Summary
Hematopoiesis is arguably one of the best understood stem cell systems; however, significant challenges remain to reach a consensus understanding of the lineage potential, heterogeneity, and relationships of hematopoietic stem and progenitor cell populations. To gain new insights, we performed quantitative analyses of mature cell production from hematopoietic stem cells (HSCs) and multiple hematopoietic progenitor populations. Assessment of the absolute numbers of mature cell types produced by each progenitor cell revealed a striking erythroid dominance of all myeloid-competent progenitors assessed, accompanied by strong platelet reconstitution. All populations with myeloid potential also produced robust numbers of red blood cells and platelets in vivo. Clonal analysis by single-cell transplantation and by spleen colony assays revealed that a significant fraction of HSCs and multipotent progenitors have multilineage potential at the single-cell level. These new insights prompt an erythroid-focused model of hematopoietic differentiation.
Keywords: lineage potential, heterogeneity, hematopoietic stem cells, quantitative analyses, erythropoiesis, multilineage differentiation, reconstitution, clonal analysis, single-cell transplantation, hematopoietic differentiation
Graphical Abstract
Highlights
-
•
RBCs are the predominant cell type produced by multipotent hematopoietic progenitors
-
•
All cell types with myeloid potential also produced RBCs and platelets in vivo
-
•
Single HSCs and MPPF cells are capable of multilineage hematopoietic reconstitution
-
•
Erythroid cell production emerges as a default hematopoietic fate
In this article, Forsberg and colleagues use quantitative reconstitution assays to demonstrate that all myeloid-competent progenitor cells also contribute to erythroid and platelet production. Single-cell in vivo analyses showed that hematopoietic stem cells and multipotent progenitors are clonally multipotent. These results suggest that erythropoiesis is a default hematopoietic fate and challenges current views on hematopoietic lineage potential and fate decisions.
Introduction
Hematopoietic stem cells (HSCs) differentiate via multiple progressively committed progenitor cell populations to maintain a balanced number of mature blood cells. Despite extensive investigation, the lineage potential, heterogeneity, and relationships of hematopoietic stem and progenitor cells (HSPCs) are under intense debate. Data from both single-cell transplantation and barcoding analyses support the existence of long-term, multilineage reconstituting clonal HSCs (Dykstra et al., 2007, Gerrits et al., 2010, Lu et al., 2011, Osawa et al., 1996, Yamamoto et al., 2013, Yamamoto et al., 2018). However, differential lineage contribution from single cells suggests heterogeneity even within strictly defined HSC compartments (Benz et al., 2012, Yamamoto et al., 2013). Similarly, the heterogeneity and physiological roles of hematopoietic progenitor cells are hotly debated. Evidence from multiple studies indicates that FLK2-positive multipotent progenitors (MPPF) serve as a developmental intermediate for all hematopoietic lineages, prior to the generation of progenitors restricted to either myeloid (common myeloid progenitors, CMPs) or lymphoid (common lymphoid progenitors, CLPs) fates (Figure S1A) (Akashi et al., 2000, Beaudin et al., 2014, Boyer et al., 2012, Boyer et al., 2011, Forsberg et al., 2006, Kondo et al., 1997, Schlenner et al., 2010). While the existence of multipotent HSCs is widely accepted and recent in situ evidence supports the existence of multilineage progenitor cells (Boyer et al., 2011, Busch et al., 2015, Sun et al., 2014), the degree of lineage commitment of hematopoietic populations remains controversial.
Several factors have made it difficult to assess the level of lineage commitment and lineage bias within hematopoietic subtypes. Tracking of mature red blood cell (RBC) and platelet (Plt) production from hematopoietic progenitor subsets in vivo was developed relatively recently; therefore, the full spectrum of mature cell types is rarely simultaneously assessed. Substitute assays, such as hematopoietic differentiation in vitro, do not always accurately reflect differentiation in situ or upon transplantation in vivo (Boyer et al., 2012, Richie Ehrlich et al., 2011, Schlenner et al., 2010). In addition, mature cell output from transplanted hematopoietic subtypes is seldom measured quantitatively, precluding accurate comparison of lineage output from specific hematopoietic subsets. Here, we use side-by-side absolute quantification of mature cell production and single-cell in vivo assays to address the lineage contribution and functional heterogeneity of HSPCs. Our new insights were combined with previous data into a model of hematopoietic differentiation that reconciles multiple longstanding controversies in HSC biology.
Results
Lineage Potential of Hematopoietic Cell Populations by Traditional Donor Chimerism
To qualitatively and quantitatively assess the differentiation potential of distinct HSPC populations (Figures S1A and S1B), we performed comprehensive analyses of mature cell production upon transplantation into sublethally irradiated mice. UBC-GFP mice allowed for the simultaneous detection of donor-derived RBCs, platelets, granulocytes/myelomonocytes (GMs), and B and T cells (Figure S1C). To enable detection of rare and transiently generated cell types, the peripheral blood (PB) of recipient mice was monitored at frequent and early time points post-transplantation.
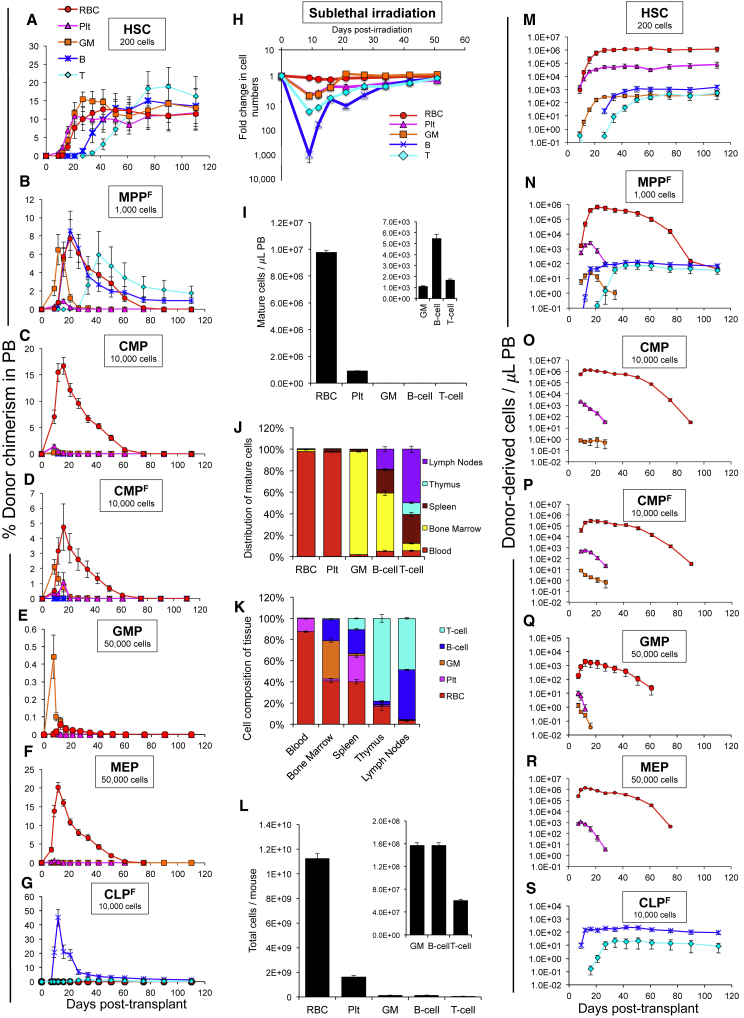
We first displayed reconstitution as donor chimerism (donor-derived cells relative to host cells), as is commonly done (Figures 1A–1G and S1D). Aside from a few notable exceptions and the addition of RBC analysis, our results largely agreed with previous reports (Akashi et al., 2000, D'Amico and Wu, 2003, Forsberg et al., 2006, Oguro et al., 2013, Yamamoto et al., 2013). Thus, HSCs gave rise to all five lineages analyzed, without evidence of decline for the duration of the experiments (16 weeks) (Figure 1A). MPPF also gave rise to all five lineages analyzed, with clear declines in chimerism 21–51 days post-transplantation (Figures 1B and S1D). Interestingly, although the Plt contribution from MPPF was lower than GM, B cell, or T cell chimerism, as reported previously (Forsberg et al., 2006, Lai and Kondo, 2006), the RBC chimerism was similar to that of nucleated white blood cells. Both FLK2− and FLK2+ CMPs produced detectable levels of RBCs, platelets, and GMs, but not B and T cells, in the PB (Figures 1C, 1D, and S1D). GM progenitors (GMPs), myeloerythroid progenitors (MEPs), and CLPF contributed primarily to GMs, RBCs, and B cells, respectively (Figures 1E–1G and S1D). Overall, these results agree with the lineage potential previously attributed to each of the HSPC populations.
Figure 1.
Reconstitution Potential of Transplanted Hematopoietic Stem and Progenitor Cell Populations
(A–G) Percentage donor chimerism over 110 days from HSCs (A), MPPF (B), CMPs (C), CMPF (D), GMPs (E), MEPs (F), or CLPF (G) upon transplantation into sublethally irradiated (500 rad) mice.
