Abstract
Small-cell lung cancer (SCLC) has the highest malignancy among all lung cancers, exhibiting aggressive growth and early metastasis to distant sites. For 30 years, treatment options for SCLC have been limited to chemotherapy, warranting the need for more effective treatments. Frequent inactivation of TP53 and RB1 as well as histone dysmodifications in SCLC suggest that transcriptional and epigenetic regulations play a major role in SCLC disease evolution. Here we performed a synthetic lethal screen using the BET inhibitor JQ1 and an shRNA library targeting 550 epigenetic genes in treatment-refractory SCLC xenograft models and identified HDAC6 as a synthetic lethal target in combination with JQ1. Combined treatment of human and mouse SCLC cell line–derived xenograft tumors with the HDAC6 inhibitor ricolinostat (ACY-1215) and JQ1 demonstrated significant inhibition of tumor growth; this effect was abolished upon depletion of NK cells, suggesting that these innate immune lymphoid cells play a role in SCLC tumor treatment response. Collectively, these findings suggest a potential new treatment for recurrent SCLC.
Significance:
These findings identify a novel therapeutic strategy for SCLC using a combination of HDAC6 and BET inhibitors.
Introduction
Small-cell lung cancer (SCLC) is a highly malignant neuroendocrine tumor in lung and accounts for 15%–20% of all primary lung cancers (1, 2). SCLC is strongly associated with cigarette smoking and displays the highest mortality among all types of lung cancer (1, 2). The treatment of SCLC continues to be a challenge; although SCLC has a relatively good initial response to chemotherapy and radiotherapy, relapse and disease progression are extremely common, leading to a 5-year survival rate of less than 2% (3).
In non-SCLC (NSCLC), oncogenic driver mutations have been identified, making molecular-targeted treatment feasible (4, 5). In contrast, SCLC is not linked to currently targetable oncogenic mutations and, instead, is predominantly associated with inactivation of TP53 and RB1, upregulation of MYC expression, as well as abnormal histone modifications (6–9). These findings suggest that epigenetic dysregulation may play a major role in this cancer. Recent strategies to target SCLC by manipulating transcription have shown some efficacy in preclinical models. For example, Christensen and colleagues reported that the transcriptional inhibitor THZ1 inhibits SCLC by targeting super enhancers of certain oncogenic transcriptional factors, including MYC, SOX2, and NFIB (10). Lenhart and colleagues reported that the BET bromodomain inhibitor JQ1 inhibits SCLC by sequestering BRD4 to prevent docking to the ASCL1 enhancer (11). In addition, Gardner and colleagues recently reported that cisplatin- and etoposide-resistant SCLC in PDX mice undergoes EZH2-mediated hypermethylation on SLFN11 (12). These studies suggest that chromatin regulators can provide manageable drug targets.
To explore the potential of epigenetic therapy in SCLC, we took advantage of the technique of synthetic lethality, which has recently contributed to the development of cancer therapeutics, especially for undrugable targets, such as Kras activation or Lkb1 deletion mutants (13–16), and developed a synthetic lethal screening strategy specifically targeting epigenetic genes in a SCLC xenograft model. As part of our screening strategy, we considered proteins of the bromodomain and extra terminal (BET) family that function as transcriptional coactivators and play roles in transcriptional elongation (17). JQ1 is a competitive inhibitor of BET proteins that blocks them from binding to acetylated histones, thus inhibiting gene transcription (18). Inhibition of BET proteins with JQ1 has shown potent antiproliferation effects in hematologic tumors through suppression of c-MYC and downstream target genes (19), in lung adenocarcinoma cells through FOSL1 and its targets (20), as well as in SCLC (11).
To maximize the impact of BET inhibition in SCLC, we screened for novel therapeutic targets using a synthetic lethal strategy with BET inhibitor JQ1 and an shRNA library specifically targeting epigenetic genes in a SCLC xenograft model. Our screen identified HDAC6, which encodes histone deacetylase 6 (HDAC6). HDACs comprise classes I, IIa, IIb, and IV of 18 members and HDAC6 belongs to class IIb (21, 22). HDAC6 is phylogenetically close to class I HDACs, but with a distinct dominant cytoplasmic localization (23, 24), although it has been reported to repress transcriptions via association with other transcriptional regulators (25–29). Our identification of HDAC6 and effective suppression of SCLC with inhibitor ACY-1215 and JQ1 shine a light on a potential new treatment for recurrent SCLC.
Materials and Methods
Cell lines and cell culture
The human SCLC NCI-H69 cell line was obtained from ATCC and the GLC-16 cell line was from our laboratory (10). Murine SCLC RP501 and RP1328 cell lines were established in our laboratory using lung tumor nodules of genetically engineered Rb/p53 mice, and murine RPP41 and RPP394 cell lines were established using lung tumor nodules of Rb/p53/p130 mice. All cell lines were authenticated by DNA fingerprinting and verified as Mycoplasma-free using Universal Mycoplasma Detection Kit (ATCC). NCI-H69 and GLC-16 were cultured in RPMI1640 supplemented with 10% FBS and 1% penicillin/streptomycin. All murine RP and RPP cell lines were cultured in RPMI1640 supplemented with 10% FBS, 1% penicillin/streptomycin, 1% insulin–transferrin–selenium (Gibco), 10 nmol/L hydrocortisone (Sigma), and 10 nmol/L β-estradiol (Sigma). Cell cultures were maintained at 37C in a humid atmosphere containing 5% CO2 and 95% air.
