Burkholderia cenocepacia is an important opportunistic pathogen which can cause life-threatening infections in susceptible individuals, particularly in cystic fibrosis and immunocompromised patients. It usually employs two types of quorum sensing (QS) systems, including the cis-2-dodecenoic acid (BDSF) system and N-acyl homoserine lactone (AHL) system, to regulate virulence. In this study, we have designed and identified an unsaturated fatty acid compound (cis-14-methylpentadec-2-enoic acid [14-Me-C16:Δ2]) that is capable of interfering with B. cenocepacia QS signaling and virulence. We demonstrate that 14-Me-C16:Δ2 reduced BDSF and AHL signal production in B. cenocepacia. It also impaired QS-regulated phenotypes in various Burkholderia species. These results suggest that 14-Me-C16:Δ2 could interfere with QS signaling in many Burkholderia species and might be developed as a new antibacterial agent.
KEYWORDS: AHL, BDSF, Burkholderia cenocepacia, quorum sensing, virulence
ABSTRACT
Quorum sensing (QS) signals are widely used by bacterial pathogens to control biological functions and virulence in response to changes in cell population densities. Burkholderia cenocepacia employs a molecular mechanism in which the cis-2-dodecenoic acid (named Burkholderia diffusible signal factor [BDSF]) QS system regulates N-acyl homoserine lactone (AHL) signal production and virulence by modulating intracellular levels of cyclic diguanosine monophosphate (c-di-GMP). Thus, inhibition of BDSF signaling may offer a non-antibiotic-based therapeutic strategy against BDSF-regulated bacterial infections. In this study, we report the synthesis of small-molecule mimics of the BDSF signal and evaluate their ability to inhibit BDSF QS signaling in B. cenocepacia. A novel structural analogue of BDSF, 14-Me-C16:Δ2 (cis-14-methylpentadec-2-enoic acid), was observed to inhibit BDSF production and impair BDSF-regulated phenotypes in B. cenocepacia, including motility, biofilm formation, and virulence, while it did not inhibit the growth rate of this pathogen. 14-Me-C16:Δ2 also reduced AHL signal production. Genetic and biochemical analyses showed that 14-Me-C16:Δ2 inhibited the production of the BDSF and AHL signals by decreasing the expression of their synthase-encoding genes. Notably, 14-Me-C16:Δ2 attenuated BDSF-regulated phenotypes in various Burkholderia species. These findings suggest that 14-Me-C16:Δ2 could potentially be developed as a new therapeutic agent against pathogenic Burkholderia species by interfering with their QS signaling.
IMPORTANCE Burkholderia cenocepacia is an important opportunistic pathogen which can cause life-threatening infections in susceptible individuals, particularly in cystic fibrosis and immunocompromised patients. It usually employs two types of quorum sensing (QS) systems, including the cis-2-dodecenoic acid (BDSF) system and N-acyl homoserine lactone (AHL) system, to regulate virulence. In this study, we have designed and identified an unsaturated fatty acid compound (cis-14-methylpentadec-2-enoic acid [14-Me-C16:Δ2]) that is capable of interfering with B. cenocepacia QS signaling and virulence. We demonstrate that 14-Me-C16:Δ2 reduced BDSF and AHL signal production in B. cenocepacia. It also impaired QS-regulated phenotypes in various Burkholderia species. These results suggest that 14-Me-C16:Δ2 could interfere with QS signaling in many Burkholderia species and might be developed as a new antibacterial agent.
INTRODUCTION
Burkholderia cenocepacia is a Gram-negative opportunistic pathogen belonging to the Burkholderia cepacia complex (Bcc), which is a group of at least 20 closely related bacterial species (1, 2). This bacterium can cause life-threatening infections in susceptible individuals, particularly in cystic fibrosis and immunocompromised patients (3). B. cenocepacia mostly utilizes two types of quorum sensing (QS) systems, the N-acylhomoserine lactone (AHL) and cis-2-dodecenoic acid (BDSF) systems, to regulate biological functions and virulence (4–6). The major AHL system is the cep system, which consists of the AHL synthase CepI and the transcriptional regulator CepR. CepI predominantly synthesizes both N-octanoyl-homoserine lactone (C8-HSL) and smaller amounts of N-hexanoyl-homoserine lactone (C6-HSL). When the cell population density reaches a threshold level, these AHL signals will bind to CepR, cause a conformational change in the regulatory protein, and result in the modulation of the target gene expression levels (7, 8). In the BDSF system, the bifunctional crotonase RpfFBc synthesizes the BDSF signal, which then binds to and stimulates the BDSF receptor protein RpfR to decrease intracellular cyclic diguanosine monophosphate (c-di-GMP) levels, resulting in the transcriptional modulation of target genes and controlling motility, biofilm formation, protease production, and virulence (9–11).
Antibiotics have been extensively used to control and prevent infectious diseases that are caused by bacterial and fungal pathogens. However, the extensive use of this live-or-die selection pressure has fostered the emergence of superbugs that are resistant to conventional antibiotics. Infections caused by antibiotic-resistant pathogens are becoming increasingly common (12, 13) and are now major health care and public concerns. The development of antibiotic resistance in B. cenocepacia and other Bcc species has also become a serious concern in the medical community (14), leading to an emergent need for new strategies and novel drugs to efficiently treat infections caused by these pathogens.
An attractive approach to avoid the emergence of superbugs is to target bacterial virulence systems rather than their essential cellular processes. This strategy can reduce selective survival pressures and slow the emergence of drug resistance (15–17). Many studies have indicated that QS inhibitors can impair QS-dependent functions in bacteria. Therefore, the design and development of novel QS inhibitors to treat infectious diseases caused by bacterial pathogens that employ QS to regulate virulence is a valuable approach to combat bacterial pathogens and prevent the emergence of drug resistance. This strategy has already been successfully utilized to develop novel drugs to target QS and other signaling systems in a number of bacterial pathogens (18–27), making anti-QS drugs attractive alternatives to antibiotics.
The BDSF system is a conserved QS system in Burkholderia species and in many other bacterial pathogens (28), regulating biological functions and playing a key role in pathogenesis. Therefore, the BDSF system is an excellent candidate target to treat diseases caused by these bacterial species. Here, we report the design of BDSF system inhibitors from unsaturated fatty acid derivatives and evaluate their activities to inhibit QS signaling and virulence. We identified an unsaturated fatty acid compound (cis-14-methylpentadec-2-enoic acid [14-Me-C16:Δ2]) that is capable of interfering with B. cenocepacia QS signaling and virulence. We demonstrate that 14-Me-C16:Δ2 reduced the QS signal production and the expression levels of their synthase-encoding genes. In other Burkholderia species, 14-Me-C16:Δ2 also inhibited motility and biofilm formation, which are BDSF-regulated phenotypes. In summary, the goal of our study was to determine the applicability of targeting the BDSF QS system to promote the development of novel anti-QS therapeutics against B. cenocepacia.
RESULTS
14-Me-C16:Δ2 is a potential inhibitor of B. cenocepacia QS.
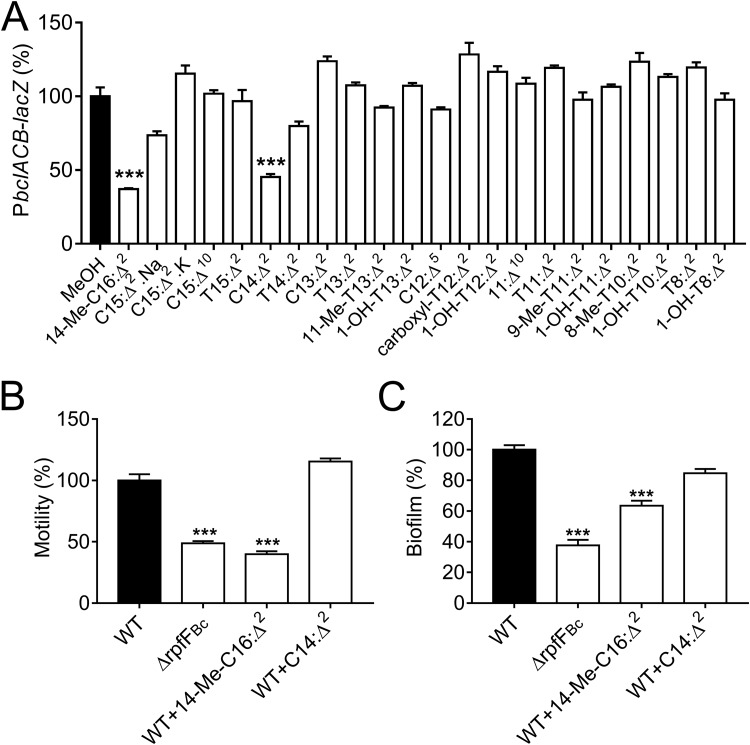
To identify potential inhibitors of QS systems in B. cenocepacia, we first tested the effect of the candidate compounds (see Table S1 in the supplemental material) on the β-galactosidase (β-Gal) activity of a B. cenocepacia strain H111 carrying a lectin-encoding bclACB operon-lacZ promoter fusion plasmid, as this operon is positively controlled by both the BDSF and AHL systems (5, 9). The reporter strain, H111(PbclACB-lacZ), was inoculated and grown in LB medium supplemented with 22 candidate compounds at a final concentration of 100 μM. β-Gal activity was measured when the cell cultures reached an optical density at 600 nm (OD600) of ∼1.5. Bioassay results showed that two candidate compounds (14-Me-C16:Δ2 and C14:Δ2) exerted significant inhibition on bclACB gene expression (Fig. 1A). We next tested the ability of 14-Me-C16:Δ2 and C14:Δ2 to interfere with the biological functions regulated by QS in B. cenocepacia H111. Data showed that swarming motility (Fig. 1B) and biofilm formation (Fig. 1C) were strongly reduced in the presence of 14-Me-C16:Δ2, but the compound C14:Δ2 did not show inhibitory activity in swarming motility and biofilm formation. Taken together, 14-Me-C16:Δ2 can act as a potential inhibitor of B. cenocepacia QS. In this study, 14-Me-C16:Δ2 was synthesized in large quantity from 11-methyl-dodecanol (Fig. S1A), and its chemical structure was confirmed via 1H and 13C nuclear magnetic resonance (NMR) spectra (Fig. S1B and C and 2A).
