Abstract
Developing novel approaches to treat skeletal disorders requires an understanding of how critical molecular factors regulate osteoblast differentiation and bone remodeling. We have reported that (1) retinoic acid receptor-related orphan receptor beta (Rorβ) is upregulated in bone samples isolated from aged mice and humans in vivo; (2) Rorβ expression is inhibited during osteoblastic differentiation in vitro; and (3) genetic deletion of Rorβ in mice results in preservation of bone mass during aging. These data establish that Rorβ inhibits osteogenesis and that strict control of Rorβ expression is essential for bone homeostasis. Because microRNAs (miRNAs) are known to play important roles in the regulation of gene expression in bone, we explored whether a predicted subset of nine miRNAs regulates Rorβ expression during both osteoblast differentiation and aging. Mouse osteoblastic cells were differentiated in vitro and assayed for Rorβ and miRNA expression. As Rorβ levels declined with differentiation, the expression of many of these miRNAs, including miR-219a-5p, was increased. We further demonstrated that miR-219a-5p was decreased in bone samples from old (24-month) mice, as compared with young (6-month) mice, concomitant with increased Rorβ expression. Importantly, we also found that miR-219a-5p expression was decreased in aged human bone biopsies compared with young controls, demonstrating that this phenomenon also occurs in aging bone in humans. Inhibition of miR-219a-5p in mouse calvarial osteoblasts led to increased Rorβ expression and decreased alkaline phosphatase expression and activity, whereas a miR-219a-5p mimic decreased Rorβ expression and increased osteogenic activity. Finally, we demonstrated that miR-219a-5p physically interacts with Rorβ mRNA in osteoblasts, defining Rorβ as a true molecular target of miR-219a-5p. Overall, our findings demonstrate that miR-219a-5p is involved in the regulation of Rorβ in both mouse and human bone.
Keywords: RorB, MicroRNA, OSTEOBLAST, OSTEOPOROSIS, AGING
Introduction
Maintenance of bone mass involves the coordinated activities of bone-forming osteoblasts and bone-resorbing osteoclasts,(1) with important signaling contributions by the matrix-embedded osteocyte.(2,3) Alterations in the activities among these cell types can lead to either excessive bone resorption or diminished bone formation, resulting in bone loss and the increased risk of fracture.(4,5) Osteoblast differentiation requires the orchestrated actions of numerous transcription factors (ie, Runx2, osterix) that function to regulate both the formation and maintenance of bone.(6) Understanding the complexity of the regulation and activity of these and other transcription factors during bone homeostasis is essential to formulate new strategies to combat osteoporotic bone loss and enhance skeletal repair following injury.
The orphan nuclear receptor, Rorβ is a transcription factor that has recently been shown to be a negative regulator of bone. Rorβ inhibits osteoblast differentiation and Rorβ expression increases with age in both mice and human bone-lineage cells,(7–9) establishing a reciprocal relation between Rorβ expression and osteogenic potential. At the molecular level, Rorβ represses both Runx2- and β-catenin-dependent transcriptional activation.(7,9) Furthermore, global deletion of Rorβ in mice results in improved bone mass, microarchitecture, and strength associated with increased bone formation, decreased resorption, and increased activation of the Wnt pathway.(9) Therefore, the identification and development of novel therapeutic approaches to downregulate Rorβ may be a new approach in osteoporosis treatment.
MicroRNAs (miRNAs) comprise a family of small noncoding RNAs (approximately 21 to 22 nucleotides in length) that perform posttranscriptional regulation of gene expression through mRNA degradation and/or translational repression.(10,11) They exert their gene regulatory function through interaction with short seed sequences usually contained within the 3’ untranslated region (UTR) of target genes,(12) and have been implicated in virtually all essential biological processes, including skeletal development and function.(13–17) Specific miRNAs have been identified that are associated with osteoporosis, aging, and skeletal fractures.(18–20) Because aberrant miRNA expression during either normal or pathological processes has the potential of affecting gene expression and altering a cellular and/or tissue phenotype, it is important to understand how these alterations affect the regulation of key transcription factors in bone, including Rorβ.
Rorβ expression drastically changes during both osteoblast differentiation and in aging bone in both mice and humans; therefore, we hypothesized that altered miRNA expression patterns control the expression levels of Rorβ, and by extension the cells/tissues in which Rorβ functions. Hence, the goal of this study was to specifically identify miRNA(s) that regulate Rorβ in osteoblastic cells, as well as in bone samples isolated from mice and humans during aging. Successful identification of these miRNA(s) would provide an alternative therapeutic strategy to downregulate Rorβ to promote bone anabolism in the context of age-related bone disorders, such as osteoporosis.
Materials and Methods
Osteoblast cell culture and differentiation
Primary mouse calvarial osteoblasts (CalOBs) were isolated as previously described(21) and maintained in alpha-minimal essential growth medium (αMEM) supplemented with 1 × antibiotic/antimycotic (ThermoFisher Scientific, Waltham, MA, USA), 1 × Glutamax, and 10% (v/v) fetal bovine serum (GE Healthcare Life Sciences HyClone Laboratories, Logan, UT, USA). For the osteoblast differentiation assays, primary CalOBs were plated at a cell density of 104 cells/cm2 in 12-well tissue culture plates and allowed to grow to confluence (typically 5 days). At confluence, the media were replaced with osteogenic differentiation medium (growth media supplemented with 50 mg/L ascorbic acid and 10mM β-glycerophosphate; Sigma-Aldrich, St. Louis, MO, USA) and allowed to differentiate for 0, 4, 7, 10, 17, and 24 days (n = 6). Osteogenic differentiation media were replaced every 3 days. Cells were lysed in QIAzol reagent (Qiagen, Valencia, CA, USA) for RNA isolation.
qPCR analyses of mRNA and miRNA assays
Total RNA was isolated from either differentiated CalOBs (n = 6) or mouse bone homogenates (n = 10) using the miRNeasy Mini Kit that included a DNase treatment step (RNase-free DNase Set) using the QIAcube instrument (all reagents from Qiagen). One μg of total RNA was used to generate cDNA using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Carlsbad, CA, USA) according to the manufacturer’s instructions. qPCR analysis for mRNA expression was performed using the ABI Prism 7900HT Real-Time System instrument (Applied Biosystems) with SYBR Green reagent (Qiagen) as previously described.(22) Data normalization was conducted using multiple endogenous reference genes (Actb, Hprt, and Tuba1a) and threshold calculations are as previously described.(23) The oligonucleotide sequences for the genes measured in this study were designed using the Primer Express program (Applied Biosystems) and are available in Supplemental Table 1.
