Some viruses that belong to the Arenaviridae family, like Lassa and Junín viruses, are notorious human pathogens, which may lead to fatal outcomes when they infect people. It is thus important to develop means to combat these viruses. For developing effective vaccines, it is vital to understand the basic mechanisms that these viruses utilize in order to evade or overcome host immune responses. It was previously noted that the GP1 receptor-binding domain from Lassa virus is shed and accumulates in the serum of infected individuals. This raised the possibility that Lassa virus GP1 may function as an immunological decoy. Here we demonstrate that mice develop nonproductive immune responses against GP1 from Lassa virus, which is in contrast to the effective neutralizing responses that GP1 from Junín virus elicits. Thus, GP1 from Lassa virus is indeed an immunological decoy and GP1 from Junín virus may serve as a constituent of a future vaccine.
KEYWORDS: arenavirus, immune response, vaccines
ABSTRACT
There are two predominant subgroups in the Arenaviridae family of viruses, the Old World and the New World viruses, that use distinct cellular receptors for entry. While New World viruses typically elicit good neutralizing antibody responses, the Old World viruses generally evade such responses. Antibody-based immune responses are directed against the glycoprotein spike complexes that decorate the viruses. A thick coat of glycans reduces the accessibility of antibodies to the surface of spike complexes from Old World viruses, but other mechanisms may further hamper the development of efficient humoral responses. Specifically, it was suggested that the GP1 receptor-binding module of the Old World Lassa virus might help with evasion of the humoral response. Here we investigated the immunogenicity of the GP1 domain from Lassa virus and compared it to that of the GP1 domain from the New World Junín virus. We found striking differences in the ability of antibodies that were developed against these immunogens to target the same GP1 receptor-binding domains in the context of the native spike complexes. Whereas GP1 from Junín virus elicited productive neutralizing responses, GP1 from Lassa virus elicited only nonproductive responses. These differences can be rationalized by the conformational changes that GP1 from Lassa virus but not GP1 from Junín virus undergoes after dissociating from the trimeric spike complex. Hence, shedding of GP1 in the case of Lassa virus can indeed serve as a mechanism to subvert the humoral immune response. Moreover, the realization that a recombinant protein may be used to elicit a productive response against the New World Junín virus may suggest a novel and safe way to design future vaccines.
IMPORTANCE Some viruses that belong to the Arenaviridae family, like Lassa and Junín viruses, are notorious human pathogens, which may lead to fatal outcomes when they infect people. It is thus important to develop means to combat these viruses. For developing effective vaccines, it is vital to understand the basic mechanisms that these viruses utilize in order to evade or overcome host immune responses. It was previously noted that the GP1 receptor-binding domain from Lassa virus is shed and accumulates in the serum of infected individuals. This raised the possibility that Lassa virus GP1 may function as an immunological decoy. Here we demonstrate that mice develop nonproductive immune responses against GP1 from Lassa virus, which is in contrast to the effective neutralizing responses that GP1 from Junín virus elicits. Thus, GP1 from Lassa virus is indeed an immunological decoy and GP1 from Junín virus may serve as a constituent of a future vaccine.
INTRODUCTION
The mammarenaviruses of the Arenaviridae family are phylogenetically composed of two main branches, the New World (NW) viruses, which are endemic to the Americas, and the Old World (OW) viruses, which are mostly found in Africa (1). Both branches are zoonotic, single-stranded, bisegmented, RNA-negative viruses that cause infectious outcomes ranging from mild symptoms to fatal hemorrhagic fevers (HF) (2–5). Junín virus (JUNV) and Lassa virus (LASV) are notorious representatives of the NW and OW branches, respectively, that may cause deadly HF in humans. Typical of their phylogenetic groups, JUNV uses transferrin receptor 1 (TfR1) and LASV uses α-dystroglycan (α-DG) as receptors for cell entry (6–9). LASV further uses a secondary receptor, the lysosome-associated membrane glycoprotein 1 (LAMP1), for exiting from the late endosome to the cytoplasm for replication (10, 11). JUNV and LASV are registered as category A pathogens by the NIAID and are select agents of the CDC due to the threat of severe illness that they pose, and no current FDA-approved vaccines and only limited therapeutic options are available.
The sole protein on the lipid membrane of arenaviruses is a class I trimeric spike complex termed the glycoprotein complex (GPC) (1). Each protomer of the GPC is made up of three components: a uniquely long and functional signal peptide termed the stable signal peptide (SSP), a receptor attachment module termed glycoprotein 1 (GP1), and a fusion protein necessary for merging host and virus membranes termed glycoprotein 2 (GP2) (1, 12, 13). GP1 is positioned on the apical region of the spike complex and is available for receptor recognition (1, 14, 15), and it further caps the GP2 fusion protein to prevent premature triggering (1, 13). Owing to its sole presence on the viral surface, GPC is an important target for the neutralizing humoral immune response against these viruses.
Solving the structure of GP1 from LASV (GP1LASV) and comparing it with GP1 from the NW Machupo virus (MACV) (GP1MACV), which was the only other known GP1 structure at the time, led to the hypothesis that soluble GP1LASV undergoes conformational changes compared with the conformation of GP1LASV in the context of the LASV GPC trimer (GPCLASV) (16). Among other reasons, the termini of GP1LASV point in the opposite direction compared to those of GP1MACV while burying a charged residue; in addition, soluble GP1LASV binds only its intracellular secondary receptor, LAMP1, and not α-DG (16). Acidic pH-induced conformational changes of the spike complex were demonstrated using electron microscopy studies (15), and these likely prime GPCLASV for engaging with LAMP1 (11). The recent important structure by Hastie et al. of the ectodomain portion of GPCLASV in its native prefusion state confirms that GP1LASV undergoes conformational changes (14). This structure helps to illustrate the conformational changes that are associated with priming as well as with the dissociation of GP1LASV from the trimeric complex. On the other hand, GP1 domains from NW mammarenaviruses, like JUNV, MACV, or Whitewater Arroyo virus, adopt structures that mostly resemble the structure of GP1LASV in the context of the prefusion trimeric spike (17–21) both as part of a complex with TfR1 and as stand-alone soluble proteins. Furthermore, so far there is no evidence that NW mammarenaviruses need to switch to secondary receptors during cell entry. Therefore, GP1 proteins from NW mammarenaviruses do not seem to undergo conformational changes when they dissociate from their trimeric spike complexes like GP1LASV does.
LASV vaccination attempts are an ongoing challenge, and recovery from LASV infection is determined by T-cell activation, while antibodies (Abs) are less of a correlate for recovery (22, 23). In contrast, JUNV can be prevented with Candid#1 vaccination (24, 25), which elicits an Ab profile similar to that elicited by live JUNV (26), and an effective means of treatment for JUNV infection is through administration of serum from convalescing patients (27). Despite the fact that GP1LASV is considered poorly immunogenic (28), Galan-Navarro et al. demonstrated that by encapsulating it in a special polymersome, nanocarrier nonneutralizing antibodies could be elicited (29). It was further elegantly demonstrated that extensive glycosylation on the GP1LASV surface masks epitopes that could otherwise serve as targets for neutralizing Abs (30) and that this masking involves underprocessed oligomannose glycans (31). However, functional sites on viral glycoproteins, like the receptor-binding surfaces, cannot be completely shielded with glycans. A known example is the CD4 binding site of the highly glycosylated gp120 receptor-binding module of HIV-1 that serves as a central epitope for neutralizing antibodies (32). Thus, additional mechanisms may hamper the elicitation of an effective humoral response against LASV. Interestingly, shedding of GP1LASV was reported in the sera of LASV-infected HF patients, which raised the possibility that GP1LASV functions as an immunological decoy (33). If GP1 is indeed a decoy, it may further impede the development of an effective humoral response. Accumulation of soluble GP1 following shedding, on the other hand, may expose epitopes that are otherwise less accessible in the context of the trimer due to its dense glycan layer (31). Shedding of GP1LASV may also increase the amount of immunogens that are available to the immune system and, thus, could further promote the development of the humoral response.
Here we present a side-by-side comparison of the immunogenicity of GP1LASV and GP1 from JUNV (GP1JUNV) in mice. We demonstrate that both proteins are highly immunogenic but that the immune response against GP1LASV is nonneutralizing and generally cannot target the native spike complex, which is in marked contrast to the immune response that GP1JUNV elicits. Based on the currently available structural information, we suggest that the altered conformation of GP1LASV specifically exposes nonnative epitopes that promote the production of Abs that cannot target the native spike. Taking this special attribute into consideration may help to improve the ongoing quest for developing an effective vaccine against LASV. Moreover, our data suggest that recombinant soluble GP1 domains from NW mammarenaviruses may serve as useful immunogens, offering an attractive path for developing a new class of safe vaccines.
