Abstract
Inflammatory caspases (caspase-1, caspase-4, caspase-5 and caspase-11 (caspase-1/–4/–5/–11)) mediate host defense against microbial infections, processing pro-inflammatory cytokines and triggering pyroptosis. However, precise checkpoints are required to prevent their unsolicited activation. Here we report that serpin family B member 1 (SERPINB1) limited the activity of those caspases by suppressing their caspase-recruitment domain (CARD) oligomerization and enzymatic activation. While the reactive center loop of SERPINB1 inhibits neutrophil serine proteases, its carboxy-terminal CARD-binding motif restrained the activation of pro-caspase-1/–4/–5/–11. Consequently, knockdown or deletion of SERPINB1 prompted spontaneous activation of caspase-1/–4/–5/–11, release of the cytokine IL-1β and pyroptosis, inducing elevated inflammation after non-hygienic co-housing with pet-store mice and enhanced sensitivity to lipopolysaccharide- or Acinetobacter baumannii–induced endotoxemia. Our results reveal that SERPINB1 acts as a vital gatekeeper of inflammation by restraining neutrophil serine proteases and inflammatory caspases in a genetically and functionally separable manner.
Inflammasomes are innate immune sensors that regulate the activation of inflammatory caspases in response to pathogen- and damage-associated molecular patterns (PAMPs and DAMPs, respectively) and, ultimately, maintain immune homeostasis. While inflammatory caspases play a fundamental role in controlling infections and responding to endogenous stressors, their unregulated activation can produce unnecessary pro-inflammatory cytokines, leading to injurious pyroptosis: an inflammatory form of cell death characterized by substantial leakage of cytosolic contents1–4. Thus, it is important that caspase activity be tightly controlled. Understanding the regulatory mechanisms that ensure the timely and beneficial activation of inflammatory caspases is therefore of particular importance.
The inflammatory caspase subfamily (caspase-1, caspase-4 and caspase-5 (collectively called ‘caspase-1/–4/–5’ here) in humans; caspase-1/–11 in mice) functions mainly during innate immune responses in various aspects of host physiology5–7. In response to cytosolic PAMPs and DAMPs, cytosolic sensors, including NOD-like receptors (NLRs), form a multi-protein inflammasome complex by recruiting the adaptor ASC (apoptosis-associated speck-like protein containing caspase-recruitment domain (CARD)) and an effector protease, caspase-18–12. This process relies on the homotypic interactions of CARD–CARD or pyrin domain (PYD)–PYD, linking NLRs either to the adaptor ASC or directly to pro-caspase-1. The inflammasome complex features prion-like structures, including PYD filaments of ASC and CARD filaments of caspase-1, which promote the coming together of component proteins for efficient activation and signal amplification13–16. The CARD of pro-caspase-4/–5/–11 directly binds to cytosolic lipopolysaccharide (LPS), triggering CARD-mediated oligomerization and inducing their enzymatic activation3,17,18.
Caspase-1 processes pro-inflammatory cytokines into their biologically active and secreted forms (for example, it processes pro-interleukin 1β (pro-IL-1β ) into IL-1β ). Caspase-4/–5/–11 are unable to cleave pro-IL-1β but mediate IL-1β maturation indirectly through the NLRP3–ASC–caspase-1 inflammasome19. Caspase-1/–4/–5/–11 cleave gasdermin D (GSDMD) to induce pyroptosis. Recent studies disclosed that caspase-4 and caspase-5 may have distinct substrate specificities and possibly have exclusive roles20,21. Pro-inflammatory cytokines and danger molecules released from pyroptotic cells, such as IL-1α and high-mobility group box 1, attract immune cells to promote rapid clearance of infection and enhance host defense systems22–24.
Serpins (serine-protease inhibitors) are a superfamily of proteins that share a conserved tertiary structure25,26. Among 16 phylogenetic clades, SERPINB1 belongs to the intracellular clade B serpins and resides mainly in the cytoplasm of neutrophils and monocytes because it lacks a secretory signal sequence27,28. SERPINB1 was originally identified as an inhibitor of serine proteases, including neutrophil elastase, proteinase-3 and cathepsin G, stored in azurophilic granules29. When the proteases leak to the cytoplasm during degranulation, SERPINB1 suppresses their enzymatic activities and prevents unwanted cellular damage. Similar to other serpins, SERPINB1 folds into a metastable structure consisting of nine α-helices and three β-sheets with a reactive center loop (RCL). In the classic serpin ‘conformational trapping’ mechanism of inhibition, cleavage of the RCL by the target protease allows structural conversion by rapid opening of β-sheet A and insertion of the cleaved RCL as an additional strand30. The target protease remains covalently bound to the RCL, which drags the protease to the opposite pole of the serpin, where the distortion inactivates its enzymatic activity.
Given that dysregulation of inflammatory caspases is associated with several diseases, various checkpoints are in place to halt accidental activation of caspase-1/–4/–5/–1131,32. Here we show a SERPINB1-mediated safeguard mechanism that prevents inflammatory caspases from undergoing unwarranted oligomerization and activation in the absence of an immunological threat, thereby curbing unsolicited inflammation.
Results
Inflammatory caspase-1/–4/–5 interact with SERPINB1.
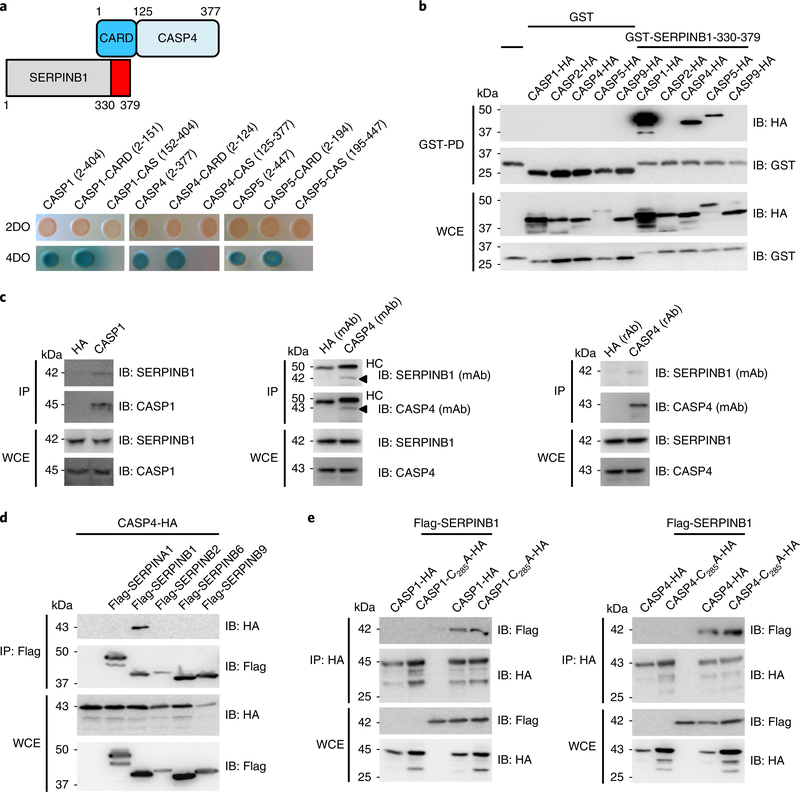
A yeast two-hybrid screening explored potential binding partners of caspase-4. Two clones encoding SERPINB1 were identified: one carrying a full-length protein (amino acids (aa) 1–379), and one with a carboxyterminal region (aa 330–379) (Fig. 1a). Further yeast binding studies revealed that SERPINB1 exhibited a specific interaction with the amino-terminal pro-domain CARD of caspase-1/–4/–5 but not with their carboxy-terminal catalytic domains (Fig. 1a). The caspase-4 CARD region is expected to carry six anti-parallel α-helices. Yeast two-hybrid mapping showed that α-helix 2 (α2) of the caspase-4 CARD seemed to be primarily responsible for SERPINB1 binding, as its binding activity was similar to that of full-length CARD, whereas α1 + α2 showed slightly reduced SERPINB1 binding (Supplementary Fig. 1a). I-TASSER-predicted structure of caspase-4 CARD indicated that residues D27 and V30 of α2 and residue E40 of α3 are potential ligand binding sites (Supplementary Fig. 1b). The D27A/V30A/E40A caspase-4 CARD mutant showed severely reduced SERPINB1 binding (Supplementary Fig. 1c).
Fig. 1 |. Interaction of inflammatory caspase-1/–4/–5 with SERPINB1.
a, Identification of SERPINB1 as a binding partner of caspase-4 CARD (aa 2–124) by yeast two-hybrid screening. For yeast co-transformation assays, full-length and truncated forms of caspase-1/–4/–5 were co-transformed with the SERPINB1 carboxy-terminal region (aa 330–379) to yeast for growth on two dropout (DO) or four dropout containing X-α-Gal plates. b, Specific interaction between SERPINB1 and inflammatory caspase family members. GST–SERPINB1–330–379 and hemagglutinin (HA)-tagged CARD-containing caspases were transfected into 293 T cells, and whole-cell extracts (WCEs) were subjected to GST pulldown (GST-PD), followed by immunoblotting (IB) using anti-HA or anti-GST. c, Endogenous interaction between caspase-1/–4 and SERPINB1. THP1 WCEs were subjected to co-immunoprecipitation (IP) with control anti-HA or anti-caspase-1. U937 WCEs were subjected to co-immunoprecipitation with control anti-HA or anti-caspase-4. mAb and rAb denote mouse and rabbit antibodies, respectively. HC, heavy chain. d, Specific binding of caspase-4 to SERPINB1. HA-tagged caspase-4 and Flag-tagged serpin family members were transfected into 293 T cells, and WCEs were subjected to co-immunoprecipitation with anti-Flag, followed by immunoblotting using anti-HA or anti-Flag. e, Interaction of enzymatically inactive caspase mutants with SERPINB1. HA-tagged caspase-1 or caspase-4 wild-type or enzymatically inactive mutants and Flag-tagged SERPINB1 were transfected into 293 T cells, and WCEs were subjected to co-immunoprecipitation with anti-HA, followed by immunoblotting with anti-Flag or anti-HA. Data in a–e are representative of two independent experiments.
We further determined binding specificity between SERPINB1 and caspase-1/–4/–5 in mammalian cells. A glutathione S-transferase (GST)-pulldown assay demonstrated that caspase-1/–4/–5 interacted with the carboxy-terminal region of SERPINB1, whereas CARD-containing apoptotic caspase-2/–9 did not (Fig. 1b). The endogenous interaction between caspase-1/–4 and SERPINB1 was also readily detected in THP1 or U937 human monocytes (Fig. 1c). Among various serpin family members, SERPINB1—but not SERPINA1, SERPINB2, SERPINB6 or SERPINB9—specifically bound caspase-4 (Fig. 1d). In addition, enzymatically inactive caspase-1-C285A and caspase-4-C258A mutants still possessed SERPINB1-binding ability, indicating that catalytic caspase activity is not required for the SERPINB1 interaction (Fig. 1e). Collectively, these results demonstrate specific interaction between SERPINB1 and caspase-1/–4/–5.
Identification of the CARD-binding motif of SERPINB1.
