The climate-active gas dimethyl sulfide (DMS) plays an important role in global sulfur cycling and atmospheric chemistry. DMS is mainly produced through the bacterial cleavage of marine dimethylsulfoniopropionate (DMSP). When released into the atmosphere from the oceans, DMS can be photochemically oxidized into DMSO or sulfate aerosols, which form cloud condensation nuclei that influence the reflectivity of clouds and, thereby, global temperature. SAR11 bacteria are the most abundant marine heterotrophic bacteria, and many of them contain DMSP lyase DddK to cleave DMSP, generating DMS. In this study, based on structural analyses and mutational assays, we revealed the catalytic mechanism of DddK, which has universal significance in SAR11 bacteria. This study provides new insights into the catalytic mechanism of DddK, leading to a better understanding of how SAR11 bacteria generate DMS.
KEYWORDS: DMS generation, DMSP, DMSP lyase DddK, SAR11, catalytic mechanism
ABSTRACT
The osmolyte dimethylsulfoniopropionate (DMSP) is produced in petagram quantities in marine environments and has important roles in global sulfur and carbon cycling. Many marine microorganisms catabolize DMSP via DMSP lyases, generating the climate-active gas dimethyl sulfide (DMS). DMS oxidation products participate in forming cloud condensation nuclei and, thus, may influence weather and climate. SAR11 bacteria are the most abundant marine heterotrophic bacteria; many of them contain the DMSP lyase DddK, and their dddK transcripts are relatively abundant in seawater. In a recently described catalytic mechanism for DddK, Tyr64 is predicted to act as the catalytic base initiating the β-elimination reaction of DMSP. Tyr64 was proposed to be deprotonated by coordination to the metal cofactor or its neighboring His96. To further probe this mechanism, we purified and characterized the DddK protein from Pelagibacter ubique strain HTCC1062 and determined the crystal structures of wild-type DddK and its Y64A and Y122A mutants (bearing a change of Y to A at position 64 or 122, respectively), where the Y122A mutant is complexed with DMSP. The structural and mutational analyses largely support the catalytic role of Tyr64, but not the method of its deprotonation. Our data indicate that an active water molecule in the active site of DddK plays an important role in the deprotonation of Tyr64 and that this is far more likely than coordination to the metal or His96. Sequence alignment and phylogenetic analysis suggest that the proposed catalytic mechanism of DddK has universal significance. Our results provide new mechanistic insights into DddK and enrich our understanding of DMS generation by SAR11 bacteria.
IMPORTANCE The climate-active gas dimethyl sulfide (DMS) plays an important role in global sulfur cycling and atmospheric chemistry. DMS is mainly produced through the bacterial cleavage of marine dimethylsulfoniopropionate (DMSP). When released into the atmosphere from the oceans, DMS can be photochemically oxidized into DMSO or sulfate aerosols, which form cloud condensation nuclei that influence the reflectivity of clouds and, thereby, global temperature. SAR11 bacteria are the most abundant marine heterotrophic bacteria, and many of them contain DMSP lyase DddK to cleave DMSP, generating DMS. In this study, based on structural analyses and mutational assays, we revealed the catalytic mechanism of DddK, which has universal significance in SAR11 bacteria. This study provides new insights into the catalytic mechanism of DddK, leading to a better understanding of how SAR11 bacteria generate DMS.
INTRODUCTION
The marine osmolyte dimethylsulfoniopropionate (DMSP) has key roles in global sulfur and carbon cycling (1) and signaling pathways (2) and is the major precursor for the climate-active gas dimethyl sulfide (DMS). Over 8 billion tonnes of DMSP are produced annually in Earth’s surface oceans (3) by many marine algae and bacteria and some corals and plants (4–8). DMSP can account for ∼10% of the total organic carbon in the surface oceans (9). Although many algae can catabolize DMSP (10, 11), heterotrophic bacteria, especially Roseobacter and SAR11 bacteria, are thought to be the major degraders of DMSP in marine environments (1, 12). These bacteria can catabolize DMSP via demethylation and/or cleavage pathways (1, 12, 13). In DMSP demethylation, a series of Dmd enzymes can generate acetaldehyde and methanethiol, providing the marine microbial food web with organic carbon and reduced sulfur (13, 14). DMSP is cleaved by various bacterial DMSP lyases, termed Ddd enzymes, to produce the climate-active volatile DMS (1, 15). Approximately 30 million tons of DMS is transferred from the oceans to the atmosphere annually (16), where it can be photochemically oxidized to form cloud condensation nuclei, which increases the backscatter of solar radiation and potentially influences the global climate (1, 17).
Eight distinct DMSP lyases, termed Alma1, DddD, DddP, DddY, DddK, DddL, DddQ, and DddW, have been identified in taxonomically diverse bacteria, fungi, and phytoplankton (11, 12, 15). DddD is a class III CoA transferase family enzyme that yields 3-hydroxypropionate and DMS from DMSP (15, 18). All the other known DMSP lyases generate acrylate and DMS from DMSP (15, 18). The algal DMSP lyase Alma1 belongs to the aspartate racemase superfamily (11), DddP is a member of the M24 peptidase family (19), and the five remaining DMSP lyases (DddY, DddK, DddL, DddQ, and DddW) belong to the cupin superfamily and contain a conserved cupin motif (12, 15, 20–22). Cupin superfamily proteins comprise functionally diverse enzymes, which contain a metal ion in their active sites (21, 23). Indeed, all cupin-containing DMSP lyases have been shown to require metal cofactors for enzyme activity (22, 24–26).
The advent of biochemical and structural studies on several DMSP lyases has led to the proposal of molecular mechanisms for many of these enzymes (20, 24–27). Structures of the DMSP lyase DddQ reveal that metal-bound DddQ undergoes a conformational change upon DMSP binding that allows a conserved tyrosine residue to act as a Lewis catalytic base (26). The catalytic mechanisms of DddP and DddY have also been proposed from studies of their crystal structures (20, 27, 28). Unsurprisingly, being a cupin enzyme, DddY adopts a catalytic mechanism similar to that of DddQ, with a specific tyrosine residue acting as the catalytic base (20). In contrast, DddP contains a di-iron core, and an ion-shift catalytic mechanism is proposed (27).
