Aspergillus oryzae is an industrially important filamentous fungus; therefore, a clear understanding of its polysaccharide metabolism and utilization is very important for its industrial utilization. In this study, we revealed that the basic-region helix-loop-helix (bHLH) transcription factor AoDevR is importantly involved in chitin and starch metabolism in A. oryzae. The overexpression of AodevR strongly suppressed the expression of amylase-related genes. The results of a yeast one-hybrid assay suggested that the DevR protein potentially interacts with the promoter of amyR, which encodes a transcription factor involved in amylase production and starch utilization. This study provides new insight for further revealing the regulation mechanism of amylase production in A. oryzae.
KEYWORDS: α-amylase production, Aspergillus oryzae, bHLH transcription factor, polysaccharide metabolism, yeast one hybrid
ABSTRACT
Basic-region helix-loop-helix (bHLH) proteins are a superfamily of transcription factors that are often involved in the control of growth and differentiation. Recently, it was reported that the bHLH transcription factor DevR is involved in both asexual and sexual development in Aspergillus nidulans and regulates the conidial melanin production in Aspergillus fumigatus. In this study, we identified and characterized an Aspergillus oryzae gene that showed high similarity with devR of A. nidulans and A. fumigatus (AodevR). In the AodevR-disrupted strain, growth was delayed and the number of conidia was decreased on Czapek-Dox (CD) minimal agar plates, but the conidiation was partially recovered by adding 0.6 M KCl. Simultaneously, the overexpression of AodevR was induced and resulted in extremely poor growth when the carbon source changed from glucose to polysaccharide (dextrin) in the CD agar plate. Scanning electron microscopy (SEM) indicated that the overexpression of AodevR resulted in extremely thin aberrant hyphal morphology. Conversely, the deletion of AodevR resulted in thicker hyphae and in more resistance to Congo red relative to the control strain. Quantitative reverse transcriptase PCR (RT-PCR) further indicated that AoDevR significantly affects chitin and starch metabolism, and importantly, the overexpression of AodevR inhibited the expression of genes related to starch degradation. A yeast one-hybrid assay suggested that the DevR protein possibly interacted with the promoter of amyR, which encodes a transcription factor involved in amylase production. Importantly, AoDevR is involved in polysaccharide metabolism and affects the growth of the A. oryzae strain.
IMPORTANCE Aspergillus oryzae is an industrially important filamentous fungus; therefore, a clear understanding of its polysaccharide metabolism and utilization is very important for its industrial utilization. In this study, we revealed that the basic-region helix-loop-helix (bHLH) transcription factor AoDevR is importantly involved in chitin and starch metabolism in A. oryzae. The overexpression of AodevR strongly suppressed the expression of amylase-related genes. The results of a yeast one-hybrid assay suggested that the DevR protein potentially interacts with the promoter of amyR, which encodes a transcription factor involved in amylase production and starch utilization. This study provides new insight for further revealing the regulation mechanism of amylase production in A. oryzae.
INTRODUCTION
Aspergillus oryzae is regarded as an industrially important filamentous fungus utilized for traditional fermentative manufacture and commercial enzyme production. Despite the significance of A. oryzae in the field of industrial production and the fact that its whole genome has been sequenced (1, 2), the function of a large number of genes encoding proteins is still unclear, which limits its industrial production and utilization.
The basic helix-loop-helix (bHLH) proteins belong to a superfamily of transcription factors widely distributed in eukaryotic organisms, with members involved in developmental processes, including cellular proliferation and differentiation. The bHLH domain of this protein family is approximately 60 amino acids, including two functionally distinct regions: the basic region and the helix-loop-helix region (3, 4). It has been reported that some bHLH proteins can form homodimers or heterodimers and bind to sequences containing a consensus core element called the E box (5′-CANNTG-3′) to affect the transcription of downstream target genes (5).
In the Aspergillus genus of filamentous fungi, some bHLH transcription factors have been described. These transcription factors are reported to be important for penicillin biosynthesis, cell cycle progression, sexual and asexual development, and other cellular regulations and physiological functions (6–9). In Aspergillus oryzae, bHLH transcription factor SclR was found to play an important role in hyphal morphology, asexual conidiospore formation, and sclerotial production (10, 11). At the same time, the deletion of sclR leads to rapid cell lysis and protein degradation under liquid culture conditions. Recently, it was reported that SclR is significantly involved in carbohydrate metabolism in A. oryzae (12). In addition, sclR overexpression may also promote the formation of heterokaryotic sclerotia and heterokaryotic fusants formed by hyphal fusion before or during sclerotial formation in A. oryzae (13). Many bHLH proteins form homodimers and heterodimers with more than one partner, some of which also exhibit bifunctional roles as activators or repressors of downstream target gene transcription to perform their own complex functions (14–16). In A. oryzae, another bHLH transcription factor, EcdR, has also been characterized. EcdR plays a major role in conidiation, especially at the early stage of conidiophore development in A. oryzae. Yeast two-hybrid assays indicated that EcdR and SclR, both bHLH proteins, competitively interacted with each other to form homo/heterodimers and resulted in a mutual inhibition of function in A. oryzae (17). Except for these, it was reported that DevR, a bHLH transcription factor, is important for both asexual and sexual development in Aspergillus nidulans (18). The devR gene deletion showed that the gene is nonessential for vegetative growth. However, the deletion caused wrinkled colonies and resulted in a yellow pigment, and the strain did not form conidia on minimal agar plates. Additionally, DevR also regulates conidial melanin production in Aspergillus fumigatus (19). As an essential component of the cell wall, melanin plays a significant role in protecting organisms from external stresses (20). Dihydroxynaphthalene (DHN)-melanin is responsible for the pigmentation of A. fumigatus conidia, and six genes are involved in its biosynthesis (21–23). Among these genes, pksP, a polyketide synthase gene, plays a core role. It has been reported that DevR and another MADS box transcription factor, RlmA, may bind to the pksP promoter to induce the transcription activity of pksP, thus regulating melanin production (19).
In this study, a gene (AO090026000797) homologous to A. nidulans devR was isolated and characterized in A. oryzae and was designated AodevR. The deletion of AodevR resulted in thicker hyphae and more resistance to Congo red than in the control strain. Conversely, the overexpression of AodevR resulted in extremely thin aberrant hyphal morphology, and the strain displayed increasing sensitivity to Congo red. Quantitative PCR (qPCR) results indicated that the expression levels of certain genes related to chitin and starch metabolisms were significantly changed in the AodevR deletion and/or overexpression strains, suggesting that AoDevR is potentially involved in polysaccharide metabolism in A. oryzae.
RESULTS
Deletion and overexpression of the AodevR gene in A. oryzae.
In this study, we searched for the gene homologous to A. nidulans devR in the A. oryzae genome database (http://www.bio.nite.go.jp/dogan/project/view/AO) using the BLAST search and found AO090026000797, which was designated AodevR. The identified AodevR gene encodes a protein of 510 amino acids, which shares relatively high similarity with DevR proteins of A. nidulans (69.6%) and A. fumigatus (76.9%). Especially as a member of the bHLH family, the bHLH domain of this protein shows even higher similarity and displays 100% and 92.3% identity with those of A. nidulans and A. fumigatus, respectively (see Fig. S1 in the supplemental material). At the same time, the protein also shares relatively high similarity with hypothetical proteins in other Aspergillus species or its related genera, such as Penicillium.
To investigate the function of DevR in A. oryzae, the AodevR gene disruptant and its overexpressing strains were constructed. First, the devR gene deletion cassette was constructed by fusion PCR as described in Materials and Methods (10). The constructed deletion cassette was transformed into the parent strain, RkuN16ptr1, and grown on selective Czapek-Dox (CD) agar medium. Genomic DNA obtained from the transformants was digested with SmaI, and the deletion of AodevR was confirmed using Southern blot analysis (see Fig. S2A and B). Additionally, the devR-overexpressing strain was constructed with pyrG as a selectable marker. The A. oryzae amyB promoter was used to induce overexpression of devR (Fig. S2C). Genomic DNA of the transformants was isolated and digested with PstI, and then a Southern blot analysis was performed using the devR open reading frame (ORF) as a probe. The change in band shift indicated that the plasmid for AodevR overexpression was integrated into the chromosome of the strain (Fig. S2D). To validate further whether the AodevR was overexpressed, the transcription level of AodevR was investigated by semiquantitative reverse transcriptase PCR (RT-PCR) analysis. Dextrin-containing CD medium was used to induce the A. oryzae amyB promoter, and the result showed that AodevR was more effectively transcribed in the AodevR-overexpressing strain (Fig. S2E).
