Background
Clozapine is an atypical antipsychotic and the gold standard for treatment refractory schizophrenia (TRS), i.e. for whom at least two traditional antipsychotics have been ineffective [1]. The reason for why guidelines only allow clozapine to be prescribed in TRS is the rare, but potentially lethal occurrence of clozapine-induced agranulocytosis (CIA)[1]. The FDA label also lists black box warnings that dose-related side effects of orthostatic hypotension, bradycardia, syncope and seizure have been observed, and warns of serious cardiac side effects and increased mortality in the elderly (see drug label). Other common side effects include sedation (mostly transient) and metabolic side effects such as weight gain. While required dosage can vary substantially among patients (~150 – 1000mg/day), clozapine has a narrow therapeutic window and therapeutic drug monitoring is recommended: serum concentrations of clozapine less than 250 ng/mL are associated with relapse and those above 750ng/mL are associated with increased risk of intoxication [PMID: 27932669][1]. While there are currently no published pharmacogenomic (PGx) guidelines for patients with variants influencing metabolism of clozapine, the FDA label cautions about CYP2D6 poor metabolizers and potential drug-drug interactions with drugs metabolized by same CYP450 enzymes than clozapine. This summary examines the pharmacokinetics (PK) of clozapine and the candidate genes involved and discusses the impact of their variants. While there are a large number of receptor that clozapine acts at, perhaps at least as many as 39 different receptors [2], the pharmacodynamics (PD) of clozapine are not within the scope of this article except where PD effects are the result of PK genes.
Metabolism
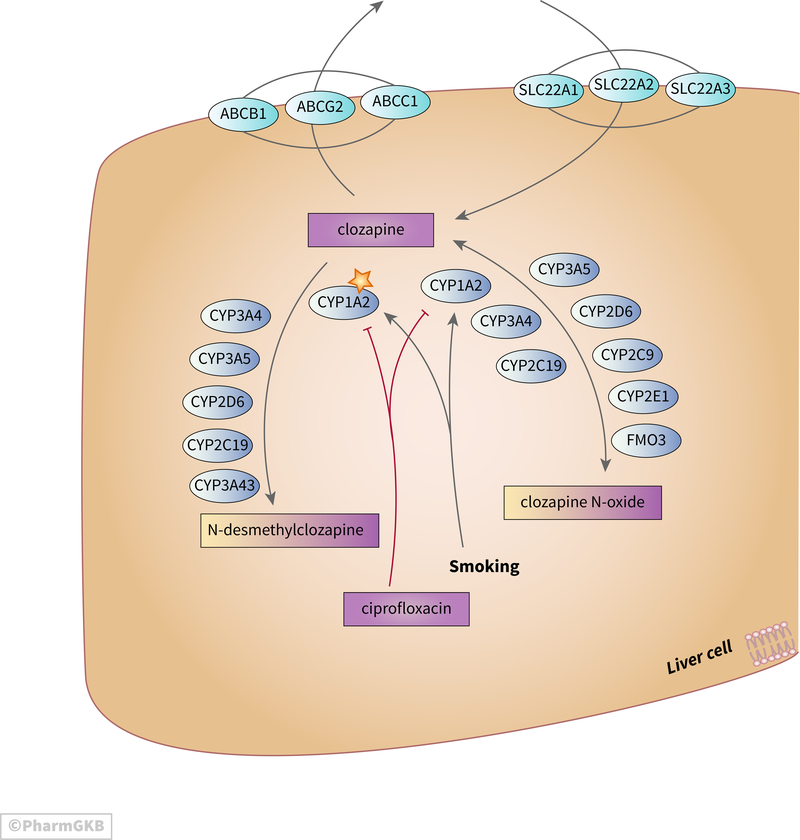
Clozapine undergoes extensive hepatic metabolism with the main routes being demethylation to N-desmethylclozapine and oxidation to clozapine n-oxide (depicted in Figure 1)[3]. In vitro experiments suggest that CYP3A4 accounts for around 70% of clozapine clearance, CYP1A2 around 15%, and 5% or less for each of CYP2C19, CYP2C8 and FMO3 [4]. In vitro studies of clozapine metabolism indicate that CYP3A4 and CYP1A2 are the major enzymes responsible for demethylation with CYP2D6 playing a very minor role [3,5]. Several enzymes were capable of generating the n-oxide metabolite of clozapine in vitro (CYP1A2, CYP2E1, CYP2C9, CYP3A4, CYP2D6, FMO3, CYP2C19) but in vivo CYP1A2 is considered to be the major catalyst [1,3,5,6]. Studies in vivo suggest that CYP3A5 and CYP3A43 may also play a role [7,8]. N-desmethylclozapine is an active metabolite, capable of effect via dopamine D2 and D3 receptors [9], histamine receptors [10], muscarinic M1 [11], and serotonin receptors [12]. Also the ratio of clozapine to N-desmethylclozapine is a strong predictor of working memory performance in patients with schizophrenia; with lower ratios associated with better working memory and executive function [11][13]. Clozapine n-oxide is considered inactive and may be metabolized back to clozapine [3,14,15]. Additional metabolites have been identified in urine from clozapine treated patients but the clinical significance of these metabolites is not clear [16].
Figure 1.
Stylized diagram showing clozapine metabolism and transport in the liver. A fully clickable version of this figure can be found at https://www.pharmgkb.org/pathway/PA166163661
While there are functional variants in CYP1A2 there is not a consensus on the impact of these particularly with respect to clozapine [17–21](discussed further below in the PGx section). Serum concentrations of clozapine are lower in smokers than non-smokers due to activation of CYP1A2 [22]. Changes in smoking behavior can significantly alter clozapine metabolism; with clozapine exposure increasing by over 50% after cessation of smoking [23]. Strong inhibitors of CYP1A2 such as the antibiotic ciprofloxacin can also significantly increase clozapine exposure, and case studies have reported fatal interactions [24]. Additionally, during infection pro-inflammatory cytokines can down-regulate expression of CYP1A2 which may exacerbate this effect [25]. The antidepressant fluvoxamine is also an inhibitor of CYP1A2 and can increase plasma concentrations of clozapine. This PK effect can be utilized as a mechanism to modulate clozapine dose; reducing the dose of clozapine in order to try to decrease side effects while maintaining efficacy [26].
Overall, dose, gender, age and smoking account for approximately 50% of variability in serum clozapine and the remaining 50% is likely composed by genetic variants in drug metabolizing enzymes and by co-medications [27,28]. Serum clozapine is lower in males compared to females despite females receiving lower doses [27–29]. Plasma clozapine is approximately 20–30% lower in smokers compared to non-smokers [27–29]. Significantly higher concentrations are observed in older patients (45+ years) compared to younger patients (18–26 years)[28]. A very small study of pregnant women receiving antipsychotics found decreased plasma concentrations although not significant for clozapine (n=4), it was significant for other drugs (aripiprazole n= 14 and quetiapine n=35) and the authors postulated that levels of CYP3A4 are induced during pregnancy and this may influence antipsychotic drug PK [30]. This, combined with alterations in behavior during pregnancy, such as change in caffeine use and smoking, may be important to monitor in women receiving clozapine.
