Genet. 25 25–29. 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakshi M., Oelmuller R. (2014). WRKY transcription factors: jack of many trades in plants. Plant Signal. Behav. 9:e27700. 10.4161/psb.27700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltruschat H., Fodor J., Harrach B. D., Niemczyk E., Barna B., Gullner G., et al. (2008). Salt tolerance of barley induced by the root endophyte Piriformospora indica is associated with a strong increase in antioxidants. New Phytol. 180 501–510. 10.1111/j.1469-8137.2008.02583.x [DOI] [PubMed] [Google Scholar]
- Bari R., Jones J. D. (2009). Role of plant hormones in plant defence responses. Plant Mol. Biol. 69 473–488. 10.1007/s11103-008-9435-0 [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57 289–300. 10.1111/j.2517-6161.1995.tb02031.x [DOI] [Google Scholar]
- Bing L. A., Lewis L. C. (1991). Suppression of Ostrinia nubilalis (Hubner) (Lepidoptera, Pyralidae) by endophytic Beauveria bassiana (Balsamo) Vuillemin. Environ. Entomol. 20 1207–1211. 10.1093/ee/20.4.1207 [DOI] [Google Scholar]
- Biswas C., Dey P., Satpathy S., Satya P. (2012). Establishment of the fungal entomopathogen Beauveria bassiana as a season long endophyte in jute (Corchorus olitorius) and its rapid detection using SCAR marker. Biocontrol 57 565–571. 10.1007/s10526-011-9424-0 [DOI] [Google Scholar]
- Bolstad B. M., Irizarry R. A., Astrand M., Speed T. P. (2003). A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19 185–193. 10.1093/bioinformatics/19.2.185 [DOI] [PubMed] [Google Scholar]
- Brotman Y., Landau U., Cuadros-Inostroza A., Takayuki T., Fernie A. R., Chet I., et al. (2013). Trichoderma-plant root colonization: escaping early plant defense responses and activation of the antioxidant machinery for saline stress tolerance. PLoS Pathog. 9:15. 10.1371/journal.ppat.1003221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown P. D., Tokuhisa J. G., Reichelt M., Gershenzon J. (2003). Variation of glucosinolate accumulation among different organs and developmental stages of Arabidopsis thaliana. Phytochemistry 62 471–481. 10.1016/S0031-9422(02)00549-6 [DOI] [PubMed] [Google Scholar]
- Brownbridge M., Reay S. D., Nelson T. L., Glare T. R. (2012). Persistence of Beauveria bassiana (Ascomycota: Hypocreales) as an endophyte following inoculation of radiata pine seed and seedlings. Biol. Control 61 194–200. 10.1016/j.biocontrol.2012.01.002 [DOI] [Google Scholar]
- Buxdorf K., Yaffe H., Barda O., Levy M. (2013). The effects of glucosinolates and their breakdown products on necrotrophic fungi. PLoS One 8:e70771. 10.1371/journal.pone.0070771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Card S., Johnson L., Teasdale S., Caradus J. (2016). Deciphering endophyte behaviour: the link between endophyte biology and efficacious biological control agents. FEMS Microbiol. Ecol. 92:fiw114. 10.1093/femsec/fiw114 [DOI] [PubMed] [Google Scholar]
- Castrillo L. A., Vandenberg J. D., Wraight S. P. (2003). Strain-specific detection of introduced Beauveria bassiana in agricultural fields by use of sequence-characterized amplified region markers. J. Invertebr. Pathol. 82 75–83. 10.1016/S0022-2011(02)00190-8 [DOI] [PubMed] [Google Scholar]
- Chen X., Liu J., Lin G., Wang A., Wang Z., Lu G. (2013). Overexpression of AtWRKY28 and AtWRKY75 in Arabidopsis enhances resistance to oxalic acid and Sclerotinia sclerotiorum. Plant Cell Rep. 32 1589–1599. 10.1007/s00299-013-1469-3 [DOI] [PubMed] [Google Scholar]