DddK is found in SAR11 bacteria, which were recently shown to cleave and demethylate DMSP under laboratory conditions, with the former pathway predominating (1, 12). SAR11 bacteria often comprise 30% of the ocean's surface microbial community, and dddK transcripts are relatively abundant in marine environments, which suggests that this DMSP lyase is environmentally important in DMS generation (1, 12, 29). Recently, a catalytic mechanism for DddK from Pelagibacter ubique strain HTCC1062 was proposed by Schnicker et al., based on structural analysis combined with mutational data (24). Tyr64 in Ni-bound DddK was proposed to act as the catalytic base to initiate the β-elimination reaction of DMSP. Tyr64 of DddK is proposed to be deprotonated by coordinating either to the metal through a conformational change or to the neighboring His96 (24). However, the distance between the hydroxyl group of Tyr64 and His96 in the Ni-DddK structure (PDB ID 5TFZ) is 3.5 Å (24), which can only allow the formation of a weak hydrogen bond. Further analysis of the structures suggests that even with the conformation changes in Tyr64 described by Schnicker et al. (24), Tyr64 is unlikely to form a coordinating bond with Ni2+. Thus, further structural work is required to understand the DddK catalytic mechanism, with emphasis on the generation of structural data on DddK/DMSP complexes.
In this study, the DddK enzyme from P. ubique HTCC1062 was overexpressed and purified in Escherichia coli cells and then characterized. The crystal structures of wild-type (WT) DddK, a Y64A mutant (bearing a change of Y to A at position 64), and a Y122A mutant complexed with DMSP were solved. Based on structural and mutational analyses, the catalytic mechanism of DddK was predicted, in which an activated water molecule plays an important role in the deprotonation of the catalytic tyrosine (Tyr64). Furthermore, the Tyr122 residue can also act as the catalytic base when Tyr64 is mutated to phenylalanine. Sequence alignment and phylogenetic analysis suggest that the proposed DddK catalytic mechanism is relevant to all known SAR11 bacteria containing DddK and that DddW may adopt a catalytic mechanism similar to that of DddK. The results provide new insights into the catalytic mechanism of DddK and bacterial DMSP cleavage.
RESULTS AND DISCUSSION
Expression and characterization of DddK of P. ubique HTCC1062.
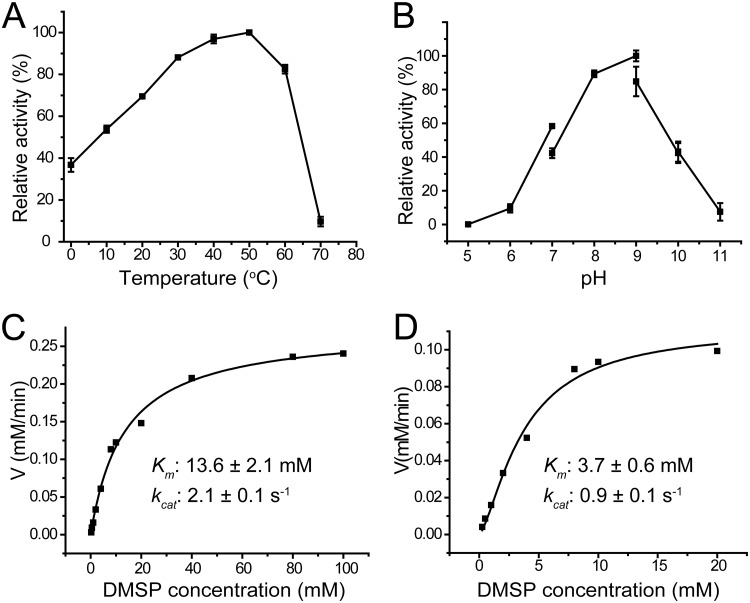
The dddK gene of P. ubique HTCC1062 contains 393 nucleotides and encodes a protein of 130 amino acid residues. Full-length dddK of P. ubique HTCC1062 was synthesized and was expressed in Escherichia coli BL21(DE3) cells, and the recombinant DddK was purified and characterized. The optimal temperature for DddK enzyme activity was ∼50°C (Fig. 1A), and the optimal pH was 9.0 (Fig. 1B). The optimal temperature and optimal pH for DddK are relatively high compared to those of marine environments. However, DddK maintained ∼70% of its highest enzymatic activity at 20°C, ∼85% at 30°C, ∼90% at pH 8.0, and 40% to 60% at pH 7.0 (Fig. 1A and B). Thus, DddK should function properly in physiological environments. The recombinant DddK exhibited a Km value of 13.6 mM for DMSP and a kcat value of 2.1 s−1 at pH 8.0 and 30°C (Fig. 1C). Its Km value at pH 7.0 was a little lower, but still in the millimolar level (3.7 mM) (Fig. 1D). The Km value of DddK determined in this study is lower than that reported previously for the same strain (82 mM) (12), which may be caused by the different buffer systems used. The relatively high Km value, in the millimolar range, is common in many DMSP catabolic enzymes, including DMSP lyases DddP, DddQ, DddY, DddW, and Alma1 and the DMSP demethylase DmdA (11, 20, 25–27, 30). The intracellular concentration of DMSP in P. ubique HTCC1062 can reach 180 mM, while the concentration of DMSP in seawater is in the nanomolar level (12), indicating that an efficient transporter for DMSP is required. In P. ubique HTCC1062, the transporter OpuAC, which belongs to the ATP binding cassette (ABC) superfamily, was speculated to import DMSP into cells (12).
FIG 1.
Characterization of recombinant DddK. (A) Effect of temperature on the enzymatic activity of DddK. (B) Effect of pH on the enzymatic activity of DddK. (C, D) Nonlinear fit curves for DMSP cleavage by DddK. The kinetic parameters were determined under pH 8.0 (C) and pH 7.0 (D) at 30°C.
Overall structure of DddK.
To elucidate the catalytic mechanism of DddK, we solved the crystal structure of WT DddK to 2.0 Å by the single-wavelength anomalous dispersion (SAD) method using a selenomethionine derivative (Se derivative) (Table 1). To further understand the DddK mechanism, it was important to obtain a structure of a DddK/DMSP complex, since the structure of DddK bound to diacrylate is the closest available (24). To obtain this, we constructed several mutants with lower enzymatic activities and cocrystalized the mutants with DMSP. Crystals were obtained for DddK containing Y64A and Y122A substitutions. After structural refinement, we found that only the structure of the Y122A mutant contains DMSP in its active site. The structures of WT DddK, the Y64A mutant, and the Y122A mutant/DMSP complex are very similar, with a root mean square deviation (RMSD) between WT DddK and the Y64A mutant of 0.27 Å and an RMSD between WT DddK and the Y122A mutant/DMSP complex of 0.19 Å. The structural analyses of DddK described below are based on the structure of WT DddK unless otherwise noted.