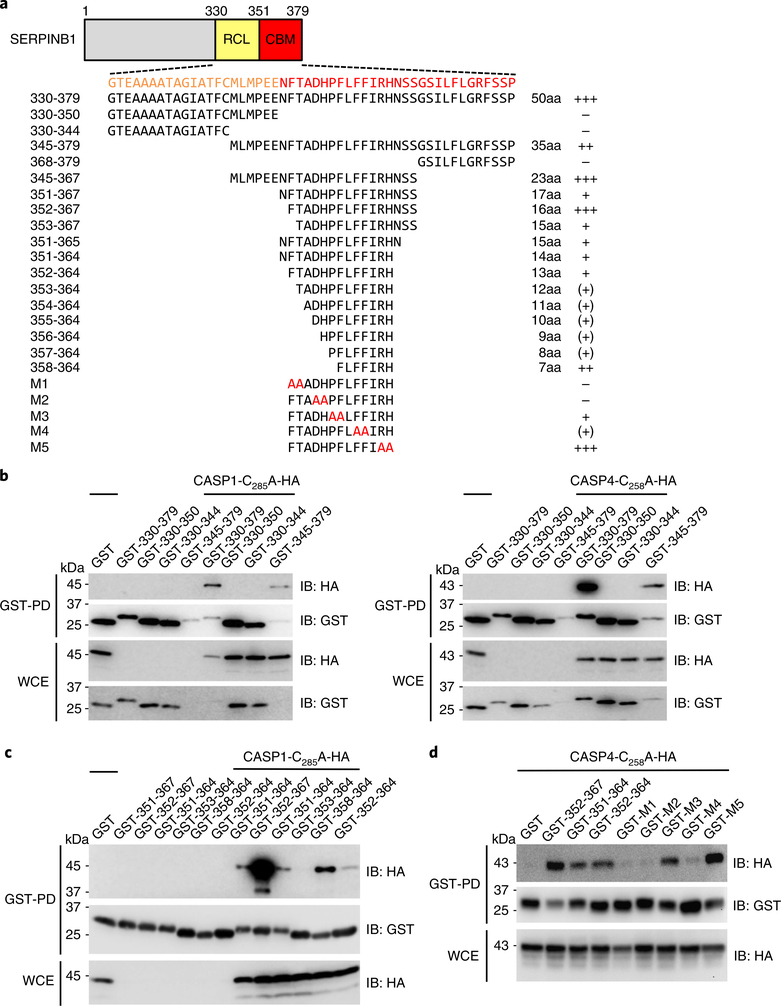
The yeast two-hybrid SERPINB1 clone carrying aa 330–379 includes the RCL region that interacts with neutrophil serine proteases. We further constructed various SERPINB1 carboxy-terminal fragments with GST fusions. GST-pulldown assays showed that the carboxy-terminal region (aa 351–379) of SERPINB1, but not the RCL region (aa 330–350), was responsible for caspase-1/–4 binding (Fig. 2a–c). We designated this carboxy-terminal 29-aa sequence as a CARD-binding motif (CBM). Within the CBM region, aa 352–367 (FTADHPFLFFIRHNSS) showed the strongest binding to caspase-1 (Fig. 2c). Obviously, the mutants with deletion of 13 aa (F352TADHPFLFFIRH364) or 7 aa (F358LFFIRH364) abolished caspase-1 CARD binding (Supplementary Fig. 2a). We also generated alanine-substitution mutagenesis of 13 aa (F352TADHPFLFFIRH364) and showed that the critical residues (F352, T353, D355, H356, F360 and F361) of the CBM were important for caspase-4 binding (Fig. 2d).
Fig. 2 |. Identification of the CBm of SERPINB1.
a, Schematic diagram of SERPINB1 truncation and alanine-substitution mutant constructs. The relative binding intensity is indicated as + ++ > ++ > + > (+ ) > −. b,c, GST-pulldown assay of SERPINB1 truncation mutants binding to caspase-1 and caspase-4. GST-SERPINB1 truncated forms and caspase-1-C285A–HA or caspase-4-C258A–HA were transfected into 293 T cells, and WCEs were subjected to GST pulldown, followed by immunoblotting using anti-HA or anti-GST. d, GST-pulldown assay of SERPINB1 alanine-substitution mutants binding to caspase-4. GST-SERPINB1 truncated or alanine-substituted forms and caspase-4-C258A–HA were transfected into 293 T cells, and WCEs were subjected to GST pulldown, followed by immunoblotting using anti-HA or anti-GST. Data in b–d are representative of two independent experiments.
While the SERPINB1 RCL mutant (F343A/C344A) lost proteinase-3-binding activity, it still effectively bound caspase-1/–4, indicating that neutrophil serine proteases and inflammatory cysteine caspases bind to disparate regions of SERPINB1 (Supplementary Fig. 2b). An in vitro enzymatic assay showed that purified SERPINB1 efficiently inhibited neutrophil elastase enzymatic activity yet did not affect the catalytic activity of caspase-1/–4 that contained only an enzymatic domain without the pro-domain CARD (Supplementary Fig. 2c,d). These results were consistent with the yeast two-hybrid data showing that the enzymatic domain of caspase-1/–4/–5 did not play a role in SERPINB1 interaction. Together, these findings demonstrate that SERPINB1 targets neutrophil serine proteases and inflammatory caspases through its RCL and CBM regions, respectively, suggesting that SERPINB1 affects two different protease families through distinct mechanisms.
SERPINB1 depletion–induced IL-1β secretion and pyroptosis.
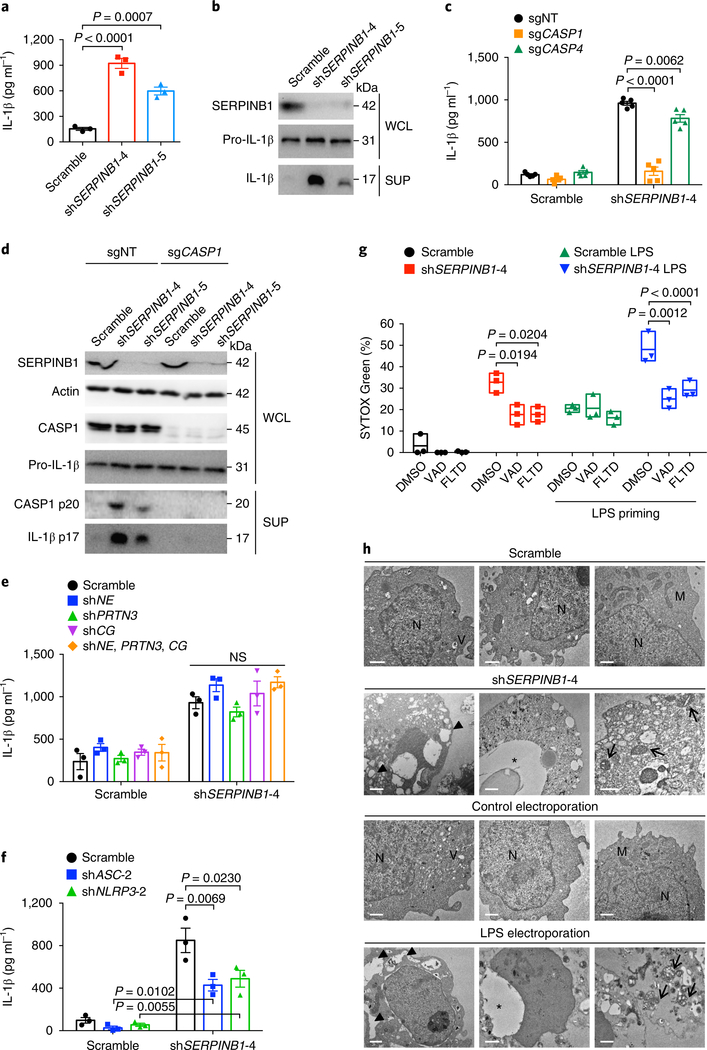
To investigate the physiological role of SERPINB1 in the regulation of inflammatory caspase activity, we depleted THP1 and U937 cells of SERPINB1 through the use of SERPINB1-specific short hairpin RNAs (shRNAs), followed by extracellular LPS priming for IL1B gene expression. Immunoblotting validated the knockdown efficiencies of three different SERPINB1-targeting shRNAs: shSERPINB1-4, –5 and –6 (Supplementary Fig. 3a and Supplementary Table 1). SERPINB1-depleted THP1 cells showed elevated IL-1β secretion compared with that of scrambled shRNA–treated cells (Fig. 3a). SERPINB1 knockdown had no considerable effect on LPS-induced secretion of tumor necrosis factor (TNF) or IL-6 concentrations (Supplementary Fig. 3b). The cleaved form of IL-1β (p17) was markedly increased after SERPINB1 depletion (Fig. 3b).
Fig. 3 |. SERPINB1 depletion induces spontaneous IL-1β secretion and cell death.
a, IL-1β secretion after SERPINB1 depletion. THP1 cells were transduced with scrambled or SERPINB1-specific shRNA (shSERPINB1) lentivirus for 48 h and were primed with LPS (1 μg ml−1) for 12 h. b, Immunoblots of the secreted p17 form of IL-1β after SERPINB1 depletion. c, Effects of deletion of the gene encoding caspase-1 or caspase-4 on SERPINB1-depletion-induced IL-1β secretion. d, Immunoblots of the cleaved p20 form of caspase-1 and processed p17 form of IL-1β in THP1-sgNT or THP1-sgCASP1 cells after SERPINB1 depletion. e,f, Effects of knockdown of genes encoding neutrophil elastase (shNE), proteinase-3 (shPRTN3), cathepsin G (shCG), ASC (shASC) or NLRP3 (shNLRP3) on SERPINB1-depletion-induced IL-1β secretion. g, Effects of caspase inhibitors on SERPINB1-depletion-induced cell death. SYTOX Green–based cell death was measured without or with LPS priming. h, Transmission electron microscopic images of U937 cells after SERPINB1 depletion. For a positive control, cells were electroporated with 1 μg LPS. Large vacuoles (filled triangles), disrupted nucleus (asterisks), swollen mitochondria (arrows). Scale bars, 1 μm. N, nucleus; V, phagocytic vacuole; M, mitochondria. Data in b,d,g,h are representative of two independent experiments. Whole-cell lysates (WCLs) and supernatants (SUPs) in b,d were immunoblotted with indicated antibodies. Data are presented as mean ± s.e.m. from n = 3 independent experiments in a,e,f and from n = 5 independent experiments in c, and as box and whiskers (min to max) from n = 3 technical replicates in g. P values were determined by one-way analysis of variance (ANOVA) with Dunnett’s comparison relative to scramble in a and by two-way ANOVA with Bonferroni’s comparison relative to non-targeting control (sgNT) in c, to scramble in e,f, or to dimethylsulfoxide (DMSO) in g. NS, not significant.
The SERPINB1-depletion-induced IL-1β secretion was completely blocked by a pan-caspase inhibitor (z-VAD-FMK) and was attenuated by caspase-1/–4/–5 inhibitors (z-YVAD-FMK, z-LEVD-FMK or z-WEHD-FMK) but was unaltered by a caspase-3 inhibitor (z-DEVD-FMK) (Supplementary Fig. 3c). We further generated THP1 cells stably expressing Cas9 and single-guide RNAs (sgRNAs) targeting CASP1 (sgCASP1) or CASP4 (sgCASP4) or a non-targeting control sgRNA (sgNT) (Supplementary Fig. 3d and Supplementary Table 2). Immunoblotting validated that caspase-1/–4 expression was completely disrupted (Supplementary Fig. 3d). Loss of CASP1 expression markedly dampened the SERPINB1-depletion-induced IL-1β secretion compared to the non-targeting control, whereas silencing CASP4 expression marginally reduced such secretion (Fig. 3c). Consistent with that, the p20 subunit of caspase-1 was readily detected in SERPINB1-depleted cells, and pro-IL-1β processing (p17) was impaired in sgCASP1 cells (Fig. 3d), indicating that caspase-1 is primarily responsible for IL-1β release after SERPINB1 knockdown.