- Contreras-Cornejo H. A., Macias-Rodriguez L., Alfaro-Cuevas R., Lopez-Bucio J. (2014). Trichoderma spp. improve growth of Arabidopsis seedlings under salt stress through enhanced root development, osmolite production, and Na+ elimination through root exudates. Mol. Plant Microbe. Interact. 27 503–514. 10.1094/MPMI-09-13-0265-R [DOI] [PubMed] [Google Scholar]
- Contreras-Cornejo H. A., Macias-Rodriguez L., Beltran-Pena E., Herrera-Estrella A., Lopez-Bucio J. (2011). Trichoderma-induced plant immunity likely involves both hormonal- and camalexin-dependent mechanisms in Arabidopsis thaliana and confers resistance against necrotrophic fungi Botrytis cinerea. Plant Signal. Behav. 6 1554–1563. 10.4161/psb.6.10.17443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das K., Roychoudhury A. (2014). Reactive oxygen species (ROS) and response of antioxidants as ROS-scavengers during environmental stress in plants. Front. Environ. Sci. 2:53 10.3389/fenvs.2014.00053 [DOI] [Google Scholar]
- Daudi A., Cheng Z., O’Brien J. A., Mammarella N., Khan S., Ausubel F. M., et al. (2012). The apoplastic oxidative burst peroxidase in Arabidopsis is a major component of pattern-triggered immunity. Plant Cell 24 275–287. 10.1105/tpc.111.093039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds P. N., Rathjen J. P. (2010). Plant immunity: towards an integrated view of plant–pathogen interactions. Nat. Rev. Genet. 11:539. 10.1038/nrg2812 [DOI] [PubMed] [Google Scholar]
- Encinas-Villarejo S., Maldonado A. M., Amil-Ruiz F., de los Santos B., Romero F., Pliego-Alfaro F., et al. (2009). Evidence for a positive regulatory role of strawberry (Fragaria x ananassa) Fa WRKY1 and Arabidopsis At WRKY75 proteins in resistance. J. Exp. Bot. 60 3043–3065. 10.1093/jxb/erp152 [DOI] [PubMed] [Google Scholar]
- Frerigmann H., Gigolashvili T. (2014). MYB34, MYB51, and MYB122 distinctly regulate indolic glucosinolate biosynthesis in Arabidopsis thaliana. Mol. Plant 7 814–828. 10.1093/mp/ssu004 [DOI] [PubMed] [Google Scholar]
- Fu M., Kang H. K., Son S. H., Kim S. K., Nam K. H. (2014). A subset of Arabidopsis RAV transcription factors modulates drought and salt stress responses independent of ABA. Plant Cell Physiol. 55 1892–1904. 10.1093/pcp/pcu118 [DOI] [PubMed] [Google Scholar]
- Ganley R. J., Newcombe G. (2006). Fungal endophytes in seeds needles of Pinus monticola. Mycol. Res. 110 318–327. 10.1016/j.mycres.2005.10.005 [DOI] [PubMed] [Google Scholar]
- Gechev T. S., Minkov I. N., Hille J. (2005). Hydrogen peroxide-induced cell death in Arabidopsis: transcriptional and mutant analysis reveals a role of an oxoglutarate-dependent dioxygenase gene in the cell death process. IUBMB Life 57 181–188. 10.1080/15216540500090793 [DOI] [PubMed] [Google Scholar]
- Gill S. S., Gill R., Trivedi D. K., Anjum N. A., Sharma K. K., Ansari M. W., et al. (2016). Piriformospora indica: potential and significance in plant stress tolerance. Front. Microbiol. 7:332 10.3389/fmicb.2016.00332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glare T., Caradus J., Gelernter W., Jackson T., Keyhani N., Kohl J., et al. (2012). Have biopesticides come of age? Trends Biotechnol. 30 250–258. 10.1016/j.tibtech.2012.01.003 [DOI] [PubMed] [Google Scholar]
- Golo P. S., Gardner D. R., Grilley M. M., Takemoto J. Y., Krasnoff S. B., Pires M. S., et al. (2014). Production of destruxins from Metarhizium spp. fungi in artificial medium and in endophytically colonized cowpea plants. PLoS One 9:e104946. 10.1371/journal.pone.0104946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Vidal S., Salinas J., Tena M., Lopez-Llorca L. V. (2009). Proteomic analysis of date palm (Phoenix dactylifera L.) responses to endophytic colonization by entomopathogenic fungi. Electrophoresis 30 2996–3005. 10.1002/elps.200900192 [DOI] [PubMed] [Google Scholar]