FIG 1.
Evaluation of QS inhibition in B. cenocepacia H111 with the candidate compounds (100 μM). (A) Effect of exogenous addition of 100 μM candidate compounds on bclACB gene expression, as determined by using a PbclACB-lacZ fusion reporter strain. These compounds are listed in Table S1. (B and C) Quantitative analysis of swarming motility (B) and biofilm formation (C) of B. cenocepacia H111 with the addition of the leading compounds (100 μM).
FIG 2.
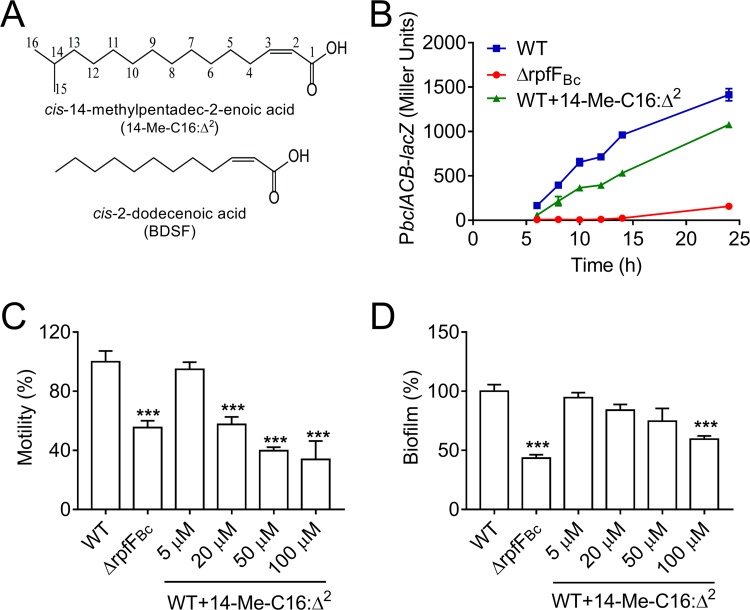
Effect of 14-Me-C16:Δ2 on QS-regulated functions of B. cenocepacia H111. (A) The chemical structure of 14-Me-C16:Δ2 compared to BDSF. (B) Effects of 14-Me-C16:Δ2 on bclACB gene expression were measured by assessing β-galactosidase activity. (C and D) Quantitative analysis of swarming motility (C) and biofilm formation (D) of B. cenocepacia H111 with the addition of 14-Me-C16:Δ2. Cells were treated with various concentrations of 14-Me-C16:Δ2 and incubated statically at 37°C, and the results are expressed as the relative values compared to those of the untreated controls. The results are based on three independent experiments. Error bars represent the means ± standard deviations (SD). Significant differences (*, P < 0.05; **, P < 0.01; ***, P < 0.001) between the tested compound and its control (n = 3) are shown.
14-Me-C16:Δ2 inhibits QS-regulated phenotypes in B. cenocepacia.
Interestingly, exogenous addition of 14-Me-C16:Δ2 at final concentrations of 5 to 100 μM did not appear to inhibit the growth of B. cenocepacia (Fig. S1D). However, 14-Me-C16:Δ2 clearly inhibited the expression of bclACB, as the addition of 100 μM 14-Me-C16:Δ2 reduced bclACB expression in the wild-type strain at all growth stages (Fig. 1A and 2B). We next tested the different concentration of 14-Me-C16:Δ2 on phenotypes controlled by the B. cenocepacia QS systems. As expected, swarming motility was inhibited by 14-Me-C16:Δ2 in a dose-dependent manner, as the addition of 5, 20, 50, and 100 μM 14-Me-C16:Δ2 reduced B. cenocepacia swarming motility by 5.2, 37.3, 42.4, and 60.2%, respectively (Fig. 2C).
Biofilm formation is a virulence trait of Burkholderia species that has been associated with the persistence of infections and the increased antibiotic tolerance of biofilm-associated cells compared with those of planktonic cells. In B. cenocepacia, QS systems regulate the expression of surface proteins, lectins, and extracellular DNA, which are all important components for biofilm matrix structures (29, 30). Therefore, we were interested to further investigate the inhibitory effect of 14-Me-C16:Δ2 on the ability of B. cenocepacia to form biofilms. After growing B. cenocepacia in microtiter dish wells in the presence of 0, 5, 20, 50, or 100 μM 14-Me-C16:Δ2, the addition of 100 μM 14-Me-C16:Δ2 was observed to cause a 37.3% reduction in biofilm formation (Fig. 2D). We also found that the addition of 14-Me-C16:Δ2 could significantly increase the antibiotic susceptibility of B. cenocepacia cells to chloramphenicol (Table S2), suggesting that 14-Me-C16:Δ2 showed an obviously synergistic effect with this antibiotic against B. cenocepacia.
14-Me-C16:Δ2 decreases BDSF signal production.
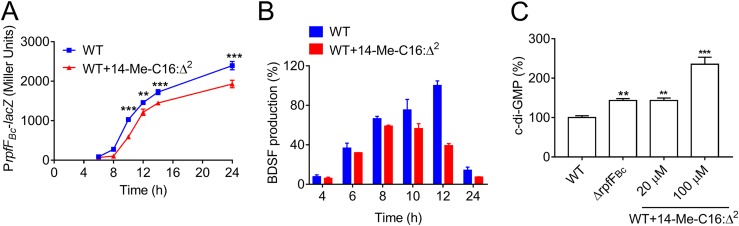
Viable QS inhibition strategies consist of the inhibition of signal sensing or signal synthesis and of signal degradation. In B. cenocepacia, the BDSF system positively regulates AHL signal production and cepI expression at the transcriptional level (9). To determine the mechanism by which 14-Me-C16:Δ2 acts upon QS signaling in B. cenocepacia, we first analyzed whether 14-Me-C16:Δ2 is involved in the competitive inhibition of the BDSF signal binding to the receptor protein RpfR using isothermal titration calorimetry (ITC). The results showed that 14-Me-C16:Δ2 did not bind to RpfR (Fig. S2A). In addition, 14-Me-C16:Δ2 did not inhibit the binding of the BDSF signal to its receptor protein RpfR (Fig. S2B and C), suggesting that 14-Me-C16:Δ2 does not interfere with the ability of RpfR to sense the BDSF signal. We next investigated whether 14-Me-C16:Δ2 exerts its inhibitory effect on BDSF signal synthesis. The data showed that treatment of cells with 14-Me-C16:Δ2 at a final concentration of 100 μM resulted in a significant decrease in the expression of rpfFBc at all growth stages, especially in the earlier growth stage (Fig. 3A).
FIG 3.