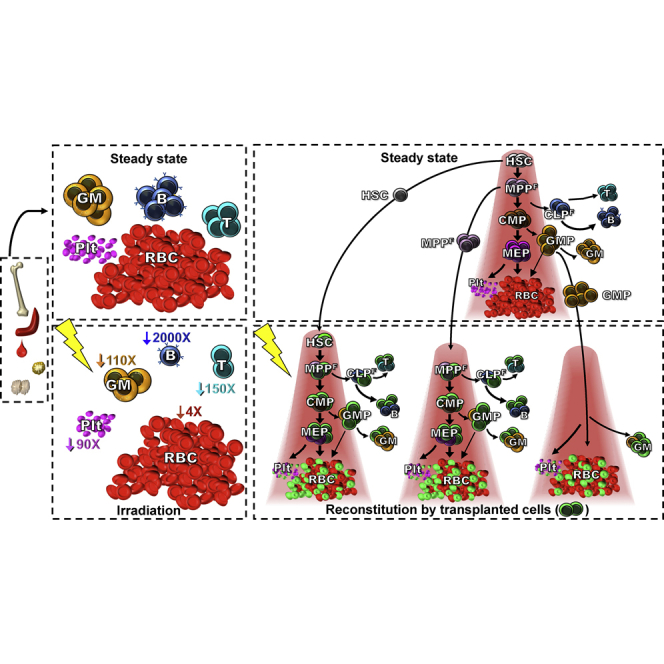
(H) B cell numbers display a rapid and more drastic decline (1,000-fold) after sublethal irradiation than other mature cell types (1.4-, 6-, 6-, and 23-fold for RBCs, platelets, GMs, and T cells, respectively). Data displayed are fold changes in mature cell numbers in the peripheral blood (PB) of sublethally irradiated (500 rad) mice over time. n ≥ 7.
(I) The number of mature hematopoietic cells in a microliter of PB at steady state. n = 10.
(J) The distribution of mature hematopoietic cells between blood, bone marrow, spleen, thymus, and lymph nodes of a mouse. n = 10.
(K) The composition of mouse blood, bone marrow, spleen, thymus, and lymph nodes displayed as a percentage of total mature hematopoietic cells. n = 10.
(L) The number of mature hematopoietic cells in a 25 g mouse at steady state. n = 10.
(M–S) Reconstitution data from (A–G) replotted as the absolute number of donor-derived cells per microliter PB. HSCs (M), MPPF (N), CMPs (O), CMPF (P), GMPs (Q), MEPs (R), and CLPF (S). Transplantation data in (A–G) and (M–S) are representative means ± SEM from at least seven recipient mice per cell type from at least two independent experiments.
See also Figures S1 and S2.
Quantifying Absolute Numbers of Mature Cells Produced by Distinct Progenitor Populations
Reconstitution displayed as chimerism depends on both donor cell production and the number of mature host cells present. To compare the effects of radiation conditioning on different types of host cells, we measured mature cell numbers at several time points post-sublethal irradiation. This analysis uncovered a dramatically cell-type-specific variation in both the magnitude and the kinetics of host cell decrease and recovery, with a rapid, greater than 1,000-fold decrease in B cell numbers and only an ∼1.4-fold, slower, decrease in RBC numbers (Figure 1H). These host cell variations affect the perceived cell generation from transplanted cells when reconstitution is displayed as donor-to-host chimerism. To remove the host variable, we determined the absolute number of each donor-derived mature cell type in the PB after transplantation of different progenitor populations, displayed as the number of donor-derived cells per microliter of PB (Figures 1M–1S). Even though these data were derived from the same transplantation experiments as for Figures 1A–1G, the absolute quantification conveyed a remarkably different perspective on the ability of different progenitors to reconstitute hematopoiesis (compare Figures 1A–1G with 1M–1S): with the exception of CLPs, RBC production exceeded all other cell types by orders of magnitude from all transplanted progenitor populations.
To determine whether this RBC dominance was apparent only in the blood, we accounted for differential tissue distribution of each cell type to convey the total number of cells produced per transplanted HSPC in the entire body of the recipient. Assessments of mature cell numbers and tissue distribution between major hematopoietic organs revealed that, as expected, RBCs far outnumbered the other cell types in PB (Figure 1I) and that the vast majority of the total RBCs present in a mouse were located in the PB (Figure 1J). Platelets had a similar distribution pattern, whereas most GMs were found in the bone marrow (BM). B cells were distributed (in order of abundance) among BM, spleen, lymph nodes, and PB, and T cells among lymph nodes, spleen, thymus, BM, and PB. Conversely, displaying each tissue based on the abundance of cell types revealed that blood is composed almost entirely of RBCs (87%) and platelets (12%) and that CD3+ T cells make up 78% of the hematopoietic cells in the thymus, whereas other tissues were less dominated by one cell type (Figure 1K). Combining the tissue distribution with PB cell counts provided an estimate of the total numbers of each cell type in a mouse (Figure 1L) that are consistent with previous reports (Kakumitsu et al., 2005, Nemzek et al., 2001).
These data enabled us to use the PB data (Figures 1M–1S) to assess the absolute number of each mature cell type generated by each transplanted cell population in each recipient mouse (Figures S2A–S2G). While the magnitude of the difference between cells that mainly distribute to the blood (RBCs and platelets) and cell types that are dispersed among other tissues (GMs, B and T cells) decreased when whole-body distribution was taken into account, the relative order of cell types produced was not altered (compare Figures 1M–1S with S2A–S2G; except for GMPs, see below). Using modified Markov birth-death modeling and published mature cell half-lives, we tested the impact of cell half-life (ranging from ∼1 day for GMs to ∼150 days for T cells) on population size. Because the identity and half-life of each intermediate population is not known, we modeled an “extreme half-life scenario” where the published half-life for each mature population was used for all progenitor intermediates giving rise to that cell type to estimate the largest possible impact of the differential half-lives (Figures S3A and S3B). We then calculated a “birth rate” to distinguish cell generation (new cells produced; Table S1 and Figure S2A′–S2G′) from cell accumulation (number of cells present; Table 1). Over time, the differential half-lives have a significant impact; thus, our estimations of cell production by HSCs are different in the short term (Tables 1 and S2) and long term (Tables S1 and S2). Importantly, because repopulation from progenitor cells is transient, half-lives have less impact on estimation of mature cell production by progenitor cells, especially during the early time points after transplantation that we used for cell quantification (compare Tables 1 and S1 and solid and dashed lines in Figures S2A′–S2G′; see Figures 2A′–2G′ for cell numbers and post-transplantation time points).
Table 1.
Approximate Proportion of the Absolute Number of Mature Cells Generated by Each Transplanted Progenitor Cell Type
| RBC | Plt | GM | B | T | |
|---|---|---|---|---|---|
| HSC | 86.2 | 7.22 | 2.87 | 2.48 | 1.25 |
| MPPF | 98.3 | 0.56 | 0.47 | 0.31 | 0.33 |
| CMP | 99.7 | 0.26 | 0.01 | nd | nd |
| CMPF | 99.4 | 0.29 | 0.31 | nd | nd |
| GMP | 90.8 | 0.79 | 8.37 | nd | nd |
| MEP | 99.9 | 0.12 | nd | nd | nd |
| CLP | nd | nd | nd | 91 | 9 |
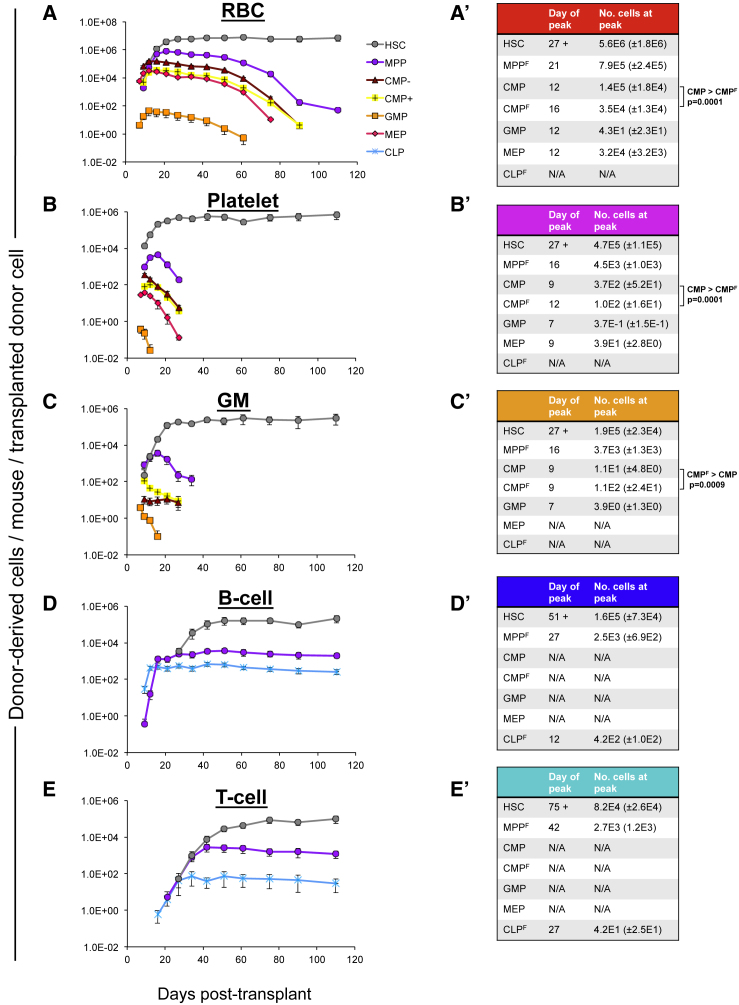
Figure 2.
Comparative Mature Cell Accumulation and Kinetics by Transplanted Hematopoietic Stem and Progenitor Cell Populations
The total numbers of RBCs (A), platelets (B), GMs (C), B cells (D), and T cells (E) present per mouse per transplanted HSPC. (A′–E′) The approximate time points and cell numbers for peak donor-derived mature cells from (A–E), respectively.