AoDevR affects the growth of the strain and conidiation.
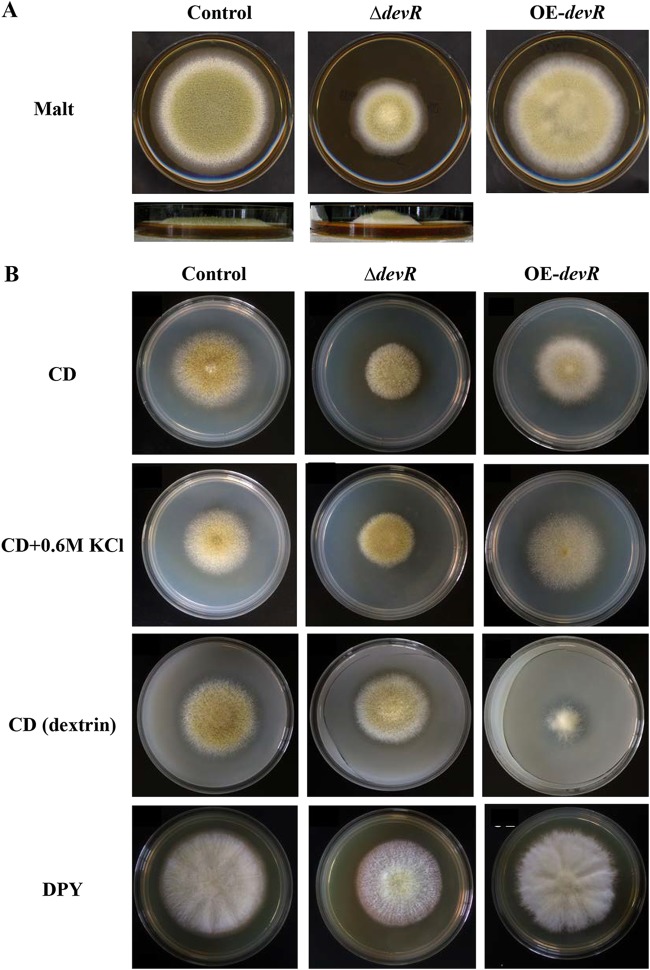
The generated AodevR-disrupted and AodevR-overexpressing strains were first spot cultivated onto malt agar medium, and their phenotypes were observed and compared. In contrast to the control strain, the AodevR-disrupted strain formed compact colonies with significantly reduced diameters. Although the deletion of AodevR caused a growth defect, the aerial hyphae grew taller (Fig. 1A). The ΔdevR strain also showed a significant decrease in conidiation compared to that of the control strain (Fig. 1A). In addition, the AodevR-overexpressing strain only displayed slightly delayed growth and reduced conidial formation compared to those of the control strain (Fig. 1A). The growth of these strains was further compared under different culture conditions. On CD minimal medium, both devR-overexpressing (OE-devR) strains showed a delay in growth and conidial formation compared to those of the control strain (Fig. 1B). However, the decreased conidiation in the ΔdevR strain was partially recovered by adding 0.6 M KCl. In addition, complementation of the AodevR deletion was performed by reintegration of the AodevR gene. The restoration of the parental phenotype suggested that the phenotype of the ΔdevR strain can be conclusively attributed to AodevR (see Fig. S3). On the CD (dextrin) plate, when the amyB promoter was induced by using dextrin as a carbon source, the OE-devR strain exhibited an extremely poor growth rate, and no conidia were observed (Fig. 1B). The result suggested that the overexpression of AodevR may strongly inhibit the growth of the strain. Interestingly, under nutrition-rich conditions, such as on the dextrin-polypeptone-yeast extract (DPY) and malt plates, the strong inhibitory effect of the OE-devR strain was undetectable (Fig. 1A and B).
FIG 1.
Phenotypic comparison. The AodevR-disrupted, AodevR-overexpressing, and control strains were spot-cultivated on malt agar medium (A) and different plates (B) at 30°C for 5 days, and their phenotypes were observed and compared. ΔdevR, AodevR-disrupted strain; OE-devR, AodevR-overexpressing strain.
AoDevR affects hyphal morphology.
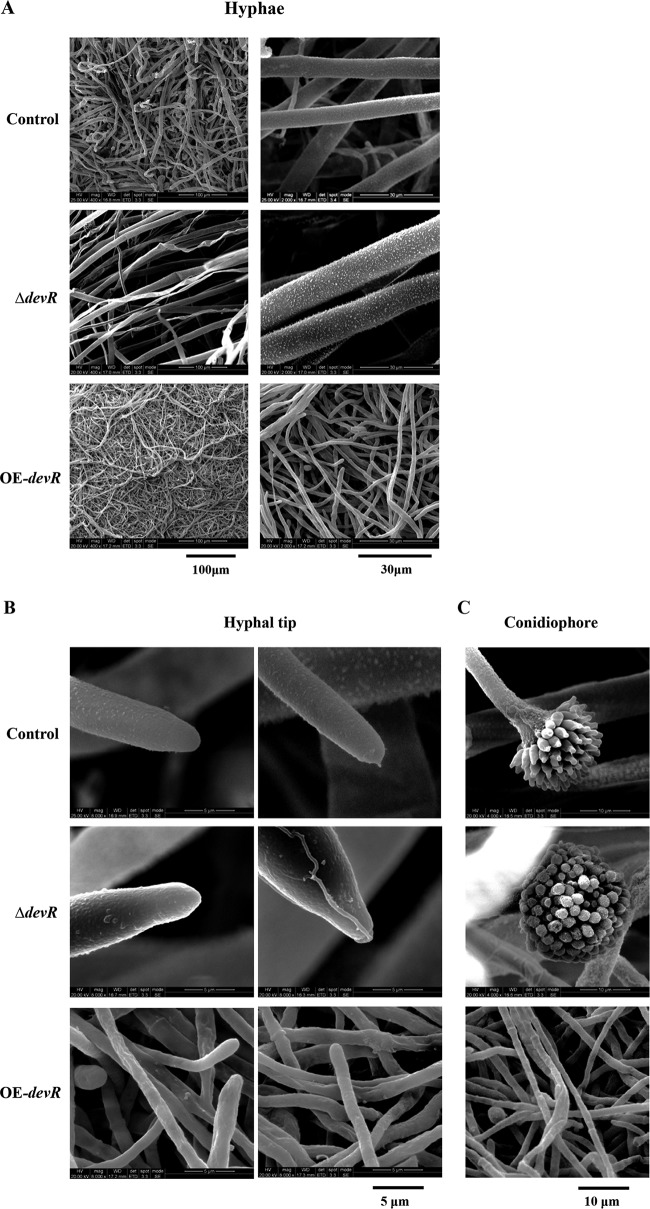
To observe hyphal morphology, the constructed AodevR-disrupted strain, the AodevR-overexpressing strain, and the control strain were inoculated on the CD (dextrin) plates at 30°C for 2 days. The marginal regions of the colonies were cut, fixed, and dehydrated. Results from observation by scanning electron microscopy (SEM) indicated that compared to the control strain, the ΔdevR strain exhibited thicker hyphae, and conversely, the OE-devR strain showed extremely thin and dense hyphae (Fig. 2A). The aberrant hyphal morphogenesis was induced by the overexpression of AodevR. We further observed the difference in the morphology at the hyphal tip. SEM indicated that the hyphal tip of the ΔdevR strain was not only thicker but also sharper than that of control strain (Fig. 2B). In addition, the ΔdevR strain produced normal conidia; however, in the OE-devR strain, almost no conidia were observed in the whole view (Fig. 2C).