- Graser G., Oldham N. J., Brown P. D., Temp U., Gershenzon J. (2001). The biosynthesis of benzoic acid glucosinolate esters in Arabidopsis thaliana. Phytochemistry 57 23–32. 10.1016/S0031-9422(00)00501-X [DOI] [PubMed] [Google Scholar]
- Haas J., Lozana E. R., Haida K. S., Mazara S. M., Souza Vismara E., Poppy G. M. (2018). Getting ready for battle: do cabbage seeds acid treated with jasmonic sap chitosan affect chewing and-feeding insects? Entomol. Exp. Appl. 166 412–419. 10.1111/eea.12678 [DOI] [Google Scholar]
- Hajek A. (2004). Natural Enemies An Introduction to Biological Control. Cambridge: Cambridge University Press; 10.1017/CBO9780511811838 [DOI] [Google Scholar]
- Hardtke C. S., Dorcey E., Osmont K. S., Sibout R. (2007). Phytohormone collaboration: zooming in on auxin–brassinosteroid interactions. Trends Cell Biol. 17 485–492. 10.1016/j.tcb.2007.08.003 [DOI] [PubMed] [Google Scholar]
- Hermosa R., Viterbo A., Chet I., Monte E. (2012). Plant-beneficial effects of Trichoderma and of its genes. Microbiology 158 17–25. 10.1099/mic.0.052274-0 [DOI] [PubMed] [Google Scholar]
- Hopkins R. J., van Dam N. M., van Loon J. J. A. (2009). Role of glucosinolates in insect-plant relationships and multitrophic interactions. Annu. Rev. Entomol. 54 57–83. 10.1146/annurev.ento.54.110807.090623 [DOI] [PubMed] [Google Scholar]
- Jaber L. R., Enkerli J. (2017). Fungal entomopathogens as endophytes: can they promote plant growth? Biocontrol Sci. Technol. 27 28–41. 10.1080/09583157.2016.1243227 [DOI] [Google Scholar]
- Jaber L. R., Ownley B. H. (2018). Can we use entomopathogenic fungi as endophytes for dual biological control of insect pests and plant pathogens? Biol. Control 116 36–45. 10.1016/j.jip.2015.07.009 [DOI] [PubMed] [Google Scholar]
- Jones J. D. G., Dangl J. L. (2006). The plant immune system. Nature 444 323–329. 10.1038/nature05286 [DOI] [PubMed] [Google Scholar]
- Kieber J. J., Schaller G. E. (2014). Cytokinins. Arabidopsis Book 12:e0168. 10.1199/tab.0168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnaswamy S., Verma S., Rahman M. H., Kav N. N. (2011). Functional characterization of four APETALA2-family genes (RAP2.6, RAP2.6L, DREB19 and DREB26) in Arabidopsis. Plant Mol. Biol. 75 107–127. 10.1007/s11103-010-9711-7 [DOI] [PubMed] [Google Scholar]
- Lacey L. A., Grzywacz D., Shapiro-Ilan D. I., Frutos R., Brownbridge M., Goettel M. S. (2015). Insect pathogens as biological control agents: back to the future. J. Invertebr. Pathol. 132 1–41. 10.1016/j.jip.2015.07.009 [DOI] [PubMed] [Google Scholar]
- Lee D. S., Kim B. K., Kwon S. J., Jin H. C., Park O. K. (2009). Arabidopsis GDSL lipase 2 plays a role in pathogen defense via negative regulation of auxin signaling. Biochem. Biophys. Res. Commun. 379 1038–1042. 10.1016/j.bbrc.2009.01.006 [DOI] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lugtenberg B. J. J., Caradus J. R., Johnson L. J. (2016). Fungal endophytes for sustainable crop production. FEMS Microbiol. Ecol. 92:fiw194. 10.1093/femsec/fiw194 [DOI] [PubMed] [Google Scholar]
- Maag D., Kandula D. R. W., Müller C., Mendoza-Mendoza A., Wratten S. D., Stewart A., et al. (2014). Trichoderma atroviride LU132 promotes plant growth but not induced systemic resistance to Plutella xylostella in oilseed rape. BioControl 59 241–252. 10.1007/s10526-013-9554-7 [DOI] [Google Scholar]
- Maleck K., Levine A., Eulgem T., Morgan A., Schmid J., Lawton K. A., et al. (2000). The transcriptome of Arabidopsis thaliana during systemic acquired resistance. Nat. Genet. 26 403–410. 10.1038/82521 [DOI] [PubMed] [Google Scholar]
