Natural products are readily isolated from cultured bacteria and exploited for useful purposes, including drug discovery. However, these compounds are rarely detected in the environments from which the bacteria are obtained, thus limiting our understanding of their ecological significance. Here, we used environmental metabolomics to directly assess chemical diversity in marine sediments. We identified numerous metabolites and, in one case, isolated strains of bacteria capable of producing one of the compounds detected. Coupling environmental metabolomics with community and metagenomic analyses provides opportunities to link compounds and producers and begin to assess the complex interactions mediated by specialized metabolites in marine sediments.
KEYWORDS: Salinispora, metabolomics, metagenomics, microbiome, natural products
ABSTRACT
Thousands of natural products have been identified from cultured microorganisms, yet evidence of their production in the environment has proven elusive. Technological advances in mass spectrometry, combined with public databases, now make it possible to address this disparity by detecting compounds directly from environmental samples. Here, we used adsorbent resins, tandem mass spectrometry, and next-generation sequencing to assess the metabolome of marine sediments and its relationship to bacterial community structure. We identified natural products previously reported from cultured bacteria, providing evidence they are produced in situ, and compounds of anthropogenic origin, suggesting this approach can be used as an indicator of environmental impact. The bacterial metabolite staurosporine was quantified and shown to reach physiologically relevant concentrations, indicating that it may influence sediment community structure. Staurosporine concentrations were correlated with the relative abundance of the staurosporine-producing bacterial genus Salinispora and production confirmed in strains cultured from the same location, providing a link between compound and candidate producer. Metagenomic analyses revealed numerous biosynthetic gene clusters related to indolocarbazole biosynthesis, providing evidence for noncanonical sources of staurosporine and a path forward to assess the relationships between natural products and the organisms that produce them. Untargeted environmental metabolomics circumvents the need for laboratory cultivation and represents a promising approach to understanding the functional roles of natural products in shaping microbial community structure in marine sediments.
IMPORTANCE Natural products are readily isolated from cultured bacteria and exploited for useful purposes, including drug discovery. However, these compounds are rarely detected in the environments from which the bacteria are obtained, thus limiting our understanding of their ecological significance. Here, we used environmental metabolomics to directly assess chemical diversity in marine sediments. We identified numerous metabolites and, in one case, isolated strains of bacteria capable of producing one of the compounds detected. Coupling environmental metabolomics with community and metagenomic analyses provides opportunities to link compounds and producers and begin to assess the complex interactions mediated by specialized metabolites in marine sediments.
INTRODUCTION
Bacterial natural products account for many of today’s most useful medicines (1). These “specialized metabolites” are traditionally isolated from bacteria cultured in the laboratory, leaving us with little understanding of when, why, and at what concentrations they are produced in nature (2). Furthermore, laboratory culture conditions typically bear little resemblance to those experienced by microbes in the habitats from which they are obtained and thus may lack environmental cues required to induce natural product biosynthesis. This possibility is supported by the large number of “orphan” biosynthetic gene clusters (BGCs) detected in bacterial genomes (3–5) and mounting evidence that interactions with other microbes induce specialized metabolite production (6–9).
While thousands of natural products have been discovered from cultured bacteria, relatively few have been detected directly in the environment. Exceptions include compounds originally isolated from marine invertebrates and subsequently shown to be of microbial origin (10, 11). Suppressive soils provide another example (12, 13), in which case metabolites produced by soil bacteria have been detected and linked to the inhibition of plant pathogens (14, 15). Microbial natural products have also been detected in the context of monitoring for toxic algal blooms (16), with polyaromatic resins used to extract waterborne toxins such as domoic acid (17, 18). While this approach has been further developed to study allelopathy among soil microorganisms (19), it has yet to be more broadly applied to the detection of bacterial natural products in marine ecosystems. Methodological advances in mass spectrometry coupled with pipelines for the visualization of complex data sets (20–22) provide new opportunities to assess environmental metabolomes and explore the ecological functions of natural products.
The diverse and highly structured bacterial communities associated with marine sediments provide ample opportunities for chemically mediated interactions to occur. While bacteria cultured from ocean sediments represent a relatively new source of natural products (23, 24) the compounds detected in the laboratory likely represent a small subset of what is produced in nature. Here, we employed adsorbent resins and mass spectrometry to analyze the metabolomes associated with marine sediment communities across a range of coastal habitats off Belize. The results reveal complex chemical landscapes that include metabolites related to those of microbial, invertebrate, and anthropogenic origin. These compounds include the bacterial metabolite staurosporine, which occurred at concentrations sufficient to affect community structure. The thousands of molecules that could not be readily identified indicate that much remains to be learned about the chemical ecology of marine sediments.
RESULTS
Sediment metabolomics.
The adsorbent resin HP20, which has been used to adsorb lipophilic toxins from the water column (16), was buried in marine sediments at five sites around the Smithsonian field station at Carrie Bow Cay, Belize. The resin deployed at site 1 was eluted with organic solvents, and mass spectral data from the crude extract was analyzed using the Global Natural Products Social Molecular Networking (GNPS) pipeline (22). The extracts from sites 2 to 5 were first fractionated prior to analysis. In total, 5,803 tandem mass spectrometry (MS/MS) spectra were collected, of which 5,242 passed the filtering requirements. Identical fragmentation spectra were clustered, resulting in the generation of 512 unique parent ions (nodes) after the removal of resin and solvent control spectra. These nodes represent parent ions with masses (m/z values) that ranged from 227 to 1,424 Da, with most falling between 350 and 900 Da. The number of parent ions detected at each site ranged from 106 (site 5, seagrass bed) to 335 (site 3, mangrove channel), with an average of 204 per site. The data were visualized as an Euler diagram, which revealed the extraordinary levels of small-molecule diversity, with only nine parent ions shared among all sites and nearly half (47%) observed at only one site (Fig. S1). The decision to combine the replicates from each site prevented a more thorough analysis of intersite variation; however, the considerable number of parent ions detected indicates that the resin technique developed for this study was effective for the extraction of a diverse set of metabolites.
Compound identification.
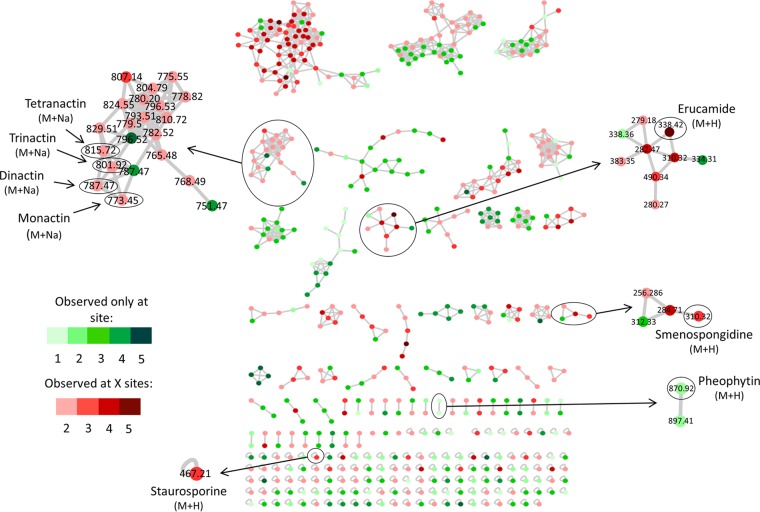
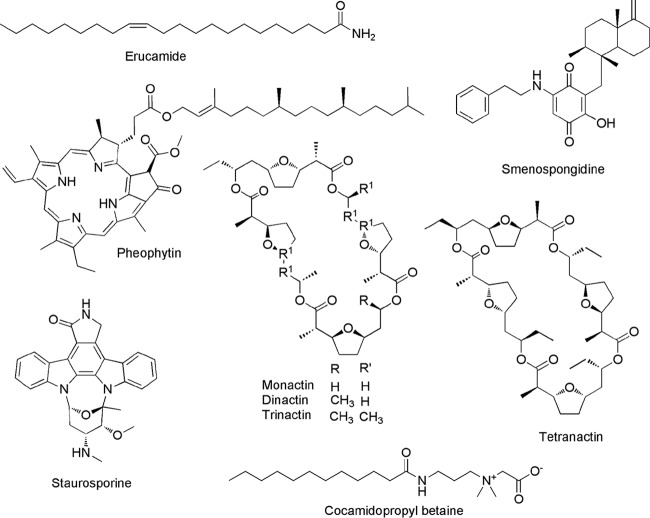
A molecular network was generated to visualize the MS/MS data. Compared to the >220,000 metabolites present in the GNPS database, the vast majority of parent ions and most molecular families had no matches (Fig. 1). While this database consists largely of spectra from the NIST 2014 ESI MS/MS library, it also includes a marine natural product library, in addition to community deposits (22). Eight parent ions showed confident database matches, including some that were detected at multiple sites (Table 1). A subsequent analysis performed using the GNPS standalone dereplication pipeline confirmed these matches and led to the additional identification of cocamidopropyl betaine. The greater sensitivity of the standalone tool is likely due to the clustering algorithm used to generate the molecular network, which can reduce match scores. Compounds were identified using a combination of approaches, including the GNPS-generated similarity (cosine) score, number of shared peaks, and a manual examination of mirror plots, in addition to m/z error. When available, authentic standards were used to further support the identifications. The nine compounds identified represent an array of structures originally reported from diverse sources, including sponges, algae, and bacteria, as well as structures of synthetic origin (Fig. 2, Table 1). Strict Chemical Analysis Working Group (CAWG) criteria (25) were applied to increase confidence in the matches. Four compounds (monactin, staurosporine, cocamidopropyl betaine, and erucamide) were assigned “level 1” classification according to CAWG recommendations based on retention time, mass, and fragmentation spectra in comparison to those of authentic standards (Fig. S2 to S5). The remaining five compounds were “putatively annotated” (level 2) based on spectral matching to library standards (Fig. S6 to S10). The m/z errors (Table 1) may indicate that the compounds detected are closely related to the library matches but may also reflect differences between the instruments on which the analyses were performed. The results demonstrate that microbial natural products can be extracted and identified directly from marine sediments, thus providing evidence for their production in situ.