FIG 2.
Electron microscopy. (A) Observation of hyphal morphology. The control, the OE-devR, and the ΔdevR strains were cultivated on CD (dextrin) plates at 30°C for 2 days. (B and C) Further observation of the hyphal tip (B) and conidiophore (C). The marginal regions of the colonies were cut, fixed, and dehydrated. The specimens were examined using an environmental scanning electron microscope. ΔdevR, AodevR-disrupted strain; OE-devR, AodevR-overexpressing strain.
AoDevR is involved in chitin metabolism.
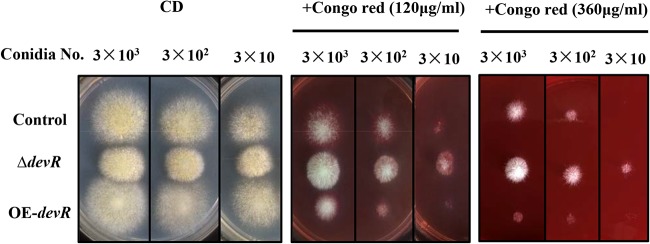
Since the AoDevR affects hyphal morphology, we further examined the role of this protein in stress tolerance. In this study, because of the poor growth of the OE-devR strain on the CD (dextrin) plate, a CD plate containing Congo red (120 μg · ml−1 or 360 μg · ml−1) was used to examine the cell wall tolerance. These strains were spot cultivated on plates with either CD or CD supplemented with Congo red at 30°C for 4 days. The results indicated that, although no obvious difference in growth rate was observed on the CD plates, the OE-devR strain still displayed increasing sensitivity to Congo red compared to that of the control strain (Fig. 3). In contrast, although the growth rate of the ΔdevR strain was delayed on the CD plate, the strain exhibited more resistance to Congo red (Fig. 3). We also performed other stress tolerance experiments (37°C, 1 M NaCl, and 5 mM H2O2), but no evident changes in growth trends were observed in the ΔdevR strain (data not shown).
FIG 3.
Cell wall stress assay. The CD plate containing Congo red (120 or 360 μg · ml−1) was used to examine cell wall tolerance. Strains were spot cultivated onto CD plates and Congo red-supplemented CD plates at 30°C for 4 days. ΔdevR, AodevR-disrupted strain; OE-devR, AodevR-overexpressing strain.
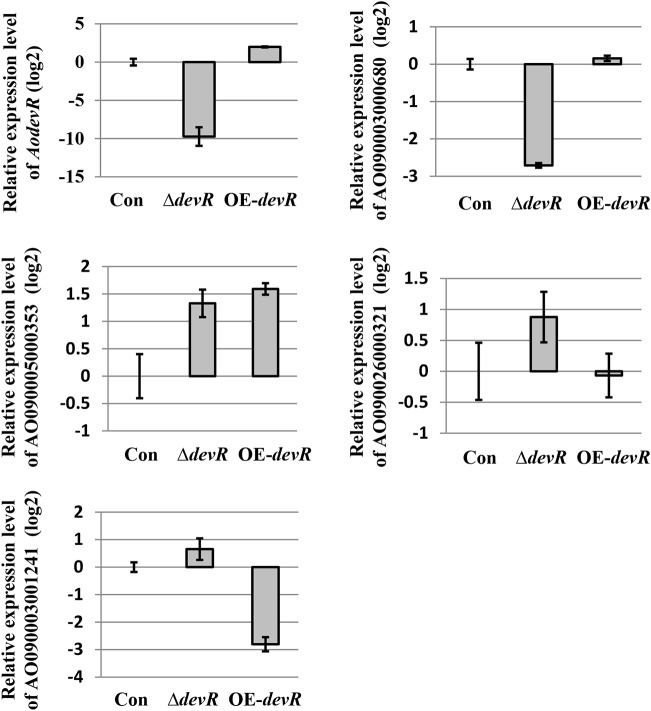
Congo red, which potently inhibits cell wall assembly by binding to chitin, inhibits chitin biosynthesis. Therefore, the expression levels of genes predicted to encode chitin synthase and chitinase in these strains were compared using qPCR. Conidia of the ΔdevR, OE-devR, and control strains were spot cultivated on CD (dextrin) plates at 30°C for 2 days. The total RNA was isolated from the strains, and quantitative real-time PCR was performed. At first, we verified whether the expression of AodevR was significantly changed in the ΔdevR and OE-devR strains. The result indicated that the expression level of AodevR was significantly reduced in the ΔdevR strain compared to that in the control strain. In contrast, the expression level of this gene was increased in the OE-devR strain, and the dextrin induction of the expression level of AodevR increased 4-fold compared to that in the control strain (Fig. 4). In this study, the expression level of the predicted chitinase-encoding gene (AO090003000680) was significantly reduced in the ΔdevR strain. This result is consistent with the results from the Congo red tolerance experiment. However, AodevR overexpression did not result in a clear change in the expression of this gene. In addition, we also examined the expression of the predicted chitin synthase genes AO090005000353 and AO090026000321. The expression level of AO090005000353 increased, and the expression of AO090026000321 was also slightly increased by the deletion of AodevR. However, the expression levels of the two genes did not have the expected results in the OE-devR strain. The expression of AO090005000353 was markedly increased in the OE-devR strain (Fig. 4). Interestingly, the AodevR overexpression resulted in a significant decrease in the expression level of AO090003001241, which encodes a putative chitin deacetylase.
FIG 4.
qPCR analysis of the genes related to chitin metabolism. The relative expression levels of the selected genes involved in cell wall integrity and chitin biosynthesis were examined and compared using qPCR. The expression levels of all genes were normalized to the expression level of the endogenous control gene, histone H2A. The expression value of each gene in the control strain was used as the baseline. For qPCR, three replicates were performed for each sample. The average values and standard deviations are represented as bars and error bars, respectively. Con, control strain; ΔdevR, AodevR-disrupted strain; OE-devR, AodevR-overexpressing strain.
AoDevR is involved in starch metabolism and utilization.
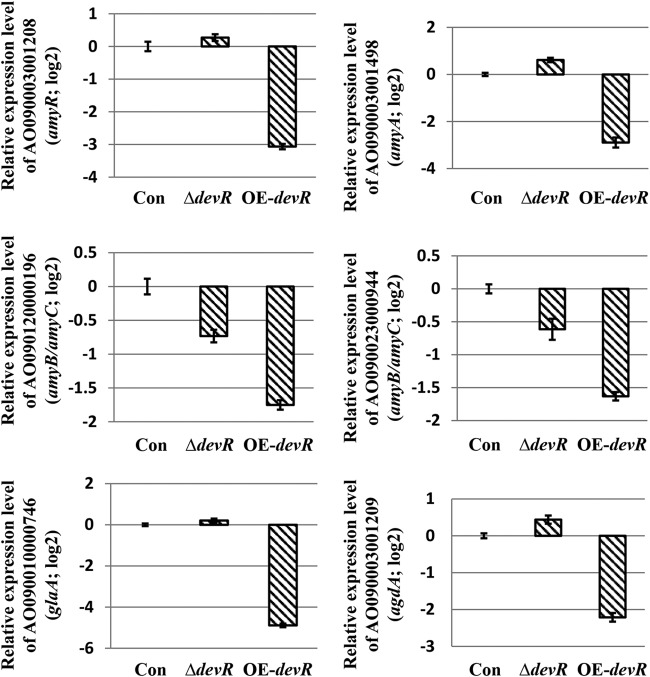
As described above, when the overexpression of AodevR was induced on the CD (dextrin) agar plate, the OE-devR strain displayed extremely poor growth (Fig. 1B). Based on this result, we considered that the overexpression of AodevR possibly inhibited starch degradation and polysaccharide utilization. Therefore, we investigated the expression level of the genes related to the starch degradation in more detail using qPCR. These strains were cultivated in CD (dextrin) liquid medium at 30°C for 2 days, and qPCR was performed as described in Materials and Methods. The results indicated that the overexpression of AodevR markedly inhibited the expression of amylase activity-related genes (Fig. 5), among which the amyR-encoded protein is a transcription factor involved in amylase production (24, 25). In addition, it has been reported that A. oryzae strains abundantly express α-amylases, and the genome reference strain RIB40 has three α-amylase genes (amyA, amyB, and amyC) (26). Among them, the DNA sequence of amyB (AO090120000196) is identical to that of amyC (AO090023000944). Although different primers were designed, the qPCR results for the two genes were consistent. Additionally, the expression of the agdA gene encoding alpha-glucosidase and the glaA gene encoding glucoamylase, which are involved in amylase activity (27), was also drastically reduced in the OE-AodevR strain. However, no evident increase in the expression levels of these genes was found in the AodevR gene-disrupted strain.