To nullify the possibility of a contribution of neutrophil serine proteases to SERPINB1-depletion-induced IL-1β secretion, we generated THP1 cells stably containing shRNA lentivirus targeting genes encoding neutrophil elastase (shNE), proteinase-3 (shPRTN3), cathepsin G (shCG), all three genes (shNE, PRTN3, CG) or none (scramble). None of the neutrophil serine proteases affected SERPINB1-depletion-induced IL-1β release (Fig. 3e and Supplementary Fig. 3e). While SERPINB1-depletion-induced IL-1β release was detectably reduced after silencing of ASC or NLRP3 expression, it was still considerably higher than that of controls (Fig. 3f and Supplementary Fig. 3f). In addition, SERPINB1 depletion did not affect IL-1β secretion induced by nigericin, ATP, muramyl dipeptide, flagellin or poly(dA:dT) (Supplementary Fig. 3g).
SERPINB1 depletion led to spontaneous cell death of THP1 and U937 cells (Supplementary Fig. 3h–k). On the other hand, treatment with a pan-caspase inhibitor (z-VAD-FMK) or a GSDMD-derived caspase-1/–4/–5 inhibitor (Ac-FLTD-CMK)33 considerably reduced SERPINB1-depletion-induced cell death (Fig. 3g). This indicates that SERPINB1-depletion-induced cell death is mediated mainly by inflammatory caspases. Moreover, transmission electron microscopy showed that SERPINB1-depleted U937 cells displayed the typical pyroptotic morphology of large vacuoles, swollen mitochondria and disrupted nucleus, similar to cytosolic LPS–treated U937 cells (Fig. 3h). Overall, these data indicate that SERPINB1 functions as a checkpoint for basal IL-1β secretion and inflammatory cell death in a caspase-1/–4/–5 expression–dependent manner after no inflammasome stimuli.
Murine SERPINB1 paralogs for caspase-1/–11 inhibition.
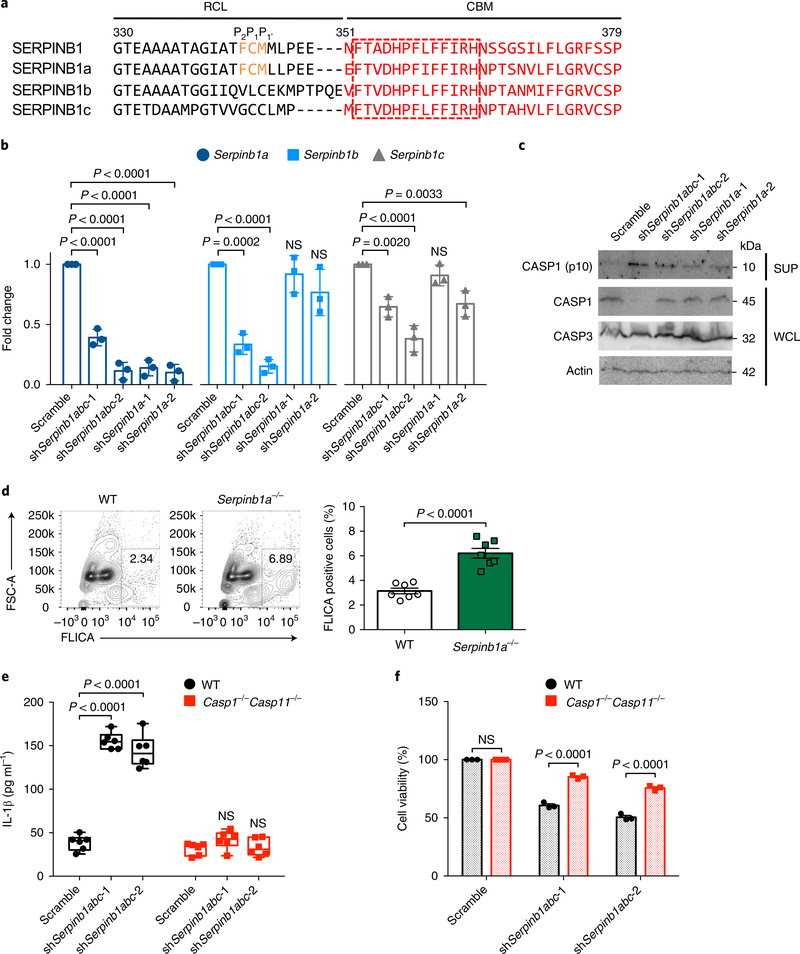
While humans have a single SERPINB1 gene, mice have three paralogs with high sequence homology: Serpinb1a, Serpinb1b and Serpinb1c. Human SERPINB1 and murine SERPINB1a show a highly conserved RCL sequence, especially the P2–P1–P1 (F343CM345) residues critical for serine-protease inhibition, whereas the putative RCLs of SERPINB1b and SERPINB1c are considerably different from that of human SERPINB1 (Fig. 4a). This suggests that SERPINB1a is the functional murine ortholog of human SERPINB1, but SERPINB1b and SERPINB1c may not replicate the serine-protease-inhibitory activity of human SERPINB1 (Fig. 4a). Conversely, all murine SERPINB1 isoforms (SERPINB1a, SERPINB1b and SERPINB1c) carry a well-conserved CBM, especially the 13-aa sequence F352TADHPFLFFIRH364 (Fig. 4a). Accordingly, those murine SERPINB1 isoforms interacted equally with murine caspase-1/–11 (Supplementary Fig. 4a). This suggests that the inhibition of inflammatory caspases may be a common function among human SERPINB1 and murine SERPINB1a, SERPINB1b and SERPINB1c, whereas the inhibition of neutrophil serine proteases may be shared primarily by human SERPINB1 and murine SERPINB1a.
Fig. 4 |. Role of murine SERPINB1 isoforms in caspase-1/–11 inhibition.
a, The RCL and CBM sequence alignment of human SERPINB1 and mouse SERPINB1a, SERPINB1b and SERPINB1c. The conserved P2–P1–P1 residues in the RCL sequence are indicated by orange font, and the CBM sequence is shown in red font. The conserved 13 aa in the CBM sequence are indicated in the red-dotted box. Specified amino acid numbers are based on human SERPINB1. b, Verification of Serpinb1-targeting shRNA’s silencing efficiency by qRT-PCR in DC2.4 cells. mRNA expression was normalized to 18S, and fold change was calculated relative to scramble. c, Immunoblots of cleaved p10 subunit of caspase-1 in DC2.4 cells after Serpinb1a, Serpinb1b and/or Serpinb1c depletion. WCLs and SUPs were obtained at 48 h post-transduction and were immunoblotted with indicated antibodies. d, Caspase-1 activation in Serpinb1a−/− pPMNs. FLICA-positive staining was analyzed by flow cytometry. e,f, IL-1β secretion and cell death after Serpinb1 depletion in BMDMs. Wild-type (WT) and Casp1−/−Casp11−/− BMDMs were transduced with scrambled or pan-Serpinb1 shRNA lentivirus. Cytokines were quantified by enzyme-linked immunosorbent assay (ELISA), and cell viability was determined by ATP-based assay. Data in c are representative of two independent experiments. Data are presented as mean ± s.e.m. from n = 3 independent experiments in b,f and from n = 7 per group, pooled from two independent experiments in d, and as box and whiskers (min to max) from n = 6 pooled from three independent experiments in e. P values were determined by one-way ANOVA with Dunnett’s comparison relative to scramble in b, by an two-tailed unpaired t-test in d and by two-way ANOVA with Bonferroni’s comparison relative to scramble in e,f.
Reverse transcription and quantitative PCR (qRT-PCR) showed that DC2.4 dendritic cells had higher expression of three Serpinb1 paralogs than that of J774A.1 or RAW264.7 macrophage cells (Supplementary Fig. 4b and Supplementary Table 3). DC2.4 cells were used for the shRNA-mediated ablation of Serpinb1 expression and the functional analysis of caspase-1 activation. Two different pan-Serpinb1 shRNAs (shSerpinb1abc-1 and shSerpinb1abc-2) efficiently abolished all expression of SERPINB1a, SERPINB1b and SERPINB1c, whereas two Serpinb1a-specific shRNAs (shSerpinb1a-1 and shSerpinb1a-2) ablated only SERPINB1a expression (Fig. 4b). Intracellular FLICA-positive staining, which labels active caspase-1, was highest in the pan-Serpinb1-depleted cells (~36%), elevated in the Serpinb1a-depleted cells (~18–20%) and low in the scrambled shRNA–treated cells (~4%) (Supplementary Fig. 4c). Consistent with that, the p10 subunit of caspase-1 was robustly detected in pan-Serpinb1-depleted cells, weakly detected in Serpinb1a-depleted cells and not detected in scrambled shRNA–treated cells (Fig. 4c). These results suggest that three Serpinb1 paralogs possess redundant functions to regulate inflammatory caspase activity. When bone marrow polymorphonuclear cells (PMNs) isolated from wild-type or Serpinb1a−/− mice were incubated in vitro overnight, Serpinb1a−/− PMNs showed consistently higher FLICA-positive staining than that of wild-type PMNs (Fig. 4d). This indicates that SERPINB1a deficiency leads to increased caspase-1 activity in primary PMNs under no-stimulation conditions.
We further examined the suppressive role of murine SERPINB1 in bone marrow–derived macrophages (BMDMs) from wild-type or Casp1−/−Casp11−/− mice. Pan-Serpinb1 depletion led to spontaneous IL-1β secretion in wild-type BMDMs but not in Casp1−/− Casp11−/− BMDMs (Fig. 4e). The silencing efficiencies were assessed by qRT-PCR (Supplementary Fig. 4d). Moreover, pan-Serpinb1-depletion-induced cell death was elevated in wild-type BMDMs, whereas it was considerably reduced in Casp1−/−Casp11−/− BMDMs (Fig. 4f). These data indicate that depletion of murine SERPINB1a, SERPINB1b and SERPINB1c induces basal IL-1β release and cell death in a caspase-1/–11 expression–dependent manner.
SERPINB1a deficiency elevates sensitivity to septic shock.
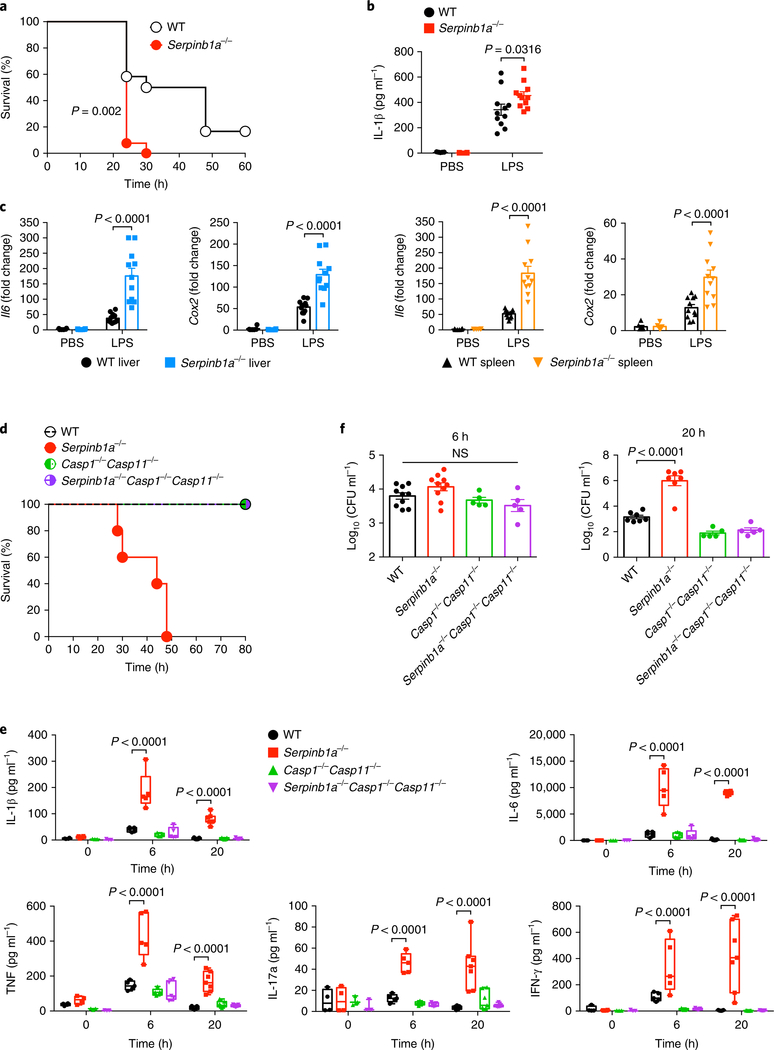
To determine the impact of SERPINB1a deficiency on LPS-induced endotoxic shock, we challeneged wild-type mice and Serpinb1a−/− mice with intraperitoneal injection of 20 mg kg−1 LPS and monitored their survival. Remarkably, 90% of Serpinb1a−/− mice succumbed to lethal LPS challenge at 1 day post-injection, whereas only 40% of wild-type mice died during the same time period (Fig. 5a). Serpinb1a−/− mice showed higher plasma IL-1β concentrations and increased liver and spleen Il6 and Cox2 mRNA expression than that of wild-type mice (Fig. 5b,c). These findings suggest that SERPINB1a deficiency elevates sensitivity to an LPS-induced systemic inflammatory response.