The miRNA expression data were generated using the same RNA as for the mRNA expression analyses; however, 1 μg of total RNA was reverse-transcribed using the miScript II RT Kit (Qiagen). The individual miRNA assays used in this study were purchased from Qiagen and used with the miScript SYBR Green PCR Kit (Qiagen) according to the manufacturer’s instructions, using the same instrumentation as for the mRNA expression analyses. Data normalization was performed using the RNU6B (U6) small nucleolar RNA (snRNA).
Bone tissue samples (mouse and human)
All mouse studies were conducted in accordance with NIH guidelines and approved by the Institutional Animal Care and Use Committee at the Mayo Clinic (#A9715–15). Validation and detailed methods of our osteocyte-enriched bone isolation protocol are described extensively elsewhere.(24) Briefly, mouse vertebrae isolated from either 6-month-old or 24-month-old male C57BL/6N mice (n = 10) were cleaned of associated connective tissue and cut into ~1-mm pieces, which then underwent two sequential 30-min digests in endotoxin-free collagenase (Liberase; Roche Diagnostics GmbH, Mannheim, Germany) and were homogenized in QIAzol (Qiagen) for RNA isolation. The remaining cell fraction has been previously shown to represent a highly enriched population of osteocytes.(24) The human bone biopsy samples used in this study were as described in previous publications(22,25); they were small-needle bone biopsies (isolated from the posterior iliac crest) procured from 10 young premenopausal women (mean age ± SD, 27 ± 3 years; range 23 to 30 years) and 10 older postmenopausal women (78 ± 5 years; range 72 to 87 years). Postmenopausal status was established by the absence of menses for >1 year and serum follicle stimulate hormone levels >20 IU/L. Extensive exclusion criteria for these patients and the study protocol are as previously described.(22) The RNA from these bone biopsy samples was reanalyzed for miRNA expression.
miR-219a-5p miRNA inhibitor cell model production
Primary CalOBs at approximately 70% confluence were treated overnight with either a negative control miRNA inhibitor lentivirus (Catalog #HLTUD002C) or a custom miR-219a-5p miRNA inhibitor lentivirus (MISSION Lenti miRNA Inhibitors; Sigma-Aldrich) at a multiplicity of infection of five virus particles per cell, in the presence of 8-μg/mL polybrene (Santa Cruz Biotechnology, Santa Cruz, CA, USA) to enhance viral infection. The cells were trypsinized and plated in growth medium supplemented with 1.5-μg/mL puromycin (ThermoFisher Scientific), which was changed every 3 days until all the non-transduced control cells were dead (approximately 7 days). The resulting cell populations were termed CalOB-Control for the negative control miRNA inhibitor cell line or CalOB-Δ219a for the miR-219a-5p miRNA inhibitor cell line; both were used for osteoblast differentiation assays as described above. The cells were then maintained in growth media supplemented with 0.5-μg/mL puromycin. The puromycin was removed during the duration of the experiments to prevent any effects of the puromycin on cell function.
Alkaline phosphatase stain
The alkaline phosphatase enzymatic activity stain was performed using the Leukocyte Alkaline Phosphatase Kit (Sigma-Aldrich), which was modified to perform in tissue culture plates. Briefly, wells containing 7-day differentiated cells were washed twice in 1 × phosphate-buffered saline (PBS), fixed in 3.75% paraformaldehyde (v/v in distilled water) for 15 min on ice, washed three times in 1 × PBS, and allowed to dry completely. In a separate tube, 500 μL of 100mM sodium nitrite solution were mixed with 500 μL of 400mM FRV-alkaline solution (fast red violet LB base; v/v in hydrochloric acid) and allowed to stand for 2 min at room temperature. The solution was diluted 1:10 with double-distilled water, and 500 μL of naphthol AS-BI solution (naphthol AS-BI phosphate; 4 mg/mL in 2M 2-amino-2-methyl-1,3-propanediol [AMPD] buffer, pH 9.5) were added. One mL of the mixed solution was added to each well and allowed to incubate for 30 min in the dark. The plates were gently rinsed with distilled water, dried, and scanned.
Transfection of miRNA mimics and siRNAs
The miR-219a-5p (Catalog #MSY0000664; 5’-UGAUUGUCCAAACGCAAUUCU-3’), miR-135a-5p (Catalog #MSY0000147; 5’-UAUGGCUUUUUAUUCCUAUGUGA-3’), and negative control (Catalog #YM00479902; 5’-UCACCGGGUGUAAAUCAGCUUG-3’) double-stranded miRNA mimics were purchased from Qiagen. For the experiment illustrated in Fig. 3E, the miRNA mimics were transfected into primary mouse CalOBs using the 4D-Nucleofector (Lonza, Basel, Switzerland) according to the manufacturer’s instructions. Specifically, 1 × 106 cells at approximately 70% confluence were trypsinized and transfected with 100nM of either mimic (n = 6) and plated into a 12-well plate. Cells were harvested in QIAzol (Qiagen) at either 6 or 24 hours following transfection for Rorβ expression analysis. For the experiment illustrated in Fig. 5, 20nM negative control or miR-219a-5p mimic (n = 6) were transfected using HiPerFect Transfection Reagent (Qiagen) according to the manufacturer’s instructions. The next day, the media were replaced with osteogenic differentiation media and cultured for 7 days (changing the media at days 3 and 6), where they were harvested in QIAzol. For the siRNA experiment illustrated in Fig. 6B to D, 20nM control or Rorβ-specific siRNAs (Dharmacon, Lafayette, CO, USA; n = 6) were transfected using HiPerFect Transfection Reagent; the cells were cultured and harvested exactly as for the miRNA mimic experiments.
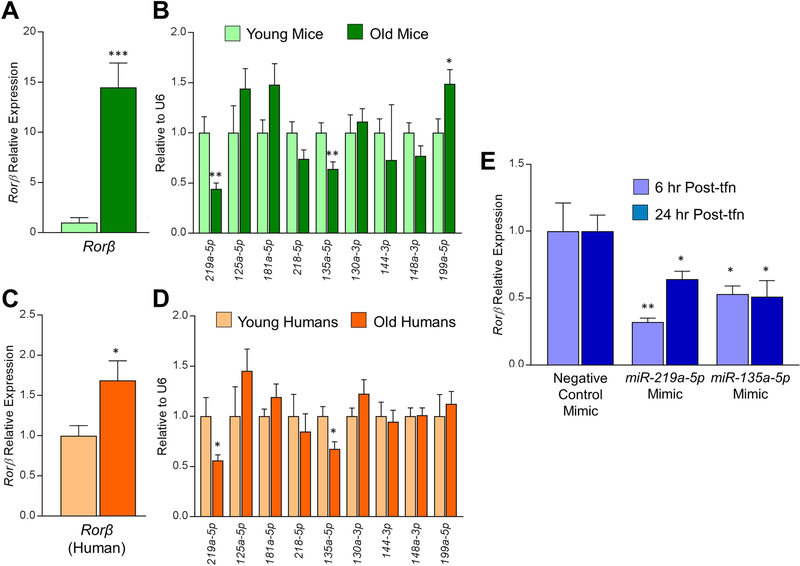
Fig. 3.