FIG 1.
Sediment metabolomes visualized as a molecular network. Parent ions from five sites are colored based on distributions. Green nodes, only found at one site (site number indicated); red nodes, found at multiple sites (total number indicated). Parent ions matching known compounds are indicated by m/z values.
TABLE 1.
Compound identification
| Site | Compound name (CAWG classification)a | Parent ion (m/z)b | Library ion (m/z) | m/z error (ppm)c | Similarity scored | No. of shared peakse | Source(s) | Reference(s) |
|---|---|---|---|---|---|---|---|---|
| 1 | Erucamide (1) | 338.341 | 338.345 | 13.3 | 0.856 | 10 | Synthetic | 47 |
| 1 | Smenospongidine (2) | 310.320 | 310.311 | 24.2 | 0.756 | 7 | Smenospongia sp., Hippospongia sp., Dysidea sp. | 26, 62 |
| 2 | Pheophytin (2) | 871.587 | 871.590 | 2.17 | 0.873 | 5 | Eukaryotic phototrophs | 39 |
| 2 | Staurosporine (1) | 467.220 | 467.210 | 32.1 | 0.872 | 5 | Streptomyces spp., Salinispora sp., ascidians, mollusks | 31, 34–37 |
| 2 | Smenospongidine (2) | 310.329 | 310.311 | 47.6 | 0.749 | 8 | Smenospongia sp., Hippospongia sp., Dysidea sp. | 26, 62 |
| 2 | Erucamide (1) | 338.361 | 338.345 | 37.3 | 0.674 | 8 | Synthetic | 47 |
| 3 | Cocamidopropyl betaine (1) | 343.302 | 343.296 | 17.7 | 0.967 | 4 | Synthetic | 44 |
| 3 | Smenospongidine (2) | 310.305 | 310.311 | 17.9 | 0.838 | 8 | Smenospongia sp., Hippospongia sp., Dysidea sp. | 26, 62 |
| 3 | Erucamide (1) | 338.350 | 338.345 | 5.5 | 0.742 | 9 | Synthetic | 47 |
| 4 | Cocamidopropyl betaine (1) | 343.299 | 343.296 | 8.1 | 0.958 | 3 | Synthetic | 44 |
| 4 | Staurosporine (1) | 467.210 | 467.210 | 1.1 | 0.883 | 6 | Streptomyces spp., Salinispora sp., ascidians, mollusks | 31, 34–37 |
| 4 | Smenospongidine (2) | 310.318 | 310.311 | 22.4 | 0.831 | 4 | Smenospongia sp., Hippospongia sp., Dysidea sp. | 26, 62 |
| 4 | Tetranactin (2) | 815.500 | 815.468 | 30.3 | 0.816 | 20 | Streptomyces sp. | 30 |
| 4 | Erucamide (1) | 338.345 | 338.345 | 14.4 | 0.807 | 8 | Synthetic | 47 |
| 4 | Dinactin (2) | 787.465 | 787.428 | 42.1 | 0.795 | 15 | Streptomyces sp. | 28 |
| 4 | Trinactin (2) | 801.480 | 801.462 | 18 | 0.780 | 10 | Streptomyces sp. | 28 |
| 4 | Monactin (1) | 773.450 | 773.458 | 10.0 | 0.688 | 8 | Streptomyces sp. | 28 |
| 5 | Cocamidopropyl betaine (1) | 343.293 | 343.296 | 8.2 | 0.963 | 3 | Synthetic | 44 |
| 5 | Staurosporine (1) | 467.208 | 467.210 | 6.0 | 0.881 | 5 | Streptomyces spp., Salinispora spp., ascidians, mollusks | 31, 34–37 |
| 5 | Trinactin (2) | 801.359 | 801.462 | 21.6 | 0.804 | 14 | Streptomyces sp. | 28 |
| 5 | Erucamide (1) | 338.351 | 338.345 | 17.2 | 0.795 | 8 | Synthetic | 47 |
| 5 | Smenospongidine (2) | 310.305 | 310.311 | 20.8 | 0.784 | 4 | Smenospongia sp., Hippospongia sp., Dysidea sp. | 31, 34–37 |
| 5 | Tetranactin (2) | 815.441 | 815.468 | 31.4 | 0.763 | 5 | Streptomyces sp. | 28 |
| 5 | Dinactin (2) | 787.392 | 787.3428 | 45.5 | 0.728 | 11 | Streptomyces sp. | 28 |
| 5 | Monactin (1) | 773.450 | 773.458 | 13.7 | 0.653 | 6 | Streptomyces sp. | 28 |
Compound names are followed by the CAWG classification (1 or 2).
Parent ions detected at the five sites were compared with standards in the GNPS library.
m/z error describes the difference between observed parent ion and library spectrum in ppm (which contains both low- and high-resolution data).
GNPS-generated similarity score is a metric from 0 to 1, with >0.65 considered a positive match.
Number of shared peaks indicates the number of MS/MS fragments that matched between library and parent ion fragmentation spectra.
FIG 2.
Compounds identified in the sediment metabolomes. Stereochemistry was not assigned as part of this study.
The compound putatively identified as the sponge metabolite smenospongidine (26) was unexpected, since the sediments did not contain any obvious sponge biomass. Sesquiterpene quinones, such as smenospongidine, belong to a well-known class of redox-active microbial metabolites that have been proposed to function as extracellular electron shuttles under hypoxic growth conditions (27). Given the m/z error (17.9 to 47.6) and lack of a standard, the compound detected could be a related metabolite. Regardless of its identity, this compound represents one of the few observed at all five sites, and the network analysis places it in a molecular family, suggesting that structurally related compounds await identification (Fig. 1).
Bacterial metabolites belonging to the nactin family of macrotetrolide cyclic esters were also identified (monactin, dinactin, trinactin, and tetranactin). These closely related ionophore polyketides are produced by Streptomyces species and exhibit a range of biological activities (28–30). Similarities in their fragmentation spectra are visualized as a large molecular family in the network analysis (Fig. 1). Comparison with an authentic standard provides strong support for the identification of these compounds (Fig. S2). Staurosporine was also identified using the GNPS dereplication protocols and supported by comparison with an authentic standard (Fig. S3). It was observed at three sites and appeared in the network as a single node (Fig. 1). Staurosporine is a protein kinase inhibitor, originally isolated from Streptomyces staurosporeus (31), with activity against fungi, bacteria, and mammalian cell lines (Table 2) (32, 33). It has subsequently been reported from marine ascidians and mollusks (34, 35), as well as from other actinomycetes (36), including the marine actinomycete Salinispora arenicola (37). When isolated from marine invertebrates, staurosporine was proposed to be of microbial origin (38); however, this has yet to be experimentally verified. While the retention time and fragmentation pattern for the compound detected closely matched those of the standard, the mass difference may indicate that it is a closely related compound. The detection of relatively lipophilic compounds, such as staurosporine, suggests that some may be particle associated and supports the utility of HP20 resin to capture a broad range of compounds. The final natural product identified was pheophytin, which was observed at site 2. This flavonoid glycoside acts as an electron carrier in photosystem II (39) and is also a breakdown product of chlorophyll (40). It is found in cyanobacteria and eukaryotic phototrophs. Pheophytin has been used to assess primary productivity in sediments (41, 42) and may be indicative of decaying seaweeds, microalgae, or meiofauna grazing (43).
TABLE 2.
Staurosporine biological activitya
| Assay | IC50 | Reference or source |
|---|---|---|
| HeLa S3 cells | 4 nM | 84 |
| Fungal mycelium | 9.6 μM | 85 |
| Fungi | 0.78–50 μM | 31 |
| Mycobacterium protein kinase | 600 nM | 32 |
| Protein kinases | 2–20 nM | 33 |
| C. elegans predation deterrence | 179 μM | This study |
| Brine shrimp toxicity | 812 nM | This study |
IC50 values are from previous studies, except for C. elegans predation deterrence and brine shrimp lethality values.
In addition to the detection of natural products, two of the parent ions matched synthetic compounds and thus are likely of anthropogenic origin. Cocamidopropyl betaine is an additive to skin care products, including sun protectants (44), and has been found in numerous habitats associated with human activity (21). It was identified using the standalone GNPS dereplication tool and supported by comparison with an authentic standard (Fig. S4). The detection of this compound at three sites raises environmental concerns, given that low levels of sunscreen have been shown to promote coral bleaching (45). Another potential contaminant is erucamide, a fatty acid derivative used as a slip agent in the manufacturing of plastics (46), waxes, and other consumer goods. Erucamide appeared in a large molecular family (Fig. 1) and was supported by comparison with an authentic standard (Fig. S5). It was detected at all five sites but not observed in methodological controls, suggesting that it was present in the environment. Since erucamide is a derivative of the natural product erucic acid (47), the possibility that the compound detected is of natural origin cannot be ruled out.
Linking compounds to community composition.