Fig. 5 |. Elevated sensitivity of Serpinb1a−/− mice to endotoxic shock.
a, Survival of wild-type (WT) and Serpinb1a−/− mice (n = 12 per group) intraperitoneally challenged with 20 mg kg−1 LPS. b,c, Plasma IL-1β and liver and spleen Il6 and Cox2 mRNAs of wild-type and Serpinb1a−/− mice (n = 6 for the PBS group and n = 11 for the LPS group). Serum and organs were collected at 3 h post-injection. IL-1β concentrations were determined by ELISA. mRNA expression was normalized to 18 S, and fold change was calculated relative to the PBS-treated wild-type. d, Survival of wild-type, Serpinb1a−/−, Casp1−/−Casp11−/− and Serpinb1a−/−Casp1−/−Casp11−/− mice (n = 5 per group) intravenously infected with 1.2 × 107 to 1.5 × 107 colony-forming units of HUMC1. e, Plasma cytokine concentrations of wild-type and Serpinb1a−/− mice (n = 3–4 per group at 0 hpi, n = 5 per group at 6 hpi, n = 7 per group at 20 hpi). Cytokine concentrations were quantified using a Bio-Plex assay. f, Blood bacterial burden of wild-type and Serpinb1a−/− mice (n = 10 for wild-type or Serpinb1a−/− at 6 hpi, n = 7 for wild-type or Serpinb1a−/− at 20 hpi, n = 5 for Casp1−/−Casp11−/− or Serpinb1a−/−Casp1−/−Casp11−/− at 6 or 20 hpi). Data are presented as Kaplan–Meier plot in a,d, as mean ± s.e.m. in b,c,f, and as box and whiskers (min to max) with line at median in e. P values were determined by log-rank test in a, by two-way ANOVA with Bonferroni’s comparison relative to wild-type in b,c,e, and by one-way ANOVA with Dunnett’s comparison relative to wild-type in f. NS, not significant.
To test if SERPINB1a deficiency affected susceptibility to bacteremia-induced sepsis, we used a highly virulent clinical isolate of Acinetobacter baumannii called ‘HUMC1’34. Wild-type, Serpinb1a−/−, Casp1−/−Casp11−/− and Serpinb1a−/−Casp1−/−Casp11−/− mice were infected intravenously with HUMC1. Wild-type mice survived the infection, but Serpinb1a−/− mice succumbed to death by 30 hours post infection (hpi) (Fig. 5d). In contrast, Serpinb1a−/− Casp1−/−Casp11−/− cleared the infection, indicating that SERPINB1a deficiency–induced lethality is dependent on caspase-1/–11 expression (Fig. 5d). Also, plasma concentrations of the cytokines IL-1β, IL-6, TNF, IL-17a and IFN-γ were markedly higher in Serpinb1a−/− mice than in wild-type, Casp1−/−Casp11−/− or Serpinb1a−/−Casp1−/− Casp11−/− mice (Fig. 5e). Although bacterial burden was similar in all four groups at 6 hpi, plasma cytokine concentrations spiked in Serpinb1a−/− mice (Fig. 5e,f). However, by 20 hpi, we detected higher bacterial loads in Serpinb1a−/− mice than in all other groups (Fig. 5f). Finally, Serpinb1a−/− mice showed enlarged spleens and germinal center disorganization (Supplementary Fig. 5). These findings demonstrate that SERPINB1a deficiency generates excess inflammatory cytokines and increases susceptibility to A. baumannii–induced sepsis. Neither Casp1−/−Casp11−/− mice nor Serpinb1a−/−Casp1−/− Casp11−/− mice showed hyper-inflammation or mortality, indicating that SERPINB1a activity is functionally linked to caspase-1/–11.
SERPINB1a deficiency increases inflammation after co-housing.
Laboratory mice live in extremely hygienic environment, as they are housed in specific pathogen-free (SPF) facilities. We examined if SERPINB1a deficiency affected basal inflammatory responses after exposure to a non-hygienic environment. Wild-type, Serpinb1a−/−, Casp1−/−Casp11−/− and Serpinb1a−/−Casp1−/−Casp11−/− mice were co-housed with retail pet-store mice that had been exposed to diverse physiological microbes35. After 2 weeks of co-housing, Serpinb1a−/− mice had significantly higher Il6 mRNA in blood leukocytes and lungs than that of wild-type, Casp1−/−Casp11−/− or Serpinb1a−/− Casp1−/−Casp11−/− mice (Supplementary Fig. 6a,b).
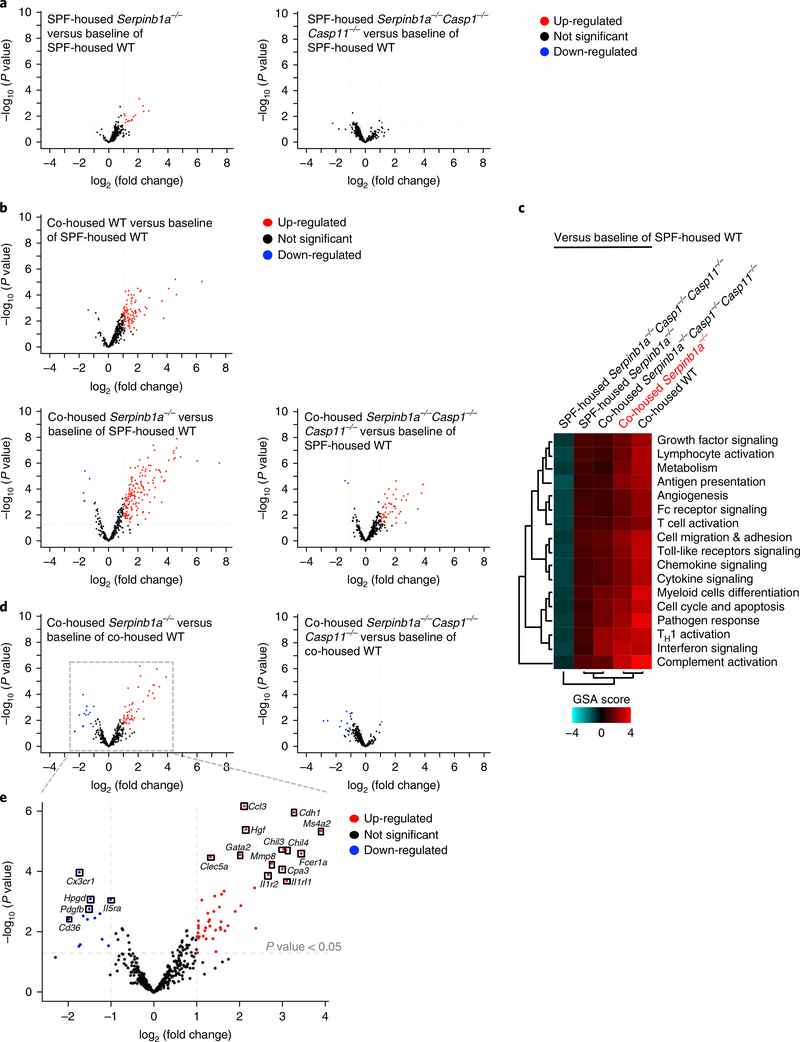
We further analyzed 754 genes encoding inflammatory immune signaling molecules in leukocytes of SPF-housed or co-housed wildtype, Serpinb1a−/− and Serpinb1a−/−Casp1−/−Casp11−/− mice. First, we observed that SPF-housed Serpinb1a−/− mice showed detectably up-regulated inflammatory gene expression compared with that of SPF-housed wild-type mice, whereas Serpinb1a−/−Casp1−/−Casp11−/− mice showed no difference from wild-type mice (Fig. 6a). When wild-type, Serpinb1a−/− and Serpinb1a−/−Casp1−/−Casp11−/− mice were co-housed with the ‘retail’ mice, co-housed Serpinb1a−/− mice exhibited the highest induction of inflammatory gene expression among three groups (Fig. 6b,c). More importantly, co-housed Serpinb1a−/− Casp1−/−Casp11−/− mice displayed considerably reduced inflammatory gene expression compared with that of co-housed Serpinb1a−/− mice and displayed similar or even slightly down-regulated inflammatory gene expression compared with that of co-housed wild-type mice (Fig. 6b–d). This indicates that SERPINB1a deficiency leads to highly elevated basal inflammation after rewilding husbandry and that this is dependent on caspase-1/–11 expression. In particular, the up-regulated inflammatory genes of co-housed Serpinb1a−/− mice encoded molecules involved mainly in cytokine and chemokine signaling and Toll-like receptor signaling, as well as the lymphocyte activation pathway (Fig. 6e and Supplementary Table 4). In conclusion, these results demonstrate that SERPINB1a deficiency lowers the threshold of inflammatory activation in natural husbandry conditions in a caspase-1/–11 expression–dependent manner, further indicating the vital role of SERPINB1a in restraining basal inflammatory caspase activity.
Fig. 6 |. Increased inflammation in non-hygienic co-housed Serpinb1a−/− mice.
a, NanoString volcano plot of SPF-housed Serpinb1a−/− mice or SPF-housed Serpinb1a−/−Casp1−/−Casp11−/− mice versus baseline of SPF-housed wild-type (WT) mice (n = 4 per group). b, NanoString volcano plot of co-housed wild-type, co-housed Serpinb1a−/− or co-housed Serpinb1a−/−Casp1−/−Casp11−/− mice versus baseline of SPF-housed wild-type mice (n = 4 per group). c, NanoString Gene Set Analysis (GSA) heatmap of directed global significance scores (n = 4 per group). Red denotes gene sets whose genes exhibit over-expression with the covariate, whereas blue denotes gene sets with under-expression with the covariate. The directed global significance score was calculated as the square root of the mean signed squared t-statistic for the genes in a gene set, with t-statistics coming from the linear regression underlying differential expression analysis. d,e, NanoString volcano plot of co-housed Serpinb1a−/− or Serpinb1a−/−Casp1−/−Casp11−/− mice versus baseline of co-housed wild-type mice (n = 4 per group). The gray-dotted box is magnified to show up- or down-regulated genes that are detailed in Supplementary Table 4. Volcano plots in a,b,d,e are presented as fold change in gene expression (log2(fold change)) against significance of change (–log10(P value)). P values were determined by two-tailed unpaired t-test in a,b,d,e.