miR-219a-5p and miR-135a-5p expression decline in aging bone in both mice and humans. (A, B) RNA was isolated from collagenase-digested vertebrae from either young (6-months-old) or old (24-months-old) male mice (n = 10) and subjected to qPCR analysis for: (A) Rorβ or (B) the panel of nine miRNAs predicted to regulate Rorβ. (C, D) Similarly, RNA isolated from needle biopsy bone core samples from younger (mean age 27 years) or older (mean age 78 years) women (n = 10) was also subjected to the same qPCR analysis for (C) Rorβ or (D) the same panel of nine miRNAs. (E) Synthetic miRNA mimics for miR-219a-5p, miR-135a-5p, or a negative control mimic were transfected into primary murine calvarial osteoblasts and RNA was isolated either 6 or 24 hours after transfection (Post-tfn) and qPCR analyses for Rorβ expression was performed. Data represent mean ± SEM. *p < 0.05; **p < 0.01; ***p < 0.001 relative to negative control mimic control transfection (independent samples t test).
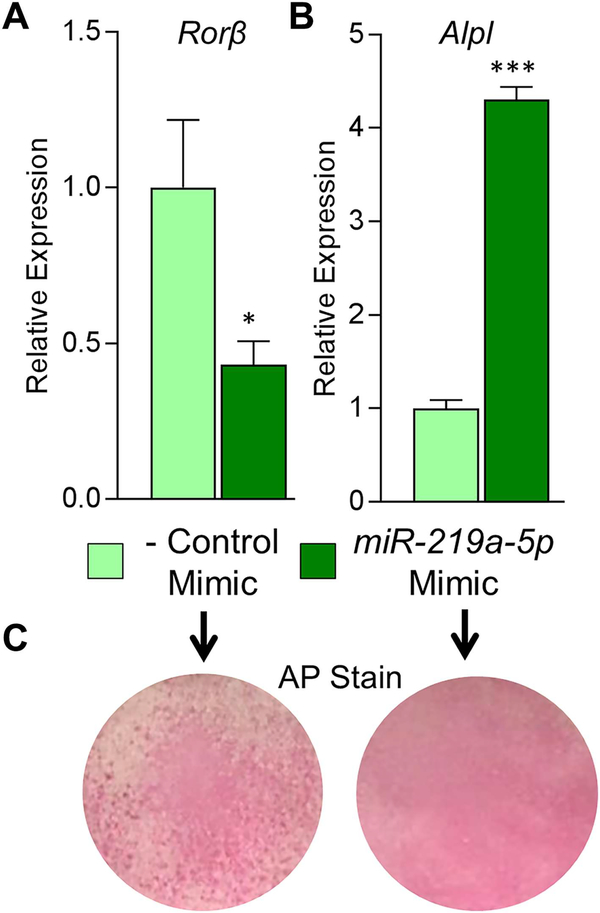
Fig. 5.
A miR-219a-5p mimic enhances the osteogenic phenotype in osteoblasts. (A) Negative control (−) or miR-219a-5p miRNA mimics were transfected into CalOB cells and assayed for Rorβ expression using qPCR. (B) The transfected cells were cultured in osteogenic differentiation medium for 7 days and Alpl expression assessed using qPCR. (C) Alkaline phosphatase enzymatic activity (AP stain) was assessed at 7 days of differentiation for both transfection conditions. Data represent mean ± SEM. *p < 0.05; ***p < 0.001 relative to negative control miRNA mimic (independent samples t test).
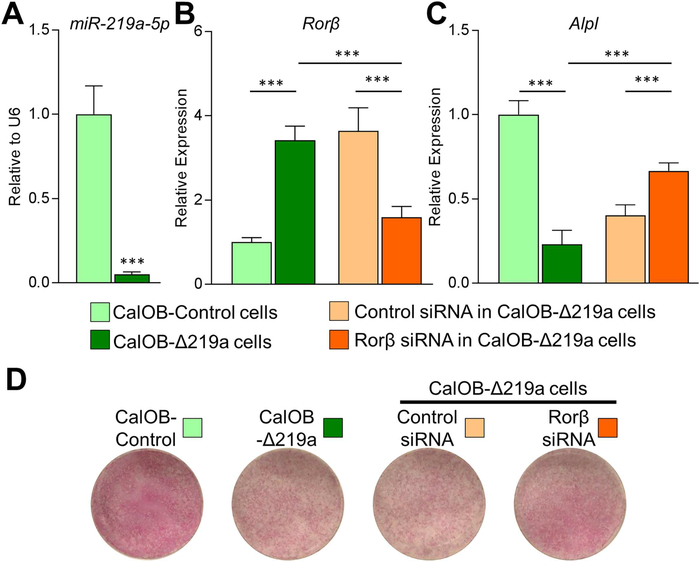
Fig. 6.
A miR-219a-5p inhibitor suppresses the osteogenic phenotype in osteoblasts. Control murine calvarial osteoblasts (CalOB-Control) and CalOB-Δ219a cells were assayed for (A) miR-219a-5p and (B) Rorβ expression using qPCR. (C) CalOB and CalOB-Δ219a cells were cultured in osteogenic differentiation medium for 7 days and Alpl expression levels were assayed using qPCR (green bars). Control or Rorβ-specific siRNAs were transfected into CalOB-Δ219a cells, cultured in osteogenic differentiation medium for 7 days, and assayed for Rorβ or Alpl expression (panels B–C, orange bars). (D) Alkaline phosphatase enzymatic activity was assessed at 7 days of differentiation for the indicated conditions. Data represent mean ± SEM. ***p < 0.001 (ANOVA followed by Tukey’s post hoc test).