Effect of 14-Me-C16:Δ2 on the BDSF QS system in B. cenocepacia H111. (A) Effects of 14-Me-C16:Δ2 on rpfFBc gene expression were measured by assessing β-galactosidase activity of the rpfFBc-lacZ transcriptional fusions in the H111 wild-type (WT) strain in the presence or absence of 100 μM 14-Me-C16:Δ2. (B) Quantitative analysis of BDSF production in B. cenocepacia H111 with the addition of 14-Me-C16:Δ2 at different time points. The relative amounts of signal molecules were calculated on the basis of their peak areas. For convenient comparison, the peak BDSF of the WT at 12 h was arbitrarily defined as 100% and used to normalize the signal ratios of the different time points. (C) Quantitative analysis of c-di-GMP levels in B. cenocepacia H111 in the presence or absence of 14-Me-C16:Δ2 (100 μM). The relative amounts of signal molecules were calculated on the basis of their peak areas. For convenient comparison, the peak c-di-GMP of the WT was arbitrarily defined as 100% and used to normalize the signal ratios of the different treatments. The results are based on three independent experiments. Error bars represent the means ± standard deviations (SD). Significant differences (*, P < 0.05; **, P < 0.01; ***, P < 0.001) between the tested compound and its control (n = 3) are shown.
The inhibitory activity of 14-Me-C16:Δ2 on BDSF signal synthesis was also assessed by liquid chromatography-mass spectrometry (LC-MS) analysis. In agreement with the above-mentioned results, 14-Me-C16:Δ2 reduced BDSF production at all growth stages (Fig. 3B). The production of BDSF in the wild-type strain supplemented with 100 μM 14-Me-C16:Δ2 was reduced to 39.2% of that observed in the untreated wild-type strain at 12 h postinoculation (Fig. 3B).
Our recent studies demonstrated that the disruption of rpfFBc or rpfR results in an increase in intracellular c-di-GMP levels (9). In addition, modified intracellular c-di-GMP levels can cause changes in the production of virulence factors and AHL signal molecules, as well as alter the biosynthesis of different biofilm matrix components (9, 31). The observed inhibition in BDSF signal synthesis by 14-Me-C16:Δ2 motivated us to investigate the effect of 14-Me-C16:Δ2 on intracellular c-di-GMP levels in B. cenocepacia H111. The LC-MS analysis showed that treatment of the wild-type strain with 14-Me-C16:Δ2 at final concentrations of 20 μM and 100 μM caused approximately a 1.4- and 2.4-fold increase in the intracellular c-di-GMP levels, respectively (Fig. 3C). Taken together, the results showed that the disruption of rpfFBc gene expression by 14-Me-C16:Δ2 resulted in a notable increase in intracellular c-di-GMP levels, consequently affecting target gene expression.
14-Me-C16:Δ2 decreases AHL signal production.
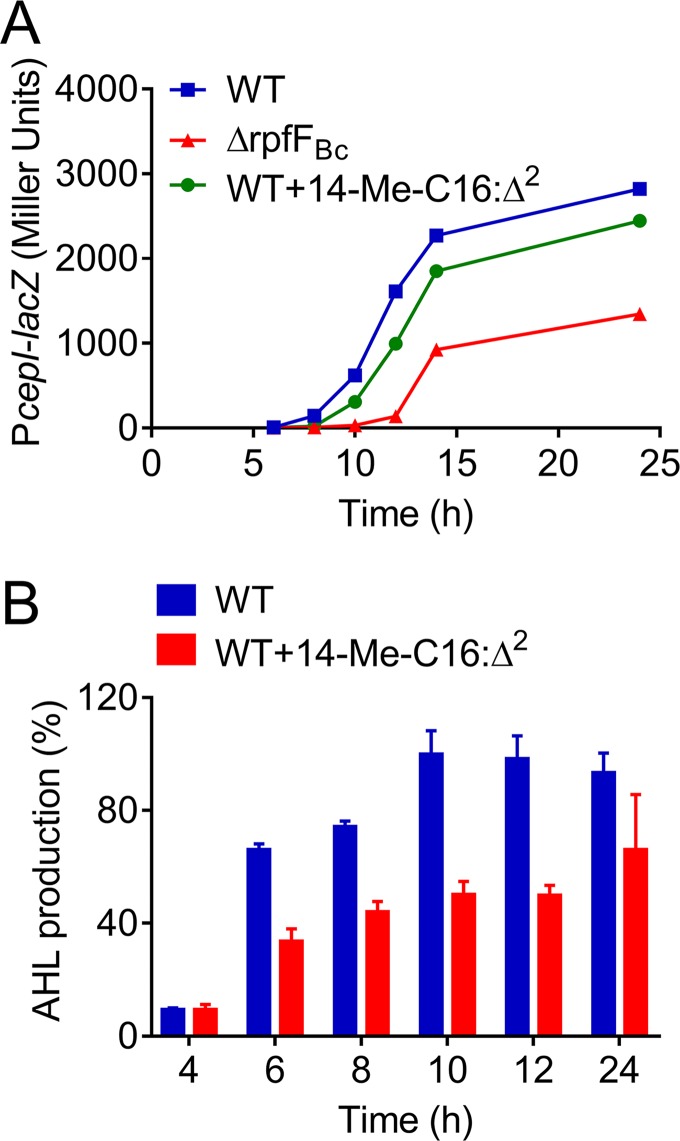
Recent studies showed that the BDSF system controls AHL signal production by regulating the expression of the AHL synthase CepI at the transcriptional level (9, 11). We next sought to measure AHL signal production in the wild-type H111 strain in the presence or absence of 14-Me-C16:Δ2. Our results revealed a notable reduction in AHL signal production in the wild-type strain when it was treated with 14-Me-C16:Δ2 (Fig. 4A). To test whether 14-Me-C16:Δ2 affected cepI (AHL synthase-encoding gene) transcription, we used cepI promoter-lacZ fusions and measured their activities in B. cenocepacia H111 strains. In agreement with the above-mentioned results, when the wild-type strain was grown in the presence of 14-Me-C16:Δ2 at a final concentration of 100 μM, a decrease in cepI expression was observed at various growth stages (Fig. 4B).
FIG 4.
Effect of 14-Me-C16:Δ2 on the AHL QS system in B. cenocepacia H111. (A) Effects of 14-Me-C16:Δ2 on cepI gene expression were measured by assessing β-galactosidase activity of the cepI-lacZ transcriptional fusions. B. cenocepacia H111 was grown in the presence or absence of 14-Me-C16:Δ2 (100 μM). Samples were taken from each culture at the OD600 values for the indicated time intervals. (B) Quantitative analysis of AHL production in B. cenocepacia H111 in the presence or absence of 100 μM 14-Me-C16:Δ2 at different time points. For convenient comparison, AHL production of the WT at 10 h was arbitrarily defined as 100% and used to normalize the signal ratios of the different time points. The results are based on three independent experiments. Error bars represent the means ± SD. Significant differences (*, P < 0.05; **, P < 0.01; ***, P < 0.001) between the tested compound and its control (n = 3) are shown.
14-Me-C16:Δ2 attenuates B. cenocepacia virulence.
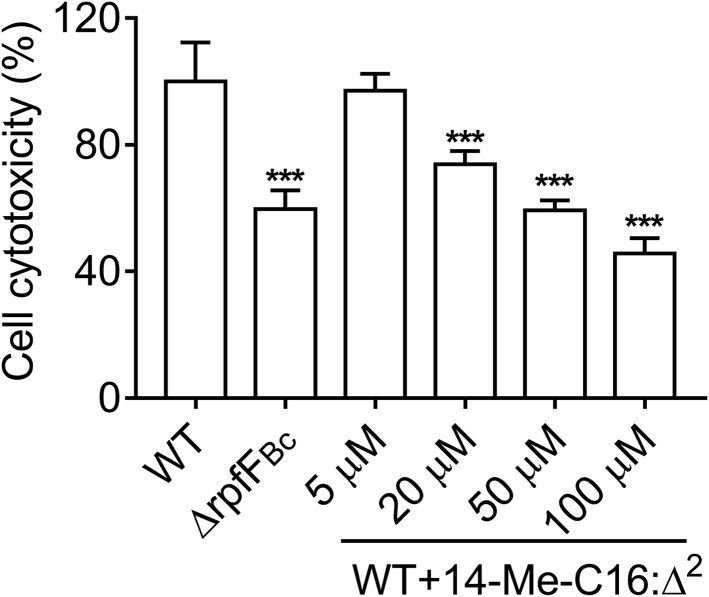
Previous studies showed that both the BDSF and AHL QS systems play vital roles in the pathogenesis of B. cenocepacia. Because the addition of 14-Me-C16:Δ2 inhibited the production of BDSF and AHL signals, we evaluated the efficacy of 14-Me-C16:Δ2 on B. cenocepacia H111 virulence using an A549 cell line infection model. Cytotoxicity was measured by quantifying the release of lactate dehydrogenase (LDH) into the supernatants of cultured cells. As expected, treatment with 14-Me-C16:Δ2 led to a reduction in bacterial virulence against this cell line (Fig. 5). When the wild-type H111 strain was incubated with A549 cells in the presence of 20, 50, and 100 μM 14-Me-C16:Δ2, the observed cytotoxicity levels were reduced to 74, 59, and 46% of that of the untreated group at 8 h postinoculation, respectively (Fig. 5).