FIG 5.
qPCR analysis of the starch degradation-related genes. The strains were cultivated in CD (dextrin) liquid medium at 30°C for 2 days. The relative expression levels of genes involved in starch degradation were examined and compared using qPCR. The expression levels of the genes were normalized to the expression level of the endogenous control gene, the gene encoding histone H2A. The expression value of each gene in the control strain was used as the baseline. For qPCR, three replicates were performed for each sample. The average values and standard deviations are represented as bars and error bars, respectively. Con, control strain; ΔdevR, AodevR disrupted strain; OE-devR, AodevR-overexpressing strain.
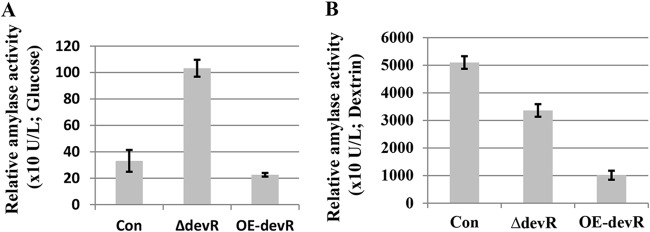
These strains were subsequently cultured in yeast extract-polypeptone-glucose (YPD; carbon source, glucose) and DPY (carbon source, dextrin) liquid media for 3 days, and the amylase activity in the supernatants was measured. Interestingly, although the deletion of AodevR did not increase the amylase activity in the supernatant of the DPY liquid medium, the amylase activity of ΔdevR strain was notably improved in the YPD liquid medium relative to that in the control strain (Fig. 6). In addition, the overexpression of AodevR inhibited the amylase activity in the DPY liquid medium, which is in accordance with the results of qPCR.
FIG 6.
Amylase activity analysis. The ΔdevR and OE-devR strains, as well as the control strains, were cultured in YPD and DPY liquid media at 30°C for 3 days. The α-amylase activity of the supernatants was then measured. Con, control strain; ΔdevR, AodevR disrupted strain; OE-devR, AodevR-overexpressing strain.
Analysis of the protein-DNA interaction.
Recent research shows that bHLH transcription factors recognize specific DNA sites named E boxes, which are characterized by a (G/C)CANNTG(G/C) sequence (19). In addition, it was reported that DevR binds to the E-box sites in the pksP promoter region (19). In this study, to verify whether the DevR protein interacts with the promoters of some chitin and starch metabolism-related genes, we first analyzed the promoter regions of these genes, some of which contain putative E-box sites within the 1.0-kb promoter regions (Table 1). Of these, the amyR-encoded protein is a transcription factor that regulates the expression of other amylase-related genes. Therefore, if DevR directly regulates the expression of amyR, it can also affect the degradation and utilization of starch.
TABLE 1.
Putative E-box sites in the promoter regions of chitin and starch metabolism-related genes
| Gene IDa | Gene name or description | E box | Position |
|---|---|---|---|
| AO090003001208 | amyR | TGCACCTGCA | −218 |
| CCCACATGCT | −720 | ||
| AO090003001498 | amyA | TGCAGCTGCG | −502 |
| AO090010000746 | glaA | GCCAGCTGCA | −259 |
| AO090003001209 | agdA | AGCATGTGGG | −943 |
| AO090005000353 | Chitin synthase | TGCAATTGCT | −16 |
| AO090026000321 | chsZ, chitin synthase | AGCAAATGGA | −711 |
| AO090003001241 | Predicted chitin deacetylase | AGCAATTGCT | −909 |
ID, identifier.
We then used the promoter of amyR as an example to verify whether it could interact directly with DevR. Since in the promoter region of amyR, there are two putative E boxes in front of the transcriptional start site, E box 1 (GCACCTGC) at −218 and E box 2 (CCACATGC) at −720 (Table 1), we selected the two target DNA sequences to conduct the yeast one-hybrid assay. To determine the specificity of DevR binding, the sequence CCACATGC of E box 2 was exchanged with CTTTAAAC as a negative control (−720m) (Fig. 7). Three tandem copies of the DNA targets were separately cloned into the pAbAi vector, and the generated plasmids [pAbAi-PamyR(−218), pAbAi-PamyR(−720), and pAbAi-PamyR(−720m)] were then efficiently integrated into the genome of the Y1HGold yeast strain by homologous recombination to generate bait-specific reporter strains. The constructed plasmid pGADT7-devR was further cotransformed into these Y1HGold strains (see Materials and Methods for detailed descriptions). The transformed cells were plated on medium lacking Leu with 50 or 100 ng/ml aureobasidin A (AbA). The positive clones showed that although both E-box sites can interact with DevR, PamyR(−218) has a stronger binding capacity (Fig. 7). The result suggested a possible interaction between the DevR protein and the promoter of amyR, implying that DevR can regulate the expression of amyR and accordingly affect the expression of other amylase-related genes.
FIG 7.
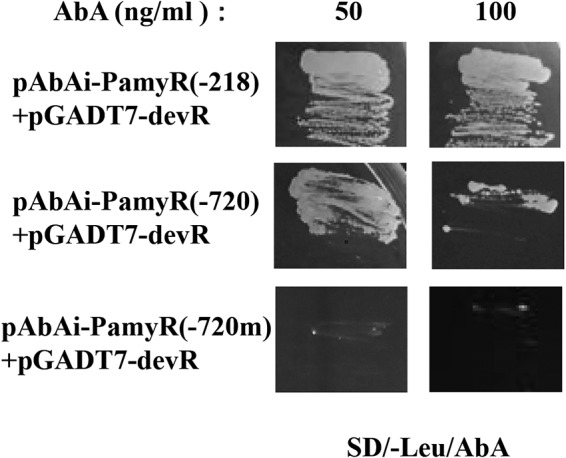
Interactions of DevR with promoter of amyR by yeast one-hybrid assay. Transformed yeast cells were grown on SD-Leu with 50 or 100 ng/ml AbA. The yeast cells that cotransformed with the indicated plasmids [pAbAi-PamyR(−218)/pAbAi-PamyR(−720)/pAbAi-PamyR(−720m) plus pGADT7-devR] were streak cultured onto the plates to examine the growth at 30°C for 3 days. The mutated sequence in the putative binding site (−720m) was used as a negative control. AbA, aureobasidin A.
DISCUSSION
In this study, we identified the AO090026000797 gene in A. oryzae, which is homologous to devR of A. nidulans and A. fumigatus (see Fig. S1 in the supplemental material). As a bHLH transcription factor, the DevR protein has been well characterized in these species. For example, DevR is required for the generation of conidia on minimal agar plates, and the deletion mutant produced wrinkled colonies in A. nidulans (18). However, in our study, AodevR deletion did not cause the loss of conidiation ability and did not produce wrinkled colonies in A. oryzae (Fig. 1B), implying that AoDevR may have some different functions or phenotypic effects from others. Except for the results just described, in our study, under osmotic stress conditions (0.6 M KCl), the reduced conidiation was partially recovered in the ΔdevR strain (Fig. 1B). However, the osmotic stress did not obviously inhibit the growth of the ΔdevR strain, which is also different from that of A. fumigatus (19). Since A. oryzae is an important filamentous fungus in the fermentation industry, characterization of the function of AoDevR is considered to still be very important. In addition, when AodevR overexpression was induced on CD (dextrin) agar plates, the OE-devR strain displayed extremely poor growth (Fig. 1B). The result indicated that AodevR overexpression results in a defect of polysaccharide utilization and strongly inhibits the growth of the strain. Further, electron microscopy indicated that AodevR overexpression also resulted in aberrant hyphal morphogenesis and that the strain formed extremely thin hyphae (Fig. 2). In contrast, deletion of this gene caused thicker hyphal formation than in the control strain (Fig. 2B).
