Abstract
Functional genomics networks are widely used to identify unexpected pathway relationships in large genomic datasets. However, it is challenging to quantitatively compare the signal-to-noise ratio of different networks, the biology they describe, and to identify the optimal network to interpret a particular genetic dataset. Via GeNets users can train a machine-learning model (Quack) to make such comparisons; and they can execute, store, and share analyses of genetic and RNA sequencing datasets.
With the significant technological advances in epigenetics, proteomics and single-cell RNA sequencing it is now possible to generate an unprecedented amount of tissue- and cell-type-specific functional genomics data that can be conveniently represented as gene networks. In these networks, genes are connected if they are functionally correlated or interacting in any of the aforementioned data types, and this representation of complicated data sets can lead to the discovery of biological relationships that would otherwise have been missed (reviewed in 1 and exemplified in 2). Importantly, combining functional genomics networks with exome-sequencing data, or genome-wide association studies, is a cost-efficient and scalable way to identify draft cellular circuits that are enriched for genetic risk in a particular disease (reviewed in 1, exemplified in 3; for a full discussion of the potential of biological networks in genome interpretation and biological discovery see Supplementary Note 1). These draft circuits can then be followed up in a targeted and cost-efficient manner both computationally and experimentally which can lead to new biological insight or focus drug-target discovery (Reviewed in 1 and exemplified in 4, 5).
However, different networks (e.g., those generated from different cell or tissue types) vary considerably in their signal-to-noise ratio as well as global and local biological signal. Here, we use the term ‘biological signal’ to describe how well a network recapitulates the functional relationships between genes that are known to be in the same pathways based on prior knowledge generally accepted by the scientific community. By global signal we denote the ability of a network to recapitulate functional relationships across hundreds of core human pathways (e.g., RNA splicing or pathways involved in the cell-cycle). Local signal refers to the signal across a subset of pathways related to a more specific user-defined biological area (e.g., neurodevelopment, osteogenesis, or blood lipid biology). At the technical level this variability means that networks diverge significantly in which genes are covered by data (i.e., which genes are connected to others in the network in question), their density (i.e., how many connections a gene has to other genes) and their topology (i.e., in the specific patterns of how genes are connected to each other).
Consider a scientist who is interested in applying a network-based approach to studying genes implicated in autism spectrum disorders (ASDs) and who has generated a RNA sequencing dataset from a specific set of neurons which can be represented as a gene network. First, this scientist should determine the global signal of the network to confirm its overall signal to noise ratio. Second, it would be valuable to know the local biological signal of that network across pathways relevant to autism (e.g., across neurodevelopmental pathways), and to make detailed comparisons of these metrics and those for other analogous networks existing in the public domain. Once both the overall quality of the network and the local signal across relevant pathways has been investigated, pathway analysis algorithms can be applied to the network to explore potentially new pathway relationships between the autism gene set. However, there is currently no technology that 1) enables users to compare the global and local biological signal of networks taking into consideration signal-to-noise, coverage, density, and unique topology; and 2) leverages the optimal network for pathway analyses that can be visualized, stored, managed, and shared with collaborators. This creates a significant bottleneck in exploiting tissue- and cell-type-specific networks for biological discovery in many areas of biomedicine.