FIG 5.
Effects of 14-Me-C16:Δ2 on the pathogenicity of B. cenocepacia were determined using an A549 cell line model system. The cell cytotoxicity of the WT was arbitrarily defined as 100% and used to normalize the signal ratios of the different treatments. The results are based on three independent experiments. Error bars represent the means ± SD. Significant differences (*, P < 0.05; **, P < 0.01; ***, P < 0.001) between the tested compound and its control (n = 3) are shown.
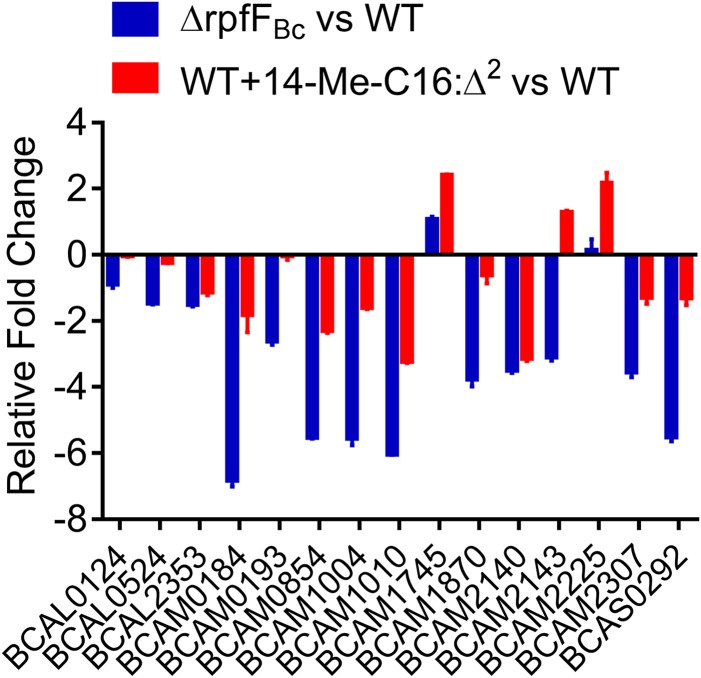
14-Me-C16:Δ2 affects the expression of a wide range of genes controlled by the BDSF system.
As the addition of 14-Me-C16:Δ2 resulted in reduced BDSF signal production, we examined whether 14-Me-C16:Δ2 affects the expression of BDSF-regulated genes. Quantitative reverse transcription-PCR (qRT-PCR) analyses revealed that cells treated with 14-Me-C16:Δ2 exhibited altered expression of many genes in the BDSF regulon (Fig. 6 and Table S3), indicating that 14-Me-C16:Δ2 inhibited BDSF signal production and affected the expression of the target genes of the BDSF system.
FIG 6.
qPCR analysis of the genes that showed differential expression between the rpfFBc mutant strain and the wild-type stain, and between the wild-type strain in the presence and absence of 14-Me-C16:Δ2 (100 μM). The results are based on three independent experiments. Error bars represent the means ± SD.
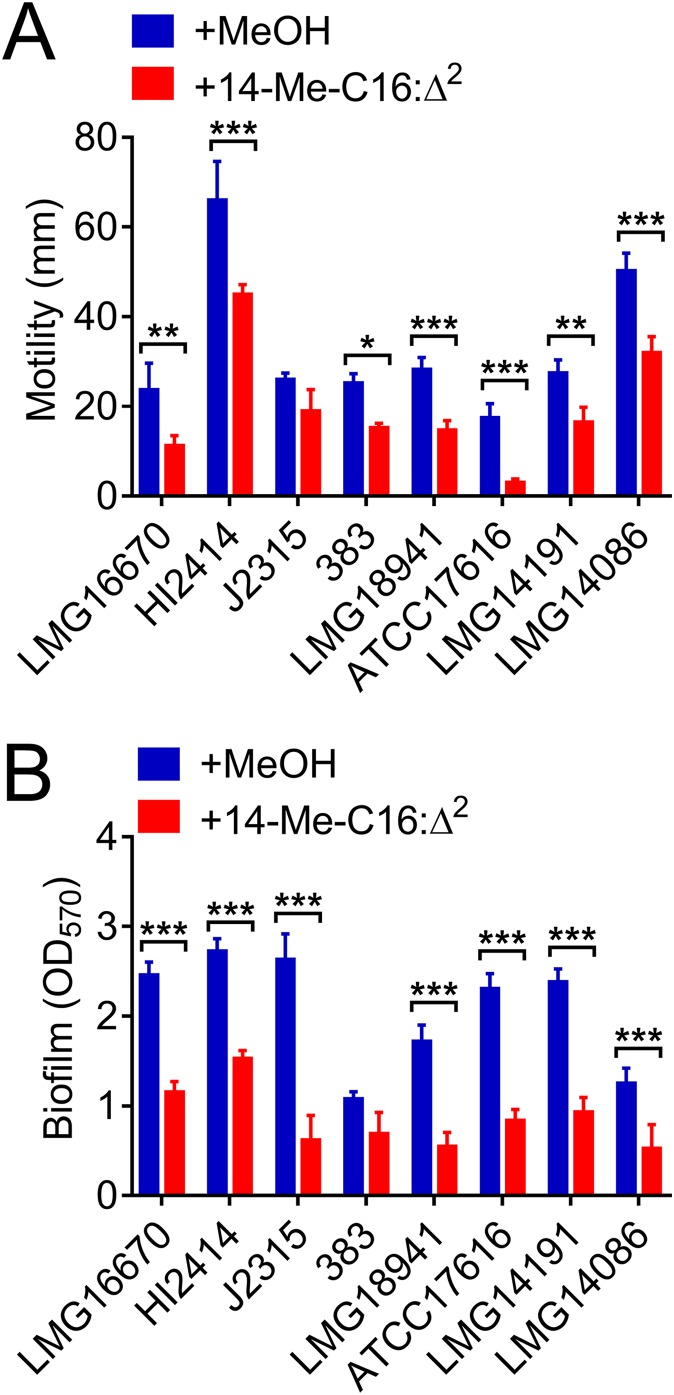
14-Me-C16:Δ2 inhibits QS-regulated phenotypes in many Burkholderia species.
The results of our previous studies indicated that many Burkholderia species may employ the BDSF system to regulate biological functions (11, 28, 32, 33). Therefore, the effect of 14-Me-C16:Δ2 on the QS-regulated phenotypes of different Burkholderia species was investigated. Swarming motility and biofilm formation were significantly inhibited in cells treated with 14-Me-C16:Δ2 for all the tested Burkholderia species, while their growth rates were unaffected (Fig. 7 and S3).
FIG 7.
(A and B) Quantitative analysis of swarming motility (A) and biofilm formation (B) of different Burkholderia species in the absence or presence of 14-Me-C16:Δ2 (100 μM). LMG16670, HI2414, J2315, 383, LMG18941, ATCC 17616, LMG14191 and LMG14086 represent B. anthina LMG16670, B. cenocepacia HI2414, B. cenocepacia J2315, B. cepacia 383, B. dolosa LMG18941, B. multivorans ATCC 17616, B. pyrrocinia LMG14191, and B. stabilis LMG14086, respectively. Cells were treated with various concentrations of 14-Me-C16:Δ2 and incubated statically at 37°C, and the results are expressed as the relative values compared to that of the untreated controls. The results are based on three independent experiments. Error bars represent the means ± SD. Significant differences (*, P < 0.05; **, P < 0.01; ***, P < 0.001) between the tested compound and its control (n = 3) are shown.
DISCUSSION
In this study, we identified cis-14-methylpentadec-2-enoic acid (14-Me-C16:Δ2) as an unsaturated fatty acid compound that is capable of interfering with the AHL and BDSF QS systems in B. cenocepacia (Fig. 3 and 4). 14-Me-C16:Δ2 reduced the production of QS signals and the expression of their synthase-encoding genes, resulting in the downregulation of factors associated with QS system-regulated biological functions in B. cenocepacia H111 (Fig. 2). As expected, because the QS system targeted by 14-Me-C16:Δ2 is a nonessential cell pathway, this molecule did not exhibit antimicrobial activity against B. cenocepacia (Fig. S1). However, promising results were obtained using the A549 cell line model, where the effect of 14-Me-C16:Δ2 was clearly demonstrated in vitro (Fig. 5). Thus, these results demonstrate that 14-Me-C16:Δ2 has efficacy against B. cenocepacia virulence.