Pooled shRNA/JQ1 screen and analysis
A pooled lentiviral shRNA library was constructed at the Broad Institute in Cambridge, MA, using a subset of The RNAi Consortium (TCR) shRNA library targeting approximately 550 epigenetic-related genes, with an average of 5–7 shRNAs per gene. Detailed pooled shRNA screen and data analysis were performed as described previously (16). In brief, target cells were infected with the pooled lentiviral shRNA library. One aliquot of shRNA-positive cells was immediately saved for analysis of the initial population, and the remaining cells were injected subcutaneously in the dorsal flank region of athymic nude mice. One day after injection, 5 mice were treated with vehicle and another 5 mice were treated with JQ1until the tumor reached approximately 1cm in diameter. Genomic DNA was extracted from the initial cell aliquot and xenograft tumors, and shRNA abundance was quantified by deep sequencing. The ratio of abundance of each shRNA in vehicle-treated and JQ1-treated versus initial shRNA was ranked on the basis of the rank of weighted second best score (25% weight for ranked top shRNAs + 75% weight for second-best shRNAs; http://www.broadinstitute.org/cancer/software/ GENE-E; ref. 30).
Mouse treatment studies
All mouse studies were conducted through Institutional Animal Care and Use Committee–approved animal protocols. Unless otherwise stated, SCLC cells were injected into the dorsal flank region of athymic nude mice (Charles River Laboratories) or NOD-scid IL2Rγnull (NSG) mice (The Jackson Laboratory) at 2 × 106 human cells or 1–2 × 106 mouse cells per implantation. Once tumors were palpable, mice were randomized into treatment arms and tumor volume was assessed by caliper 1–2 times per week, depending on tumor growth rate.
ACY-1215 was provided by Acetylon Pharmaceuticals, Inc. and JQ1 was provided by the James Bradner laboratory at Dana Farber Cancer Institute (Boston, MA). Each drug was prepared in the following solvents: ACY-1215, 10% DMSO and 4.5% dextrose in H2O; JQ1, 10% DMSO and 9% 2-hydroxylpropyl β-cyclodextrin in H2O; vehicle, 10% DMSO and 4.5% dextrose, or 9% 2-hydroxylpropyl β-cyclodextrin in H2O. All drugs were administered intraperitoneally, and, when combinations were administered, we allowed a 30-minute interval between administration of different drugs.
Both anti-asialo GM1 and control rabbit sera were diluted 1:10 in PBS and then intraperitoneally injected at 100 μL per mouse.
qRT-PCRs
Total RNA was extracted from cultured cells using TRIzol (Invitrogen). To generate cDNA, 1 mg total RNA was reverse transcribed using the ImProm-II RT system (Promega) as per the manufacturer’s instructions. Real-time quantitative PCR (qRT-PCR) reactions were performed in a final volume of 20 μL, containing 10 μL of 2 × SYBR Green PCR Master Mix (Applied Biosystems), 1 μL of 10 μmol/L forward primer, 1 μL of 10 μmol/L reverse primer, and cDNA corresponding to 45 ng RNA using StepOnePlus Real-Time PCR System (Applied Biosystems) as instructed in the manufacturer’s protocol. All reactions were performed in triplicate. qPCR primers were designed using Primer3 software (http://bioinfo.ut.ee/primer3/): HDAC6 (forward/reverse) 5′-ATGCCCAGACTATCAGTGGG/ATAGCACACTGGGGTCATCC-3′; ACTB (forward/reverse) 5′-GTCTTCCCCTCCATCGTG/TACTTCAGGG TGAGGATGCC-30. All qPCR reactions were performed in triplicate.
Antibodies
In vivo use rabbit anti-mouse asialo GM1 antibody was from Wako Chemicals and rabbit sera were from Sigma. IHC use biotinylated anti-cleaved caspase-3 antibody was from Abcam. Flow cytometry use PerCP/Cy5.5-conjugated mAbs to CD45; PerCP/Cy5.5 or APC/Cy7-conjugated mAbs to CD3 (17A2); AF488-conjugated mAbs to CD49b (DX5); BV421 or PE/Cy7-conjugated mAbs to CD335 (Nkp46); PE/Cy7-conjugated mAbs to CD103; FITC-conjugated mAbs to H2; AF700-conjugated mAbs to IA/IE; AF488-conjugated anti-human CD326 (EpCAM) antibody; PE/Cy7-conjugated anti-human HLA-A,B,C antibody; and APC/Cy7-conjugated anti-human HLA-DR antibody were from Biolegend. Aqua live/dead dye was from Life Technologies.