Data were generated from the transplantation and cell distribution experiments of Figure 1. The mature cell production capacity was always in the same order: HSCs > MPPF > CLPF (for B and T cells) and HSCs > MPPF > CMP > CMPF > MEP > GMP (for RBCs, platelets, and GMs). The only exception was that significantly more GMs accumulated after CMPF than after CMP transplantation (C and C′; p < 0.0009 by Student's two-tailed t test). All other comparisons were also significant. See also Figure S3.
The absolute quantification revealed that RBCs were by far the most abundant cell type produced by each progenitor (Figures 1M–1S and S2A–S2G and Table 1). The only exception was CLPF, which stayed true to their reported lymphoid commitment by producing only B and T cells (Figure 1S) (Forsberg et al., 2006, Kondo et al., 1997, Schlenner et al., 2010). Of note, T cell production by CLPF was more readily detectable when displayed as absolute numbers (Figures 1S and S2G) than as chimerism (Figure 1G). After RBCs, platelets were the next most abundant mature cell type produced. Despite the low Plt chimerism after transplantation of MPPF, CMPs, CMPF, MEPs, and GMPs (Figures 1B–1F), donor-derived platelets outnumbered or equaled the GMs, B cells, and T cells produced from each population (Figures 1N–1R and S2B–S2F, Table 1). Surprisingly, GMPs, previously considered committed to myelomonocytic cell production (Akashi et al., 2000, Forsberg et al., 2006, Na Nakorn et al., 2002), produced more RBCs than GMs and also contributed to platelets (Figures 1Q, S2E, S4G, and S4H, Table 1). HSCs, CMPs, and MEPs displayed a more expected reconstitution pattern. Notably, CMPF gave rise to far greater numbers of RBCs than GMs, with Plt production roughly equaling that of GM generation (Figures 1P and S2D, Table 1). Similarly, although MPPF displayed complete multipotency, they produced RBCs and platelets in much greater abundance than they produced nucleated mature cells (Figures 1N and S2B, Table 1), defying reports that FLK2 expression signifies loss of MegE potential. Overall, the proportions of different mature cell types produced by all erythromyeloid-competent cells were strikingly similar (Figures 1M–1R and S2A–S2F, Table 1).
Direct Comparison of the Mature Cell Production Capacity of Progenitor Populations
To assess the relative reconstitution capacity of each progenitor cell type, we then compared the total output of each mature cell type per transplanted HSPC. Each HSC generated more of each mature cell type than any progenitor tested (Figures 2A–2E). In addition, the time between transplantation and the peak of mature cell production (time to peak) was the longest for HSCs, and mature cells persisted without evidence of decline (Figures 2A–2E and 2A′–2E′). These properties are consistent with self-renewal and with HSCs existing at the top of the hematopoietic hierarchy (Figure S1A). Per transplanted cell, MPPF gave rise to more RBCs, platelets, GMs, B cells, and T cells than any lineage-restricted progenitor (Figures 2A–2E). In addition, the time to peak for mature cell production from transplanted MPPF was shorter than for HSCs, but longer compared with lineage-restricted progenitors (Figures 2A–2E and 2A′–2E′). Likewise, the timing and total cell production from both CMP populations were in between those of the MPPF and MEPs. Relative to CMPF, CMPs displayed more robust RBC and Plt generation (Figures 2A and 2B; 2A′–2B′), while CMPF excelled in GM production (Figures 2C and 2C′). While the capacity of CMPF to produce RBCs and platelets in vivo contrasts with in vitro data where MegE output from CMPF was not observed (Nutt et al., 2005), our results are consistent with the relative lineage preferences from previously described in vitro data, with CMPF exhibiting a relative preference for GM production compared with CMP (Figures 2A′–2C′ and Table 1). GMPs produced the same cell types as CMPs and CMPF, but in fewer numbers and with the shortest time to peak for any HSPC (Figures 2A–2E and 2A′–2E′). Overall, these data are consistent with the developmental relationship between HSPCs displayed in Figure S1A.
MPPF Give Rise to Myeloid Progenitors In Vivo
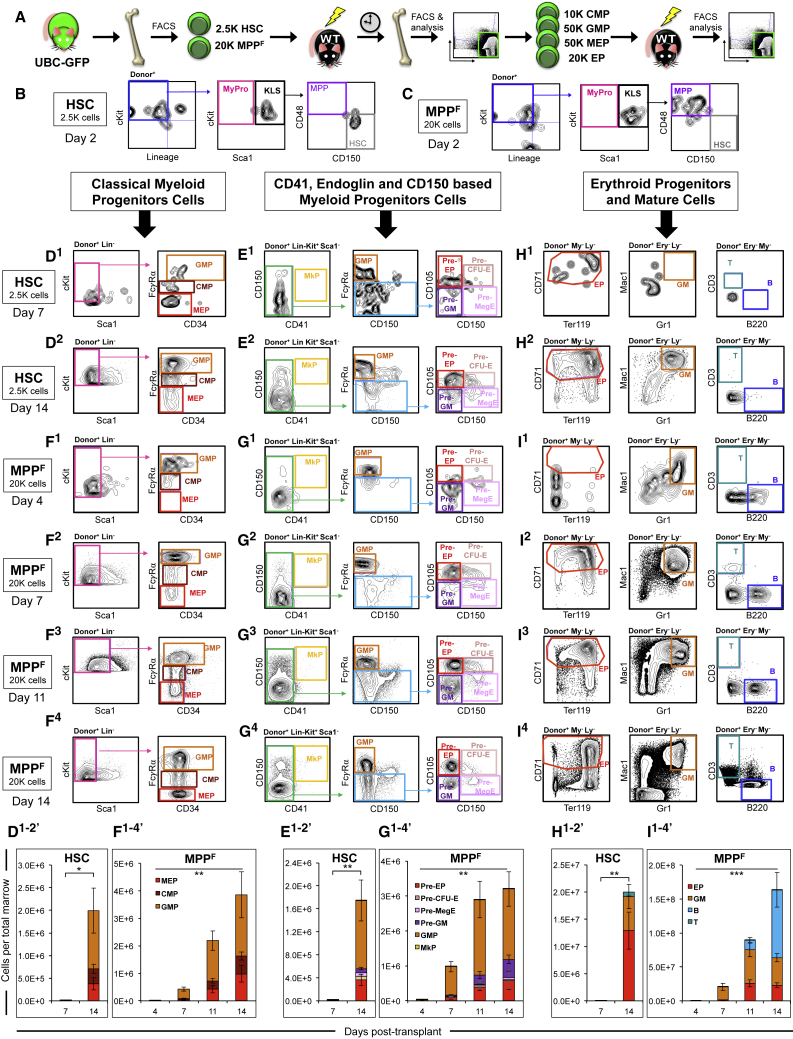
The lineage potential and absolute number of cells produced can provide insights into the relative hierarchy of cell populations, but not direct mother-daughter relationships. To directly determine if MPPF can give rise to erythro- and erythromyeloid-restricted progenitors, and to assess the extent of expansion of various progenitor populations, we performed phenotypic, quantitative, and functional analysis of donor-derived cells shortly after transplantation (Figure 3A). Two days after transplantation of HSCs or MPPF, the phenotype of the donor cells in the recipient BM remained similar to their respective phenotype prior to transplantation (Figures 3B and 3C). By days 7 and 14, transplanted HSCs maintained the self-renewing HSC pool while also repopulating phenotypic MPPs, “classical” (Figures 3D1 and 3D2) and “alternative” myeloid progenitors (Figures 3E1 and 3E2), as well as erythroid progenitors (EPs) and mature GM cells (Figures 3H1 and H2). Few HSC-derived B and T cells were detected within the 14 day analysis period. BM analyses at days 4, 7, 11, and 14 revealed that MPPF repopulated all types of erythromyeloid progenitors and mature cells (Figures 3F1–3I4). Quantification of the absolute donor-derived cell numbers showed substantial expansion of donor-derived cells during the analysis period (Figures 3D1−2′–3I1−4′ and S4A–S4D). By day 14, the transplanted HSCs had given rise to ∼2 million and MPPF to nearly 4 million myeloid progenitor cells (Figures 3D1−2′–3G1−4′). While MPPF clearly produced more B cells compared with HSCs within this time frame (Figures 3H1′–3I4′), the proportions of progenitor cells generated by HSCs and MPPF were similar (Figures 3D1′–3I4′). For example, comparing CMP/GMP/MEP proportions from HSCs at day 14 with CMP/GMP/MEP from MPPF at day 11 revealed that distributions were not statistically significantly different (Figures 3D1−2′ versus 3F1−4′). Indeed, GMPs were the second most abundantly produced progenitor population from both HSCs (∼1.3 million GMPs by day 14) and MPPF (∼2.2 million GMPs; approximately 2-fold more than from HSCs), after EPs (∼13 million from HSCs and, as with GMP generation, ∼2-fold more [∼26 million] EPs from MPPF). Interestingly, these experiments did not reveal a clear hierarchy between CMPs and their presumed GMP and MEP descendants, as GMPs outnumbered CMPs at all time points from both HSCs and MPPF. Secondary transplantation of HSC- and MPPF-derived CMPs, GMPs, MEPs, and EPs confirmed that these populations have the same lineage potential as the corresponding cell population in primary transplantation (Figures S4E–S4L). MPPF were also capable of producing phenotypic CLPs, although in significantly lower numbers compared with erythromyeloid-restricted progenitors (∼35- and ∼220-fold differences at days 7 and 14, respectively; Figures S4M and S4N). Quantitatively, these experiments demonstrate that there is substantial expansion in progenitor cell numbers in the BM soon after transplantation, that HSCs and MPPF produce various erythromyeloid progenitors in roughly the same proportions, and that MPPF produce more erythromyeloid- than lymphoid-restricted progenitor cells. Qualitatively, these data provide direct evidence that transplanted MPPF produce all types of erythromyeloid- and lymphoid-competent progenitor cells, consistent with full multipotency.