B. cenocepacia infections are often associated with poor clinical outcomes and high mortality due to declines in lung function that lead to fatal pneumonia (1, 3, 34). These infections are often refractory to treatment with common antibiotics because of the emergence of multidrug-resistant (MDR) B. cenocepacia strains (14). This serious fact suggests that there is an urgent need to develop alternative strategies. Targeting bacterial virulence rather than cell growth is one attractive strategy. The significant advances of such “antivirulence” therapies include the inhibition of biofilm formation, motility, virulence, and QS signaling (12, 15–17, 35). In particular, the development of QS inhibitors as novel drugs to treat bacterial infections has attracted significant attention over the past 20 years (18–21, 23–27, 36). For example, a number of compounds have been screened and identified or synthesized as antagonists of QS systems, including diketopiperazines (37) and baicalein (38). The compounds with activities that interfere with the BDSF QS significantly inhibit B. cenocepacia virulence, suggesting that interference in QS signaling can be used as favorable therapeutic method to treat B. cenocepacia infections.
Diverse strategies have been explored to control QS signaling, including inhibition of signal synthesis and signal sensing and the promotion of signal degradation (39). However, specific Burkholderia species produce multiple types of QS signals and sense them using different cognate receptors. Thus, simultaneous inhibition of all signal-specific QS pathways in Burkholderia spp. requires targeting the upstream region of the QS signals. Among these factors, RpfFR homologues function as the master QS regulators within Burkholderia QS pathways (11, 32, 40). Moreover, these homologs display high sequence similarity and structural similarity (28), making them attractive targets for Burkholderia QS inhibition. In this study, 14-Me-C16:Δ2 inhibited RpfFBc activity and markedly affected the QS-regulated phenotypes in all the Burkholderia species examined (Fig. 7). The potent and broad activity of this molecule would be especially important in practical settings, since multiple Burkholderia species can cause pulmonary infections in hospital environments. In fact, multiple Burkholderia species, rather than a single species, are responsible for Burkholderia pathogenesis in naturally infected CF patients.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The Escherichia coli strain and Burkholderia strains used in this work are listed in Table 1 and were grown at 28 or 37°C, as indicated previously (4, 9), with shaking at 250 rpm in Luria-Bertani (LB) broth. B. cenocepacia H111 and its rpfFBc deletion mutant were described previously (32). The following antibiotics were used to supplement media when necessary: ampicillin, 100 μg ml−1; gentamicin, 50 μg ml−1; kanamycin, 100 μg ml−1; and tetracycline, 10 μg ml−1. A549 human lung carcinoma cells (American Type Culture Collection #CCL-185) were grown in Dulbecco’s modified Eagle medium (DMEM; Gibco) supplemented with 10% fetal bovine serum and 1× penicillin-streptomycin (Pen-Strep; Sigma) at 37°C.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Phenotypes and/or characteristicsa | Reference or sourceb |
|---|---|---|
| Strains | ||
| Burkholderia spp. | ||
| B. cenocepacia H111 | Wild-type strain, genomovar III of the B. cepacia complex | 42 |
| H111(PcepI-lacZ) | H111 harboring reporter construct PcepI-lacZ | 9 |
| H111(PbclACB-lacZ) | H111 harboring reporter construct PbclACB-lacZ | 11 |
| H111(PrpfFBc-lacZ) | H111 harboring reporter construct PrpfFBc-lacZ | 11 |
| ΔrpfFBc mutant | BDSF-deficient mutant derived from H111 with rpfFBc being deleted | 32 |
| ΔrpfFBc(PcepI-lacZ) mutant | ΔrpfFBc mutant harboring reporter construct PcepI-lacZ | 9 |
| ΔrpfFBc(PbclACB-lacZ) mutant | ΔrpfFBc mutant harboring reporter construct PbclACB-lacZ | 11 |
| B. multivorans ATCC 17616 | Soil, USA | E. Mahenthiralingam laboratory |
| B. cepacia 383 | Soil, Trinidad | E. Mahenthiralingam laboratory |
| B. cenocepacia J2315 | Cystic fibrosis isolate, UK | ATCC |
| B. stabilis LMG 14086 | Respirator, UK | BccMtm |
| B. cenocepacia HI2414 | Isolated from agricultural soil, USA | BccMtm |
| B. dolosa LMG 18941 | Cystic fibrosis isolate, USA | BccMtm |
| B. anthina LMG 16670 | Rhizosphere, UK | BccMtm |
| B. pyrrocinia LMG 14191 | Soil, Japan | BccMtm |
| E. coli | ||
| DH5α | supE44 lacU169(80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 pir | Laboratory collection |
| BL21 | F− ompT hsdS (rB− mB−) dcm+ Tetr gal (DE3) endA | Stratagene |
| Plasmids | ||
| pET-28a | Expression vector, Kanr | Novagen |
| pET-rpfR | pET-28a containing rpfR | 7 |
Tetr tetracycline resistance; Kanr, kanamycin resistance.
BccMtm, Belgian Coordinated Collections of Microorganisms.
Synthesis of 14-Me-C16:Δ2.
14-Me-C16:Δ2 was synthesized using 11-methyl-dodecanol as a starting material. 11-Methyl-dodecanol was used to synthesize 1-bromo-11-methyl-dodecane by bromination, which together with propiolic acid was subsequently used to synthesize 14-methylpentadec-2-ynoic acid under the catalytic action of hexamethylphosphoric triamide (HMPT) and n-butyllithium (n-BuLi). Finally, cis-14-methylpentane-2-enoic acid was synthesized by a one-step Pd-BaSO4 catalytic hydrogenation reaction.
Construction of reporter strains and measurement of β-galactosidase activity.
The bclACB, cepI, and rpfFBc reporters were introduced into the B. cenocepacia H111 wild-type and rpfFBc mutant strains by electroporation. Transconjugants were then selected on LB agar plates containing tetracycline. Measurements of β-galactosidase activity were made according to previously described methods (41). Bacteria were cultured at 37°C, and the cells were harvested to measure β-galactosidase activity.
Swarming motility assay.
Burkholderia strains treated with candidate compounds were used for the swarming motility assay. Swarming motility was determined on semisolid agar (0.3%). Bacteria were inoculated into the center of plates containing 0.8% tryptone, 0.5% glucose, and 0.3% agarose. The plates were incubated at 30°C for 18 h before the diameter of the colony was measured.
Biofilm formation assays.
Biofilm formation assays were performed as described previously, with minor modifications (42). Overnight cultures of Burkholderia strains were inoculated into 200 μl of LB broth in 96-well microtiter plates at an OD600 of 0.05. Candidate compounds were added as indicated below, and the planktonic cells and medium were removed after culturing at 24 h. The crystal violet staining method was used to quantify biofilm mass, as described by Huber et al. (42). The crystal violet from stained biofilms was dissolved in 200 μl of 95% (vol/vol) ethyl alcohol, and the absorbance at 570 nm was measured.
Detection of MIC values.
The MICs of 12 antibiotics (ampicillin, kanamycin, gentamicin, tetracycline, chloramphenicol, penicillin, trimethoprim, streptomycin, rifampin, tobramycin, apramycin sulfate, and cefotaxime) and the combinations of antibiotics with 14-Me-C16:Δ2 (100 μM) against B. cenocepacia were determined using broth microdilution according to the CLSI 2015 guidelines (43). Overnight culture of B. cenocepacia was diluted 1,000-fold for the subsequent tests. The cultures were placed in a 96-well plate and incubated at 37°C supplemented with different concentrations of antibiotics for 24 h. The MIC was defined as the lowest concentration of antibiotic in which bacterial growth in the well was not measurable by determination of the turbidity at 600 nm.
Quantification of BDSF and AHL signals.
Bacteria were cultured overnight in LB broth with agitation at 37°C, inoculated into 1 liter of fresh LB medium, and incubated for 20 h. Cells were removed by centrifugation, and the supernatants were mixed with an equal volume of ethyl acetate. The ethyl acetate was collected and evaporated to dryness and dissolved in 1 ml of methanol. BDSF signals were measured by liquid chromatography-mass spectrometry (LC-MS) (33).
Quantification of AHL signals was performed using the β-galactosidase assay with the aid of the AHL reporter strain CF11, as described previously (44). Briefly, the reporter strain CF11 was grown in minimal medium at 28°C with shaking at 220 rpm for 12 h. The cultures were inoculated into the same medium supplemented with extracts containing AHL signals. After bacterial cells were harvested, β-galactosidase activity was assayed as described previously (41).
Quantification of c-di-GMP.