Immunostainings and flow cytometry
For immunostaining, xenograft tumors were cut into small pieces, dissociated in RPMI1640 containing 100 IU/mL collagenase type IV (Invitrogen) and 50 μg/mL DNase I (Roche) for 45 minutes at 37C, and then gently squeezed through 70-mm cell strainer to generate single-cell suspension. Spleens were mashed in PBS/2% FBS and then filtered with 70-μm cell strainer to generate single-cell suspension. After centrifugation, cells were resuspended into red blood cell lysis butter (Gibco/Thermo Fisher Scientific) for 3 minutes at room temperature and then resuspended in PBS/2% FBS. A total of 5 × 106tumor cells in 100 μL PBS or 1 × 106 spleen cells in 50 mL PBS were stained with fluorophore-conjugated antibodies (diluted at 1:100 for mouse cell staining or at 1:20 for human cell staining) for 20 minutes. Cells were fixed with 2% paraformaldehyde (eBioscience) for 1 hour at 4C in the dark, washed with PBS/2% FBS, resuspended in 150 μL PBS. Cells were acquired using a BD LSRFortessa flow cytometer and analyzed using BD FACSDiva software.
For IHC, graft tumors were fixed with 10% formalin for overnight and then stored in 70% ethanol. Tumor samples were embedded in paraffin, sectioned at 5 mm, and then stained for cleaved caspase 3.
Statistical analysis
Statistical analyses were carried out using GraphPad Prism 7. All numerical data are presented as mean ± SEM. Grouped analysis was performed using two-way ANOVA. Column analysis was performed using one-way ANOVA or Student t test. P < 0.05 was considered statistically significant.
Results
Identification of HDAC6 as a potential target for combination therapy with JQ1 in SCLC xenograft model
Knowing that SCLC is predominantly driven by epigenetic dysregulation (6), we performed an in vivo synthetic lethal screen using a pooled shRNA library targeting approximately 550human epigenetic genes (Supplementary Table S1) in the presence or absence of BET inhibitor JQ1 to identify novel therapeutic targets (Fig. 1A). To set up in vivo screening conditions, eight individual SCLC cell lines with similar genetic characteristics but varying treatment histories (Supplementary Table S2) were examined for their ability to form xenograft tumors and sensitivity of the resulting tumors to JQ1 in athymic nude mice. Among the cell lines tested, most “prior treatment” SCLC cell lines, including NCI-H69, NCI-H82, GLC-16, and GLC-19, formed xenograft tumors, but with variable sensitivities to JQ1, from moderately resistant (GLC-16) to moderately sensitive (GLC-19 and NCIH69; Supplementary Fig. S1). To identify combination strategies independent of JQ1 sensitivity, we chose two cell lines, JQ1-resistent GLC-16 and JQ1-sensitive NCI-H69, for screening use.
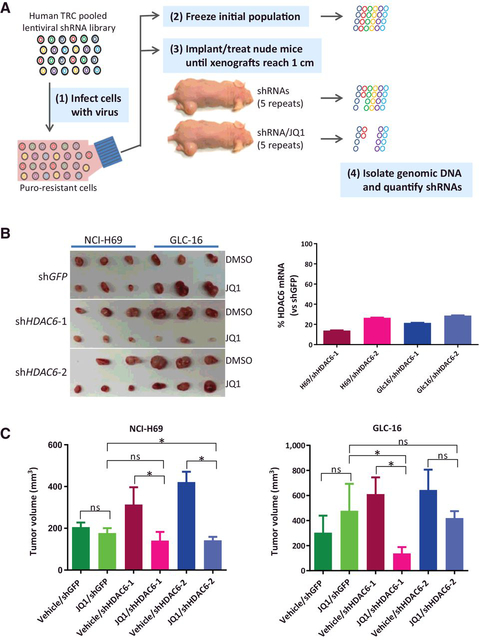
Figure 1.
Identification of HDAC6 as a potential target for combination therapy with the BET bromodomain inhibitor JQ1 in SCLC. A, Schematic overview of the pooled shRNA/JQ1 screen. 1, Target cells are infected with a pooled lentiviral shRNA library targeting approximately 550 human epigenetic genes and shRNA-positive cells are selected with puromycin. 2, One aliquot of the infected cells was immediately frozen for analysis of the initial population. 3, Another aliquot of the cells was subcutaneously injected into the right flank of athymic nude mice. Mice were then treated daily with vehicle or JQ1 at 25 mg/kg until xenograft tumors reached a size of approximately 1 cm in diameter. 4, Xenograft tumors were collected and genomic DNA was extracted from tumors, lentiviral shRNA cassettes were PCR-amplified, and individual shRNA abundance was quantified by deep sequencing. B, Confirmation of shHDAC6/JQ1 synthetic lethal interaction in SCLC xenograft tumors. SCLC NCI-H69 and GLC-16 cells were infected with lentiviral-shHDAC6–1, shHDAC6–2, or shGFP and selected with puromycin for 2 days. Cells were collected, confirmed for HDAC6 knocking down by qRT-PCR (graph on right), and then subcutaneously implanted into athymic nude mice. Mice were treated with vehicle or JQ1 until xenograft tumors in the control group reached approximately 1 cm in diameter. C, Quantification of tumor volume in B. Error bars, SD. ns, not significant (*, P < 0.05).
There were 13 merged hits in the top 10% of ranked genes from NCI-H69 and GLC-16 xenograft tumors (Supplementary Fig. S2A and S2B; Supplementary Table S3). We selected HDAC6, PAX5, STAG1, and YEATS4 for gene-specific validation for their shRNA ranking being greatly enhanced by JQ1 (Supplementary Fig. S2C). We used two shRNAs per hit (Supplementary Fig. S3A) to validate the synergistic lethal interaction of shRNA and JQ1 in vivo. Our data confirmed that JQ1 synergized with individual shRNAs of HDAC6 and YEATS4, but of STAG1 and PAX5 (Fig. 1B and C; Supplementary Fig. S3B and S3C).Therefore, HDAC6 andYEATS4 were validated hits of the screen.