TABLE 1.
Crystallographic data collection and refinement parameters of DddK
| Parameter | Value(s)a for: |
|||
|---|---|---|---|---|
| Se derivative of DddK | WT DddK | Y64A mutant | Y122A mutant | |
| Diffraction data | ||||
| Space group | P21 | P21 | P21 | P21 |
| Unit cell dimensions | ||||
| a, b, c (Å) | 34.9, 56.9, 49.8 | 35.3, 57.4, 55.8 | 35.1, 57.4, 55.4 | 36.8, 49.1, 76.2 |
| α, β, γ (°) | 90.0, 91.0, 90.0 | 90.0, 94.2, 90.0 | 90.0, 94.1, 90.0 | 90.0, 100.1, 90.0 |
| Resolution range (Å) | 50.0–2.7 (2.75–2.70) | 50.0–2.0 (2.03–2.00) | 50.0–2.3 (2.34–2.30) | 50.0–1.6 (1.63–1.60) |
| Redundancy | 13.0 (10.9) | 12.8 (13.4) | 5.7 (5.8) | 6.4 (6.0) |
| Completeness (%) | 99.9 (98.8) | 99.7 (99.9) | 95.4 (98.9) | 98.9 (94.0) |
| Rmergeb | 0.1 (0.4) | 0.1 (0.3) | 0.1 (0.2) | 0.1 (0.3) |
| I/σI | 38.6 (14.7) | 49.1 (13.4) | 29.5 (7.0) | 37.6 (5.3) |
| Refinement statistics | ||||
| R factor | 0.17 | 0.22 | 0.16 | |
| Free R factor | 0.21 | 0.29 | 0.22 | |
| RMSD from ideal geometry | ||||
| Bond lengths (Å) | 0.006 | 0.007 | 0.005 | |
| Bond angles (°) | 1.0 | 1.0 | 0.9 | |
| Ramachandran plot (%) | ||||
| Favored | 96.4 | 93.5 | 97.9 | |
| Allowed | 3.6 | 6.5 | 2.1 | |
| Outliers | 0 | 0 | 0 | |
| Overall B factors (Å2) | 34.9 | 54.4 | 29.5 | |
Numbers in parentheses refer to data in the highest-resolution shell.
Rmerge = ∑hkl∑i|I(hkl)i − <I(hkl)>|/∑hkl∑iI(hkl)i, where I is the observed intensity, <I(hkl)> represents the average intensity, and I(hkl)i represents the observed intensity of each unique reflection.
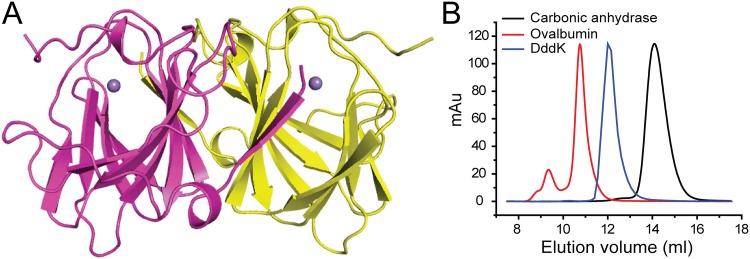
The crystals of DddK belong to the P21 space group, with two molecules arranged as a dimer in an asymmetric unit (Fig. 2A). Gel filtration analysis indicated that DddK is also a dimer in solution (Fig. 2B), which is further supported by the PISA server prediction (http://www.ebi.ac.uk/pdbe/prot_int/pistart.html). The overall structure of DddK is similar to that reported for Ni-DddK (24). DddK is composed of mainly beta strands that adopt a beta-barrel fold typical of cupin superfamily members, including DMSP lyases DddQ and DddY (20, 21, 26). The electron density map indicates that DddK contains a metal ion. Inductively coupled plasma atomic emission spectrometry measurements suggested that the metal ion is a Mn2+, occupying ∼34% of the DddK molecules. The element iron was estimated to occupy ∼24% of DddK molecules, and elements Co, Zn, and Ni were estimated to occupy less than 5% of the DddK molecules. It has been reported that DddK can utilize Ni2+, Mn2+, Co2+, Fe2+, Zn2+, or Cu2+ as a cofactor to catalyze DMSP cleavage (24). When the metal ion is fully replaced by Mn2+, DddK presents a relatively high enzymatic activity (24). Furthermore, SAR11 bacteria possess a manganese transporter, MntX, to take up Mn2+ (31), and thus, it is possible that DddK utilizes Mn2+ as a cofactor in natural environments. In addition to DddK, DMSP lyases DddQ and DddW can also utilize different metal ions for catalysis (25, 26, 32), suggesting that this might be an adaptation strategy of bacteria to survive in different environments.
FIG 2.
Overall structural analysis of DddK. (A) Overall structure of DddK. There are two DddK molecules arranged as a dimer in an asymmetric unit. One DddK molecule is colored in magenta, and the other is colored in yellow. The metal ion in DddK is shown as a purple sphere. (B) Gel filtration analysis of DddK. Carbonic anhydrase (13,700 Da; GE Healthcare) and ovalbumin (43,000 Da; GE Healthcare) were used as markers. The predicted molecular mass of DddK is 14,423 Da (https://web.expasy.org/protparam/).
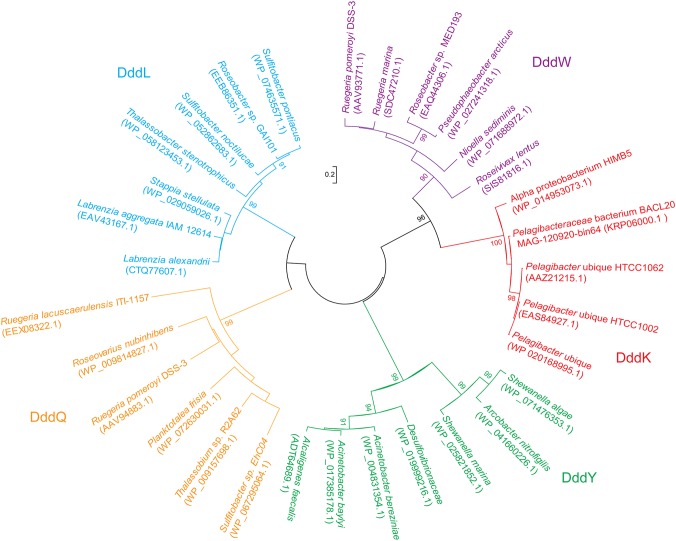
Of DMSP lyases belonging to the cupin superfamily, only DddW and DddK exist as dimers in solution (24, 25), while DddQ and DddY function as monomers (20, 26). The association state of DddL is currently unknown. Our phylogenetic analysis also suggested that DddK is more closely related to DddW, while DddL is more closely related to DddQ, and DddY forms a separate clade (Fig. 3).