Transport
Overall experiments on clozapine transport have been in a variety of cell types and have not given a concerted picture about which transporters are involved in influx or export. The scattered evidence is presented below and all reasonable candidates depicted on Figure 1: Clozapine is transported efficiently across the blood-brain barrier and into the brain as shown by PET imaging of C11 labeled clozapine [31]. However, this study used intravenous clozapine whereas most patients receive the drug orally and the transporters responsible were not identified.
For oral clozapine, transport across the gut is important. There is evidence that clozapine metabolism in the gut is different than in the liver since grapefruit juice (an inhibitor of gut CYP3A4 but not liver CYP3A4) does not impact plasma clozapine concentrations [32,33].
Uptake in the liver may be catalyzed by SLC22A1, since clozapine was able to inhibit SLC22A1 transport of a model substrate (MPP+) in vitro [34]. In the same in vitro experiments clozapine also interacted with SLC22A2 and SLC22A3 [34].
The relationship of clozapine to the ABCB1 encoded transporter (also known as PgP) is conflicting in the literature. Early in vitro experiments with CaCo2 cells (an intestinal cell line) showed clozapine and N-desmethylclozapine as low affinity substrates of ABCB1 and inhibitors of ABCB1-mediated talinolol transport [35]. However, although ABCB1 variants are associated with differential PK or outcomes [36,37], more recent articles report clozapine is not substrate or effective inhibitor of ABCB1 [38].
Clozapine inhibits ABCG2 transport of mitoxantrone in vitro [39]. Furthermore variants in the ABCG2 transporter are associated with clozapine exposure and variants in ABCC1 are associated with outcomes but it has not been shown definitively that clozapine is a substrate for either (discussed further in PGx section)[1,37].
Drug-Drug Interactions, DDIs
There are many publications concerning clozapine drug-drug interactions but most studies are very small or individual case reports and very few consider whether PGx variants impacted the interaction (reviewed in [40]).
The FDA label for clozapine categorizes the DDIs into 4 types with recommendations for each:
Strong CYP1A2 inhibitors e.g., fluvoxamine and ciprofloxacin the recommendation is to reduce the clozapine dose to one third.
Moderate or weak CYP1A2 inhibitors such as oral contraceptives or caffeine the recommendation is to monitor for adverse reactions and consider dose reduction.
CYP2D6 or CYP3A4 inhibitors such as fluoxetine the recommendation is to monitor for adverse reactions and consider dose reduction.
Strong CYP3A4 inducers are not recommended e.g., carbamazepine, rifampicin, phenytoin and St Johns Wort.
Studies of DDIs of each type are discussed below and summarized in Table 1:
Table 1.
Summary of clozapine DDIs reported in the literature and the candidate genes involved.
| Interacting drug | Type DDI | Probable gene interaction(s) |
Effect compared to clozapine alone |
Reference |
|---|---|---|---|---|
| ciprofloxacin | Type 1 | Inhibition of CYP1A2 |
Higher plasma clozapine concentrations |
[PMID:9167522] [PMID:17624521] [PMID:19067475] [PMID:27872784] [PMID:22929408] |
| fluvoxamine | Type 1 | Inhibition of CYP1A2 and CYP219 |
Higher plasma clozapine concentration |
[PMID:7962687] [PMID:7782489] |
| caffeine | Type 2 | Inhibition of CYP1A2 |
Higher plasma clozapine concentrations |
[PMID: 8601555] [PMID:22926611] [PMID:23104241] [PMID: 17515710] [PMID:14725610] |
| oral contraceptives |
Type 2 | Inhibition of CYP1A2 |
Higher plasma clozapine concentrations |
PMID:12454563, PMID:25890012, PMID:24717251 |
| antidepressants | Type 3 | Inhibition of CYP2D6 |
Higher plasma clozapine concentrations |
PMID: 17214606 PMID: 24494611 |
| erythromycin | Type 3 | Inhibition of CYP3A4 |
Higher plasma clozapine concentration |
[PMID: 7977898] |
| rifampicin | Type 4 | Induction of CYP1A2 and CYP3A4 |
Lower plasma clozapine concentrations |
[PMID:9472849] |
| phenobarbital | Type 4 | Induction of CYP1A2 and CYP3A4 |
Lower plasma clozapine concentrations |
[PMID:9541158] [PMID:9853978] |
| omeprazole | Not classified | Induction of CYP1A2 |
Lower plasma clozapine concentrations |
[PMID:12806570] PMID:24419309 |
Type 1 DDIs : Strong CYP1A2 inhibitors
There are multiple reports in the literature of serious, sometimes fatal interactions between clozapine and ciprofloxacin supporting the recommendation to modulate the clozapine dose [24,41–47]. There is also a large body of literature on fluvoxamine and clozapine (reviewed in [40]): several papers discuss the use of fluvoxamine as adjunct to avoid high doses clozapine; with lower clozapine doses reducing the incidence of side effects including weight gain [48], glucose/metabolic dysfunction [49], and hematotoxicity [50].
An in vitro study using human liver microsomes investigated production of N-desmethylclozapine and clozapine n-oxide in response to cotreatment with fluvoxamine [51]. The authors concluded that caffeine phenotyping was not reliable in predicting CYP1A2-related clozapine-fluvoxamine DDI [51].
Type 2 DDIs : Moderate or weak CYP1A2 inhibitors
Caffeine consumption is a significant predictor of serum clozapine levels [52]. Various case studies have shown that caffeine intake can influence clozapine plasma levels and clozapine efficacy [53,54]. A randomized study that alternated patients to drink either regular coffee or decaffeinated coffee demonstrated that caffeine was associated with plasma clozapine [55]. Caffeine intake can also be from cola soda, where 6 glasses a day increased plasma clozapine to toxic levels in one case study [56]. Intraindividual variation in plasma clozapine responses to caffeine were influenced by CYP1A2 [55].
There are a few case reports of DDI with clozapine and oral contraceptives [57–59]. The recommendation to monitor and consider dose alterations can be more complicated in real world situations since the clozapine-prescribing physician may be unaware of a patient’s contraceptive prescription history and the patient may not realize that sharing the information is important.
Type 3 DDIs: CYP2D6 or CYP3A4 inhibitors
Co-treatment of clozapine with antidepressants metabolized by CYP2D6 including sertraline, paroxetine and fluoxetine, is reported to lead to increases of plasma clozapine by 20–40% (reviewed in [60]). While the study by Centorrino examined 40 patients co-treated with SSRIs and 40 with clozapine alone and showed increased serum clozapine and N-desmethylclozapine, there were very large standard deviations serum concentrations (for example, the mean clozapine concentration in paroxetine co-treated patients was 417ng/ml with SD 373ng/ml)[61]. Another study of 14 patients on paroxetine and clozapine showed no change in clozapine or major metabolites [62,63]. In a single case report, a patient on clozapine and paroxetine had a toxic increase in serum clozapine [64]. While a study of 9 patients co-treated with paroxetine and 8 co-treated with sertraline found increased clozapine and N-desmethyl clozapine in the paroxetine group only [65]. However, since paroxetine is also metabolized by CYP1A2 and CYP3A4 the relationship may not all be due to CYP2D6 inhibition [see Paroxetine Pathway https://www.pharmgkb.org/pathway/PA166121347].