Combination therapy with HDAC6 inhibitor ACY-1215 and JQ1 synergistically suppresses SCLC growth in preclinical mouse models
Although our shRNA library covered all members of the HDAC family, only HDAC6 knockdown resulted in enhanced sensitivity to JQ1 (Supplementary Fig. S4A). Next, we performed a pilot treatment study in athymic nude mice carrying NCI-H69 xenograft tumors using JQ1 in combination with the HDAC6 inhibitors ACY-1215, ACY-1083, or tubastatin. Although these HDAC6 inhibitors have different selectivities for HDAC6 over class I HDACs, varying from 12- to 15-fold (ACY-1215) to 300- to 1,000-fold (ACY-1083 and tubastatin; Supplementary Fig. S4B), they displayed similar inhibitory effects on NCI-H69 tumor growth when cotreated with JQ1 (Supplementary Fig. S4C), indicating that the different selectivities of HDAC6 inhibitors do not affect their combined inhibitory effect with JQ1 on SCLC. Because the synthetic lethal effect is independent of the selectivity of HDAC6 inhibitors, we chose ACY-1215 in subsequent treatment studies as it is currently being tested in multiple clinical trials (31).
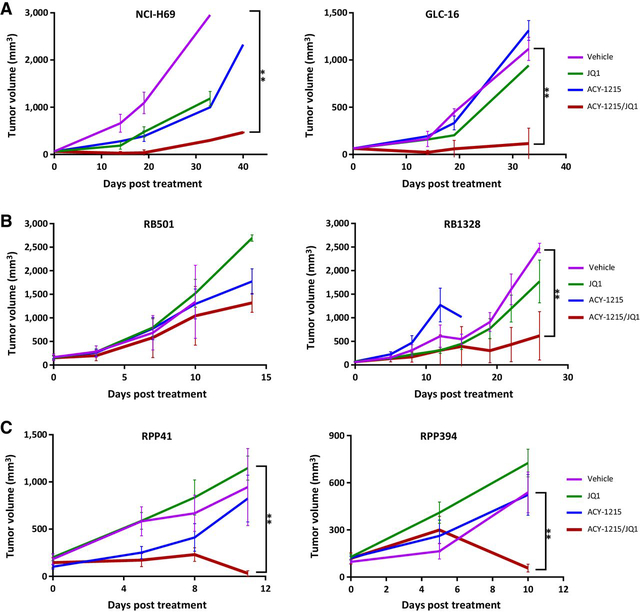
We next evaluated the treatment efficacy of ACY-1215/JQ1 on multiple human/mouse SCLC cell line–derived xenograft/allograft tumors. Consistent with the above shHDAC6/JQ1 results, the ACY-1215/JQ1 combinational treatment significantly suppressed growth of two human SCLC xenograft tumors (Fig. 2A), one of two Rb/p53 mouse SCLC allograft tumors (Fig. 2B), and two Rb/p53/p130 mouse SCLC allograft tumors (Fig. 2C). ACY-1215 or JQ1 treatment alone demonstrated either no or moderate suppression of tumor growth relative to vehicle-treated controls (Fig. 2A–C).
Figure 2.
In vivo efficacy of HDCA6 inhibitor ACY-1215 and JQ1 combination treatment. Athymic nude mice carrying SCLC NCI-H69 or GLC-16 xenograft tumors (A), mouse SCLC Rb/p53 RB501 or RB1328 allograft tumors (B), and mouse SCLC Rb/p53/p130 RPP41 or RPP394 allograft tumors (C) were treated with vehicle, JQ1 (25 mg/kg daily), ACY-1215 (50 mg/kg daily), or both drugs in combination. Tumor volume (mm3) was calculated as (length × width2)/2. Data represent mean ± SD for 5–7 mice (**, P < 0.001).
NK cells mediate ACY-1215/JQ1’s synergistic inhibitory effects on SCLC xenograft tumor growth
To investigate the mechanism underlying ACY-1215/JQ1 suppression of SCLC, we performed RNA-seq and proteomics analyses. Gene expression clustering pattern analysis revealed that JQ1 had a strong effect while ACY-1215 had limited impact on transcription as compared with the vehicle control (Supplementary Fig. S5A). Next, we sorted out the gene lists of synergistic up-/downregulation upon ACY-1215/JQ1 treatment (Supplementary Fig. S5B), followed by gene ontology (GO) analysis of the enriched genes using GOrilla software. All these analyses only revealed one relative significant pathway (GO: 0045670 with a false discovery rate q value of 0.102) associated with upregulation of osteoclast differentiation (Supplementary Table S4), which has been linked to positive regulation of myeloid leukocyte differentiation (32).
