Coconut is an important crop for both industry and small stakeholders in many intertropical countries. Phytoplasma-associated lethal yellowing-like diseases have become one of the major pests that limit coconut cultivation as they have emerged in different parts of the world. We developed a multilocus sequence typing scheme (MLST) for tracking epidemics of “Ca. Phytoplasma palmicola,” which is responsible for coconut lethal yellowing disease (CLYD) on the African continent. MLST analysis applied to diseased coconut samples collected in western and eastern African countries also showed the existence of three distinct populations of “Ca. Phytoplasma palmicola” with low intrapopulation diversity. The reasons for the observed strong geographic patterns remain to be established but could result from the lethality of CLYD and the dominance of short-distance insect-mediated transmission.
KEYWORDS: phytoplasma, coconut lethal yellowing disease, multilocus sequence typing
ABSTRACT
To sustain epidemiological studies on coconut lethal yellowing disease (CLYD), a devastating disease in Africa caused by a phytoplasma, we developed a multilocus sequence typing (MLST) scheme for “Candidatus Phytoplasma palmicola” based on eight housekeeping genes. At the continental level, eight different sequence types were identified among 132 “Candidatus Phytoplasma palmicola”-infected coconuts collected in Ghana, Nigeria, and Mozambique, where CLYD epidemics are still very active. “Candidatus Phytoplasma palmicola” appeared to be a bacterium that is subject to strong bottlenecks, reducing the fixation of positively selected beneficial mutations into the bacterial population. This phenomenon, as well as a limited plant host range, might explain the observed country-specific distribution of the eight haplotypes. As an alternative means to increase fitness, bacteria can also undergo genetic exchange; however, no evidence for such recombination events was found for “Candidatus Phytoplasma palmicola.” The implications for CLYD epidemiology and prophylactic control are discussed. The usefulness of seven housekeeping genes to investigate the genetic diversity in the genus “Candidatus Phytoplasma” is underlined.
IMPORTANCE Coconut is an important crop for both industry and small stakeholders in many intertropical countries. Phytoplasma-associated lethal yellowing-like diseases have become one of the major pests that limit coconut cultivation as they have emerged in different parts of the world. We developed a multilocus sequence typing scheme (MLST) for tracking epidemics of “Ca. Phytoplasma palmicola,” which is responsible for coconut lethal yellowing disease (CLYD) on the African continent. MLST analysis applied to diseased coconut samples collected in western and eastern African countries also showed the existence of three distinct populations of “Ca. Phytoplasma palmicola” with low intrapopulation diversity. The reasons for the observed strong geographic patterns remain to be established but could result from the lethality of CLYD and the dominance of short-distance insect-mediated transmission.
INTRODUCTION
Molecular epidemiology requires easy-to-use tools that can sufficiently discriminate at a population level. Both multilocus sequence typing (MLST) and multilocus variable number tandem repeat (VNTR) analysis (MLVA) have been developed to fulfill this objective and then were applied to the surveillance of human, animal, and plant pathogens (1, 2). Next-generation sequencing (NGS) allows the study of bacterial diversity at the genomic level through whole-genome sequencing (WGS). WGS considerably increased the resolution of subpopulations of Staphylococcus aureus (3), Mycobacterium abscessus (4), and Mycoplasma pneumoniae (5). Mycoplasma-like phytoplasmas are bacteria of the class Mollicutes (6). Mycoplasmas are restricted to vertebrate hosts, while phytoplasmas are plant pathogens vectored through circulative-propagative transmission by phloem-feeding insects of the order Hemiptera (7, 8).
The inability to grow phytoplasmas in axenic culture remains a major constraint in developing WGS strategies, as it limits access to adequate amounts of pure phytoplasma DNA. Only six full-chromosome sequences of phytoplasmas have been deciphered to date (9), and genome surveys depend on fastidious and complex molecular strategies (10–13). However, metagenomic approaches through NGS have opened new opportunities, and 17 additional phytoplasma genome draft sequences have been deposited in GenBank (NCBI). Such genome-wide sequence analyses have provided easy access to genetic markers, allowed unbiased definitions of species boundaries in the genus “Candidatus Phytoplasma,” and have revealed the horizontal transfer of potential mobile units and effectors (14, 15).
MLST schemes have been developed for many Mollicutes, almost exclusively Mycoplasma and Ureaplasma, because of their impacts on humans and animals (16–18). For phytoplasmas, most of the MLST schemes that have been developed for “Candidatus Phytoplasma” species or 16S taxonomic groups target rRNAs, housekeeping genes, protein coding genes, and also positively selected genes involved in the interaction with the host (19, 20). A single study following the classical requirement of an MLST scheme that includes at least seven housekeeping genes and targets a small number of samples is currently available (2, 21). Other studies that targeted three to five housekeeping genes have addressed taxonomic or epidemiological questions (22, 23). Nevertheless, primers tend to be species specific most of the time, and MLST schemes are not transferable to other phytoplasma species or taxonomic groups.
Among the numerous diseases caused by phytoplasmas, coconut lethal yellowing diseases (CLYD), also known as lethal yellowing type syndromes (LYTS), are among the most destructive, with significant economic and social impact (24). CLYD are present in Africa, the Caribbean, Central America, and Oceania, where they are associated with different “Ca. Phytoplasma” species according to the area considered (25).
In Africa, CLYD are responsible for the loss of millions of coconut trees since the beginning of the 20th century. In east Africa, CLYD were first described in 1905 in Tanzania (26) and then in Kenya (27). The first description in Mozambique was in 1958 in Cabo Delgado (28), and that in Zambezia was in 1972 (29), growing to an epidemic in the 1990s. In West Africa, CLYD was described for the first time in Nigeria as Awka wilt disease in 1917 (30). From the 1930s, similar diseases were described in the eastern Volta region of Ghana (31), in Togo (32), and in Cameroon (33). In 1964, coconut lethal yellowing was observed in the western region of Ghana (31) and in the 2000s in Ivory Coast (34). Molecular characterization of the phytoplasmas associated with these different syndromes was performed only in a few countries (see Fig. S1 in the supplemental material), and two different “Candidatus Phytoplasma” species described exclusively on the African continent were identified: “Ca. Phytoplasma cocostanzania” in Kenya and Tanzania (35) and North Mozambique (36) and “Ca. Phytoplasma palmicola” in Ghana, Nigeria (37), Ivory Coast (38), and Mozambique (39).
Based on the sequence and restriction fragment length polymorphism (RFLP) pattern of the central part of its 16S rRNA gene, “Ca. Phytoplasma palmicola” is classified in group 16SrXXII (40) and is divided into two subgroups that can be differentiated by HaeIII enzymatic restriction. Subgroup 16SrXXII-A includes “Ca. Phytoplasma palmicola” phytoplasmas from Mozambique and Nigeria, and subgroup 16SrXXII-B corresponds to “Ca. Phytoplasma palmicola” from Ghana and Ivory Coast (37). A few studies have been conducted to differentiate between populations of “Ca. Phytoplasma palmicola” at the national level by using different genes. The sequence of the secA gene differentiated between “Ca. Phytoplasma palmicola” samples from Ivory Coast, Ghana, and Mozambique (41, 42), while the rplV gene was able to distinguish “Ca. Phytoplasma palmicola” isolates from different Ghanaian regions (43).
The propagation of “Ca. Phytoplasma palmicola” depends on its insect vectors, dissemination potential, alternative host plants, and a still unproved seed transmission. In practice, sampling CLYD and insects potentially vectoring the disease is also challenging due to the difficulty in accessing the several-meters-high canopy of coconut trees. Despite years of epidemiological research, the insect vectors are still unidentified. The rationale of this study is based on the assumption that deciphering the genetic structure of “Ca. Phytoplasma palmicola” populations at regional, national, and continental levels and mapping the distribution of its genetic variants will give insights into the origin and pathways of the spread of “Ca. Phytoplasma palmicola.” We therefore developed an eight-gene MLST scheme that follows the general requirements for genotyping bacterial agents and applied this scheme to a set of 132 “Ca. Phytoplasma palmicola” isolates collected from three African countries where active CLYD epidemics had been previously associated with the two “Ca. Phytoplasma palmicola” taxonomic subgroups, 16SrXXII-A and 16SrXXII-B.
RESULTS
Housekeeping gene selection and properties.
Eight housekeeping genes, namely, dnaC, gyrB, leuS, lpd, secA, recA, rsmI, and rplV, were selected to investigate the genetic diversity among 132 samples of “Ca. Phytoplasma palmicola” originating from three of the main African countries affected by this phytoplasma, i.e., Ghana, Nigeria, and Mozambique. The primers designed for this study successfully amplified the respective gene targets for all 132 DNA samples of “Ca. Phytoplasma palmicola” of both the 16SrXXII-A and 16SrXXII-B groups, irrespective of their collection dates or geographic origins. The PCR products observed on agarose gels each showed a unique and clear DNA band from 553 to 983 bp depending on the target gene (Table 1). None of the 1,056 double-strand-sequenced PCR products displayed double peaks or ambiguous bases, thus demonstrating the specificity of the primers and the absence of mixed infections.
TABLE 1.
PCR primer pairs used to amplify each “Ca. Phytoplasma palmicola” MLST locus
| Target gene | Primer | Sequence (5′ to 3′) | Ta (°C)a | Amplicon (bp)b | Reference or source |
|---|---|---|---|---|---|
| lpd | lpd_CPpml-F | AGTTCAATTAGATGTTTGTCCTCGT | 58 | 833 | This study |
| lpd_CPpml-R | TCAGATAAAGTTGGATGAGGATGA | ||||
| dnaC | dnaC_CPpml-F | CTGCTCGTCCTTCTATGGGA | 58 | 670 | This study |
| dnaC_CPpml-R | AGCCACAATTAATTCTATATTACCTG | ||||
| leuS | leuS_CPpml-F | CAGAACAATATGCTTTACAAACAGG | 58 | 783 | This study |
| leuS_CPpml-R | TCACAAGCAGGAACAGACATAA | ||||
| gyrB | gyrB_CPpml-F | TGGAAAAATGTTTGTTAGCTGT | 54 | 732 | This study |
| gyrB_CPpml-R | CGAGCAGTTACTTCTTCGCC | ||||
| secA | secA_CPpml-F | AAAAACCTCAAACCACAACATT | 54 | 750 | This study |
| secA_CPpml-R | TATCAGTACCACGACCAGCC | ||||
| rsmI | rsmI_CPpml-F | ATATATCAGATATTAGTTTTCGAGCT | 54 | 553 | This study |
| rsmI_CPpml-R | TTCACCATGAATAATAGTTTCGAA | ||||
| recA | recA_CPpml-F | TTCCCACTGGTTCTTTGTCTTT | 54 | 832 | This study |
| recA_CPpml-R | ATCAGCTATGTTTGGGTTTTGT | ||||
| rplV and rpsC | rpLYF1 | TTTAAAGAAGGTATTAACATGA | 51 | 983 | 50 |
| rpLYR1 | TAATACCTATAACTCCGTG | ||||
| 16S rRNA gene | P1 | AAGAGTTTGATCCTGGCTCAGGATT | 56 | 1,756 | 48 |
| P7 | CAGAACAATATGCTTTACAAACAGG |
Ta, annealing temperature used in PCRs.