RESULTS
GP1 proteins from both JUNV and LASV are immunogenic in mice.
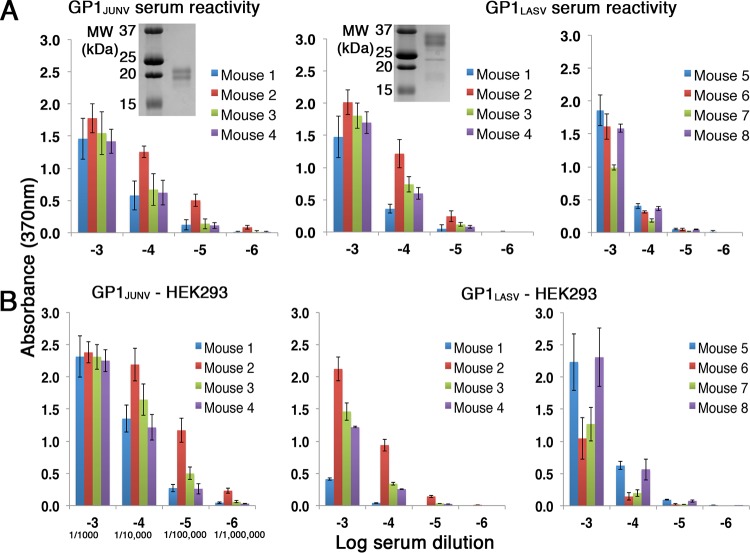
We set out to test the immunogenicity of the GP1 proteins from mammarenaviruses of the OW and NW phylogenetic branches causing HF. As immunogens, we constructed portions of GP1 from JUNV (amino acids 77 to 246 of GPC) and LASV (amino acids 75 to 237 of GPC) fused to a 6×His tag at their C termini. We expressed both GP1JUNV and GP1LASV in insect cells and purified them using immobilized metal affinity chromatography followed by size exclusion chromatography (SEC) (Fig. 1A). Purified proteins were injected into two sets of four mice by the Antibody Laboratory of the Core Biological Facilities at the Weizmann Institute of Science following a protocol that was approved by the Institutional Animal Care and Use Committee (IACUC), and serum samples were collected after a set of boosts. We used an immunization protocol that is typically used by the core facility to maximize the Ab response prior to isolating monoclonal Abs (MAbs). This protocol is based on a series of sequential injections followed by a double boost (see Materials and Methods). Before sacrificing the animals, bleeding was used to evaluate the Ab response that was developed. To test the reactivity of sera from immunized mice, we used an enzyme-linked immunosorbent assay (ELISA). The plates were coated with the same purified GP1s that were used for the mouse immunizations. For each serum sample, we tested a series of serial dilutions (Fig. 1A). All mice developed robust immune responses. The responses were specific to the injected immunogens, as no cross-reactivity against the reciprocal immunogen was observed (data not shown). Using ELISA, a positive response was observed up to dilutions of 10−6 and up to dilutions of 10−5 for sera from mice immunized with GP1JUNV and GP1LASV, respectively (Fig. 1A).
FIG 1.
Both GP1JUNV and GP1LASV are immunogenic in mice. (A) ELISA using serial dilutions of sera from mice immunized with GP1JUNV and GP1LASV. Plates were coated with the same immunogen that was produced in insect cells and that was injected into mice. (Left and middle) The response of two groups of 4 mice each that were immunized with GP1JUNV and GP1LASV, respectively. Colors correspond to sera from different mice. (Right) Data for a second group of 4 mice that were immunized with GP1LASV. Bars represent average values from three independent experiments, each consisting of several technical replicates. Error bars represent standard deviations. (Insets) Analyses of the corresponding GP1JUNV and GP1LASV that were used for immunization using SDS-PAGE and Coomassie staining. MW, molecular weight. (B) ELISA using the indicated serum dilution series and plates coated with the GP1 domains produced in HEK293F cells. Bars represent average values from three independent experiments, each consisting of several technical replicates. Error bars represent standard deviations. For convenience, the serum dilutions used are further indicated using a serology standard format in the lower left plot.
The GP1 immunogens that we used were produced in insect cells, which incorporate high-mannose-type glycans instead of complex-type glycans like mammalian cells do. To test for a potential influence of glycans on the recognition of the immunogens, we produced GP1JUNV and GP1LASV in HEK293F cells and tested the serum reactivity toward plates coated with these proteins (Fig. 1B). Sera both from GP1JUNV-immunized mice and from GP1LASV-immunized mice were reactive (Fig. 1B), indicating that the Ab responses that were obtained are not high-mannose-type glycan dependent. The absolute response levels of sera against GP1 that was produced in insect cells and against GP1 that was produced in HEK293F cells could not be directly compared due to potential variations of the coating efficiencies of the proteins. Nevertheless, the relative response of sera from different mice should be the same if the type of glycans present does not affect recognition. Comparing the relative responses of the sera (Fig. 1A and B) indicated that they were generally maintained but that small variations were visible. For instance, the response of mouse 1 from the GP1LASV-immunized mouse cohort was lower than expected in the case of HEK293F cell-produced GP1LASV when the response to mouse 2 was used as a reference (Fig. 1B). In the absence of measurable cross-reactivity between the two immunogens, it is not likely that such differences are due to recognition of the high-mannose-type glycans. Instead, the complex-type glycans may, in some cases, sterically restrict access to epitopes on the GP1 surface due to their overall larger size than the high-mannose-type glycans. Regardless, the GP1 immunogens elicited humoral responses in all of the injected mice.
GP1JUNV but not GP1LASV elicits a neutralizing immune response.
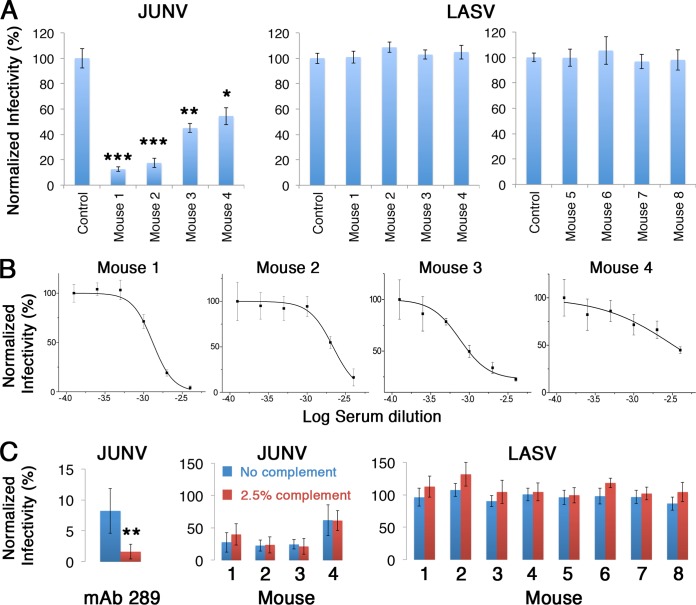
To test whether sera from GP1-immunized mice contain Abs that have neutralization capacity, we performed a viral neutralization assay using a murine leukemia virus (MLV)-based pseudotyped virus reporter system. For this purpose, we generated pseudotyped viruses carrying a luciferase reporter gene and bearing the spike complexes of either JUNV or LASV on their surfaces. Pseudotyped viruses were applied on HEK293T cells after preincubation with a 1:250 dilution of serum from immunized mice, and infectivity was determined by following the luciferase levels at 48 h postinfection (Fig. 2A). The infectivity of pseudotyped viruses bearing the JUNV spike complexes was significantly reduced in the presence of sera from all four mice that were immunized with GP1JUNV (Fig. 2A). On the other hand, none of the serum samples from GP1LASV-immunized mice were able to reduce the infectivity of pseudotyped viruses bearing the LASV spike complexes (Fig. 2A). Prompted by this result, we decided to include a second group of four mice to further substantiate the putative inability of GP1LASV to elicit a neutralizing response. For that, we immunized an additional cohort of four mice (i.e., mice 5 to 8) with GP1LASV and confirmed that reactive serum responses against GP1LASV were indeed developed both from insect cells (Fig. 1A) and from HEK293F cells (Fig. 1B), similar to the findings for the first group of mice. We repeated the infectivity assays with these serum samples and could not observe any neutralization activity (Fig. 2A). Thus, GP1JUNV but not GP1LASV elicits a neutralizing humoral response in mice.
FIG 2.