- Mathys J., De Cremer K., Timmermans P., Van Kerckhove S., Lievens B., Vanhaecke M., et al. (2012). Genome-wide characterization of ISR induced in Arabidopsis thaliana by Trichoderma hamatum T382 against Botrytis cinerea infection. Front. Plant Sci. 3:108. 10.3389/fpls.2012.00108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinnon A. C., Saari S., Moran-Diez M. E., Meyling N. V., Raad M., Glare T. R. (2017). Beauveria bassiana as an endophyte: a critical review on associated methodology and biocontrol potential. BioControl 62 1–17. 10.1007/s10526-016-9769-5 [DOI] [Google Scholar]
- Mi H., Huang X., Muruganujan A., Tang H., Mills C., Kang D., et al. (2017). PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 45 D183–D189. 10.1093/nar/gkw1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molitor A., Zajic D., Voll L. M., Pons K. H. J., Samans B., Kogel K. H., et al. (2011). Barley leaf transcriptome and metabolite analysis reveals new aspects of compatibility and Piriformospora indica-mediated systemic induced resistance to powdery mildew. Mol. Plant Microbe Interact. 24 1427–1439. 10.1094/MPMI-06-11-0177 [DOI] [PubMed] [Google Scholar]
- Mukherjee P. K., Horwitz B. A., Herrera-Estrella A., Schmoll M., Kenerley C. M. (2013). Trichoderma research in the genome era. Annu. Rev. Phytopathol. 51 105–129. 10.1146/annurev-phyto-082712-102353 [DOI] [PubMed] [Google Scholar]
- Muvea A. M., Meyhöfer R., Subramanian S., Poehling H.-M., Ekesi S., Maniana N. K. (2014). Colonisation of onions by endophytic fungi and their impacts on the biology of Thrips tabaci. PLoS One 9:e108242. 10.1371/journal.pone.0108242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Návarová H., Bernsdorff F., Döring A.-C., Zeier J. (2012). Pipecolic acid, an endogenous mediator of defense amplification and priming, is a critical regulator of inducible plant immunity. Plant Cell 24 5123–5141. 10.1105/tpc.112.103564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndamukong I., Abdallat A. A., Thurow C., Fode B., Zander M., Weigel R., et al. (2007). SA-inducible Arabidopsis glutaredoxin interacts with TGA factors and suppresses JA-responsive PDF1.2 transcription. Plant J. 50 128–139. 10.1111/j.1365-313X.2007.03039.x [DOI] [PubMed] [Google Scholar]
- Nongbri P., Vahabi K., Mrozinska A., Seebald E., Sun C., Sherameti I., et al. (2012). Balancing defense and growth—Analyses of the beneficial symbiosis between Piriformospora indica and Arabidopsis thaliana. Symbiosis 58 17–28. 10.1007/s13199-012-0209-8 [DOI] [Google Scholar]
- Oh I. S., Park A. R., Bae M. S., Kwon S. J., Kim Y. S., Lee J. E., et al. (2005). Secretome analysis reveals an Arabidopsis lipase involved in defense against Alternaria brassicicola. Plant Cell 17 2832–2847. 10.1105/tpc.105.034819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ownley B. H., Griffin M. R., Klingeman W. E., Gwinn K. D., Moulton J. K., Pereira R. M. (2008). Beauveria bassiana: endophytic colonization and plant disease control. J. Invertebr. Pathol. 98 267–270. 10.1016/j.jip.2008.01.010 [DOI] [PubMed] [Google Scholar]
- Park C. J., Seo Y. S. (2015). Heat shock proteins: a review of the molecular chaperones for plant immunity. Plant Pathol. J. 31 323–333. 10.5423/PPJ.RW.08.2015.0150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfalz M., Vogel H., Kroymann J. (2009). The gene controlling the Indole Glucosinolate Modifier1 quantitative trait locus alters indole glucosinolate structures and aphid resistance in Arabidopsis. Plant Cell 21 985–999. 10.1105/tpc.108.063115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse C. M., Zamioudis C., Berendsen R. L., Weller D. M., Van Wees S. C., Bakker P. A. (2014). Induced systemic resistance by beneficial microbes. Annu. Rev. Phytopathol. 52 347–375. 10.1146/annurev-phyto-082712-102340 [DOI] [PubMed] [Google Scholar]