Rorβ−3’UTR luciferase report construct and assay
To construct the 3’UTR reporter vector, the luciferase reporter gene luc2P was excised from the pGL4.11[luc2P] vector (Promega, Madison, WI, USA) as a HindIII/XbaI fragment. The luc2P reporter gene was used because it contains a C-terminal hPEST domain, a protein destabilization sequence, which allows for quicker and greater magnitude responses. The luc2P fragment was subcloned into the HindIII/XbaI sites of pcDNA/4TO (Promega) to make the 3’UTR reporter vector, Luc2P-4TO. The entire mouse Rorβ 3’UTR sequence (7279 nucleotide bases in length; Genbank Accession NM_001043354), was synthesized as four approximately equal-sized gBLOCKs (Integrated DNA Technologies, Coralville, IA, USA) and assembled into the XbaI site of Luc2P-4TO using Gibson Assembly Master Mix (New England BioLabs, Ipswich, MA, USA) to produce Rorβ−3’UTR-Luc. To assess the effects of miR-219a-5p on the Rorβ 3’UTR, 100 ng of Rorβ−3’UTR-Luc were cotransfected with 100nM of either negative control or miR-219a-5p miRNA mimics (Qiagen) into primary mouse CalOBs (n = 6) at approximately 70% confluence using DharmaFECT DUO transfection reagent (Dharmacon), a reagent specifically made for cotransfection of DNA and miRNAs. Twenty-four hours later, protein and luciferase assays were performed as previously described.(26) The data are presented as luciferase activity per μg protein and normalized to the negative control miRNA mimic.
miRNA/mRNA pulldown assays
The miRNA pulldown assay was modified from Phatak and Donahue.(27) Briefly, custom miRIDIAN 3’-end biotinylated (*) double-stranded miRNA mimics for mmu-miR-219a-5p (5’-UGAUUGUCCAAACGCAAUUCU-3’*) and the same negative control miRNA sequence (Dharmacon), as described in the Transfection of miRNA mimics and siRNAs subsection (5’-UCACCGGGUGUAAAUCAGCUUG-3’*), were prepared. One million cells at approximately 70% confluence were trypsinized and transfected, using the 4D-Nucleofector (Lonza), with 100nM of either of the biotinylated mimics (n = 3) for 6 hours. Fifty μL of streptavidin-dyna beads (M-280; ThermoFisher Scientific) were sequentially washed twice using a tube magnet with solution A (0.1M NaOH [w/v] and 0.05M [w/v] NaCl in nuclease-free double-distilled water [ddH2O]), twice with solution B (0.1M NaCl [w/v] and 70% ethanol [v/v] in ddH2O) and three times with lysis buffer (20mM Tris-HCl [pH 7.5] [w/v], 100mM KCl [w/v], 5mM MgCl2 [w/v], and 0.3% IGEPAL [v/v] in ddH2O). All chemicals were purchased from Sigma-Aldrich unless otherwise stated. The beads were then pretreated with 10-μL yeast tRNA (10 mg/mL; ThermoFisher Scientific) in lysis buffer for 2 hours at 4°C and washed once with lysis buffer. Following the 6-hour transfection procedure, the cells were washed twice in ice-cold 1 × PBS and lysed in 550 μL of lysis buffer (supplemented with Halt Protease Inhibitor and Riboblock RNase Inhibitor, both from Thermo-Fisher Scientific). Following a 10-min lysis on ice, the samples were centrifuged at 14,000 rpm and the supernatant saved. Fifty μL of each sample were saved as an “input” control; the remaining 500 μL were mixed with the prepared streptavidin beads overnight at 4°C. The bead-sample mixture was placed on the tube magnet and the beads washed three times with lysis buffer. The RNA was extracted by the addition of QIAzol (Qiagen). RNA isolation and cDNA synthesis were performed as described above on the whole RNA sample for the experimental pulldown, with 1 μg of the input control for each replicate. The data were calculated by first normalizing each pulldown to its own input control using 2Input-Pulldown. The three replicate samples for the miR-219a-5p and a negative control miRNA pulldowns were averaged and normalized to the negative control miRNA pulldown. These calculations gave enrichment values for the miR-219a-5p pulldown over the negative control miRNA pulldown.
Statistics
All values are expressed as mean ± SEM. Mean values were compared between two groups and performed by an independent samples t test. Differences were considered significant at p < 0.05 (two-tailed test). For comparison among more than two groups, ANOVA was performed, followed by the Tukey’s post hoc test, to adjust for multiple comparisons. Analyses were performed using the Statistical Package for the Social Sciences for Windows (Version 22.0; SPSS, Chicago, IL, USA); figures were created using GraphPad Prism (Version 5.03; GraphPad Software, San Diego, CA, USA).
Results
Identification of miRNAs predicted to regulate Rorβ
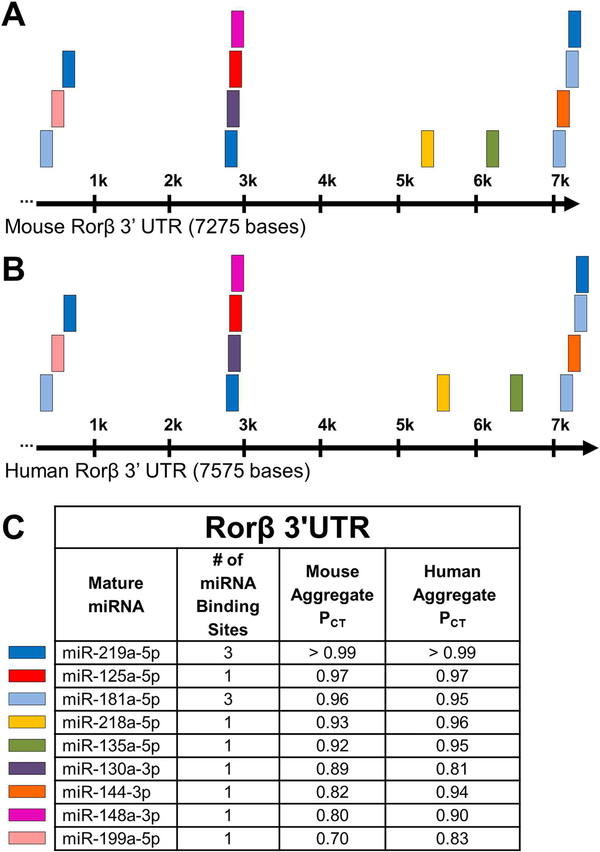
miRNAs typically regulate gene expression through recognition and interaction with short seed sequences (typically seven to eight nucleotide bases) contained within the 3’UTRs of their cognate target genes.(10–12) Because both mouse and human Rorβ mRNAs contain relatively large 3’UTRs of approximately 7.3 kb and 7.6 kb, respectively (Fig. 1A, B), many miRNAs would be predicted to recognize the Rorβ 3’UTR with varying efficiencies. To reduce the list down to the most highly predicted miRNA/Rorβ mRNA interactions, we used the TargetScanMouse (Release 7.1; laboratory of David Bartel [Massachusetts Institute of Technology, Cambridge, MA, USA] in conjunction with the Whitehead Institute for Biomedical Research [Cambridge, MA, USA]) online prediction tool(28) to identify miRNAs that have the highest probability of conserved targeting (PCT score).(29) We chose those nine miRNAs with the highest prediction scores (using a PCT cutoff ≥0.70) and ranked them separately in mice and humans (Fig. 1C). The predicted miRNA target sites were remarkably conserved between both species in terms of miRNA identity, number of miRNA binding sites, and location within the 3’UTR, suggesting a strong evolutionary conservation and regulatory function of Rorβ expression.