In an attempt to link compounds identified in the sediment metabolomes to candidate producers, bacterial diversity was assessed by analyzing 16S rRNA gene amplicon libraries at a rarefied depth of 95,000 reads for three to five replicates from each of the five sites. The number of observed OTUs (99% sequence identity) was high, ranging from 16,063 ± 344 (site 1) to 17,917 ± 222 (site 4). We searched the sediment libraries for the genus Streptomyces, since it is known to produce staurosporine and nactins; however, no streptomycete OTUs were identified. We also searched for the marine actinomycete genus Salinispora, which includes the known staurosporine producer Salinispora arenicola (37), and identified this taxon at all five sites (Fig. 3A). The relative Salinispora abundance was low, ranging from 0.001% ± 0.001% (site 5) to 0.010% ± 0.004% (site 2) of the community. Assuming bacterial concentrations in marine sediments of 108 to 109/ml (48), the culture-independent abundances detected correspond well with previous reports of 103 Salinispora CFU per ml sediment (49). Although among-site differences in Salinispora abundance were not significant (analysis of variance [ANOVA], F4,16 = 1.285; P = 0.317), sites 2 and 4 had the highest average Salinispora relative abundance. Due to the region of the 16S rRNA gene sequenced, it was not possible to distinguish among the three named Salinispora species. We therefore used culture-dependent methods to test for the presence of S. arenicola at sites 2 and 4 and successfully isolated strains belonging to this species (Table S1). Metabolomic analyses revealed that these strains produced staurosporine (Fig. S11), thus establishing the presence of bacteria capable of producing a compound detected in the sediment metabolome.
FIG 3.
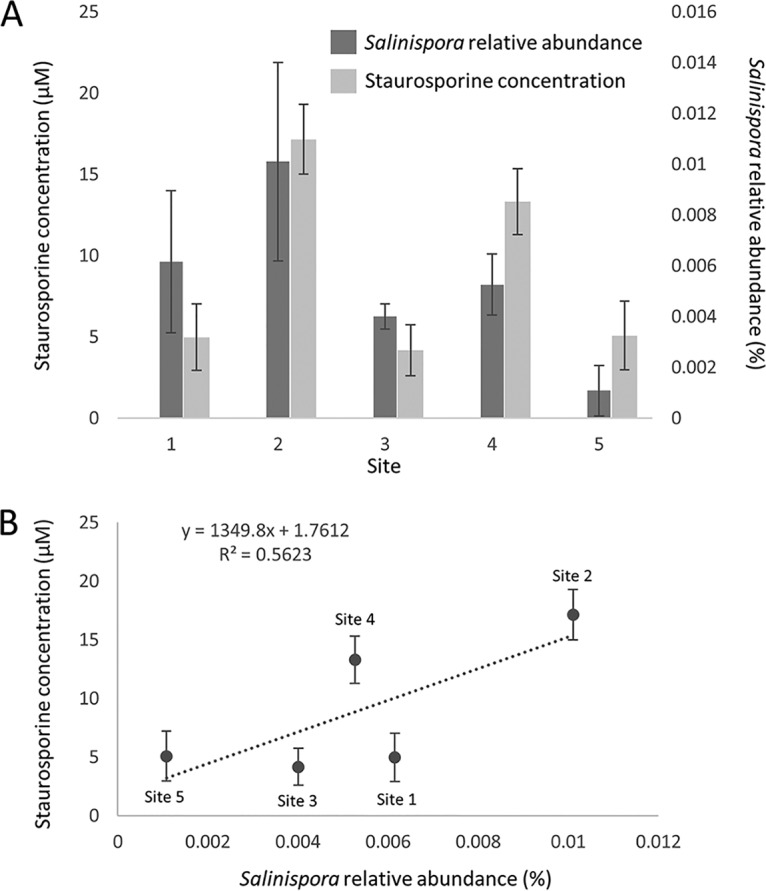
Correlation between Salinispora and staurosporine abundance in marine sediments. (A) Salinispora relative abundance (%) and staurosporine abundance (μM) at five sites. (B) Linear regression correlating Salinispora abundance with staurosporine concentration. Error bars are standard deviations.
Staurosporine quantification and biological activity.
Given that the resin capture method developed for the metabolomic analyses is not quantitative, the relationships between Salinispora relative abundance and staurosporine concentration was determined using volumetric sediment extraction protocols. Surprisingly, staurosporine was identified by high-performance liquid chromatography with UV detection (HPLC-UV) in extracts generated from all five sites, thus allowing concentrations to be calculated (Fig. S12 to S13). The inorganic component of the sediment was determined to be 32% to 41% at the various sites and was factored into the concentration calculations by dividing the total volumetric concentration by the percent organic material (Table S2) to establish “biologically relevant” staurosporine concentrations. These ranged from 5 to 17 μM and established the highest concentrations at sites 2 and 4 (Table S2), which represent two of the three sites at which staurosporine was detected by mass spectroscopy. Salinispora relative abundance was correlated with staurosporine concentration at the five sites (Fig. 3B), supporting the hypothesis that S. arenicola contributes to the staurosporine pool identified in marine sediments.
Staurosporine is a low-nanomolar kinase inhibitor that is highly toxic to mammalian cell lines, bacteria, and fungi (Table 2). The staurosporine concentrations at all five sites were above the 50% inhibitory concentration (IC50) values for HeLa cell (4 nM), protein kinase (2 to 20 nM), and mycobacterium protein kinase inhibition (600 nM). Sites 2 and 4 contained staurosporine levels above those shown to inhibit fungal growth (9.6 μM). The staurosporine concentrations also exceeded brine shrimp IC50 values, suggesting that it may function as a feeding deterrent, although this concentration did not deter feeding by Caenorhabditis elegans (Table 2 and Fig. S14). These results suggest that staurosporine can achieve allelopathic concentrations in marine sediments.
Metagenomic analyses.
Metagenomics represents an increasingly feasible approach to linking natural products to their producers (50). This is facilitated by the growing number of experimentally characterized biosynthetic gene clusters (BGCs) and by databases such as the Minimum Information about a Biosynthetic Gene cluster (MiBIG) repository (51). In an effort to link compounds detected in the metabolomics data to Salinispora spp., a 37-million-read metagenomic library generated from site 2 was mapped to the complete genome sequences of Salinispora tropica CNB-440 and Salinispora arenicola CNS-205. At a BLAST E value threshold of 1 × 10−50 and a DNA sequence identity of ≥96%, 14 reads mapped to S. arenicola, 22 mapped to S. tropica, and four mapped to both. Half of the S. arenicola reads and one-third of the S. tropica reads are >99% identical to the genomes, providing strong support for linkage to these species. These results suggest an overall Salinispora abundance of 0.5 to 1 ppm of the DNA in the sediment microbiome, which is in general agreement with the culture-independent results (0.01% to 0.001%). The low relative abundance of Salinispora sequences provides an explanation for our inability to detect Salinispora-derived staurosporine biosynthesis genes despite our ability to culture staurosporine-producing Salinispora strains from the same sediment samples. Given these results, it can be estimated that ca. 1,000× more reads would be needed to confidently obtain 1× coverage of a Salinispora genome, thus demonstrating current challenges associated with detecting biosynthetic gene clusters from rare members of the microbial community using metagenomics.
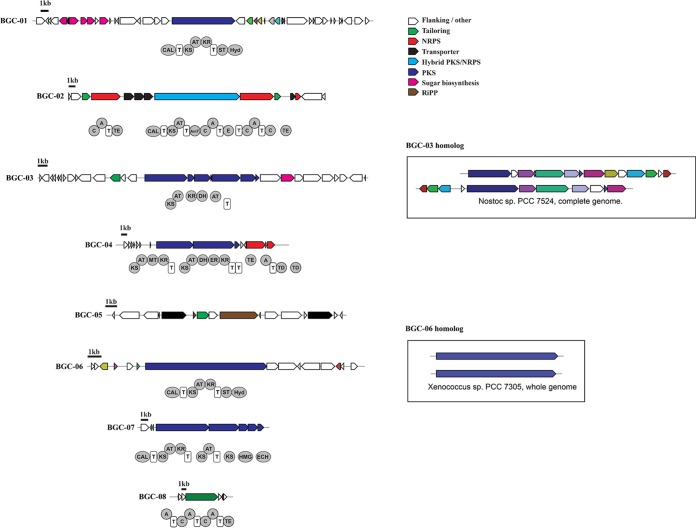
We next queried the metagenome for other potential sources of staurosporine using the staD gene, which is integral to staurosporine biosynthesis (52). A tBLASTn analysis against the 1,147,515 contig (>300 bp) metagenomic assembly identified 14 hits (E value cutoff of 1 × 10−10) that ranged in length from 277 to 7,834 nucleotides and coverage from 0.9× to 6.3×. The closest NCBI matches originated in cyanobacteria, a phylum known to produce staurosporine-related indolocarbazoles (53). Moreover, the longest contig contained a flavin adenine dinucleotide (FAD)-dependent amino acid oxidase (l-tryptophan oxidase) known to be involved in indolocarbazole biosynthesis, supporting our partial recovery of a canonical indolocarbazole BGC from the metagenome. Cyanobacteria were in considerably greater relative abundance (6%) than Salinispora at site 2, suggesting that they represent another potential source of the staurosporine or staurosporine-like compound detected, although the community analysis did not reveal any known cyanobacterial indolocarbazole producers. Similarly, BGCs associated with macrotetrolide (nactin) biosynthesis were not identified in the metagenomic analysis despite the level 1 identification of these compounds, suggesting that the producers were below the detection limit or similar compounds are produced by different biosynthetic routes.
In total, 83 different BGCs were assembled from the site 2 metagenome (Fig. 4). These encompass a wide range of biosynthetic paradigms, including 23 ribosomally synthesized and posttranslationally modified peptides (RiPPs; predicted to encode bacteriocins, lantipeptides, cyanobactins, and lasso peptides), 26 polyketide synthases (PKSs), 10 nonribosomal peptide synthetases (NRPSs), 1 PKS-NRPS hybrid, 14 terpenes, and 9 classified as “other” based on antiSMASH analyses. BLAST analyses linked 75% of these to cyanobacteria; however, others were linked to diverse taxonomic groups, including Proteobacteria (15%), Bacteroidetes (5%), Aquificae (1%), Firmicutes (1%), and Planctomycetes (2%). To further explore the identity of the host strains, we identified housekeeping genes flanking eight of the assembled BGCs. All eight housekeeping genes were most closely related to those of cyanobacteria, with percent identities from 67 to 85% (Table S3). Phylogenetic trees generated with the 100 top NCBI BLAST matches revealed that some formed a clade with the natural product-rich genus Moorea (Fig. S15), while others formed clades with less-studied genera, such as Myosarcina and Pleurocapsa (Fig. S16 to S17). Interestingly, five of the housekeeping genes did not cluster with those of any known cyanobacteria (Fig. S18), suggesting that they may belong to previously undescribed cyanobacterial lineages.