These changes in morphology are possibly related to cell wall biosynthesis. Therefore, our next step was to examine whether the deletion or overexpression of AodevR affected the cell wall tolerance. The results from stress tolerance experiments suggested that overexpression of AodevR increased the sensitivity to Congo red, and deletion of this gene resulted in more resistance to Congo red relative to that of the control strain, as expected (Fig. 3). No important changes in growth were observed in other stress tolerance experiments, such as heat shock, osmotic stress, and oxidative stress (data not shown). Congo red potently inhibits cell wall assembly by binding to chitin and inhibiting chitin biosynthesis. Therefore, we compared the expression of several genes in these strains predicted to participate in chitin biosynthesis or chitin degradation by qPCR (Fig. 4). According to the results, the expression level of a predicted chitinase-encoding gene (AO090003000680) was significantly reduced and those of two chitin synthase genes (AO090005000353 and AO090026000321) were increased in the ΔdevR strain, as expected (Fig. 4). The result implied that excessive chitin biosynthesis or a decrease in chitin degradation possibly resulted in the thickening of the cell wall of the ΔdevR strain, therefore resulting in delayed growth and increased Congo red tolerance. Simultaneously, the expression of a putative chitin deacetylase gene (AO090003001241) was significantly decreased in the OE-devR strain. Cell wall metabolism and cell wall modification are very important processes that microorganisms use to adjust to various environmental conditions. The AO090003001241 protein-encoding sequence was also similar to that of PgdA, which was shown to participate in cell wall modification, and its deletion mutant was sensitive to lysozyme treatment (28, 29). However, the expression of the chitin synthase gene (AO090005000353) was significantly increased in the OE-devR strain. It was considered that the excessive defects in cell wall biosynthesis could be related to the feedback effect.
In addition, since overexpression of AodevR resulted in extremely poor growth when dextrin was used as a carbon source in the CD (dextrin) plate (Fig. 1B), we concluded that overexpression of AodevR possibly inhibited the expression of amylase-related genes and polysaccharide utilization. qPCR results confirmed that AodevR overexpression actually inhibited the expression of amylase-related genes (Fig. 5). α-Amylase, which hydrolyzes starch into dextrin and short oligosaccharides, is one of the most abundantly expressed proteins in A. oryzae. It is widely used in industry and has been well characterized in academic studies (26, 30). Therefore, as an industrial fermentative strain, it is very important to reveal the regulation of amylase production in A. oryzae. Interestingly, the amylase assay showed that although the deletion of AodevR did not increase amylase activity in the supernatant of the DPY liquid medium with dextrin as the carbon source, the amylase activity was notably improved in the YPD liquid medium with glucose as the carbon source (Fig. 6). At the same time, compared to that of the control strain, no obvious changes occurred in the biomass in the ΔdevR and OE-devR strains (data not shown). In addition, during strain growth in DPY liquid medium, AodevR overexpression markedly inhibited amylase activity, which is consistent with the qPCR result. As mentioned above, α-amylase is one of the most abundantly expressed proteins in A. oryzae; therefore, amyB, as one of the amylase-encoding genes, exhibits strong expression under the conditions of starch-induced culture. According to our results, although AodevR overexpression partly inhibited and reduced expression of amyR and even amyB (Fig. 5), the amyB promoter still induced AodevR overexpression compared to that in the control strain during the early stages of cultivation (Fig. 4). However, with the extension of cultivation time, when DevR accumulated to a certain level, the inducing effect of the amyB promoter disappeared, possibly because of negative feedback. The results of a yeast one-hybrid assay suggested that the DevR protein possibly interacted with the amyR promoter (Fig. 7), implying that AodevR overexpression may directly inhibit the expression of amyR and accordingly affect the expression of other amylase-related genes. However, the question remains whether the amyB promoter can be used to induce AodevR overexpression to investigate amylase activity. Therefore, we also tried another carbon source-independent thiA promoter to replace the amyB promoter to induce AodevR overexpression (see Fig. S4). thiA encodes an enzyme involved in thiamine biosynthesis and is regulated transcriptionally by thiamine (31). The phenotypic analysis showed that the number of conidia in and the growth of the PthiA-devR strain were slightly suppressed on CD (dextrin) agar plates (Fig. S4B), but the inhibition effect of growth caused by the thiA promoter was not as strong as that by the amyB promoter. The qPCR result indicated that the expression level of AodevR was increased in the PthiA-devR strain relative to that in the control strain when cultured in liquid medium. At the same time, the expression levels of some amylase-related genes were clearly decreased in the PthiA-devR strain, which showed a trend similar to that with amyB as the promoter (Fig. S4C).
In summary, in this study, we consider that AoDevR is significantly involved in chitin and starch metabolism and thus affects starch utilization by and growth of A. oryzae. In our previous research, we also found that another bHLH transcription factor, SclR, is critically involved in carbohydrate metabolism in A. oryzae (12). These studies may provide a greater understanding of the function and mechanism of the bHLH transcription factor family, especially, carbohydrate metabolism, and a theoretical basis for more effective industrial utilization of A. oryzae.
MATERIALS AND METHODS
Strains, media, and cultivation.
The Aspergillus oryzae wild-type strain RIB40 was used as the DNA donor strain. Strain RkuN16ptr1 (Δku70::ptrA ΔpyrG) (32) was used for the disruption and overexpression of the devR gene in A. oryzae. Escherichia coli DH5α was used for DNA manipulations. Potato-dextrose (PD) agar medium was used to grow the A. oryzae strains for the collection of conidia. DPY medium (2% dextrin, 1% polypeptone, 0.5% yeast extract, 0.5% KH2PO4, and 0.05% MgSO4·7H2O; pH 5.8) was used to culture the A. oryzae strains. Czapek-Dox (CD) medium (0.3% NaNO3, 0.2% KCl, 0.1% KH2PO4, 0.05% MgSO4·7H2O, 0.002% FeSO4·7H2O, and 2% glucose; pH 5.5) was used to select the A. oryzae transformants. CD (dextrin) medium (0.3% NaNO3, 0.2% KCl, 0.1% KH2PO4, 0.05% MgSO4·7H2O, 0.002% FeSO4·7H2O, and 2% dextrin; pH 5.5) was used to induce the amyB promoter and detect the growth. CD plus Congo red (120 μg · ml−1 or 360 μg · ml−1) was used for the stress culture. Malt medium (per liter: malt extract, 80 g; CuSO4, 2.0 mg; Na2B4O7, 0.04 mg; FePO4, 0.87 mg; MnSO4, 0.95 mg; Na2MoO4, 0.8 mg; ZnSO4, 8.0 mg; agar, 15 g; pH 6.0) was used for the growth experiment. YPD (2% glucose, 1% polypeptone, 0.5% yeast extract, 0.5% KH2PO4, and 0.05% MgSO4·7H2O; pH 5.8) and DPY were used for the production and amylase activity test. A. oryzae transformation was performed as previously described (10).
Construction of the devR gene deletion cassette by fusion PCR.