GP1JUNV as opposed to GP1LASV elicits a neutralizing response in mice. (A) The infectivity of pseudotyped viruses bearing the spike complexes of JUNV and LASV was tested in the presence of serum samples at 1:250 dilutions from the various GP1JUNV- and GP1LASV-immunized mice. Sera from unimmunized mice were used as controls, and the data were normalized to the values for those sera. Bars represent average values from three independent experiments, each consisting of several technical replicates. Error bars represent standard deviations. Statistically significant reductions in infectivity were determined by a two-tailed Student's t test. *, P < 0.05; **, P < 0.005; ***, P < 0.0005. (B) Dose-response neutralization by GP1JUNV-immunized mouse sera. Graphs show representative results from dose-response experiments using a serum dilution range of 1:250 to 1:8,000 for each of the four mice immunized with GP1JUNV. Mean values from technical replicates are shown, and error bars show standard deviations. Each experiment was repeated at least twice. (C) Complement does not enhance serum neutralization potentials. (Left) Enhanced neutralization of pseudotyped viruses by anti-JUNV Ab289 at 1 μg/ml in the presence of 2.5% complement (red bar) compared to neutralization without the addition of complement (blue bar) as a positive control. The difference in neutralization was significant (two-tailed Student's t test, P < 0.005). Sera from GP1JUNV-immunized mice (middle) and from GP1LASV-immunized mice (right) were tested for neutralization of the corresponding pseudotyped viruses at 1:1,000 and 1:500 dilutions of sera (for GP1JUNV- and GP1LASV-immunized mice, respectively) in the presence or absence of 2.5% complement. None of the differences in neutralization was statistically significant. Results are the averages from three independent experiments, each one of which included several technical replicates.
Although sera from all four GP1JUNV-immunized mice exhibited significant neutralization activity at a 1:250 dilution, the level of neutralization varied from about 50% to more than 85% (Fig. 2A). To better characterize the individual response in each mouse, we performed neutralization experiments using serial dilution series of sera (Fig. 2B). These experiments indicated that in mice 1 and 2, the neutralization responses reached almost 100% at the lowest dilutions used (i.e., 1:250), and from the behavior of the fitted dose-response curves, it looks like 100% neutralization may have been achieved at lower serum dilutions. The neutralization responses of mice 3 and 4 were less potent. Nevertheless, extrapolating from the neutralization curve for the serum from mouse 3, it is reasonable to assume that complete neutralization could be achieved using undiluted serum. In mouse 4, it is, however, less clear if undiluted serum would achieve 100% neutralization, but higher than 50% neutralization is certain. Given these observations, immunizing mice with GP1JUNV elicited at least transiently strong neutralizing immune responses that in some mice might actually be sterilizing.
Differential recognition of the trimeric spike complexes by GP1-immunized mouse sera.
Albeit GP1LASV-immunized mouse serum was unable to neutralize pseudotyped viruses bearing the GPCLASV spike complex, the GP1LASV-immunized sera may still contain antibodies that can recognize and bind to the native spike. Merely binding to the trimeric spike complex on the surface of infected cells may be beneficial for promoting Fc-mediated effector functions, like antibody-dependent cell-mediated cytotoxicity or activation of the complement system (34). To test for the potential ability of sera to mediate complement-dependent neutralization, we repeated the neutralization assays in the presence of complement. First, we verified that we could measure complement-enhanced neutralization using a purified monoclonal Ab against JUNV (Ab289 [see below]) as a control. Supplementing Ab289 with complement significantly augmented its neutralization activity (Fig. 2C). Next, we tested sera both from GP1JUNV-immunized mice and from GP1LASV-immunized mice in the presence of complement. We did not observe enhanced neutralization of JUNV-pseudotyped viruses or a gain of neutralization of LASV-pseudotyped viruses in the presence of complement (Fig. 2C). These observations suggest that in the case of GP1JUNV-immunized mice, if complement-compatible Abs are present, they do not achieve effective concentrations. In sera from GP1LASV-immunized mice, either such Abs exist at concentrations too low to be detected or these serum samples are deprived of Abs that can recognize the native spike.
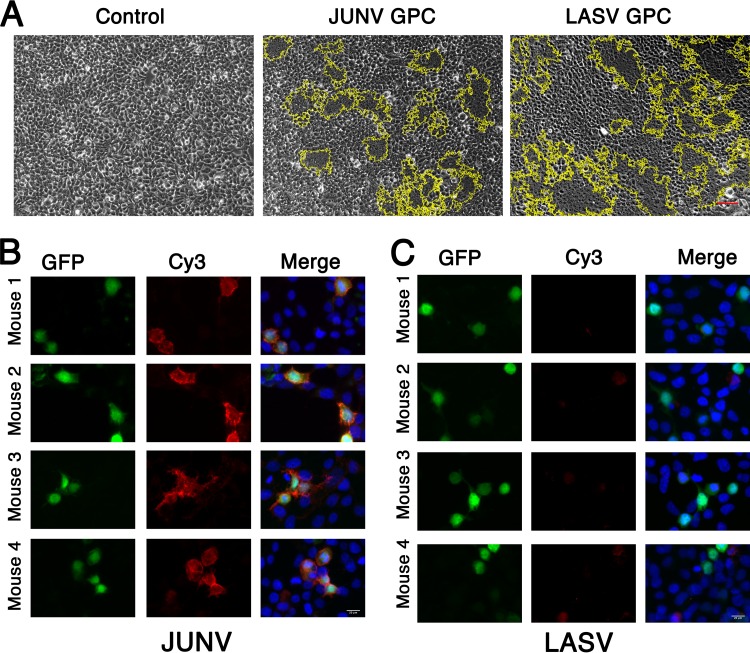
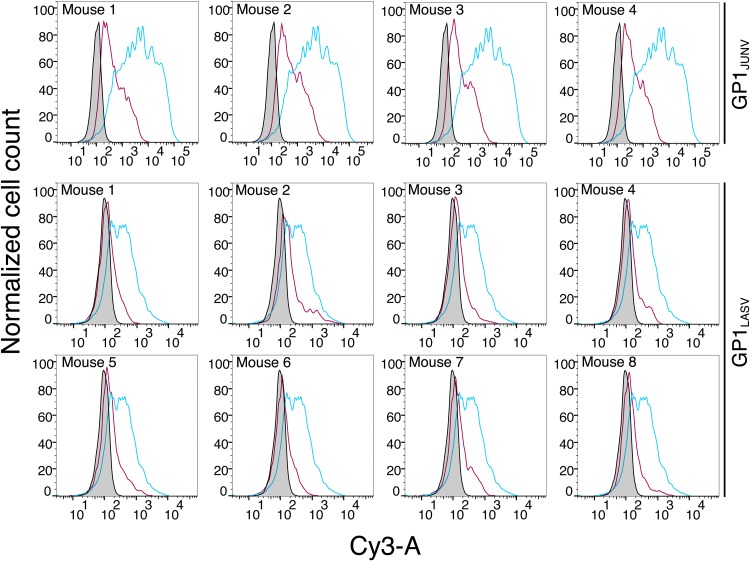
In order to test whether GP1-immunized sera can indeed recognize the GP1 immunogen in the context of the full GPC, we stained HEK293T cells expressing either GPCJUNV or GPCLASV with the serum samples. To identify cells that were successfully transfected with the GPC expression vector, we replaced the neomycin resistance cassette of the pcDNA3.1 plasmid with a green fluorescent protein (GFP) reporter gene. First, we verified that both GPCs were properly produced and processed from this modified vector to give functional spike complexes on cell surfaces by performing a cell fusion assay. GPCJUNV and GPCLASV were able to efficiently induce the formation of syncytia upon exposure to acidic pH (Fig. 3A), indicating that, indeed, both GPCJUNV and GPCLASV appeared in their functional forms on the surfaces of the cell. Next, we fixed cells that were transiently transfected with GPCJUNV and GPCLASV on microscope slides and stained them with the corresponding GP1-immunized sera, followed by a fluorescently labeled secondary anti-mouse Ab. Sera from all GP1JUNV-immunized mice effectively recognized the native spike expressed on HEK293T cells (Fig. 3B). On the other hand, sera from GP1LASV-immunized mice failed to stain GPCLASV-expressing cells (Fig. 3C). Furthermore, flow cytometry analyses of unfixed cells that were stained by the various serum samples indicated robust recognition of GPCJUNV by sera from GP1JUNV-immunized mice (Fig. 4). In contrast to that, sera from GP1LASV-immunized mice showed only traces of binding to GPCLASV that was visible on only a small subset of cells (Fig. 4). The inability of sera from GP1LASV-immunized mice to effectively recognize the native GPCLASV is in marked contrast to the observed reactivity toward the isolated GP1LASV (Fig. 1A and B), indicating that isolated GP1LASV mostly displays nonnative epitopes. Thus, GP1JUNV but not GP1LASV elicits antibody responses that are capable of robustly recognizing the native spike complex.
FIG 3.