B. cenocepacia H111 and its derivatives were grown in 400 ml of LB medium at 37°C to an OD600 of 2.5 to ∼3.0 with shaking at 200 rpm. Formaldehyde (final concentration, 0.18%) was added to block the degradation of c-di-GMP. Cells were collected by centrifugation at 8,000 × g for 10 min at 4°C. The cell pellets were washed with 40 ml of phosphate-buffered saline (pH 7.0) containing 0.18% formaldehyde and were centrifuged at 8,000 × g for 10 min at 4°C. Next, the cell pellets were dissolved in water, boiled for 10 min, and cooled on ice for 10 min. Nucleotides were extracted using 65% ethanol. The supernatants were retained, and the extractions were repeated. The supernatants were concentrated and lyophilized, and the pellets were dissolved in 1 ml H2O and filtered using polyvinylidene difluoride (PVDF) filters (0.22-μm pore size). c-di-GMP levels were measured by LC-MS (32).
Protein expression and purification.
rpfR was fused to the expression vector pET-28a. The fusion gene construct was transformed into E. coli strain BL21. Affinity purification of the HIS-RpfR fusion protein was performed following the method described previously (32). Fusion protein was cleaved with PreScission protease (GE Healthcare; 2 units/100 μl of bound proteins) at 4°C overnight. The cleaved fusion proteins were eluted and analyzed by SDS-PAGE.
ITC analysis.
The isothermal titration calorimetry (ITC) measurements were performed using an ITC-200 microcalorimeter following the manufacturer’s protocol (MicroCal, Northampton, MA). In brief, titrations began with one injection of 1.5 μl of 14-Me-C16:Δ2 (500 μM) solution into the sample cell containing 350 μl of the RpfR solution (25 μM) in the ITC-200 microcalorimeter. The volume of 14-Me-C16:Δ2 injection was changed to 2 μl in the subsequent 18 injections. The heat changes accompanying injections were recorded. The titration experiment was repeated at least three times, and the data were calibrated with the final injections and fitted with the one-site model to determine the binding constant (Kd) using the MicroCal ORIGIN version 7 software.
Quantitative real-time PCR assays.
B. cenocepacia H111 was cultured at 37°C in LB broth in the presence or absence of 14-Me-C16:Δ2 at a final concentration of 100 μM to the logarithmic-growth phase. B. cenocepacia H111 cultured without 14-Me-C16:Δ2 was used as the control. Total RNA was extracted with TRIzol reagent (Invitrogen), according to the manufacturer’s instructions. RNA quality was determined by measuring the A260/A280 and A260/A230 values and by gel electrophoresis. Reverse transcription-PCR was performed using a cDNA synthesis kit (Promega), according to the manufacturer’s instructions. Specific qRT-PCR primers (listed in Table 2) were used to amplify central fragments of approximately 200 bp in length from different genes. qRT-PCR was performed using SYBR green qPCR mastermixes (Thermo Scientific) and a 7300Plus real-time PCR system (Applied Biosystems). The recA gene was used as an internal reference (33). The relative expression levels of the target genes were calculated using the quantitation-comparative CT (ΔΔCT) method according to Livak and Schmittgen (45).
TABLE 2.
PCR primers used in this study
| Primer | Sequence (5′–3′)a |
|---|---|
| For protein expression | |
| rpfR-His-F | CGCGGATCCATGGATGACGAAAACGATAGCGCGG |
| rpfR-His-R | CCCAAGCTTTCAGGCGATCAGCCTGAGCTTTCTC |
| For qPCR assay | |
| BCAL0017-F | AATGAAGACCCTCGCACAGG |
| BCAL0017-R | TCGATATCCGCGTAAAGCCC |
| BCAL0111-F | TCAGCGTAGTGGTTGAACCC |
| BCAL0111-R | TCGGTAATGACGGCGATCAG |
| BCAL0113-F | TTGATCGTGTTCAGCGTCGA |
| BCAL0113-R | AACTTCGCGACGCTCTACAA |
| flhD-F | CTACCAGCGAAATGCTCAGT |
| flhD-R | CATACCCATCGCCTTGTCTT |
| flhC-F | GAAGGAAATCACCCTCGCCA |
| flhC-R | AACAGCGACGAGTGGATGTT |
| flhF-F | ACATTCGCAAATTCACCGGC |
| flhF-R | CGAGTGCAACGATTTCGACG |
| BCAL0248-F | CAATCGCGCAAGTCCTGAAG |
| BCAL0248-R | ATGACCTGCGGAATGTCGTT |
| BCAL0251-F | GTGAAGGGTGGCCGTATTCT |
| BCAL0251-R | TTCTTGAGCGGCACCTTGAA |
| secY-F | CTAACAGCCCGAGTCTTGCA |
| secY-R | AGTTGATCCGGATCGATGCC |
| filI-F | CTCGATCACCGCGTTCTACA |
| filI-R | GATGCGTTTCGTCGATCAGC |
| BCAL0524-F | CAGATGGTGCTCAAGGAAGT |
| BCAL0524-R | GACATGTTCGCGAGGAACT |
| BCAL0566-F | CGGATCCTGGTTCTGCAACT |
| BCAL0566-R | TGACTTCCTCCAACACGACG |
| BCAL0571-F | TGTTGATCGAGATGCCGAGG |
| BCAL0571-R | CTGAAGGATCTCGCGCAGAT |
| BCAL0831-F | TTGCAGGTTGAGTTCGACGA |
| BCAL0831-R | TCCGTATTTGCCCCCGAAAA |
| BCAL0990-F | GCAAAACAAGAAGTCGCCGT |
| BCAL0990-R | ACGACTTTCTTGCCGCGATA |
| BCAL1059-F | CACGCTTCGGCATCTTCAAG |
| BCAL1059-R | CAGGTCGAACGGAATCACCA |
| BCAL1060-F | ATGATGTACATCGCCGCGAA |
| BCAL1060-R | GCATCACTTCCGCGATGAAC |
| leuE-F | TGAGCCTGCTGAATCCGAAG |
| leuE-R | GAAGATCAGCGTGCTCAGGT |
| gtrR-F | GTGAAGGAGGGACTGTTCCG |
| gtrR-R | GACGAACGACTTGAACAGCG |
| zmpB-F | GGAAGGCTTGTCGGAAGG |
| zmpB-R | CCAGTTGTAGACCCAGTGATAC |
| BCAM2374-F | CAATCTGCTCTCCGTCTCCG |
| BCAM2374-R | AATCGGTTCGTAGTGCGTGT |
| BCAM2627-F | CTATCGTCACTGGGTGTCGG |
| BCAM2627-R | CCTTGTGGATCGCATGACCT |
| BCAS0263-F | CGGACGAATTGCTGCACAAG |
| BCAS0263-R | TCGTACTGGTGCAACACGAT |
| BCAS0292-F | GCCGATCGAAGCGGAAAT |
| BCAS0292-R | CAAAGAGCCGGTTGTCGTTG |
| BCAS0638-F | TTGCCGAAGAGAACCTGGTC |
| BCAS0638-R | CCGAGCAGGGTGAAGTGATT |
| BCAL2337-F | TCCACGACAGGAAGTTCACG |
| BCAL2337-R | AAATCTCGATGGGCTTCGCA |
| BCAL2353-F | GTCGTTTCTGGGCAAGGTA |
| BCAL2353-R | CACGTCGTGATCGATGTAGTC |
| BCAL2946-F | GAGCGCACGATCTACATGGA |
| BCAL2946-R | AGGAAGTGATAGCCGATGCG |
| BCAL2978-F | GAACGTGTCGATGTGCATGG |
| BCAL2978-R | TCGGCTACGACATGCACTAC |
| BCAL3041-F | CGATGGTCATCAACGGCAAG |
| BCAL3041-R | GTTCTTCTCGCCCTTCTGCT |
| BCAL3405-F | TACAACATCGGCAAGCAGGT |
| BCAL3405-R | GGCAACTGCTCGTAGGTGAT |
| bclB-F | CTTTACCCACGACGACCTTTAC |
| bclB-R | TCGTATTGCGGCAGTTTCTC |
| BCAM0193-F | TATCGGGAAGGCAGCTACT |
| BCAM0193-R | CCGTAGTAATGGATTTCGAGCA |
| BCAM0196-F | GGAGCCTGATCCATACGTCG |
| BCAM0196-R | CATCGCCATGTCGAGAAAGC |
| BCAM0854-F | GGGACGATGGCGATTTCTT |
| BCAM0854-R | GGTTCCATCACCGCATAGTC |
| BCAM0859-F | TCGCTACAACGATCTCGACG |
| BCAM0859-R | CTTGCCTTCGAGCGAGATCT |
| BCAM0862-F | GCAACAAGTTCTACGCGACC |
| BCAM0862-R | GTCGAAGCTGCCGTATTTGC |
| BCAM0948-F | GCATGATGATGAGCAAGCCG |
| BCAM0948-R | CCCAGTAGTCCGGATAACGC |
| BCAM1004-F | TGAACTACCGTGAATCGTATGG |
| BCAM1004-R | CAACGGTGTCCGTGATCTT |
| BCAM1005-F | GCGTCAGGTAGTAGATCGGC |
| BCAM1005-R | CGGTCGAGGAGCAGTTCTAC |
| BCAM1010-F | GTGGGAAGAGGACATCATCAAG |
| BCAM1010-R | CGAAGATCGTCGGCATGAATA |
| BCAM1745-F | GCGATGTCGATCAGGTTCTT |
| BCAM1745-R | TGGCCGTCATGTTCAGGTAC |
| BCAM1870-F | AGTTCGATCGCGACGATACC |
| BCAM1870-R | AGCGACTTCAGCAGATACGG |
| BCAM1871-F | CTCGAACGACAGGTTGACGA |
| BCAM1871-R | GTATTTGCTGCGCATCTCCG |
| BCAM2044-F | CGGTCACGTAAAGCTCGGTA |
| BCAM2044-R | ACGTTTACGATCGGCTTCCA |
| BCAM2060-F | GTTGAGCAGGAACAGGTCGA |
| BCAM2060-R | GTGCTGTACGTGAACCAGGA |
| BCAM2140-F | GCGACATCGCATTCATTCATC |
| BCAM2140-R | ATCGTGTCGGGCGAAATC |
| bapA-F | CTGTTGTTGGTGCGATCATTT |
| bapA-R | CAACGTCGTGCCGTCATA |
| BCAM2215-F | AATCGGTCGTTGACGGGAAA |
| BCAM2215-R | CTTCAGCGTGAACGTGTAGC |
| BCAM2225-F | TCGCTCAAACGTTTGCAGTG |
| BCAM2225-R | GAACCATGCGCGGATCAAC |
| aidA-F | GACGTTGTCCTGGTTGGTCA |
| aidA-R | CGCGTTACCGATGTACTCGT |
Restriction enzyme sites are underlined.