The deletion cassette was constructed using the fusion PCR method described previously (33). Using A. oryzae RIB40 genomic DNA as a template, the 2.2-kb pyrG gene (with primer pair pyrG-F/pyrG-R) and two approximately 1.0-kb DNA fragments outside the targeted AodevR gene [left region with primer pair DH797-F(−980)/DH797-R(-1F) and right region with primer pair DH797-F(1670R)/DH797-R(2700) listed in Table 2] were amplified by PCR. PCR amplification was carried out in a GeneAmp 9700 system (Applied Biosystems, Foster City, CA) with PrimeSTAR HS DNA polymerase (TaKaRa, Shiga, Japan). The sequences of the left and right regions of the AodevR gene were designed to overlap the sequences of the primer pair of the pyrG gene. The three DNA fragments amplified were then mixed, and fusion PCR was performed using primer pair DH797-F(−900)/DH797-R(2620) (Table 2). The PCR cycle was as follows: denaturation at 98°C for 10 s followed by 35 cycles of 98°C for 10 s, 55°C for 15 s, and 72°C for 5 min. The deletion cassette generated was used for A. oryzae transformation to delete the devR gene from the genome (Fig. S2A and B).
TABLE 2.
Primers designed for this study
| Primer | Sequence (5′→3′) | Description |
|---|---|---|
| DH797-F(-980) | GTTCTACTGGCCATCATCGGTCG | Construction of devR gene deletion cassette |
| DH797-R(-1F) | CATCACAGGGTACGTCTGTTGTGATGTGTAAGTGTGGCTTCCATG | |
| pyrG-F | ACAACAGACGTACCCTGTGATG | |
| pyrG-R | AACTGCACCTCAGAAGAAAAGGAT | |
| DH797-F(1670R) | ATCCTTTTCTTCTGAGGTGCAGTTCAATGCAAGGCGTGCAGTACAC | |
| DH797-R(2700) | CAGAATGCTAAATACCCGTGCTC | |
| DH797-F(-900) | GATTTTGTGCCACGTTCCGCTG | |
| DH797-R(2620) | CATTGGACGAGAAGCTCTGGTGAC | |
| OE797-F(IF) | ATTTGAATTCGAATCGATGGAAGCCACACTTACACATCG | Construction of plasmid for devR overexpression |
| OE797-R(IF) | CACGTCGTAGGGGTACCGTCGCTCATCGGTGTACTG | |
| HA-F | TACCCCTACGACGTGCCCGACTAC | |
| Pamy-R | CGATTCGAATTCAAATGCCTTCTGTGG | |
| 797-F(RT) | CCAAACCCCTACAGTATCTTCA | qPCR analysis of AodevR |
| 797-R(RT) | GAGACGGCTGAGAGGACGTAAG | |
| 680-F(RT) | CTCGCTCTTCTTCTCTCTCGAT | qPCR of genes related to chitin metabolism |
| 680-R(RT) | TCGGACATTGGCGAATGCATAG | |
| 353-F(RT) | CCTCCCCGTGTACTACTTTATC | |
| 353-R(RT) | TGGGAGACATGAGAGCCTTTTC | |
| 321-F(RT) | GACGTTGCTGAATGCGTTGCAT | |
| 321-R(RT) | CTCTTCTCCTTGCATGTTCCCA | |
| 1241-F(RT) | CATGTCTTGTCCGCACCATTGA | |
| 1241-R(RT) | ATGAAGAAGGTAGCCTTGGCTC | |
| 944-F | CATGGCTACTGGCAGCAGGATA | qPCR of genes related to starch degradation |
| 944-R | CATATGGTTAGCAACCACATCGAC | |
| 1028-F | TCGGAGTCGTTCACTCGACTAC | |
| 1028-R | CAGGTACTTGAGGAAGACATTGAC | |
| 196-F | GCATCATCGACAAGTTGGAC | |
| 196-R | CGTTCAGAGAGTATATATCCTGC | |
| 746-F | CCTATCATCGAGGAGTTCATTAGC | |
| 746-R | GCTGTCTCGTCGACATTGAACT | |
| 1209-F | GCTCTACCTTAGTCTACGAGA | |
| 1209-R | GTCCATAGATGTTATCGTCAATG | |
| 1498-F | GGTCATCAACAACATGGCATAT | |
| 1498-R | GGTGTAATTGTTCCAGTCAG | |
| AD797-F | GGAGGCCAGTGAATTCATGGAAGCCACACTTACACATCGG | Yeast one-hybrid assay |
| AD797-R | CGAGCTCGATGGATCCCTACCGTCGCTCATCGGTGTACTG | |
| PthiA-F(IF) | AGGAAACAGCTATGAC-TTCGGTAAATACACTATCACA | Construction of plasmid of thiA promoter inducing devR overexpression |
| PthiA-R(FP) | CGATGTGTAAGTGTGGCTTCCAT-GTTTCAAGTTGCAATGACTAT | |
| 797-F(FP) | ATAGTCATTGCAACTTGAAAC-ATGGAAGCCACACTTACACATCG | |
| 797-R(IF) | TACCGAGCTCGAATTC-CTACCGTCGCTCATCGGTGTAC | |
| pUPA-F1 | GAATTCGAGCTCGGTACCCG | |
| pUPA-R1 | GTCATAGCTGTTTCCTGTGTG |
Construction of the plasmid for the overexpression of the AodevR gene.
To construct the plasmid for AodevR overexpression, the AodevR ORF was amplified by PCR using the A. oryzae RIB40 genomic DNA and the primer pair OE797-F(IF) and OE797-R(IF). The PCR product was inserted into the linearized plasmid pamy902 (17) using PCR with the primer pair HA-F and Pamy-R listed in Table 2. PrimeSTAR HS DNA polymerase (TaKaRa, Shiga, Japan) was used for PCR amplification. An In-fusion PCR cloning kit (Clontech, USA) was used to fuse the AodevR ORF into the linearized plasmid, and the ecdR ORF in the plasmid pamy902 was replaced by the AodevR ORF. The generated plasmid (pOE-devR) (Fig. S2C and D), which contains the amyB promoter and the selectable marker pyrG gene, was introduced into the RkuN16ptr1 strain to obtain the AodevR overexpressing strain.
Southern blot hybridization analysis.
AodevR-disrupted and AodevR-overexpressing strains were identified by PCR and Southern blot analysis. The genomic DNA of the A. oryzae strains was extracted as previously described (10). After electrophoresis, the digested genomic DNA was transferred onto a Hybond-N+ membrane (Amersham Biosciences, Amersham, UK). Southern hybridization was performed as described (10). A digoxigenin (DIG)-labeled probe was constructed using a PCR DIG labeling kit (Roche Diagnostics, Mannheim, Germany). The hybridization and detection of signals with the DIG system were performed according to the manufacturer’s instructions (Roche Diagnostics). A GeneGnome XRQ chemiluminescent imaging system (Syngene, UK) was used for detection. The probe was generated using the genomic DNA of A. oryzae strain RIB40 as a template. The primer pair DH797-F(1670R)/DH797-R(2620) was used for the amplification of the probe examining the devR gene deletion, and AodevR ORF was amplified as a probe to check its overexpression.
Complementation of the AodevR deletion by reintegration of AodevR gene.
For reintegration of AodevR into the ΔdevR strain, an approximately 3.7-kb DNA fragment including AodevR was amplified by PCR with primer pair DH797-F(−980)/DH797-R(2700) (Table 2). The PCR product was used to transform the ΔdevR strain, and transformants were grown on CD medium with 15 mM uridine. The obtained conidia (106 to 108) were placed on CD medium containing 5-fluoroorotic acid (5-FOA) and uridine to select pyrG-deficient transformants. The pyrG-deficient strains were isolated 4 days later. The obtained transformants were confirmed by PCR and Southern blot analysis (Fig. S3A).
RNA isolation and quantitative real-time PCR analysis.