Reactivity of GP1-immunized mouse sera against GP1 in the context of the full GPC. (A) Syncytium formation assay for cells expressing either GPCJUNV or GPCLASV. Untransfected cells were used as a negative control. Both GPCJUNV and GPCLASV induced cell fusion upon exposure to acidic pH, marked by dark syncytium patches within the yellow boundaries, indicating functional membrane expression of the corresponding GPCs. These are representative images from three independent repeats. (B) Staining of HEK293T cells expressing GPCJUNV on the membrane surface and cytosolic GFP as a transfection control. Images are shown for staining with 1:1,000 dilutions of sera from 4 mice. GFP is shown as an indicator for transfection (green, left column), GPC staining is visualized using a Cy3-conjugated secondary Ab (red, middle column), and a merge between the channels (right column) with DAPI (4′,6-diamidino-2-phenylindole) staining (blue) is shown. (C) Cell staining as described in the legend to panel B using cells expressing GPCLASV that were stained with GP1LASV-immunized mouse sera. Images obtained using a 1:1,000 dilution are shown. Lower serum dilutions up to 1:10 were also tested for GP1LASV-immunized mouse sera and gave similar results (data not shown). Bars, 50 μm (A) and 20 μm (B and C).
FIG 4.
Recognition of native unfixed spike complexes by GP1-immunized mouse sera. The results of fluorescent-activated cell sorting analyses of live unfixed cells expressing the GPCJUNV or GPCLASV spike complexes (top row and bottom two rows, respectively) are shown. Each graph shows staining by serum from the indicated mouse in a red curve. Control staining using serum from an unimmunized mouse is shown in gray. For GPCJUNV-bearing cells, staining with purified Ab289 is shown in cyan. For GPCLASV-bearing cells, staining with purified Ab37.7 is shown in cyan.
GP1LASV displays nonnative epitopes due to conformational changes.
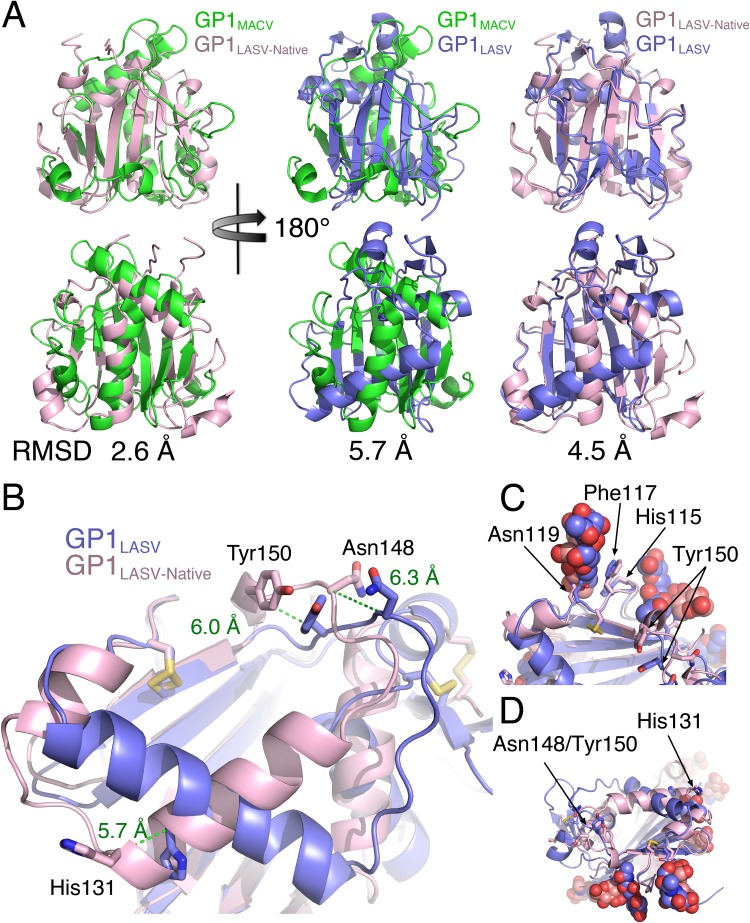
The structure of the ectodomain portion from GPCLASV in its native conformation, which was recently solved by Hastie et al. (14), confirmed that GP1LASV as a stand-alone protein undergoes substantial conformational changes, which allows us to analyze these changes. Interestingly, beside the loops that vary substantially among GP1 domains from different mammarenaviruses, the native conformation of GP1LASV is more similar to that of GP1s from NW mammarenaviruses than to the conformation of itself in its primed conformation (Fig. 5A). Albeit this primed conformation of GP1 is needed for LAMP1 binding in the case of LASV, it is also seen in other OW mammarenaviruses, like Morogoro virus (35), which does not switch to LAMP1. Hence, such conformational switches may serve to form immunological decoys, in addition to priming the spike complexes for switching to secondary receptors. Receptor-binding sites are known vulnerabilities of viral glycoproteins (19, 32, 36–38). In the case of GP1LASV, a complete map of the α-DG binding site is still not available, but several residues that contribute to α-DG binding were previously identified (39, 40). These residues are found at two distinct regions of GP1LASV that cluster together between two adjacent GP1LASV proteins in the context of the native trimer to form the α-DG binding site (14). His131, which is one of the α-DG binding residues, is located on a C-terminal edge of an α-helix in the native conformation of GP1LASV (Fig. 5B). The α-helix that harbors His131 is positioned parallel to a short succeeding α-helix, and a loop connects these two helices (Fig. 5B). In the native trimeric spike complex, this loop is fully exposed on the surface. In the primed conformation of GP1LASV, these two α-helices rearrange together with the loop region to form a single elongated α-helix (Fig. 5B). This completely alters the position of His131 (a change of 5.7 Å in the position of its C-α atom) and the secondary structure context as well as shifts the surrounding residues on the loop that are likely to contribute to the formation of the α-DG binding site. Asn148 and Tyr150, which make up the second cluster, are located at a loop region which is also affected by the rearrangements of GP1LASV (Fig. 5B). In the primed conformation of GP1LASV, these residues alter their positions (6.3-Å and 6.0-Å movements of the C-α atoms of Asn148 and Tyr150, respectively), as well as their orientations (Fig. 5B). Thus, the α-DG binding surfaces that exist in the context of the native trimeric spike complex are completely deformed in the primed monomeric GP1LASV. Importantly, it was previously demonstrated that glycans on the surface of the spike complex of LASV mask prominent epitopes that can otherwise serve as targets for neutralizing antibodies if they are exposed by mutagenesis (30). One such glycan is at Asn119 of LASV (previously referred to as glycan 9). Asn119 is close enough to the α-DG binding site (Fig. 5C) that a MAb bound to Asn119 would likely interfere with α-DG binding. Examining the region near Asn119 reveals some structural elements that do not change conformation upon priming of GP1LASV, like Phe117 and His115 (Fig. 5C). On the other hand, the two regions that make the putative α-DG binding site and display altered conformations at the soluble GP1LASV are devoid of shielding glycans (Fig. 5D). Hence, in a soluble GP1LASV, parts that preserve the native-like conformations are concealed by glycans, and exposed regions display nonnative conformations.
FIG 5.
Structural comparison of soluble GP1LASV with GP1LASV of GPC in the native conformation (GP1LASV-Native). (A) Superpositions of GP1LASV-Native (pink; PDB accession number 5VK2) with soluble GP1LASV (blue; PDB accession number 4ZJF) and GP1MACV (green; PDB accession number 2WFO). The views in the lower row are rotated 180° compared to the views in the upper row. Root mean square deviation (RMSD) values that were calculated using the TM-align server (45) are indicated for each pair of aligned structures. (B) A close-up view of GP1LASV-Native superimposed with GP1LASV showing His131, Asn148, and Tyr150 as sticks. Green dotted lines illustrate the relative C-α atom movements of the indicated residues in the two aligned structures, and the distances are indicated in ångströms. (C) A glycan attached to Asn119 masks a conformation-stable region. Phe117 and His115, which maintain the same conformation in both GP1LASV-Native and GP1LASV, are shown as sticks. Glycans are shown using spheres. The color scheme is the same as that in panel A. (D) The regions that make the α-DG binding site are devoted from glycans. The two regions that form the α-DG binding site are indicated, as are the observed glycans. The color scheme is the same as that described in the legend to panel A.
Isolation and characterization of an anti-JUNV neutralizing MAb.