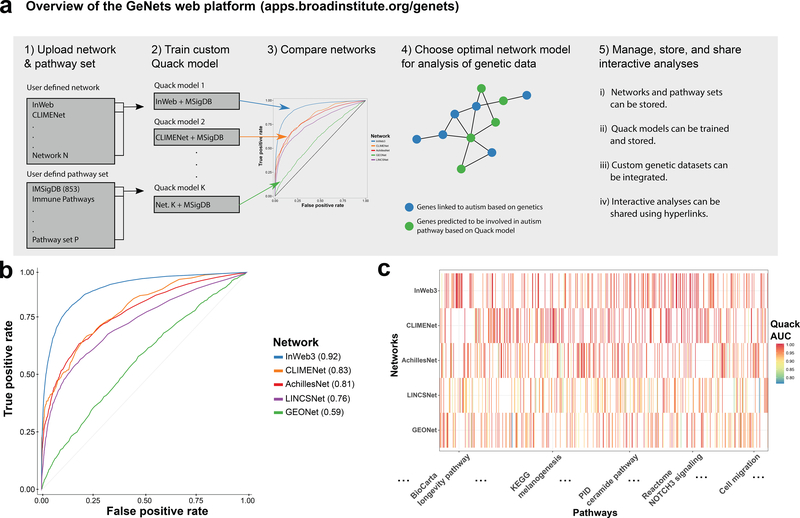
To address these key barriers, we have developed the ‘Broad Institute Web Platform for Genome Networks’ (GeNets, http://apps.broadinstitute.org/genets), with some of its uniquely enabling features listed in Fig. 1a. To make detailed comparisons of the global and local biological signals of any user-defined network, we designed a fast and efficient machine learning method (Quack) that can learn the topological patterns of pathway sets in any network defined by the user. For example, using the InWeb6 protein-protein interaction network and 853 expertly curated pathways from the Molecular Signatures Database (http://software.broadinstitute.org/gsea/msigdb/, Supplementary Data 1), Quack tests 18 different topological properties of how genes in each of these pathways are connected to each other in the protein-protein interaction data (for all details on the 18 topological properties and how Quack is trained see Online Methods, Supplementary Notes 2-6, Supplementary Table 1, Supplementary Figures 1-11). We train Quack on 70% of the data from these pathways and then, for a given pathway set and evaluate its ability to predict the 30% of the genes we hold out. The InWeb-specific Quack model gives an area under the receiver operating characteristics curve (AUC) of 0.92 across the 853 MSigDB pathways.
Figure 1 |. Features of the GeNets web platform.
a) GeNets overview. b) AUCs of five heterogeneous networks as determined by the Quack machine-learning algorithm. Each model was trained on N = 597 pathways (70% of the 853 curated MSigDB pathways). c) Local biological signal of five networks (rows) across 730 pathways (columns). Colors as indicated in the color key and cells are blank if genes in a pathway were not covered by enough connections in the network in question for Quack to determine an AUC. Interactive view with all pathway names and more details is available from the GeNets Dashboard.
To compare the global signal of InWeb to other networks - and to exemplify the broad utility of Quack across different types of functional genomics networks commonly used in biomedicine - we used the same 853 MSigDB pathways to train Quack models for another four networks based on i] mRNA expression patterns in tissue samples from the Gene Expression Omnibus7 (GEONet), ii] cancer co-dependency relationships from project Achilles8 (AchillesNet), iii] phylogenetic patterns from inferred models of evolution9 (CLIMENet), and iv] cell perturbation profiles of eight cell lines from the LINCS project10 (LINCSNet, see Online Methods and Supplementary Note 3 for details on the networks). The network-specific Quack models reveal that the five networks generally have a good global biological signal (median AUC = 0.81, Fig. 1b, where the local biological signal across individual pathways can be seen in Fig. 1c).
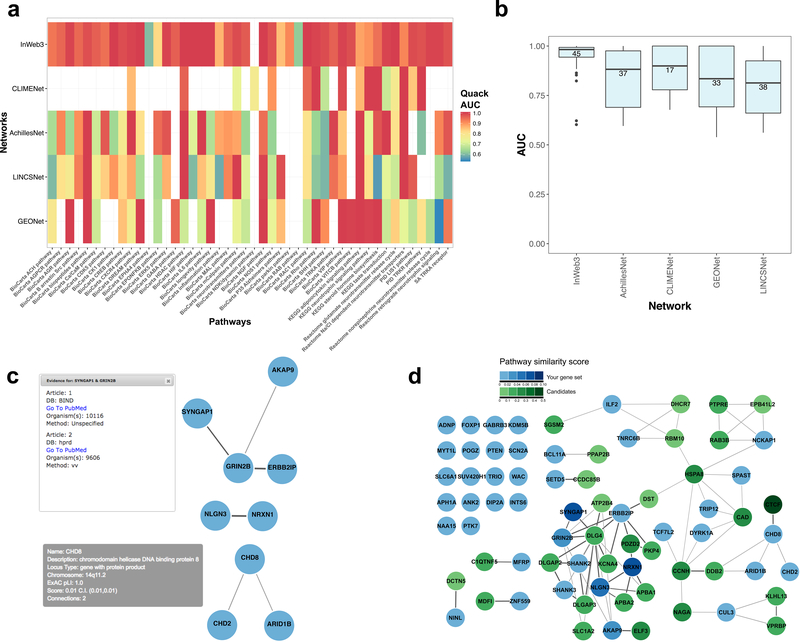
To explore which of the five networks is optimal for exploring pathway relationships between 65 genes involved in ASD11, we extracted information on the local biological signals across a set of neurological and neurodevelopmental pathways from the five Quack models we had trained previously. This analysis shows that InWeb has particularly high AUCs across this set of pathways (Figs. 2a and b). In addition to testing global and local biological signals of networks, another feature of Quack is its ability to explore pathways of user-defined gene sets by mapping and evaluating their topological connections to a seed set of genes (Online Methods, Supplementary Notes 2-6, Supplementary Table 1, Supplementary Figures 1-11). Specifically, using the 65 ASD genes as a seed set, the InWeb-specific Quack model predicts 31 ASD candidates because they topologically connect in a way that, based on the training procedure of Quack, suggests the 31 genes and the seed set coalesce into pathways together (Figs. 2 c and d).