PCR product length in base pairs observed by electrophoresis on agarose gel.
The GC content of “Ca. Phytoplasma palmicola” housekeeping genes (cGC) was between 26.3% (gyrB) and 30.4% (recA) (Table 2). The average cGC for the eight genes was 27.80%.
TABLE 2.
Genetic parameters calculated for each individual locus and concatenated sequences from the “Ca. Phytoplasma palmicola” MLST schemea
| Parameter | Population | n | Value for: |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| lpd | dnaC | leuS | gyrB | secA | rsmI | recA | rplV | Concat | |||
| Length (bp) | 630 | 543 | 657 | 606 | 627 | 417 | 708 | 324 | 4,512 | ||
| cG+C (%) | 29.0 | 26.7 | 27.3 | 26.3 | 26.5 | 27.8 | 30.4 | 28.3 | 27.8 | ||
| Ka/Ks | 0.172 | 0.082 | 0.103 | 0.010 | 0.168 | 0.067 | 0.037 | 0.077 | 0.132 | ||
| H/ST (SS) | GHA | 96 | 3 (2) | 1 (0) | 2 (2) | 1 (0) | 1 (0) | 2 (1) | 1 (0) | 2 (1) | 4 (6) |
| NGA | 4 | 1 (0) | 1 (0) | 1 (0) | 1 (0) | 1 (0) | 1 (0) | 1 (0) | 1 (0) | 1 (0) | |
| MOZ | 32 | 2 (1) | 2 (1) | 2 (1) | 2 (1) | 1 (0) | 2 (3) | 2 (3) | 2 (1) | 3 (9) | |
| AFR | 132 | 6 (36) | 4 (20) | 5 (28) | 4 (25) | 3 (23) | 5 (23) | 4 (36) | 5 (14) | 8 (205) | |
| π | GHA | 96 | 0.0008 | 0.00 | 0.0010 | 0.00 | 0.00 | 0.0005 | 0.00 | 0.0010 | 0.0004 |
| MOZ | 32 | 0.0001 | 0.0001 | 0.0008 | 0.0001 | 0.00 | 0.0002 | 0.0003 | 0.0002 | 0.0002 | |
| AFR | 132 | 0.0213 | 0.0114 | 0.0153 | 0.0147 | 0.0117 | 0.0195 | 0.0167 | 0.0112 | 0.0154 | |
| Hd | GHA | 96 | 0.455 | 0.321 | 0.189 | 0.321 | 0.475 | ||||
| MOZ | 32 | 0.063 | 0.063 | 0.516 | 0.063 | 0.063 | 0.063 | 0.063 | 0.546 | ||
| AFR | 132 | 0.658 | 0.418 | 0.613 | 0.418 | 0.415 | 0.518 | 0.418 | 0.587 | 0.696 | |
| Tajima's D | GHA | 96 | 0.4105 | 1.0039 | –0.0362 | 0.7491 | 0.9207 | ||||
| MOZ | 32 | –1.1424 | –1.1424 | 1.6467 | –1.1424 | –1.1424 | –1.7295 | –1.1424 | –1.6703 | ||
| AFR | 132 | 2.8042c | 1.9326 | 2.6036b | 2.3302b | 2.1193b | 2.6459b | 2.0925b | 1.1031 | 2.5782b | |
| Fu & Li's D | GHA | 96 | 0.6886 | 0.0 | 0.6886 | 0.00 | 0.00 | 0.4949 | 0.00 | 0.4949 | 1.1233 |
| MOZ | 32 | –1.7034 | –1.7034 | 0.5871 | –1.7036 | 0.00 | –1.7034 | –2.7326b | –1.7034 | –3.3707c | |
| AFR | 132 | 2.1032c | 1.7462c | 1.9567c | 1.9168c | 1.8261c | 1.8261c | 1.2041 | 0.9461 | 2.4606c | |
| Fu & Li's F | GHA | 96 | 0.7049 | 0.00 | 0.9183 | 0.00 | 0.00 | 0.3910 | 0.00 | 0.6651 | 1.2465 |
| MOZ | 32 | –1.7820 | –1.7820 | 1.0149 | –1.7034 | 0.00 | –1.7820 | –2.8304 | –1.7820 | –3.3301 | |
| AFR | 132 | 2.8710 | 2.1773 | 2.6581 | 2.4961 | 2.3260 | 2.5722 | 1.8837 | 1.1993 | 3.0119 | |
Parameters were determined for Ghanaian (GHA), Nigerian (NGA), and Mozambican (MOZ) populations and at the African continent (AFR) level. The lengths of the trimmed gene and concatenated sequences (Concat) in base pairs considered for MLSA and their coding G+C percentages (cG+C) are presented. The synonymous/nonsynonymous substitution (Ka/Ks) ratio was calculated using the MEGA 7.0 program. The number of haplotypes (H) or sequence types (ST) with the number of segregating sites (SS), haplotype diversity (Hd), and nucleotide diversity (π) were calculated using DnaSP 6.1, as well as the neutrality tests using Tajima's D, Fu and Li's D, and Fu and Li's F statistics. Most parameters were not calculated for the Nigerian population (NGA) because of the occurrence of a single haplotype.
P < 0.05.
P < 0.02.
The Ka/Ks values, measuring the ratio of the number of nonsynonymous substitutions per nonsynonymous site (Ka) to the number of synonymous substitutions per synonymous site (Ks), are presented in Table 2. Low Ka/Ks values ranging from 0.01 (gyrB) to 0.17 (lpd) indicated that all the selected genes underwent purifying selection.
Genetic diversity of “Ca. Phytoplasma palmicola.”
The number of haplotypes (H/ST), the number of segregating sites (SS), the nucleotide diversity (π), and the haplotype diversity (Hd) for each housekeeping gene and its concatenated sequence are presented in Table 2 according to the infected-coconut country of origin.
The less discriminating gene, secA, revealed three haplotypes, HsecA1 to HsecA3, each corresponding to a different country of origin. The more discriminating gene, lpd, differentiated into six haplotypes, Hlpd1 to Hlpd6.
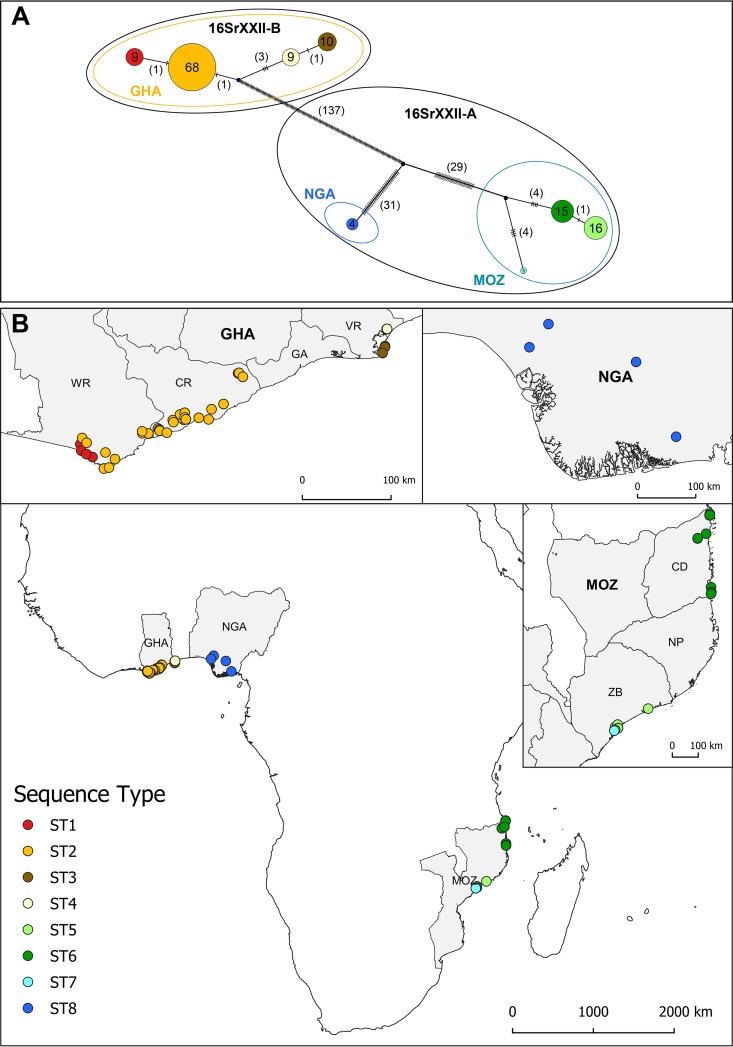
The eight genes were concatenated to obtain a sequence of 4,512 bp for each isolate. With 205 segregating nucleotides, the concatenated sequences allowed the differentiation of eight sequence types (STs) (Table 3; Fig. 1). This combination of housekeeping gene haplotypes showed that four STs are present in Ghana (ST1 to ST4), three different STs in Mozambique (ST5 to ST7), and a unique ST, ST8, in Nigeria (Table 3; Fig. 1). All but ST7 were represented by several isolates.
TABLE 3.
Combination of the 8 housekeeping gene haplotypes allowing discrimination of the 8 STs of “Ca. Phytoplasma palmicola”
| Sequence type | Geographic population | n | Housekeeping gene haplotype |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| H lpd | H dnaC | H leuS | H gyrB | H secA | H rsmI | H recA | H rplV | |||
| ST1 | Ghana | 9 | Hlpd1 | HdnaC1 | HleuS1 | HgyrB1 | HsecA1 | HrsmI1 | HrecA1 | HrplV1 |
| ST2 | Ghana | 68 | Hlpd2 | HdnaC1 | HleuS1 | HgyrB1 | HsecA1 | HrsmI1 | HrecA1 | HrplV1 |
| ST3 | Ghana | 10 | Hlpd3 | HdnaC1 | HleuS2 | HgyrB1 | HsecA1 | HrsmI2 | HrecA1 | HrplV2 |
| ST4 | Ghana | 9 | Hlpd3 | HdnaC1 | HleuS2 | HgyrB1 | HsecA1 | HrsmI1 | HrecA1 | HrplV2 |
| ST5 | Mozambique | 16 | Hlpd4 | HdnaC2 | HleuS3 | HgyrB2 | HsecA2 | HrsmI3 | HrecA2 | HrplV3 |
| ST6 | Mozambique | 15 | Hlpd4 | HdnaC2 | HleuS4 | HgyrB2 | HsecA2 | HrsmI3 | HrecA2 | HrplV3 |
| ST7 | Mozambique | 1 | Hlpd5 | HdnaC3 | HleuS4 | HgyrB3 | HsecA2 | HrsmI4 | HrecA3 | HrplV4 |
| ST8 | Nigeria | 4 | Hlpd6 | HdnaC4 | HleuS5 | HgyrB4 | HsecA3 | HrsmI5 | HrecA4 | HrplV5 |
FIG 1.