The total RNA was isolated from strains grown on CD (dextrin) agar or liquid medium at 30°C for 2 days using a TaKaRa MiniBEST Universal RNA extraction kit (TaKaRa, Otsu, Japan) according to the manufacturer’s protocol. The total RNA was treated with DNase I (TaKaRa Bio, Shiga, Japan) to remove the chromosomal DNA. RT-PCR was performed using 0.5 μg total RNA and a PrimeScript RT Master Mix (TaKaRa, Shiga, Japan) according to the manufacturer’s instructions. Quantitative real-time PCR was performed using the designed primer pairs listed in Table 2, and histone H2A was used as a reference gene (12). Gene expression was analyzed with 300 nM primers using an SYBR Premix Ex Taq II (TaKaRa, Shiga, Japan) and an Applied Biosystems StepOnePlus Real-Time PCR system (Thermo Fisher Scientific, Waltham, USA). The PCR cycle conditions were 30 s at 95°C and 40 cycles of 5 s at 95°C and 30 s at 60°C. The relative expression gene level was determined by the ratio of the target gene to the reference gene after amplification. For qPCR, three replicates were performed for each sample. The relative gene expression levels were calculated using the comparative cycle threshold (CT) method, and the mixture included a negative no-reverse transcription (RT) control.
Scanning electron microscopy.
Conidia were cultivated on CD (dextrin) agar medium at 30°C for 2 days. Samples were cut, fixed with 2.5% (vol/vol) glutaraldehyde in 0.1 M phosphate-buffered saline (pH 7.4) for 3 h, and postfixed with 1% osmium tetroxide in the same buffer for 1.5 h. They were then dehydrated through a graded alcohol series (30%, 50%, 70%, 90%, and 100% ethanol) and dried in a critical point dryer using liquid CO2 as the drying agent. The specimens were examined using an environmental scanning electron microscope (Quanta 200; FEI Co., Hillsboro, OR, USA).
Assay of amylase activity.
Conidia were cultivated in 40 ml YPD or DPY liquid medium at 30°C for 3 days. The medium was centrifuged at 13,000 × g at 4°C for 10 min. α-Amylase activity in the supernatant was measured using an amylase assay kit (Jining, Shanghai, China) per the manufacturer’s instructions.
Yeast one-hybrid assay.
Three copies of the DNA target elements (PamyR(−218), TCGAGCTCGGTACCCTTTTGCACCTGCAGCCTTTTGCACCTGCAGCCTTTTGCACCTGCAGCCGGGGATCTGTCGACC; PamyR (−720), TCGAGCTCGGTACCCCATCCCCACATGCTGGATCATCCCCACATGCTGGATCATCCCCACATGCTGGATGGGGATCTGTCGACC; and the mutated sequence in the putative binding site PamyR(-720m), TCGAGCTCGGTACCCCATCCCTTTAAACTGGATCATCCCTTTAAACTGGATCATCCCTTTAAACTGGATGGGGATCTGTCGACC) were synthesized and inserted into the linearized pAbAi vector (Clontech), which was digested with SmaI using the In-fusion PCR cloning kit. A yeast one-hybrid system kit was used to construct the plasmids [pAbAi-PamyR(−218), pAbAi-PamyR(−720), and pAbAi-PamyR(-720m)], which were linearized with BstBI and then transformed into Saccharomyces cerevisiae Y1HGold (Clontech). Colonies were grown in the absence of uracil on SD−Ura agar plates. Next, AodevR cDNA was amplified by PCR with primer pair AD797-F and AD797-R (Table 2). The PCR product was inserted into the linearized pGADT7 AD cloning vector (Clontech), which was digested with EcoRI and BamHI using the In-fusion PCR cloning kit. The generated plasmid pGADT7-devR was further transformed into Y1HGold [pAbAi-PamyR(−218), pAbAi-PamyR(−720), and pAbAi-PamyR(−720m)]. The transformed cells were plated on medium lacking Leu with 50 or 100 ng/ml AbA to identify the interactions of DevR with the amyB promoter. Y1HGold[pAbAi-PamyR(−218)/pAbAi-PamyR(−720) plus pGADT7] was used as a negative control (data not shown).
Construction of the PthiA-containing AodevR overexpression strain.
The 1.3-kb thiA promoter region (31) and AodevR ORF were amplified by PCR using the A. oryzae RIB40 genomic DNA as a template and the primer pairs PthiA-F(IF)/PthiA-R(FP) and 797-F(FP)/797-R(IF) listed in Table 2. The two PCR fragments were fused by fusion PCR, and the generated PCR product was inserted into the plasmid pOE-devR, which was linearized by PCR with the primer pair pUPA-F1/pUPA-R1 (Table 2). PrimeSTAR HS DNA polymerase (TaKaRa, Shiga, Japan) was used for PCR amplification. An In-fusion PCR cloning kit (Clontech, USA) was used to fuse the PthiA-AodevR into the linearized plasmid, and the amyB promoter was replaced by the PthiA in the plasmid. The generated plasmid (pthiA-devR) (Fig. S4A) was introduced into the RkuN16ptr1 strain to obtain the AodevR-overexpressing strain.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the Natural Science Foundation of China (31570107) and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
We declare no conflict of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00089-19.
REFERENCES
- 1.Machida M, Asai K, Sano M, Tanaka K, Kumagai T, Terai G, Kusumoto K, Arima T, Akita O, Kashiwagi Y, Abe K, Gomi K, Horiuchi H, Kitamoto K, Kobayashi T, Takeuchi M, Denning DW, Galagan JE, Nierman WC, Yu J, Archer DB, Bennett JW, Bhatnagar D, Cleveland TE, Fedorova ND, Gotoh O, Horikawa H, Hosoyama A, Ichinomiya M, Igarashi R, Iwashita K, Juvvadi PR, Kato M, Kato Y, Kin T, Kokubun A, Maeda H, Maeyama N, Maruyama J, Nagasaki H, Nakajima T, Oda K, Okada K, Paulsen I, Sakamoto K, Sawano T, Takahashi M, Takase K, Terabayashi Y, Wortman JR, et al. 2005. Genome sequencing and analysis of Aspergillus oryzae. Nature 438:1157–1161. doi: 10.1038/nature04300. [DOI] [PubMed] [Google Scholar]
- 2.Kobayashi T, Abe K, Asai K, Gomi K, Juvvadi PR, Kato M, Kitamoto M, Takeuchi M, Machida M. 2007. Genomics of Aspergillus oryzae. Biosci Biotechnol Biochem 71:646–670. doi: 10.1271/bbb.60550. [DOI] [PubMed] [Google Scholar]
- 3.Murre C, McCaw PS, Baltimore D. 1989. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell 56:777–783. doi: 10.1016/0092-8674(89)90682-X. [DOI] [PubMed] [Google Scholar]
- 4.Li FM, Liu WY. 2017. Genome-wide identification, classification, and functional analysis of the basic helix-loop-helix transcription factors in the cattle, Bos Taurus. Mamm Genome 28:176–197. doi: 10.1007/s00335-017-9683-x. [DOI] [PubMed] [Google Scholar]
- 5.Martinez-Garcia JF, Huq E, Quail PH. 2000. Direct targeting of light signals to a promoter element-bound transcription factor. Science 288:859–863. [DOI] [PubMed] [Google Scholar]
- 6.Caruso ML, Litzka O, Martic G, Lottspeich F, Brakhage AA. 2002. Novel basic-region helix-loop-helix transcription factor (AnBH1) of Aspergillus nidulans counteracts the CCAAT-binding complex AnCF in the promoter of a penicillin biosynthesis gene. J Mol Biol 323:425–439. doi: 10.1016/S0022-2836(02)00965-8. [DOI] [PubMed] [Google Scholar]
- 7.Miller KY, Wu J, Miller BL. 1992. StuA is required for cell pattern formation in Aspergillus. Genes Dev 6:1770–1782. doi: 10.1101/gad.6.9.1770. [DOI] [PubMed] [Google Scholar]