FIG 3.
Neighbor-joining phylogenetic tree of DddK, DddW, DddL, DddQ, and DddY. Phylogenetic analysis was performed using MEGA version 6.0 (37).
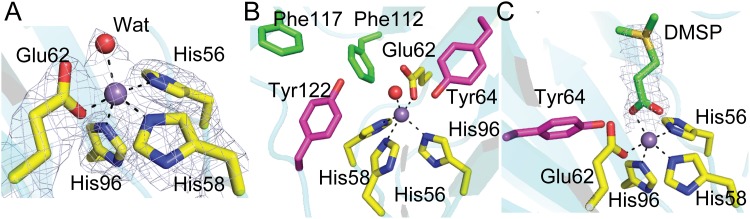
Key residues in the active site of DddK.
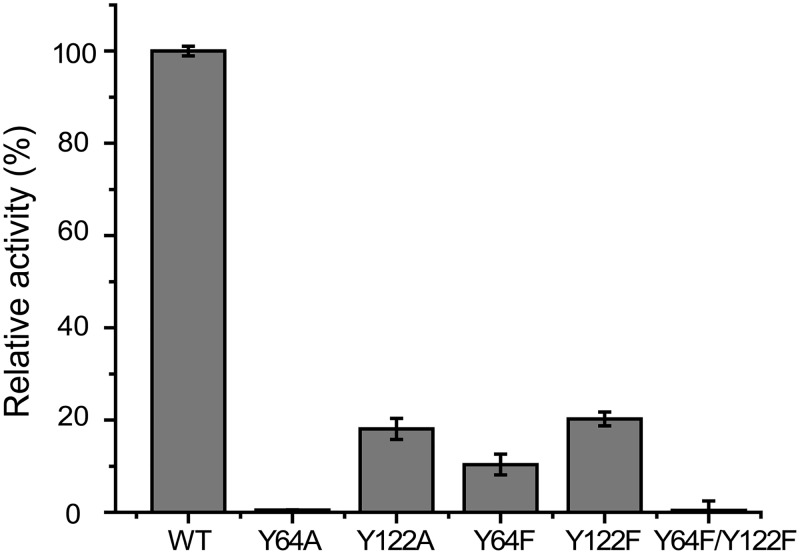
In the DddK crystal structure, residues His56, His58, Glu62, and His96 and a water molecule participate in coordinating Mn2+ (Fig. 4A). There are also two tyrosine residues and two phenylalanine residues in the active site (Fig. 4B). The overall B factor of DddK is moderate (Table 1), and the B factors of the active-site residues and Mn2+ are similar to those of other residues. Also, the electron densities of the active-site residues and Mn2+ are clear (Fig. 4A), indicating that the conformation of these residues is relatively stable. In the crystal structure of the Y122A mutant/DMSP complex, DMSP replaces the water molecule in the active site and coordinates with Mn2+ (Fig. 4C). Compared to the overall B factor of the Y122A mutant/DMSP complex (29.5 Å2), the B factor of DMSP (44.5 Å2) is relatively high. Moreover, the electron density of the sulfonium group of DMSP is weak (Fig. 4C), suggesting that the binding of DMSP is not strong. These structural observations are consistent with the biochemical results in which DddK exhibited low affinity for DMSP (Fig. 1C and D). Structural analyses indicated that Tyr64 and Tyr122 are close to DMSP, and both residues can act as a catalytic base to attack DMSP. To further ascertain the potential role of these residues in catalysis, we generated four mutants (with Y64A, Y122A, Y64F, and Y122F mutations) and studied their enzymatic activities (Fig. 5). Mutation of Tyr64 to alanine abolished the activity of DddK, while the Y122A mutant still maintained ∼20% enzymatic activity (Fig. 5), indicating that Tyr64 is likely the catalytic base of DddK, as was proposed by Schnicker et al. (24). However, it is surprising that the Y64F mutant maintained ∼10% enzymatic activity, because phenylalanine lacks the hydroxyl group necessary to act as a catalytic base. Therefore, there should be another residue which acts as a compensating catalytic base when Tyr64 is mutated to phenylalanine, contributing the residual 10% enzymatic activity. Based on structural analysis, Tyr122 is the only possible compensating catalytic base (Fig. 4B), which is supported by the data showing that the Y64F Y122F double mutation completely abolished the enzymatic activity of DddK (Fig. 5).
FIG 4.
Structural analyses of important residues in the active site of DddK. The Mn2+ in DddK is shown as a purple sphere, and the water molecule as a red sphere. (A) Residues and molecules involved in coordinating Mn2+ in DddK. The 2Fo-Fc densities for Mn2+, the water molecule, and selected DddK residues are contoured in blue-white (“bluewhite” per PyMOL software) at 1.0 σ. (B) Residues and molecules involved in DMSP cleavage. The possibly catalytic residues of DddK are colored in magenta. (C) Residues and molecules involved in DMSP cleavage in the Y122A mutant/DMSP complex. The 2Fo-Fc densities for DMSP are contoured in blue-white at 1.5 σ.
FIG 5.
Mutational analysis of residues of DddK that may participate in catalyzing DMSP cleavage. The enzymatic activity of WT DddK was defined as 100%.