(A) Integer neighbor-joining network calculated from eight housekeeping gene (dnaC, gyrB, leuS, lpd, recA, rplV, rsmI, secA) concatenated sequences (4,512 bp) of 132 “Candidatus Phytoplasma palmicola” samples originating from three African countries, Ghana (GHA), Mozambique (MOZ), and Nigeria (NGA). Each circle represents a sequence type (ST). The numbers inside each circle correspond to the numbers of samples presenting identical STs, and the numbers in parentheses represent the numbers of single-nucleotide polymorphism (SNPs) between STs. Small ellipses describe the three different geographic populations. Large ellipses represent the 16Sr RFLP subgroup deduced from 16S rRNA gene sequences. (B) Geographical distribution of the 132 “Ca. Phytoplasma palmicola” samples according to their STs at the African continent level, in Ghana (96 samples), in Mozambique (32 samples), and in Nigeria (4 samples). The Ghanaian coastal regions of western region (WR), central region (CR), greater Accra (GA), and Volta region (VR) and the Mozambican provinces of Cabo Delgado (CD), Nampula (NP), and Zambezia (ZB) are delimited.
The 16S rRNA of 41 samples representing each of the eight STs was sequenced (6 isolates of ST1, 9 of ST2, 4 of ST3, 5 of ST4, 6 of ST5, 6 of ST6, 1 of ST7, 4 of ST8). Two types of sequences were identified. Seventeen samples corresponding to ST5 to ST8 originating from Nigeria and Mozambique were all identical to the 16S rRNA sequence of the LDN strain from Nigeria (accession number Y14175) (44, 45) and were therefore classified as members of subgroup 16SrXXII-A. Twenty-four samples of ST1 to ST4 from Ghana were all identical to the 16S rRNA of CSPW-dna19 and were therefore assigned to subgroup 16SrXXII-B (accession number KF364359) (37). Sequences from subgroups 16SrXXII-A and -B differed by six single-nucleotide polymorphisms (SNPs) over 1,505 bp (99.6% homology).
Geographic distribution of the “Ca. Phytoplasma palmicola” sequence types.
All the DNA samples were spatially referenced, allowing the mapping of the distribution of their corresponding STs. The geographic distribution of the STs illustrates their aggregation in each country (Fig. 1B). Each ST presents a continuous distribution, each constituting a well-defined focus, except for ST7, which was represented by the single isolate MZ12-187. The geographic origins of the 132 samples and their corresponding STs are available (see Data Set S1 in the supplemental material). In central Ghana (CR), ST2 was the most abundant and only type present, while in the western region (WR), it coexisted with ST1, a single SNP variant of ST2 (Fig. 1). ST3 and ST4 constituted two foci in the eastern Volta region of Ghana (VR). In Nigeria, the second western African country surveyed, only ST8 was found, but only four DNA samples were analyzed. In Mozambique in East Africa, ST5 and ST6 were prevalent. ST5 was restricted to the province Zambezia, while ST6 appeared only along the coast of the northern province Cabo Delgado. A unique ST7 sample was also localized to Zambezia. ST7 exhibited 8 or 9 SNPs compared to the two other Mozambican sequence types, ST5 and ST6 (Fig. 1A).
Population structure of “Ca. Phytoplasma palmicola.”
Tajima's D and Fu and Li's D and F were calculated for each housekeeping gene, and the concatenated sequences were calculated for each country and at the African continent level (Table 2). Tajima's D was positive but not significant for the concatenated sequences and sequences of all the genes except rsmI from Ghana. Even though they were not significant, the calculated Tajima's D values were negative using most of the Mozambican sequences, probably because ST7 was represented by a unique sample. With the exception of dnaC and rplV, all Tajima's D and Fu and Li's D and F values were significantly positive at the continental level, suggesting bottleneck effects.
An integer neighbor-joining network was calculated using the concatenated sequences of the 132 isolates (Fig. 1A). Of over 205 segregating sites, 202 were parsimony informative. The structure of the network illustrates both strong regional differentiation and the clonal structure of the “Ca. Phytoplasma palmicola” populations. From the network, we observed three distinct populations. Multilocus sequence analysis (MLSA)-concatenated sequences of samples from Ghana are similarly distant from the closer Nigerian and Mozambican genotypes, from which they differed by 169 (3.75%) and 171 (3.79%) SNPs, respectively. While from the same 16SrXXII-A subgroup, Mozambican and Nigerian samples differed by at least 64 SNPs (1.42%). The within-country genetic diversity was low, with no SNPs for Nigeria, 6 SNPs for Ghana, and 9 SNPs for Mozambique.
Within the 16Sr subgroups, the genetic diversity was higher for subgroup 16SrXXII-A than for subgroup 16SrXXII-B. In detail, the STs of subgroup 16srXXII-B split into two divergent branches (Fig. 1A). Only 1 SNP differentiated STs within the same subregion, whereas interregional diversity ranged from 4 to 6 SNPs. Within subgroup 16SrXXII-A, the two main STs, ST5 and ST6, differed by only one mutation, while ST7 differed by eight mutations compared to the more closely related ST6 in Mozambique. In this subgroup, the Nigerian ST8 differed from the Mozambican STs by a minimum of 64 SNPs but was, however, more distant from the Ghanaian STs of the other subgroup by a minimum of 138 SNPs.
Absence of recombination among “Ca. Phytoplasma palmicola” sequence types.
Split networks were calculated from “Ca. Phytoplasma palmicola” STs for each housekeeping gene (Fig. S2). The split-graph structures observed for lpd and rsmI did not appear to exclude recombination phenomena, and phi test results with P values of 0.3405 and 0.1417, respectively, were not significant irrespective of the gene being considered. The phi test result calculated for the concatenated sequences was also not significant (P = 0.0748), and the Recombination Detection Program (RDP; http://web.cbio.uct.ac.za/~darren/rdp.html) did not identify any recombination signals.
Interspecific comparison using the developed scheme.
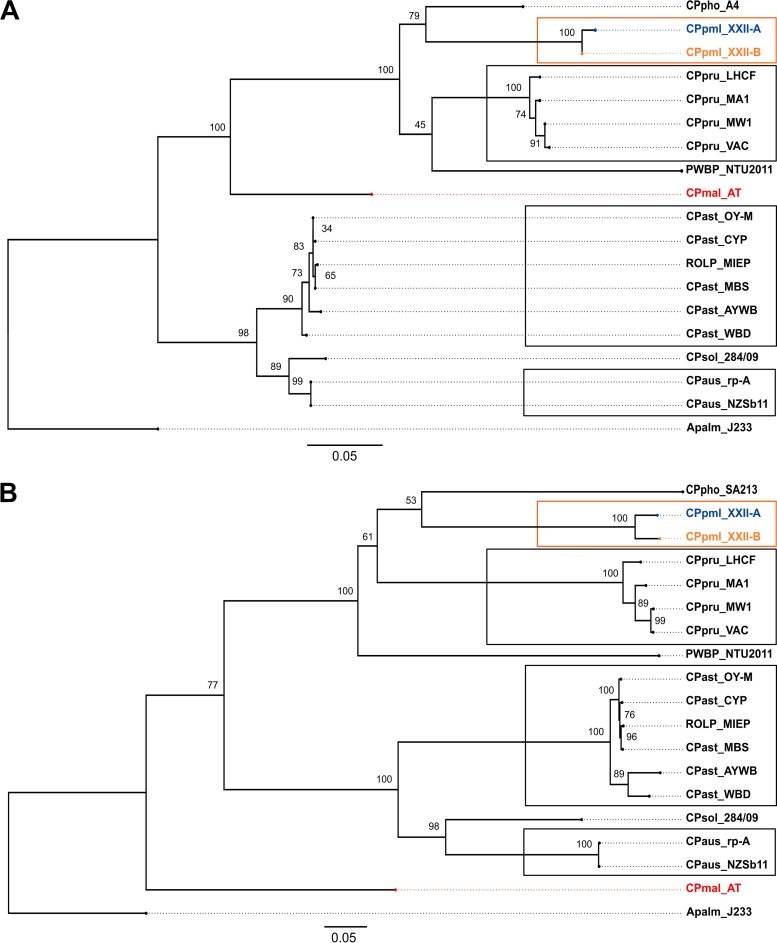
Seven housekeeping genes (dnaC, gyrB, leuS, lpd, secA, rsmI, and rplV) and 16S rRNA gene were found for 16 of 23 phytoplasma genomes or draft genomes. The 16S rRNA sequences of the corresponding phytoplasmas were used to construct a maximum likelihood (ML) phylogenetic tree after removal of a recombinant region of 155 bp. The ML phylogenetic tree presents a topology apparently similar to that of the housekeeping genes but increased the strength of the phylogenetic tree (Fig. 2). However, the congruency index (Icong) of 1.1039 was not significant (P = 0.35), indicating that the two calculated trees were not more congruent than what is expected by chance (46).
FIG 2.
Phylogenetic relationship of 18 Candidatus phytoplasmas. A rooted maximum likelihood tree was calculated for the 16S rRNA gene sequences (A) and for 3,756-bp concatenated DNA sequences of the dnaC, leuS, gyrB, rsmI, lpd, secA, and rplV housekeeping genes with the recombinant region removed (B). The codes used are as follows: CPmal_AT (“Ca. Phytoplasma mali” strain AT [CU469464.1]), CPaus_rp-A (“Ca. Phytoplasma australiense” [NC_010544.1]), CPast_OY-M (“Ca. Phytoplasma asteris” strain onion yellows phytoplasma OY-M [NC_005303.2]), CPpru_MW1 (“Ca. Phytoplasma pruni” strain milkweed yellows-MW1 [AKIL00000000.1]). CPpru_MA1 (“Ca. Phytoplasma pruni” strain Italian clover phyllody MA1 [AKIM00000000.1]), CPpru_VAC (“Ca. Phytoplasma pruni” strain vaccinium witches'-broom VAC [AKIN00000000.1]), PWBP_NTU2011 (peanut witches'-broom phytoplasma NTU2011 [AMWZ00000000.1]), CPaus_NZSb11 (“Ca. Phytoplasma australiense” strain strawberry lethal yellows phytoplasma NZSb11 [NC_021236.1]), CPsol_284/09 (“Ca. Phytoplasma solani” strain 284/09 [FO393427.1]), CPast_WBD (“Ca. Phytoplasma asteris” strain wheat blue dwarf [AVAO00000000.1]), CPpru_LHCF (“Ca. Phytoplasma pruni” strain CX [LHCF00000000.1]), and CPast_CYP (“Ca. Phytoplasma asteris” chrysanthemum yellows strain CYP [JSWH00000000.1]). CPast_AYWB (“Ca. Phytoplasma asteris” strain aster yellows witches broom [NC_007716.1]), CPpho_SA213 (“Ca. Phytoplasma phoenicum” [JPSQ00000000.1]) without recA, and CPho_A4 (AF515636), CPast_MBS (“Ca. Phytoplasma asteris” strain maize bushy stunt [NZ_CP015149.1]), and ROLP_MIEP (rice orange leaf phytoplasma LD1 [MIEP00000000.1]). Apalm_J233 (Acholeplasma palmae [NC_022538]) was used as an outgroup. “Candidatus Phytoplasma palmicola” is represented by one isolate (MZ11-005) from the 16SrXXII-A subgroup CPpml_XXII-A and by one isolate (GH04-009) from the 16SrXXII-B subgroup CPpml_XXII-B. Node values represent bootstrap test results with 500 replicates.