In an in vitro microsome experiment, the CYP2D6 inhibitors quinidine and dextromethorphan influenced the metabolism of clozapine and reduced the generation of minor metabolic products as shown by HPLC, but did not change the amount of N-desmethylclozapine or clozapine n-oxide [66].
The CYP3A4 inhibitor ketoconazole has been shown to strongly inhibit production of N-desmethylclozapine, clozapine N-oxide and other minor metabolites in vitro [3]. However, in a study of 21 patients receiving clozapine and ketoconazole, no significant changes in clozapine and metabolites were observed [33]. Also in a series of human liver microsome experiments using CYP3A4 phenotyping with testosterone 6β‐hydroxylation, the CYP3A4 DDI of clozapine with ketoconazole was not predictable [51].
Type 4 DDIs: strong CYP3A4 inducers
Although co-prescription of clozapine and strong CYP3A4 inducers is not recommended, the label lists examples as phenytoin and carbamazepine but not oxcarbazepine. Case reports of DDI with oxcarbazepine suggest that it may be especially likely to interact with clozapine via both CYP3A4 and CYP1A2 [67,68].
Other DDIs
One of the side effects of clozapine is gastroesophageal reflux disease, GERD [69]. Treatment for GERD is proton pump inhibitors (PPIs), the majority of which are metabolized by CYP2C19. There are several possible routes of DDIs between clozapine and PPIs; induction of CYP1A2, induction of FMO3, and competitive inhibition of CYP2C19. Any or all of these interactions may lead to increased N-desmethylclozapine and clozapine n-oxide and increased risk for hematological toxicity [69]. Clozapine n-oxide inhibits the CYP2C19 mediated metabolism of s-mephenytoin in vitro [70]. Potentially then clozapine n-oxide may inhibit CYP2C19 metabolism of PPIs.
Moderate or weak CYP1A2 or CYP3A4 inducers are also discussed in the drug label, with the suggestion to increase dose if lack of efficacy is observed. No specific drugs are mentioned in the label.
The drug label also cautions on the use of clozapine in those already at risk for long QT and mentions many QT-prolonging drugs where concomitant use may increase the risk of long QT syndrome. Several of the drugs listed are also contraindicated as CYP3A4 inhibitors eg. erythromycin or CYP2D6 inhibitors eg quinidine, chlorpromazine or CYP1A2 inhibitors eg amiodarone.
Pharmacogenomics
While the majority of studies of clozapine PGx have examined PD candidate genes, this paper is focused on PK candidate genes and their effects on PK or on PD/clinical outcomes (summarized in table 2). The candidate genes CYP1A2, CYP3A4, CYP2C19, CYP2D6 and ABCB1 are all well known pharmacogenes and detailed summaries on their variants are described on the PharmGKB website [https://www.pharmgkb.org/vips]. In addition there are detailed descriptions of each PGx relationship paper that are combined into interactive table (see the clozapine Variant Annotations) on the PharmGKB website [https://www.pharmgkb.org/chemical/PA449061/variantAnnotation].
Table 2.
Summary of PK gene variants and their associated phenotypes.
| Gene Variant(s) |
Allele/ Genotype |
Association | Study size |
p-value | Reference |
|---|---|---|---|---|---|
| ABCB1 rs1045642 | AA | increased plasma clozapine | 75 | 0.046 | 19593168 |
| ABCB1 rs1045642 | AA | increased risk of Agranulocytosis and Neutropenia with clozapine |
310 | 0.05 | 27168101 |
| ABCB1 rs1045642 | AG+GG | increased weight gain and risk for hypertension with clozapine in men |
65 | 0.026 0.006 |
28919802 |
| ABCB1 rs7787082 | G | nonresponse | 93 | 0.036 | 22722500 |
| ABCB1 rs10248420 | A | nonresponse | 89 | 0.046 | 22722500 |
| ABCC1 rs212090T | AT+TT | increased weight gain and risk for hypertension in men |
65 | 0.022 0.031 |
28919802 |
| ABCG2 rs2231142 | GT+TT | increased dose-adjusted trough concentrations of clozapine |
45 | 0.01 | 27932669 |
| CYP1A2 rs762551 (*1F) |
AA | decreased likelihood of Metabolic Syndrome in non-smokers |
38 | Not given |
27681143 |
| CYP1A2 rs762551 (*1F) |
AA | Increased likelihood of Metabolic Syndrome in smokers |
21 | 0.0213 | 27681143 |
| CYP1A2 rs762551 (*1F) |
AA | nonresponse | 4 | Not given |
15206669 |
| CYP1A2 rs35694136 and/or rs2069514 (*1C) |
DEL/A | increased likelihood of elevated insulin levels |
17 | 0.04 | 17503978 |
| CYP1A2 *1F | *1F/*1F | increased risk of seizures | 108 | 0.033 | 23601795 |
| CYP1A2 *1C | *1C | increased severity of Confusion, Drug Toxicity, Headache, Muscle Rigidity, sedation and Tachycardia |
5 | Not given |
21481946 |
| CYP2C19 | *17/*17 | higher serum N-desmethylclozapine, lower prevalence of diabetes, increased improvement of Schizophrenia symptoms |
17 45 125 |
0.049 0.042 0.012 |
28664816 |
| CYP2C19 | *2/*2 | higher serum clozapine | 75 | 0.036 | 19593168 |
| CYP2C19 | *2 | Increased likelihood of metabolic syndrome |
59 | 0.033 | 27681143 |
| CYP3A5 | *1/*3 | decreased concentrations of clozapine compared to *3/*3 |
92 | < 0.0001 | 28340122 |
| CYP3A43 rs680055 | CG | increased response compared to CC | 152 | 0.013 | 25150845 |
All alleles/genotypes stated as on plus chromosomal strand. The ABCB1 gene is on the minus strand therefore alleles may be shown complementary to in other publications.
CYP1A2
There are no coding sequence variants of CYP1A2 associated with clozapine metabolism, likely due to very low frequencies in most populations (reviewed in [71] and summarized at PharmGKB in the CYP1A2 VIP summary). The most commonly studied variants are in the upstream promoter region : CYP1A2: (−163)C>A (rs762551), CYP1A2: (−3860)G>A (rs2069514) and CYP1A2: (−729)C>T (rs12720461) which form the haplotypes *1F, *1C, *1D and *1K [71]. Since there has been some historical confusion about which variant at −164 (rs762551) represents the *1F allele [71], the specific variants are given where reported. The haplotype *1F is considered an inducible variant that has high activity, haplotypes *1C, *1D and *1K are considered low activity [71].