Cytotoxicity assays.
Cytotoxicity was assessed by measuring the release of LDH from A549 cells, which were routinely grown in DMEM with the addition of 10% fetal bovine serum (FBS) in 96-well tissue culture plates with 1 × 104 cells per well. Confluent A549 cells were washed and incubated in DMEM supplemented with 1% FBS before infection. The B. cenocepacia H111 wild-type strain and its derivatives were grown in LB medium at 37°C, centrifuged, and resuspended in culture medium to an OD600 of 1.0. A549 cells were infected with bacterial cells at 109 CFU/ml for 8 h, and then the culture supernatants were collected by centrifugation for 4 min at 250 × g. The LDH in the supernatant was measured, and the cytotoxicity was calculated relative to that of an uninfected control.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the Guangdong Natural Science Funds for Distinguished Young Scholars (grant 2014A030306015), the National Key Project for Basic Research of China (973 Project, grant 2015CB150600), and the National Natural Science Foundation of China (grant 31571969).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00105-19.
REFERENCES
- 1.Mahenthiralingam E, Urban TA, Goldberg JB. 2005. The multifarious, multireplicon Burkholderia cepacia complex. Nat Rev Microbiol 3:144–156. doi: 10.1038/nrmicro1085. [DOI] [PubMed] [Google Scholar]
- 2.Holden MTG, Seth-Smith HMB, Crossman LC, Sebaihia M, Bentley SD, Cerdeno-Tarraga AM, Thomson NR, Bason N, Quail MA, Sharp S, Cherevach I, Churcher C, Goodhead I, Hauser H, Holroyd N, Mungall K, Scott P, Walker D, White B, Rose H, Iversen P, Mil-Homens D, Rocha EPC, Fialho AM, Baldwin A, Dowson C, Barrell BG, Govan JR, Vandamme P, Hart CA, Mahenthiralingam E, Parkhill J. 2009. The genome of Burkholderia cenocepacia J2315, an epidemic pathogen of cystic fibrosis patients. J Bacteriol 191:261–277. doi: 10.1128/JB.01230-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mahenthiralingam E, Vandamme P, Campbell ME, Henry DA, Gravelle AM, Wong LTK, Davidson AGF, Wilcox PG, Nakielna B, Speert DP. 2001. Infection with Burkholderia cepacia complex genomovars in patients with cystic fibrosis: virulent transmissible strains of genomovar III can replace Burkholderia multivorans. Clin Infect Dis 33:1469–1475. doi: 10.1086/322684. [DOI] [PubMed] [Google Scholar]
- 4.Deng Y, Boon C, Eberl L, Zhang LH. 2009. Differential modulation of Burkholderia cenocepacia virulence and energy metabolism by the quorum-sensing signal BDSF and its synthase. J Bacteriol 191:7270–7278. doi: 10.1128/JB.00681-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmid N, Pessi G, Deng Y, Aguilar C, Carlier AL, Grunau A, Omasits U, Zhang L, Ahrens CH, Eberl L. 2012. The AHL- and BDSF-dependent quorum sensing systems control specific and overlapping sets of genes in Burkholderia cenocepacia H111. PLoS One 7:e49966. doi: 10.1371/journal.pone.0049966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suppiger A, Schmid N, Aguilar C, Pessi G, Eberl L. 2013. Two quorum sensing systems control biofilm formation and virulence in members of the Burkholderia cepacia complex. Virulence 4:400–409. doi: 10.4161/viru.25338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lewenza S, Conway B, Greenberg EP, Sokol PA. 1999. Quorum sensing in Burkholderia cepacia: Identification of the LuxRI homologs CepRI. J Bacteriol 181:748–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gotschlich A, Huber B, Geisenberger O, Tögl A, Steidle A, Riedel K, Hill P, Tümmler B, Vandamme P, Middleton B, Camara M, Williams P, Hardman A, Eberl L. 2001. Synthesis of multiple N-acylhomoserine lactones is wide-spread among the members of the Burkholderia cepacia complex. Syst Appl Microbiol 24:1–14. doi: 10.1078/0723-2020-00013. [DOI] [PubMed] [Google Scholar]
- 9.Deng Y, Lim A, Wang J, Zhou T, Chen S, Lee J, Dong Y, Zhang L. 2013. cis-2-dodecenoic acid quorum sensing system modulates N-acyl homoserine lactone production through RpfR and cyclic di-GMP turnover in Burkholderia cenocepacia. BMC Microbiol 13:148. doi: 10.1186/1471-2180-13-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Udine C, Brackman G, Bazzini S, Buroni S, Van Acker H, Pasca MR, Riccardi G, Coenye T. 2013. Phenotypic and genotypic characterisation of Burkholderia cenocepacia J2315 mutants affected in homoserine lactone and diffusible signal factor-based quorum sensing systems suggests interplay between both types of systems. PLoS One 8:e55112. doi: 10.1371/journal.pone.0055112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang C, Cui C, Ye Q, Kan J, Fu S, Song S, Huang Y, He F, Zhang L, Jia Y, Gao Y, Harwood CS, Deng Y. 2017. Burkholderia cenocepacia integrates cis-2-dodecenoic acid and cyclic dimeric guanosine monophosphate signals to control virulence. Proc Natl Acad Sci U S A 114:13006–13011. doi: 10.1073/pnas.1709048114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Livermore DM. 2004. The need for new antibiotics. Clin Microbiol Infect 10:1–9. doi: 10.1111/j.1465-0691.2004.1004.x. [DOI] [PubMed] [Google Scholar]
- 13.Pfaller MA, Jones RN, Doern GV, Kugler K. 1998. Bacterial pathogens isolated from patients with bloodstream infection: frequencies of occurrence and antimicrobial susceptibility patterns from the SENTRY antimicrobial surveillance program (United States and Canada, 1997). Antimicrob Agents Chemother 42:1762–1770. doi: 10.1128/AAC.42.7.1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rhodes KA, Schweizer HP. 2016. Antibiotic resistance in Burkholderia species. Drug Resist Updat 28:82–90. doi: 10.1016/j.drup.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Njoroge J, Sperandio V. 2009. Jamming bacterial communication: new approaches for the treatment of infectious diseases. EMBO Mol Med 1:201–210. doi: 10.1002/emmm.200900032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Escaich S. 2010. Novel agents to inhibit microbial virulence and pathogenicity. Expert Opin Ther Pat 20:1401–1418. doi: 10.1517/13543776.2010.511176. [DOI] [PubMed] [Google Scholar]
- 17.Clatworthy AE, Pierson E, Hung DT. 2007. Targeting virulence: a new paradigm for antimicrobial therapy. Nat Chem Biol 3:541–548. doi: 10.1038/nchembio.2007.24. [DOI] [PubMed] [Google Scholar]
- 18.Dobretsov S, Teplitski M, Alagely A, Gunasekera SP, Paul VJ. 2010. Malyngolide from the cyanobacterium Lyngbya majuscula interferes with quorum sensing circuitry. Environ Microbiol Rep 2:739–744. doi: 10.1111/j.1758-2229.2010.00169.x. [DOI] [PubMed] [Google Scholar]