FIG 4.
Biosynthetic gene clusters assembled from the site 2 metagenome. BGCs were identified from the SPAdes-assembled metagenome using antiSMASH (scaffolds of >5,000 bp), and further annotated manually through BLASTp searches against the NCBI database. For BGC-03 and BGC-06, homologs identified in sequenced cyanobacterial genomes are shown for reference.
DISCUSSION
Thousands of natural products have been isolated from cultured bacteria, yet the biosynthetic potential detected in microbial communities remains far from being realized (50). While new approaches to cultivation have helped gain access to this potential (54), the isolation of compounds from individual strains cultured in the laboratory provides little insight into the diversity of compounds that occur in nature. Environmental metabolomics (55) provides a new window from which to view this complexity by circumventing the need for laboratory cultivation. Allowing the environment to provide the myriad conditions required for growth and compound production alleviates the need to reproduce these conditions in the lab. When applied to the human microbiome, metabolomics led to the recognition that human-associated microbes produce antibiotics and other biologically active compounds (56) that play important roles in human health (57). These approaches have yet to be applied more broadly to ocean sediments, a major global biome where the effects of natural products on microbial community structure have yet to be recognized.
Here, we demonstrate that untargeted metabolomics can be used to detect a vast array of compounds in marine sediments, the majority of which could not be readily identified. Given the inherent limitations of the capture (HP20) and detection (LC-MS) techniques employed, the results likely represent only a portion of the compounds present in the sites sampled. The ability to directly detect unknown metabolites in marine sediments draws parallels with the first culture-independent assessments of bacterial diversity, which revealed extensive levels of diversity that had yet to be cultured or described (58). Environmental metabolomics has enormous potential to reveal community-level chemical interactions and establish correlations between compounds and community composition. As data for additional standards are deposited into GNPS and other libraries, these analyses will increasingly include compound identification.
Although most compounds detected in this study could not be identified, the bacterial metabolite staurosporine was observed in both sediment metabolomes and extracts generated from strains of the marine actinomycete S. arenicola cultured from the same sediments. While anthropogenic antibiotics have been detected in various environmental samples, including river sediments (59), microbial natural products have seldom been detected in nature (2). The low relative abundance of Salinispora spp. in marine sediments supports the concept that members of the “rare biosphere” (60) play important roles in structuring microbial communities (61), in this case via the production of biologically active natural products. The high concentrations of staurosporine detected suggest that it may accumulate over time or originate from alternative biogenic sources. Support for the latter hypothesis comes from the detection of indolocarbazole biosynthetic genes in metagenomic assemblies assigned to cyanobacteria. Regardless of biosynthetic origin, the detection of biologically active concentrations of staurosporine at all five sites provides evidence that it functions as an allelochemical in nature. In the future, it will be important to better understand the fine-scale spatial dynamics of this and other biologically active compounds in marine sediments. It remains to be seen how effectively metagenomic BGC assembly can be used to connect rare community members with the compounds they produce.
The detection of a compound tentatively identified as smenospongidine provides another potentially interesting application of the resin capture technique. This sesquiterpene quinone was originally reported from a marine sponge (62). Given that there was no obvious sponge tissue in the sediments sampled, environmental metabolomics may provide a method to help identify compounds of microbial origin that were originally ascribed to marine invertebrates (11). Other interesting observations include the possibility that compounds such as the nactin family of macrotetrolide antibiotics (63) and staurosporine originate from organisms other than those from which they were originally reported. Finally, the detection of synthetic compounds indicates that sediment metabolomics can be used to assess anthropogenic influences, even in relatively pristine sites.
Environmental metabolomics provides a unique window with which to assess specialized metabolite diversity in nature. When coupled with community analyses, links between compounds and their producers can be established and subsequently tested. This approach can be used to establish baseline metabolomic fingerprints, links to microbiome composition, and opportunities to monitor baseline shifts over time or across environmental gradients as an indicator of sediment community “health.” The ability to detect and quantify natural products in marine sediments provides unique opportunities to assess their impact on community structure and opens new opportunities for natural product discovery.
MATERIALS AND METHODS
Locations and samples.
Five sites near the Smithsonian field station at Carrie Bow Cay, Belize, were sampled between 3 and 7 September 2015. The sites were characterized as (1) shallow reef (depth, 8 m; 16°47.901′N, 88°04.995′W), (2) reef slope (depth, 20 m; 16°48.191′N, 88°04.691′W), (3) mangrove channel (depth, 1 m; 16°49.546′N, 88°06.408′W), (4) lagoon reef (depth, 6 m; 16°47.726′N, 88°07.349′W), and (5) seagrass bed (depth, 1 m; 16°48.174′N, 88°04.925′W). For sediment metabolomes, nine hand-made Miracloth (EMD Millipore) bags, each containing ca. 80 g of MeOH-activated HP20 adsorbent resin (Diaion), were buried just below the surface of the sediment over an area ca. 1 m2 at each site. After 24 h, the bags were retrieved and stored frozen (−20°C) prior to elution. One similarly prepared resin bag was not deployed and was used as a negative control. Five replicate sediment samples (ca. 80 g each) were collected from each site in sterile Whirl-pak bags (Sigma-Aldrich) for bacterial cultivation and additional metabolomic analyses. A subsample from each site was transferred into a 50-ml falcon tube with RNAlater (Thermo Fisher) for molecular (DNA) analysis. All samples were stored frozen (−20°C) prior to processing.
Resin extraction and analysis.
The HP20 resin samples from each site were removed from the Miracloth bags, combined, and suspended in 500 ml of 1:1 dichloromethane (DCM):methanol (MeOH) for 30 min in a 2.8-liter Fernbach flask while shaking at 190 rpm on a G10 gyratory shaker (New Brunswick Scientific). The solvent was collected by filtration through qualitative P8 coarse fluted filter paper (Fisher Scientific, Hampton, NH), dried by rotary evaporation, and reextracted with ethyl acetate (EtOAc) followed by MeOH. The combined extracts were filtered (0.2 μm) to remove particulates, and those from sites 2 to 5 were subjected to C18 reversed phase flash chromatography using an acetonitrile (ACN)/water gradient (100% H2O, 20% ACN, 40% ACN, 60% ACN, 80% ACN, and 100% ACN). The fractions were dried by rotary evaporation, dissolved in MeOH at 1 mg/ml, and subjected to liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis by injecting 5 μl into an Agilent 1290 ultraperformance liquid chromatography (UHPLC) system. The site 1 crude extract was similarly analyzed. Chromatographic separation was achieved at 40°C, using a flow rate of 0.5 ml/min and a 1.7-μm C18 (50 × 2.1 mm) Kinetex UHPLC column (Phenomenex). The following linear gradient of solvents A (water and 0.1% formic acid [vol/vol]) and B (acetonitrile and 0.1% formic acid [vol/vol]) was used: 0 to 0.5 min (5% B), 0.5 to 8 min (5% to 95% B), and 8 to 9 min (95% B). The column eluent was introduced directly into a Bruker Daltonics microTOF focus II quadrupole time of flight (qTOF) mass spectrometer equipped with an Apollo II electrospray ionization source and controlled via otofControl v3.4 (build 16) and Hystar v3.2 software packages (Bruker Daltonics). The instrument was first externally calibrated using ESI-L low-concentration tuning mix (Agilent Technologies) prior to sample analysis and the internal calibrant (lock mass) hexakis(1H,1H,2H-difluoroethoxy)phosphazene (Synquest Laboratories), m/z 622.0295089613, was continuously introduced during the entirety of each LC-MS run. Data were collected in positive-ion mode, scanning from 100 to 2,000 m/z. Instrument source parameters were set as follows: nebulizer gas (nitrogen) pressure, 300 kPa; capillary voltage, 4,500 V; ion source temperature, 200°C; and dry gas flow, 9 liters/min. The initial mass spectral acquisition (MS1) rate was set at 2 Hz, and the MS/MS acquisition rate varied (5 to 10 Hz) depending on precursor intensity. Data-dependent MS/MS acquisition was programmed to the top ten most intense precursors per MS1 scan, and precursors were actively excluded for 0.5 min after being fragmented twice. Each MS/MS scan was the average of 4 collision energies, paired optimally with specific collision radio frequency (RF) (or “ion cooler RF”) voltages and transfer times in order to maximize the qualitative structural information from each precursor. The MS/MS data were converted to mzXML and uploaded to the MassIVE server (https://massive.ucsd.edu/) via the FTP client FileZilla (https://filezilla-project.org). Data are publicly available at GNPS (http://gnps.ucsd.edu) under accession number MSV000082961.
MS data were analyzed using the Global Natural Products Social Molecular Networking (GNPS) pipeline (22) using a modification of previously validated parameters (64). Only parent ions that fragmented at least twice were included. Networks were visualized using Cytoscape (http://www.cytoscape.org/cy3.html) with the built-in solid layout (65). Nodes with a cosine score of >0.95 were combined into consensus spectra. The algorithm parameters included mass tolerance for fragment peaks (0.3 Da), parent mass tolerance (1.0 Da), a minimum number of matched peaks per spectral alignment (4), a maximum component size of 1, and a minimum cosine score of 0.6. Nodes in the top ten cosine scores (K parameter) in both directions were connected by an edge. MS/MS spectra from “no injection blanks” and negative controls were subtracted from the network to remove both instrument noise and parent ions associated with the resin extraction protocol.