- Pieterse C. M. J., Van der Does D., Zamioudis C., Leon-Reyes A., Van Wees S. C. (2012). Hormonal modulation of plant immunity. Annu. Rev. Cell. Dev. Biol. 28 489–521. 10.1146/annurev-cellbio-092910-154055 [DOI] [PubMed] [Google Scholar]
- Pieterse C. M. J., Van Pelt J. A., Ton J., Parchmann S., Mueller M. J., Buchala A. J., et al. (2000). Rhizobacteria-mediated induced systemic resistance (ISR) in Arabidopsis requires sensitivity to jasmonate and ethylene but is not accompanied by an increase in their production. Physiol. Mol. Plant Pathol. 57 123–134. 10.1006/pmpp.2000.0291 [DOI] [Google Scholar]
- Pre M., Atallah M., Champion A., De Vos M., Pieterse C. M. J., Memelink J. (2008). The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiol. 147 1347–1357. 10.1104/pp.108.117523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qayyum M. A., Wakil W., Arif M. J., Sahi S. T., Dunlap C. A. (2015). Infection of Helicoverpa armigera by endophytic Beauveria bassiana colonizing tomato plants. Biol. Control 90 200–207. 10.1016/j.biocontrol.2015.04.005 [DOI] [Google Scholar]
- Queitsch C., Hong S. W., Vierling E., Lindquist S. (2000). Heat shock protein 101 plays a crucial role in thermotolerance in Arabidopsis. Plant Cell 12 479–492. 10.1105/tpc.12.4.479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos Y., Portal O., Lysoe E., Meyling N. V., Klingen I. (2017). Diversity and abundance of Beauveria bassiana in soils, stink bugs and plant tissues of common bean from organic and conventional fields. J. Invertebr. Pathol. 150 114–120. 10.1016/j.jip.2017.10.003 [DOI] [PubMed] [Google Scholar]
- Ratzka A., Vogel H., Kliebenstein D. J., Mitchell-Olds T., Kroymann J. (2002). Disarming the mustard oil bomb. Proc. Natl. Acad. Sci. U.S.A. 99 11223–11228. 10.1073/pnas.172112899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reay S. D., Brownbridge M., Gicquel B., Cummings N. J., Nelson T. L. (2010). Isolation and characterization of endophytic Beauveria spp. (Ascomycota: Hypocreales) from Pinus radiata in New Zealand forests. Biol. Control 54 52–60. 10.1016/j.biocontrol.2010.03.002 [DOI] [Google Scholar]
- Ren X., Chen Z., Liu Y., Zhang H., Zhang M., Liu Q., et al. (2010). ABO3, a WRKY transcription factor, mediates plant responses to abscisic acid and drought tolerance in Arabidopsis. Plant J. 63 417–429. 10.1111/j.1365-313X.2010.04248.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robatzek S., Somssich I. E. (2002). Targets of AtWRKY6 regulation during plant senescence and pathogen defense. Genes Dev. 16 1139–1149. 10.1101/gad.222702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rostás M., Ton J., Mauch-Mani B., Turlings T. C. (2006). Fungal infection reduces herbivore-induced plant volatiles of maize but does not affect naive parasitoids. J. Chem. Ecol. 32 1897–1909. 10.1007/s10886-006-9147-3 [DOI] [PubMed] [Google Scholar]
- Roy H. E., Steinkraus D. C., Eilenberg J., Hajek A. E., Pell J. K. (2006). Bizarre interactions and endgames: entomopathogenic fungi and their arthropod hosts. Annu. Rev. Entomol. 51 331–357. 10.1146/annurev.ento.51.110104.150941 [DOI] [PubMed] [Google Scholar]
- Rushton P. J., Somssich I. E., Ringler P., Shen Q. J. (2010). WRKY transcription factors. Trends Plant Sci. 15 247–258. 10.1016/j.tplants.2010.02.006 [DOI] [PubMed] [Google Scholar]