Figure 3.
Both HSCs and MPPF Generate all Types of Erythromyeloid Progenitors after Transplantation
(A) Schematic of HSC and MPPF transplantation from UBC-GFP mice, short-term analysis of donor-derived progenitor cells, and subsequent functional analysis of the same donor-derived progenitor cells by secondary transplantation.
(B–I) Analysis of donor-derived cells in the BM of recipients shortly after transplantation of HSCs and MPPF. Cells were pre-gated on GFP+ to display only donor-derived cells. (B and C) At day 2 post-transplantation, the phenotype of GFP+ cells in the BM of sublethally irradiated (500 rad) recipient mice was predominantly the same as the phenotype of the input cell type. (D, E, and H) Transplanted HSCs gave rise to phenotypic classical CMPs, GMPs, and MEPs (D1 and D2); progenitors defined by alternative markers as MkP, “GMP,” pre-GM, pre-EP, pre-CFU-E, and pre-MegE (E1 and E2); and EP and GM cells (H1 and H2) by days 7 and 14 post-transplantation into lethally irradiated (1,000 rad) recipients. Few HSC-derived B and T cells were observed at these time points (H1 and H2). (F, G, and I) Transplanted MPPF gave rise to phenotypic classical CMPs, GMPs, and MEPs (F1–F4); MkP, “GMP,” pre-GM, pre-EP, pre-CFU-E, and pre-MegE (G1–G4); and EP, GM, and B and T cells (I1–I4) by days 4, 7, 11, and 14 post-transplantation. (D1′–I4′) Quantification of results displayed in (D–I), respectively, demonstrated substantial expansion in donor-derived cell numbers during the course (4–14 days) of the experiments. Most comparisons of the absolute cell numbers between time points and between HSCs and MPPF were significantly different; however, the proportions of progenitor cells generated were similar. For example, both HSCs and MPPF generated more GMPs than MEPs and CMPs, and the proportions of CMP/GMP/MEP generated by HSCs at day 11 and MPPF at day 14 were not significantly different from each other (Student's two-tailed t test for HSCs; one-way ANOVA with multiple post hoc comparisons for MPPF; D1′–I4′). n = 3 or more recipients in two or more independent experiments for each donor cell type and analysis time point.
Data are displayed as means ± SEM. Abbreviations for pre-gating of (H1–I4): Ery, TER-119; Ly, CD3 and B220; My, Gr1 and Mac1. FACS, fluorescence-activated cell sorting; WT, wild-type. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. See also Figure S4.
Transplantation of Single MPPF Provides Multilineage Reconstitution
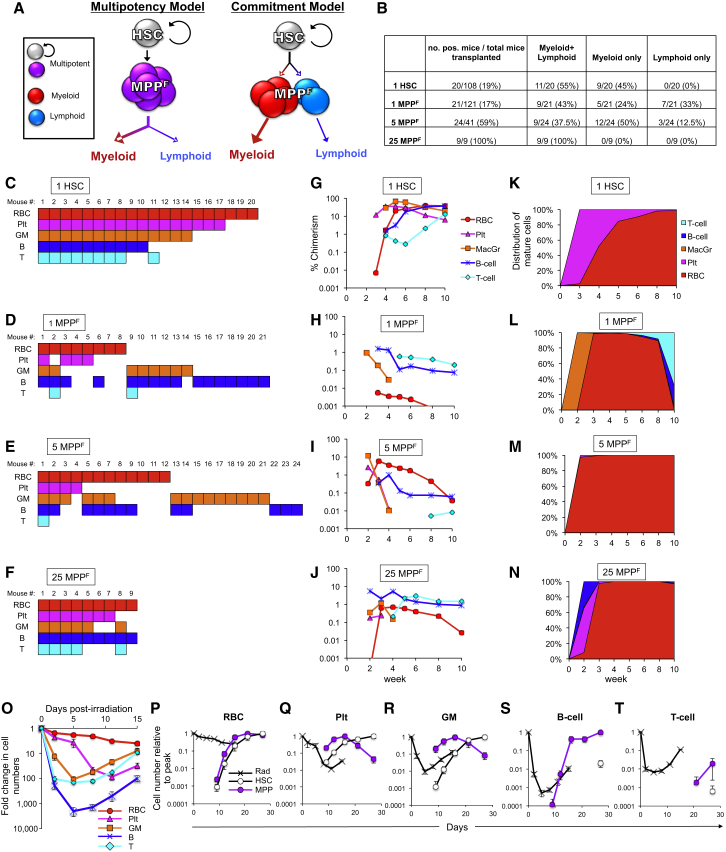
It is clear from the data presented in Figures 1M, 1N, 3F–3I, S2A, and S2B that transplantation of multiple MPPF results in a cell production profile similar to that of HSCs. However, population data cannot determine if the MPPF compartment is functionally homogeneous, with each cell being multipotent (Figure 4A, multipotency model; Boyer et al., 2012), if MPPF are a heterogeneous population of committed progenitors that share a common phenotype (Figure 4A, commitment model), or a combination of the two. To differentiate between these possibilities, we implemented two in vivo clonal approaches: single-cell transplantation and analysis of single-cell-derived colony-forming units of the spleen (CFU-S).
Figure 4.
Single HSCs and MPPF Reconstituted Both Myeloid and Lymphoid Lineages In Vivo
(A) Alternative models of MPPF multipotency. The multilineage reconstitution by MPPF transplanted in bulk (Figures 1B and 1N) may be derived from clonally multipotent MPPF (left) or from the combined contribution of lineage-committed cells with a shared MPPF surface phenotype (right).
(B) Summary of the results from single transplanted HSCs and from single and multiple MPPF. p = 0.05 for 1 HSC versus 25 MPPF; p > 0.1 for 1 HSC versus 5 MPPF; p < 0.05 for 1 HSC versus 1 MPPF by chi-square test of independence in reconstitution patterns.
(C–F) Lineages detected in individual lethally irradiated (1,000 rad) recipients receiving either single HSCs (C) or 1 (D), 5 (E), or 25 (F) MPPF. Mature cell detection by flow cytometry is indicated for each recipient and cell type by a filled square (RBCs, red; platelets, pink; GMs, orange; B cells, blue; T cells, teal).
(G–N) (G–J) Percentage donor chimerism and (K–N) distribution of mature cells from single HSCs (G and K) and 25 (J and N), 5 (I and M), or single (H and L) MPPF. Representative recipients that displayed multilineage reconstitution are shown.
(O) B cell numbers display a rapid and more drastic decline (2,000-fold) after lethal irradiation than other mature cell types (4- to 150-fold for RBCs and T cells, respectively). Data displayed are fold change in mature cell numbers in the PB of lethally irradiated (1,000 rad) mice over time. n = 4.
(P–T) The magnitude and timing of host cell depletion and recovery affect the ability to detect reconstitution from transplanted progenitor cells. Numbers from (O) were superimposed with the kinetics of mature cell generation by HSCs and MPPF for each lineage, and displayed relative to the peak reconstitution (set at 1.00) for each population. RBCs (P), platelets (Q), GMs (R), B cells (S), and T cells (T). All data are from at least three independent experiments.
See also Figures S5 and S6.
Single-cell transplantation of one HSC or one MPPF led to detectable levels of donor-derived cells in 19% and 17% of mice, respectively (Figures 4B and S5). Eight of the 20 reconstituted recipients of a single HSC had donor-derived cells of all five lineages investigated (Figures 4C and S5). This may be an underestimation of their true capacity, as some quiescent HSCs do not produce progeny until many weeks after transplantation or until secondary transfer (Yamamoto et al., 2013). Fifty-five percent of mice reconstituted with a single HSC produced cells of at least one myeloid lineage (RBCs, platelets, and/or GMs) and one lymphoid lineage (B cells and/or T cells) and were therefore categorized as multipotent (Figures 4B and 4C). The remaining positive recipients of single HSCs produced only myeloid cells, whereas none produced only lymphoid cells (Figures 4B and 4C). None of the single MPPF gave rise to all five cell types analyzed. Importantly, however, some single MPPF gave rise to four different types of mature cells, and a considerable fraction of single MPPF (43%; 9/21) generated both myeloid and lymphoid cells (Figures 4B and 4D). Increasing the number of transplanted MPPF to 5 or 25 cells led to a higher frequency of recipients with detectable donor cells (Figure 4B), an increased number of cell types detected (Figures 4D–4F), and an increase in donor chimerism levels (Figures 4H–4J; average overall reconstitution from 1, 5, and 25 MPPF was 0.9%, 1.5%, and 2.2%, respectively). Twenty-five MPPF were sufficient for combined myeloid and lymphoid detection in 100% of recipients, whereas mice transplanted with 5 or 1 MPPF or a single HSC were multilineage reconstituted at lower frequencies (37.5%, 43%, and 55%, respectively; Figures 4B–4F). Although a single MPPF did not contribute as robustly to recipient chimerism as a single HSC (Figures 4G and 4H; average overall reconstitution levels were 0.9% and 8%, respectively), more RBCs than any other mature cell type were generated from both HSCs and MPPF (Figures 4K–4N).