Compounds were identified using the GNPS reference library, which contains over 220,000 curated spectra (22). To minimize false positives, a false-discovery rate (FDR) analysis was conducted using the Passatutto test on GNPS. This test generates a decoy library to assess the rate at which a user’s submitted spectra match spectra in the decoy library for a range of minimum matched peaks and cosine (similarity) scores (66). Parameters were set to maintain a false discovery rate below 1% (Fig. S18). Library matches were assigned if they shared at least four MS/MS peaks, were within one dalton, and had a similarity score above 0.65. Mirror spectra plots generated in GNPS were used to manually investigate all spectral matches. Positive matches to short amino acid sequences or primary metabolites were excluded.
Four of the nine compounds identified were compared to authentic standards (staurosporine [NETA Scientific], erucamide [VWR], cocamidopropyl betaine [Spectrum], and monactin [Santa Cruz Biotechnology]) using the Chemical Analysis Working Group’s established criteria (25). These included retention time, fragmentation spectrum, and mass matching using a Dionex UltiMate 3000 high-performance liquid chromatograph (ThermoFisher Scientific, Waltham, MA) coupled to a Bruker Impact high-definition (HD) qTOF mass spectrometer. Chromatographic separation was achieved at 40°C, using a flow rate of 0.5 ml/min and a Kinetex C18 (1.7-μm, 100-Å) column (50 mm × 2.1 mm) (Phenomenex). The following linear gradient of solvents A (water and 0.1% formic acid [vol/vol]) and B (acetonitrile and 0.1% formic acid [vol/vol]) was used: 0-1 min (5% B), 1 to 2 min (5% to 40% B), 2 to 8 min (40% to 100% B), and 8 to 9 min (100% B).
For the mass spectrometry analysis, data were collected in positive ion mode, scanning from 800 to 2,000 m/z. Instrument source parameters were set as follows: nebulizer gas (nitrogen) pressure, 300 kPa; capillary voltage, 4,500 V; ion source temperature, 200°C; dry gas flow, 9 liters/min. MS1 spectral acquisition rate was set at 3 Hz and MS/MS acquisition rate was variable depending on precursor intensity. Data-dependent MS/MS acquisition was programmed to the top ten most intense precursors per MS1 scan and precursors were actively excluded for 0.5 min after being fragmented twice. Each MS/MS scan was subjected to five collision energies, starting with 3 eV and stepped 50%, 75%, 150%, and 200%, in order to maximize the qualitative structural information from each precursor. HP-921 lock mass was introduced during the entirety of the mass spectrometry run.
Staurosporine quantification.
Three replicate sediment samples ranging from 6 to 11 ml from each site were extracted with 50 ml of 1:1 DCM:MeOH. The extracts were filtered, the solvent removed under a stream of nitrogen, and the remaining water dried via lyophilization. The dried extracts were solubilized in MeOH and adsorbed onto 100 mg C18, loaded onto 2-g Isolute SPE C18 columns (Biotage), and subjected to flash chromatography using a 20% acetonitrile:water step gradient. The fractions were dried by rotary evaporation and dissolved in 2 ml MeOH, and 30 μl was injected into an Agilent 1100 series high-performance liquid chromatograph with a Phenomenex Kinetex C18 reversed-phase column (2.6 mm, 100 × 4.6 mm) using a flow rate of 0.7 ml/min. A linear gradient of solvents A (water and 0.1% formic acid [vol/vol]) and B (acetonitrile and 0.1% formic acid [vol/vol]) was used with 0 to 2 min (5% B), 2 to 14 min (5% to 100% B), and 14 to 15 min (100% B). The diverter valve was set to waste for the first 2 min. Staurosporine was identified based on retention time and UV-spectral matching in comparison with an authentic standard. The average area under the curve for the staurosporine peak (350 nm) was calculated for triplicate extracts from each location and converted to concentration using the best-fit line equation generated in R (67) for a standard curve generated from an authentic standard.
Biologically relevant sediment volumes were calculated by subtracting the inorganic volume from the total sediment volume of each sample. Inorganic volumes were determined at each site using standard protocols (68) by first oven drying (60°C for 48 h) triplicate 5-ml volumes of wet sediment. The dried sediments were then treated with excess 20% H2O2, heated (60°C) for 30 min to oxidize organics, and redried, and the volume of remaining inorganic material calculated via volumetric displacement and subtracted from the initial wet sediment volume. Molar staurosporine concentrations were calculated by converting mg/ml concentrations calculated from the HPLC-UV data to μM using the biologically relevant sediment volumes.
To assess C. elegans feeding deterrence, staurosporine was dissolved in 50 μl dimethyl sulfoxide (DMSO) to generate concentrations of 210 μM, 160 μM, and 110 μM (corresponding to 0.1 mg/ml, 0.075 mg/ml, and 0.05 mg/ml, respectively) in sterile 10-ml glass culture tubes. These concentrations were picked because they bracketed the MIC at which staurosporine deterred C. elegans feeding. Escherichia coli strain OP-50 (Troemel laboratory, University of California, San Diego [UCSD]) was grown in 50 ml of Miller's LB broth (Fisher Scientific) for 3 days, autoclaved, and 950 μl added to the staurosporine dilutions and DMSO controls. Twenty-five μl of the treated and control E. coli cultures were then placed 1 cm apart in a 60-cm diameter petri dish containing 4 ml of nematode growth medium (Research Products International Corp., Mount Prospect, IL). Sterile C. elegans embryos were generated as previously described (69), and ca. 75 were placed equidistant from treatment and control foods, with an average of 6 plates per trial. The number of living C. elegans individuals per food source was counted under a dissecting scope 24 h after embryo addition, converted to percentage C. elegans per food source, and differences between treatments and controls tested using a nonparametric t test (Wilcoxon rank-sum test) in R version 3.4.1.
Brine shrimp assay.
The brine shrimp assay was performed as previously described (70). Briefly, Artemia eggs (Fisher Scientific) were hatched in brine (1 liter deionized [DI] water, 40 g instant ocean) for 48 h at room temperature with excess airflow. Approximately 25 Artemia individuals were then transferred into each well of a 24-well plate containing 1.95 ml of brine, after which serially diluted staurosporine dissolved in 50 μl DMSO was added to the wells (0.16 to 16 μM final concentrations) in triplicate along with DMSO controls. After 24 h, the numbers of dead and living brine shrimp were counted in each well under a dissecting microscope.
For bacterial community analysis, environmental DNA (eDNA) was extracted from 24 sediment samples (5 replicates per site for sites 1 to 4 and 4 replicates for site 5). Approximately 1 g of sediment per sample was added into FastPrep tubes and subjected to a physical (bead beating) and chemical (phenol-chloroform) DNA extraction protocol (71). All extractions were done in duplicate and combined during the purification process. The v4 region of the 16S rRNA gene was PCR amplified in triplicate for each sample from 1 μl of 5 ng/μl DNA template using the Phusion Hot Start Flex 2× master mix, an annealing temperature of 60°C, and the primers 515F (TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGGTGYCAGCMGCCGCGGTAA) and 806Rb (GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGGACTACNVGGGTWTCTAAT) (72). Replicate reactions were pooled and the products cleaned using ExoSap-IT. Illumina barcodes were added using the following program: 98°C for 1 min followed by five cycles of 98°C for 10 s, 65°C for 20 s, and 72°C for 30 s, with a final extension at 72°C for 2 min. Libraries were constructed following Illumina protocols and PCR products cleaned with ExoSap-IT and pooled in equimolar concentrations, cleaned using AMPure beads, and sequenced on a MiSeq v2 PE250 instrument at the Institute for Genomic Medicine (IGM), University of California, San Diego.
Sequences were trimmed using the paired-end command in Trimmomatic (http://www.usadellab.org/cms/?page=trimmomatic) with a quality control sliding window of 4 bases, a minimum quality of 25, and a minimum length of 50 bases. All samples were standardized to a rarefication depth of 95,000 reads, resulting in three replicates (one each from sites 3, 4, and 5) being removed due to inadequate sequencing depth. The remaining reads were clustered into OTUs based on 99% similarity and taxonomy assigned using the Silva rRNA Database (73) in Qiime v1.9.1 (74). To assess Salinispora relative abundance, replicates were analyzed with chloroplast sequences excluded (75).
Actinomycete cultivation and identification were performed following standard protocols (49). Briefly, 1 g frozen sediment from each of the five sites was air dried for 48 h and blotted onto petri dishes containing medium A1 (10 g starch, 4 g yeast extract, 2 g peptone, 16 g agar, 750 ml 0.2 μM filtered seawater, and 250 ml deionized water). The petri dishes were incubated at room temperature for 2 weeks, after which Salinispora-like colonies were purified by dilution streaking. Colony PCR was performed by suspending a single colony in 10 μl DMSO and using 1 μl as the PCR template. Each PCR mixture consisted of 10× PCR buffer (Applied Biosciences, Foster City, CA), 2.5 mM MgCl2 (Applied Biosciences), 0.7% DMSO, 10 mM deoxynucleoside triphosphates (dNTPs), 1.5 U AmpliTaq Gold DNA polymerase (Applied Biosciences) and 10 μmol of each 16S rRNA primer (FC27 [5′-AGAGTTTGATCCTGGCTCAG-3′] forward and RC1492 [5′TACGGCTACCTTGTTACGACTT-3′] reverse). PCR conditions were as follows: 5 min of initial denaturation at 95°C, followed by 30 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 1 min. The resulting amplicons were sequenced in the forward and reverse directions (Eton Biosciences, San Diego, CA), assembled, and trimmed in Geneious v10.2.3 (76), yielding ca. 1,300 bp products. Strains were identified based on NCBI BLAST analysis (77).