- Salas-Marina M. A., Silva-Flores M. A., Uresti-Rivera E. E., Castro-Longoria E., Herrera-Estrella A., Casas-Flores S. (2011). Colonization of Arabidopsis roots by Trichoderma atroviride promotes growth and enhances systemic disease resistance through jasmonic acid/ethylene and salicylic acid pathways. Eur. J. Plant Pathol. 131 15–26. 10.1007/s10658-011-9782-6 [DOI] [Google Scholar]
- Schmelz E. A., Engelberth J., Tumlinson J. H., Block A., Alborn H. T. (2004). The use of vapor phase extraction in metabolic profiling of phytohormones and other metabolites. Plant J. 39 790–808. 10.1111/j.1365-313X.2004.02168.x [DOI] [PubMed] [Google Scholar]
- Schulz B., Boyle C. (2005). The endophytic continuum. Mycol. Res. 109 661–686. 10.1017/S095375620500273X [DOI] [PubMed] [Google Scholar]
- Song J. T., Lu H., McDowell J. M., Greenberg J. T. (2004). A key role for ALD1 in activation of local and systemic defenses in Arabidopsis. Plant J. 40 200–212. 10.1111/j.1365-313X.2004.02200.x [DOI] [PubMed] [Google Scholar]
- Steyaert J. M., Ridgway H. J., Elad Y., Stewart A. (2003). Genetic basis of mycoparasitism: a mechanism of biological control by species of Trichoderma. N. Z. J. Crop Hortic. Sci. 31 281–291. 10.1080/01140671.2003.9514263 [DOI] [Google Scholar]
- Stotz H. U., Sawada Y., Shimada Y., Hirai M. Y., Sasaki E., Krischke M., et al. (2011). Role of camalexin, indole glucosinolates, and side chain modification of glucosinolate-derived isothiocyanates in defense of Arabidopsis against Sclerotinia sclerotiorum. Plant J. 67 81–93. 10.1111/j.1365-313X.2011.04578.x [DOI] [PubMed] [Google Scholar]
- Sun B. T., Akutse K. S., Xia X. F., Chen J. H., Ai X., Tang Y., et al. (2018) Endophytic effects of Aspergillus oryzae on radish (Raphanus sativus) and its herbivore, Plutella xylostella. Planta 248 705–714. 10.1007/s00425-018-2928-4 [DOI] [PubMed] [Google Scholar]
- Suzuki N., Bajad S., Shuman J., Shulaev V., Mittler R. (2008). The transcriptional co-activator MBF1c is a key regulator of thermotolerance in Arabidopsis thaliana. J. Biol. Chem. 283 9269–9275. 10.1074/jbc.M709187200 [DOI] [PubMed] [Google Scholar]
- Sword G. A., Tessnow A., Ek-Ramos M. J. (2017). Endophytic fungi alter sucking bug responses to cotton reproductive structures. Insect Sci. 24 1003–1014. 10.1111/1744-7917.12461 [DOI] [PubMed] [Google Scholar]
- Tall S., Meyling N. V. (2018). Probiotics for plants? growth promotion by the entomopathogenic fungus beauveria bassiana depends on nutrient availability. Microbe. Ecol. 76 1002–1008. 10.1007/s00248-018-1180-6 [DOI] [PubMed] [Google Scholar]
- Tang D., Wang G., Zhou J.-M. (2017). Receptor kinases in plant-pathogen interactions: more than pattern recognition. Plant Cell 29 618–637. 10.1105/tpc.16.00891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gene Ontology Consortium (2017). Expansion of the gene ontology knowledgebase and resources. Nucleic Acids Res. 45 D331–D338. 10.1093/nar/gkw1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O., Blasing O., Gibon Y., Nagel A., Meyer S., Kruger P., et al. (2004). MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37 914–939. 10.1111/j.1365-313X.2004.02016.x [DOI] [PubMed] [Google Scholar]
- Umezawa T., Okamoto M., Kushiro T., Nambara E., Oono Y., Seki M., et al. (2006). CYP707A3, a major ABA 8’-hydroxylase involved in dehydration and rehydration response in Arabidopsis thaliana. Plant J. 46 171–182. 10.1111/j.1365-313X.2006.02683.x [DOI] [PubMed] [Google Scholar]
- Usadel B., Poree F., Nagel A., Lohse M., Czedik-Eysenberg A., Stitt M. (2009). A guide to using MapMan to visualize and compare Omics data in plants: a case study in the crop species, Maize. Plant Cell Environ. 32 1211–1229. 10.1111/j.1365-3040.2009.01978.x [DOI] [PubMed] [Google Scholar]