DISCUSSION
Multilocus sequence analysis highlights a genetic structure congruent with 16S rRNA subgroups.
To date, molecular characterization of “Ca. Phytoplasma palmicola” has been based mainly on the amplification of the 16S-23S rRNA gene operon by using the universal phytoplasma 16S rRNA gene primers, P1/P7 (47, 48), or specific primers (49), or, alternatively, on the amplification of the ribosomal protein gene (50) or the secA gene (51, 52). The MLST scheme we report is based on eight housekeeping genes and allows the deciphering of “Ca. Phytoplasma palmicola” genetic diversity as it could be applied to all CLYD-infected coconut samples from the different countries of origin and sampling dates. The protocol has been optimized to avoid a nested-PCR step, thereby reducing the risk of false positives. Because of their specificity for “Ca. Phytoplasma palmicola” and their efficiency in amplifying our entire sample set, each of the seven primer pairs developed in this study could be used independently to confirm “Ca. Phytoplasma palmicola” diagnostically.
Intraspecific diversity among Mollicutes observed by MLST is variable, from very low for Mycoplasma mycoides subsp. mycoides (53, 54) to high for Mycoplasma agalactiae and Mycoplasma bovis (18). Even though MLST is available for some phytoplasmas, it has not yet been applied on a large scale for phytoplasmas (55, 56). Although the present MLSA scheme allowed the differentiation of only 8 STs of “Ca. Phytoplasma palmicola,” it clearly differentiated three distinct populations distributed into two existing 16SrXXII subgroups. The current division of “Ca. Phytoplasma palmicola” into two distinct 16SrXXII subgroups (37) is consistent with the MLSA data, because the two distinct subgroups correspond to the lowest similarity value of the concatenated sequence of 96.21 to 96.25%, depending on the ST considered. The 16S rRNA sequences do not allow differentiation between the populations from Mozambique and Nigeria (16SrXXII-A), while MLSA shows a dissimilarity of 1.4%. The complementarity of the two methods, 16S rRNA sequencing and MLSA, for assigning strains to species or lineages has been described previously (57).
“Ca. Phytoplasma palmicola” is subject to bottlenecks leading to a strong geographic structure.
Intracountry diversity of “Ca. Phytoplasma palmicola” measured by MLST is low, with the detection of 4 and 3 STs in Ghana and Mozambique, respectively. However, the identification of 3 STs in Mozambique contrasts drastically with the findings of Bila et al. (42), who observed high diversity and expanding populations of “Ca. Phytoplasma palmicola” in Mozambique based on both the secA and 16S rRNA genes, whereas those genes appear to be the most conserved in this study.
Except for the rare ST7, we observed four genetically related STs in Ghana versus two STs in Mozambique. A unique ST8 was observed in Nigeria, the first African country in which the disease was described, in 1917 (30). While only four Nigerian samples could be found and analyzed in this study, those samples were collected from coconuts separated by distances of up to 300 km. This contrasts to the situation in Ghana, where three or four STs were observed at similar distances. Only genotyping of a larger set of samples can confirm the lack of diversity in Nigeria.
The occurrence of strong bottlenecks might explain the very low diversity at the national or regional scales. Coconut lethal yellowing phytoplasmas lead to the rapid and inescapable death of the infected coconut in about 1 year. A new genetic phytoplasma variant has a short span of time to appear and emerge from the initial plant host. If such a variant appears, the probability for it to be acquired by the insect vectors would be low compared to that of the prevalent original genotype, unless this new variant acquires a higher multiplicative capacity in the coconut or a greater ability to be acquired by the insect vector.
Geographic isolation of “Ca. Phytoplasma palmicola” puts potential intraspecific recombination at a disadvantage.
Phytoplasmas are considered to have a high level of plasticity through genome rearrangement and recombination (58–60), and both recA, when present, and PMU (potential mobile unit) genes would play a role in adapting to different environments (61, 62). Despite the presence of the recA gene in “Ca. Phytoplasma palmicola,” no signs of recombination were observed among its different populations. Since multiple infections have been observed for “Ca. Phytoplasma mali” (63), spatial aggregation of the different “Ca. Phytoplasma palmicola” STs and low diversity at the regional level could explain why we detected neither coinfection nor recombination. This distribution reduces the probability that two STs would be in the same plant and hence reduces the probability of observing signals of recombination. Detecting the occurrence of such a rare event of intraspecific recombination would require extensive sampling in places where genetically distant STs share the same geographic area, for example, in the Zambezia province in Mozambique with ST6 and ST7.
A second phytoplasma that infects coconut trees in Africa and that is responsible for lethal decline (LD) in Tanzania, “Ca. Phytoplasma cocostanzania,” has been described in both the Cabo Delgado (36) and Zambezia (42) provinces of Mozambique, where “Ca. Phytoplasma palmicola” is present. Interspecific coinfection has not yet been reported. The seven new primer pairs developed for the “Ca. Phytoplasma palmicola” MLST scheme were designed to be specific for “Ca. Phytoplasma palmicola,” and they are therefore not appropriate for the detection of interspecific coinfections. However, interspecific coinfection could not be detected through 16S rRNA or rplV gene amplification and sequencing, while the corresponding primers can detect both phytoplasmas (50, 64).
New clues about “Ca. Phytoplasma palmicola” epidemiology.
Genotyping old historical samples would have been the best way to redraw the routes of introduction and the evolution of “Ca. Phytoplasma palmicola.” However, since phytoplasmas cannot be cultivated and since coconut cannot be grafted, historical “Ca. Phytoplasma palmicola” strains have not been maintained. Until now, we have been unable to find old frozen CLYD plant material or nucleic acids.
“Ca. Phytoplasma palmicola” STs are geographically clustered and distributed as small or large foci, suggesting a gradual spread. This observation is consistent with the previous observation of aggregated disease patterns and predominant dissemination within short distances (65). A low dispersion capacity of the unidentified “Ca. Phytoplasma palmicola” insect vector or poor acquisition or transmission efficiencies could explain this clustered geographic pattern. A longer-distance dissemination pattern would imply extreme weather events or human-assisted long-distance movements of nut, seedling, alternative plant hosts, or plant-carrying insect vectors.
Regarding alternative plant hosts for palm phytoplasmas, Bahder et al. (66) present the hypothesis that “Ca. Phytoplasma palmae” (group 16SrIV) could have been introduced in one plot in Florida by the planting of St. Augusta grass, on which Haplaxus crudus, identified as an insect vector of this phytoplasma, could achieve part of its life cycle as it does on many grasses (67). Studies are conflicting about possible “Ca. Phytoplasma palmicola” alternative plant hosts. No alternative hosts were identified in Ghana (68), whereas “Ca. Phytoplasma palmicola”-related strains were detected in different weed plant families in Ivory Coast (69). Surveys must be undertaken in other African countries to determine if these new findings are or are not restricted to this particular strain of “Ca. Phytoplasma palmicola” and to evaluate the potential role of these plant species as reservoirs for the short- and long-distance propagation of the disease. “Ca. Phytoplasma palmicola” presents a strong geographical pattern, in contrast to that of other phytoplasmas, such as fruit tree phytoplasmas in Europe (19). In fruit trees, both grafting and vegetative multiplication enhance dissemination and genetic uniformity; however, these practices do not exist for coconut trees. In general, human dissemination of coconut varieties is achieved through nuts or seedlings disseminated at the local or regional level. Dissemination at the continental or intercontinental scale is achieved through nuts or, more recently, coconut embryos. Phytoplasmas have been detected in coconut embryos (70, 71), and transmission to the seedling has been observed through the in vitro germination of embryos (72). However, the natural germination of phytoplasma-contaminated seeds and transmission of the phytoplasma to a viable seedling have not yet been demonstrated. Seed transmission may theoretically explain the occurrence of “Ca. Phytoplasma palmicola” of the subgroup 16SrXXII-A in both eastern and western Africa, because some nuts may have been introduced from West to East Africa (73). However, since at least 60 SNPs differentiate Nigerian and Ghanaian “Ca. Phytoplasma palmicola” strains from those detected in Mozambique, this excludes the recent introduction in Mozambique of a strain originating from areas we sampled in West Africa. It cannot be ruled out that the three genetically distinct African populations of “Ca. Phytoplasma palmicola” might have emerged independently due to a local plant host shift of an unidentified African phytoplasma. However, our data do not allow us to confirm such a possibility.
In Mozambique, ST7 is represented by a single sample and differs by at least 8 SNPs from the other Mozambican “Ca. Phytoplasma palmicola” STs. It is presently unknown whether several distinct introductions or a single introduction, followed by a local diversification, occurred in this country.
“Ca. Phytoplasma palmicola” genetic diversity has implications for LYD management.
The only method for the large-scale control of LYD is early eradication (74–76) and/or deployment of genetic resistance (77, 78). During the last decades, many trials have been performed for the identification of promising coconut varieties with resistance to CLYD in Ghana (78), Mozambique, or, more recently, Ivory Coast, where resistance trials are ongoing. In Ghana, trials that were conducted in different locations and coconut varieties showed different disease levels between trials in Agona Junction and in Axim, where ST2 and ST1 are present, respectively (78). Geographical differences in disease incidence could be explained by differences in the aggressiveness or the epidemic propensity of the considered ST. Trials aimed to improve coconut germplasm must include the precise identification of the strain and a multisite establishment.
An MLST scheme for “Ca. Phytoplasma palmicola”: a first step toward improved control of CLYD epidemics.
The proposed MLST scheme will enhance the resolution for better tracking of CLYD epidemics at the continental, regional, or provincial level. It could be implemented to identify the “Ca. Phytoplasma palmicola” strain that recently emerged in the Mozambican province of Inhambane. It could also confirm or refute the recently suggested introduction of “Ca. Phytoplasma palmicola” in Ivory Coast from western Ghana (41).