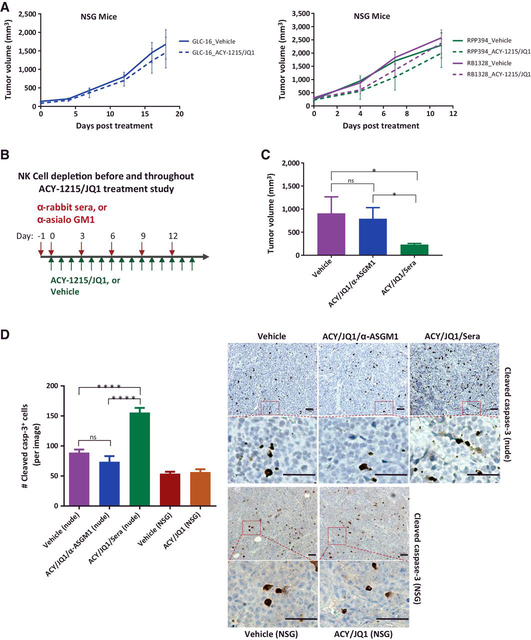
Proteomic analysis of human proteins identified mostly cytoskeletal proteins. After expanding the analysis to include mouse proteins coexisting with xenograft tumors, we identified a group of proteins, including granzymes, functionally involved in innate immune responses (Supplementary Table S5). To investigate whether the residual immunity in athymic nude mice mediated ACY-1215/JQ1 effect, we first utilized NSG mice as these mice lack NK cells compared with athymic nude mice (33, 34). Indeed, there was almost no difference in human and mouse SCLC xenograft/allograft tumor growth comparing mice treated with ACY-1215/JQ1 and vehicle (Fig. 3A; Supplementary Fig. S6), suggesting that NK cells in athymic nude mice may be involved in the suppression of tumor growth. Next, athymic nude mice carrying GLC-16 xenograft tumors were treated with ACY-1215/ JQ1 plus α-asialo GM1(ASGM1) to deplete NK cells (35, 36) or rabbit sera (Sera) for IgG control (Fig. 3B). Consistent with the findings in NSG mice, ACY-1215/JQ1/α-ASGM1 treatment only moderately suppressed tumor growth, whereas ACY-1215/JQ1/Sera treatment significantly suppressed GLC-16 xenograft tumor growth (Fig. 3C). Depletion of NK cells after α-ASGM1 administration was confirmed by flow cytometry (Supplementary Fig. S7A and S7B). Finally, GLC-16 cells were cocultured with purified NK cells of nude mice in the presence of ACY-1215/JQ1. The results of this study showed that NK-cell population negatively correlated with cell viability (Supplementary Fig. S8A). However, without ACY-1215/JQ1, a positive correlation was observed (Supplementary Fig. S8B). Collectively, these results confirmed that NK cells in athymic nude mice mediate ACY-1215/JQ1 antitumor effect.
Figure 3.
NK cells mediate ACY-1215/JQ1’s synergistic inhibitory effect. A, NSG mice carrying human SCLC NCI-H69 xenograft tumors or mouse SCLC RPP349 or RB1328 allograft tumors were treated with vehicle or ACY-1215/JQ1. Tumor volume (mm3) was calculated as (length × width2)/2. Data represent mean ± SD for 5–7 mice. B, Schematic overview of antibody-mediated NK-cell depletion before and during ACY-1215/JQ1 treatment in athymic nude mice. C, Tumor volumes from athymic nude mice carrying GLC-16 xenograft tumors treated with vehicle, ACY-1215 (ACY)/JQ1/α-asialo GM1 (ASGM1), or ACY-1215 (ACY)/JQ1/rabbit sera (Sera), as indicated, for 14 days. Data represent mean ± SD for 5 mice (**, P < 0.005). D, GLC-16 xenograft tumors of NSG (A) and nude (C) mice were submitted for IHC staining of cleaved caspase-3 on FFPE slides. Each column in the bar graph of cleaved caspase-3 represents mean ± SD for 50 images per treatment group. ns, not significant (**, P ≤ 0.0001). Representative images of cleaved caspase-3 IHC stain per treatment group are shown. Scale bar, 500 μm.