Despite high frequency of combined myeloid and lymphoid reconstitution by MPPF, we noted that MPPF led to only GM, only B cell, or combined GM/B cell reconstitution of some recipients, whereas HSCs did not. In addition, some myelo/lympho reconstitution occurred in unexpected combinations, such as only RBCs and B cells (mouse 6 for single MPPF, mouse 9 for 25 MPPF, and mice 8 and 9 for 5 MPPF). GM and B cells occurred together in several mice, without detection of T cells. Indeed, T cell readout was the most difficult lineage to detect from both HSC and MPPF transplants (Figures 4C–4F). As different mature cell types in the host are depleted to different extents upon sublethal (Figure 1H) and lethal (Figure 4O) irradiation, and donor-derived progeny are produced with different kinetics from different HSCs and MPPF (Figures 2A′–2E′), we hypothesized that the kinetics of host and donor cells differentially affects donor-derived cell detection. Superimposing the decline and recovery of host cells with the production of mature cells by HSCs or MPPF revealed that RBC production from both HSCs and MPPF largely coincided with the reduction in host RBCs (Figure 4P), facilitating detection of donor-derived RBCs in recipient mice. In contrast, neither HSCs nor MPPF produce large numbers of T cells until after host T cell numbers have significantly recovered (Figure 4T), contributing to poor detection of T cells in mice transplanted with a single cell (Figures 4C and 4D). Similarly, the timing of GM and B cell production from MPPF occurs near the low point of host GMs and B cells, whereas the main contribution of HSCs to GMs and B cells occurs after host cell recovery (Figures 4R and 4S). The fact that B cells are the host cell type most affected by irradiation (Figures 1H and 4O) and that MPPF produce B cells prior to host recovery, whereas HSCs do not (Figure 4S), contributes to the relatively high frequency of B cell detection in mice transplanted by a single MPPF, as well as the more apparent contribution of CLPF to B cells than to T cells (Figures 1G, 1S, and S6).
Multilineage Reconstitution from Single HSCs and MPPF in Spleen Colony Assays
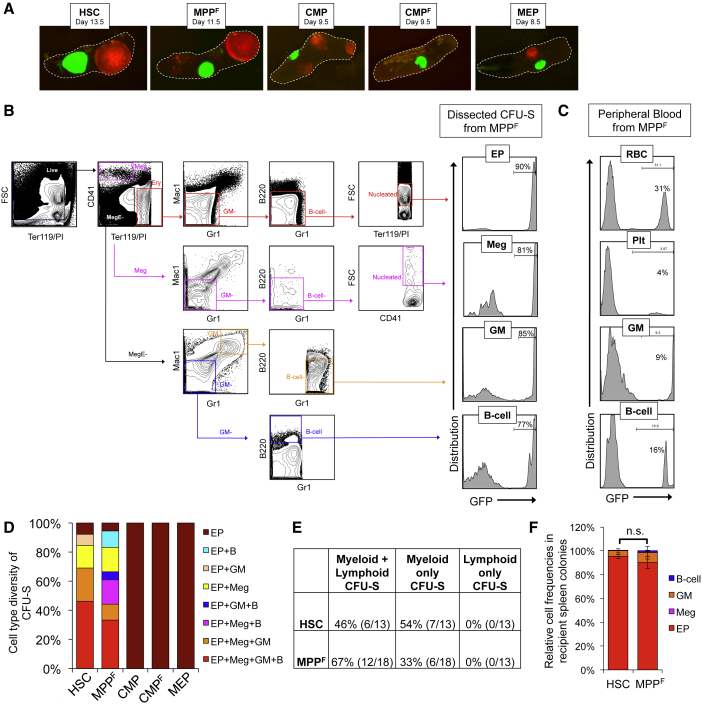
To test the multipotency of clonal HSCs and MPPF by an independent method, we analyzed donor-derived CFU-S. Like single-cell transplants, this assay measures clonal lineage potential, as each colony consists of progeny from a single cell (Becker et al., 1963, Weber et al., 2011). In support of the clonal origin of CFU-S, we observed only single-color colonies when a mixture of Tomato+ cells and GFP+ cells was transplanted into the same recipient (Figure 5A). To test for multilineage potential of single cells, we examined individually dissected CFU-S for EPs, megakaryocytes (Megs), GMs, and B cells (Figure 5B). It is highly improbable that CFU-S colonies were contaminated with circulating, donor-derived mature cells, as donor contribution within individual CFU-S was substantially higher than the donor cell chimerism in the PB (compare Figures 5B and 5C). T cell output was not assessed, as T cells require extended development in the thymus and are not produced in the spleen or in the time frame of this assay.
Figure 5.
Multilineage Reconstitution by Single HSCs and MPPF in In Vivo Spleen Colony Assays
(A) In support of the clonal origin of spleen colonies, CFU-S were composed of only a single color, either red or green, after transplantation of a mixture of Tomato+ and GFP+ cells. Representative fluorescence microscopy images are shown.
(B–F) Spleen colonies derived from single HSCs or MPPF contain erythroid, megakaryocytic, GM, and B cell lineages. Donor cells from UBC-GFP mice were transplanted into lethally irradiated (1,000 rad) mice. CFU-S were dissected and analyzed by flow cytometry at the time point post-transplantation of largest colony size; HSCs on day 13.5 (n = 13), MPPF on day 11.5 (n = 18), CMPs on day 9.5 (n = 16), CMPF on day 9.5 (n = 8), and MEPs on day 8.5 (n = 9). (B) Gating strategy and analysis of a representative MPPF-derived CFU-S. The percentages of donor-derived (GFP+) cells for each cell type within representative colonies are shown in the histograms on the right, with gating strategies shown on the left. (C) Substantially lower donor contribution was observed in the PB than within CFU-S (B), indicating that the detection of multiple lineages within a colony is not due to contamination of circulating cells. Of note, there were no detectable EPs or Megs in the PB. Representative flow cytometry plots are from a recipient of MPPF. (D) Proportion of individual CFU-S containing detectable EPs, Megs, GMs, and/or B cells. (E) Summary of the CFU-S data shown in (D). The colony type distribution was not significantly different for HSCs versus MPPF (p > 0.1; chi-square test of independence). (F) The frequency of EPs, Megs, GMs, and B cells in dissected CFU-S colonies from (D). Percentages are shown as donor-derived (GFP+) cells only. Data represent four independent experiments with a total of 12–17 mice per group.
Of 13 HSC-derived CFU-S, six (46%) contained all four lineages that can be detected in this assay (EPs, Megs, GMs, and B cells; Figure 5D, red bar; Figure 5E). The remaining colonies lacked B cells and were thus a mixture of myeloid lineages. Similarly, six of the 18 (33%) single-MPPF-derived colonies contained all four lineages (Figures 5D and 5E). Other MPPF-derived CFU-S contained various combinations of myeloid only or myeloid plus B cells. The proportions of myelo/lympho, myeloid only, or lymphoid only colonies produced by HSCs versus MPPF were not significantly different (Figure 5E). All colonies visible to the eye contained erythroid cells. In fact, the vast majority of total cells produced were erythroid cells (Figure 5F), and CFU-S derived from CMP, CMPF, and MEP contained only EPs (Figure 5D). The absence of additional lineages from myeloid progenitors is likely due to their reduced total cell production compared with HSCs and MPPF (Figures 2A–2C), limiting our ability to detect mature cells that are not produced in as high abundance as erythrocytes. The CFU-S data, like the single-cell transplantations, demonstrate that a substantial fraction of both HSCs and MPPF is multipotent at the single-cell level.
Discussion
A Quantitative Perspective of Progenitor Repopulation Capacity
By simply quantifying the absolute number of mature cells generated per transplanted stem and progenitor cell, we provide a new perspective of the regenerative capacity of HSCs and several progenitor populations. Our strategy revealed that all progenitor cells with myeloid potential produce far more RBCs than any other cell type (Table 1 and Figures 1M–1S, 2A–2E, 2A′–2E′, 3A–3F, 4K–4N, and 5D–5F). As illustrated by the concordance with previous findings when the same results were displayed as traditional donor chimerism (Figures 1A–1G), our results are not due to differences in gating strategies, cell population purity, or transplantation methods. Rather, the new insights were reached by eliminating the drastically variable fluctuations in host cell disappearance and recovery (Figures 1H and 4O) and by taking into account the absolute numbers of each mature cell type, including RBCs and platelets, produced by each stem and progenitor cell (Figures 2A–2E, 2A′–2E′, and S2). As transplanted donor cells differ not only in the types of cells generated, but also in the number, timing, and duration of mature cell production, each progenitor cell has a distinct reconstitution pattern. In addition, the half-lives of mature cell types vary considerably. Collectively, these dynamics differentially affect the ability to detect and quantify the contributions of different donor populations to each mature lineage, as exemplified in Figures 4P–4T and S6. Absolute quantification of donor-derived cells removes host variables and therefore facilitates comparative assessment of reconstitution both across lineages and between transplanted populations.