SERPINB1 suppresses CARD oligomerization of caspase-1/–4.
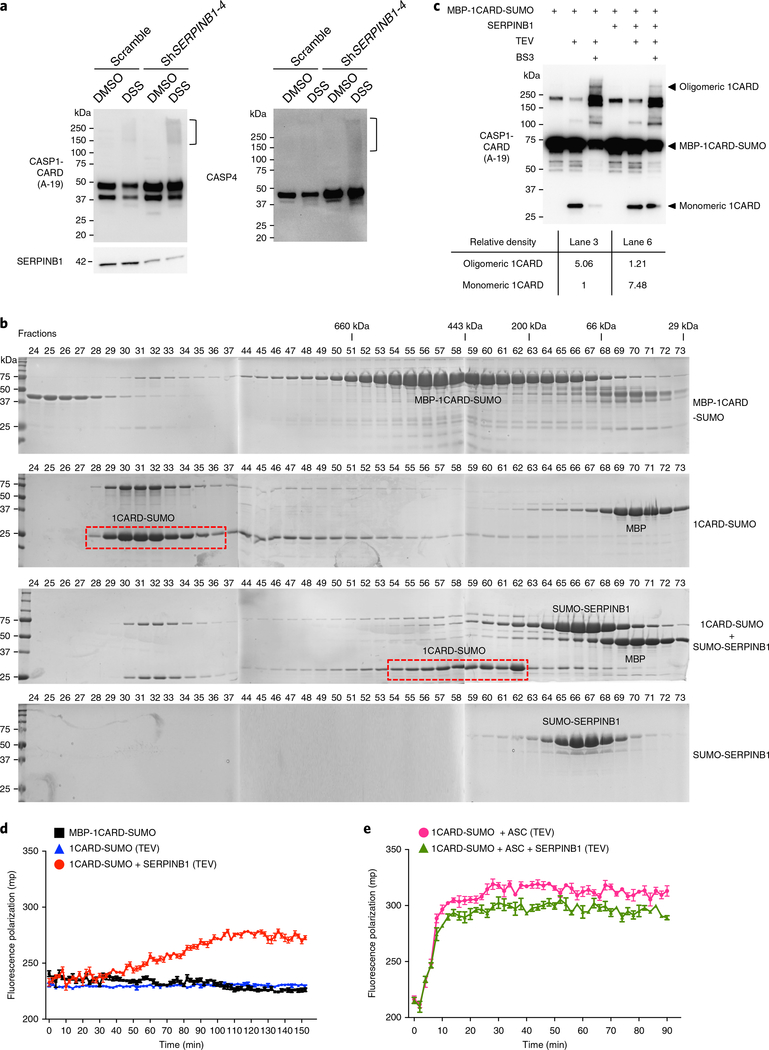
The key structural signature of inflammasome activation is the filamentous CARD oligomerization of caspase-1/–4. To address whether SERPINB1 inhibited caspase-1/–4 oligomerization, we first detected endogenous caspase-1/–4 oligomerization after SERPINB1 depletion by crosslinking with disuccinimidyl suberate (DSS). SERPINB1 depletion alone considerably increased the oligomerization of endogenous caspase-1/–4 compared to scramble control without inflammatory stimulation (Fig. 7a).
Fig. 7 |. SERPINB1 suppresses CARD oligomerization of caspase-1/–4.
a, Increased oligomerization of endogenous caspase-1/–4 after SERPINB1 depletion. Scramble or shSERPINB1 lentivirus–infected THP1 cells were pre-incubated with z-VAD-FMK and LPS for 6 h before DSS crosslinking. Crosslinked cells were lysed, and WCEs were immunoblotted with indicated antibodies. Square bracket (right margin) indicates oligomerized caspase-1 or caspase-4. b, Size-exclusion chromatography assay of caspase-1 CARD oligomerization. MBP-1CARD-SUMO was incubated with or without SUMO-SERPINB1 at 4 °C for 12 h and subjected to TEV cleavage. The indicated samples were analyzed by size-exclusion columns. SDS-PAGE of each fraction was stained with Coomassie blue. The red-dotted box indicates the fractions containing 1CARD-SUMO proteins. c, SERPINB1-mediated inhibition of caspase-1 CARD oligomerization. MBP-1CARD-SUMO (100 nM) was incubated with or without SERPINB1 (100 nM) at 4 °C for 12 h and was subjected to TEV cleavage for 2 h, followed by BS3 crosslinking. Crosslinked proteins were immunoblotted with indicated antibodies. Relative density was calculated by Image Lab Software. d,e, Fluorescence polarization assay of caspase-1 CARD oligomerization. SERPINB1 was incubated with FITC-conjugated MBP-1CARD-SUMO or together with MBP-ASC at 4 °C for 12 h and was subjected to TEV cleavage. The fluorescence polarization signals were collected at the indicated time point with an Envision plate reader. Data in a–e are representative of two independent experiments. Data in d,e are presented as mean ± s.e.m. from n = 3 technical replicates.
To further test the effect of SERPINB1 on CARD oligomerization in vitro, we obtained a sandwich-tagged caspase-1 CARD (1CARD) protein generated by conjugation with an amino-terminal maltose-binding protein (MBP) tag and a carboxy-terminal small ubiquitin-related modifier (SUMO) tag and acquired the soluble and monomeric forms of the MBP-1CARD-SUMO fusion protein. The addition of tobacco etch virus (TEV) protease resulted in cleavage of the amino-terminal His-MBP from the fusion protein, leading to oligomerization of the 1CARD-SUMO protein (Supplementary Fig. 7a). SERPINB1 was also purified as an amino-terminal SUMO fusion protein (SUMO-SERPINB1) to increase solubility. The oligomerization of 1CARD-SUMO protein after TEV treatment was determined by size-exclusion chromatography, whereby the oligomerized 1CARD-SUMO protein was eluted in void fractions 29–34 in the form of large aggregates (Fig. 7b). Pre-incubation with SUMO-SERPINB1 resulted in a considerable reduction in 1CARD-SUMO oligomerization after cleavage by TEV protease; most 1CARD-SUMO protein eluted in fractions 54–62 (Fig. 7b). Antibody to SUMO (antiSUMO) corroborated the location of SUMO-conjugated proteins in size-exclusion chromatography (Supplementary Fig. 7b).
We captured and visualized the oligomeric state of 1CARD protein by crosslinking with bis(sulfosuccinimidyl) suberate (BS3) and immunoblotting with antibody to caspase-1 CARD. TEV treatment produced a monomeric 1CARD, whereas subsequent BS3 crosslinking effectively induced 1CARD oligomerization, resulting a severe reduction in monomeric 1CARD (Fig. 7c, lanes 2 and 3). Conversely, the addition of SERPINB1 retained most of 1CARD in a monomeric form and considerably reduced the oligomerization of 1CARD (Fig. 7c, lane 6). Densitometry showed that the presence of SERPINB1 led to a 7.48-fold increase in 1CARD monomer and a 4.18-fold decrease in 1CARD oligomer compared to the absence of SERPINB1 (Fig. 7c, bottom), demonstrating that SERPINB1 suppresses in vitro CARD oligomerization of caspase-1.
To quantitatively assess the degree of 1CARD oligomerization, we performed an in vitro fluorescence polarization assay using fluorescein isothiocyanate (FITC)-conjugated MBP-1CARD-SUMO and SERPINB1 proteins (Supplementary Fig. 7c–e). Subsequent removal of MBP by TEV protease initiated the oligomerization of 1CARD-SUMO, as indicated by a gradual increase in fluorescence polarization values (Fig. 7d). Pre-incubation with SERPINB1 completely abolished the increased fluorescence polarization of 1CARD-SUMO after TEV treatment (Fig. 7d). As the adaptor ASC associates with caspase-1 through CARD–CARD homotypic interactions13,16, rapid oligomerization of 1CARD-SUMO was detected after incubation with MBP-ASC, in contrast to the slow oligomerization of 1CARD-SUMO alone (Fig. 7d,e). However, pre-incubation with SERPINB1 showed little or no effect on the ASC-mediated rapid oligomerization of 1CARD-SUMO (Fig. 7e). Collectively, these results demonstrate that SERPINB1 interaction effectively suppresses the spontaneously induced caspase-1 CARD oligomerization, which is overridden by ASC nucleation–mediated caspase-1 CARD oligomerization.
Overall, our study reveals that SERPINB1 has two separate sites for protease binding and inhibition (Supplementary Fig. 7f). The RCL is cleaved by and then covalently attaches to neutrophil elastase, proteinase-3 and cathepsin G, leading to irreversible protease inhibition. The CBM, consisting of β-sheet strand 1C (s1C) and β-sheet strands 4 and 5 (s4B and s5B), restrains the CARD oligomerization of caspase-1/–4/–5/–11, leading to reversible protease inhibition.
Discussion
The identification of regulatory mechanisms to limit adventitious inflammation is an important goal with the potential for many clinically relevant implications. Genetic mutations that cause dysregulated inflammasome signaling result in not only numerous inflammatory diseases, including cryopyrin-associated periodic syndromes and familial Mediterranean fever, but also chronic illnesses, including type 2 diabetes and neurodegenerative diseases36–38. CARD-only proteins, including COP1, INCA and ICEBERG, have been identified as caspase-1 decoys16,39,40. In particular, INCA prevents the elongation of caspase-1 CARD filaments at post-activation step. Cowpox virus homolog of serpin (CrmA) interrupts the p20–p10 dimerization of caspase-1, and the cellular serpin-protease inhibitor PI-9 inhibits caspase-1 (p20/p10) enzymatic activity41–44. While INCA, CrmA and PI-9 function as negative-feedback regulators of active caspase-1, SERPINB1 acts as a gatekeeper of inflammatory pro-caspases to avoid self-damage from inadvertent activation without stimulation. SERPINB1 deficiency increases the CARD–CARD self-association of inflammatory caspases, leading to the spontaneous dimerization and activation of their protease domains. It should be noted that this is independent of stimulation-induced inflammasome activation, in which inflammatory caspases cluster by binding ASC polymers after inflammasome assembly.
Unlike the exposed RCL region, the CBM of SERPINB1 (s4B and s5B) is masked by α-helices (αH, αG and αA), raising accessibility issues for CARD binding. This suggests that the surrounding α-helices and β-strands might be flexible or adopt a conformational change. Hormone-carrier serpins SERPINA6 (also known as corticosteroid-binding globulin) and SERPINA7 (also known as thyroxine-binding globulin) have a surface pocket between β-strands (s3B, s4B and s5B) and α-helices (αH and αA) for hormone binding45. The CARD-binding mode of SERPINB1 emulates, although not identically, the hormone-binding mode of SERPINA6 and SERPINA7. The CBM of SERPINB1 may cooperate with surrounding secondary structures to generate the CARD-binding pocket with high affinity and selectivity.
While the CARD forms helix–helix oligomerization in a heptad repeat fashion16, the SERPINB1 CBM does not have an α-helix but does have a β-sheet. This indicates that SERPINB1 CBM and caspase-4 CARD do not interact as a heptad repeat but hydrophobic interactions are rather important for the interaction. Furthermore, apoptotic caspase-2 and caspase-9 have amino acids at positions V30 and E40 different from those of caspase-4, whereas the residue D27 is conserved. Also, at the equivalent positions of the D27, V30 and E40 residues of caspase-4, caspase-1 and caspase-5 have D27, L30 and M40 residues and N85, A88 and E98 residues, respectively. This suggests that the specific residues and conserved structure of inflammatory caspase CARD are required for SERPINB1 interaction. Further investigation is needed to determine whether the D27A/V30A/E40A caspase-4 mutant is appropriately folded and functional.
