Summary
Astrocytes tile the central nervous system, but their functions in neural microcircuits in vivo and their roles in mammalian behavior remain incompletely defined. We used 2-photon laser scanning microscopy, electrophysiology, MINIscopes, RNA-seq and a genetic approach to explore the effects of reduced striatal astrocyte Ca2+ signaling in vivo. In wild type mice, reducing striatal astrocyte Ca2+-dependent signaling increased repetitive self-grooming behaviors by altering medium spiny neuron (MSN) activity. The mechanism involved astrocyte-mediated neuromodulation mediated by ambient GABA and was corrected by blocking astrocyte GABA transporter 3 (GAT-3). Furthermore, in a mouse model of Huntington’s disease, dysregulation of GABA and astrocyte Ca2+ signaling accompanied excessive self-grooming, which was relieved by blocking GAT-3. Assessments with RNA-seq revealed astrocyte genes and pathways regulated by Ca2+ signaling in a cell autonomous and non-cell autonomous manner, including Rab11a, a regulator of GAT-3 functional expression. Thus, striatal astrocytes contribute to neuromodulation controlling mouse obsessive-compulsive-like behavior.
ETOC/ “In brief”
The Khakh laboratory evaluated the consequences of genetically attenuating astrocyte calcium signaling in the adult mouse striatum in vivo. They discovered excessive self-grooming phenotypes, the mechanisms of which were explored at molecular, cellular and in vivo levels.
Introduction
Astrocytes tile the entire central nervous system and serve multiple roles including ion homeostasis, synapse formation, synapse removal, synaptic regulation and contributions to neurovascular coupling (Khakh and Sofroniew, 2015). Astrocytes are also proposed to regulate neuronal function via intracellular Ca2+-dependent signaling that has been documented in vitro and in vivo in diverse species (Bazargani and Attwell, 2016; Shigetomi et al., 2016). Furthermore, astrocyte Ca2+ signaling is known to be dysfunctional in mouse models of neurological disease (Khakh and Sofroniew, 2015). However, despite progress, the functional significance of astrocyte Ca2+-dependent signaling for the function(s) of neural microcircuits in vivo, for vertebrate behavior and for brain diseases remains incompletely understood.
A barrier to exploring astrocyte roles in vivo has been the inability to reduce Ca2+ signaling specifically within astrocytes in the fully developed adult brain in a nucleus specific manner. Previous studies addressed the functions of astrocyte Ca2+ signaling in vivo by employing IP3 receptor type 2 (IP3R2) deletion mice, which attenuated Ca2+ signals, but produced no behavioural consequences (Agulhon et al., 2010; Petravicz et al., 2014). However, the approach was not brain area specific, the mice lacked IP3R2s throughout the body, throughout development, and some astrocyte Ca2+ signaling persisted (Rungta et al., 2016; Srinivasan et al., 2015; Stobart et al., 2016). These are important considerations and the use of IP3R2 deletion mice has led to a confusing picture on the relevance of astrocyte Ca2+ signaling (Khakh and McCarthy, 2015; Shigetomi et al., 2016). The suggestion that astrocyte Ca2+ signaling has little role in regulating neurons or behavior is at odds with in vivo imaging and slice electrophysiology data (Nimmerjahn and Bergles, 2015; Volterra et al., 2014). An additional consideration is the fact that astrocytes change during development, display brain-area specialisations and exhibit diverse types of Ca2+ signals (Shigetomi et al., 2016). Furthermore, behavior derives from the output of microcircuits located in distinct brain nuclei. Therefore, brain area specific methods to attenuate astrocyte Ca2+-dependent signaling in adult mice in vivo are needed. In these regards, some studies have used overexpression of IP3 binding proteins as a “sponge” to decrease IP3-dependent signaling (Xie et al., 2010). The uncertainty on the functions of astrocyte Ca2+ signaling in vivo provided the impetus to explore complementary approaches in vertebrates.