To evaluate the apoptotic status upon the treatment, nude and NSG xenograft tumors were IHC stained for cleaved caspase 3. In agreement, ACY-1215/JQ1/Sera treatment in nude tumors significantly increased the numbers of cleaved caspase 3–positive cells as compared with vehicle treatment (Fig. 3D), whereas the nude tumors treated with ACY-1215/JQ1/α-ASGM1 or NSG tumors treated with ACY-1215/JQ1 displayed similar numbers on cleaved caspase 3–positive cells as their vehicle control (Fig. 3D). Collectively, these data confirmed the role of NK cells in mediating ACY-1215/JQ1 antitumor effect
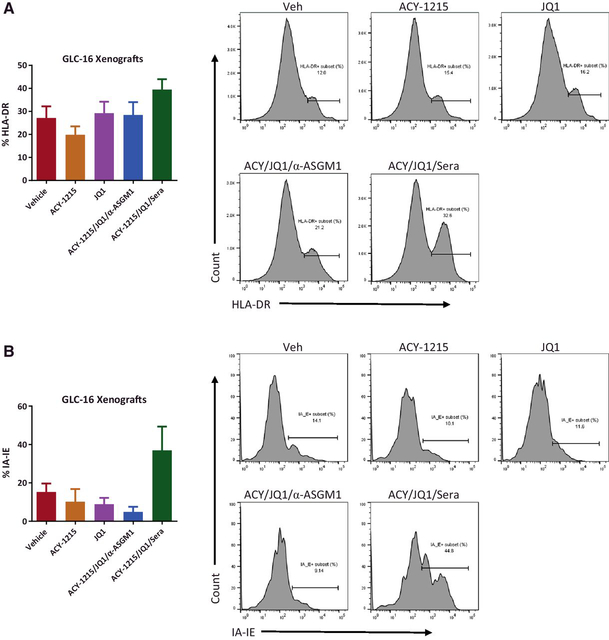
ACY-1215/JQ1 combination therapy increases MHC II expression in both SCLC xenograft tumor cells and tumor-resident myeloid cells
To determine the potential interaction between tumor cells and tumor-resident NK cells, GLC-16 xenograft tumors from nude mice pretreated with vehicle, ACY-1215 and/or JQ1 plus α-ASGM1, or Sera for 7 days were processed into single-cell suspensions and then immunostained for PD-L1, MHCs, and NK-cell–activating ligands (MICA/MICB and B7-H6) on GLC-16 cells, and NK-cell–activating receptors (NKp46 and NKG2D) on tumor-resident NK cells. Flow cytometry analysis of NK-related markers on both tumor and tumor-resident NK cells did not bring useful information for either none or low signals. Instead, we observed moderately increased HLA-DR expression on tumor cells upon ACY-1215/JQ1/Sera but not ACY-1215/JQ1/α-ASGM1 treatment (Fig. 4A; Supplementary Fig. S9A), suggesting the upregulation of MHC II on tumor cells relies on NK cells. We further examined the expression of MHC II in the tumor-resident myeloid cells and observed a similar NK-cell–relied moderately increased IA/IE expression (Fig. 4B; Supplementary Fig. S9B). Depletions of NK cells in spleen and xenograft tumors were confirmed (Supplementary Fig. S9C and S9D).
Figure 4.
NK cells mediate ACY-1215/JQ1’s synergistic upregulation of MHC class II molecules. GLC-16 xenograft nude tumors pretreated for 7 days with ACY-1215 (ACY) and/or JQ1 plus a-asialo GM1 (ASGM1) or rabbit sera (Sera), as indicated, were submitted for flow cytometry analysis. A, Percentage of HLA-DR subset in CD45-negative SCLC single-cell population of GLC-16 xenograft tumors. B, Percentage of IA-IE subset in tumor-infiltrating myeloid cells of GLC-16 xenograft tumors.
Discussion
Over the past three decades, standard chemotherapy in combination with radiotherapy has been the typical treatment option for SCLC (37–39). SCLC patients respond well to initial therapy; however, most patients eventually die of recurrent disease (40). Therefore, new treatment strategies, especially for recurrent SCLC, are urgently needed.
Among all HDACs being covered in the library, HDAC6 was the only hit showing synthetic lethal interaction with JQ1 in our shRNA/JQ1 in vivo screen. We also performed shRNA/JQ1 in vitro screen by culturing the shRNA-library infected NCI-H69 and GLC-16 cells in the presence or absence of JQ1 (Supplementary Fig. S10A). Unlike the in vivo results, HDAC6 in vitro neither ranked in the top 10% hits nor was synthetic lethal interaction with JQ1 (Supplementary Fig. S10B). The effective suppression of SCLC with shHDAC6 (or ACY-1215)/JQ1 in athymic nude mice but not in cell culture or in NSG mice would suggest that the remaining immune responses in nude mice likely mediate ACY-1215/JQ1’s suppressive effect.
In this study, we demonstrated that NK cells mediated ACY-1215/JQ1’s antitumor effect. In addition, NK cells also mediated ACY-1215/JQ1’s treatment to upregulate MHC II expression on both tumor cells and tumor-infiltrating myeloid cells. The presence of MHC II molecules in tumor cells has been associated with a favorable prognosis in triple-negative breast cancer and colon cancer (41–43). Therefore, our finding fits the notion. The elevated expression of MHC II would suggest a potential involvement of antigen-specific helper T-lymphocyte receptors. In a recent treatment study of mouse SCLC allograft tumors in immunocompetence mice with ACY-1215/JQ1 plus depleting NK, CD4, or CD8 T cells, the preliminary results of this study showed that depleting either NK or CD4 T cells blocked ACY-1215/JQ1’s antitumor effect to a similar extent. Further studies will focus on how NK cells sense ACY-1215/JQ1 treatment and then deliver stimulating signals to downstream effectors.
Consistent with the reported cytotoxic effects, ACY-1215/JQ1 killed SCLC cells in cell culture (44, 45). Even if our in vivo study suggested that ACY-1215/JQ1 provoked NK-cell–mediated immunity, we cannot rule out the possibility that tumor debris from the cytotoxic effect of ACY-1215/JQ1 may serve as a primary immunogenic antigen. Of note, ACY-1215 is currently being tested in several clinical studies for the treatment of multiple myeloma and lymphoma, while the use of JQ1 in clinical studies is limited because of its toxicity. Combining ACY-1215 and JQ1 enables us to use a lower dose of JQ1 in the current treatments of SCLC and previous NSCLC (46). It is likely that optimized doses of ACY-1215 and/or JQ1 in combination with different methods of drug administration to mice can achieve similar or better treatment results in mice with reduced toxicity, and these findings can be translated to SCLC patients.