In patients receiving clozapine, CYP1A2*1F has been associated with treatment non-response requiring higher doses [17–19,72]. Homozygotes of CYP1A2 *1F (rs762551 AA) who are smokers have lower plasma clozapine, higher metabolite concentrations and faster clearance than non-smokers. In a case study of 4 non-responders to clozapine, who were all heavy smokers (30 or more cigarettes/day), found all were homozygous *1F (described as −164C > A, all were AA)[17]. All of the patients experienced a marked or a moderate improvement of their clinical state after the increase of clozapine plasma levels above the therapeutic threshold either by increased of clozapine doses to very high values or by the introduction of fluvoxamine [17]. Another case report of two patients ceasing smoking and experiencing severe adverse effects: one who smoked more than 40 cigarettes/day and quit abruptly and had extremely high plasma clozapine and adverse effects; one who reported smoking 3 or 4 cigarettes/day who was hospitalized and not allowed to smoke and had many co-medications. Both patients were homozygous CYP1A2*1F and defined specifically as −164AA [19]. Additionally, a study of 58 patients with schizophrenia receiving clozapine found decreased concentration dose (C/D) ratios in smokers with CYP1A2*1F (AA) but this was not significant [18]. Non-smokers who were *1F hetero or homozygotes had increased C/D ratios. The smokers were described as smoking at least 15 cigarettes/day but the range was not reported [18,73].
The risk for seizures with patients receiving clozapine tends to be at low doses (< 300 mg/d) during the titration phase, and at high doses (> or = 600 mg/d) during the maintenance phase [74]. The homozygous CYP1A2*1F genotype is associated with increased risk for seizures in patients receiving clozapine (n=108)[75], perhaps reflecting the association CYP1A2*1F has with higher doses.
Patients with low expression alleles of CYP1A2 were more likely to have adverse events related to increased plasma clozapine, decreased metabolites and decreased clearance. A case study of three individuals with clozapine intolerance and tardive dyskinesia (TD) on low/normal dose with no other confounding drugs found their genotypes to be CYP1A2*1C (2 heterozygous, 1 homozygous and all without *1F) which is a low expression allele. The CYP1A2-mRNA expression levels in these patients’ lymphocytes were less than 1/30 of those found in controls [20]. Another case series of Schizophrenia hospital inpatients where a portion were receiving clozapine (n=18 out of 209), and found low activity CYP1A2 (no inducers CBZ, VPA or smoking, CYP1A2*1D delT or CYP1A2*1F C) was associated with decreased clinical outcomes as measured by PSP-P and CGI-2 scores [21].
In general, PGx studies of clozapine and BMI and metabolic syndrome-related side effects have focused on PD genes and not included measurements of PK gene variants. There is some variability of opinion about if metabolic side effects are dose dependent [76]. One small study (n=17) showed that low activity variants CYP1A2 *1C and *1D were associated with higher serum clozapine concentrations and an increased risk of developing insulin and lipid elevations and insulin resistance on a given dose of clozapine [77].
CYP3A
Low CYP3A4 expression (as measured by leukocyte mRNA levels) is associated with high serum clozapine [7]. In those low expressors of CYP3A4, the expression of CYP3A5 ie. CYP3A5*1 resulted in decreased serum clozapine compared to those who were CYP3A5*3 homozygotes and did not express functional CYP3A5 [7]. Another study showed no effect of CYP3A5 with CYP3A4 contributing only in patients with reduced CYP1A2 activity [36].
CYP3A43 is another gene in the CYP3A locus [78] and although its exact involvement in the clozapine pathway is as yet unclear, variants rs680055 and rs472660 in CYP3A43 are associated with increased response to clozapine [8].
CYP2D6
An early study of patients receiving clozapine showed no association between PM and EM genotypes and response [79]. Several other studies concluded CYP2D6 variants are not significantly associated with clozapine/N-desmethylclozapine ratio or metabolic effects [36,77,79,80]. The FDA drug label does not give references supporting the inclusion of wording about CYP2D6 variants.
CYP2C19
One study of patients receiving clozapine showed that the CYP2C19*17/*17 genotype, who are considered ultrarapid metabolizers, had higher serum N-desmethylclozapine, was associated with lower prevalence of diabetes, and had increased improvement of their Schizophrenia symptoms compared to CYP2C19*1/*1 [15]. In another study, the poor metabolizer genotype CYP2C19*2/*2 was associated with higher serum clozapine [36]. CYP2C19*17/*17 was not associated with significant changes in serum clozapine or N-desmethylclozapine in the full cohort but was significant in small group not receiving fluvoxamine [36]. In a large study of 91 patients, neither *2 or *17 was associated with alterations in serum clozapine [6].
Transporter variants
A few studies have examined variants in transporters although they have not been reproduced. The ABCB1 variant rs1045642 AA genotype (3435G>A) is associated with increased plasma clozapine [36]. The same genotype of ABCB1 rs1045642 was also associated with agranulocytosis and neutropenia [81] but another study found that this genotype, when part of a haplotype of ABCB1, might be protective against agranulocytosis [82]. In general clozapine induced agranulocytosis is not thought to be dose dependent but increased drug concentrations specifically in leukocytes could be a factor and may be influenced by genetic variants. Variants rs1045642G in ABCB1 and rs212090T in ABCC1 were associated with increased weight gain and risk for hypertension in men but not women treated with clozapine [37].
ABCB1 rs7787082 G and rs10248420 A alleles were more frequently observed in nonresponders to clozapine [83]. Variant rs2231142 in ABCG2 is associated with increased dose-adjusted trough concentrations of clozapine [1].
Conclusions
We have presented a short summary of the candidates involved in clozapine metabolism and their involvement in DDIs and PGx responses. There is still a large part of PK variability for clozapine that is not adequately explained. The evidence from DDI experiments in vitro and in vivo does not align perfectly [13,61–63]. There are many possible reasons for this including differences in metabolism in different cell types, consideration of different metabolites and pathway branches and PGx influences. The in vitro experiments were done with expressed proteins or liver microsomes and whereas in vivo studies may involve many cell and organ systems. In vitro systems do not allow for the transcriptional modulation of CYP1A2 or CYP3A4 which can be a big part of DDIs.
Both types of DDI studies also mostly focused on the parent drug and its two main serum metabolites, N-desmethylclozapine and clozapine n-oxide, and little is known about the relevance of minor metabolites and pathways downstream of N-desmethylclozapine [6,66]. Very few studies considered a what the implication of a PGx variant might be on a DDI.