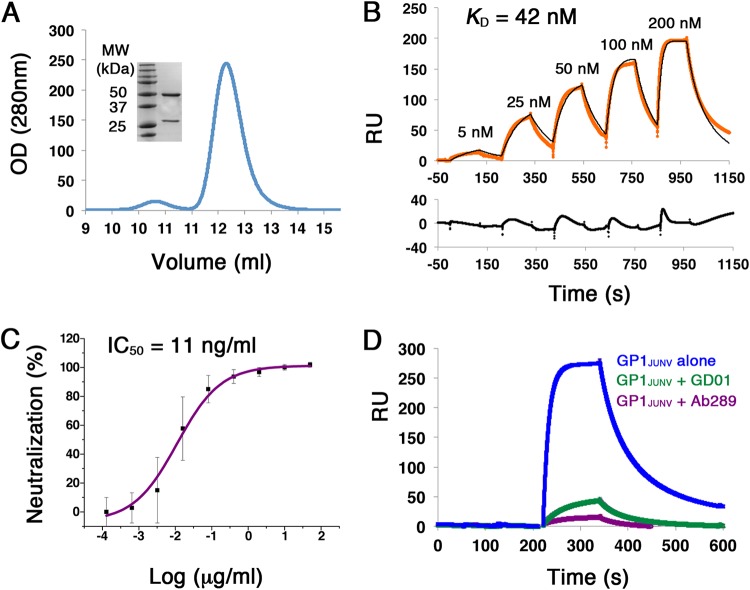
An important observation from the mouse immunization experiments is the ability of GP1JUNV to elicit neutralizing responses in mice. To gain insights regarding the mechanism underlying such a neutralization capacity, we decided to examine a representative MAb. We thus generated hybridomas and cloned the heavy chain variable region (VH) and the light chain variable region (VL) of Ab289, a strong binder of GP1JUNV in ELISA, into an IgG scaffold. We expressed Ab289 in HEK293F cells and purified it using protein A affinity chromatography followed by SEC (Fig. 6A). By capturing Ab289 on a surface plasmon resonance protein A sensor chip and injecting soluble GP1JUNV as an analyte in a single-cycle kinetic experiment, we measured the binding affinity (KD) of this MAb without avidity. Ab289 has a KD of 42 nM toward GP1JUNV (Fig. 6B). Next, we tested the neutralization capacity of Ab289. For that, MLV-pseudotyped viruses bearing GPCJUNV were incubated with a serial dilution series of Ab289 prior to applying them on HEK293T cells, and the levels of the luciferase reporter gene were subsequently determined. Ab289 effectively neutralizes JUNV-pseudotyped viruses with a 50% inhibitory concentration (IC50) of 11 ng/ml (Fig. 6C). To gain insights into the mechanism by which Ab289 neutralizes the viruses, we made use of GD01, a previously characterized MAb against JUNV that targets the TfR1 receptor-binding site on the surface of GP1JUNV (19). Using competition SPR, we tested if Ab289 competes with GD01 for binding to the receptor-binding site. For that, we immobilized the Ab289 IgG on a protein A sensor chip and used GP1JUNV as an analyte either alone or in the presence of the Fab portions from GD01 or Ab289 in excess (Fig. 6D). When supplementing GP1JUNV with either GD01 Fab or Ab289 Fab, the binding to the immobilized Ab289 IgG was greatly reduced (Fig. 6D). Performing the reciprocal experiment by immobilizing GD01 IgG instead of Ab289 IgG gave very similar results (data not shown). Hence, Ab289 competes with GD01 and thus uses the same overall neutralization mechanism of blocking the binding to the cellular receptor.
FIG 6.
Isolation and characterization of Ab289 from GP1JUNV-immunized mouse serum. (A) Chromatogram showing the elution profile of Ab289 using SEC. OD, optical density. (Inset) SDS-PAGE analysis of Ab289 stained with Coomassie blue. (B) SPR sensorgram of GP1JUNV as the analyte injected over immobilized Ab289 in a single-cycle kinetic experiment. The recorded sensorgram is shown in orange, and the fitted graph is shown in black. KD was calculated by fitting a 1:1 binding model to the experimental data. A residual plot showing the quality of the fitted models is included below the sensorgram. The binding experiment was repeated twice, and a representative sensorgram is shown. (C) Neutralization IC50 plot of JUNV-pseudotyped viruses by Ab289. The neutralization was tested using a 5-fold dilution series of Ab289. No Ab control was used to determine 100% infectivity. The IC50 was calculated to be 11 ng/ml using a four-parameter sigmoidal function. The indicated values are averages from three independent experiments, each containing several technical replicates. Error bars show standard deviations. (D) Competition SPR experiments. GP1JUNV was injected as the analyte at 200 nM over immobilized Ab289 either alone (blue) or in the presence of 1 μM Fab 289 (purple) or 1 μM Fab GD01 (green) as competitors.
DISCUSSION
Viruses belong to the Arenaviridae family share many fundamental biological attributes but also exhibit critical differences. Considering the GP1 receptor-binding modules of viruses from the NW and OW groups, it is becoming apparent that they do not only differ in the type of cellular receptors that they have evolved to engage with. Owing to novel structural information that became available in recent years, we now know that GP1 from the OW LASV and likely from other OW viruses undergoes conformational changes due to pH-induced priming or as a result of dissociating from the trimeric spike complexes. Based on these conformational changes and due to the fact that elevated levels of GP1LASV were identified in sera from LASV-infected individuals, it was previously postulated that GP1LASV could serve as an immunological decoy (16, 33). Here we compared the humoral immune responses that were developed in mice following immunization by GP1LASV and GP1JUNV. Both proteins were found to be immunogenic, and Abs that can bind these immunogens were easily detected in sera using ELISA. However, we found a striking difference in the reactivity of sera from mice immunized with GP1LASV and that of sera from mice immunized with GP1JUNV to the respective native spike complexes. While sera from mice that were injected with GP1JUNV readily recognized GPCJUNV on cell surfaces and were further able to neutralize pseudotyped viruses bearing the GPCJUNV, sera from mice immunized with GP1LASV showed very little capacity to bind and were practically functionally inert toward native GPCLASV. Neutralization of GPCJUNV-bearing pseudotyped viruses was achieved in part by blocking the TfR1 receptor-binding site, as exemplified by Ab289, which we isolated. The inability of GP1LASV to elicit a neutralizing humoral response could be rationalized by the conformational changes that it undergoes upon dissociating from the trimer, which allows it to display nonnative conformations at the critically exposed receptor-binding surfaces.
For testing the immunogenicity of the GP1 domains, we used a standard immunization protocol that included the use of complete Freund’s adjuvant. A previous study by Galan-Navarro et al. demonstrated that encapsulation of a GP1LASV protein spanning residues 92 to 256 in polymersome nanocarriers together with monophosphoryl lipid A, which is a Toll-like receptor 4 agonist, greatly enhances the humoral immune response in mice (29). Albeit it does not provide neutralizing Abs, this immunization scheme was able to elicit the production of Abs that recognize the native GPCLASV spike complex (29). Thus, the presentation of the GP1 domains to the immune system is a critical aspect that is influenced by the delivery method, perhaps having the capacity to expose more concealed epitopes that are otherwise less available.
The marked differences in the immune responses against GP1LASV and GP1JUNV have important implications for fighting pathogenic OW and NW mammarenaviruses. First, our data verify that shed GP1LASV is immunogenic but elicits a nonneutralizing humoral response that cannot target the native spike complex. Hence, GP1LASV could indeed serve as an immunological decoy. It is not clear what the mechanisms that allow shedding of GP1LASV are and what the extent of this phenomenon in infected individuals is. Nevertheless, there are ongoing efforts to develop vaccines against LASV that use various platforms to display the spike complex of LASV. Potentially, undesired shedding of the GP1LASV portion from these vaccines may lead to suboptimal results and, hence, may hamper their development. In such cases, structure-based design could be used to stabilize the trimeric spike complex to prevent shedding and to mitigate this problem.
Equally important is our observation that administering recombinant GP1JUNV leads to the elicitation of productive neutralizing Ab response in mice. The Candid#1 vaccine that is used to fight JUNV is based on attenuated virus (24), which raises safety concerns, and this vaccine has not been approved for use by the FDA so far. A detergent-solubilized protein fraction that corresponds to GP1 from live JUNV was previously shown to elicit a protective response in rabbits (41). Our data corroborate this insight and further suggest that the recombinant GP1 portion of GPCJUNV that is produced in a heterologous expression system is sufficient to elicit the production of neutralizing Abs that target the TfR1 receptor-binding site. Moreover, since GP1 domains from other NW mammarenaviruses seem to maintain their native conformations as soluble proteins (17–21), we postulate that using such recombinant GP1 domains as immunogens may also elicit a productive immune response. Therefore, the development of vaccines based on these recombinant proteins seems to be feasible. Such putative vaccines are not anticipated to raise any special safety concerns.
MATERIALS AND METHODS
Materials.