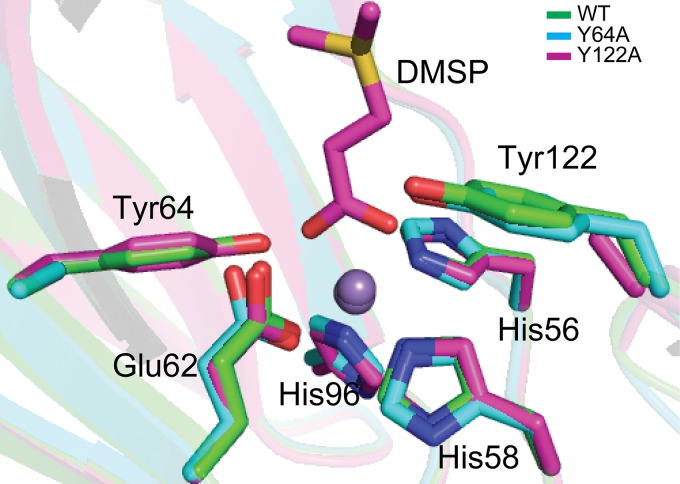
Tyrosine exhibits a high pKa in solution. To act as a catalytic base, it is necessary for a tyrosine to be deprotonated. Structural analysis suggested that Tyr64 of DddK can form a hydrogen bond with the activated water molecule which coordinates with Mn2+, with the distance between the hydroxyl group of Tyr64 and the activated water molecule being 2.6 Å. Interaction with the activated water molecule should help the deprotonation of Tyr64. A previous study of DddK suggested that Tyr64 might undergo a conformational change to coordinate with the metal to be deprotonated (24). However, the conformations of the key residues identified in the active sites of structures of WT DddK, the Y64F mutant, and the Y122F mutant/DMSP complex are nearly identical (Fig. 6), indicating that Tyr64 is unlikely to generate a conformational change during catalysis sufficient to allow metal coordination. There are two phenylalanine residues (Phe112 and Phe117) which are close to Tyr122 (Fig. 4C). The benzene ring of these two phenylalanine residues may attract the proton of the Tyr122 hydroxyl group via cation-π interactions, leading to the deprotonation of Tyr122.
FIG 6.
Structural alignment of important residues from WT DddK, the Y64A mutant, and the Y122A mutant/DMSP complex. The structure of WT DddK is colored in green, the structure of the Y64A mutant is in cyan, and the structure of the Y122A mutant/DMSP complex is in magenta. The Mn2+ in DddK is shown as a purple sphere.
Mechanism of DMSP cleavage by DddK to generate DMS and acrylate.
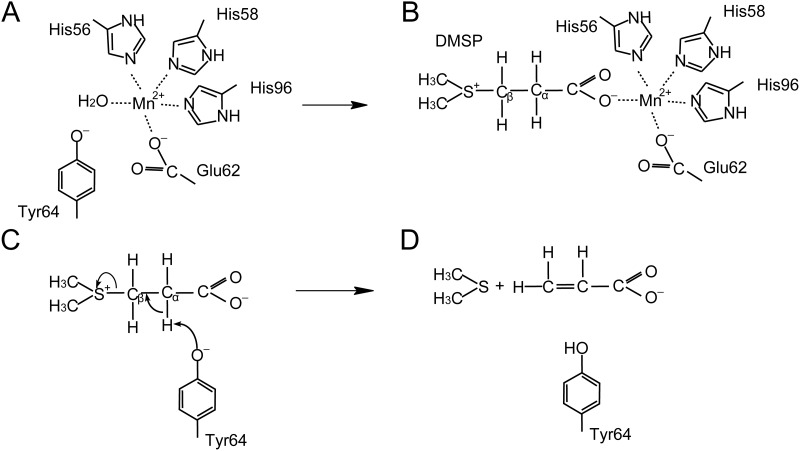
Although Tyr122 may facilitate DddK catalysis when Tyr64 is mutated to phenylalanine, there is no doubt that Tyr64 is the major catalytic residue utilized during DMSP cleavage, based on mutational analyses (Fig. 5). This is in agreement with the study by Schnicker et al. (24). Thus, the catalytic mechanism of DddK proposed below is based on Tyr64 acting as the catalytic base. In the absence of DMSP, residues His56, His58, Glu62, and His96 and a water molecule coordinate Mn2+ in the active site of DddK (Fig. 7A). After DMSP enters the active site of DddK, it replaces the water molecule and forms a new coordination bond with Mn2+ (Fig. 7B). Subsequently, Tyr64 likely attacks the Cα-H proton of DMSP, leading to the formation of a Cα carbanion, which attacks the Cβ of DMSP, breaking the Cβ-S bond of DMSP (Fig. 7C). Consequently, DMSP is cleaved into DMS and acrylate, which are released from the active site of DddK (Fig. 7D).
FIG 7.
Catalytic mechanism by which DddK cleaves DMSP. (A) In the absence of DMSP, Mn2+ is coordinated by residues His56, His58, Glu62, and His96 and a water molecule. (B) DMSP displaces the water molecule and forms a new coordination bond with Mn2+. (C) The residue Tyr64 acts as the catalytic base to initiate the reaction. (D) DMS and acrylate are generated from DMSP cleavage.
Considering that dddK homologues have been found in 8 of 12 SAR11 Ia genomes (12), it is important to elucidate the catalytic mechanism in detail. Based on our structural and mutational data, during the catalysis of DddK, it is an activated water molecule that likely allows Tyr64 to be deprotonated and gain the ability to be a catalytic base, which is different from the previous report that it is the conformational change of Tyr64 that performs this (24).
Universality of the catalytic mechanism of DddK.
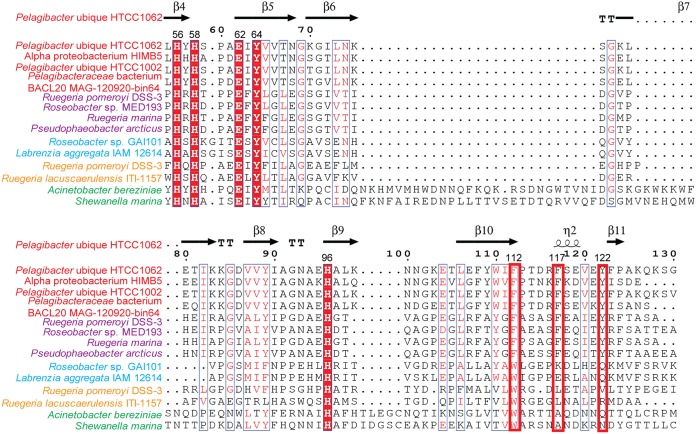
To investigate the ubiquity of the DddK catalytic mechanism, we performed sequence alignment of DddK proteins and other DMSP lyases that belong to the cupin superfamily (Fig. 8). The residues coordinating the metal ion (His56, His58, Glu62, and His96) and the major catalytic residue (Tyr64) are highly conserved in all DMSP lyases that belong to the cupin superfamily (Fig. 8). Residues involved in facilitating the catalysis when Tyr64 is mutated to phenylalanine (Tyr122, Phe112, and Phe117) are only conserved in DddK and DddW proteins (Fig. 8). Phylogenetic analysis also indicated that DddK is more closely related to DddW (Fig. 3). These results suggest that the proposed catalytic mechanism of DddK has universal significance in SAR11 bacteria containing DddK and that DddW may adopt a catalytic mechanism similar to that of DddK.