At present the PGx evidence is very scattered and few associations have been replicated but CYP1A2, CYP2C19 and CYP3A family genes are all good candidates. The evidence for CYP2D6 variation on clozapine PGx is lacking, although the DDI studies show that CYP2D6 is involved in clozapine PK and there may yet be a role in generation of minor metabolites [3,65,29,69]. Larger in vivo studies are needed that include mechanisms that quantify the influence of environmental effects of caffeine and smoking as well as multiple PGx candidates. While one small study concluded the effect of smoking on clozapine/N-desmethylclozapine plasma concentrations was not related to the number of cigarettes smoked per day (>20, 11–20, 6–10, <=5), this study only examined 45 smokers [36]. A more recent study combining caffeine metabolism and smoking data for 863 healthy individuals, including 385 smokers, to model clozapine metabolism suggests the number of cigarettes is related to clearance. The model was able to predict clozapine clearance in agreement with published data for ranges of cigarettes per day (>20, 11–20, 6–10, <=5)[84]. Difficulties in generating such a model involve factoring the induction coffee consumption may have on CYP1A2 expression [85]. Passive smoking may also need to be considered as there is evidence that it influences phenacetin metabolism, another substrate of CYP1A2. Passive smokers had an intermediate metabolism of phenacetin between that of smokers and non-smokers [86]. In addition, changes in housing status of a patient should be considered in any predictive model since changes from home-based care, hospital emergency care and in-patient facility care can affect smoking behavior.
The ability to predict the size of effects from a change in care arrangement may aid in avoiding adverse events. Other factors to consider are other inducer/inhibitors including environmental toxins (which may be encountered if patient is in unstable housing situation), prescription drugs, drugs of abuse, alternative medicines, alcohol, and diet. There is evidence of inflammation induced downregulation of CPY1A2 and CYP3A4 resulting in phenoconversion from a EM genotype to a PM phenotype [25].
More data is needed on the contribution of CYP3A5 and CYP3A43. While genetic variants appear to have a limited effect on CYP3A4, there are variants that have large effects on CYP3A5 and CYP3A43 [7,8]. In addition the impact of transporter variation both hepatically and at the blood-brain barrier needs more consistent examination. The combination of all these factors into an algorithm along with those for PD candidate genes is the ultimate aim.
Acknowledgements
PharmGKB is supported by the NIH/NIGMS (R24GM61374).
Footnotes
Conflict of interest:
RBA is a stockholder in Personalis Inc. and 23andMe, and a paid advisor for Karius.
References
- 1.Akamine Y, Sugawara-Kikuchi Y, Uno T, Shimizu T, Miura M. Quantification of the steady-state plasma concentrations of clozapine and N-desmethylclozapine in Japanese patients with schizophrenia using a novel HPLC method and the effects of CYPs and ABC transporters polymorphisms. Ann Clin Biochem. 2017; 54 (6):677–685. [DOI] [PubMed] [Google Scholar]
- 2.Roth BD,J PDSP Ki Database”. Psychoactive Drug Screening Program (PDSP). University of North Carolina at Chapel Hill and the United States National Institute of Mental Health. Available at: https://pdsp.unc.edu/databases/pdsp.php?knowID=0&kiKey=&receptorDD=&receptor=&speciesDD=&species=&sourcesDD=&source=&hotLigandDD=&hotLigand=&testLigandDD=&testFreeRadio=testFreeRadio&testLigand=clozapine&referenceDD=&reference=&KiGreater=&KiLess=&kiAllRadio=all&doQuery=Submit+Query [Accessed February 2018.
- 3.Pirmohamed M, Williams D, Madden S, Templeton E, Park BK. Metabolism and bioactivation of clozapine by human liver in vitro. J Pharmacol Exp Ther. 1995; 272 (3):984–990. [PubMed] [Google Scholar]
- 4.Wagmann L, Meyer MR, Maurer HH. What is the contribution of human FMO3 in the N-oxygenation of selected therapeutic drugs and drugs of abuse? Toxicol Lett. 2016; 258:55–70. [DOI] [PubMed] [Google Scholar]
- 5.Zhang WV, D’Esposito F, Edwards RJ, Ramzan I, Murray M. Interindividual variation in relative CYP1A2/3A4 phenotype influences susceptibility of clozapine oxidation to cytochrome P450-specific inhibition in human hepatic microsomes. Drug Metab Dispos. 2008; 36 (12):2547–2555. [DOI] [PubMed] [Google Scholar]
- 6.Dragovic S, Gunness P, Ingelman-Sundberg M, Vermeulen NP, Commandeur JN. Characterization of human cytochrome P450s involved in the bioactivation of clozapine. Drug Metab Dispos. 2013; 41 (3):651–658. [DOI] [PubMed] [Google Scholar]
- 7.Toth K, Csukly G, Sirok D, Belic A, Kiss A, Hafra E, et al. Potential Role of Patients’ CYP3A-Status in Clozapine Pharmacokinetics. Int J Neuropsychopharmacol. 2017; 20 (7):529–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brandl EJ, Chowdhury NI, Tiwari AK, Lett TA, Meltzer HY, Kennedy JL, et al. Genetic variation in CYP3A43 is associated with response to antipsychotic medication. J Neural Transm (Vienna). 2015; 122 (1):29–34. [DOI] [PubMed] [Google Scholar]
- 9.Mendoza MC, Lindenmayer JP. N-desmethylclozapine: is there evidence for its antipsychotic potential? Clin Neuropharmacol. 2009; 32 (3):154–157. [DOI] [PubMed] [Google Scholar]
- 10.Humbert-Claude M, Davenas E, Gbahou F, Vincent L, Arrang JM. Involvement of histamine receptors in the atypical antipsychotic profile of clozapine: a reassessment in vitro and in vivo. Psychopharmacology (Berl). 2012; 220 (1):225–241. [DOI] [PubMed] [Google Scholar]
- 11.Rajji TK, Mulsant BH, Davies S, Kalache SM, Tsoutsoulas C, Pollock BG, et al. Prediction of working memory performance in schizophrenia by plasma ratio of clozapine to N-desmethylclozapine. Am J Psychiatry. 2015; 172 (6):579–585. [DOI] [PubMed] [Google Scholar]
- 12.Lameh J, Burstein ES, Taylor E, Weiner DM, Vanover KE, Bonhaus DW. Pharmacology of N-desmethylclozapine. Pharmacol Ther. 2007; 115 (2):223–231. [DOI] [PubMed] [Google Scholar]