The development of an expanded MLST (eMLST) by including genes involved in the pathogenicity and insect transmission to the present MLST could increase its resolution. Using eMLST that had been expanded by adding two putative virulence genes (ureG and mba-np1) improved the discrimination of Ureaplasma urealyticum strains (79). Phytoplasma genes coding for surface proteins under positive selection, such as AMP/STAMP, IMP, or VMP, which were proven to participate in various steps of insect vector colonization, are particularly effective in phytoplasma molecular epidemiology (80–87). Only an IMP homolog was found to be encoded in the “Ca. Phytoplasma palmicola” genome draft. Such genes evolve faster than housekeeping genes and therefore do not fulfill the requirement defined by Maiden for the MLST scheme (2). The imp gene will be evaluated in a new eMLST scheme in further investigations.
On a smaller scale, MLST is clearly not sufficiently discriminating to trace “Ca. Phytoplasma palmicola” populations on a local or field scale. VNTRs and single nucleotide repeats (SNRs) are widely used to investigate monomorphic or poorly diversified bacterial species, such as Bacillus anthracis (88). If plastic Mollicutes genomes harbor large quantities of DNA repeats (89), their types and distributions are variable among phytoplasmas (11). Investigating the occurrence of VNTRs in phytoplasmas revealed that the repetition of short motifs is overrepresented in phytoplasmas compared to other prokaryotes, while long motifs are underrepresented (90). Although difficult to achieve, the identification of VNTRs applicable for inoculum tracing would benefit from a larger number of sequenced phytoplasma genomes. The ultimate approach is whole-genome sequencing (WGS). WGS of maize bushy stunt phytoplasma isolates from the same field successfully revealed genetic polymorphisms associated with functional traits (9).
In conclusion, the current MLST scheme will be valuable for assigning future outbreaks to existing or presently unreported genetic clusters of “Ca. Phytoplasma palmicola.” The interspecific comparisons presented in the present paper were based on available draft genomes. Generalization of this MLST scheme would require new genome sequencing to develop new sets of primers for each group or “Candidatus Phytoplasma” species. However, it would be a pertinent approach to compare the genetic diversities and to identify evolutionary constraints acting on the small genome of these insect-transmitted plant pathogens.
MATERIALS AND METHODS
Housekeeping gene selection and primer design.
Highly fragmented preliminary genome drafts obtained for one Mozambican isolate of the “Ca. Phytoplasma palmicola” subgroup 16SrXXII-A and one Ghanaian isolate of the “Ca. Phytoplasma palmicola” subgroup 16SrXXII-B (unpublished data) were used to select housekeeping genes and to design the corresponding PCR primers.
Housekeeping genes were selected for their availabilities on both drafts and their locations on different contigs to avoid contiguity. The two sequences of each selected gene from the two “Ca. Phytoplasma palmicola” genome drafts were aligned with BioEdit 7.0 software (http://www.mbio.ncsu.edu/BioEdit/bioedit.html) to produce a consensus sequence. Consensus sequences served as the templates to define specific PCR primers with predicted product sizes of 450 to 850 bp using Primer-BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast). The seven housekeeping genes selected, primer sequences, annealing temperature, and PCR product sizes are shown in Table 1. An eighth gene, rplV, coding for the 50S ribosomal protein L22, was included in the MLSA scheme because of its capacity to differentiate two strains in Ghana (43).
DNA phytoplasma: origin of the samples and DNA extraction.
In West Africa, Ghana and Nigeria are the only countries where the CLYD epidemics associated with “Ca. Phytoplasma palmicola” were active during the period of sampling (2004 to 2013). Sampling was extensively performed in Ghana during the 2004-2009 period, as the CLYD epidemics were very active. There was no restriction of sampling in any country. Ghana and Nigeria were subjected to extensive surveys in 2009 and 2012, respectively. In Nigeria, where the disease was first reported in 1932, four samples, collected in four different areas, were found positive for “Ca. Phytoplasma palmicola.” In Ivory Coast, “Ca. Phytoplasma palmicola” had not yet been reported at the time of the survey. A survey performed in Togo in 2006 was unsuccessful, and the disease was not reported during the 2004-2013 period. Similar feedback was obtained from Cameroon. In East Africa, LYTS is associated with “Ca. Phytoplasma cocostanzania” except in Mozambique, where both “Ca. Phytoplasma palmicola” and “Ca. Phytoplasma cocostanzania” are associated with distinct disease cases. In Mozambique, a survey in Cabo Delgado Province was done proposedly with systematic sampling of symptomatic coconut trees. Samples from the Zambezia Province of Mozambique were collected during a time-limited survey.
The MLST scheme was applied to the 132 “Ca. Phytoplasma palmicola” DNA samples (Table 4; Data Set S1 in the supplemental material). The DNA samples were extracted from stem sawdust or inflorescences of LYD-infected coconuts collected between 2004 and 2013 in Ghana (96 samples), Nigeria (4 samples), and Mozambique (32 samples). DNA was extracted using a modified CTAB protocol (39) or the DNeasy plant mini kit (Qiagen, Hilden, Germany).
TABLE 4.
Origin of “Ca. Phytoplasma palmicola”-infected coconut trees analyzed
| Country and region or province | No. of coconut trees sampled by yr |
Total no. | |||||||
|---|---|---|---|---|---|---|---|---|---|
| 2004 | 2005 | 2006 | 2007 | 2009 | 2011 | 2012 | 2013 | ||
| Ghana | 2 | 5 | 6 | 11 | 72 | 96 | |||
| Central region | 3 | 2 | 8 | 39 | 52 | ||||
| Volta region | 3 | 16 | 19 | ||||||
| Western region | 2 | 2 | 1 | 3 | 17 | 25 | |||
| Mozambique | 13 | 4 | 15 | 32 | |||||
| Cabo Delgado | 15 | 15 | |||||||
| Zambezia | 13 | 4 | 17 | ||||||
| Nigeria | 4 | 4 | |||||||
| Total | 2 | 5 | 6 | 11 | 72 | 13 | 8 | 15 | 132 |
Housekeeping genes and 16S rRNA gene amplification and sequencing.
PCR amplification was performed for each of the seven selected housekeeping genes (i.e., genes encoding DNA recombinase A [recA], DNA gyrase subunit B [gyrB], leucyl-tRNA synthetase [leuS], dihydrolipoyl dehydrogenase [lpd], DNA replication protein [dnaC], protein translocase subunit [secA], and rRNA small subunit methyltransferase I [rsmI]) plus the rplV gene for the 132 isolates. The rplV gene was amplified with the rpLYF/rpLYR primer pair (50). In a subset of 41 “Ca. Phytoplasma palmicola” samples, the 16S rRNA gene was amplified for comparison. Amplification of the locus sequences was obtained by a single PCR performed in 25 μl containing a final concentration of 1× PCR buffer, 1.5 mM MgCl2, 0.2 mM each deoxynucleoside triphosphate, 0.25 μM each forward and reverse primer, and 1.25 U of GoTaq Flexi DNA polymerase (Promega, Madison, WI, USA). The 16S rRNA gene was amplified with the classic P1/P7 (48) primers. Both phytoplasma-free coconut tree DNA and water controls were systematically included as negative controls in all tests. Amplifications were performed with the GeneAmpPCR system 9700 (Thermo Fisher Scientific, Waltham, MA, USA) with an initial denaturation at 95°C for 2 min, followed by 35 cycles of 30 s at 94°C, 50 s at the optimal annealing temperature (Ta) presented in Table 1, 60 s at 72°C, and a final 6-min extension at 72°C. For P1/P7 amplification, the annealing temperature was 58°C for 50 s with an extension step of 1 min 30 s at 72°C. The quality and concentrations of the amplified products were estimated on 1% agarose gels. PCR primers were used directly for double-strand sequencing of the PCR products by Genewiz (Takeley, United Kingdom). Two internal primers were designed for each strand to complete the P1/P7 sequencing.
Forward and reverse chromatograms were assembled using Geneious R8 (Biomatters Ltd., Auckland, New Zealand), extremities were trimmed to identical lengths and to start at the first base of a codon, and finally consensus sequences were exported for phylogenetic analyses. Because rpLYF/rpLYR primers amplify both the rplV sequence coding for the 50S ribosomal protein L22 and the rpsC genes coding for the 30S ribosomal protein, S3, the sequences were trimmed to eliminate the short intergenic and rpsC regions. The P1/P7 16S rRNA sequences were trimmed to eliminate the ITS1-tRNA-Ile-ITS2 and 23S rRNA gene sequences.
“Ca. Phytoplasma palmicola” diversity analysis.
The number of polymorphic sites, nucleotide diversity (π), and average percent cGC were calculated by using DnaSP version 6.10. The Ka/Ks ratio, measuring the level of selection based on the ratio of the number of nonsynonymous substitutions per nonsynonymous site (Ka) to the number of synonymous substitutions per synonymous site (Ks), was calculated for each housekeeping gene by using MEGA version 7.0.26 (https://www.megasoftware.net). An integer neighbor-joining network (reticulation = 0.5) of the 132 concatenated sequences was constructed using PopART v1.7 software (91).
Split network and recombination analysis.
A split network of the sequence types observed for each housekeeping gene was generated by using the neighbor-net method (92) of SplitTree4 v4.14 (93), and recombination was tested for each gene and for the concatenated sequences by performing the pairwise homoplasy index (phi) test (94). Recombination was also estimated for concatenated sequences by RDP4 (95).
Interspecific analysis.
To conduct interspecific analysis, a total of 23 full and incomplete phytoplasma chromosomes available in GenBank were screened for the availability of all eight MLST-selected housekeeping genes and the 16SrRNA operon. Since some of the drafts lack complete recA sequences, the analysis was restricted to the seven other housekeeping genes (i.e., dnaC, gyrB, leuS, lpd, secA, rsmI, and rplV).
The data set included “Ca. Phytoplasma asteris” strain onion yellows OY-M (NC_005303.2), wheat blue dwarf (AVAO00000000.1), chrysanthemum yellows strain CYP (JSWH00000000.1), rice orange leaf phytoplasma (MIEP00000000.1), maize bushy stunt (NZ_CP015149.1), and aster yellow witches broom (NC_007716.1), “Ca. Phytoplasma mali” strain AT (CU469464.1); “Ca. Phytoplasma australiense” strains Australian grapevine yellows (NC_010544.1) and strawberry lethal yellows NZSb11 from New Zealand (NC_021236.1); “Ca. Phytoplasma pruni” strains milkweed yellows-MW1 (AKIL00000000.1), Italian clover phyllody MA1 (AKIM00000000.1), and vaccinium witches'-broom VAC (AKIN00000000.1); 16SrII-A phytoplasma strain peanut witches'-broom phytoplasma NTU2011 (AMWZ00000000.1); “Ca. Phytoplasma solani” strain 284/09 (FO393427.1); “Ca. Phytoplasma pruni” (LHCF00000000.1); and “Ca. Phytoplasma phoenicum” (JPSQ00000000.1). Acholeplasma palmae (NC_022538) was used as an outgroup.