What triggers the release of inflammatory caspases from SERPINB1 inhibition? It has been shown that the protease-mediated RCL cleavage of SERPINA6 induces conformational transition, generating an additional β-strand (s4A) and unwinding α-helix D46. The allosteric modulation of the cortisol-binding site allows cortisol release at sites of inflammation. Similarly, the elastase- or other protease-mediated RCL cleavage of SERPINB1 may perturb the configuration of the CARD-binding site, liberating inflammatory caspases from SERPINB1-mediated restraint in response to PAMPs and DAMPs. Thus, understanding the structural plasticity and flexibility of SERPINB1 is key to revealing the molecular details of SERPINB1-mediated CARD entrapment and to dissecting potential coordinated action between neutrophil serine proteases and inflammatory caspases.
As IL-1β rapidly induces NLRP3, ASC and CASP1 expression to amplify IL-1β response in an autocrine or paracrine manner47, we observed that NLRP3 or ASC silencing partially lowered SERPINB1-depletion-induced IL-1β secretion. This potential indirect role of the NLRP3–ASC–caspase-1 inflammasome in positive-feedback amplification of IL-1β signaling needs further investigation. In addition, non-canonical inflammasome-mediated GSDMD cleavage has been shown to induce neutrophil extracellular traps21,48. Future studies are warranted to understand the role of SERPINB1 and caspase-4/–5/–11 in neutrophil extracellular trap formation.
Although the complexity of sepsis makes it difficult to understand specific mechanisms in vivo, uncontrolled and excessive inflammatory cytokines ultimately result in immune paralysis, tissue damage and, finally, organ failure49. This suggests that SERPINB1a deficiency–mediated inflammatory caspase-1/–11 activation enhances the inflammatory response to LPS challenge or bacterial infection, thereby exacerbating mortality and morbidity. A. baumannii burdens were similar in wild-type, Serpinb1a−/−, Casp1−/−Casp11−/− and Serpinb1a−/−Casp1−/−Casp11−/− mice at 6 hpi, indicating that the initial abundant production of inflammatory cytokines in Serpinb1a−/− mice was not due to a large bacterial load. These results underscore the idea that the outcome of infection is driven as much by the host response to the pathogen as by bacterial burden. Furthermore, rewilding of laboratory mice allowed us to examine the gatekeeper function of SERPINB1a in vivo, which was not obvious when mice were housed in a SPF facility. This is an important observation, given the growing interest in different immunological events of ‘clean’ laboratory mice versus those of ‘dirty’wild mice with diverse microbial experience35,50.
In summary, our study has revealed that a new checkpoint mechanism, SERPINB1, targets neutrophil serine proteases and inflammatory caspases in a genetically and functionally separable manner, restricting unwanted inflammation. This also represents an important function of serpin-family proteins that minimizes damage to the host from detrimental inflammation by achieving a delicate balance between latent versus active inflammatory caspases.
Methods
Yeast two-hybrid screen.
Complementary DNA encoding the human caspase-4 CARD (aa 2–124) was cloned into the bait vector pGBKT7. The reporter AH109 yeast strain (Saccharomyces cerevisiae, Clontech) expressing caspase-4 CARD was transformed with a human leukocyte matchmaker cDNA library in accordance with Matchmaker GAL4 Two-Hybrid System 3 instructions (Clontech). For yeast co-transformation assay, the bait vector containing indicated caspase constructs and the prey vector pACT2 harboring SERPINB1 were co-transformed and screened in four dropout plates with 5 mM 3-amino-1,2,4-triazole.
Co-immunoprecipitation and GST pulldown.
293T cells were transfected with the indicated DNA plasmids using polyethylenimine transfection (Polysciences). Cells were collected after 48 h post-transfection, washed by PBS (Sigma) and resuspended in 1% NP40 lysis buffer containing 50 mM Tris-HCl, pH 8.0 (Invitrogen), 150 mM NaCl (Sigma) and 1% Nonidet P-40 (Sigma) supplemented with EDTA-free complete protease inhibitor cocktail (Roche). After sonication or three freeze/thaw cycles, WCL were centrifuged for 10 min at 16,000g. The supernatants, referred to as ‘WCEs’, were precleared with Sepharose beads (GE) rotating at 4 °C for 1 h and filtered through a 0.45 μm polyethersulfone filter (Thermo Fisher). For co-immunoprecipitation, WCEs were incubated with indicated antibodies at 4 °C for 3–12 h, followed by incubation with protein A/G agarose beads (Thermo Fisher) at 4 °C for 3 h. For GST pulldown, WCEs were incubated with glutathione-conjugated Sepharose beads (GE) at 4 °C for 1 h. The immobilized immunocomplexes or GST complexes containing beads were washed five times using 1% NP40 lysis buffer with various concentrations of NaCl (150–500 mM). Beads were eluted in 2× Laemmli dye (Sigma) by heating at 95 °C for 5 min and subjecting to immunoblotting.
Immunoblotting.
Primary antibodies include antibodies to human SERPINB1 (3B4, Origene), human caspase-1 (14F468, Santa Cruz), human caspase-1 (EPR16883, Abcam), human caspase-1 p20 (Bally-1, Adipogen), human caspase-1 CARD (A-19, Santa Cruz), mouse caspase-1 (M-20, Santa Cruz), human caspase-4 (4B9, Santa Cruz), caspase-2 (611022, BD), caspase-3 (8G10, Cell Signaling), IL-1β (2002, Cell Signaling), neutrophil elastase (C-17, Santa Cruz), proteinase-3 (684042, R&D Systems), cathepsin G (N-19, Santa Cruz), NLRP3 (Cryo-2, Adipogen), ASC (F-9, Santa Cruz), SMT3 (y-84, Santa Cruz), His (H-15, Santa Cruz), GST (B-14, Santa Cruz), Actin (C4, Santa Cruz), rabbit-Flag (F7425, Sigma), mouse-Flag (F1804, Sigma), rabbit-HA (PRB-101P, Covance), mouse-HA (16B12, BioLegend), mouse IgG-HRP (7076, Cell Signaling), rabbit IgG-HRP (7074, Cell Signaling) and goat IgG-HRP (sc-2020, Santa Cruz).
In vitro enzymatic assays.
Neutrophil elastase enzymatic assay (2 μM, Sigma) was incubated with SERPINB1 (20 μM or 40 μM) at 37 °C for 5 min and the reactions were diluted in 500 μl assay buffer containing MeO-SucAAPV-pNA (0.8 mM; 160 nmol, Sigma) chromogenic substrates. The 200 μl reaction mixtures were used to measure free p-nitroanilide (pNA) cleaved from substrate by spectrophotometric detection at 400 nm. For the caspase-1 and caspase-4 enzymatic assay, caspase-1 and caspase-4 (10 μM, Sigma) were incubated with SERPINB1 (40 μM) at 4 °C for 1 h and the reactions were diluted in assay buffer containing the following chromogenic substrates: caspase-1 substrate, Ac-YVAD-pNA (Sigma); caspase-4 substrate, Ac-LEVD-pNA (Sigma); 0.1 mM, 20 nmol final concentration. The reaction mixtures were incubated at 37 °C for hydrolysis of substrates. At the indicated time points, residual protease activity was quantified with PerkinElmer Envision plate reader (15 s, 100 times for neutrophil elastase; every 10 min for caspase-1 and caspase-4).
Plasmids.
All constructs for transient and stable expression in mammalian cells include pEF-MCS-IRES-puro (made in-house), pEBG-GST-MCS (made inhouse), pLKO.1-TRC (Addgene) and pL-CRISPR.EFS.PAC (Addgene) vectors. All caspase-1, –2, –4, –5, –9, –11 and proteinase-3 expression plasmids contain a carboxy-terminal hemagglutinin epitope tag, and all SERPINA1, B1, B2, B6, B9, B1a, B1b and B1c expression plasmids have an amino-terminal Flag epitope tag. Caspase-1-C285A and caspase-4-C258A expression plasmids were kindly provided by H.D. Beer. SERPINB1 truncation, deletion and point mutants, the caspase-4 CARD point mutant and caspase-11-C254A were constructed by a standard PCR cloning strategy or a GeneArt Site-Directed mutagenesis system (Invitrogen).
Cell culture.
The 293 T, THP1, U937, J774A.1 and Raw264.7 cell lines were purchased from ATCC. DC2.4 cells were kindly provided by K.L. Rock. 293 T, J774A.1 and RAW264.7 cell lines were grown in DMEM (Gibco) supplemented with 10% fetal bovine serum (FBS, Seradigm), 100 U per ml penicillin and 100 μg ml−1 streptomycin (1% Pen/Strep, Gibco). THP1 and U937 cells were maintained in RPMI-1640 (Gibco) with 10% FBS and 1% Pen/Strep. DC2.4 cells were cultured in RPMI-1640 supplemented with 10% FBS, 2 mM l-glutamine (Gibco), 1× non-essential amino acids (Gibco MEM NEAA × 100), 10 mM HEPES, pH 7.5 (Gibco), 55 μM β-mercaptoethanol (Gibco) and 1% Pen/Strep. All cells were maintained at 37 °C with 5% CO2.
Lentivirus-mediated shRNA knockdown.
The targeting sense strand sequences are listed in Supplementary Table 1. Recombinant shRNA pseudotyped viral particles were produced by packaging plasmids, including pGag/Pol, pRev and pVSV-G. THP1 cells (1 × 106 cells per well in a six-well plate), U937 cells (5 × 105 cells per well in a six-well plate), and DC2.4 cells (3 × 105 cells per well in a 12-well plate) were transduced with lentivirus by centrifugation at 524g for 45 min in media containing 4 μg ml−1 polybrene (Sigma).
CRISPR/Cas9 system.
CASP1-specific and CASP4-specific sgRNA sequences are listed in Supplementary Table 2. THP1 cells were constructed to stably express Cas9 and sgRNAs by selection with puromycin (0.5 μg ml−1). Single clones were obtained by serial dilution.
ELISA.
Human IL-1β ELISA Set II, human TNF ELISA Set, human IL-6 ELISA Set and mouse IL-1β ELISA Set (BD OptEIA). THP1 cells (1 × 106 cells per well in a six-well plate) were transduced with scramble or shSERPINB1 lentivirus for 48 h and primed with 1 μg ml−1 LPS (0127:B8, Sigma) for 12 h. For NLRP3 inflammasome activation, THP1 cells were differentiated with 100 ng ml−1 phorbol 12-myristate 13-acetate (Calbiochem) for 72 h, transduced by scramble or shSERPINB1 lentivirus for 48 h, and primed with LPS (0.5–1 μg ml−1) overnight. Cells were washed by PBS and stimulated with control media for 3 h, 2 μM nigericin (Sigma) for 3 h, 10 μg ml−1 muramyl dipeptide (Sigma) for 6 h, 5 mM ATP (Sigma) for 1 h, 2.5 μg ml−1 flagellin (Invivogen) for 6 h, or 1 μg ml−1 poly(dA:dT)/LyoVec (Invivogen) for 6 h. For caspase inhibitor treatment, THP1 cells were transduced with scramble or shSERPINB1 lentivirus for 48 h, treated with indicated caspase inhibitors (20 μM) 1 h before LPS (1 μg ml−1, 12 h) priming. Caspase inhibitors include z-YVAD-FMK (Santa Cruz), z-LEVD-FMK (BioVision), z-WEHD-FMK (BioVision), z-DEVD-FMK (Santa Cruz) and z-VAD-FMK (Santa Cruz).
SYTOX Green-based cell death.