Unexpected Lineage Potential of Hematopoietic Progenitor Populations
The lineage potential that we uncovered was in some cases unexpected. For example, GMPs have been reported to express few erythroid-associated genes and to lack MegE potential (Akashi et al., 2000, Forsberg et al., 2006, Paul et al., 2015, Pronk et al., 2007), and CMPF produce few Meg/erythroid colonies in vitro (Nutt et al., 2005), whereas our experiments here revealed that CMPF and GMPs produce RBCs and platelets (Figures 1D, 1E, 1P, 1Q, 2A–2C, 2A′–2C′, 3A–3J, S2, and S4E–S4H, Table 1). Significant heterogeneity of myeloid progenitor populations, especially classical CMPs, has been reported (Miyawaki et al., 2015, Paul et al., 2015, Perié et al., 2015), confounding interpretation of lineage potential. While the purity of bulk populations is not absolute, contamination cannot explain the differences between the lineage potential apparent in Figures 1A–1G and 1M–1S, as these data are derived from the exact same experiments. Other possible contributors to the contradictory findings include differences between the in vitro and the in vivo assay conditions, limits of detection, and the relatively recent development of mice that make it possible to directly track RBCs and platelets in vivo. The cell production capacity varies drastically between different progenitors: our estimates revealed that each MPPF produces, on average, ∼800,000 cells, whereas one GMP gives rise to ∼47 progeny (43 RBCs, 1 Plt, and 4 GMs; Figures 2A′–2E′). The low burst size of GMPs precludes detection of progeny in single-cell transplants and in CFU-S; thus, the data presented here do not conclusively rule out that RBC and/or Plt generation from presumed GMPs is from “contaminating” cells. Notably, whereas we readily detected GFP+ RBCs and platelets in every recipient of GMPs in both primary and secondary transplantation experiments (Figures 1E, 1Q, S4G, and S4H), we did not observe RBC or Plt production from CLPs (Figures 1G and 1S) or platelets from EPs (Figures S4K–S4L), indicating that the technical purity of our transplanted cell populations is good. In future studies we will test whether GMPs, like MPPF, fit the “default” model proposed later by displaying significant in vivo MegE potential that is not readily detected in vitro. In contrast, the numbers of RBCs produced by CMPs and CMPF (averaging ∼140,000 and 35,000, respectively) were sufficient for detection of EPs in CFU-S (Figure 5D), whereas detection of platelets from CMPs, CMPF, GMPs, and MEPs required transplantation of higher numbers of progenitors. We previously demonstrated that Flk2+ MPPs give rise to all lineages, including erythroid cells, in situ both at steady state and under stress, as well as upon transplantation (Boyer et al., 2012, Boyer et al., 2011, Forsberg et al., 2006). Collectively, our results indicate that it is difficult to separate functional RBC and Plt potential from GM potential in vivo, reinforcing the existence of functional CMPs and the retention of RBC capability across several phenotypically distinct populations.
CLPF Appear Restricted to B and T Cell Generation
The only population in our study that did not produce RBCs and platelets was CLPF. CLPF also lacked detectable GM potential and therefore appear committed to the lymphoid fate, in agreement with their initial designation as CLPs (Kondo et al., 1997) and with in vivo lineage tracing experiments (Schlenner et al., 2010). Interestingly, while B cell production was readily detectable, T cell capacity was more apparent by absolute quantification than by donor chimerism data (compare Figures 1G–1S). The relative difficulty in detecting CLPF-derived T cells can be attributed, in part, to host T cells being less affected by irradiation compared with host B cells (∼20-fold versus ∼1,000-fold, respectively; Figure 1H) and because host T cell numbers recover before CLPF-derived T cells start accumulating (approximately day 19; Figures 1S and S6). In contrast, CLPF-derived B cells are detected as early as day 9 after transplantation, closely coinciding with the sharp reduction in host B cells (Figure S6). Their more limited lineage potential and lower expansion capacity are consistent with CLPF residing hierarchically downstream of MPPF. However, our calculations estimated that one MPPF gives rise to equal numbers of B cells (∼2,500) and T cells (∼2,700), whereas one CLPF gives rise to 10-fold more B cells than T cells (420 versus 42) (Figures 2D′ and 2E′). This appears inconsistent with a direct and exclusive MPPF-to-CLPF transition, but rather evokes additional intermediate populations and/or flexibility in differentiation pathways from upstream progenitors.
Multilineage Differentiation Potential of Single HSCs and MPPF
It is clear from both the chimerism (Figure 1B) and the absolute cell number data (Figures 1N and 2A–2E), as well as previous publications (Forsberg et al., 2006, Miyawaki et al., 2015, Yamamoto et al., 2013), that despite downregulation of erythroid-associated genes (Forsberg et al., 2006, Moignard et al., 2013), MPPF are multipotent at the population level. Here, we also showed that a substantial fraction of MPPF is multipotent at the single-cell level. In CFU-S assays, MPPF produced both myeloid and lymphoid cells in 67% of the colonies, with the remaining colonies consisting of myeloid cells only (Figures 5D and 5E). While the CFU-S frequency from MPPF is lower than that from HSCs (∼1/78 versus 1/33; Beaudin et al., 2014, Forsberg et al., 2006), CFU-S capability is clearly an underestimation of the MegE potential of a cell, as far more than 1/33 HSCs are multipotent in other assays. Indeed, the CFU-S frequency increases ∼10-fold when HSCs or MPPF are injected directly into the spleen as opposed to intravenously (Beaudin et al., 2014). Collectively, these data demonstrate that the ability of MPPF to generate MegE cells is substantially greater than estimated by CFU-S alone.
Similar proportions of HSCs and MPPF displayed combined myelo/lymphoid potential at the clonal level upon single-cell transplantation (55% of HSCs and 43% of MPPF; Figures 4B and 4C). Our numbers are similar to those of Yamamoto et al., who found that 56% (9/16 mice) of “LMPPs” (in their report defined as KLS CD34+ FLK2+, a population significantly overlapping with our MPPF) displayed combined myelo/lymphoid readout (Yamamoto et al., 2013). In vivo evidence for the existence of clonal MPP was also provided by in situ barcoding studies (Rodriguez-Fraticelli et al., 2018, Sun et al., 2014). The exact proportion of uncommitted cells within the HSC and MPPF compartment is difficult to estimate, as variable lineage outputs could be a result of detection limits (progeny were produced but not in sufficient numbers to detect), stochasticity (the transplanted cell happened to encounter a particular combination of cytokines), population heterogeneity (a proportion of the cells are multipotent, whereas some are not), or a combination of the three. Few studies have tested the full lineage potential of MPPF, but the heterogeneity of HSCs has been tested and debated extensively (Ema et al., 2014, Hock, 2010, Kokkaliaris et al., 2016, Schroeder, 2010). The HSC results are highly relevant for the heterogeneity of MPPF: a lineage-restricted cell (a committed “HSC”) cannot give rise to a cell with greater lineage potential (a multipotent MPP). Thus, the proportion of multipotent cells within the MPPF fraction must be equal to or smaller than the fraction of HSCs that are multipotent (Figure 4A; Boyer et al., 2012). Conversely, if some MPPF are uncommitted, an equal or greater proportion of HSCs and other upstream populations should also be uncommitted. In our experiments, the frequency of single cells with combined myelo/lympho potential is similar for HSCs and MPPF (55% and 43% for singly transplanted HSCs and MPPF, respectively, and 46% and 67% for HSCs and MPPF in CFU-S). Given that it is harder to detect progeny from MPPF than from HSCs, the extent of lineage restriction upon the transition from HSCs to MPPF appears quite low. The existence of fully multipotent progenitor cells is consistent with the notion that steady-state hematopoiesis is largely sustained by MPPs rather than HSCs (Busch et al., 2015, Sun et al., 2014).