The seven housekeeping gene sequences were aligned using Muscle (Codon) implemented in MEGA version 7.0.26 and then concatenated. The maximum likelihood (ML) phylogenetic tree was constructed by using the best-fitting model option of RDP4 (95), with recombinant regions removed when detected by more than three methods. Bootstrap values were calculated from 1,000 replications. The analysis was repeated with one of the two 16Sr RNA gene sequences of each genome. The congruence of the phylogenetic trees from the two sets of sequences (MLSA and 16S) were calculated by using the congruency index Icong (46) (http://max2.ese.u-psud.fr/icong/index.help.html).
Accession number(s).
The 1,056 sequences obtained are available in the European Nucleotide Archive (ENA) (https://www.ebi.ac.uk/ena/) under project no. PRJEB27044 (study ERP109081). The sequences can be found under accession numbers LR028084 to LR029139. The 16S rRNA sequences were deposited in the ENA under project no. PRJEB27044 with accession numbers LR028043 to LR028083.
Supplementary Material
ACKNOWLEDGMENTS
We thank O. Pruvost for helpful discussions during manuscript preparation and C. Boyer, K. Boyer, S. Javegny, and S. Sisteron at the Plant Protection Platform (3P, IBISA) as well as N. Munguambe for technical support. We thank S. Hogenhout, M. Kube, C. Siewert, D. Severac, and S. Carrere for their contributions toward obtaining the first LY phytoplasma genome drafts under the Phytoplasma Sequencing Initiative of the European COST action FA0807.
This work was cofunded by the European Union under the European Regional Development Fund (grant ERDF GURDT I2016-1731-0006632), the Conseil Régional de La Réunion, and the Centre de Coopération Internationale en Recherche Agronomique pour le Développement (CIRAD).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02716-18.
REFERENCES
- 1.Achtman M. 2008. Evolution, population structure, and phylogeography of genetically monomorphic bacterial pathogens. Annu Rev Microbiol 62:53–70. doi: 10.1146/annurev.micro.62.081307.162832. [DOI] [PubMed] [Google Scholar]
- 2.Maiden MCJ. 2006. Multilocus sequence typing of bacteria. Annu Rev Microbiol 60:561–588. doi: 10.1146/annurev.micro.59.030804.121325. [DOI] [PubMed] [Google Scholar]
- 3.Aanensen DM, Feil EJ, Holden MTG, Dordel J, Yeats CA, Fedosejev A, Goater R, Castillo-Ramírez S, Corander J, Colijn C, Chlebowicz MA, Schouls L, Heck M, Pluister G, Ruimy R, Kahlmeter G, Åhman J, Matuschek E, Friedrich AW, Parkhill J, Bentley SD, Spratt BG, Grundmann H. 2016. Whole-genome sequencing for routine pathogen surveillance in public health: a population snapshot of invasive Staphylococcus aureus in Europe. mBio 7:e00444-16. doi: 10.1128/mBio.00444-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bryant JM, Grogono DM, Greaves D, Foweraker J, Roddick I, Inns T, Reacher M, Haworth CS, Curran MD, Harris SR, Peacock SJ, Parkhill J, Floto RA. 2013. Whole-genome sequencing to identify transmission of Mycobacterium abscessus between patients with cystic fibrosis: a retrospective cohort study. Lancet 381:1551–1560. doi: 10.1016/S0140-6736(13)60632-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Diaz MH, Desai HP, Morrison SS, Benitez AJ, Wolff BJ, Caravas J, Read TD, Dean D, Winchell JM. 2017. Comprehensive bioinformatics analysis of Mycoplasma pneumoniae genomes to investigate underlying population structure and type-specific determinants. PLoS One 12:e0174701. doi: 10.1371/journal.pone.0174701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Razin S, Yogev D, Naot Y. 1998. Molecular biology and pathogenicity of mycoplasmas. Microbiol Mol Biol Rev 62:1094–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen L-L, Chung W-C, Lin C-P, Kuo C-H. 2012. Comparative analysis of gene content evolution in phytoplasmas and mycoplasmas. PLoS One 7:e34407. doi: 10.1371/journal.pone.0034407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strauss E. 2009. Phytoplasma research begins to bloom. Science 325:388–390. doi: 10.1126/science.325_388. [DOI] [PubMed] [Google Scholar]
- 9.Orlovskis Z, Canale MC, Haryono M, Lopes JRS, Kuo C-H, Hogenhout SA. 2017. A few sequence polymorphisms among isolates of Maize bushy stunt phytoplasma associate with organ proliferation symptoms of infected maize plants. Ann Bot 119:869–884. doi: 10.1093/aob/mcw213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia-Chapa M, Batlle A, Rekab D, Rosquete MR, Firrao G. 2004. PCR-mediated whole genome amplification of phytoplasmas. J Microbiol Methods 56:231–242. doi: 10.1016/j.mimet.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 11.Cimerman A, Arnaud G, Foissac X. 2006. Stolbur phytoplasma genome survey achieved using a suppression subtractive hybridization approach with high specificity. Appl Environ Microbiol 72:3274–3283. doi: 10.1128/AEM.72.5.3274-3283.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tran-Nguyen LTT, Gibb KS. 2007. Optimizing phytoplasma DNA purification for genome analysis. J Biomol Tech 18:104–112. [PMC free article] [PubMed] [Google Scholar]
- 13.Liefting LW, Kirkpatrick BC. 2003. Cosmid cloning and sample sequencing of the genome of the uncultivable mollicute, Western X‐disease phytoplasma, using DNA purified by pulsed‐field gel electrophoresis. FEMS Microbiol Lett 221:203–211. doi: 10.1016/S0378-1097(03)00183-6. [DOI] [PubMed] [Google Scholar]
- 14.Chung W-C, Chen L-L, Lo W-S, Lin C-P, Kuo C-H. 2013. Comparative analysis of the peanut witches'-broom phytoplasma genome reveals horizontal transfer of potential mobile units and effectors. PLoS One 8:e62770. doi: 10.1371/journal.pone.0062770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Firrao G, Martini M, Ermacora P, Loi N, Torelli E, Foissac X, Carle P, Kirkpatrick BC, Liefting L, Schneider B, Marzachi C, Palmano S. 2013. Genome wide sequence analysis grants unbiased definition of species boundaries in “Candidatus Phytoplasma.” Syst Appl Microbiol 36:539–548. doi: 10.1016/j.syapm.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 16.Brown RJ, Holden MTG, Spiller OB, Chalker VJ. 2015. Development of a multilocus sequence typing scheme for molecular typing of Mycoplasma pneumoniae. J Clin Microbiol 53:3195–3203. doi: 10.1128/JCM.01301-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghanem M, El-Gazzar M. 2016. Development of multilocus sequence typing (MLST) assay for Mycoplasma iowae. Vet Microbiol 195:2–8. doi: 10.1016/j.vetmic.2016.08.013. [DOI] [PubMed] [Google Scholar]
- 18.Manso-Silván L, Dupuy V, Lysnyansky I, Ozdemir U, Thiaucourt F. 2012. Phylogeny and molecular typing of Mycoplasma agalactiae and Mycoplasma bovis by multilocus sequencing. Vet Microbiol 161:104–112. doi: 10.1016/j.vetmic.2012.07.015. [DOI] [PubMed] [Google Scholar]
- 19.Danet JL, Balakishiyeva G, Cimerman A, Sauvion N, Marie-Jeanne V, Labonne G, Laviňa A, Batlle A, Križanac I, Škorić D, Ermacora P, Serçe ÇU, Çağlayan K, Jarausch W, Foissac X. 2011. Multilocus sequence analysis reveals the genetic diversity of European fruit tree phytoplasmas and supports the existence of inter-species recombination. Microbiology 157:438–450. doi: 10.1099/mic.0.043547-0. [DOI] [PubMed] [Google Scholar]
- 20.Quaglino F, Kube M, Jawhari M, Abou-Jawdah Y, Siewert C, Choueiri E, Sobh H, Casati P, Tedeschi R, Lova MM, Alma A, Bianco PA. 2015. ‘Candidatus Phytoplasma phoenicium’ associated with almond witches’-broom disease: from draft genome to genetic diversity among strain populations. BMC Microbiol 15:148. doi: 10.1186/s12866-015-0487-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yong L, Chun‐Gen P, Guo‐Zhong T, Zhi‐Xin L, Min‐Wei G, Cai‐Li L, Xi‐Zhuo W. 2014. Multilocus sequences confirm the close genetic relationship of four phytoplasmas of peanut witches'‐broom group 16SrII‐A. J Basic Microbiol 54:818–827. doi: 10.1002/jobm.201300140. [DOI] [PubMed] [Google Scholar]
- 22.Arnaud G, Malembic-Maher S, Salar P, Bonnet P, Maixner M, Marcone C, Boudon-Padieu E, Foissac X. 2007. Multilocus sequence typing confirms the close genetic interrelatedness of three distinct flavescence dorée phytoplasma strain clusters and group 16SrV phytoplasmas infecting grapevine and alder in Europe. Appl Environ Microbiol 73:4001–4010. doi: 10.1128/AEM.02323-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malembic-Maher S, Salar P, Filippin L, Carle P, Angelini E, Foissac X. 2011. Genetic diversity of European phytoplasmas of the 16SrV taxonomic group and proposal of ‘Candidatus Phytoplasma rubi.’ Int J Syst Evol Microbiol 61:2129–2134. doi: 10.1099/ijs.0.025411-0. [DOI] [PubMed] [Google Scholar]
- 24.Gurr GM, Johnson AC, Ash GJ, Wilson BAL, Ero MM, Pilotti CA, Dewhurst CF, You MS. 2016. Coconut lethal yellowing diseases: a phytoplasma threat to palms of global economic and social significance. Front Plant Sci 7:1521. doi: 10.3389/fpls.2016.01521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yankey EN, Bila J, Rosete YA, Oropeza C, Pilet F. 2018. Phytoplasma diseases of palms, p 267–285. In Rao GP, Bertaccini A, Fiore N, Liefting LW (ed), Phytoplasmas: plant pathogenic bacteria. I. Characterisation and epidemiology of phytoplasma-associated diseases. Springer Singapore, Singapore. [Google Scholar]
- 26.Stein H. 1905. Die Kokosnuss und deren Bearbeitung in Deutsch-Ostafrika. Der Tropenpflanzer 9:195–201. [Google Scholar]
- 27.Dowson W. 1921. Some problems of economic biology in East Africa (Kenya Colony). Ann Appl Biol 8:85–87. [Google Scholar]
- 28.de Carvalho T, Mendes O.1958. Doenças de plantas em Moçambique. Direcção de Agricultura e Florestas, Repartição de Sanidade Vegetal, Secçaõ de Patologia Vegetal, Lourenço Marques, Provincia de Moçambique.