Fig. 1.
Predicted miRNA target sites in the 3’UTR of mouse and human Rorβ mRNA. (A, B) Schematic representation of both mouse and human Rorβ 3’UTRs, with the length of each denoted in nucleotide bases. (C) Table of the top nine miRNAs with a PCT score ≥0.70 against both mouse and human Rorβ mRNA 3’UTR. The mature miRNA name, number of binding sites, and the aggregate PCT scores are listed. On the left of the table, color-coded boxes represent the location of each miRNA within the mouse and human Rorβ mRNA 3’UTR from the panels above.
miRNA expression and activity in osteoblasts and aging
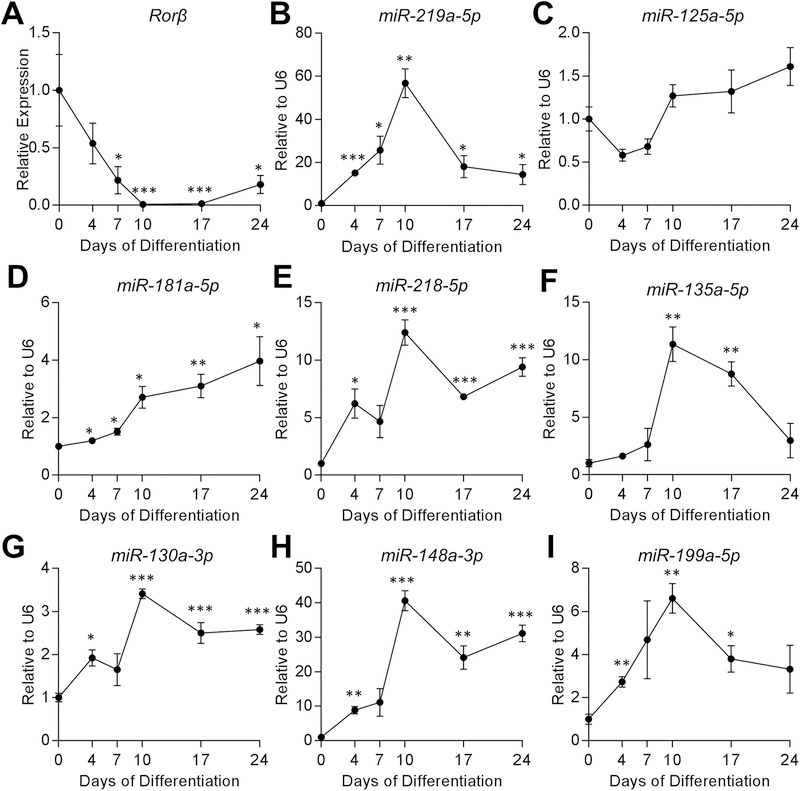
To begin to understand the regulatory function of these miRNAs with Rorβ expression and in osteoblast differentiation, we first conducted an osteoblast differentiation assay using WT primary murine CalOBs. The mRNA transcripts for Alpl (Supplemental Fig. 1A), Bglap (Supplemental Fig. 1B), Col1a1 (Supplemental Fig. 1C), and Runx2 (Supplemental Fig. 1D) increased throughout the time course, demonstrating a normal osteoblast-differentiation gene-expression phenotype. As anticipated based on previous reports,(8) Rorβ transcripts dramatically declined throughout the osteoblast differentiation time course (Fig. 2A). If any of these miRNAs do have a role in the regulation of Rorβ mRNA stability, which is one of the possible outcomes of miRNA-mediated silencing of gene expression,(10,11) then the miRNA would be hypothesized to increase. Consistent with this hypothesis, miR-219a-5p (Fig. 2B), miR-181a-5p (Fig. 2D), miR-218–5p (Fig. 2E), miR-135a-5p (Fig. 2F), miR-130a-3p (Fig. 2G), miR-148a-3p (Fig. 2H), and miR-199a-5p (Fig. 2I) significantly increase to varying levels throughout the time course (compared with day 0), whereas miR-125a-5p expression (Fig. 2C) was unchanged and miR-144–3p was not expressed in CalOBs (data not shown). Remarkably, these data indicate that seven of the nine (78%) highest predicted miRNAs for Rorβ targeting follow a pattern consistent with Rorβ expression during osteoblast differentiation (eg, miRNA expression increased and Rorβ expression decreased).
Fig. 2.
Rorβ expression decreases, whereas expression of multiple miRNAs predicted to target Rorβ increases during osteoblastic differentiation. (A) Primary murine CalOBs were cultured in osteogenic differentiation medium and qPCR performed for Rorβ mRNA expression on samples collected at 0, 4, 7, 10, 17, and 24 days of differentiation (n = 6). (B–I) Similarly, expression analyses of the following miRNAs were also performed on the same osteoblast differentiation time course; (B) miR-219a-5p, (C) miR-125a-5p, (D) miR-181a-5p, (E) miR-218–5p, (F) miR-135a-5p, (G) miR-130a-3p, (H) miR-148a-3p, and (I) miR-199a-5p. Data represent mean ± SEM. *p < 0.05; **p < 0.01; ***p < 0.001 relative to day 0 (independent samples t test).
To further refine the number of possible miRNAs that may be involved in the regulation of Rorβ levels, we took advantage of the fact that Rorβ expression dramatically increases in the bone cortex (containing mostly osteocytes and osteoblasts) during aging in both mice(9) and humans.(7) Following the same logic as in Fig. 2, if these miRNAs have a role in the regulation of Rorβ levels during aging, then they would be hypothesized to decrease in aged bone samples. Therefore, we profiled all nine of the miRNAs in young (6-month-old) and old (24-month-old) male mice, along with Rorβ expression. Although Rorβ levels in bone samples increased approximately 14-fold with age (Fig. 3A), only miR-219a-5p and miR-135a-5p decreased in old mice (Fig. 3B). Similarly, in primary bone core samples isolated from younger premenopausal women (mean age 27 years) and older postmenopausal women (mean age 78 years), where Rorβ levels increased ~1.6-fold (Fig. 3C), again only miR-219a-5p and miR-135a-5p decreased significantly in aging bone (Fig. 3D). The striking congruency of the downregulation of miR-219a-5p and miR-135a-5p expression in aging bone in both humans and mice, where Rorβ expression is increased, strongly suggests that these two miRNAs regulate Rorβ expression in bone. Indeed, transient transfection of synthetic miR-219a-5p and miR-135a-5p miRNA mimics into CalOBs resulted in significant suppression of Rorβ mRNA expression after 6 and 24 hours following transfection (Fig. 3E). It is important to note that although miR-135a-5p expression also inversely correlated with Rorβ expression during osteoblast differentiation and in aging bone, we chose to focus the next set of experiments on miR-219a-5p because it was the top predicted miRNA that regulates Rorβ in both mice and humans.