A GSDMD-derived peptide inhibitor (Ac-FLTD-CMK) was provided by T.S. Xiao. Scramble or shSERPINB1 lentivirus–transduced THP1 cells were continuously treated with Ac-FLTD-CMK (5 μM) or z-VAD-FMK (5 μM) for 72 h. In addition, 5 × 104 cells per well in a 96-well black clear-bottomed plate were incubated with 0.5 μM SYTOX Green cell-impermeable dye (Thermo Fisher) for 3 h without or with LPS (1 μg ml−1).
Cell viability and cytotoxicity.
Cell viability was measured by ATP-based CellTiter-Glo Luminescent Cell Viability assay (Promega). Lactate dehydrogenase release was determined by CytoTox96 Non-Radioactive Cytotoxicity assay (Promega).
Mice.
Serpinb1a−/− mice (C57BL/6) have been previously described51. Casp1−/− Casp11−/− mice were purchased from The Jackson Laboratory. Serpinb1a−/−Casp1−/− Casp11−/− mice were generated by breeding of Serpinb1a−/− mice with Casp1−/− Casp11−/− mice. All mice were maintained in pathogen-free University of Southern California (USC) animal facilities and used in accordance with protocols and ethical regulations approved by the USC Institutional Animal Care and Use.
LPS-induced endotoxemia.
6- to 12-week-old age- and sex-matched wild-type and Serpinb1a−/− inbred mice were administered intraperitoneally with 20 mg kg−1 ultrapure Escherichia coli 0111:B4 LPS (Sigma) suspended in PBS. Mice were euthanized if they were moribund; i.e., they did not respond, were immobile or lost >25 % body mass. For plasma IL-1β, mice were euthanized 3 h after injection, and blood was obtained through cardiac puncture. For Il6 and Cox2 transcripts, the liver and spleen were isolated 3 h after injection and were subjected to TRI reagent (Sigma) and TissueLyser II (Qiagen) for 30 s and 30 frequency per s.
A. baumannii infection.
For infection with a hypervirulent strain of A. baumannii (HUMC1), 8- to 12-week-old age- and sex-matched mice were challenged intravenously via the tail vein with 1.2 × 107 to 1.5 × 107 colony-forming units. HUMC1 were grown overnight in tryptic soy broth at 37 °C with shaking. The bacteria were passaged to mid-log growth phase in tryptic soy broth at 37 °C with shaking. Cells were washed three times with PBS and resuspended at the appropriate concentration for infection. For survival studies, mice were monitored twice daily and were euthanized when they appeared moribund. Blood was obtained by retro-orbital bleeding or cardiac puncture. For quantifying bacterial burden, blood was serially diluted in PBS and plated on tryptic soy agar plates. Following overnight incubation at 37 °C, colony-forming units were counted. Plasma cytokine concentrations were quantified using a custom Bio-Plex mouse cytokine assay (Bio-Rad) and were analyzed on a Bio-Plex 200 plate reader using Bio-Plex Manager software 6.1 (Bio-Rad) at the USC Immune Monitoring Core.
Co-housing.
8- to 12-week-old wild-type, Serpinb1a−/− and Serpinb1a−/−Casp1−/− Casp11−/− inbred mice were co-housed with pet-store mice within a Biosafety Level-3 facility. Blood was obtained through retro-orbital bleeding, red blood cells were lysed with RBC lysis buffer (BioLegend), and total RNA was isolated using an RNeasy Mini kit (Qiagen). Isolated lung was subjected to TRI reagent and lysed by TissueLyser II, then 100 ng RNA (20 ng μl−1) was subjected to NanoString gene expression analysis at the USC Molecular Genomics Core and 754 myeloid innate immunity panel genes were analyzed using nSolver software v.4.0.
Primary BMDM and PMN isolation.
Bone marrow cells were resuspended in DMEM supplemented with 20% L929 supernatants containing macrophage colony-stimulating factor (prepared in-house), 10% FBS, 2 mM l-glutamine, 1 mM sodium pyruvate (Gibco) and 1% Pen/Strep and was plated in a 10-cm untreated plate (1.5 × 107 cells per plate). On day 3, fresh media containing 10% L929 supernatants containing macrophage colony-stimulating factor was added. On day 4, BMDMs were transduced with scrambled or pan-Serpinb1 shRNA lentivirus. On day 6, cells (3 × 105 cells per well of a 24-well plate) were primed with LPS (1 μg ml−1) for 12 h. On day 7, cells and supernatants were used for experiment. Cold DPBS (Gibco) was used to harvest cells instead of trypsin. For PMN preparation, bone marrow cells isolated from 6- to 12-week-old sex-matched littermate mice were resuspended in 1 ml Hank’s Balanced Salt Solution without calcium and magnesium (HBSS, Gibco) and were laid on two-layer of Histopaque 1119 (3 ml) and Histopaque 1077 (3 ml) (Sigma). After centrifugation at 800g for 30 min at 23 °C without braking, PMNs were collected from interface between the two layers. For in vitro culture, PMNs were cultured for 16–20 h in RPMI-1640 containing 10% FBS.
FLICA staining.
DC2.4 cells (3 × 105 cells per well in a 12-well plate) were transduced with scrambled or shSerpinb1 shRNA lentivirus for 48 h. PMNs of wild-type or Serpinb1a−/− mice were cultured for 16–20 h. Cells were washed by PBS and resuspended in 300 μl PBS. In addition, 5 × 105 cells were incubated with 10 μl of × 30 FLICA, FAM-YVAD-FLICA (ImmunoChemistry Technologies) at 37 °C for 30 min. Cells were fixed and analyzed on a FACSCanto II flow cytometer (BD) and data were obtained using FlowJo software (VWR).
qRT-PCR.
A total of 1 μg RNA was extracted using TRI reagent or RNeasy Mini, digested with DNase I (Sigma) and reverse-transcribed into cDNA using an iScript cDNA synthesis kit (Bio-Rad). cDNA was quantified using an iQ SYBR Green Supermix kit (Bio-Rad) and a CFX96 thermocycler (Bio-Rad). Gene-specific probes for qRT–PCR are listed in Supplementary Table 3.
Immunohistochemistry.
Spleens were fixed in 4% paraformaldehyde and embedded in paraffin. Sectioning and hematoxylin–eosin staining were performed by the USC Immunohistochemistry Core facility. Images were captured using a BZ-X710 series microscope (Keyence).
Negative stained transmission electron microscopy
U937 cells (1 × 106) were harvested after 48 h post-transduction with scramble or shSERPINB1 lentivirus. For the LPS control, U937 cells (1 × 106) were transfected with 1 μg LPS using Neon electroporation (Thermo Fisher) and were harvested 2 h later. The cell pellets were fixed overnight in half-strength Karnovsky’s Fixative (2% paraformaldehyde and 2.5% gluteraldehyde) in 0.1 M cacodylate buffer, pH 7.0, were rinsed in buffer and were post-fixed for 1 h in 1% osmium tetroxide buffer. They were stained overnight en bloc with 1% uranyl acetate. Following this, the pellets were soaked in 10% BSA in H2O for 2 h and the BSA was hardened to a gelatin with the half-strength Karnovsky’s Fixative to prevent pellet dispersion in subsequent steps. The pellets were dehydrated in a graded series of ethanol, infiltrated with Epon over several days, placed in Epon blocks and polymerized at 60 °C overnight. The blocks were sectioned at 70 nm, and the sections were placed on copper grids. Sections were examined and photographed using a JEOL JEM-2100 Transmission Electron Microscope at 80 kV employing Gatan Micrograph software v.1.84.
Recombinant protein expression and purification.
The sandwich-tagged caspase-1 CARD plasmid (modified pDB-His-MBP vector) contains an amino-terminal MBP tag with 6× His tag and a carboxy-terminal SUMO tag. SERPINB1 was cloned into a modified pET-28 vector containing an amino-terminal SUMO tag. These constructs were transformed and expressed in E. coli BL21 (DE3) RIPL strain (Novagen). Cells were grown at 37 °C until OD600 reached 0.8, and protein expression was induced with 0.5 mM isopropyl β-D-1-thiogalactoside (GoldBio) at 18 °C for 18–20 h. Bacteria were lysed in buffer containing 50 mM Tris-HCl, pH 8.0, 150 mM NaCl and 1% Nonidet P-40 supplemented with complete protease inhibitor EDTA-free cocktail. After sonication, cell lysates were centrifuged at 16,000g for 1 h and the supernatants containing His-tagged proteins were purified by affinity chromatography using HisPur Cobalt resin (Thermo Fisher) by gravity. The beads were washed with 20 column-volumes of buffer containing 50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 10 mM imidazole (Sigma) and 3 mM β-mercaptoethanol (Sigma). The protein was eluted with 200 mM imidazole and dialyzed in buffer containing 50 mM Tris-HCl, pH 8.0, and 150 mM NaCl to remove imidazole. When needed, the eluted proteins were passed through size-exclusion chromatography to achieve monomeric fractions. All purification steps were carried out at 4 °C to avoid protein aggregation.
Size-exclusion chromatography.
Sandwich-tagged caspase-1 CARD was incubated with SUMO-SERPINB1 at a molar ratio of 1:1 at 4 °C for 12 h, and nucleation of caspase-1 CARD was initiated by removal of the MBP tag by TEV (Sigma, His- and GST-tagged) in buffer containing 1 mM DL-dithiothreitol (DTT, Sigma) at 4 °C for 12 h. The TEV enzyme was removed by incubation with glutathione-conjugated Sepharose beads at 4 °C for 1 h. The protein mixtures were passed through a pre-equilibrated Superose 6 10/300 GL size-exclusion column (GE) with a Biologic Duo Flow system (Bio-Rad). Size-exclusion chromatography buffer contains 20 mM Tris-HCl, pH 8.0, 150 mM NaCl and 1 mM DTT. Chromatography was operated with 0.2 ml min−1 flow rate, 217 psi max pressure and 250 μl fractionation volume. Each fraction (20 μl) was separated by SDS-PAGE, followed by Coomassie Brilliant Blue staining (Brilliant blue R, Sigma). The molecular mass of eluted proteins was estimated by pre-running of standard markers (MWGF1000, Sigma).
Fluorescence polarization assay.
In vitro Sortase A labeling was used for site-specific incorporation. A Sortase A recognition motif (LPETGG) was inserted into the carboxyl terminus of the sandwich-tagged caspase-1 CARD plasmid (MBP-1CARD-SUMO). Then, 2 mg monomeric MBP-1CARD-SUMO protein was incubated with purified Sortase A (Addgene, plasmid) and Gly-Gly-Gly-Lys-FITC (GGG-K-FITC) fluorescence probe (Biomatik service) in sortase buffer (10× buffer: 500 mM Tris-HCl, pH 7.5, 1.5 M NaCl and 100 mM CaCl2) at 4 °C overnight. The ratio was protein:Sortase A:GGG-K-FITC = 50 μM:30 μM:500 μM. The mixtures were subjected to size-exclusion chromatography to remove excess free fluorophore-peptide. The FITC-conjugated monomeric MBP-1CARD-SUMO was diluted to 4 μM for the fluorescence polarization assay. His-ULP1 protease (LifeSensors) was used to cleave the amino-terminal SUMO from SERPINB1. The cleaved His-SUMO and His-ULP1 were removed by passage through HisPur Cobalt resin. The labeled MBP-1CARD-SUMO protein was incubated with SERPINB1 at a molar ratio of 1:2 at 4 °C for 12 h and was subjected to TEV cleavage. The amino-terminal MBP-fused ASC was incubated with MBP-1CARD-SUMO at a molar ratio of 10:1. The 20 μl mixtures were transferred into a black, round-bottomed, 384-well Corning plate in triplicate, on ice. Fluorescence polarization data were collected with an Envision plate reader (excitation channel, 104 FITC FP480; two emission channels, 208 FITC fluorescence polarization Spol 535 and 209 FITC fluorescence polarization Ppol 535; FITC fluorescence polarization, single; 2100–4090 Optical Module, excitation light; reduced 1%).