- Vadassery J., Tripathi S., Prasad R., Varma A., Oelmuller R. (2009). Monodehydroascorbate reductase 2 and dehydroascorbate reductase 5 are crucial for a mutualistic interaction between Piriformospora indica and Arabidopsis. J. Plant Physiol. 166 1263–1274. 10.1016/j.jplph.2008.12.016 [DOI] [PubMed] [Google Scholar]
- Van der Ent S., Van Wees S. C. M., Pieterse C. M. J. (2009). Jasmonate signaling in plant interactions with resistance-inducing beneficial microbes. Phytochemistry 70 1581–1588. 10.1016/j.phytochem.2009.06.009 [DOI] [PubMed] [Google Scholar]
- Vega F. E. (2018). The use of fungal entomopathogens as endophytes in biological control: a review. Mycologia 110 4–30. 10.1080/00275514.2017.1418578 [DOI] [PubMed] [Google Scholar]
- Vega F. E., Goettel M. S., Blackwell M., Chandler D., Jackson M. A., Keller S., et al. (2009). Fungal entomopathogens: new insights on their ecology. Fungal Ecol. 2 149–159. 10.1016/j.funeco.2009.05.001 [DOI] [Google Scholar]
- Vega F. E., Simpkins A., Aime M. C., Posada F., Peterson S. W., Rehner S. A., et al. (2010). Fungal endophyte diversity in coffee plants from Colombia, Hawai’i, Mexico and Puerto Rico. Fungal Ecol. 3 122–138. 10.1016/j.funeco.2009.07.002 16187260 [DOI] [Google Scholar]
- Vidal S., Jaber L. R. (2015). Entomopathogenic fungi as endophytes: plant-endophyte-herbivore interactions and prospects for use in biological control. Curr. Sci. 109 46–54. [Google Scholar]
- Vos C. M. F., De Cremer K., Cammue B. P. A., De Coninck B. (2015). The toolbox of Trichoderma spp. in the biocontrol of Botrytis cinerea disease. Mol. Plant Pathol. 16 400–412. 10.1111/mpp.12189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waller F., Achatz B., Baltruschat H., Fodor J., Becker K., Fischer M., et al. (2005). The endophytic fungus Piriformospora indica reprograms barley to salt-stress tolerance, disease resistance, and higher yield. Proc. Natl. Acad. Sci. U.S.A. 102 13386–13391. 10.1073/pnas.0504423102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J., Zhang X. C., Neece D., Ramonell K. M., Clough S., Kim S. Y., et al. (2008). A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell 20 471–481. 10.1105/tpc.107.056754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Withers J., Dong X. (2017). Post-translational regulation of plant immunity. Curr. Opin. Plant Biol. 38 124–132. 10.1016/j.pbi.2017.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittstock U., Burow M. (2010). Glucosinolate breakdown in Arabidopsis: mechanism, regulation and biological significance. Arabidopsis Book 8:e0134. 10.1199/tab.0134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Deng F., Ramonell K. M. (2012). Receptor-like kinases and receptor-like proteins: keys to pathogen recognition and defense signaling in plant innate immunity. Front. Biol. 7:155–166. 10.1007/s11515-011-1185-8 [DOI] [Google Scholar]
- Yoon H. S., Lee H., Lee I. A., Kim K. Y., Jo J. (2004). Molecular cloning of the monodehydroascorbate reductase gene from Brassica campestris and analysis of its mRNA level in response to oxidative stress. Biochim. Biophys. Acta 1658 181–186. 10.1016/j.bbabio.2004.05.013 [DOI] [PubMed] [Google Scholar]
- Yoshioka Y., Ichikawa H., Naznin H. A., Kogure A., Hyakumachi M. (2012). Systemic resistance induced in Arabidopsis thaliana by Trichoderma asperellum SKT-1, a microbial pesticide of seedborne diseases of rice. Pest Manag. Sci. 68 60–66. 10.1002/ps.2220 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated for this study can be found in Lincoln University Research Archive, https://researcharchive.lincoln.ac.nz/handle/10182/8097.