FIG 8.
Sequence alignment of DddK, DddW, DddL, DddQ, and DddY proteins. Numbers in parentheses refer to the amino acid numbers in the DddK sequence from P. ubique HTCC1062. Bacteria containing the different proteins are colored as follows: red for DddK, purple for DddW, cyan for DddL, yellow for DddQ, and green for DddY.
Conclusion.
The cleavage of DMSP to generate DMS is an important step in the global sulfur cycle. SAR11 bacteria are the most abundant bacteria in the ocean and are very likely important environmental DMS producers via the DddK DMSP lyase pathway, since dddK homologues are found in 8 of 12 SAR11 Ia genomes and dddK transcripts are relatively abundant in seawater samples. In this study, the crystal structures of WT DddK, the Y64A mutant, and the Y122A mutant/DMSP complex were solved and the catalytic mechanism of DddK is explained based on structural and mutational analyses. The results provide important new data on the structure of DddK and the molecular mechanism of DddK catalysis for DMSP cleavage. Sequence alignment indicated that the catalytic mechanisms of DddK proteins from different SAR11 bacteria may be similar, thereby having universal significance. This study enriches our understanding of DMSP cleavage into DMS in SAR11 bacteria.
MATERIALS AND METHODS
Gene cloning, point mutation, and protein expression and purification.
The 393-bp full-length dddK gene from P. ubique HTCC1062 was synthesized by the Beijing Genomics Institute (China). The gene was then subcloned into the pET22b (Novagen, USA) vector with a C-terminal His tag. All of the point mutations in DddK were introduced using PCR-based methods and were verified by DNA sequencing. The DddK protein and all of its mutants were expressed in E. coli BL21(DE3). The cells were cultured at 37°C in lysogeny broth medium to an optical density at 600 nm (OD600) of 0.8 to 1.0 and then induced at 18°C for 14 h with 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG). The proteins were first purified with Ni2+-nitrilotriacetic acid (NTA) resin (Qiagen, Germany) and then fractionated by gel filtration on a Superdex-75 column (GE Healthcare, USA). The selenomethionine derivative of DddK was overexpressed in E. coli BL21(DE3) under 0.5 mM IPTG induction in M9 minimal medium supplemented with selenomethionine, lysine, valine, threonine, leucine, isoleucine, and phenylalanine. The recombinant selenomethionine derivative was purified in a manner similar to that used for WT DddK.
Enzyme assays.
The enzymatic activity of DddK was measured by detecting the production of acrylate as previously described (27). DddK (at a final concentration of 1.5 μM) and DMSP (at a final concentration of 20 mM) were mixed with reaction buffer containing 100 mM Tris-HCl (pH 9.2) in a total volume of 200 μl. After the mixture was incubated at 50°C for 15 min, the reaction was stopped by adding perchloric acid, and the amount of acrylate in the reaction mixture was detected by high-performance liquid chromatography (HPLC) on a Sunfire C18 column (Waters, Ireland). To determine the optimal temperature for DddK, reaction mixtures were incubated at 0°C, 10°C, 20°C, 30°C, 40°C, 50°C, 60°C, or 70°C for 15 min. The optimum pH for DddK was examined at 50°C (the optimal temperature for DddK enzymatic activity) using Bis-Tris buffer for pH 5 to 7, Tris buffer for pH 7 to 9, glycine buffer for pH 9 to 10, and Na2CO3 buffer for pH 10 to 11. The kinetic parameters of DddK were determined by nonlinear analysis based on the initial rates determined with 2.2 μM DddK and 0.25 to 100 mM DMSP. The measurement was performed under conditions of pH 7.0 and pH 8.0 at 30°C.
Crystallization and data collection.
The purified DddK protein was concentrated to ∼20 mg/ml in 10 mM Tris-HCl (pH 8.0) and 100 mM NaCl. Initial crystallization trials for DddK were performed at 18°C using the sitting-drop vapor diffusion method. Diffraction-quality crystals of DddK were obtained in hanging drops containing 0.2 M sodium chloride, 0.1 M HEPES (pH 7.5), and 25% (wt/vol) polyethylene glycol (PEG) 3350 at 18°C after a 2-week incubation. Crystals of the DddK Se derivative were obtained in hanging drops containing 0.2 M sodium chloride, 0.1 M HEPES (pH 7.5), and 25% (wt/vol) PEG 4000 at 18°C after a 2-week incubation. To obtain the structure of the DddK/DMSP complex, DddK Y64A and Y122A mutants were cocrystallized with DMSP (1 mM). After structural refinement, however, we found that only the structure of the Y122A mutant contains DMSP in its active site. Crystals of the Y64A mutant were obtained in hanging drops containing 0.1 M Tris (pH 8.5) and 25% PEG 3350 at 18°C after a 2-week incubation. Crystals of the Y122A mutant/DMSP complex were obtained in hanging drops containing 0.1 M HEPES (pH 7.5) and 25% PEG 3350 at 18°C after a 2-week incubation. X-ray diffraction data were collected on the BL18U1 and BL19U1 beamlines at the Shanghai Synchrotron Radiation Facility. The initial diffraction data sets were processed using the HKL3000 program with its default settings (33).
Structure determination and refinement.
The crystals of WT DddK, the Y64A mutant, and the Y122A mutant/DMSP complex all belong to the P21 space group. The structure of the DddK Se derivative was determined by single-wavelength anomalous dispersion (SAD) phasing. The crystal structures of WT DddK, the Y64A mutant, and the Y122A mutant/DMSP complex were determined by molecular replacement using the CCP4 program Phaser (34) with the structure of the DddK Se derivative as the search model. The refinements of these structures were performed using Coot (35) and Phenix (36). The default parameters in CCP4, Coot, and Phenix were used. All the structure figures were processed using the program PyMOL (http://www.pymol.org/).
Detection of metal ions.
Inductively coupled plasma atomic emission spectrometry measurement was performed using an IRIS Intrepid II XSP (Thermo Electron, USA). To detect the metal ions in DddK, 1 ml of DddK protein (20 mg/ml) was mixed with 10 ml nitric acid. The sample was incubated at 140°C overnight until the digestion was complete. The sample was then diluted to 5 ml with distilled water and filtered through a 0.22-μm filter membrane before detection.