- 8.Twumasi-Boateng K, Yu Y, Chen D, Gravelat FN, Nierman WC, Sheppard DC. 2009. Transcriptional profiling identifies a role for BrlA in the response to nitrogen depletion and for StuA in the regulation of secondary metabolite clusters in Aspergillus fumigatus. Eukaryot Cell 8:104–115. doi: 10.1128/EC.00265-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park BC, Park YH, Yi S, Choi YK, Kang EH, Park HM. 2014. Transcriptional regulation of fksA, a β-1,3-glucan synthase gene, by the APSES protein StuA during Aspergillus nidulans development. J Microbiol 52:940–947. doi: 10.1007/s12275-014-4517-y. [DOI] [PubMed] [Google Scholar]
- 10.Jin FJ, Takahashi T, Machida M, Koyama Y. 2009. Identification of a basic helix-loop-helix-type transcription regulator gene in Aspergillus oryzae by systematically deleting large chromosomal segments. Appl Environ Microbiol 75:5943–5951. doi: 10.1128/AEM.00975-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jin FJ, Takahashi T, Matsushima K, Hara S, Shinohara Y, Maruyama J, Kitamoto K, Koyama Y. 2011. SclR, a basic helix-loop-helix transcription factor, regulates hyphal morphology and promotes sclerotial formation in Aspergillus oryzae. Eukaryot Cell 10:945–955. doi: 10.1128/EC.00013-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jin FJ, Han P, Zhuang M, Zhang ZM, Jin L, Koyama Y. 2018. Comparative proteomic analysis: SclR is importantly involved in carbohydrate metabolism in Aspergillus oryzae. Appl Microbiol Biotechnol 102:319–332. doi: 10.1007/s00253-017-8588-7. [DOI] [PubMed] [Google Scholar]
- 13.Wada R, Jin FJ, Koyama Y, Maruyama J, Kitamoto K. 2014. Efficient formation of heterokaryotic sclerotia in the filamentous fungus Aspergillus oryzae. Appl Microbiol Biotechnol 98:325–334. doi: 10.1007/s00253-013-5314-y. [DOI] [PubMed] [Google Scholar]
- 14.Hoshizaki D, Hill J, Henry S. 1990. The Saccharomyces cerevisiae INO4 gene encodes a small, highly basic protein required for derepression of phospholipid biosynthetic enzymes. J Biol Chem 265:4736–4745. [PubMed] [Google Scholar]
- 15.Nikoloff D, McGraw P, Henry S. 1992. The INO2 gene of Saccharomyces cerevisiae encodes a helix-loop-helix protein that is required for activation of phospholipid synthesis. Nucleic Acids Res 20:3253. doi: 10.1093/nar/20.12.3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Endoh-Yamagami S, Hirakawa K, Morioka D, Fukuda R, Ohta A. 2007. Basic helix-loop-helix transcription factor heterocomplex of Yas1p and Yas2p regulates cytochrome P450 expression in response to alkanes in the yeast Yarrowia lipolytica. Eukaryot Cell 6:734–743. doi: 10.1128/EC.00412-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin FJ, Nishida M, Hara S, Koyama Y. 2011. Identification and characterization of a putative basic helix-loop-helix transcription factor involved in the early stage of conidiophore development in Aspergillus oryzae. Fungal Genet Biol 48:1108–1115. doi: 10.1016/j.fgb.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 18.Tüncher A, Reinke H, Martic G, Caruso ML, Brakhage AA. 2004. A basic-region helix-loop-helix protein-encoding gene (devR) involved in the development of Aspergillus nidulans. Mol Microbiol 52:227–241. doi: 10.1111/j.1365-2958.2003.03961.x. [DOI] [PubMed] [Google Scholar]
- 19.Valiante V, Baldin C, Hortschansky P, Jain R, Thywißen A, Straßburger M, Shelest E, Heinekamp T, Brakhage AA. 2016. The Aspergillus fumigatus conidial melanin production is regulated by the bifunctional bHLH DevR and MADS-box RlmA transcription factors. Mol Microbiol 102:321–335. doi: 10.1111/mmi.13462. [DOI] [PubMed] [Google Scholar]
- 20.Langfelder K, Streibel M, Jahn B, Haase G, Brakhage AA. 2003. Biosynthesis of fungal melanins and their importance for human pathogenic fungi. Fungal Genet Biol 38:143–158. doi: 10.1016/S1087-1845(02)00526-1. [DOI] [PubMed] [Google Scholar]
- 21.Tsai HF, Wheeler MH, Chang YC, Kwon-Chung KJ. 1999. A developmentally regulated gene cluster involved in conidial pigment biosynthesis in Aspergillus fumigatus. J Bacteriol 181:6469–6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nosanchuk JD, Stark RE, Casadevall A. 2015. Fungal melanin: what do we know about structure? Front Microbiol 6:1463. doi: 10.3389/fmicb.2015.01463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsai HF, Chang YC, Washburn RG, Wheeler MH, Kwon-Chung KJ. 1998. The developmentally regulated alb1 gene of Aspergillus fumigatus: its role in modulation of conidial morphology and virulence. J Bacteriol 180:3031–3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ito T, Tani S, Itoh T, Tsukagoshi N, Kato M, Kobayashi T. 2004. Mode of AmyR binding to the CGGN8AGG sequence in the Aspergillus oryzae taaG2 promoter. Biosci Biotechnol Biochem 68:1906–1911. doi: 10.1271/bbb.68.1906. [DOI] [PubMed] [Google Scholar]
- 25.Suzuki K, Tanaka M, Konno Y, Ichikawa T, Ichinose S, Hasegawa-Shiro S, Shintani T, Gomi K. 2015. Distinct mechanism of activation of two transcription factors, AmyR and MalR, involved in amylolytic enzyme production in Aspergillus oryzae. Appl Microbiol Biotechnol 99:1805–1815. doi: 10.1007/s00253-014-6264-8. [DOI] [PubMed] [Google Scholar]
- 26.Nemoto T, Maruyama J, Kitamoto K. 2012. Contribution ratios of amyA, amyB, amyC genes to high-level α-amylase expression in Aspergillus oryzae. Biosci Biotechnol Biochem 76:1477–1483. doi: 10.1271/bbb.120142. [DOI] [PubMed] [Google Scholar]
- 27.Minetoki T, Gomi K, Kitamoto K, Kumagai C, Tamura G. 1995. Characteristic expression of three amylase-encoding genes, agdA, amyB, and glaA in Aspergillus oryzae transformants containing multiple copies of the agdA gene. Biosci Biotechnol Biochem 59:2251–2254. doi: 10.1271/bbb.59.2251. [DOI] [PubMed] [Google Scholar]
- 28.Kobayashi K, Sudiarta IP, Kodama T, Fukushima T, Ara K, Ozaki K, Sekiguchi J. 2012. Identification and characterization of a novel polysaccharide deacetylase C (PdaC) from Bacillus subtilis. J Biol Chem 287:9765–9776. doi: 10.1074/jbc.M111.329490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rismondo J, Wamp S, Aldridge C, Vollmer W, Halbedel S. 2018. Stimulation of PgdA-dependent peptidoglycan N-deacetylation by GpsB-PBP A1 in Listeria monocytogenes. Mol Microbiol 107:472–487. doi: 10.1111/mmi.13893. [DOI] [PubMed] [Google Scholar]
- 30.Matsuura Y, Kusunoki M, Harada W, Kakudo M. 1984. Structure and possible catalytic residues of Taka-amylase A. J Biochem 95:697–702. doi: 10.1093/oxfordjournals.jbchem.a134659. [DOI] [PubMed] [Google Scholar]
- 31.Shoji J, Maruyama J, Arioka M, Kitamoto K. 2005. Development of Aspergillus oryzae thiA promoter as a tool for molecular biological studies. FEMS Microbiol Lett 244:41–46. doi: 10.1016/j.femsle.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 32.Takahashi T, Jin FJ, Sunagawa M, Machida M, Koyama Y. 2008. Generation of large chromosomal deletions in koji molds Aspergillus oryzae and Aspergillus sojae via a loop-out recombination. Appl Environ Microbiol 74:7684–7693. doi: 10.1128/AEM.00692-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jin FJ, Maruyama J, Juvvadi PR, Arioka M, Kitamoto K. 2004. Development of a novel quadruple auxotrophic host transformation system by argB gene disruption using adeA gene and exploiting adenine auxotrophy in Aspergillus oryzae. FEMS Microbiol Lett 239:79–85. doi: 10.1016/j.femsle.2004.08.025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.