- 29.Santana Quadros A. 1972. A “doença desconhecida” do coqueiro na Zambézia. Rev Agric 14:33–34. [Google Scholar]
- 30.Johnson WH.1918. Annual report of the agricultural department of southern provinces Nigeria for the year 1917, Ibadan, Nigeria. Government Publication 14.
- 31.Chona BL, Addoh PG. 1970. Cape St. Paul wilt: the present position, p 3–8. In Chona BL, Adansi MA (ed), Coconut in Ghana. Crops Research Institute bulletin no. 3, Council for Scientific and Industrial Research Kwadaso, Kumasi, Ghana. [Google Scholar]
- 32.Meiffren M. 1951. Note préliminaire sur l’étude de la maladie des cocotiers au Togo. Agron Trop 6:163–174. [Google Scholar]
- 33.Grimaldi J, Monveiller G. 1965. La Maladie des cocotiers de Kribi. Cameroun Agric Pastoral Forest 90:18–27. [Google Scholar]
- 34.Konan Konan J, Allou K, Atta Diallo H, Saraka Yao D, Koua B, Kouassi N, Benabid R, Michelutti R, Scott J, Arocha-Rosete Y. 2013. First report on the molecular identification of the phytoplasma associated with a lethal yellowing-type disease of coconut palms in Cote d'Ivoire. New Dis Rep 28:3. doi: 10.5197/j.2044-0588.2013.028.003. [DOI] [Google Scholar]
- 35.IRPCM Phytoplasma/Spiroplasma Working Group. 2004. 'Candidatus Phytoplasma', a taxon for the wall-less, non-helical prokaryotes that colonize plant phloem and insects. Int J Syst Evol Microbiol 54:1243–1255. doi: 10.1099/ijs.0.02854-0. [DOI] [PubMed] [Google Scholar]
- 36.Dollet M, Lourenço E, Macome F, Vaz A, Fabre S. 2012. In Mozambique, at least two different phytoplasmas induce lethal yellowing type syndromes in coconut palms. 19th Congress of the International Organization for Mycoplasmology, Toulouse, France, 15 to 20 July 2012. [Google Scholar]
- 37.Harrison NA, Davis RE, Oropeza C, Helmick EE, Narváez M, Eden-Green S, Dollet M, Dickinson M. 2014. ‘Candidatus Phytoplasma palmicola’, associated with a lethal yellowing-type disease of coconut (Cocos nucifera L.) in Mozambique. Int J Syst Evol Microbiol 64:1890–1899. doi: 10.1099/ijs.0.060053-0. [DOI] [PubMed] [Google Scholar]
- 38.Arocha-Rosete Y, Konan Konan JL, Diallo AH, Allou K, Scott JA. 2014. Identification and molecular characterization of the phytoplasma associated with a lethal yellowing-type disease of coconut in Côte d’Ivoire. Can J Plant Pathol 36:141–150. doi: 10.1080/07060661.2014.899275. [DOI] [Google Scholar]
- 39.Mpunami AA, Tymon A, Jones P, Dickinson MJ. 1999. Genetic diversity in the coconut lethal yellowing disease phytoplasmas of East Africa. Plant Pathol 48:109–114. doi: 10.1046/j.1365-3059.1999.00314.x. [DOI] [Google Scholar]
- 40.Wei W, Davis RE, Lee I-M, Zhao Y. 2007. Computer-simulated RFLP analysis of 16S rRNA genes: identification of ten new phytoplasma groups. Int J Syst Evol Microbiol 57:1855–1867. doi: 10.1099/ijs.0.65000-0. [DOI] [PubMed] [Google Scholar]
- 41.Rosete YA, Diallo HA, Konan Konan JL, Yankey N, Saleh M, Pilet F, Contaldo N, Paltrinieri S, Bertaccini A, Scott J. 2017. Detection and differentiation of the coconut lethal yellowing phytoplasma in coconut‐growing villages of Grand‐Lahou, Côte d'Ivoire. Ann Appl Biol 170:333–347. doi: 10.1111/aab.12333. [DOI] [Google Scholar]
- 42.Bila J, Mondjana A, Samils B, Högberg N. 2015. High diversity, expanding populations and purifying selection in phytoplasmas causing coconut lethal yellowing in Mozambique. Plant Pathol 64:597–604. doi: 10.1111/ppa.12306. [DOI] [Google Scholar]
- 43.Pilet F, Poulin L, Nkansah-Poku J, Quaicoe RN. 2011. Ribosomal protein gene sequences reveal a geographical differentiation between CSPWD phytoplasmas in Ghana. Bull Insectol 64:S219–S220. [Google Scholar]
- 44.Osagie IJ, Ojomo EE, Pilet F. 2015. Occurrence of Awka wilt disease of coconut in Nigeria for one century. Phytopathogenic Mollicutes 5:S61–S62. doi: 10.5958/2249-4677.2015.00025.0. [DOI] [Google Scholar]
- 45.Tymon AM, Jones P, Harrison NA. 1998. Phylogenetic relationships of coconut phytoplasmas and the development of specific oligonucleotide PCR primers. Ann Appl Biol 132:437–452. doi: 10.1111/j.1744-7348.1998.tb05220.x. [DOI] [Google Scholar]
- 46.de Vienne DM, Giraud T, Martin OC. 2007. A congruence index for testing topological similarity between trees. Bioinformatics 23:3119–3124. doi: 10.1093/bioinformatics/btm500. [DOI] [PubMed] [Google Scholar]
- 47.Deng S, Hiruki C. 1991. Amplification of 16S rRNA genes from culturable and nonculturable Mollicutes. J Microbiol Methods 14:53–61. doi: 10.1016/0167-7012(91)90007-D. [DOI] [Google Scholar]
- 48.Schneider B, Cousin MT, Klinkong S, Seemüller E. 1995. Taxonomic relatedness and phylogenetic positions of phytoplasmas associated with diseases of faba bean, sunnhemp, sesame, soybean, and eggplant. Z Pflanzenkr Pflanzenschutz 102:225–232. [Google Scholar]
- 49.Dollet M, Fabre S, Pilet F, De Almeida Marinho VL, Quaicoe RN. 2006. Diagnosis and variability in coconut lethal yellowing phytoplasmas in Africa. Annual Meeting of the American Phytopathological Society, Cartegena, Colombia, 12 to 16 September 2006. [Google Scholar]
- 50.Marinho VLA, Fabre S, Dollet M. 2006. Diagnóstico do “Coconut lethal yellowing phytoplasma”, praga de quarentena para o Brasil, baseado no estudo de genes da proteína ribosomal. Fitopatol Bras 31:S363–S364. [Google Scholar]
- 51.Obeng‐Darko SA, Quaicoe RN, Yankey EN, Twumasi P. 2018. Comparative PCR analyses for the detection of the Cape St. Paul wilt disease phytoplasma in coconut palms in Ghana. J Phytopathol 166:477–483. doi: 10.1111/jph.12707. [DOI] [Google Scholar]
- 52.Yankey EN, Swarbrick PJ, Nipah JO, Quaicoe RN, Dickinson MJ. 2014. Detection of the Cape St. Paul wilt phytoplasma in coconut palms in Ghana through the combination of visual symptoms assessment and molecular diagnosis using a secA gene based assay. J Plant Pathol 96:281–285. [Google Scholar]
- 53.Dupuy V, Manso-Silvan L, Barbe V, Thebault P, Dordet-Frisoni E, Citti C, Poumarat F, Blanchard A, Breton M, Sirand-Pugnet P, Thiaucourt F. 2012. Evolutionary history of contagious bovine pleuropneumonia using next generation sequencing of Mycoplasma mycoides subsp. mycoides “Small Colony”. PLoS One 7:e46821. doi: 10.1371/journal.pone.0046821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yaya A, Manso-Silván L, Blanchard A, Thiaucourt F. 2008. Genotyping of Mycoplasma mycoides subsp. mycoides SC by multilocus sequence analysis allows molecular epidemiology of contagious bovine pleuropneumonia. Vet Res 39:14. doi: 10.1051/vetres:2007052. [DOI] [PubMed] [Google Scholar]
- 55.El-Sisi Y, Omar AF, Sidaros SA, Elsharkawy MM, Foissac X. 2018. Multilocus sequence analysis supports a low genetic diversity among ‘Candidatus Phytoplasma australasia’ related strains infecting vegetable crops and periwinkle in Egypt. Eur J Plant Pathol 150:779–784. doi: 10.1007/s10658-017-1311-9. [DOI] [Google Scholar]
- 56.Kakizawa S, Kamagata Y. 2014. A multiplex-PCR method for strain identification and detailed phylogenetic analysis of AY-group phytoplasmas. Plant Dis 98:299–305. doi: 10.1094/PDIS-03-13-0216-RE. [DOI] [PubMed] [Google Scholar]
- 57.Gevers D, Cohan FM, Lawrence JG, Spratt BG, Coenye T, Feil EJ, Stackebrandt E, de Peer YV, Vandamme P, Thompson FL, Swings J. 2005. Re-evaluating prokaryotic species. Nat Rev Microbiol 3:733. doi: 10.1038/nrmicro1236. [DOI] [PubMed] [Google Scholar]
- 58.Andersen MT, Liefting LW, Havukkala I, Beever RE. 2013. Comparison of the complete genome sequence of two closely related isolates of ‘Candidatus Phytoplasma australiense’ reveals genome plasticity. BMC Genomics 14:529. doi: 10.1186/1471-2164-14-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Siampour M, Izadpanah K, Marzachi C, Salehi Abarkoohi M. 2015. Identification and characterization of conserved and variable regions of lime witches’ broom phytoplasma genome. Microbiology 161:1741–1751. doi: 10.1099/mic.0.000133. [DOI] [PubMed] [Google Scholar]
- 60.Wei W, Davis RE, Jomantiene R, Zhao Y. 2008. Ancient, recurrent phage attacks and recombination shaped dynamic sequence-variable mosaics at the root of phytoplasma genome evolution. Proc Natl Acad Sci U S A 105:11827–11832. doi: 10.1073/pnas.0805237105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bai X, Zhang J, Ewing A, Miller SA, Jancso Radek A, Shevchenko DV, Tsukerman K, Walunas T, Lapidus A, Campbell JW, Hogenhout SA. 2006. Living with genome instability: the adaptation of phytoplasmas to diverse environments of their insect and plant hosts. J Bacteriol 188:3682–3696. doi: 10.1128/JB.188.10.3682-3696.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tran-Nguyen LTT, Kube M, Schneider B, Reinhardt R, Gibb KS. 2008. Comparative genome analysis of 'Candidatus Phytoplasma australiense' (subgroup tuf-Australia I; rp-A) and “Ca. Phytoplasma asteris” strains OY-M and AY-WB. J Bacteriol 190:3979–3991. doi: 10.1128/JB.01301-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Seemüller E, Kiss E, Sule S, Schneider B. 2010. Multiple infection of apple trees by distinct strains of ‘Candidatus Phytoplasma mali’ and its pathological relevance. Phytopathology 100:863–870. doi: 10.1094/PHYTO-100-9-0863. [DOI] [PubMed] [Google Scholar]