Human embryonic kidney (HEK293T) cells were maintained in Dulbecco's modified Eagle medium (DMEM; Gibco) supplemented with minimal essential medium nonessential amino acids (NEAA; Biological Industries), 1% (vol/vol) l-glutamine solution 200 mM (Biological Industries), 1% (vol/vol) penicillin-streptomycin solution (penicillin at 10,000 units/ml, streptomycin at 10 mg/ml) (Biological Industries), and 10% (vol/vol) fetal bovine serum (FBS; Gibco), here referred to as 293 cell full medium (FM). The GP2-293 retroviral packaging cell line (TaKaRa, Clontech) was maintained in DMEM with the same supplements described above, except for the addition of 1% (vol/vol) 100 mM sodium pyruvate (Biological Industries) instead of NEAA. HEK293F cells were maintained in FreeStyle medium (Gibco). SF9 and TNi insect cells were grown in ESF921 medium (Expression Systems). All transfections for mammalian cells mentioned here were performed with the 25-kDa polyethyleneimine (PEI) reagent (Polysciences). Secondary anti-mouse immunoglobulin-horseradish peroxidase (HRP) was purchased from Jackson (catalog number 115-035-003), and secondary anti-mouse immunoglobulin and human immunoglobulin conjugated to Cy3 were purchased from Jackson (catalog numbers 115-165-146 and 109-165-008, respectively).
Construction of expression vectors.
Sequences coding the GPC proteins of JUNV and LASV were chemically synthesized (GenScript) and subcloned into the pcDNA3.1 expression vector, using the BamHI-NotI restriction sites. The addition of soluble GFP to this vector was performed by replacing the neomycin resistance gene with a gene encoding GFP by restriction-free cloning while keeping the neomycin resistance gene promoter. The GP1JUNV (amino acids 77 to 246) and GP1LASV (amino acids 75 to 237) proteins were generated on pAcGP67b plasmids for expression with insect cells using a baculovirus transfection system with a construct fused to a C-terminal His tag. Constructs for expressing GP1 in mammalian cells were generated by fusing GP1JUNV (amino acids 77 to 247) and GP1LASV (amino acids 75 to 237) upstream of a fragment encoding a portion of the human Fc (IgG1) and a tobacco etch virus (TEV) protease site in a modified pHLsec plasmid. Cloning was performed using BamHI-KpnI restriction sites introducing a V5H8 signal peptide at the N termini.
Protein expression and purification.
To produce the GP1 immunogens, we used the same protocol that we previously described (20). Briefly, we expressed the proteins in insect cells and purified them using Ni2+-affinity chromatography, followed by size exclusion chromatography. To produce recombinant Abs, we transfected HEK293F cells with the plasmids encoding the heavy and light chains, using 25-kDa polyethyleneimine (Polysciences) at a DNA/polyethyleneimine ratio of 1 μg/2 μl. Cells were maintained for 5 days in suspension before harvesting the supernatants. After clarifying the supernatants by centrifugation, Abs were captured using protein A affinity chromatography (GE Lifesciences). Abs were eluted using 0.1 M citric acid, and the buffer was adjusted to pH 8.0 using Tris-HCl. For obtaining the Fab portions, we used papain to digest the Abs in a cutting buffer made of 20 mM cysteine-HCl (Sigma-Aldrich) and 10 mM EDTA adjusted to pH 7.0 with Tris-HCl buffer, pH 8.0. The amount of papain was calibrated for each Ab. Cutting was performed for 90 min in 37°C. Negative protein A was performed to remove Fc fragments by capturing them on the column and collecting the flow-through, followed by size exclusion chromatography on a Superdex200 10/300 column (GE Lifesciences). Expression and purification of GP1-Fc fusion proteins from HEK293F cells were done as described above for the recombinant Abs. Following transfection and protein A column purification, protein was digested with TEV protease to remove the Fc fraction. The cleaved product was isolated using size exclusion chromatography on a Superdex200 10/300 column (GE Lifesciences).
Mouse immunization.
To immunize mice, we followed a protocol for generating monoclonal antibodies described by Eshhar (42). Briefly, BALB/c mice were immunized with emulsions of 50 μg His-tagged GP1JUNV or GP1LASV with complete Freund’s adjuvant (Difco Laboratories) 3 times at intervals of 3 weeks. Blood samples were collected 10 days after the 3rd boost and were subjected to ELISA to confirm an immune response. For GP1JUNV, another boost was performed, and for GP1LASV, an additional 3 boosts were performed. At 4 and 5 days prior to final serum collection and the generation of a hybridoma, mice were given two additional boosts. Hybridomas were selected by screening supernatants via ELISA. The protocol of the study with mice was approved by the Institutional Animal Care and Use Committee of the Weizmann Institute of Science.
Selecting MAbs from single-cell hybridomas.
Spleen cells from immunized mice were fused with NSO myeloma cells via polyethylene glycol (PEG) utilization. Hybridoma cells were then selected with hypoxanthine-aminopterin-thymidine medium. Hybridoma clone supernatants were screened for GP1 binding by performing ELISA. Top hits were selected for single-cell subcloning and further testing with ELISA.
Generating a cDNA library and cloning of MAbs.
RNA from positive ELISA hits of hybridoma cells was extracted using an RNeasy kit (Qiagen) and was used to generate a cDNA library using a New England Biolabs ProtoScript first-strand cDNA kit according to the protocol described by the manufacturer. A cleaning process using New England Biolabs DNase was performed according to the manufacturer’s protocol. Degenerative primers described by Wang et al. (43) were used in a standard PCR to amplify VH and VL. Cloning was performed using AgeI and SalI restriction enzymes to insert VH into a human heavy-chain IgG1 scaffold. Restriction-free cloning was performed to insert VL upstream of a human light chain of the constant region (CL) and downstream of the signal peptide.
Indirect ELISA.
Ninety-six-well plates (Greiner Bio-One) were coated with 100 μl of 1 μg/ml GP1 antigen in phosphate-buffered saline (PBS), and blocking was with 1% bovine serum albumin (BSA) in PBS with 0.05% (vol/vol) Tween 20. Sera from GP1-immunized mice were added at various dilutions as the primary Ab in blocking buffer, followed by the addition of the secondary Ab conjugated to HRP. Incubations were for 1 h at 37°C for all Abs except the secondary Ab, which was incubated for 30 min. Between each step, wells were washed using PBS with 0.05% (vol/vol) Tween 20. For enzymatic detection, 3,3′,5,5′-tetramethylbenzidine detection reagent (Sigma-Aldrich) was added to each well, and the plates were immediately read using the Tecan Infinite M200 Pro plate reader at a wavelength of 370 nm.
Pseudoviral particle production.
MLV virus-like particles (VLPs) pseudotyped with GPC from Junín and Lassa viruses were produced by transfecting retroviral transfer vector pLXIN-Luc encoding luciferase as the reporter gene (pLXIN-Luc [Addgene plasmid 60683] was a gift from Alice Wong), along with the envelope of Junín or Lassa virus GPC-pcDNA3.1 plasmid into the GP2-293 retroviral packaging cell line (Clontech). Briefly, GP2-293 cells were seeded at 4 × 106 to 5 × 106 in 100-mm culture dishes. On the next day, the cells were transfected with the retroviral vector and the envelope plasmid at a 1:1 ratio using the calcium phosphate transfection method. After 5 h, the transfection medium was replaced with GP2-293 cell FM, and the supernatants containing viral particles were harvested 48 h later. VLPs were concentrated by the addition of 40% (wt/vol) PEG 6000 (Sigma-Aldrich) in phosphate-buffered saline (PBS) into the VLP supernatant to obtain a final concentration of 8%. The solution was kept at 4°C for 18 h, followed by centrifugation at 10,000 × g for 20 min. The supernatant was then removed, and the pseudotyped viruses were resuspended in GP2-293 cell FM and stored at −80°C until use.
Serum endpoint dilution and Ab neutralization assay.
For neutralization assays, the HEK293T human TfR1-stable cell line for Junín virus VLPs and HEK293T cells for Lassa virus VLPs were seeded on a poly-l-lysine-precoated white, chimney 96-well plate (Greiner Bio-One). Cells were left to adhere for 4 h, followed by the addition of pseudoviruses concentrated 20 times for Junín virus and 10 times for Lassa virus. The VLPs were then preincubated with serum samples. For the neutralization assay, 2-fold descending concentrations starting with 1:250 μl of GP1-immunized mouse serum was performed. For the neutralization assay with Ab289, a 5-fold descending dilution was performed starting from 50 μg/ml. Cells were washed from the viruses at 18 h postinfection, and luminescence from the activity of luciferase was measured at 48 h postinfection using a Tecan Infinite M200 Pro plate reader after applying Bright-Glo reagent (Promega) to the cells.
Complement assay.
Viral neutralization was performed as described above. JUNV VLPs were used in a serum dilution of 1:1,000, and LASV VLPs were used in a serum dilution of 1:500. Assays with triplicates of each sample were performed in parallel with either 2.5% standard rabbit complement (Cedarlane) or DMEM. As a positive control, Ab289 was used at a final concentration of 1 μg/ml with the JUNV VLPs.
Cell fusion assay.