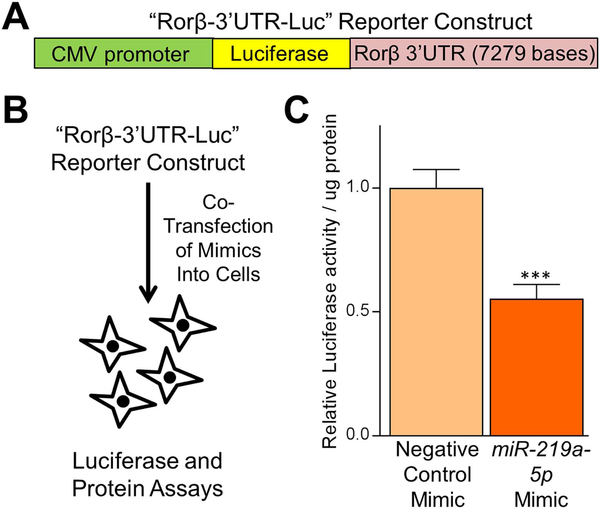
To assess the activity of miR-219a-5p in Rorβ 3’UTR regulation, we utilized a reporter assay where the 3’UTR of Rorβ was inserted 3’ of a luciferase reporter gene (Fig. 4A). In this assay (Fig. 4B), if the cotransfected miRNA mimic regulates the 3’UTR and causes mRNA degradation and/or translational repression, then luciferase activity should decline. Indeed, cotransfection of the Rorβ 3’UTR reporter construct with miR-219a-5p miRNA mimic resulted in a significant suppression of luciferase activity (Fig. 4C).
Fig. 4.
A miR-219a-5p miRNA mimic regulates a Rorβ−3’UTR luciferase reporter construct. (A) Schematic diagram of the Rorβ−3’UTR-Luc reporter construct and (B) the miRNA/reporter assay in primary mouse CalOBs. (C) Relative luciferase activity per μg protein was measured 24 hours following cotransfection of the Rorβ−3’UTR-Luc reporter with either negative control or miR-219a-5p miRNA mimics. Data represent mean ± SEM. ***p < 0.001 relative to negative control miRNA mimic (independent samples t test).
MiR-219a-5p enhances osteoblastic differentiation
We continued our analysis of miR-219a-5p and Rorβ regulation by determining whether transfection of a miR-219a-5p mimic into WT CalOB cells enhances osteoblast activity. CalOBs were transfected with either a negative control miRNA mimic or a miR219a-5p mimic, where Rorβ mRNA levels decreased by approximately 60% (Fig. 5A). The cells were then cultured in osteogenic differentiation medium for 7 days, where the miR-219a-5p mimic increased Alpl mRNA levels by 4.3-fold (Fig. 5B) and increased alkaline phosphatase staining (Fig. 5C).
We then performed the converse experiment, where either a negative control miRNA inhibitor or a miR-219a-5p inhibitor was introduced into WT CalOBs, producing the CalOB-Control or CalOB-Δ219a cell lines, respectively. As shown in Fig. 6A, miR219a-5p expression was significantly decreased by 95% in the CalOB-Δ219a cell model when compared with CalOB-Control cells, whereas Rorβ mRNA levels significantly increased by 3.4-fold (Fig. 6B, green bars). Additionally, Alpl mRNA expression was decreased by 77% (Fig. 6C, green bars) and alkaline phosphatase enzymatic activity was also noticeably decreased in the CalOB-Δ219a cell model as compared with CalOB-Control cells (Fig. 6D). To determine whether increased Rorβ levels in CalOB-Δ219a cells contribute to the suppressed osteogenic phenotype, control or Rorβ-specific siRNAs were transfected into CalOB-Δ219a cells. Rorβ mRNA levels decreased by 60% with the Rorβ specific siRNA (Fig. 6B, orange bars) and Alpl mRNA levels increased significantly (Fig. 6C, orange bars), relative to the control siRNA. The alkaline phosphatase staining in these conditions confirmed these observations (Fig. 6D). It should be noted that Alpl expression was not rescued to the same levels as observed in CalOB-Control cells, suggesting that factors other than Rorβ also contribute to the CalOB-Δ219a osteoblastic phenotype. Taken together, these results demonstrate that inhibition of miR-219a-5p suppresses the osteogenic phenotype, which is partially based on increased Rorβ.
Interaction of miR-219a-5p with Rorβ mRNA in osteoblasts
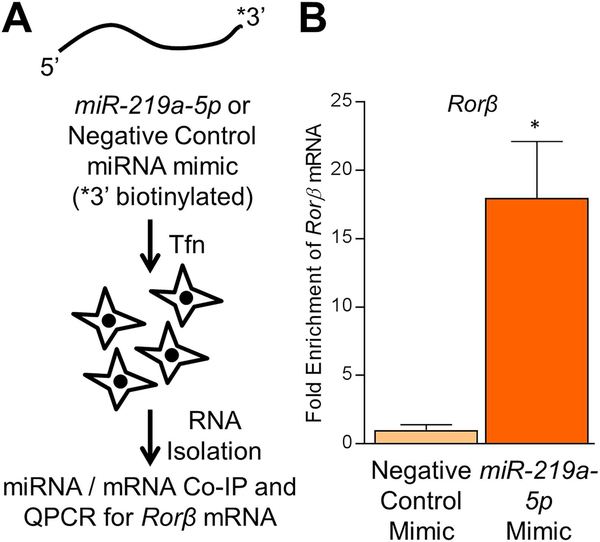
Finally, to confirm that miR-219a-5p directly targets Rorβ mRNA through direct interaction, we conducted a miRNA/mRNA pulldown assay using a transfected biotinylated miR-219a-5p mimic or biotinylated negative control mimic into WT CalOBs, followed by qPCR for the presence of the Rorβ mRNA (Fig. 7A). As shown in Fig. 7B, pulldown with the biotinylated miR-219a-5p mimic resulted in a 17-fold enrichment of Rorβ mRNA over the biotinylated negative control miRNA mimic. This demonstrates that Rorβ mRNA is a bona fide target of miR-219a-5p in osteoblasts through direct physical interaction.
Fig. 7.
miR-219a-5p interacts with Rorβ mRNA in osteoblasts. (A) Schematic diagram of the miRNA/mRNA pulldown assay used in this study. (B) Fold enrichment of Rorβ mRNA by the biotinylated miR-219a-5p mimic (over the biotinylated negative control mimic) was determined by qPCR. Data represent mean ± SEM. *p < 0.05 relative to the negative control mimic (independent samples t test).