- 13.Molins C, Carceller-Sindreu M, Navarro H, Carmona C, Pineiro M, Martinez E, et al. Plasma ratio of clozapine to N-desmethylclozapine can predict cognitive performance in treatment-resistant psychotic patients. Psychiatry Res. 2017; 258:153–157. [DOI] [PubMed] [Google Scholar]
- 14.Murray M Role of CYP pharmacogenetics and drug-drug interactions in the efficacy and safety of atypical and other antipsychotic agents. J Pharm Pharmacol. 2006; 58 (7):871–885. [DOI] [PubMed] [Google Scholar]
- 15.Piatkov I, Caetano D, Assur Y, Lau SL, Coelho M, Jones T, et al. CYP2C19*17 protects against metabolic complications of clozapine treatment. World J Biol Psychiatry. 2017; 18 (7):521–527. [DOI] [PubMed] [Google Scholar]
- 16.Schaber G, Stevens I, Gaertner HJ, Dietz K, Breyer-Pfaff U. Pharmacokinetics of clozapine and its metabolites in psychiatric patients: plasma protein binding and renal clearance. Br J Clin Pharmacol. 1998; 46 (5):453–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eap CB, Bender S, Jaquenoud Sirot E, Cucchia G, Jonzier-Perey M, Baumann P, et al. Nonresponse to clozapine and ultrarapid CYP1A2 activity: clinical data and analysis of CYP1A2 gene. J Clin Psychopharmacol. 2004; 24 (2):214–219. [DOI] [PubMed] [Google Scholar]
- 18.Kootstra-Ros JE, Smallegoor W, van der Weide J. The cytochrome P450 CYP1A2 genetic polymorphisms *1F and *1D do not affect clozapine clearance in a group of schizophrenic patients. Ann Clin Biochem. 2005; 42 (Pt 3):216–219. [DOI] [PubMed] [Google Scholar]
- 19.Bondolfi G, Morel F, Crettol S, Rachid F, Baumann P, Eap CB. Increased clozapine plasma concentrations and side effects induced by smoking cessation in 2 CYP1A2 genotyped patients. Ther Drug Monit. 2005; 27 (4):539–543. [DOI] [PubMed] [Google Scholar]
- 20.Bolla E, Bortolaso P, Ferrari M, Poloni N, Callegari C, Marino F, et al. Are CYP1A2*1F and *1C associated with clozapine tolerability?: a preliminary investigation. Psychiatry Res. 2011; 189 (3):483. [DOI] [PubMed] [Google Scholar]
- 21.Czerwensky F, Leucht S, Steimer W. CYP1A2*1D and *1F polymorphisms have a significant impact on olanzapine serum concentrations. Ther Drug Monit. 2015; 37 (2):152–160. [DOI] [PubMed] [Google Scholar]
- 22.Ozdemir V, Kalow W, Posner P, Collins EJ, Kennedy JL, Tang BK, et al. CYP1A2 activity as measured by a caffeine test predicts clozapine and active metabolite steady-state concentrationin patients with schizophrenia. J Clin Psychopharmacol. 2001; 21 (4):398–407. [DOI] [PubMed] [Google Scholar]
- 23.Meyer JM. Individual changes in clozapine levels after smoking cessation: results and a predictive model. J Clin Psychopharmacol. 2001; 21 (6):569–574. [DOI] [PubMed] [Google Scholar]
- 24.Meyer JM, Proctor G, Cummings MA, Dardashti LJ, Stahl SM. Ciprofloxacin and Clozapine: A Potentially Fatal but Underappreciated Interaction. Case Rep Psychiatry. 2016; 2016:5606098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shah RR, Smith RL. Inflammation-induced phenoconversion of polymorphic drug metabolizing enzymes: hypothesis with implications for personalized medicine. Drug Metab Dispos. 2015; 43 (3):400–410. [DOI] [PubMed] [Google Scholar]
- 26.Lu ML, Lane HY, Chen KP, Jann MW, Su MH, Chang WH. Fluvoxamine reduces the clozapine dosage needed in refractory schizophrenic patients. J Clin Psychiatry. 2000; 61 (8):594–599. [DOI] [PubMed] [Google Scholar]
- 27.Perry PJ, Bever KA, Arndt S, Combs MD. Relationship between patient variables and plasma clozapine concentrations: a dosing nomogram. Biol Psychiatry. 1998; 44 (8):733–738. [DOI] [PubMed] [Google Scholar]
- 28.Haring C, Meise U, Humpel C, Saria A, Fleischhacker WW, Hinterhuber H. Dose-related plasma levels of clozapine: influence of smoking behaviour, sex and age. Psychopharmacology (Berl). 1989; 99 Suppl:S38–40. [DOI] [PubMed] [Google Scholar]
- 29.Centorrino F, Baldessarini RJ, Kando JC, Frankenburg FR, Volpicelli SA, Flood JG. Clozapine and metabolites: concentrations in serum and clinical findings during treatment of chronically psychotic patients. J Clin Psychopharmacol. 1994; 14 (2):119–125. [PubMed] [Google Scholar]
- 30.Westin AA, Brekke M, Molden E, Skogvoll E, Castberg I, Spigset O. Treatment With Antipsychotics in Pregnancy: Changes in Drug Disposition. Clin Pharmacol Ther. 2018; 103 (3):477–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park HS, Kim E, Moon BS, Lim NH, Lee BC, Kim SE. In Vivo Tissue Pharmacokinetics of Carbon-11-Labeled Clozapine in Healthy Volunteers: A Positron Emission Tomography Study. CPT Pharmacometrics Syst Pharmacol. 2015; 4 (5):305–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lane HY, Jann MW, Chang YC, Chiu CC, Huang MC, Lee SH, et al. Repeated ingestion of grapefruit juice does not alter clozapine’s steady-state plasma levels, effectiveness, and tolerability. J Clin Psychiatry. 2001; 62 (10):812–817. [DOI] [PubMed] [Google Scholar]
- 33.Lane HY, Chiu CC, Kazmi Y, Desai H, Lam YW, Jann MW, et al. Lack of CYP3A4 inhibition by grapefruit juice and ketoconazole upon clozapine administration in vivo. Drug Metabol Drug Interact. 2001; 18 (3–4):263–278. [DOI] [PubMed] [Google Scholar]
- 34.Haenisch B, Drescher E, Thiemer L, Xin H, Giros B, Gautron S, et al. Interaction of antidepressant and antipsychotic drugs with the human organic cation transporters hOCT1, hOCT2 and hOCT3. Naunyn Schmiedebergs Arch Pharmacol. 2012; 385 (10):1017–1023. [DOI] [PubMed] [Google Scholar]
- 35.El Ela AA, Hartter S Schmitt, U. et al. Identification of P-glycoprotein substrates and inhibitors among psychoactive compounds--implications for pharmacokinetics of selected substrates. J Pharm Pharmacol. 2004; 56 (8): 967–75. [DOI] [PubMed] [Google Scholar]
- 36.Jaquenoud Sirot E, Knezevic B, Morena GP, Harenberg S, Oneda B, Crettol S, et al. ABCB1 and cytochrome P450 polymorphisms: clinical pharmacogenetics of clozapine. J Clin Psychopharmacol. 2009; 29 (4):319–326. [DOI] [PubMed] [Google Scholar]