Figure 2 |. Using GeNets to explore pathways implicated in autism spectrum disorders (ASD).
a) Heat map of the local biological signal of the five networks across neurological and neurodevelopmental pathways [determined by training network-specific Quack models]. b) AUC distributions of neurological pathways represented in the five networks. Only pathways with enough connections for Quack to determine an AUC are included, and their numbers are indicated in each network. Center line, median; box limits, upper and lower quartiles; whiskers, 1.5× interquartile range; points, outliers. c) Direct protein-protein interactions (from InWeb) between 65 genes implicated in ASD (where only genes with interactions are shown). Upon clicking the edges in GeNets, there is a direct link to the publication supporting the relevant data exemplified here with the SYNGAP1-GRIN2B protein interaction. The bottom box illustrates information available upon mouse over of genes in the network exemplified with CHD8. d) Thirty-one potential autism candidate proteins (green) based on protein-protein interactions to 65 ASD input genes (light blue) after training of a neurodevelopmental-specific Quack Model. Darker green means higher confidence candidate as indicated.
We further explored these 31 candidates, by using a GeNets visualization feature to cross-reference proteins the network that have been genetically linked to psychiatric and neurodevelopmental phenotypes through GWAS or exome sequencing 12–13 (Supplementary Figure 12a). The overlap of the 31 candidates and these independent genetic datasets is significant (P < 0.05 using a hypergeometric distribution, overlapping genes from three different loci are CTCF, NAGA, SGSM2). This provides some support that the InWeb-specific Quack model predicts candidates linked to ASDs. We annotated the genes in the network that are under brain-specific regulation using expression quantitative trait loci (eQTL) from the GTEx project (Version 6, http://www.gtexportal.org/home/) (Supplementary Figure 12b) and together, both the independent genetic data and the brain-specific eQTL data converge on NAGA suggesting it could be an interesting autism candidate gene. A targeted literature analysis of NAGA shows that mutations in this gene have been implicated in Schindler disease14, which has overlapping symptoms with ASDs.
The five networks highlighted in this manuscript are available in GeNets and users can upload and test any network they choose by training network-specific Quack models. Furthermore, we provide pre-trained analyses and visualizations of 853 MSigDB pathways and 168 GWAS datasets from the NIH GWAS catalog (https://www.genome.gov/gwastudies/ ) and a number of predefined gene annotations that help users interpret genetic data. For example, upon hovering over network nodes users are presented with descriptions of the genes, the Quack-determined probabilities of belonging to a pathway with the user-defined seed set, and also a metric of their genetic intolerance to loss of function mutations as determined in large population genetic studies from the Exome Aggregation Consortium15. This is a useful metric to have available directly from the browser when interpreting clinical exome-sequencing data through the GeNets framework. More information on specific applications of GeNets can be found in the ‘how GeNets can help you’ guide in the web platform.