Supplementary Material
Acknowledgments
This work was supported by the National Cancer Institute R01 CA195740, CA163896, CA166480, CA122794, and CA140594 (all to K.K. Wong). K.K. Wong was also supported by a Stand Up To Cancer – American Cancer Society Lung Cancer Dream Team Translational Research Grant (Grant Number: SU2C-AACR-DT1715). Stand Up To Cancer is a division of the Entertainment Industry Foundation. Research grants are administered by the American Association for Cancer Research, the scientific partner of SU2C. We thank the Histopathology Core Facility in BWH for assistance in IHC.
Footnotes
Disclosure of Potential Conflicts of Interest
J.E. Bradner is the president at NIBR. S.N. Quayle is the director of pharmacology at Cue Biopharma. No potential conflicts of interest were disclosed by the other authors.
Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
References
- 1.Rodriguez E, Lilenbaum RC. Small cell lung cancer: past, present, and future. Curr Oncol Rep 2010;12:327–34. [DOI] [PubMed] [Google Scholar]
- 2.Califano R, Abidin AZ, Peck R, Faivre-Finn C, Lorigan P. Management of small cell lung cancer: recent developments for optimal care. Drugs 2012;72:471–90. [DOI] [PubMed] [Google Scholar]
- 3.Jackman DM, Johnson BE. Small-cell lung cancer. Lancet 2005;366: 1385–96. [DOI] [PubMed] [Google Scholar]
- 4.Vadakara J, Borghaei H. Personalized medicine and treatment approaches in non-small-cell lung carcinoma. Pharmgenomics Pers Med 2012;5:113–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu K, House L, Liu W, Cho WC. Personalized targeted therapy for lung cancer. Int J Mol Sci 2012;13:11471–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peifer M, Fernandez-Cuesta L, Sos ML, George J, Seidel D, Kasper LH, et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet 2012;44:1104–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rudin CM, Durinck S, Stawiski EW, Poirier JT, Modrusan Z, Shames DS, et al. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat Genet 2012; 44:1111–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim DW, Wu N, Kim YC,Cheng PF, Basom R, Kim D, et al. Genetic requirement for Mycl and efficacy of RNA Pol I inhibition in mouse models of small cell lung cancer. Genes Dev 2016;30: 1289–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Augert A, Zhang Q, Bates B, Cui M, Wang X, Wildey G, et al. Small cell lung cancer exhibits frequent inactivating mutations in the histone methyltransferase KMT2D/MLL2: CALGB 151111 (Alliance). J Thorac Oncol 2017;12:704–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christensen CL, Kwiatkowski N, Abraham BJ, Carretero J, Al-Shahrour F, Zhang T, et al. Targeting transcriptional addictions in small cell lung cancer with a covalent CDK7 inhibitor. Cancer Cell 2014;26: 909–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lenhart R, Kirov S, Desilva H, Cao J, Lei M, Johnston K, et al. Sensitivity of small cell lung cancer to BET inhibition is mediated by regulation of ASCL1 gene expression. Mol Cancer Ther 2015;14:2167–74. [DOI] [PubMed] [Google Scholar]
- 12.Gardner EE, Lok BH, Schneeberger VE, Desmeules P, Miles LA, Arnold PK, et al. Chemosensitive relapse in small cell lung cancer proceeds through an EZH2-SLFN11 Axis. Cancer Cell 2017;31:286–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Whitehurst AW, Bodemann BO, Cardenas J, Ferguson D, Girard L, Peyton M, et al. Synthetic lethal screen identification of chemosensitizer loci in cancer cells. Nature 2007;446:815–9. [DOI] [PubMed] [Google Scholar]
- 14.Luo J, Emanuele MJ, Li D, Creighton CJ, Schlabach MR, Westbrook TF, et al. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell 2009;137:835–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corcoran RB, Cheng KA, Hata AN, Faber AC, Ebi H, Coffee EM, et al. Synthetic lethal interaction of combined BCL-XL and MEK inhibition promotes tumor regressions in KRAS mutant cancer models. Cancer Cell 2013;23:121–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Y, Marks K, Cowley GS, Carretero J, Liu Q, Nieland TJ, et al. Metabolic and functional genomic studies identify deoxythymidylate kinase as a target in LKB1-mutant lung cancer. Cancer Discov 2013;3:870–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang Z, Yik JH, Chen R, He N, Jang MK, Ozato K, et al. Recruitment of PTEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell 2005;19:535–45. [DOI] [PubMed] [Google Scholar]
- 18.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, et al. Selective inhibition of BET bromodomains. Nature 2010;468:1067–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 2011;146:904–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lockwood WW, Zejnullahu K, Bradner JE, Varmus H. Sensitivity of human lung adenocarcinoma cell lines to targeted inhibition of BET epigenetic signaling proteins. Proc Natl Acad Sci U S A 2012;109: 19408–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grozinger CM, Hassig CA, Schreiber SL. Three proteins define a class of human histone deacetylases related to yeast Hda1p. Proc Natl Acad Sci U S A 1999;96:4868–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seto E, Yoshida M. Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harb Perspect Biol 2014;6:a018713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Ruijter AJ, van Gennip AH, Caron HN, Kemp S, van Kuilenburg AB. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem J 2003;370:737–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Valenzuela-Fernandez A, Cabrero JR, Serrador JM, Sanchez-Madrid F. HDAC6: a key regulator of cytoskeleton, cell migration and cell-cell interactions. Trends Cell Biol 2008;18:291–7. [DOI] [PubMed] [Google Scholar]