Discussion
The goal of this study was to identify potential miRNAs that regulate Rorβ levels, both in an in vitro osteoblastic cell model and in vivo in both mouse and human bone fractions during physiological aging. We found that miR-219a-5p fits these criteria, as its expression increased during osteoblastic differentiation and decreased in both mouse and human bone during aging, exhibiting a reciprocal pattern to Rorβ expression. Furthermore, a miR-219a-5p mimic decreased Rorβ expression and enhanced osteoblast differentiation, whereas a miR-219a-5p inhibitor increased Rorβ expression and suppressed osteoblast differentiation. These data suggest that miR-219a-5p is bone anabolic, partially through suppression of Rorβ. Finally, we demonstrate that miR-219a-5p and Rorβ mRNA physically interact using a RNA-based pulldown assay. Collectively, these data demonstrate that miR-219a-5p regulates Rorβ levels in osteoblasts and bone, and that miR-219a-5p could represent a novel tool to suppress Rorβ activity and promote bone anabolism in the context of age-related bone disorders, such as osteoporosis.
It is estimated that over 1500 distinct miRNAs exist in both mice and humans, which regulate expression of approximately 60% of all genes in nearly every tissue,(10,29,30) including bone.(13–18) MiRNAs exert their regulatory function by triggering mRNA degradation and/or translational repression through the recognition of short miRNA seed sequences contained within the 3’UTR of target genes.(11,12) In general, genes with longer 3’UTRs are more likely to be regulated by miRNAs simply based on their size.(31) For example, expression of the isoform of Nurr1 with the longer 3’UTR is under more miRNA control than the shorter isoforms.(32) Therefore, it is not surprising that Rorβ, with its long 3’UTR lengths of approximately 7.3 kb and approximately 7.6 kb in mice and humans, respectively, would likely be highly controlled by miRNAs. Using the TargetScan miRNA site prediction software,(28) we showed that many miRNAs are predicted to target Rorβ with varying efficiencies in both mice and humans. Interestingly, the identity and approximate location of the highly predicted miRNAs between mice and humans are also well conserved, providing evolutionary evidence of the importance of miRNA-mediated regulation of Rorβ expression. Of these sets of potential Rorβ-regulating miRNAs, one particular miRNA, miR-219a-5p, not only displayed the highest PCT score (≥0.99) of all miRNAs for Rorβ 3’UTR recognition, but also recognizes three independent seed sequences in the Rorβ 3’UTR. This lends credence to the notion that miR-219a-5p would be predicted to have a significant function in the regulation of Rorβ expression levels.
The regulation of Rorβ expression is an important aspect of osteoblast biology, as its expression naturally decreases during differentiation and its constitutive expression of Rorβ inhibits differentiation.(8) Our data suggest that miR-219a-5p, as well as a number of other miRNAs (Fig. 2), might have involvement in the regulation of Rorβ mRNA levels during osteoblast differentiation, as they increase in expression while Rorβ decreases. These other miRNAs may work in concert with miR-219a-5p to achieve this expression pattern, resulting in a cumulative effect on Rorβ mRNA levels. However, the effects of these same miRNAs on Rorβ mRNA levels in bone may be quite different in the context of aging. Contrary to what we observed in differentiating osteoblasts, the expression of only miR-219a-5p and miR-135a-5p changed in a manner consistent with increased Rorβ mRNA levels (eg, decreased miRNA expression) in both mice and humans, suggesting that the complete array of miRNAs controlling Rorβ expression during osteoblast differentiation and in aging bone are different.
Only a few reports have examined miR-219a-5p function in other systems, which may provide clues to its function in bone. Chang and colleagues demonstrated that miR-219a-5p is involved in vestibular compensation and that the predicted targets of miR-219a-5p were related to the processes of intracellular signal transmission, calcium/phosphate signaling, and cell–extracellular matrix interactions,(30) processes also known to be involved in bone metabolism. Kocerha et al. showed that miR-219a-5p negatively regulates the physiological function of N-methyl-D-aspartate glutamate receptors in the brain, which regulates neurotransmission and synaptic plasticity by downregulation of calcium/calmodulin-dependent protein kinase II γ subunit(33) that also has an important regulatory function in endochondral bone growth.(34) Interestingly, Dugas and colleagues demonstrated that miR-219a-5p is required for normal oligodendrocyte differentiation in the brain and is an inhibitor of cellular proliferation.(35) Because we know that Rorβ inhibits osteoblast differentiation and increases cellular proliferation,(8) it is possible that miR-219a-5p may play a similar role in osteoblasts by promoting osteoblast differentiation through the downregulation of Rorβ levels. This is supported by our data that demonstrate that miR-219a-5p plays an anabolic role in osteoblast differentiation. Based on these observations, we propose that one function of miR-219a-5p is to suppress Rorβ levels and therefore promote differentiation. The function of miR-219a-5p in the context of aging bone may be similar; however, further experimentation (eg, miR-219a-5p transgenic mouse models) would be needed to clarify this role in vivo.
A limitation of this study is that only miR-219a-5p function was tested in calvarial osteoblasts, and not miR-135a-5p. Granted that miR-135a-5p levels also increased during osteoblast differentiation and decreased during aging, we chose to focus on miR-219a-5p for the following reasons. First, miR-219a-5p was the top predicted Rorβ-regulating miRNA in both mice and humans, with a PCT score of ≥0.99. Second, three independent seed sequences for miR-219a-5p exist in the Rorβ 3’UTR, where only one exists for miR-135a-5p. Therefore, we considered miR-219a-5p as the prime candidate for further study. However, this does not preclude the possibility that miR-135a-5p may also be involved in Rorβ regulation during differentiation and in aging, although future studies would be necessary to characterize its role and whether it functions in unison with miR-219a-5p in Rorβ regulation. Another limitation is that we were not able to examine correlations between the expression of miR-219a-5p in aging mouse and human bone fractions, as we did not obtain assessments of bone mass on these cohorts. Although bone loss in both mice and humans at these ages is well characterized,(4,36) future studies would be needed to directly correlate miR-219a-5p expression with bone mass.
One of the most interesting aspects of this study is the striking congruency of miR-219a-5p expression during physiological aging in bone of both mice and humans. This is exemplified by the observation that Rorβ levels increase in osteocyte-enriched bone fractions isolated both from mice(8,9) and in bone core biopsies in humans(7) during aging. Thus, Rorβ and miR-219a-5p levels change similarly between mice and humans. Collectively, these findings strongly suggest that an identifiable Rorβ/miR-219a-5p axis exists that is conserved in mammalian evolution. This opens up the possibility that clinical therapies involving the delivery of miR-219a-5p to osteoblast lineage cells in the context of human aging, where Rorβ levels are high and promote bone catabolism,(8) may be a useful approach in attenuating bone loss during aging.