HEK293T cells were seeded in a 24-well plate precoated with poly-l-lysine (Sigma). Seeded cells were transfected 24 h later with 0.2 μg DNA of Junín or Lassa virus GPC pcDNA3.1 plasmids with a GFP reporter gene under the control of a separate promoter using PEI transfection. At 24 h after transfection, cells were rinsed once with FM supplemented with 20 mM MES (morpholineethanesulfonic acid; Acros Organics) and titrated to pH 5. Cells were then incubated with the same medium for 10 min, followed by a wash and incubation with 293 cell FM for 2 h at 37°C. Following incubation, cells were fixed in prewarmed 3.7% (vol/vol) formaldehyde solution (J. T. Baker) in phosphate-buffered saline (PBS; Biological Industries) for 10 min. Phase images of syncytia were taken at a ×10 magnification using a phase microscope. Syncytia were manually spotted and selected on the images. The boundaries and coverage area were automatically traced and calculated using the versatile wand tool of ImageJ software (44).
Cell staining and fluorescence microscopy imaging.
HEK293T cells were seeded on poly-l-lysine-precoated coverslips in 24-well plates and transfected with the above-described plasmids encoding JUNV and LASV GPCs with soluble GFP under the control of a simian virus 40 promoter using the PEI reagent. At 24 h posttransfection, cells were fixed with prewarmed 3.7% (vol/vol) formaldehyde (PFA) solution in PBS and blocked with 3% (wt/vol) BSA in PBS. Cells were stained by applying the corresponding GP1-activated sera at dilutions of 1:1,000, followed by Cy3-conjugated anti-human Fc. Cells were imaged at a ×40 magnification using a fluorescence microscope. Images were processed using ImageJ software (44).
Flow cytometry.
HEK293T cells were transfected using the Lipofectamine 2000 reagent (Thermo Fisher) with GPCLASV or GPCJUNV on pcDNA3.1. At 48 h posttransfection, the cells were harvested in PBS with 0.5% BSA and were incubated on ice for 10 min for blocking. The cells were then incubated with serum diluted in incubation buffer to 1:1,000 for 1 h, washed twice, and incubated with secondary Ab conjugated to Cy3 at a dilution of 1:800 in incubation buffer for 30 min. Cells were washed, resuspended in PBS, and subjected to flow cytometry on an LSRII flow cytometer (BD Biosciences). Excitation was performed at a wavelength of 445 nm to detect Cy3. Data were analyzed by gating the population of live single cells, counting the fluorescence levels of Cy3 using FlowJo LLC software.
SPR measurements.
All surface plasmon resonance (SPR) measurements were performed using a Biacore T200 system (GE Healthcare) at 25°C. Abs were immobilized to a protein A chip from a stock of 5 μg/ml to a surface density of ∼1,500 response units (RU). GP1JUNV (amino acids 77 to 247 with a C-terminal 6×His tag) in a series of increasing concentrations of 5, 25, 50, 100, and 200 nM in Tris-buffered saline (TBS) buffer with 0.02% (wt/vol) azide was used as the analyte, and a single-cycle kinetics experiment was performed. For regeneration, we used 10 mM glycine-HCl, pH 1.5. Sensorgrams were analyzed with Biacore T200 evaluation software using a 1:1 binding model.
ACKNOWLEDGMENTS
We thank Michal Katz for critical reading of our manuscript.
Ron Diskin is the incumbent of the Tauro Career Development Chair in Biomedical Research. The Ron Diskin lab is supported by research grants from the Enoch Foundation, from the Abramson Family Center for Young Scientists, from Rudolfine Steindling, and from the Israel Science Foundation (grant no. 682/16). This research was further supported by a grant from the Minerva Foundation with funding from the Federal German Ministry for Education and Research.
R.D. and A.B.-K. are listed as inventors on a patent application titled Use of GP1 Domain from New World Arenaviruses as a Vaccine.
REFERENCES
- 1.Nunberg JH, York J. 2012. The curious case of arenavirus entry, and its inhibition. Viruses 4:83–101. doi: 10.3390/v4010083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moraz ML, Kunz S. 2011. Pathogenesis of arenavirus hemorrhagic fevers. Expert Rev Anti Infect Ther 9:49–59. doi: 10.1586/eri.10.142. [DOI] [PubMed] [Google Scholar]
- 3.Geisbert TW, Jahrling PB. 2004. Exotic emerging viral diseases: progress and challenges. Nat Med 10:S110–S121. doi: 10.1038/nm1142. [DOI] [PubMed] [Google Scholar]
- 4.Oldstone MB. 2002. Arenaviruses. II. The molecular pathogenesis of arenavirus infections. Introduction. Curr Top Microbiol Immunol 263:V–XII. [PubMed] [Google Scholar]
- 5.Oldstone MB. 2002. Arenaviruses. I. The epidemiology molecular and cell biology of arenaviruses. Introduction. Curr Top Microbiol Immunol 262:V–XII. [PubMed] [Google Scholar]
- 6.Radoshitzky SR, Abraham J, Spiropoulou CF, Kuhn JH, Nguyen D, Li W, Nagel J, Schmidt PJ, Nunberg JH, Andrews NC, Farzan M, Choe H. 2007. Transferrin receptor 1 is a cellular receptor for New World haemorrhagic fever arenaviruses. Nature 446:92–96. doi: 10.1038/nature05539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao W, Henry MD, Borrow P, Yamada H, Elder JH, Ravkov EV, Nichol ST, Compans RW, Campbell KP, Oldstone MB. 1998. Identification of alpha-dystroglycan as a receptor for lymphocytic choriomeningitis virus and Lassa fever virus. Science 282:2079–2081. doi: 10.1126/science.282.5396.2079. [DOI] [PubMed] [Google Scholar]
- 8.Kunz S, Borrow P, Oldstone MB. 2002. Receptor structure, binding, and cell entry of arenaviruses. Curr Top Microbiol Immunol 262:111–137. [DOI] [PubMed] [Google Scholar]
- 9.Kunz S, Rojek JM, Perez M, Spiropoulou CF, Oldstone MB. 2005. Characterization of the interaction of Lassa fever virus with its cellular receptor alpha-dystroglycan. J Virol 79:5979–5987. doi: 10.1128/JVI.79.10.5979-5987.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jae LT, Raaben M, Herbert AS, Kuehne AI, Wirchnianski AS, Soh TK, Stubbs SH, Janssen H, Damme M, Saftig P, Whelan SP, Dye JM, Brummelkamp TR. 2014. Virus entry. Lassa virus entry requires a trigger-induced receptor switch. Science 344:1506–1510. doi: 10.1126/science.1252480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cohen-Dvashi H, Israeli H, Shani O, Katz A, Diskin R. 2016. Role of LAMP1 binding and pH sensing by the spike complex of Lassa virus. J Virol 90:10329–10338. doi: 10.1128/JVI.01624-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burri DJ, da Palma JR, Kunz S, Pasquato A. 2012. Envelope glycoprotein of arenaviruses. Viruses 4:2162–2181. doi: 10.3390/v4102162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harrison SC. 2008. Viral membrane fusion. Nat Struct Mol Biol 15:690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hastie KM, Zandonatti MA, Kleinfelter LM, Heinrich ML, Rowland MM, Chandran K, Branco LM, Robinson JE, Garry RF, Saphire EO. 2017. Structural basis for antibody-mediated neutralization of Lassa virus. Science 356:923–928. doi: 10.1126/science.aam7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li S, Sun Z, Pryce R, Parsy ML, Fehling SK, Schlie K, Siebert CA, Garten W, Bowden TA, Strecker T, Huiskonen JT. 2016. Acidic pH-induced conformations and LAMP1 binding of the Lassa virus glycoprotein spike. PLoS Pathog 12:e1005418. doi: 10.1371/journal.ppat.1005418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cohen-Dvashi H, Cohen N, Israeli H, Diskin R. 2015. Molecular mechanism for LAMP1 recognition by Lassa virus. J Virol 89:7584–7592. doi: 10.1128/JVI.00651-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bowden TA, Crispin M, Graham SC, Harvey DJ, Grimes JM, Jones EY, Stuart DI. 2009. Unusual molecular architecture of the Machupo virus attachment glycoprotein. J Virol 83:8259–8265. doi: 10.1128/JVI.00761-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abraham J, Corbett KD, Farzan M, Choe H, Harrison SC. 2010. Structural basis for receptor recognition by New World hemorrhagic fever arenaviruses. Nat Struct Mol Biol 17:438–444. doi: 10.1038/nsmb.1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mahmutovic S, Clark L, Levis SC, Briggiler AM, Enria DA, Harrison SC, Abraham J. 2015. Molecular basis for antibody-mediated neutralization of New World hemorrhagic fever mammarenaviruses. Cell Host Microbe 18:705–713. doi: 10.1016/j.chom.2015.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shimon A, Shani O, Diskin R. 2017. Structural basis for receptor selectivity by the Whitewater Arroyo mammarenavirus. J Mol Biol doi: 10.1016/j.jmb.2017.07.011. [DOI] [PubMed] [Google Scholar]