We explored the functions of astrocytes within the microcircuitry of the striatum, which is the largest nucleus of the basal ganglia, a group of interconnected subcortical nuclei. We define brain microcircuits as comprising neurons and associated cells such as glia, organized to carry out specific operations within a region of the nervous system (Shepherd and Grillner, 2010).
Results
Reducing striatal astrocyte calcium-dependent signaling
To reduce astrocyte Ca2+ signaling we tested isoforms of the plasma membrane Ca2+ pump PMCA, which functions to constitutively extrude cytosolic Ca2+ (Strehler, 2015). We hypothesised this would reduce astrocyte Ca2+ signaling, because most such signals last seconds. Such an approach would also conceivably be general with regards to the many types of slow Ca2+ signals. We focussed on PMCA2, because of its high pump rate and because it is unlikely expressed by astrocytes (Srinivasan et al., 2016; Zhang et al., 2014), which we confirmed for adult striatal astrocytes (Figure S1A–D). After screening and testing (Figure S1A–H), we focused on human PMCA isoform type 2 with the w/b splice variant that lacks cytosolic interaction domains, and modified to carry a mCherry tag. We abbreviated the construct as hPMCA2w/b and used “CalEx” for the approach to reflect Calcium Extrusion. hPMCA2w/b significantly reduced by >95% 50 μM ATP-evoked P2Y1 receptor-mediated Ca2+ signals in HEK-293 cells (Figure S1E–H; n = 33, P < 0.001, One-way ANOVA).
To deliver hPMCA2w/b to astrocytes in vivo, we used local microinjections of adeno-associated viruses (AAV2/5) with an astrocyte specific GfaABC1D promoter (Shigetomi et al., 2013). We confirmed that microinjection of AAV2/5-delivered cargo was astrocyte selective and restricted to the striatum, although there was a little expression along the needle tract in the cortex (Figure S2A, n = 3 mice): our studies need to be interpreted with this in mind. We microinjected hPMCA2w/b and the genetically-encoded calcium indicator GCaMP6f in vivo, resulting in expression of both proteins in ~90% of astrocytes in the dorsolateral striatum (Figure 1A–C; Figure S2B,C). Neuronal expression was not observed (n = 4 mice; Figure S2D). In controls that expressed tdTomato instead of hPMCA2w/b, we measured frequent, large amplitude spontaneously occurring Ca2+ elevations that we call Ca2+ signals: previously they have also been called transients, events, fluctuations, and oscillations. Such signals lasted seconds in striatal astrocyte somata, branches and territories (Figure 1D–G; n = 14 cells, n = 4 mice). In hPMCA2w/b-expressing astrocytes, Ca2+ signals were reduced by ~70% in terms of amplitude (Figure 1D P < 0.001, un-paired Mann-Whitney tests; Movie S1), and were also significantly shorter in duration in striatal astrocyte somata, branches and territories (Figure S2E, un-paired Mann-Whitney tests), but the effects on frequency were variable between compartments (Figure S2F). We interpret this to indicate that, by virtue of its Ca2+ extrusion ability, hPMCA2w/b significantly reduced the amplitude and shortened the duration of Ca2+ signals, but not their occurrence.