Supplementary Material
Acknowledgments
This work was supported by NIH Grants R01 AR068275 (DGM), P01 AG004875 (SK/DGM), R01 AG048792 (SK/DGM), K01 AR070241 (JNF), K01 AR070281 (MMW), R01 AR049069 (AJV), and career development awards (JNF, MMW) from the Mayo Clinic Robert and Arlene Kogod Center on Aging. We would like to thank Mr. James Peterson for data analysis and Drs. John Hawse and Malayannan Subramaniam (Mayo Clinic) for the primary CalOBs used in this study.
Footnotes
Disclosure
All authors state that they have no conflicts of interest.
Additional Supporting Information may be found in the online version of this article.
References
- 1.Almeida M, O’Brien CA. Basic biology of skeletal aging: role of stress response pathways. J Gerontol A Biol Sci Med Sci. 2013;68(10): 1197–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonewald LF. The amazing osteocyte. J Bone Miner Res. 2011;26(2): 229–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jilka RL, O’Brien CA. The role of osteocytes in age-related bone loss. Curr Osteoporos Rep. 2016;14(1):16–25. [DOI] [PubMed] [Google Scholar]
- 4.Khosla S Pathogenesis of age-related bone loss in humans. J Gerontol A Biol Sci Med Sci. 2013;68(10):1226–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wright NC, Looker AC, Saag KG, et al. The recent prevalence of osteoporosis and low bone mass in the United States based on bone mineral density at the femoral neck or lumbar spine. J Bone Miner Res. 2014;29(11):2520–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Long F Building strong bones: molecular regulation of the osteoblast lineage. Nat Rev Mol Cell Biol. 2011;13(1):27–38. [DOI] [PubMed] [Google Scholar]
- 7.Roforth MM, Khosla S, Monroe DG. Identification of Rorβ targets in cultured osteoblasts and in human bone. Biochem Biophys Res Comm. 2013;440(4):768–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roforth MM, Liu G, Khosla S, Monroe DG. Examination of nuclear receptor expression in osteoblasts reveals Rorβ as an important regulator of osteogenesis. J Bone Miner Res. 2012;27:891–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farr JN, Weivoda MM, Nicks KM, et al. Osteoprotection through the deletion of the transcription factor Rorβ in mice. J Bone Miner Res. 2018;33(4):720–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–97. [DOI] [PubMed] [Google Scholar]
- 11.Kim VN, Nam JW. Genomics of microRNA. Trends Genet. 2006;22(3): 165–73. [DOI] [PubMed] [Google Scholar]
- 12.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He X, Eberhart JK, Postlethwait JH. MicroRNAs and micromanaging the skeleton in disease, development and evolution. J Cell Mol Med. 2009;13(4):606–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kobayashi T, Lu J, Cobb BS, et al. Dicer-dependent pathways regulate chondrocyte proliferation and differentiation. Proc Natl Acad Sci U S A. 2008;105(6):1949–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kronenberg HM. PTHrP and skeletal development. Ann N Y Acad Sci.2006;1068:1–13. [DOI] [PubMed] [Google Scholar]
- 16.Gaur T, Hussain S, Mudhasani R, et al. Dicer inactivation in osteoprogenitor cells compromises fetal survival and bone formation, while excision in differentiated osteoblasts increases bone mass in the adult mouse. Dev Biol. 2010;340(1):10–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y, Xie RL, Gordon J, et al. Control of mesenchymal lineage progression by microRNAs targeting skeletal gene regulators Trps1 and Runx2. J Bio Chem. 2012;287(26):21926–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davis C, Dukes A, Drewry M, et al. MicroRNA-183–5p increases with age in bone-derived extracellular vesicles, suppresses bone marrow stromal (stem) cell proliferation, and induces stem cell senescence. Tissue Eng Part A. 2017;23(21–22):1231–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dole NS, Delany AM. MicroRNA variants as genetic determinants of bone mass. Bone. 2016;84:57–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lian JB, Stein GS, van Wijnen AJ, et al. MicroRNA control of bone formation and homeostasis. Nat Rev Endocrinol. 2012;8(4): 212–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fujita K, Xing Q, Khosla S, Monroe DG. Mutual enhancement of differentiation of osteoblasts and osteocytes occurs through direct cell–cell contact. J Cell Biochem. 2014;115(11):2039–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farr JN, Roforth MM, Fujita K, et al. Effects of age and estrogen on skeletal gene expression in humans as assessed by RNA sequencing. PLoS One. 2015;10:e0138347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Modder UI, Oursler MJ, Khosla S, Monroe DG. Wnt10b activates the Wnt, notch, and NFkappaB pathways in U2OS osteosarcoma cells. J Cell Biochem. 2011;112(5):1392–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Farr JN, Fraser DG, Wang H, et al. Identification of senescent cells in the bone microenvironment. J Bone Miner Res. 2016;31(11): 1920–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fujita K, Roforth MM, Demaray S, et al. Effects of estrogen on bone mRNA levels of sclerostin and other genes relevant to bone metabolism in postmenopausal women. J Clin Endocrinol Metab. 2014;99:E81–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Modder UI, Rudnik V, Liu G, Khosla S, Monroe DG. A DNA binding mutation in estrogen receptor-a leads to supression of Wnt signaling via b-catenin destabilization in osteoblasts. J Cell Biochem. 2012;113(7):2248–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Phatak P, Donahue JM. Biotinylated micro-RNA pull down assay for identifying miRNA targets. Bio-protocol. 2017;7(9):e2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19(1): 92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chang MY, Park S, Choi JJ, et al. MicroRNAs 218a-5p, 219a-5p, and 221–3p regulate vestibular compensation. Sci Rep. 2017;7(1): 8701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheng C, Bhardwaj N, Gerstein M. The relationship between the evolution of microRNA targets and the length of their UTRs. BMC Genomics. 2009;10:431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pereira LA, Munita R, Gonzalez MP, Andres ME. Long 3’UTR of Nurr1 mRNAs is targeted by miRNAs in mesencephalic dopamine neurons. PLoS One. 2017;12(11):e0188177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kocerha J, Faghihi MA, Lopez-Toledano MA, et al. MicroRNA-219 modulates NMDA receptor-mediated neurobehavioral dysfunction. Proc Natl Acad Sci U S A. 2009;106(9):3507–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Y, Ahrens MJ, Wu A, Liu J, Dudley AT. Calcium/calmodulin-dependent protein kinase II activity regulates the proliferative potential of growth plate chondrocytes. Development. 2011;138(2):359–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dugas JC, Cuellar TL, Scholze A, et al. Dicer1 and miR-219 Are required for normal oligodendrocyte differentiation and myelination. Neuron. 2010;65(5):597–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Glatt V, Canalis E, Stadmeyer L, Bouxsein ML. Age-related changes in trabecular architecture differ in female and male C57BL/6J mice. J Bone Miner Res. 2007;22(8):1197–207. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.