Other excellent and very successful network-analysis packages and tools like Cytoscape16, STRING17, GeneMANIA18, SANTA19, and IMP20 exist. However, a unique strength of GeNets is that it enables users to train a custom machine learning model on any network, to compare the signal of networks (both globally and locally) and to manage, store, and share results of analyses that we illustrate above, in Fig. 1a, and in the online tutorial: http://apps.broadinstitute.org/genets#users/userguide. See also Supplementary Notes 7-10 and Supplementary Figures 13-15 for a more detailed comparison of GeNets to other methods and a discussion of the platforms strengths and weaknesses. Furthermore, the Quack algorithm is available as an open source software package from https://github.com/lagelab/quack so it can be seamlessly incorporated in any functional genomics analysis pipeline.
Overall, GeNets enables a very broad group of expert and non-expert users alike to i) upload networks, ii) train network-specific machine learning models, iii) make detailed comparisons of the global and local biological signal of the many biological networks that are now emerging with the ongoing revolution in large-scale functional genomics approaches. The technology also provides a framework for pathway analyses of genetic data. We believe that as more and more genetic and network datasets become available the value of GeNets will continue to increase.
Data Availability
GeNets visualizations and pre-loaded network data can be accessed on http://apps.broadinstitute.org/genets. Quack algorithm is detailed in Supplementary Note 6 and provided as a R package on https://github.com/lagelab/quack. The 853 canonical pathways used to train Quack models in this manuscript are obtained from MSigDB and provided as Supplementary Data 1. Further data that support the findings of this study are available from the corresponding author upon request.
Online Methods
Designing and training the Quack algorithm to compare networks.
Hypothesis:
For a given network, we hypothesized that genes in a common pathway would share pathway-specific topological properties that systematically distinguish them from genes that are not part of the pathway in question.
Exemplification of Quack:
Using the InWeb network and the Biocarta AKT pathway to exemplify our approach, we defined six topological metrics that describe the relationships of a gene (e.g., AKT1) to other genes in the same pathway (i.e., betweenness centrality in pathway, weighted degree in pathway, clustering coefficient in pathway, closeness centrality in pathway, eigenvector centrality in pathway, and degree in pathway; see below for a detailed description of these metrics). The analogous six metrics for AKT1 in the overall InWeb network (e.g., the betweenness centrality in the overall network) were also computed and a ratio between the pathway-specific metric and the overall network metric was derived (e.g., betweenness centrality in pathway / betweenness centrality in overall network). Expanding this calculation to all genes in the AKT pathway resulted in a total of 18 metrics being calculated for each of the 21 AKT pathway genes. To look for topological properties that systematically distinguished AKT pathway genes from other genes in InWeb, we also computed these metrics for 2,449 genes that are in the context of the AKT pathway. Hereafter, we define the context of a specific pathway (e.g., the AKT pathway) in a specific network (e.g., InWeb) as all genes that are not part of that pathway set, but have least one connection to a gene in the pathway under investigation. This resulted in a set of 21 data points for each topological metric for the AKT pathway genes and 2,449 data points for each topological metric for the AKT context genes. This data was then used to show the topological differences between the AKT pathway members and context genes (see Supplementary Figure 1 for conceptual exemplification and Supplementary Figures 2-7 for full datasets). To systematically map the topologies of many pathways in InWeb, we repeated the analysis above for 853 pathways from the MSigDB database.
Topological signature distinguish genes in a common pathway from their context:
A univariate analysis of the distributions of scores for pathway genes versus context genes for each of the 18 metrics (Supplementary Figure 1b) confirms our hypothesis that there are topological signatures that clearly distinguish genes that together form a pathway in InWeb, from genes that are not part of the pathway in question. Expanding this analysis to all five networks revealed two pathway topological principles: First, in all networks, the distributions of these metrics are generally different between pathway genes and context genes (6 of 18 metrics illustrated in Supplementary Figure 1b-c and the complete set for each network shown in Supplementary Figures 2-7). This means that when considered on the background of a complex set of network properties, genes in a common pathway have a topological signature that distinguishes them from other genes in the network. Second, we observe differential pathway topologies in the five networks, meaning that for each network, the distributions of topological metrics for pathway members form a network-specific signature (partial signature with 6 of 18 metrics illustrated in Supplementary Figure 1d and complete signatures in Supplementary Fig 2-7).