Metabolite analyses were performed on single actinomycete colonies removed from A1 agar plates and extracted with 5 ml of MeOH in 15 ml Falcon tubes. Extracts were dried by rotary evaporation, weighed, redissolved at a concentration of 5 mg/ml in MeOH, and subjected to HPLC analysis on the Agilent 1100 series instrument using previously described parameters.
Metagenomics.
Mechanically sheared metagenomic DNA from site 2 (average size, ca. 500 bp) was used to prepare an Illumina sequencing library using the Apollo 324 next-generation sequencing (NGS) library prep system and the PrepX DNA library kit (Wafergen, CA). This library was then sequenced on an Illumina HiSeq 2500 rapid flow cell to yield 36,894,038 paired-end reads (2 × 175 bp). PRINSEQ was used to filter and trim the resulting raw Illumina reads, discarding reads that had an average quality score less than 30 or a length of less than 87 bp (78). SPAdes (default parameters) was used to assemble reads that passed quality filtering (79). Metagenomic BGCs were detected in the resulting scaffolds (>5,000 bp) using antiSMASH 3.0 (80) and were further analyzed manually.
Phylogenetic analyses.
Housekeeping genes found flanking BGCs of interest were subjected to a BLAST search against the NCBI database. FASTA sequences for the top 100 hits were downloaded, and each set was aligned with the corresponding query sequence using the MUSCLE 3.8.31 (81) alignment option in Mesquite 3.04 (82). RaxmlGUI v1.5 was used with default parameters and no set outgroup to construct phylogenetic trees with 50 bootstrap replicates (83).
Supplementary Material
ACKNOWLEDGMENTS
E. coli strain OP-50 was kindly provided by the Troemel laboratory, UCSD. The protocol for the brine shrimp assay was provided by Christopher Leber, UCSD.
This research was supported by the National Science Foundation under grant OCE-1235142.
Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02830-18.
REFERENCES
- 1.Berdy J. 2005. Bioactive microbial metabolites. A personal view. J Antibiot 58:1–26. doi: 10.1038/ja.2005.1. [DOI] [PubMed] [Google Scholar]
- 2.Davies J. 2006. Are antibiotics naturally antibiotics? J Ind Microbiol Biotechnol 33:496–499. doi: 10.1007/s10295-006-0112-5. [DOI] [PubMed] [Google Scholar]
- 3.Letzel AC, Jing L, Amos G, Millán‐Aguiñaga N, Ginigini J, Abdelmohsen UR, Gaudêncio SP, Ziemert N, Moore BS, Jensen PR. 2017. Genomic insights into specialized metabolism in the marine actinomycete Salinispora. Environ Microbiol 19:3660–3673. doi: 10.1111/1462-2920.13867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Milshteyn A, Schneider JS, Brady SF. 2014. Mining the metabiome: identifying novel natural products from microbial communities. Chem Biol 21:1211–1223. doi: 10.1016/j.chembiol.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rutledge PJ, Challis GL. 2015. Discovery of microbial natural products by activation of silent biosynthetic gene clusters. Nat Rev Microbiol 13:509–523. doi: 10.1038/nrmicro3496. [DOI] [PubMed] [Google Scholar]
- 6.Burgess JG, Jordan EM, Bregu M, Mearns-Spragg A, Boyd KG. 1999. Microbial antagonism: a neglected avenue of natural products research. Prog Ind Microbiol 35:27–32. doi: 10.1016/S0168-1656(99)00054-1. [DOI] [PubMed] [Google Scholar]
- 7.Patin NV, Floros DJ, Hughes CC, Dorrestein PC, Jensen PR. 2018. The role of inter-species interactions in Salinispora specialized metabolism. Microbiol 164:946–955. doi: 10.1099/mic.0.000679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Traxler MF, Watrous JD, Alexandrov T, Dorrestein PC, Kolter R. 2013. Interspecies interactions stimulate diversification of the Streptomyces coelicolor secreted metabolome. mBio 4:e00459-13. doi: 10.1128/mBio.00459-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trischman JA, Oeffner RE, de Luna MG, Kazaoka M. 2004. Competitive induction and enhancement of indole and a diketopiperazine in marine bacteria. Mar Biotechnol (NY) 6:215–220. doi: 10.1007/s10126-003-0010-z. [DOI] [PubMed] [Google Scholar]
- 10.König GM, Kehraus S, Seibert SF, Abdel‐Lateff A, Müller D. 2006. Natural products from marine organisms and their associated microbes. Chembiochem 7:229–238. doi: 10.1002/cbic.200500087. [DOI] [PubMed] [Google Scholar]
- 11.Wilson MC, Mori T, Ruckert C, Uria AR, Helf MJ, Takada K, Gernert C, Steffens UAE, Heycke N, Schmitt S, Rinke C, Helfrich EJN, Brachmann AO, Gurgui C, Wakimoto T, Kracht M, Crusemann M, Hentschel U, Abe I, Matsunaga S, Kalinowski J, Takeyama H, Piel J. 2014. An environmental bacterial taxon with a large and distinct metabolic repertoire. Nature 506:58–62. doi: 10.1038/nature12959. [DOI] [PubMed] [Google Scholar]
- 12.Raaijmakers JM, Vlami M, De Souza JT. 2002. Antibiotic production by bacterial biocontrol agents. Antonie Van Leeuwenhoek 81:537–547. doi: 10.1023/A:1020501420831. [DOI] [PubMed] [Google Scholar]
- 13.Mazzola M. 2002. Mechanisms of natural soil suppressiveness to soilborne diseases. Antonie Van Leeuwenhoek 81:557–564. doi: 10.1023/A:1020557523557. [DOI] [PubMed] [Google Scholar]
- 14.Raaijmakers JM, Bonsall RF, Weller DM. 1999. Effect of population density of Pseudomonas fluorescens on production of 2,4-diacetylphloroglucinol in the rhizosphere of wheat. Phytopathology 89:470–475. doi: 10.1094/PHYTO.1999.89.6.470. [DOI] [PubMed] [Google Scholar]
- 15.Thrane C, Harder Nielsen T, Neiendam Nielsen M, Sørensen J, Olsson S. 2000. Viscosinamide-producing Pseudomonas fluorescens DR54 exerts a biocontrol effect on Pythium ultimum in sugar beet rhizosphere. FEMS Microbiol Ecol 33:139–146. doi: 10.1111/j.1574-6941.2000.tb00736.x. [DOI] [PubMed] [Google Scholar]
- 16.Lane JQ, Roddam CM, Langlois GW, Kudela RM. 2010. Application of solid phase adsorption toxin tracking (SPATT) for field detection of the hydrophilic phycotoxins domoic acid and saxitoxin in coastal California. Limnol Oceanogr Methods 8:645–660. doi: 10.4319/lom.2010.8.0645. [DOI] [Google Scholar]
- 17.Kudela RM. 2011. Characterization and deployment of solid phase adsorption toxin tracking (SPATT) resin for monitoring of microcystins in fresh and saltwater. Harmful Algae 11:117–125. doi: 10.1016/j.hal.2011.08.006. [DOI] [Google Scholar]
- 18.Takahashi E, Yu Q, Eaglesham G, Connell DW, McBroom J, Costanzo S, Shaw GR. 2007. Occurrence and seasonal variations of algal toxins in water, phytoplankton and shellfish from North Stradbroke Island, Queensland, Australia. Mar Environ Res 64:429–442. doi: 10.1016/j.marenvres.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 19.Weidenhamer JD, Mohney BK, Shihada N, Rupasinghe M. 2014. Spatial and temporal dynamics of root exudation: how important is heterogeneity in allelopathic interactions? J Chem Ecol 40:940–952. doi: 10.1007/s10886-014-0483-4. [DOI] [PubMed] [Google Scholar]
- 20.Dunn WB, Erban A, Weber RJM, Creek DJ, Brown M, Breitling R, Hankemeier T, Goodacre R, Neumann S, Kopka J, Viant MR. 2013. Mass appeal: metabolite identification in mass spectrometry-focused untargeted metabolomics. Metabolomics 9:44–66. doi: 10.1007/s11306-012-0434-4. [DOI] [Google Scholar]