- 64.Marinho VLA, Fabre S, Dollet M. 2008. Genetic variability among isolates of coconut lethal yellowing phytoplasmas determined by heteroduplex mobility assay (HMA). Trop Plant Pathol 33:377–380. doi: 10.1590/S1982-56762008000500006. [DOI] [Google Scholar]
- 65.Bonnot F, de Franqueville H, Lourenço E. 2010. Spatial and spatiotemporal pattern analysis of coconut lethal yellowing in Mozambique. Phytopathology 100:300–312. doi: 10.1094/PHYTO-100-4-0300. [DOI] [PubMed] [Google Scholar]
- 66.Bahder BW, Helmick EE, Chakrabarti S, Osorio S, Soto N, Chouvenc T, Harrison NA. 2018. Disease progression of a lethal decline caused by the 16SrIV-D phytoplasma in Florida palms. Plant Pathol 67:1821–1828. doi: 10.1111/ppa.12882. [DOI] [Google Scholar]
- 67.Ramos Hernández E, Magaña Alejandro MA, Ortiz García CF, Oropeza Salín C, Lesher Gordillo JM, Sánchez Soto S. 2018. The coconut pathosystem: weed hosts of nymphs of the American palm Cixiid Haplaxius crudus (Hemiptera: Fulgoroidea). J Nat Hist 52:255–268. doi: 10.1080/00222933.2017.1420832. [DOI] [Google Scholar]
- 68.Yankey EN, Pilet F, Quaicoe RN, Dery SK, Dollet M, Dzogbefia VP. 2009. Search for alternate hosts of the coconut Cape Saint Paul wilt disease pathogen. OCL 16:123–126. doi: 10.1051/ocl.2009.0250. [DOI] [Google Scholar]
- 69.Rosete YA, Diallo HA, Konan Konan JL, Kouamé AEP, Séka K, Kra KD, Toualy MN, Kwadjo KE, Daramcoum WAMP, Beugré NI, Ouattara BWM, Kouadjo Zaka CG, Allou K, Fursy-Rodelec ND, Doudjo-Ouattara ON, Yankey N, Dery S, Maharaj A, Saleh M, Summerbell R, Contaldo N, Paltrinieri S, Bertaccini A, Scott J. 2016. Detection and identification of the coconut lethal yellowing phytoplasma in weeds growing in coconut farms in Côte d’Ivoire. Can J Plant Pathol 38:164–173. doi: 10.1080/07060661.2016.1191044. [DOI] [Google Scholar]
- 70.Cordova I, Jones P, Harrison NA, Oropeza C. 2003. In situ PCR detection of phytoplasma DNA in embryos from coconut palms with lethal yellowing disease. Mol Plant Pathol 4:99–108. doi: 10.1046/j.1364-3703.2003.00152.x. [DOI] [PubMed] [Google Scholar]
- 71.Nipah J, Jones P, Dickinson M. 2007. Detection of lethal yellowing phytoplasma in embryos from coconut palms infected with Cape St Paul wilt disease in Ghana. Plant Pathol 56:777–784. doi: 10.1111/j.1365-3059.2007.01623.x. [DOI] [Google Scholar]
- 72.Oropeza C, Cordova I, Puch-Hau C, Castillo R, Chan JL, Sáenz L. 2017. Detection of lethal yellowing phytoplasma in coconut plantlets obtained through in vitro germination of zygotic embryos from the seeds of infected palms. Ann Appl Biol 171:28–36. doi: 10.1111/aab.12351. [DOI] [Google Scholar]
- 73.Krain E, Issa JA, Kullaya A, Harries HC. 1994. The coconut palm in East Africa. 2. The Pemba Dwarf in Zanzibar. Principes 38:138–141. [Google Scholar]
- 74.Munguambe N, Timbrine R, Freire M, Gadaga S, Dias P, Do Rosario B, Pudivitr J, Pilet F. 2013. Large scale management of coconut lethal yellowing disease in Mozambique. Acta Phytopathol Sin 43(Suppl):205. [Google Scholar]
- 75.Nkansah Poku J, Philippe R, Quaicoe RN, Dery SK, Ransford A. 2009. Cape Saint Paul wilt disease of coconut in Ghana: surveillance and management of disease spread. OCL 16:111–115. doi: 10.1051/ocl.2009.0247. [DOI] [Google Scholar]
- 76.Philippe R, Dery SK, Nkansah Poku J. 2004. New data on a cultural control method against coconut lethal yellowing in Ghana. CORD Coconut Res Dev 20:21–27. [Google Scholar]
- 77.Baudouin L, Philippe R, Quaicoe R, Dery S, Dollet M. 2009. General overview of genetic research and experimentation on coconut varieties tolerant/resistant to lethal yellowing. OCL 16:127–131. doi: 10.1051/ocl.2009.0244. [DOI] [Google Scholar]
- 78.Dery SK, Philippe R, Baudouin L, Quaicoe RN, Nkansah-Poku J, Owusu-Nipah J, Arthur R, Dare D, Yankey N, Dollet M. 2008. Genetic diversity among coconut varieties for susceptibility to Cape St Paul wilt disease. Euphytica 164:1–11. doi: 10.1007/s10681-008-9691-8. [DOI] [Google Scholar]
- 79.Zhang J, Kong Y, Ruan Z, Huang J, Song T, Song J, Jiang Y, Yu Y, Xie X. 2014. Correlation between Ureaplasma subgroup 2 and genitourinary tract disease outcomes revealed by an expanded multilocus sequence typing (eMLST) scheme. PLoS One 9:e104347. doi: 10.1371/journal.pone.0104347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Arricau-Bouvery N, Duret S, Dubrana M-P, Batailler B, Desqué D, Béven L, Danet J-L, Monticone M, Bosco D, Malembic-Maher S, Foissac X. 2018. Variable membrane protein A of flavescence dorée phytoplasma binds the midgut perimicrovillar membrane of Euscelidius variegatus and promotes adhesion to its epithelial cells. Appl Environ Microbiol 84:e02487-17. doi: 10.1128/AEM.02487-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cimerman A, Pacifico D, Salar P, Marzachì C, Foissac X. 2009. Striking diversity of vmp1, a variable gene encoding a putative membrane protein of the stolbur phytoplasma. Appl Environ Microbiol 75:2951–2957. doi: 10.1128/AEM.02613-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fabre A, Balakishiyeva G, Ember I, Omar A, Acs Z, Koelber M, Kauzner L, Della Bartola M, Danet J-L, Foissac X. 2011. StAMP encoding the antigenic membrane protein of stolbur phytoplasma is useful for molecular epidemiology. Bull Insectol 64:S21–S22. [Google Scholar]
- 83.Fabre A, Danet J-L, Foissac X. 2011. The stolbur phytoplasma antigenic membrane protein gene stamp is submitted to diversifying positive selection. Gene 472:37–41. doi: 10.1016/j.gene.2010.10.012. [DOI] [PubMed] [Google Scholar]
- 84.Kakizawa S, Oshima K, Jung H-Y, Suzuki S, Nishigawa H, Arashida R, Miyata S-i, Ugaki M, Kishino H, Namba S. 2006. Positive selection acting on a surface membrane protein of the plant-pathogenic phytoplasmas. J Bacteriol 188:3424–3428. doi: 10.1128/JB.188.9.3424-3428.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Neriya Y, Sugawara K, Maejima K, Hashimoto M, Komatsu K, Minato N, Miura C, Kakizawa S, Yamaji Y, Oshima K, Namba S. 2011. Cloning, expression analysis, and sequence diversity of genes encoding two different immunodominant membrane proteins in poinsettia branch-inducing phytoplasma (PoiBI). FEMS Microbiol Lett 324:38–47. doi: 10.1111/j.1574-6968.2011.02384.x. [DOI] [PubMed] [Google Scholar]
- 86.Pacifico D, Alma A, Bagnoli B, Foissac X, Pasquini G, Tessitori M, Marzachi C. 2009. Characterization of Bois noir isolates by restriction fragment length polymorphism of a stolbur-specific putative membrane protein gene. Phytopathology 99:711–715. doi: 10.1094/PHYTO-99-6-0711. [DOI] [PubMed] [Google Scholar]
- 87.Suzuki S, Oshima K, Kakizawa S, Arashida R, Jung H-Y, Yamaji Y, Nishigawa H, Ugaki M, Namba S. 2006. Interaction between the membrane protein of a pathogen and insect microfilament complex determines insect-vector specificity. Proc Natl Acad Sci U S A 103:4252–4257. doi: 10.1073/pnas.0508668103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Keim P, Van Ert MN, Pearson T, Vogler AJ, Huynh LY, Wagner DM. 2004. Anthrax molecular epidemiology and forensics: using the appropriate marker for different evolutionary scales. Infect Genet Evol 4:205–213. doi: 10.1016/j.meegid.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 89.Rocha EPC, Blanchard A. 2002. Genomic repeats, genome plasticity and the dynamics of Mycoplasma evolution. Nucleic Acids Res 30:2031–2042. doi: 10.1093/nar/30.9.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wei W, Davis RE, Suo X, Zhao Y. 2015. Occurrence, distribution and possible functional roles of simple sequence repeats in phytoplasma genomes. Int J Syst Evol Microbiol 65:2748–2760. doi: 10.1099/ijs.0.000273. [DOI] [PubMed] [Google Scholar]
- 91.Leigh JW, Bryant D. 2015. Popart: full‐feature software for haplotype network construction. Methods Ecol Evol 6:1110–1116. doi: 10.1111/2041-210X.12410. [DOI] [Google Scholar]
- 92.Bryant D, Moulton V. 2004. Neighbor-Net: an agglomerative method for the construction of phylogenetic networks. Mol Biol Evol 21:255–265. doi: 10.1093/molbev/msh018. [DOI] [PubMed] [Google Scholar]
- 93.Huson DH, Bryant D. 2006. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- 94.Bruen TC, Philippe H, Bryant D. 2006. A simple and robust statistical test for detecting the presence of recombination. Genetics 172:2665–2681. doi: 10.1534/genetics.105.048975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Martin DP, Murrell B, Golden M, Khoosal A, Muhire B. 2015. RDP4: detection and analysis of recombination patterns in virus genomes. Virus Evol 1:vev003. doi: 10.1093/ve/vev003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.