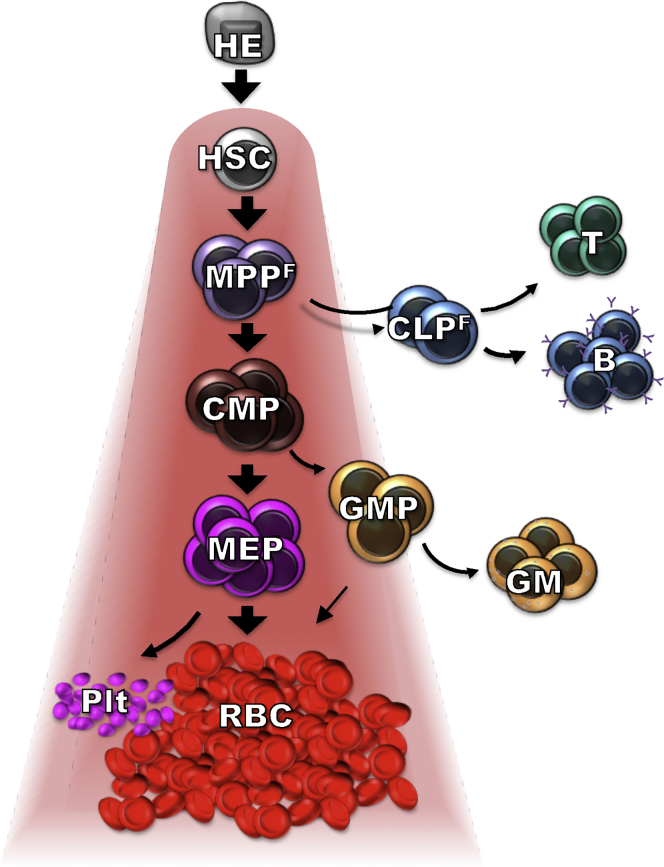
RBC Production as the Default Hematopoietic Fate
Previous studies have shown that substantial amplification of RBC production occurs at the level of erythroid-committed progenitors and precursors. While the overall dominance of RBC production should not be unexpected, models of hematopoietic differentiation rarely take the vastly different numbers of mature cells into account. The fact that cell populations like MPPF and GMPs, despite their poor ability to generate MegE cells in vitro and clear downregulation of erythroid-driving genes, generate RBCs not only in the largest absolute numbers, but also in roughly similar relative proportions compared with other HSPCs, was particularly surprising. This discordance of gene expression and functional differentiation potential may hamper efforts to construct lineage maps based on transcriptome data. To reconcile these apparent discrepancies, we propose a model based on functional lineage potential where RBC production is the default pathway (Figure 6). In this model, MegE potential is gained upon specification of HSCs from a hematovascular precursor. HSCs, at all stages of development, thus have inherent capability to generate RBCs and platelets, but not necessarily GMs, T cells, and B cells. This notion makes sense through ontogeny and as a mechanism to ensure survival, and fits with newly proposed views of human hematopoiesis (Notta et al., 2016). Our model proposes that differentiation into alternative fates is accomplished downstream of HSCs by the combination of two mechanisms: increased expression of proteins promoting GM, B cell, and/or T cell differentiation and a concurrent decrease in expression of genes driving RBC and Plt generation. In vivo, continuous expression of MegE-promoting genes does not appear necessary for retention of RBC and Plt capability, conceivably because MegE differentiation has already been initiated. Unless exposed to sufficient concentrations of factors to alter this path, most hematopoietic progenitors will therefore produce primarily RBCs and platelets. Conversely, downregulation of MegE-promoting genes may be necessary for deviation from this default pathway to allow for the relatively rare production of GMs, B cells, and T cells. When the default pathway is interrupted (removal of cells from their natural environment), progenitors that have downregulated MegE-specifying receptors (MPPF, CMPF, and GMPs) are unable to reinitiate MegE production. Thus, they perform relatively poorly in MegE assays in vitro (Adolfsson et al., 2005, Nutt et al., 2005, Pronk et al., 2007). As MPPF, CMPF, and GMPs have acquired expression of receptors that promote GM or lymphoid fates, they are reactive to the corresponding cytokines and consequently readily differentiate in vitro into cell types that normally (in vivo) represent alternative fates. The proposed model influences our understanding of erythroid-specific versus pan-hematopoietic disorders, and also provides explanations for the discordant lineage potential by in vivo and in vitro strategies and for the disconnection between gene expression patterns and functional cell production in vivo. Collectively, MPPF may be viewed more accurately as cells that have gained GM, B, and T potential than as cells that have lost MegE potential. This view is supported by our previous demonstration that all hematopoietic lineages are derived via an FLK2+ stage in situ and upon transplantation (Boyer et al., 2011, Boyer et al., 2012), by FLK2 promoting expansion of all mature blood cell types (Beaudin et al., 2014), and by recent reports pointing to MPPs, rather than HSCs, as the major source of mature hematopoietic cells during steady-state hematopoiesis in situ (Busch et al., 2015, Sun et al., 2014).
Figure 6.
Hematopoietic Differentiation Model in which Erythroid Production Represents the Default Fate of HSCs
Both functional experiments and gene expression data indicate that the capacity to generate RBCs and platelets is acquired upon specification of HSCs from a hematovascular precursor. Despite downregulation of genes that drive MegE development, RBC and Plt production remains the predominant fate of MPPF and other non-lymphoid committed progenitors. Relatively rare production of GMs and B and T cells occurs through combined downregulation of MegE drivers and a gain of gene products promoting the alternative fates. Initiation of non-MegE fates in MPPF may thus be viewed as gain of GM and lymphoid potential, rather than loss of capacity to generate RBCs and/or platelets.
Experimental Procedures
Transplantation Assays
Hematopoietic cells were isolated from BM isolated from murine femurs and tibias from wild-type (C57BL/6) or UBC-GFP mice (Schaefer et al., 2001) in accordance with UCSC IACUC guidelines, as described in the Supplemental Experimental Procedures and previously (Beaudin et al., 2016, Beaudin et al., 2014, Smith-Berdan et al., 2015, Smith-Berdan et al., 2011, Ugarte et al., 2015).
Mature Cell Quantification
A known volume of PB was mixed with an antibody solution containing a known quantity of Calibrite APC beads prior to flow cytometry analysis. For tissues, a known quantity of beads was added to each tissue preparation prior to antibody staining and analysis. The number of beads counted by flow cytometry was used to calculate the number of mature cells per microliter of blood or within each tissue. The distribution of mature hematopoietic cells in a mouse was measured in the blood obtained by perfusion, in BM by analysis of two femurs and tibias, in spleen, in thymus, and in lymph nodes (inguinal, axillary, and superficial cervical).
Single-Cell Transplants
Individual HSCs and MPPF were double-sorted into separate wells on Terasaki plates using a FACSAria III from lineage-depleted BM cells from UBC-GFP mice, similar to our previous reports (Byrne et al., 2017, Cole et al., 2018) and as detailed in the Supplemental Experimental Procedures.
Author Contributions
S.W.B. and E.C.F. conceived the study, designed, analyzed, and interpreted experiments, and co-wrote the manuscript. S.W.B., S.R., and A.E.B. designed, performed, analyzed, and interpreted experiments. S.S.B., J.P.C., P.K.M., E.W.M., C.C., H.T., and M.L. assisted with experiments and data analysis. E.C.F. oversaw the study and obtained funding. All authors reviewed the manuscript.
Acknowledgments
We thank Forsberg lab members for comments on the manuscript. This work was supported by an NIH/NHLBI award (R01HL115158) to E.C.F.; by CIRM training grant TG2-01157 to S.W.B. and A.E.B.; by NIH/NIGMS T32-GM008646 to S.W.B. and J.P.C.; by an HHMI Gilliam pre-doctoral award to J.P.C.; by CIRM SCILL grant TB1-01195 to E.W.M. via San Jose State University; and by CIRM Shared Stem Cell Facilities (CL1-00506) and CIRM Major Facilities (FA1-00617-1) awards to the University of California, Santa Cruz.
Published: March 21, 2019
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stemcr.2019.02.007.
Supplemental Information
References
- Adolfsson J., Månsson R., Buza-Vidas N., Hultquist A., Liuba K., Jensen C.T., Bryder D., Yang L., Borge O.-J., Thoren L.A.M. Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential. Cell. 2005;121:295–306. doi: 10.1016/j.cell.2005.02.013. [DOI] [PubMed] [Google Scholar]
- Akashi K., Traver D., Miyamoto T., Weissman I.L. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404:193–197. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- Beaudin A.E., Boyer S.W., Forsberg E.C. Flk2/Flt3 promotes both myeloid and lymphoid development by expanding non-self-renewing multipotent hematopoietic progenitor cells. Exp. Hematol. 2014;42:218–229.e4. doi: 10.1016/j.exphem.2013.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudin A.E., Boyer S.W., Perez-Cunningham J., Hernandez G.E., Derderian S.C., Jujjavarapu C., Aaserude E., MacKenzie T., Forsberg E.C. A transient developmental hematopoietic stem cell gives rise to innate-like B and T cells. Cell Stem Cell. 2016;19:768–783. doi: 10.1016/j.stem.2016.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker A.J., McCulloch E.A., Till J.E. Cytological demonstration of the clonal nature of spleen colonies derived from transplanted mouse marrow cells. Nature. 1963;197:452–454. doi: 10.1038/197452a0. [DOI] [PubMed] [Google Scholar]
- Benz C., Copley M.R., Kent D.G., Wohrer S., Cortes A., Aghaeepour N., Ma E., Mader H., Rowe K., Day C. Hematopoietic stem cell subtypes expand differentially during development and display distinct lymphopoietic programs. Cell Stem Cell. 2012;10:273–283. doi: 10.1016/j.stem.2012.02.007. [DOI] [PubMed] [Google Scholar]
- Boyer S.W., Schroeder A.V., Smith-Berdan S., Forsberg E.C. All hematopoietic cells develop from hematopoietic stem cells through Flk2/Flt3-positive progenitor cells. Cell Stem Cell. 2011;9:64–73. doi: 10.1016/j.stem.2011.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer S.W., Beaudin A.E., Forsberg E.C. Mapping differentiation pathways from hematopoietic stem cells using Flk2/Flt3 lineage tracing. Cell Cycle. 2012;11:3180–3188. doi: 10.4161/cc.21279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch K., Klapproth K., Barile M., Flossdorf M., Holland-Letz T., Schlenner S.M., Reth M., Höfer T., Rodewald H.-R. Fundamental properties of unperturbed haematopoiesis from stem cells in vivo. Nature. 2015;518:542–546. doi: 10.1038/nature14242. [DOI] [PubMed] [Google Scholar]