- 19.Hentzer M, Givskov M. 2003. Pharmacological inhibition of quorum sensing for the treatment of chronic bacterial infections. J Clin Invest 112:1300–1307. doi: 10.1172/JCI20074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ishida T, Ikeda T, Takiguchi N, Kuroda A, Ohtake H, Kato J. 2007. Inhibition of quorum sensing in Pseudomonas aeruginosa by N-acyl cyclopentylamides. Appl Environ Microbiol 73:3183–3188. doi: 10.1128/AEM.02233-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jakobsen TH, Bjarnsholt T, Jensen PØ, Givskov M, Høiby N. 2013. Targeting quorum sensing in Pseudomonas aeruginosa biofilms: current and emerging inhibitors. Future Microbiol 8:901–921. doi: 10.2217/fmb.13.57. [DOI] [PubMed] [Google Scholar]
- 22.Jakobsen TH, Bragason SK, Phipps RK, Christensen LD, van Gennip M, Alhede M, Skindersoe M, Larsen TO, Høiby N, Bjarnsholt T, Givskov M. 2012. Food as a source for quorum sensing inhibitors: iberin from horseradish revealed as a quorum sensing inhibitor of Pseudomonas aeruginosa. Appl Environ Microbiol 78:2410–2421. doi: 10.1128/AEM.05992-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim C, Kim J, Park H, Park H, Lee JH, Kim CK, Yoon J. 2008. Furanone derivatives as quorum-sensing antagonists of Pseudomonas aeruginosa. Appl Microbiol Biotechnol 80:37–47. doi: 10.1007/s00253-008-1474-6. [DOI] [PubMed] [Google Scholar]
- 24.Norizan SN, Yin WF, Chan KG. 2013. Caffeine as a potential quorum sensing inhibitor. Sensors (Basel) 13:5117–5129. doi: 10.3390/s130405117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Majik MS, Naik D, Bhat C, Tilve S, Tilvi S, D’Souza L. 2013. Synthesis of (R)-norbgugaine and its potential as quorum sensing inhibitor against Pseudomonas aeruginosa. Bioorg Med Chem Lett 23:2353–2356. doi: 10.1016/j.bmcl.2013.02.051. [DOI] [PubMed] [Google Scholar]
- 26.O’Loughlin CT, Miller LC, Siryaporn A, Drescher K, Semmelhack MF, Bassler BL. 2013. A quorum-sensing inhibitor blocks Pseudomonas aeruginosa virulence and biofilm formation. Proc Natl Acad Sci U S A 110:17981–17986. doi: 10.1073/pnas.1316981110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smyth AR, Cifelli PM, Ortori CA, Righetti K, Lewis S, Erskine P, Holland ED, Givskov M, Williams P, Camara M, Barrett DA, Knox A. 2010. Garlic as an inhibitor of Pseudomonas aeruginosa quorum sensing in cystic fibrosis-a pilot randomized controlled trial. Pediatr Pulmonol 45:356–362. doi: 10.1002/ppul.21193. [DOI] [PubMed] [Google Scholar]
- 28.Deng Y, Wu J, Eberl L, Zhang LH. 2010. Structural and functional characterization of diffusible signal factor family quorum-sensing signals produced by members of the Burkholderia cepacia complex. Appl Environ Microbiol 76:4675–4683. doi: 10.1128/AEM.00480-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fazli M, Almblad H, Rybtke ML, Givskov M, Eberl L, Tolker-Nielsen T. 2014. Regulation of biofilm formation in Pseudomonas and Burkholderia species. Environ Microbiol 16:1961–1981. doi: 10.1111/1462-2920.12448. [DOI] [PubMed] [Google Scholar]
- 30.Inhülsen S, Aguilar C, Schmid N, Suppiger A, Riedel K, Eberl L. 2012. Identification of functions linking quorum sensing with biofilm formation in Burkholderia cenocepacia H111. Microbiologyopen 1:225–242. doi: 10.1002/mbo3.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmid N, Suppiger A, Steiner E, Pessi G, Kaever V, Fazli M, Tolker-Nielsen T, Jenal U, Eberl L. 2017. High intracellular c-di-GMP levels antagonize quorum sensing and virulence gene expression in Burkholderia cenocepacia H111. Microbiology 163:754–764. doi: 10.1099/mic.0.000452. [DOI] [PubMed] [Google Scholar]
- 32.Deng Y, Schmid N, Wang C, Wang J, Pessi G, Wu D, Lee J, Aguilar C, Ahrens CH, Chang C, Song H, Eberl L, Zhang LH. 2012. cis-2-dodecenoic acid receptor RpfR links quorum-sensing signal perception with regulation of virulence through cyclic dimeric guanosine monophosphate turnover. Proc Natl Acad Sci U S A 109:15479–15484. doi: 10.1073/pnas.1205037109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cui C, Yang C, Song S, Fu S, Sun X, Yang L, He F, Zhang L, Zhang Y, Deng Y. 2018. A novel two-component system modulates quorum sensing and pathogenicity in Burkholderia cenocepacia. Mol Microbiol 108:32–44. doi: 10.1111/mmi.13915. [DOI] [PubMed] [Google Scholar]
- 34.Depoorter E, Bull MJ, Peeters C, Coenye T, Vandamme P, Mahenthiralingam E. 2016. Burkholderia: an update on taxonomy and biotechnological potential as antibiotic producers. Appl Microbiol Biotechnol 100:5215–5229. doi: 10.1007/s00253-016-7520-x. [DOI] [PubMed] [Google Scholar]
- 35.Cegelski L, Marshall GR, Eldridge GR, Hultgren SJ. 2008. The biology and future prospects of antivirulence therapies. Nat Rev Microbiol 6:17–27. doi: 10.1038/nrmicro1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bobadilla Fazzini RA, Skindersoe ME, Bielecki P, Puchałka J, Givskov M, Martins Dos Santos VAP. 2013. Protoanemonin: a natural quorum sensing inhibitor that selectively activates iron starvation response. Environ Microbiol 15:111–120. doi: 10.1111/j.1462-2920.2012.02792.x. [DOI] [PubMed] [Google Scholar]
- 37.Scoffone VC, Chiarelli LR, Makarov V, Brackman G, Israyilova A, Azzalin A, Forneris F, Riabova O, Savina S, Coenye T, Riccardi G, Buroni S. 2016. Discovery of new diketopiperazines inhibiting Burkholderia cenocepacia quorum sensing in vitro and in vivo. Sci Rep 6:32478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brackman G, Hillaert U, Van Calenbergh S, Nelis HJ, Coenye T. 2009. Use of quorum sensing inhibitors to interfere with biofilm formation and development in Burkholderia multivorans and Burkholderia cenocepacia. Res Microbiol 160:144–151. doi: 10.1016/j.resmic.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 39.Kalia VC. 2013. Quorum sensing inhibitors: an overview. Biotechnol Adv 31:224–245. doi: 10.1016/j.biotechadv.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 40.Boon C, Deng Y, Wang L, He Y, Xu J, Fan Y, Pan SQ, Zhang L. 2008. A novel DSF-like signal from Burkholderia cenocepacia interferes with Candida albicans morphological transition. ISME J 2:27–36. doi: 10.1038/ismej.2007.76. [DOI] [PubMed] [Google Scholar]
- 41.Zhou L, Wang J, Zhang L. 2007. Modulation of bacterial type III secretion system by a spermidine transporter dependent signaling pathway. PLoS One 2:e1291. doi: 10.1371/journal.pone.0001291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huber B, Riedel K, Hentzer M, Heydorn A, Gotschlich A, Givskov M, Molin S, Eberl L. 2001. The cep quorum-sensing system of Burkholderia cepacia H111 controls biofilm formation and swarming motility. Microbiology 147:2517–2528. doi: 10.1099/00221287-147-9-2517. [DOI] [PubMed] [Google Scholar]
- 43.Clinical and Laboratory Standards Institute. 2015. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 10th ed. CLSI document M07-A10. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 44.Zhang L, Murphy PJ, Kerr A, Tate ME. 1993. Agrobacterium conjugation and gene regulation by N-acyl-l-homoserine lactones. Nature 362:446–448. doi: 10.1038/362446a0. [DOI] [PubMed] [Google Scholar]
- 45.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.