Accession numbers.
The structures of WT DddK, the Y64A mutant, and the Y122A mutant/DMSP complex have been deposited in PDB under accession codes 6A53, 6A54, and 6A55, respectively.
ACKNOWLEDGMENTS
We thank the staffs from BL18U1 and BL19U1 beamlines of National Facility for Protein Sciences Shanghai (NFPS) and Shanghai Synchrotron Radiation Facility for assistance during data collection.
This work was supported by the National Key Research and Development Program of China (grants number 2016YFA0601303 and 2018YFC1406700), the National Science Foundation of China (grants number 31630012, 31728001, U1706207, 41706152, and 91851205), the AoShan Talents Cultivation Program supported by Qingdao National Laboratory for Marine Science and Technology (grant number 2017ASTCP-OS14), the Program of Shandong for Taishan Scholars (grant number TS20090803), funding from the Key Laboratory of Global Change and Marine-Atmospheric Chemistry of the State Oceanic Administration (grant number 2018GCMAC16), the National Postdoctoral Program for Innovative Talents (grant number BX201600095), the China Postdoctoral Science Foundation (grant number 2017M610420), the Natural Science Foundation of Jiangsu Province (grant number BK20170397), the Natural Science Foundation of Shandong Province (grant number ZR2017BC079), and the Shandong Province Postdoctoral Innovation Projects (grant number 201701002).
We declare that we have no conflicts of interest with the contents of this article.
REFERENCES
- 1.Curson AR, Todd JD, Sullivan MJ, Johnston AW. 2011. Catabolism of dimethylsulphoniopropionate: microorganisms, enzymes and genes. Nat Rev Microbiol 9:849–859. doi: 10.1038/nrmicro2653. [DOI] [PubMed] [Google Scholar]
- 2.Seymour JR, Simo R, Ahmed T, Stocker R. 2010. Chemoattraction to dimethylsulfoniopropionate throughout the marine microbial food web. Science 329:342–345. doi: 10.1126/science.1188418. [DOI] [PubMed] [Google Scholar]
- 3.Ksionzek KB, Lechtenfeld OJ, McCallister SL, Schmitt-Kopplin P, Geuer JK, Geibert W, Koch BP. 2016. Dissolved organic sulfur in the ocean: biogeochemistry of a petagram inventory. Science 354:456–459. doi: 10.1126/science.aaf7796. [DOI] [PubMed] [Google Scholar]
- 4.Curson ARJ, Williams BT, Pinchbeck BJ, Sims LP, Martinez AB, Rivera PPL, Kumaresan D, Mercade E, Spurgin LG, Carrion O, Moxon S, Cattolico RA, Kuzhiumparambil U, Guagliardo P, Clode PL, Raina JB, Todd JD. 2018. DSYB catalyses the key step of dimethylsulfoniopropionate biosynthesis in many phytoplankton. Nat Microbiol 3:430–439. doi: 10.1038/s41564-018-0119-5. [DOI] [PubMed] [Google Scholar]
- 5.Kiene RP, Linn LJ, Bruton JA. 2000. New and important roles for DMSP in marine microbial communities. J Sea Res 43:209–224. doi: 10.1016/S1385-1101(00)00023-X. [DOI] [Google Scholar]
- 6.Raina JB, Tapiolas DM, Foret S, Lutz A, Abrego D, Ceh J, Seneca FO, Clode PL, Bourne DG, Willis BL, Motti CA. 2013. DMSP biosynthesis by an animal and its role in coral thermal stress response. Nature 502:677–680. doi: 10.1038/nature12677. [DOI] [PubMed] [Google Scholar]
- 7.Stefels J, Steinke M, Turner S, Malin G, Belviso S. 2007. Environmental constraints on the production and removal of the climatically active gas dimethylsulphide (DMS) and implications for ecosystem modelling. Biogeochemistry 83:245–275. doi: 10.1007/s10533-007-9091-5. [DOI] [Google Scholar]
- 8.Curson AR, Liu J, Bermejo Martinez A, Green RT, Chan Y, Carrion O, Williams BT, Zhang SH, Yang GP, Bulman Page PC, Zhang XH, Todd JD. 2017. Dimethylsulfoniopropionate biosynthesis in marine bacteria and identification of the key gene in this process. Nat Microbiol 2:17009. doi: 10.1038/nmicrobiol.2017.9. [DOI] [PubMed] [Google Scholar]
- 9.Simo R, Archer SD, Pedros-Alio C, Gilpin L, Stelfox-Widdicombe CE. 2002. Coupled dynamics of dimethylsulfoniopropionate and dimethylsulfide cycling and the microbial food web in surface waters of the North Atlantic. Limnol Oceanogr 47:53–61. doi: 10.4319/lo.2002.47.1.0053. [DOI] [Google Scholar]
- 10.Stefels J. 2000. Physiological aspects of the production and conversion of DMSP in marine algae and higher plants. J Sea Res 43:183–197. doi: 10.1016/S1385-1101(00)00030-7. [DOI] [Google Scholar]
- 11.Alcolombri U, Ben-Dor S, Feldmesser E, Levin Y, Tawfik DS, Vardi A. 2015. Marine sulfur cycle. Identification of the algal dimethyl sulfide-releasing enzyme: a missing link in the marine sulfur cycle. Science 348:1466–1469. doi: 10.1126/science.aab1586. [DOI] [PubMed] [Google Scholar]
- 12.Sun J, Todd JD, Thrash JC, Qian Y, Qian MC, Temperton B, Guo J, Fowler EK, Aldrich JT, Nicora CD, Lipton MS, Smith RD, De Leenheer P, Payne SH, Johnston AW, Davie-Martin CL, Halsey KH, Giovannoni SJ. 2016. The abundant marine bacterium Pelagibacter simultaneously catabolizes dimethylsulfoniopropionate to the gases dimethyl sulfide and methanethiol. Nat Microbiol 1:16065. doi: 10.1038/nmicrobiol.2016.65. [DOI] [PubMed] [Google Scholar]
- 13.Moran MA, Reisch CR, Kiene RP, Whitman WB. 2012. Genomic insights into bacterial DMSP transformations. Annu Rev Mar Sci 4:523–542. doi: 10.1146/annurev-marine-120710-100827. [DOI] [PubMed] [Google Scholar]
- 14.Reisch CR, Stoudemayer MJ, Varaljay VA, Amster IJ, Moran MA, Whitman WB. 2011. Novel pathway for assimilation of dimethylsulphoniopropionate widespread in marine bacteria. Nature 473:208–211. doi: 10.1038/nature10078. [DOI] [PubMed] [Google Scholar]