DSS or BS3 non-cleavable crosslinking.
At 3 days post-transduction of scramble or shSERPINB1 lentivirus, THP1 cells (1 × 107) were treated with LPS (0.5 μg ml−1) and z-VAD-FMK (20 μM) for 6 h. Cells were washed by PBS and resuspended in 200 μl of 2.5 mM DSS (Abcam) in PBS for 30 min at 23 °C with constant mixing. Reactions were quenched by incubation with 50 mM Tris-HCl pH 7.5 buffer for 10 min. Crosslinked cells were centrifuged at 300g for 5 min and lysed in 50 μl of 1% NP40 lysis buffer containing 50 mM Tris-HCl, pH 8.0, 150 mM NaCl and 1% Nonidet P-40 supplemented with complete protease inhibitor cocktail. WCEs were subjected to immunoblotting. For BS3 crosslinking of in vitro–purified proteins, sandwich caspase-1 CARD protein was incubated with SERPINB1 protein at a molar ratio of 1:1 at 4 °C for 12 h and was subjected to TEV treatment in buffer containing 1 mM DTT at 4 °C for 2 h. The protein mixtures were then incubated with or without 2.5 mM BS3 (Abcam) at 4 °C for 10 min, mixed with 2× Laemmli buffer (without boiling) and resolved on a 4–12% gradient SDS-PAGE gel (NuPAGE Bis-Tris gel, Thermo Fisher).
Statistics and reproducibility.
All experiments were repeated at least twice with similar results. Results are presented as mean ± s.e.m., Kaplan–Meier plot, or box and whiskers (min to max) with line at median or floating bars (min to max) with line at mean. For data analysis, log-rank test was used for survival; one-way ANOVA with Dunnett’s comparison and two-way ANOVA with Bonferroni’s comparison for multi-component comparisons; two-tailed unpaired Student’s t-test for two component comparisons in GraphPad Prism v.6.0.
Reporting Summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The data that support the findings of this study are available from the corresponding author upon request.
Supplementary Material
Acknowledgements
We thank E. Remold-O’Donnell (Boston Children’s Hospital) for Serpinb1a−/− mice, J. Yuan (Harvard Medical School) for Casp11−/− mice, T.S. Xiao (Case Western Reserve University), H.D. Beer (University of Zurich) and K.L. Rock (University of Massachusetts Medical School) for reagents, M.J. Kwak and B.H. Oh for protein purification, and J.W. Bowman and R. Amatya for manuscript preparation. This work was partly supported by grant nos. CA180779, CA200422, AI073099, AI116585, AI129496, AI140718, AI140705, DE023926, DE027888 and DE028521 (J.U.J.), NRF-2016R1D1A1B03931761 (H.-R.L.), Al124491 and HD087988 (H.W.), and AI081719, AI117211, AI127954 and AI106375 (B.S.).
Footnotes
Competing interests
The authors declare no competing interests.
Additional information
Supplementary information is available for this paper at https://doi.org/10.1038/s41590-018-0303-z.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, statements of data availability and associated accession codes are available at https://doi.org/10.1038/s41590-018-0303-z.
References
- 1.Kayagaki N et al. Non-canonical inflammasome activation targets caspase-11. Nature 479, 117–121 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Kayagaki N et al. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature 526, 666–671 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Shi J et al. Inflammatory caspases are innate immune receptors for intracellular LPS. Nature 514, 187–192 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Shi J et al. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature 526, 660–665 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Martinon F & Tschopp J Inflammatory caspases: linking an intracellular innate immune system to autoinflammatory diseases. Cell 117, 561–574 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Franchi L, Munoz-Planillo R & Nunez G Sensing and reacting to microbes through the inflammasomes. Nat. Immunol 13, 325–332 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lamkanfi M & Dixit VM Mechanisms and functions of inflammasomes. Cell 157, 1013–1022 (2014). [DOI] [PubMed] [Google Scholar]
- 8.Mariathasan S et al. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature 430, 213–218 (2004). [DOI] [PubMed] [Google Scholar]
- 9.Mariathasan S et al. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature 440, 228–232 (2006). [DOI] [PubMed] [Google Scholar]
- 10.Miao EA et al. Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1β via Ipaf. Nat. Immunol 7, 569–575 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Munoz-Planillo R et al. K+ efflux is the common trigger of NLRP3 inflammasome activation by bacterial toxins and particulate matter. Immunity 38, 1142–1153 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen J & Chen ZJ PtdIns4P on dispersed trans-Golgi network mediates NLRP3 inflammasome activation. Nature. 564, 71–76 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu A et al. Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes. Cell 156, 1193–1206 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cai X et al. Prion-like polymerization underlies signal transduction in antiviral immune defense and inflammasome activation. Cell 156, 1207–1222 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang L et al. Cryo-EM structure of the activated NAIP2-NLRC4 inflammasome reveals nucleated polymerization. Science 350, 404–409 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu A et al. Molecular basis of caspase-1 polymerization and its inhibition by a new capping mechanism. Nat. Struct. Mol. Biol 23, 416–425 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hagar JA, Powell DA, Aachoui Y, Ernst RK & Miao EA Cytoplasmic LPS activates caspase-11: implications in TLR4-independent endotoxic shock. Science 341, 1250–1253 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kayagaki N et al. Noncanonical inflammasome activation by intracellular LPS independent of TLR4. Science 341, 1246–1249 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Ruhl S & Broz P Caspase-11 activates a canonical NLRP3 inflammasome by promoting K+ efflux. Eur. J. Immunol 45, 2927–2936 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Bitto NJ, et al. Membrane vesicles from Pseudomonas aeruginosa activate the noncanonical inflammasome through caspase-5 in human monocytes. Immunol. Cell Biol 96, 1120–1130 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Lagrange B et al. Human caspase-4 detects tetra-acylated LPS and cytosolic francisella and functions differently from murine caspase-11. Nat. Commun 9, 242 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ng TM & Monack DM Revisiting caspase-11 function in host defense. Cell. Host. Microbe 14, 9–14 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miao EA et al. Caspase-1-induced pyroptosis is an innate immune effector mechanism against intracellular bacteria. Nat. Immunol 11, 1136–1142 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Broz P Getting rid of the bad apple: inflammasome-induced extrusion of Salmonella-infected enterocytes. Cell. Host. Microbe 16, 153–155 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Carrell R & Travis J α1-Antitrypsin and the serpins: variation and countervariation. Trends. Biochem. Sci 10, 20–24 (1985). [Google Scholar]
- 26.Silverman GA et al. The serpins are an expanding superfamily of structurally similar but functionally diverse proteins. Evolution, mechanism of inhibition, novel functions, and a revised nomenclature. J. Biol. Chem 276, 33293–33296 (2001). [DOI] [PubMed] [Google Scholar]
- 27.Benarafa C & Remold-O’Donnell E The ovalbumin serpins revisited: perspective from the chicken genome of clade B serpin evolution in vertebrates. Proc. Natl Acad. Sci. USA 102, 11367–11372 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Remold-O’Donnell E, Chin J & Alberts M Sequence and molecular characterization of human monocyte/neutrophil elastase inhibitor. Proc. Natl Acad. Sci. USA 89, 5635–5639 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cooley J, Takayama TK, Shapiro SD, Schechter NM & Remold-O’Donnell E The serpin MNEI inhibits elastase-like and chymotrypsin-like serine proteases through efficient reactions at two active sites. Biochemistry 40, 15762–15770 (2001). [DOI] [PubMed] [Google Scholar]
- 30.Law RH et al. An overview of the serpin superfamily. Genome. Biol 7, 216 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rathinam VA, Vanaja SK & Fitzgerald KA Regulation of inflammasome signaling. Nat. Immunol 13, 333–342 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Latz E, Xiao TS & Stutz A Activation and regulation of the inflammasomes. Nat. Rev. Immunol 13, 397–411 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang J et al. Mechanism of gasdermin D recognition by inflammatory caspases and their inhibition by a gasdermin D-derived peptide inhibitor. Proc. Natl Acad. Sci. USA 115, 6792–6797 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bruhn KW et al. Host fate is rapidly determined by innate effector-microbial interactions during Acinetobacter baumannii bacteremia. J. Infect. Dis 211, 1296–1305 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beura LK et al. Normalizing the environment recapitulates adult human immune traits in laboratory mice. Nature 532, 512–516 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Strowig T, Henao-Mejia J, Elinav E & Flavell R Inflammasomes in health and disease. Nature 481, 278–286 (2012). [DOI] [PubMed] [Google Scholar]
- 37.Robbins GR, Wen H & Ting JP Inflammasomes and metabolic disorders: old genes in modern diseases. Mol. Cell 54, 297–308 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo H, Callaway JB & Ting JP Inflammasomes: mechanism of action, role in disease, and therapeutics. Nat. Med 21, 677–687 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Le HT & Harton JA Pyrin- and CARD-only proteins as regulators of NLR functions. Front. Immunol 4, 275 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pedraza-Alva G, Perez-Martinez L, Valdez-Hernandez L, Meza-Sosa KF & Ando-Kuri M Negative regulation of the inflammasome: keeping inflammation under control. Immunol. Rev 265, 231–257 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Dobo J, Swanson R, Salvesen GS, Olson ST & Gettins PG Cytokine response modifier a inhibition of initiator caspases results in covalent complex formation and dissociation of the caspase tetramer. J. Biol. Chem 281, 38781–38790 (2006). [DOI] [PubMed] [Google Scholar]
- 42.Young JL et al. The serpin proteinase inhibitor 9 is an endogenous inhibitor of interleukin 1β-converting enzyme (caspase-1) activity in human vascular smooth muscle cells. J. Exp. Med 191, 1535–1544 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Annand RR et al. Caspase-1 (interleukin-1beta-converting enzyme) is inhibited by the human serpin analogue proteinase inhibitor 9. Biochem. J 342(Pt 3), 655–665 (1999). [PMC free article] [PubMed] [Google Scholar]
- 44.Simonovic M, Gettins PGW & Volz K Crystal structure of viral serpin crmA provides insights into its mechanism of cysteine proteinase inhibition. Protein Sci. 9, 1423–1427 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou A, Wei Z, Read RJ & Carrell RW Structural mechanism for the carriage and release of thyroxine in the blood. Proc. Natl Acad. Sci. USA 103, 13321–13326 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou A et al. The S-to-R transition of corticosteroid-binding globulin and the mechanism of hormone release. J. Mol. Biol 380, 244–251 (2008). [DOI] [PubMed] [Google Scholar]
- 47.Weber A, Wasiliew P & Kracht M Interleukin-1 (IL-1) pathway. Sci. Signal 3, cm1 (2010). [DOI] [PubMed] [Google Scholar]
- 48.Sollberger G et al. Gasdermin D plays a vital role in the generation of neutrophil extracellular traps. Sci. Immunol 3, eaar6689 (2018). [DOI] [PubMed] [Google Scholar]
- 49.Buras JA, Holzmann B & Sitkovsky M Animal models of sepsis: setting the stage. Nat. Rev. Drug. Discov 4, 854–865 (2005). [DOI] [PubMed] [Google Scholar]
- 50.Reese TA et al. Sequential infection with common pathogens promotes human-like immune gene expression and altered vaccine response. Cell. Host. Microbe 19, 713–719 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 51.Benarafa C, Priebe GP & Remold-O’Donnell E The neutrophil serine protease inhibitor serpinb1 preserves lung defense functions in Pseudomonas aeruginosa infection. J. Exp. Med 204, 1901–1909 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon request.