- Byrne A., Beaudin A.E., Olsen H.E., Jain M., Cole C., Palmer T., DuBois R.M., Forsberg E.C., Akeson M., Vollmers C. Nanopore long-read RNAseq reveals widespread transcriptional variation among the surface receptors of individual B cells. Nat. Commun. 2017;8:16027. doi: 10.1038/ncomms16027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole C., Byrne A., Beaudin A.E., Forsberg E.C., Vollmers C. Tn5Prime, a Tn5 based 5' capture method for single cell RNA-seq. Nucleic Acids Res. 2018;46:e62. doi: 10.1093/nar/gky182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amico A., Wu L. The early progenitors of mouse dendritic cells and plasmacytoid predendritic cells are within the bone marrow hemopoietic precursors expressing Flt3. J. Exp. Med. 2003;198:293–303. doi: 10.1084/jem.20030107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykstra B., Kent D., Bowie M., McCaffrey L., Hamilton M., Lyons K., Lee S.-J., Brinkman R., Eaves C. Long-term propagation of distinct hematopoietic differentiation programs in vivo. Cell Stem Cell. 2007;1:218–229. doi: 10.1016/j.stem.2007.05.015. [DOI] [PubMed] [Google Scholar]
- Ema H., Morita Y., Suda T. Heterogeneity and hierarchy of hematopoietic stem cells. Exp. Hematol. 2014;42:74–82.e2. doi: 10.1016/j.exphem.2013.11.004. [DOI] [PubMed] [Google Scholar]
- Forsberg E.C., Serwold T., Kogan S., Weissman I.L., Passegué E. New evidence supporting megakaryocyte-erythrocyte potential of Flk2/Flt3+ multipotent hematopoietic progenitors. Cell. 2006;126:415–426. doi: 10.1016/j.cell.2006.06.037. [DOI] [PubMed] [Google Scholar]
- Gerrits A., Dykstra B., Kalmykowa O.J., Klauke K., Verovskaya E., Broekhuis M.J.C., de Haan G., Bystrykh L.V. Cellular barcoding tool for clonal analysis in the hematopoietic system. Blood. 2010;115:2610–2618. doi: 10.1182/blood-2009-06-229757. [DOI] [PubMed] [Google Scholar]
- Hock H. Some hematopoietic stem cells are more equal than others. J. Exp. Med. 2010;207:1127–1130. doi: 10.1084/jem.20100950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakumitsu H., Kamezaki K., Shimoda K., Karube K., Haro T., Numata A., Shide K., Matsuda T., Oshima K., Harada M. Transgenic mice overexpressing murine thrombopoietin develop myelofibrosis and osteosclerosis. Leuk. Res. 2005;29:761–769. doi: 10.1016/j.leukres.2004.12.009. [DOI] [PubMed] [Google Scholar]
- Kokkaliaris K.D., Lucas D., Beerman I., Kent D.G., Perié L. Understanding hematopoiesis from a single-cell standpoint. Exp. Hematol. 2016;44:447–450. doi: 10.1016/j.exphem.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo M., Weissman I.L., Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 1997;91:661–672. doi: 10.1016/s0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- Lai A.Y., Kondo M. Asymmetrical lymphoid and myeloid lineage commitment in multipotent hematopoietic progenitors. J. Exp. Med. 2006;203:1867–1873. doi: 10.1084/jem.20060697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Neff N.F., Quake S.R., Weissman I.L. Tracking single hematopoietic stem cells in vivo using high-throughput sequencing in conjunction with viral genetic barcoding. Nat. Biotechnol. 2011;29:928–933. doi: 10.1038/nbt.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyawaki K., Arinobu Y., Iwasaki H., Kohno K., Tsuzuki H., Iino T., Shima T., Kikushige Y., Takenaka K., Miyamoto T. CD41 marks the initial myelo-erythroid lineage specification in adult mouse hematopoiesis: redefinition of murine common myeloid progenitor. Stem Cells. 2015;33:976–987. doi: 10.1002/stem.1906. [DOI] [PubMed] [Google Scholar]
- Moignard V., MacAulay I.C., Swiers G., Buettner F., Schütte J., Calero-Nieto F.J., Kinston S., Joshi A., Hannah R., Theis F.J. Characterization of transcriptional networks in blood stem and progenitor cells using high-throughput single-cell gene expression analysis. Nat. Cell Biol. 2013;15:363–372. doi: 10.1038/ncb2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na Nakorn T., Traver D., Weissman I.L., Akashi K. Myeloerythroid-restricted progenitors are sufficient to confer radioprotection and provide the majority of day 8 CFU-S. J. Clin. Invest. 2002;109:1579–1585. doi: 10.1172/JCI15272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemzek J.A., Bolgos G.L., Williams B.A., Remick D.G. Differences in normal values for murine white blood cell counts and other hematological parameters based on sampling site. Inflamm. Res. 2001;50:523–527. doi: 10.1007/PL00000229. [DOI] [PubMed] [Google Scholar]
- Notta F., Zandi S., Takayama N., Dobson S., Gan O.I., Wilson G., Kaufmann K.B., McLeod J., Laurenti E., Dunant C.F. Distinct routes of lineage development reshape the human blood hierarchy across ontogeny. Science. 2016;351:aab2116. doi: 10.1126/science.aab2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutt S.L., Metcalf D., D’Amico A., Polli M., Wu L. Dynamic regulation of PU.1 expression in multipotent hematopoietic progenitors. J. Exp. Med. 2005;201:221–231. doi: 10.1084/jem.20041535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oguro H., Ding L., Morrison S.J. SLAM family markers resolve functionally distinct subpopulations of hematopoietic stem cells and multipotent progenitors. Cell Stem Cell. 2013;13:102–116. doi: 10.1016/j.stem.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa M., Hanada K., Hamada H., Nakauchi H. Long-term lymphohematopoietic reconstitution by a single CD34-low/negative hematopoietic stem cell. Science. 1996;273:242–245. doi: 10.1126/science.273.5272.242. [DOI] [PubMed] [Google Scholar]
- Paul F., Arkin Y., Giladi A., Jaitin D.A., Kenigsberg E., Keren-Shaul H., Winter D., Lara-Astiaso D., Gury M., Weiner A. Transcriptional heterogeneity and lineage commitment in myeloid progenitors. Cell. 2015;163:1663–1677. doi: 10.1016/j.cell.2015.11.013. [DOI] [PubMed] [Google Scholar]
- Perié L., Duffy K.R., Kok L., De Boer R.J., Schumacher T.N. The branching point in erythro-myeloid differentiation. Cell. 2015;63:1655–1662. doi: 10.1016/j.cell.2015.11.059. [DOI] [PubMed] [Google Scholar]
- Pronk C.J.H., Rossi D.J., Månsson R., Attema J.L., Norddahl G.L., Chan C.K.F., Sigvardsson M., Weissman I.L., Bryder D. Elucidation of the phenotypic, functional, and molecular topography of a myeloerythroid progenitor cell hierarchy. Cell Stem Cell. 2007;1:428–442. doi: 10.1016/j.stem.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Richie Ehrlich L.I., Serwold T., Weissman I.L. In vitro assays misrepresent in vivo lineage potentials of murine lymphoid progenitors. Blood. 2011;117:2618–2624. doi: 10.1182/blood-2010-05-287102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Fraticelli A.E., Wolock S.L., Weinreb C.S., Panero R., Patel S.H., Jankovic M., Sun J., Calogero R.A., Klein A.M., Camargo F.D. Clonal analysis of lineage fate in native haematopoiesis. Nature. 2018;553:212–216. doi: 10.1038/nature25168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer B.C., Schaefer M.L., Kappler J.W., Marrack P., Kedl R.M. Observation of antigen-dependent CD8+ T-cell/dendritic cell interactions in vivo. Cell. Immunol. 2001;214:110–122. doi: 10.1006/cimm.2001.1895. [DOI] [PubMed] [Google Scholar]
- Schlenner S.M., Madan V., Busch K., Tietz A., Läufle C., Costa C., Blum C., Fehling H.J., Rodewald H.R. Fate mapping reveals separate origins of T cells and myeloid lineages in the thymus. Immunity. 2010;32:426–436. doi: 10.1016/j.immuni.2010.03.005. [DOI] [PubMed] [Google Scholar]
- Schroeder T. Hematopoietic stem cell heterogeneity: subtypes, not unpredictable behavior. Cell Stem Cell. 2010;6:203–207. doi: 10.1016/j.stem.2010.02.006. [DOI] [PubMed] [Google Scholar]
- Smith-Berdan S., Nguyen A., Hassanein D., Zimmer M., Ugarte F., Ciriza J., Li D., García-Ojeda M.E., Hinck L., Forsberg E.C. Robo4 cooperates with CXCR4 to specify hematopoietic stem cell localization to bone marrow niches. Cell Stem Cell. 2011;8:72–83. doi: 10.1016/j.stem.2010.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith-Berdan S., Nguyen A., Hong M.A., Forsberg E.C. ROBO4-mediated vascular integrity regulates the directionality of hematopoietic stem cell trafficking. Stem Cell Reports. 2015;4:255–268. doi: 10.1016/j.stemcr.2014.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Ramos A., Chapman B., Johnnidis J.B., Le L., Ho Y.J., Klein A., Hofmann O., Camargo F.D. Clonal dynamics of native haematopoiesis. Nature. 2014;514:322–327. doi: 10.1038/nature13824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugarte F., Sousae R., Cinquin B., Martin E.W., Krietsch J., Sanchez G., Inman M., Tsang H., Warr M., Passegué E. Progressive chromatin condensation and H3K9 methylation regulate the differentiation of embryonic and hematopoietic stem cells. Stem Cell Reports. 2015;5:728–740. doi: 10.1016/j.stemcr.2015.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber K., Thomaschewski M., Warlich M., Volz T., Cornils K., Niebuhr B., Täger M., Lütgehetmann M., Pollok J.-M., Stocking C. RGB marking facilitates multicolor clonal cell tracking. Nat. Med. 2011;17:504–509. doi: 10.1038/nm.2338. [DOI] [PubMed] [Google Scholar]
- Yamamoto R., Morita Y., Ooehara J., Hamanaka S., Onodera M., Rudolph K.L., Ema H., Nakauchi H. Clonal analysis unveils self-renewing lineage-restricted progenitors generated directly from hematopoietic stem cells. Cell. 2013;154:1112–1126. doi: 10.1016/j.cell.2013.08.007. [DOI] [PubMed] [Google Scholar]
- Yamamoto R., Wilkinson A.C., Ooehara J., Lan X., Lai C.Y., Nakauchi Y., Pritchard J.K., Nakauchi H. Large-scale clonal analysis resolves aging of the mouse hematopoietic stem cell compartment. Cell Stem Cell. 2018;5:600–607. doi: 10.1016/j.stem.2018.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.