- 37.Piatkov I, Caetano D, Assur Y, Lau SL, Jones T, Boyages SC, et al. ABCB1 and ABCC1 single-nucleotide polymorphisms in patients treated with clozapine. Pharmgenomics Pers Med. 2017; 10:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moons T, de Roo M, Claes S, Dom G. Relationship between P-glycoprotein and second-generation antipsychotics. Pharmacogenomics. 2011; 12 (8):1193–1211. [DOI] [PubMed] [Google Scholar]
- 39.Wang JS, Zhu HJ, Markowitz JS, Donovan JL, Yuan HJ, Devane CL. Antipsychotic drugs inhibit the function of breast cancer resistance protein. Basic Clin Pharmacol Toxicol. 2008; 103 (4):336–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spina E, de Leon J. Metabolic drug interactions with newer antipsychotics: a comparative review. Basic Clin Pharmacol Toxicol. 2007; 100 (1):4–22. [DOI] [PubMed] [Google Scholar]
- 41.Shuman M, Lee Demler T, Trigoboff E, Opler LA. Hematologic impact of antibiotic administration on patients taking clozapine. Innov Clin Neurosci. 2012; 9 (11–12):18–30. [PMC free article] [PubMed] [Google Scholar]
- 42.Espnes KA, Heimdal KO, Spigset O. A puzzling case of increased serum clozapine levels in a patient with inflammation and infection. Ther Drug Monit. 2012; 34 (5):489–492. [DOI] [PubMed] [Google Scholar]
- 43.Brouwers EE, Sohne M, Kuipers S, van Gorp EC, Schellens JH, Koks CH, et al. Ciprofloxacin strongly inhibits clozapine metabolism: two case reports. Clin Drug Investig. 2009; 29 (1):59–63. [DOI] [PubMed] [Google Scholar]
- 44.Brownlowe K, Sola C. Clozapine toxicity in smoking cessation and with ciprofloxacin. Psychosomatics. 2008; 49 (2):176. [DOI] [PubMed] [Google Scholar]
- 45.Sambhi RS, Puri R, Jones G. Interaction of clozapine and ciprofloxacin: a case report. Eur J Clin Pharmacol. 2007; 63 (9):895–896. [DOI] [PubMed] [Google Scholar]
- 46.Sandson NB, Cozza KL, Armstrong SC, Eckermann G, Fischer BA, Phillips B. Clozapine case series. Psychosomatics. 2007; 48 (2):170–175. [DOI] [PubMed] [Google Scholar]
- 47.Raaska K, Neuvonen PJ. Ciprofloxacin increases serum clozapine and N-desmethylclozapine: a study in patients with schizophrenia. Eur J Clin Pharmacol. 2000; 56 (8):585–589. [DOI] [PubMed] [Google Scholar]
- 48.Whitney Z, Procyshyn RM, Fredrikson DH, Barr AM. Treatment of clozapine-associated weight gain: a systematic review. Eur J Clin Pharmacol. 2015; 71 (4):389–401. [DOI] [PubMed] [Google Scholar]
- 49.Lu ML, Chen TT, Kuo PH, Hsu CC, Chen CH. Effects of adjunctive fluvoxamine on metabolic parameters and psychopathology in clozapine-treated patients with schizophrenia: A 12-week, randomized, double-blind, placebo-controlled study. Schizophr Res. 2017. [DOI] [PubMed] [Google Scholar]
- 50.Legare N, Gregoire CA, De Benedictis L, Dumais A. Increasing the clozapine: norclozapine ratio with co-administration of fluvoxamine to enhance efficacy and minimize side effects of clozapine therapy. Med Hypotheses. 2013; 80 (6):689–691. [DOI] [PubMed] [Google Scholar]
- 51.Murray M, Zhang WV, Edwards RJ. Variation in the Response of Clozapine Biotransformation Pathways in Human Hepatic Microsomes to CYP1A2- and CYP3A4-selective Inhibitors. Basic Clin Pharmacol Toxicol. 2017. [DOI] [PubMed] [Google Scholar]
- 52.Rajkumar AP, Poonkuzhali B, Kuruvilla A, Jacob M, Jacob KS. Clinical predictors of serum clozapine levels in patients with treatment-resistant schizophrenia. Int Clin Psychopharmacol. 2013; 28 (1):50–56. [DOI] [PubMed] [Google Scholar]
- 53.Odom-White A, de Leon J. Clozapine levels and caffeine. J Clin Psychiatry. 1996; 57 (4):175–176. [PubMed] [Google Scholar]
- 54.Dratcu L, Grandison A, McKay G, Bamidele A, Vasudevan V. Clozapine-resistant psychosis, smoking, and caffeine: managing the neglected effects of substances that our patients consume every day. Am J Ther. 2007; 14 (3):314–318. [DOI] [PubMed] [Google Scholar]
- 55.Raaska K, Raitasuo V, Laitila J, Neuvonen PJ. Effect of caffeine-containing versus decaffeinated coffee on serum clozapine concentrations in hospitalised patients. Basic Clin Pharmacol Toxicol. 2004; 94 (1):13–18. [PubMed] [Google Scholar]
- 56.Al Hadithy A, Leeffers E, Bruggeman R. Clozapine levels might be affected by excessive cola consumption. J Clin Psychopharmacol. 2012; 32 (5):717–719. [DOI] [PubMed] [Google Scholar]
- 57.Gabbay V, O’Dowd MA, Mamamtavrishvili M, Asnis GM. C lozapine and oral contraceptives: a possible drug interaction. J Clin Psychopharmacol. 2002; 22 (6):621–622. [DOI] [PubMed] [Google Scholar]
- 58.Cadeddu G, Deidda A, Stochino ME, Velluti N, Burrai C, Del Zompo M. Clozapine toxicity due to a multiple drug interaction: a case report. J Med Case Rep. 2015; 9:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bookholt DE, Bogers JP. Oral contraceptives raise plasma clozapine concentrations. J Clin Psychopharmacol. 2014; 34 (3):389–390. [DOI] [PubMed] [Google Scholar]
- 60.Spina E, de Leon J. Clinically relevant interactions between newer antidepressants and second-generation antipsychotics. Expert Opin Drug Metab Toxicol. 2014; 10 (5):721–746. [DOI] [PubMed] [Google Scholar]
- 61.Centorrino F, Baldessarini RJ, Frankenburg FR, Kando J, Volpicelli SA, Flood JG. Serum levels of clozapine and norclozapine in patients treated with selective serotonin reuptake inhibitors. Am J Psychiatry. 1996; 153 (6):820–822. [DOI] [PubMed] [Google Scholar]
- 62.Anghelescu I, Szegedi A, Schlegel S, Weigmann H, Hiemke C, Wetzel H. Combination treatment with clozapine and paroxetine in schizophrenia: safety and tolerability data from a prospective open clinical trial. Eur Neuropsychopharmacol. 1998; 8 (4):315–320. [DOI] [PubMed] [Google Scholar]
- 63.Wetzel H, Anghelescu I, Szegedi A, Wiesner J, Weigmann H, Harter S, et al. Pharmacokinetic interactions of clozapine with selective serotonin reuptake inhibitors: differential effects of fluvoxamine and paroxetine in a prospective study. J Clin Psychopharmacol. 1998; 18 (1):2–9. [DOI] [PubMed] [Google Scholar]