- 21.Petras D, Nothias L-F, Quinn RA, Alexandrov T, Bandeira N, Bouslimani A, Castro-Falcón G, Chen L, Dang T, Floros DJ, Hook V, Garg N, Hoffner N, Jiang Y, Kapono CA, Koester I, Knight R, Leber CA, Ling T-J, Luzzatto-Knaan T, McCall L-I, McGrath AP, Meehan MJ, Merritt JK, Mills RH, Morton J, Podvin S, Protsyuk I, Purdy T, Satterfield K, Searles S, Shah S, Shires S, Steffen D, White M, Todoric J, Tuttle R, Wojnicz A, Sapp V, Vargas F, Yang J, Zhang C, Dorrestein PC. 2016. Mass spectrometry-based visualization of molecules associated with human habitats. Anal Chem 88:10775–10784. doi: 10.1021/acs.analchem.6b03456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang M, Carver JJ, Phelan VV, Sanchez LM, Garg N, Peng Y, Nguyen DD, Watrous J, Kapono CA, Luzzatto-Knaan T, Porto C, Bouslimani A, Melnik AV, Meehan MJ, Liu W-T, Crüsemann M, Boudreau PD, Esquenazi E, Sandoval-Calderón M, Kersten RD, Pace LA, Quinn RA, Duncan KR, Hsu C-C, Floros DJ, Gavilan RG, Kleigrewe K, Northen T, Dutton RJ, Parrot D, Carlson EE, Aigle B, Michelsen CF, Jelsbak L, Sohlenkamp C, Pevzner P, Edlund A, McLean J, Piel J, Murphy BT, Gerwick L, Liaw C-C, Yang Y-L, Humpf H-U, Maansson M, Keyzers RA, Sims AC, Johnson AR, Sidebottom AM, Sedio BE, Klitgaard A, Larson CB, P CAB, Torres-Mendoza D, Gonzalez DJ, Silva DB, Marques LM, Demarque DP, Pociute E, O'Neill EC, Briand E, Helfrich EJN, Granatosky EA, Glukhov E, Ryffel F, Houson H, Mohimani H, Kharbush JJ, Zeng Y, Vorholt JA, Kurita KL, Charusanti P, McPhail KL, Nielsen KF, Vuong L, Elfeki M, Traxler MF, Engene N, Koyama N, Vining OB, Baric R, Silva RR, Mascuch SJ, Tomasi S, Jenkins S, Macherla V, Hoffman T, Agarwal V, Williams PG, Dai J, Neupane R, Gurr J, Rodríguez AMC, Lamsa A, Zhang C, Dorrestein K, Duggan BM, Almaliti J, Allard P-M, Phapale P, Nothias L-F, Alexandrov T, Litaudon M, Wolfender J-L, Kyle JE, Metz TO, Peryea T, Nguyen D-T, VanLeer D, Shinn P, Jadhav A, Müller R, Waters KM, Shi W, Liu X, Zhang L, Knight R, Jensen PR, Palsson BO, Pogliano K, Linington RG, Gutiérrez M, Lopes NP, Gerwick WH, Moore BS, Dorrestein PC, Bandeira N. 2016. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat Biotechnol 34:828–837. doi: 10.1038/nbt.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fenical W, Jensen P. 2006. Developing a new resource for drug discovery: marine actinomycete bacteria. Nat Chem Biol 2:666–673. doi: 10.1038/nchembio841. [DOI] [PubMed] [Google Scholar]
- 24.Bull AT, Stach JEM. 2007. Marine actinobacteria: new opportunities for natural product search and discovery. Trends Microbiol 15:491–499. doi: 10.1016/j.tim.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 25.Sumner LW, Amberg A, Barrett D, Beale MH, Beger R, Daykin CA, Fan TW-M, Fiehn O, Goodacre R, Griffin JL, Hankemeier T, Hardy N, Harnly J, Higashi R, Kopka J, Lane AN, Lindon JC, Marriott P, Nicholls AW, Reily MD, Thaden JJ, Viant MR. 2007. Proposed minimum reporting standards for chemical analysis. Metabolomics 3:211–221. doi: 10.1007/s11306-007-0082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kondracki M-L, Guyot M. 1989. Biologically active quinone and hydroquinone sesquiterpenoids from the sponge Smenospongia sp. Tetrahedron 45:1995–2004. doi: 10.1016/S0040-4020(01)80062-4. [DOI] [Google Scholar]
- 27.Gallagher KA, Wanger G, Henderson J, Llorente M, Hughes CC, Jensen PR. 2017. Ecological implications of hypoxia‐triggered shifts in secondary metabolism. Environ Microbiol 19:2182–2191. doi: 10.1111/1462-2920.13700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Graven SN, Lardy HA, Johnson D, Rutter A. 1966. Antibiotics as tools for metabolic studies. V. Effect of nonactin, monactin, dinactin, and trinactin on oxidative phosphorylation and adenosine triphosphatase induction. Biochemistry 5:1729–1735. doi: 10.1021/bi00869a040. [DOI] [PubMed] [Google Scholar]
- 29.Callewaert DM, Radcliff G, Tanouchi Y, Shichi H. 1988. Tetranactin, a macrotetrolide antibiotic, suppresses in vitro proliferation of human lymphocytes and generation of cytotoxicity. Immunopharmacology 16:25–32. doi: 10.1016/0162-3109(88)90047-1. [DOI] [PubMed] [Google Scholar]
- 30.Ando K, Oishi H, Hirano S, Okutomi T, Suzuki K, Okazaki H, Sawada M, Sagawa T. 1971. Tetranactin, a new miticidal antibiotic. J Antibiot (Tokyo) 24:347–352. doi: 10.7164/antibiotics.24.347. [DOI] [PubMed] [Google Scholar]
- 31.Omura S, Iwai Y, Hirano A, Nakagawa A, Awaya J, Tsuchiya H, Takahashi Y, Asuma R. 1977. A new alkaloid AM-2282 of Streptomyces origin taxonomy, fermentation, isolation and preliminary characterization. J Antibiot 30:275–282. doi: 10.7164/antibiotics.30.275. [DOI] [PubMed] [Google Scholar]
- 32.Fernandez P, Saint-Joanis B, Barilone N, Jackson M, Gicquel B, Cole ST, Alzari PM. 2006. The Ser/Thr protein kinase PknB is essential for sustaining mycobacterial growth. J Bacteriol 188:7778–7784. doi: 10.1128/JB.00963-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meggio F, Deana AD, Ruzzene M, Brunati AM, Cesaro L, Guerra B, Meyer T, Mett H, Fabbro D, Furet P, Dobrowolska G, Pinna LA. 1995. Different susceptibility of protein kinases to staurosporine inhibition: kinetic studies and molecular bases for the resistance of protein kinase CK2. Eur J Biochem 234:317–322. doi: 10.1111/j.1432-1033.1995.317_c.x. [DOI] [PubMed] [Google Scholar]
- 34.Schupp P, Eder C, Proksch P, Wray V, Schneider B, Herderich M, Paul V. 1999. Staurosporine derivatives from the ascidian Eudistoma toealensis and its predatory flatworm Pseudoceros sp. J Nat Prod 62:959–962. doi: 10.1021/np980527d. [DOI] [PubMed] [Google Scholar]
- 35.Cantrell CL, Groweiss A, Gustafson KR, Boyd MR. 1999. A new staurosporine analog from the prosobranch mollusk Coriocella nigra. Nat Prod Lett 14:39–46. doi: 10.1080/10575639908045433. [DOI] [Google Scholar]
- 36.Williams DE, Bernan VS, Ritacco FV, Maiese WM, Greenstein M, Andersen RJ. 1999. Holyrines A and B, possible intermediates in staurosporine biosynthesis produced in culture by a marine actinomycete obtained from the North Atlantic Ocean. Tetrahedron Lett 40:7171–7174. doi: 10.1016/S0040-4039(99)01495-1. [DOI] [Google Scholar]
- 37.Jensen PR, Williams PG, Oh DC, Zeigler L, Fenical W. 2007. Species-specific secondary metabolite production in marine actinomycetes of the genus Salinispora. Appl Environ Microbiol 73:1146–1152. doi: 10.1128/AEM.01891-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Steinert G, Taylor MW, Schupp PJ. 2015. Diversity of actinobacteria associated with the marine ascidian Eudistoma toealensis. Mar Biotechnol (NY) 17:377–385. doi: 10.1007/s10126-015-9622-3. [DOI] [PubMed] [Google Scholar]
- 39.Klimov VV. 2003. Discovery of pheophytin function in the photosynthetic energy conversion as the primary electron acceptor of photosystem II. Photosynthesis Res 76:247–253. doi: 10.1023/A:1024990408747. [DOI] [PubMed] [Google Scholar]
- 40.Lorenzen CJ. 1967. Determionation of chlorophyll and pheo-pigments: spectrophotometric equations. Limnol Oceanogr 12:343–346. doi: 10.4319/lo.1967.12.2.0343. [DOI] [Google Scholar]
- 41.Brown LM, Hargrave BT, MacKinnon MD. 1981. Analysis of chlorophyll a in sediments by high-pressure liquid chromatography. Can J Fish Aquat Sci 38:205–214. doi: 10.1139/f81-027. [DOI] [Google Scholar]
- 42.Bunt J, Lee C, Lee E. 1972. Primary productivity and related data from tropical and subtropical marine sediments. Mar Biol 16:28–36. doi: 10.1007/BF00347844. [DOI] [Google Scholar]
- 43.Cartaxana P, Jesus B, Brotas V. 2003. Pheophorbide and pheophytin a-like pigments as useful markers for intertidal microphytobenthos grazing by Hydrobia ulvae. Estuar Coast Shelf Sci 58:293–297. doi: 10.1016/S0272-7714(03)00081-7. [DOI] [Google Scholar]
- 44.Jacob SE, Amini S. 2008. Cocamidopropyl betaine. Dermatitis 19:157–160. [PubMed] [Google Scholar]
- 45.Danovaro R, Bongiorni L, Corinaldesi C, Giovannelli D, Damiani E, Astolfi P, Greci L, Pusceddu A. 2008. Sunscreens cause coral bleaching by promoting viral infections. Environ Health Persp 116:441. doi: 10.1289/ehp.10966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bhunia K, Sablani SS, Tang J, Rasco B. 2013. Migration of chemical compounds from packaging polymers during microwave, conventional heat treatment, and storage. Comp Rev Food Sci Food Safety 12:523–545. doi: 10.1111/1541-4337.12028. [DOI] [PubMed] [Google Scholar]
- 47.Molnar N. 1974. Erucamide. J Am Oil Chem Soc 51:84–87. doi: 10.1007/BF00000019. [DOI] [Google Scholar]
- 48.Schmidt JL, Deming JW, Jumars PA, Keil RG. 1998. Constancy of bacterial abundance in surficial marine sediments. Limnol Oceanogr 43:976–982. doi: 10.4319/lo.1998.43.5.0976. [DOI] [Google Scholar]
- 49.Mincer TJ, Jensen PR, Kauffman CA, Fenical W. 2002. Widespread and persistent populations of a major new marine actinomycete taxon in ocean sediments. Appl Environ Microbiol 68:5005–5011. doi: 10.1128/AEM.68.10.5005-5011.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Crits-Christoph A, Diamond S, Butterfield CN, Thomas BC, Banfield JF. 2018. Novel soil bacteria possess diverse genes for secondary metabolite biosynthesis. Nature 558:440–444. doi: 10.1038/s41586-018-0207-y. [DOI] [PubMed] [Google Scholar]