- 21.Hastie KM, Saphire EO. 2018. Lassa virus glycoprotein: stopping a moving target. Curr Opin Virol 32:52–58. doi: 10.1016/j.coviro.2018.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCormick JB, Mitchell SW, Kiley MP, Ruo S, Fisher-Hoch SP. 1992. Inactivated Lassa virus elicits a non protective immune response in rhesus monkeys. J Med Virol 37:1–7. doi: 10.1002/jmv.1890370102. [DOI] [PubMed] [Google Scholar]
- 23.Baize S, Marianneau P, Loth P, Reynard S, Journeaux A, Chevallier M, Tordo N, Deubel V, Contamin H. 2009. Early and strong immune responses are associated with control of viral replication and recovery in Lassa virus-infected cynomolgus monkeys. J Virol 83:5890–5903. doi: 10.1128/JVI.01948-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McKee KT Jr, Oro JG, Kuehne AI, Spisso JA, Mahlandt BG. 1992. Candid no. 1 Argentine hemorrhagic fever vaccine protects against lethal Junin virus challenge in rhesus macaques. Intervirology 34:154–163. doi: 10.1159/000150276. [DOI] [PubMed] [Google Scholar]
- 25.Maiztegui JI, McKee KT Jr, Barrera Oro JG, Harrison LH, Gibbs PH, Feuillade MR, Enria DA, Briggiler AM, Levis SC, Ambrosio AM, Halsey NA, Peters CJ. 1998. Protective efficacy of a live attenuated vaccine against Argentine hemorrhagic fever. AHF Study Group. J Infect Dis 177:277–283. doi: 10.1086/514211. [DOI] [PubMed] [Google Scholar]
- 26.del Carmen Saavedra M, Sottosanti JM, Riera L, Ambrosio AM. 2003. IgG subclasses in human immune response to wild and attenuated (vaccine) Junin virus infection. J Med Virol 69:447–450. doi: 10.1002/jmv.10308. [DOI] [PubMed] [Google Scholar]
- 27.Enria DA, Briggiler AM, Fernandez NJ, Levis SC, Maiztegui JI. 1984. Importance of dose of neutralising antibodies in treatment of Argentine haemorrhagic fever with immune plasma. Lancet ii:255–256. [DOI] [PubMed] [Google Scholar]
- 28.Robinson JE, Hastie KM, Cross RW, Yenni RE, Elliott DH, Rouelle JA, Kannadka CB, Smira AA, Garry CE, Bradley BT, Yu H, Shaffer JG, Boisen ML, Hartnett JN, Zandonatti MA, Rowland MM, Heinrich ML, Martinez-Sobrido L, Cheng B, de la Torre JC, Andersen KG, Goba A, Momoh M, Fullah M, Gbakie M, Kanneh L, Koroma VJ, Fonnie R, Jalloh SC, Kargbo B, Vandi MA, Gbetuwa M, Ikponmwosa O, Asogun DA, Okokhere PO, Follarin OA, Schieffelin JS, Pitts KR, Geisbert JB, Kulakoski PC, Wilson RB, Happi CT, Sabeti PC, Gevao SM, Khan SH, Grant DS, Geisbert TW, Saphire EO, Branco LM, Garry RF. 2016. Most neutralizing human monoclonal antibodies target novel epitopes requiring both Lassa virus glycoprotein subunits. Nat Commun 7:11544. doi: 10.1038/ncomms11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Galan-Navarro C, Rincon-Restrepo M, Zimmer G, Ollmann Saphire E, Hubbell JA, Hirosue S, Swartz MA, Kunz S. 2017. Oxidation-sensitive polymersomes as vaccine nanocarriers enhance humoral responses against Lassa virus envelope glycoprotein. Virology 512:161–171. doi: 10.1016/j.virol.2017.09.013. [DOI] [PubMed] [Google Scholar]
- 30.Sommerstein R, Flatz L, Remy MM, Malinge P, Magistrelli G, Fischer N, Sahin M, Bergthaler A, Igonet S, Ter Meulen J, Rigo D, Meda P, Rabah N, Coutard B, Bowden TA, Lambert PH, Siegrist CA, Pinschewer DD. 2015. Arenavirus glycan shield promotes neutralizing antibody evasion and protracted infection. PLoS Pathog 11:e1005276. doi: 10.1371/journal.ppat.1005276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Watanabe Y, Raghwani J, Allen JD, Seabright GE, Li S, Moser F, Huiskonen JT, Strecker T, Bowden TA, Crispin M. 2018. Structure of the Lassa virus glycan shield provides a model for immunological resistance. Proc Natl Acad Sci U S A 115:7320–7325. doi: 10.1073/pnas.1803990115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.West AP Jr, Scharf L, Scheid JF, Klein F, Bjorkman PJ, Nussenzweig MC. 2014. Structural insights on the role of antibodies in HIV-1 vaccine and therapy. Cell 156:633–648. doi: 10.1016/j.cell.2014.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Branco LM, Grove JN, Moses LM, Goba A, Fullah M, Momoh M, Schoepp RJ, Bausch DG, Garry RF. 2010. Shedding of soluble glycoprotein 1 detected during acute Lassa virus infection in human subjects. Virol J 7:306. doi: 10.1186/1743-422X-7-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu LL, Suscovich TJ, Fortune SM, Alter G. 2018. Beyond binding: antibody effector functions in infectious diseases. Nat Rev Immunol 18:46–61. doi: 10.1038/nri.2017.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Israeli H, Cohen-Dvashi H, Shulman A, Shimon A, Diskin R. 2017. Mapping of the Lassa virus LAMP1 binding site reveals unique determinants not shared by other Old World arenaviruses. PLoS Pathog 13:e1006337. doi: 10.1371/journal.ppat.1006337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou T, Georgiev I, Wu X, Yang ZY, Dai K, Finzi A, Kwon YD, Scheid JF, Shi W, Xu L, Yang Y, Zhu J, Nussenzweig MC, Sodroski J, Shapiro L, Nabel GJ, Mascola JR, Kwong PD. 2010. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science 329:811–817. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Diskin R, Scheid JF, Marcovecchio PM, West AP Jr, Klein F, Gao H, Gnanapragasam PN, Abadir A, Seaman MS, Nussenzweig MC, Bjorkman PJ. 2011. Increasing the potency and breadth of an HIV antibody by using structure-based rational design. Science 334:1289–1293. doi: 10.1126/science.1213782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clark LE, Mahmutovic S, Raymond DD, Dilanyan T, Koma T, Manning JT, Shankar S, Levis SC, Briggiler AM, Enria DA, Wucherpfennig KW, Paessler S, Abraham J. 2018. Vaccine-elicited receptor-binding site antibodies neutralize two New World hemorrhagic fever arenaviruses. Nat Commun 9:1884. doi: 10.1038/s41467-018-04271-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sullivan BM, Emonet SF, Welch MJ, Lee AM, Campbell KP, de la Torre JC, Oldstone MB. 2011. Point mutation in the glycoprotein of lymphocytic choriomeningitis virus is necessary for receptor binding, dendritic cell infection, and long-term persistence. Proc Natl Acad Sci U S A 108:2969–2974. doi: 10.1073/pnas.1019304108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smelt SC, Borrow P, Kunz S, Cao W, Tishon A, Lewicki H, Campbell KP, Oldstone MB. 2001. Differences in affinity of binding of lymphocytic choriomeningitis virus strains to the cellular receptor alpha-dystroglycan correlate with viral tropism and disease kinetics. J Virol 75:448–457. doi: 10.1128/JVI.75.1.448-457.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cresta B, Padula P, de Martinez Segovia M. 1980. Biological properties of Junin virus proteins. I. Identification of the immunogenic glycoprotein. Intervirology 13:284–288. doi: 10.1159/000149136. [DOI] [PubMed] [Google Scholar]
- 42.Eshhar Z. 1985. Monoclonal antibody strategy and techniques, hybridoma technology in the biosciences and medicine. Springer, New York, NY. [Google Scholar]
- 43.Wang Z, Raifu M, Howard M, Smith L, Hansen D, Goldsby R, Ratner D. 2000. Universal PCR amplification of mouse immunoglobulin gene variable regions: the design of degenerate primers and an assessment of the effect of DNA polymerase 3' to 5' exonuclease activity. J Immunol Methods 233:167–177. doi: 10.1016/S0022-1759(99)00184-2. [DOI] [PubMed] [Google Scholar]
- 44.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Y, Skolnick J. 2005. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res 33:2302–2309. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]