- 25.Westendorf JJ, Zaidi SK, Cascino JE, Kahler R, van Wijnen AJ, Lian JB, et al. Runx2 (Cbfa1, AML-3) interacts with histone deacetylase 6 and represses the p21(CIP1/WAF1) promoter. Mol Cell Biol 2002;22: 7982–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Palijan A, Fernandes I, Bastien Y, Tang L, Verway M, Kourelis M, et al. Function of histone deacetylase 6 as a cofactor of nuclear receptor coregulator LCoR. J Biol Chem 2009;284:30264–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Villagra A, Cheng F, Wang HW, Suarez I, Glozak M, Maurin M, et al. The histone deacetylase HDAC11 regulates theexpression of interleukin 10 and immune tolerance. Nat Immunol 2009;10:92–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, et al. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell 2009;138:1019–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Y, Peng L, Seto E, Huang S, Qiu Y. Modulation of histone deacetylase 6 (HDAC6) nuclear import and tubulin deacetylase activity through acetylation. J Biol Chem 2012;287:29168–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luo B, Cheung HW, Subramanian A, Sharifnia T, Okamoto M, Yang X, et al. Highly parallel identification of essential genes in cancer cells. Proc Natl Acad Sci U S A 2008;105:20380–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Acetylon Pharmaceuticals. Available from: http://www.acetylon.com/ricolinostat_in_multiple_myeloma.php.
- 32.QuickGO. Available from: https://www.ebi.ac.uk/QuickGO/term/GO:0045670.
- 33.Flanagan SP. ‘Nude’, a new hairless gene with pleiotropic effects in the mouse. Genet Res 1966;8:295–309. [DOI] [PubMed] [Google Scholar]
- 34.Nehls M, Pfeifer D, Schorpp M, Hedrich H, Boehm T. New member of the winged-helix protein family disrupted in mouse and rat nude mutations. Nature 1994;372:103–7. [DOI] [PubMed] [Google Scholar]
- 35.Nishikado H, Mukai K, Kawano Y, Minegishi Y, Karasuyama H. NK celldepleting anti-asialo GM1 antibody exhibits a lethal off-target effect on basophils in vivo. J Immunol 2011;186:5766–71. [DOI] [PubMed] [Google Scholar]
- 36.Monnier J, Zabel BA. Anti-asialo GM1 NK cell depleting antibody does not alter the development of bleomycin induced pulmonary fibrosis. PLoS One 2014;9:e99350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kalemkerian GP, Gadgeel SM. Modern staging of small cell lung cancer. J Natl Compr Canc Netw 2013;11:99–104. [DOI] [PubMed] [Google Scholar]
- 38.Rossi A, Di Maio M, Chiodini P, Rudd RM, Okamoto H, Skarlos DV, et al. Carboplatin- or cisplatin-based chemotherapy in first-line treatment of small-cell lung cancer: the COCIS meta-analysis of individual patient data. J Clin Oncol 2012;30:1692–8. [DOI] [PubMed] [Google Scholar]
- 39.Goldberg SB, Willers H, Heist RS. Multidisciplinary management of small cell lung cancer. Surg Oncol Clin N Am 2013;22:329–43. [DOI] [PubMed] [Google Scholar]
- 40.Kalemkerian GP, Akerley W, Bogner P, Borghaei H, Chow LQ, Downey RJ, et al. Small cell lung cancer. J Natl Compr Canc Netw 2013;11:78–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ruiz-Cabello F, Klein E, Garrido F. MHC antigens on human tumors. Immunol Lett 1991;29:181–9. [DOI] [PubMed] [Google Scholar]
- 42.Sconocchia G, Eppenberger-Castori S, Zlobec I, Karamitopoulou E, Arriga R, Coppola A, et al. HLA class II antigen expression in colorectal carcinoma tumors as a favorable prognostic marker. Neoplasia 2014;16:31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Forero A, Li Y, Chen D, Grizzle WE, Updike KL, Merz ND, et al. Expression of the MHC class II pathway in triple-negative breast cancer tumor cells is associated with a good prognosis and infiltrating lymphocytes. Cancer Immunol Res 2016;4:390–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Santo L, Hideshima T, Kung AL, Tseng JC, Tamang D, Yang M, et al. Preclinical activity, pharmacodynamic, and pharmacokinetic properties of a selective HDAC6 inhibitor, ACY-1215, in combination with bortezomib in multiple myeloma. Blood 2012;119:2579–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garcia PL, Miller AL, Kreitzburg KM, Council LN, Gamblin TL, Christein JD, et al. The BET bromodomain inhibitor JQ1 suppresses growth of pancreatic ductal adenocarcinoma in patient-derived xenograft models. Oncogene 2016;35:833–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adeegbe DO, Liu Y, Lizotte PH, Kamihara Y, Aref AR, Almonte C, et al. Synergistic immunostimulatory effects and therapeutic benefit of combined histone deacetylase and bromodomain inhibition in non-small cell lung cancer. Cancer Discov 2017;7:852–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.