- 51.Medema MH, Kottmann R, Yilmaz P, Cummings M, Biggins JB, Blin K, de Bruijn I, Chooi YH, Claesen J, Coates RC, Cruz-Morales P, Duddela S, Düsterhus S, Edwards DJ, Fewer DP, Garg N, Geiger C, Gomez-Escribano JP, Greule A, Hadjithomas M, Haines AS, Helfrich EJN, Hillwig ML, Ishida K, Jones AC, Jones CS, Jungmann K, Kegler C, Kim HU, Kötter P, Krug D, Masschelein J, Melnik AV, Mantovani SM, Monroe EA, Moore M, Moss N, Nützmann H-W, Pan G, Pati A, Petras D, Reen FJ, Rosconi F, Rui Z, Tian Z, Tobias NJ, Tsunematsu Y, Wiemann P, Wyckoff E, Yan X, Yim G, Yu F, Xie Y, Aigle B, Apel AK, Balibar CJ, Balskus EP, Barona-Gómez F, Bechthold A, Bode HB, Borriss R, Brady SF, Brakhage AA, Caffrey P, Cheng Y-Q, Clardy J, Cox RJ, De Mot R, Donadio S, Donia MS, van der Donk WA, Dorrestein PC, Doyle S, Driessen AJM, Ehling-Schulz M, Entian K-D, Fischbach MA, Gerwick L, Gerwick WH, Gross H, Gust B, Hertweck C, Höfte M, Jensen SE, Ju J, Katz L, Kaysser L, Klassen JL, Keller NP, Kormanec J, Kuipers OP, Kuzuyama T, Kyrpides NC, Kwon H-J, Lautru S, Lavigne R, Lee CY, Linquan B, Liu X, Liu W, Luzhetskyy A, Mahmud T, Mast Y, Méndez C, Metsä-Ketelä M, Micklefield J, Mitchell DA, Moore BS, Moreira LM, Müller R, Neilan BA, Nett M, Nielsen J, O'Gara F, Oikawa H, Osbourn A, Osburne MS, Ostash B, Payne SM, Pernodet J-L, Petricek M, Piel J, Ploux O, Raaijmakers JM, Salas JA, Schmitt EK, Scott B, Seipke RF, Shen B, Sherman DH, Sivonen K, Smanski MJ, Sosio M, Stegmann E, Süssmuth RD, Tahlan K, Thomas CM, Tang Y, Truman AW, Viaud M, Walton JD, Walsh CT, Weber T, van Wezel GP, Wilkinson B, Willey JM, Wohlleben W, Wright GD, Ziemert N, Zhang C, Zotchev SB, Breitling R, Takano E, Glöckner FO. 2015. Minimum information about a biosynthetic gene cluster. Nat Chem Biol 11:625–631. doi: 10.1038/nchembio.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Freel KC, Nam S-J, Fenical W, Jensen PR. 2011. Evolution of secondary metabolite genes in three closely related marine actinomycete species. Appl Environ Microbiol 77:7261–7270. doi: 10.1128/AEM.05943-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sánchez C, Méndez C, Salas JA. 2006. Indolocarbazole natural products: occurrence, biosynthesis, and biological activity. Nat Prod Rep 23:1007–1045. doi: 10.1039/b601930g. [DOI] [PubMed] [Google Scholar]
- 54.Ling LL, Schneider T, Peoples AJ, Spoering AL, Engels I, Conlon BP, Mueller A, Schäberle TF, Hughes DE, Epstein S, Jones M, Lazarides L, Steadman VA, Cohen DR, Felix CR, Fetterman KA, Millett WP, Nitti AG, Zullo AM, Chen C, Lewis K. 2015. A new antibiotic kills pathogens without detectable resistance. Nature 517:455–459. doi: 10.1038/nature14098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Viant MR. 2008. Recent developments in environmental metabolomics. Mol Biosyst 4:980–986. doi: 10.1039/b805354e. [DOI] [PubMed] [Google Scholar]
- 56.Donia MS, Fischbach MA. 2015. Small molecules from the human microbiota. Science 349:1254766. doi: 10.1126/science.1254766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mazmanian SK, Round JL, Kasper DL. 2008. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 453:620. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 58.Hugenholtz P, Goebel BM, Pace NR. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J Bacteriol 180:4765–4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pei R, Kim S-C, Carlson KH, Pruden A. 2006. Effect of river landscape on the sediment concentrations of antibiotics and corresponding antibiotic resistance genes (ARG). Water Res 40:2427–2435. doi: 10.1016/j.watres.2006.04.017. [DOI] [PubMed] [Google Scholar]
- 60.Sogin ML, Morrison HG, Huber JA, Welch DM, Huse SM, Neal PR, Arrieta JM, Herndl GJ. 2006. Microbial diversity in the deep sea and the underexplored “rare biosphere.” Proc Natl Acad Sci U S A 103:12115–12120. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ramirez KS, Knight CG, De Hollander M, Brearley FQ, Constantinides B, Cotton A, Creer S, Crowther TW, Davison J, Delgado-Baquerizo M. 2017. Detecting macroecological patterns in bacterial communities across independent studies of global soils. Nat Microbiol 3:189–196. doi: 10.1038/s41564-017-0062-x. [DOI] [PubMed] [Google Scholar]
- 62.Iguchi K, Sahashi A, Yamada Y, Kohno J. 1990. New sesquiterpenoid hydroquinone and quinones from the Okinawan marine sponge (Dysidea sp.). Chem Pharm Bull 38:1121–1123. doi: 10.1248/cpb.38.1121. [DOI] [Google Scholar]
- 63.Walczak RJ, Woo AJ, Strohl WR, Priestley ND. 2000. Nonactin biosynthesis: the potential nonactin biosynthesis gene cluster contains type II polyketide synthase-like genes. FEMS Microbiol Lett 183:171–175. doi: 10.1111/j.1574-6968.2000.tb08953.x. [DOI] [PubMed] [Google Scholar]
- 64.Duncan KR, Crüsemann M, Lechner A, Sarkar A, Li J, Ziemert N, Wang M, Bandeira N, Moore BS, Dorrestein PC, Jensen PR. 2015. Molecular networking and pattern-based genome mining improves discovery of biosynthetic gene clusters and their products from Salinispora species. Chem Biol 22:460–471. doi: 10.1016/j.chembiol.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Smoot ME, Ono K, Ruscheinski J, Wang P-L, Ideker T. 2011. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Scheubert K, Hufsky F, Petras D, Wang M, Nothias L-F, Dührkop K, Bandeira N, Dorrestein P, Boecker S. 2017. Significance estimation for large scale metabolomics annotations by spectral matching. Nat Commun 8:1494. doi: 10.1038/s41467-017-01318-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.R Development Core Team. 2011. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: http://www.R-project.org. [Google Scholar]
- 68.Parker J. 1983. A comparison of methods used for the measurement of organic matter in marine sediment. Chem Ecol 1:201–209. doi: 10.1080/02757548308070802. [DOI] [Google Scholar]
- 69.Girard LR, Fiedler TJ, Harris TW, Carvalho F, Antoshechkin I, Han M, Sternberg PW, Stein LD, Chalfie M. 2007. WormBook: the online review of Caenorhabditis elegans biology. Nucleic Acids Res 35:D472–D475. doi: 10.1093/nar/gkl894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Solis PN, Wright CW, Anderson MM, Gupta MP, Phillipson JD. 1993. A microwell cytotoxicity assay using Artemia salina (brine shrimp). Planta Med 59:250–252. doi: 10.1055/s-2006-959661. [DOI] [PubMed] [Google Scholar]
- 71.Patin NV, Kunin V, Lidström U, Ashby MN. 2013. Effects of OTU clustering and PCR artifacts on microbial diversity estimates. Microb Ecol 65:709–719. doi: 10.1007/s00248-012-0145-4. [DOI] [PubMed] [Google Scholar]
- 72.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R. 2012. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hanshew AS, Mason CJ, Raffa KF, Currie CR. 2013. Minimization of chloroplast contamination in 16S rRNA gene pyrosequencing of insect herbivore bacterial communities. J Microbiol Methods 95:149–155. doi: 10.1016/j.mimet.2013.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TL. 2008. NCBI BLAST: a better web interface. Nucleic Acids Res 36:W5–W9. doi: 10.1093/nar/gkn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schmieder R, Edwards R. 2011. Quality control and preprocessing of metagenomic datasets. Bioinformatics 27:863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Weber T, Blin K, Duddela S, Krug D, Kim HU, Bruccoleri R, Lee SY, Fischbach MA, Müller R, Wohlleben W. 2015. antiSMASH 3.0—a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res 43:W237–W243. doi: 10.1093/nar/gkv437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Maddison WP, Maddison DR. 2015. Mesquite: a modular system for evolutionary analysis. Version 3.04 http://mesquiteproject.org/.
- 83.Silvestro D, Michalak I. 2012. raxmlGUI: a graphical front-end for RAxML. Org Divers Evol 12:335–337. doi: 10.1007/s13127-011-0056-0. [DOI] [Google Scholar]
- 84.Tamaoki T, Nomoto H, Takahashi I, Kato Y, Morimoto M, Tomita F. 1986. Staurosporine, a potent inhibitor of phospholipid/Ca++ dependent protein kinase. Biochem Biophys Res Commun 135:397–402. [DOI] [PubMed] [Google Scholar]
- 85.Magae Y, Magae J. 1993. Effect of staurosporine on growth and hyphal morphology of Pleurotus ostreatus. Microbiology 139:161–164. doi: 10.1099/00221287-139-1-161. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.