Ensuring non-redundancy in pathway datasets.
We ensured that the training pathway data was non redundant using the following approach: Among 1,329 C2:CP gene sets in MSigDB, we calculated pairwise Jaccard index, and in cases where Jaccard index > 0.5, we randomly selected one pathway from the two and removed it. Repeating this procedure, we obtained 853 pathways with pairwise Jaccard index <=0.5. Below is the distribution of pairwise Jaccard Index in the resulting 853 pathways, where 99.1% of pathway pairs have Jaccard Index <=0.15 and 93.3% have a Jaccard Index <= 0.05.
Network topological metrics used by Quack.
Let G=(V,E) be a graph with vertex set V and edge set E. |V| = N is the number of vertices in the graph and |E| = M is the number of edges. Let A be defined as the adjacency matrix of G, i.e., the N × N matrix such that non-diagonal entries a_vw are positive real numbers (which depends on the network, see Supplementary Note 5 for interpreting edge weights), and the diagonal elements are all zero (in all networks edges between the same gene [self interactions or self loops] are disregarded). Degree: the degree of a vertex v is defined as the number of vertices directly connected to v (i.e., direct neighbors or just “neighbors”). Weighted degree: the weighted degree, also called the “strength”, is defined as the sum of the weights of the edges which connect the neighbors to v. Clustering coefficient: the clustering coefficient of a vertex v relates to the tendency of its first order interactors to also interact with each other. Technically it is defined as C_v = 1/(s_v*(k_v-1))*sum((wgt_vw+wgt_vu)/2 *a_vw * a_vu * a_wu) across w, u. Here, s_v is the strength of vertex v, 1/(s_v*(k_v-1)) is the normalization factor, a_vw is an adjacency indicator a_vw={0: no edge; 1: edge exists, k_v is the vertex degree, wgt_vw are the weights. C_v is continuous on [0,1]. As C_v approaches 1, the neighbors of v are becoming fully connected to one another. As C_v approaches 0, the neighbors of v are not well connected (i.e., a star with v in the middle has C=0). Closeness centrality: the closeness centrality of vertex v is a measure of how close it is to all other vertices in the network. It is defined as (N-1)/sum( shortest_path(v,w), v != w), the inverse of the average shortest path length all the other vertices w in the graph. Betweenness centrality: the betweenness of vertex v is a measure of how many shortest paths between the graphs vertices go through v. It is defined as sum( spath_uvw / spath_uw, u!=w,u!=v,w!=v), where spath_uw is total number of shortest paths from node u to node w and spath_uvw is the number of those paths that pass through v. Eigenvector centrality: the eigenvector centrality of the vertex v is defined as x_v = 1/lambda * sum(a_vw*x_w) where lambda is the eigenvalue corresponding to the principal eigenvector (the eigenvector for which all entries are positive), a_vw is the value of the adjacency matrix corresponding to vertices v and w, and x_w is the component of the principal eigenvector corresponding to vertex w.
Computing topological metrics for pathway and context genes.
The six topological metrics are computed for genes (i.e., vertices) both: 1) within a pathway using only the sub-network formed by the pathway genes, 2) for the genes using the entire functional network. Additionally, the ratios of (within pathway / entire network) are computed for these metrics as well. When the denominator is zero the ratio is set to zero, otherwise, the natural logarithm ln(ratio) is computed. Therefore in total, 6 × 3 = 18 metrics are calculated for each gene. The full list of metrics can be seen in Supplementary Table 2.
Networks used in this work.
We created and used pre-existing networks from the following sources (see all details in Supplementary Notes 2 and 3): 1. gene-gene correlations based on mRNA expression patterns in 19,019 tissue samples from the Gene Expression Omnibus7 (GEONet, hereafter); 2. cancer codependency relationships across 216 cancer cell lines from project Achilles8 (AchillesNet, hereafter); 3. phylogenetic relationships from ‘clustering of inferred models of evolution’ between genes in 502 species9 (CLIMENet, hereafter); 4. cell perturbation profiles from eight cell lines from the LINCS project10 (LINCSNet, hereafter); and 5. 428,429 protein-protein interactions between 12,509 human proteins6 (from the InWeb database). Supplementary Table 1 provides a summary of final network sizes after pre-processing. For details of the pre-processing, including the removal of indirect edges from matrix data21–22, thresholding edge scores and optimizing sparse network sizes (see Supplementary Notes 2 and 3).