- 64.Joos AA, Konig F, Frank UG, Kaschka WP, Morike KE, Ewald R. Dose-dependent pharmacokinetic interaction of clozapine and paroxetine in an extensive metabolizer. Pharmacopsychiatry. 1997; 30 (6):266–270. [DOI] [PubMed] [Google Scholar]
- 65.Spina E, Avenoso A, Salemi M, Facciola G, Scordo MG, Ancione M, et al. Plasma concentrations of clozapine and its major metabolites during combined treatment with paroxetine or sertraline. Pharmacopsychiatry. 2000; 33 (6):213–217. [DOI] [PubMed] [Google Scholar]
- 66.Fischer V, Vogels B, Maurer G, Tynes RE. The antipsychotic clozapine is metabolized by the polymorphic human microsomal and recombinant cytochrome P450 2D6. J Pharmacol Exp Ther. 1992; 260 (3):1355–1360. [PubMed] [Google Scholar]
- 67.Yousra H, Pierrick L, Laurent L, Daniele D. Interaction between clozapine and oxcarbazepine: a case report. Ther Adv Psychopharmacol. 2017; 7 (2):95–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sugiyama I, Murayama N, Kuroki A, Kota J, Iwano S, Yamazaki H, et al. Evaluation of cytochrome P450 inductions by anti-epileptic drug oxcarbazepine, 10-hydroxyoxcarbazepine, and carbamazepine using human hepatocytes and HepaRG cells. Xenobiotica. 2016; 46 (9):765–774. [DOI] [PubMed] [Google Scholar]
- 69.Wicinski M, Weclewicz MM, Mietkiewicz M, Malinowski B, Grzesk E, Klonowska J. Potential Mechanisms of Hematological Adverse Drug Reactions in Patients Receiving Clozapine in Combination With Proton Pump Inhibitors. J Psychiatr Pract. 2017; 23 (2):114–120. [DOI] [PubMed] [Google Scholar]
- 70.Giri P, Naidu S, Patel N, Patel H, Srinivas NR. Evaluation of In Vitro Cytochrome P450 Inhibition and In Vitro Fate of Structurally Diverse N-Oxide Metabolites: Case Studies with Clozapine, Levofloxacin, Roflumilast, Voriconazole and Zopiclone. Eur J Drug Metab Pharmacokinet. 2017; 42 (4):677–688. [DOI] [PubMed] [Google Scholar]
- 71.Thorn CF, Aklillu E, Klein TE, Altman RB. PharmGKB summary: very important pharmacogene information for CYP1A2. Pharmacogenet Genomics. 2012; 22 (1):73–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Maric NP, Nikolic SP, Buzadzic I, Jovicic M, Andric S, Mihaljevic M, et al. >>Treatment Resistance<< Enigma Resolved by Pharmacogenomics - A Case Study of Clozapine Therapy in Schizophrenia. J Med Biochem. 2015; 34 (2):223–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.van der Weide J, Steijns LS, van Weelden MJ. The effect of smoking and cytochrome P450 CYP1A2 genetic polymorphism on clozapine clearance and dose requirement. Pharmacogenetics. 2003; 13 (3):169–172. [DOI] [PubMed] [Google Scholar]
- 74.Pacia SV, Devinsky O. Clozapine-related seizures: experience with 5,629 patients. Neurology. 1994; 44 (12):2247–2249. [DOI] [PubMed] [Google Scholar]
- 75.Kohlrausch FB, Severino-Gama C, Lobato MI, Belmonte-de-Abreu P, Carracedo A, Hutz MH. The CYP1A2 −163C>A polymorphism is associated with clozapine-induced generalized tonic-clonic seizures in Brazilian schizophrenia patients. Psychiatry Res. 2013; 209 (2):242–245. [DOI] [PubMed] [Google Scholar]
- 76.Sriretnakumar V, Huang E, Muller DJ. Pharmacogenetics of clozapine treatment response and side-effects in schizophrenia: an update. Expert Opin Drug Metab Toxicol. 2015; 11 (11):1709–1731. [DOI] [PubMed] [Google Scholar]
- 77.Melkersson KI, Scordo MG, Gunes A, Dahl ML. Impact of CYP1A2 and CYP2D6 polymorphisms on drug metabolism and on insulin and lipid elevations and insulin resistance in clozapine-treated patients. J Clin Psychiatry. 2007; 68 (5):697–704. [DOI] [PubMed] [Google Scholar]
- 78.Chen X, Wang H, Zhou G, Zhang X, Dong X, Zhi L, et al. Molecular population genetics of human CYP3A locus: signatures of positive selection and implications for evolutionary environmental medicine. Environ Health Perspect. 2009; 117 (10):1541–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Arranz MJ, Dawson E, Shaikh S, Sham P, Sharma T, Aitchison K, et al. Cytochrome P4502D6 genotype does not determine response to clozapine. Br J Clin Pharmacol. 1995; 39 (4):417–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dahl ML, Llerena A, Bondesson U, Lindstrom L, Bertilsson L. Disposition of clozapine in man: lack of association with debrisoquine and S-mephenytoin hydroxylation polymorphisms. Br J Clin Pharmacol. 1994; 37 (1):71–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.van der Weide K, Loovers H, Pondman K, Bogers J, van der Straaten T, Langemeijer E, et al. Genetic risk factors for clozapine-induced neutropenia and agranulocytosis in a Dutch psychiatric population. Pharmacogenomics J. 2017; 17 (5):471–478. [DOI] [PubMed] [Google Scholar]
- 82.Anil Yagcioglu AE, Yoca G, Ayhan Y, Karaca RO, Cevik L, Muderrisoglu A, et al. Relation of the Allelic Variants of Multidrug Resistance Gene to Agranulocytosis Associated With Clozapine. J Clin Psychopharmacol. 2016; 36 (3):257–261. [DOI] [PubMed] [Google Scholar]
- 83.Lee ST, Ryu S, Kim SR, Kim MJ, Kim S, Kim JW, et al. Association study of 27 annotated genes for clozapine pharmacogenetics: validation of preexisting studies and identification of a new candidate gene, ABCB1, for treatment response. J Clin Psychopharmacol. 2012; 32 (4):441–448. [DOI] [PubMed] [Google Scholar]
- 84.Plowchalk DR, Rowland Yeo K. Prediction of drug clearance in a smoking population: modeling the impact of variable cigarette consumption on the induction of CYP1A2. Eur J Clin Pharmacol. 2012; 68 (6):951–960. [DOI] [PubMed] [Google Scholar]
- 85.Djordjevic N, Ghotbi R, Bertilsson L, Jankovic S, Aklillu E. Induction of CYP1A2 by heavy coffee consumption in Serbs and Swedes. Eur J Clin Pharmacol. 2008; 64 (4):381–385. [DOI] [PubMed] [Google Scholar]
- 86.Dong SX, Ping ZZ, Xiao WZ, Shu CC, Bartoli A, Gatti G, et al. Effect of active and passive cigarette smoking on CYP1A2-mediated phenacetin disposition in Chinese subjects. Ther Drug Monit. 1998; 20 (4):371–375. [DOI] [PubMed] [Google Scholar]