- 15.Johnston AWB, Green RT, Todd JD. 2016. Enzymatic breakage of dimethylsulfoniopropionate—a signature molecule for life at sea. Curr Opin Chem Biol 31:58–65. doi: 10.1016/j.cbpa.2016.01.011. [DOI] [PubMed] [Google Scholar]
- 16.Kettle AJ, Andreae MO. 2000. Flux of dimethylsulfide from the oceans: a comparison of updated data seas and flux models. J Geophys Res 105:26793–26808. doi: 10.1029/2000JD900252. [DOI] [Google Scholar]
- 17.Vallina SM, Simó R. 2007. Strong relationship between DMS and the solar radiation dose over the global surface ocean. Science 315:506–508. doi: 10.1126/science.1133680. [DOI] [PubMed] [Google Scholar]
- 18.Todd JD, Rogers R, Li YG, Wexler M, Bond PL, Sun L, Curson AR, Malin G, Steinke M, Johnston AW. 2007. Structural and regulatory genes required to make the gas dimethyl sulfide in bacteria. Science 315:666–669. doi: 10.1126/science.1135370. [DOI] [PubMed] [Google Scholar]
- 19.Todd J, Curson A, Dupont C, Nicholson P, Johnston A. 2009. The dddP gene, encoding a novel enzyme that converts dimethylsulfoniopropionate into dimethyl sulfide, is widespread in ocean metagenomes and marine bacteria and also occurs in some Ascomycete fungi. Environ Microbiol 11:1376–1385. doi: 10.1111/j.1462-2920.2009.01864.x. [DOI] [PubMed] [Google Scholar]
- 20.Li CY, Zhang D, Chen XL, Wang P, Shi WL, Li PY, Zhang XY, Qin QL, Todd JD, Zhang YZ. 2017. Mechanistic insights into dimethylsulfoniopropionate lyase DddY, a new member of the cupin superfamily. J Mol Biol 429:3850–3862. doi: 10.1016/j.jmb.2017.10.022. [DOI] [PubMed] [Google Scholar]
- 21.Dunwell JM, Purvis A, Khuri S. 2004. Cupins: the most functionally diverse protein superfamily? Phytochemistry 65:7–17. doi: 10.1016/j.phytochem.2003.08.016. [DOI] [PubMed] [Google Scholar]
- 22.Lei L, Cherukuri KP, Alcolombri U, Meltzer D, Tawfik DS. 2018. The dimethylsulfoniopropionate (DMSP) lyase and lyase-like cupin family consists of bona fide DMSP lyases as well as other enzymes with unknown function. Biochemistry 57:3364–3377. doi: 10.1021/acs.biochem.8b00097. [DOI] [PubMed] [Google Scholar]
- 23.Dunwell JM. 1998. Cupins: a new superfamily of functionally diverse proteins that include germins and plant storage proteins. Biotechnol Genet Eng Rev 15:1–32. doi: 10.1080/02648725.1998.10647950. [DOI] [PubMed] [Google Scholar]
- 24.Schnicker NJ, De Silva SM, Todd JD, Dey M. 2017. Structural and biochemical insights into dimethylsulfoniopropionate cleavage by cofactor-bound DddK from the prolific marine bacterium Pelagibacter. Biochemistry 56:2873–2885. doi: 10.1021/acs.biochem.7b00099. [DOI] [PubMed] [Google Scholar]
- 25.Brummett AE, Schnicker NJ, Crider A, Todd JD, Dey M. 2015. Biochemical, kinetic, and spectroscopic characterization of Ruegeria pomeroyi DddW—a mononuclear iron-dependent DMSP lyase. PLoS One 10:e0127288. doi: 10.1371/journal.pone.0127288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li CY, Wei TD, Zhang SH, Chen XL, Gao X, Wang P, Xie BB, Su HN, Qin QL, Zhang XY, Yu J, Zhang HH, Zhou BC, Yang GP, Zhang YZ. 2014. Molecular insight into bacterial cleavage of oceanic dimethylsulfoniopropionate into dimethyl sulfide. Proc Natl Acad Sci U S A 111:1026–1031. doi: 10.1073/pnas.1312354111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang P, Chen XL, Li CY, Gao X, Zhu DY, Xie BB, Qin QL, Zhang XY, Su HN, Zhou BC, Xun LY, Zhang YZ. 2015. Structural and molecular basis for the novel catalytic mechanism and evolution of DddP, an abundant peptidase-like bacterial dimethylsulfoniopropionate lyase: a new enzyme from an old fold. Mol Microbiol 98:289–301. doi: 10.1111/mmi.13119. [DOI] [PubMed] [Google Scholar]
- 28.Hehemann JH, Law A, Redecke L, Boraston AB. 2014. The structure of RdDddP from Roseobacter denitrificans reveals that DMSP lyases in the DddP-family are metalloenzymes. PLoS One 9:e103128. doi: 10.1371/journal.pone.0103128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morris RM, Rappe MS, Connon SA, Vergin KL, Siebold WA, Carlson CA, Giovannoni SJ. 2002. SAR11 clade dominates ocean surface bacterioplankton communities. Nature 420:806–810. doi: 10.1038/nature01240. [DOI] [PubMed] [Google Scholar]
- 30.Reisch CR, Moran MA, Whitman WB. 2008. Dimethylsulfoniopropionate-dependent demethylase (DmdA) from Pelagibacter ubique and Silicibacter pomeroyi. J Bacteriol 190:8018–8024. doi: 10.1128/JB.00770-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Green RT, Todd JD, Johnston AW. 2013. Manganese uptake in marine bacteria; the novel MntX transporter is widespread in Roseobacters, Vibrios, Alteromonadales and the SAR11 and SAR116 clades. ISME J 7:581–591. doi: 10.1038/ismej.2012.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brummett AE, Dey M. 2016. New mechanistic insight from substrate- and product-bound structures of the metal-dependent dimethylsulfoniopropionate lyase DddQ. Biochemistry 55:6162–6174. doi: 10.1021/acs.biochem.6b00585. [DOI] [PubMed] [Google Scholar]
- 33.Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. 2006. HKL-3000: the integration of data reduction and structure solution—from diffraction images to an initial model in minutes. Acta Crystallogr D Biol Crystallogr 62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 34.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AGW, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. 2011. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]