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported in part by grants from National Institutes of Health: HHSN268201000033C and R01HL096738 from NHLBI and U24CA160034 from NCI Clinical Proteomics Tumor Analysis Consortium initiative to SAC. HH was supported by a Fund for Medical Discovery Award from the Executive Committee On Research at Massachusetts General Hospital. HH and KL are supported by the MGH IRG American Cancer Society. KL, AK, TL and HH are supported by a grant from the Stanley Center at the Broad Institute, a Broadnext10 grant from the Broad Institute, 1R01MH109903, U01-DK078616, 5P01HD068250-07, a Large Thematic Project Grant from the Lundbeck Foundation, and a Research Award from the Simons Foundation (SFARI).
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
REFERENCES
- 1.Lage K Protein-protein interactions and genetic diseases: The interactome. Biochim. Biophys. Acta - Mol. Basis Dis. 1842, 1971–1980 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li T et al. A scored human protein-protein interaction network to catalyze genomic interpretation. Nat. Methods 14, 61–64 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Greene CS et al. Understanding multicellular function and disease with human tissue-specific networks. Nat. Genet. 47, 569–76 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lundby A et al. Annotation of loci from genome-wide association studies using tissue-specific quantitative interaction proteomics. Nat. Methods 11, 868–74 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Okada Y et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature 506, 376–381 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lage K et al. A human phenome-interactome network of protein complexes implicated in genetic disorders. Nat. Biotechnol. 25, 309–16 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Edgar R, Domrachev M & Lash AE Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–10 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cowley GS et al. Parallel genome-scale loss of function screens in 216 cancer cell lines for the identification of context-specific genetic dependencies. Sci. data 1, 140035 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Y, Calvo SE, Gutman R, Liu JS & Mootha VK Expansion of biological pathways based on evolutionary inference. Cell 158, 213–25 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lamb J The Connectivity Map: a new tool for biomedical research. Nat. Rev. Cancer 7, 54–60 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Sanders SJ et al. Insights into Autism Spectrum Disorder Genomic Architecture and Biology from 71 Risk Loci. Neuron 87, 1215–1233 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schizophrenia Working Group of the Psychiatric Genomics, C. Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McRae JF et al. Prevalence and architecture of de novo mutations in developmental disorders. Nature (2017). doi: 10.1038/nature21062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clark NE & Garman SC The 1.9 a structure of human alpha-N-acetylgalactosaminidase: The molecular basis of Schindler and Kanzaki diseases. J. Mol. Biol. 393, 435–47 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lek M et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shannon P et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Szklarczyk D et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43, D447–52 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zuberi K et al. GeneMANIA prediction server 2013 update. Nucleic Acids Res. 41, W115–22 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cornish AJ & Markowetz F SANTA: quantifying the functional content of molecular networks. PLoS Comput. Biol. 10, e1003808 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wong AK, Krishnan A, Yao V, Tadych A & Troyanskaya OG IMP 2.0: a multi-species functional genomics portal for integration, visualization and prediction of protein functions and networks. Nucleic Acids Res. 43, W128–33 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barzel B & Barabási A-L Network link prediction by global silencing of indirect correlations. Nat. Biotechnol. 31, 720–5 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feizi S, Marbach D, Médard M & Kellis M Network deconvolution as a general method to distinguish direct dependencies in networks. Nat. Biotechnol. 31, 726–33 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
GeNets visualizations and pre-loaded network data can be accessed on http://apps.broadinstitute.org/genets. Quack algorithm is detailed in Supplementary Note 6 and provided as a R package on https://github.com/lagelab/quack. The 853 canonical pathways used to train Quack models in this manuscript are obtained from MSigDB and provided as Supplementary Data 1. Further data that support the findings of this study are available from the corresponding author upon request.