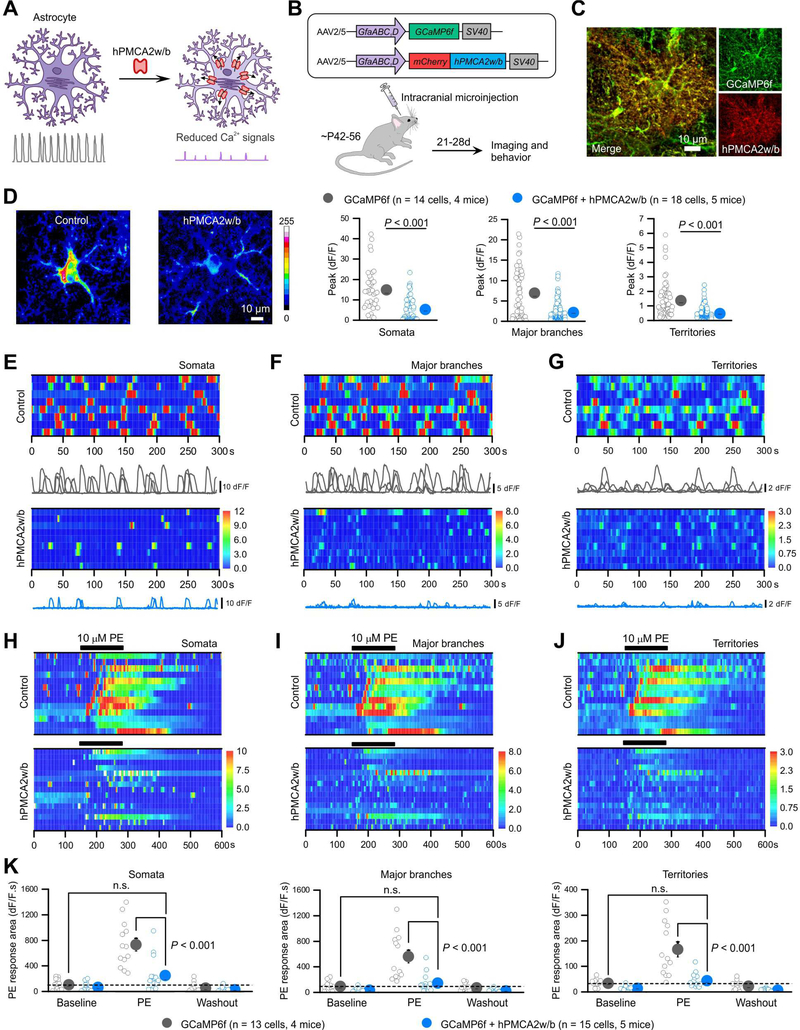
Figure 1: Reducing Ca2+-dependent signaling in striatal astrocytes with hPMCA2w/b.
(A) Schematic of hPMCA2w/b to attenuate Ca2+ signals. (B) AAVs for expressing mCherryh-PMCA2w/b with GCaMP6f in astrocytes. (C) IHC images showing colocalization of GCaMP6f and hPMCA2w/b in striatal astrocytes (Figure S2). (D) Z-stack images of GCaMP6f-expressing striatal astrocytes for control and hPMCA2w/b mice. Bar graphs: peak amplitude of spontaneous Ca2+ signals in somata, major branches and territories of striatal astrocytes (Mann-Whitney tests). (E-G) Kymographs and ΔF/F traces of Ca2+ signals in somata (E), major branches (F) and territories (G) of control and hPMCA2w/b-expressing astrocytes. (H-J) Kymographs of cells with Ca2+ responses evoked by phenylephrine (PE, 10 μM) in somata (H), major branches (I) and territories (J) of control and hPMCA2w/b-expressing astrocytes. (K) Summary plots for experiments in H-J. Average data are shown as mean ± SEM. In some cases the SEM symbol is smaller than the symbol for the mean.
Astrocytes respond with global Ca2+ elevations to a variety of G-protein coupled receptor (GPCR) agonists. We next explored if hPMCA2w/b affected pharmacologically-evoked Ca2+ elevations in striatal slices. hPMCA2w/b significantly reduced 10 μM phenylephrine (PE)-evoked GPCR–mediated astrocyte Ca2+ signals in all astrocyte compartments such that PE failed to evoke significant Ca2+ elevations (Figure 1H–K). hPMCA2w/b also attenuated global Ca2+ elevations triggered by intracellular Ca2+ store depletion (Figure S2G–I). Recent data emphasize the relevance of astrocyte basal Ca2+ levels (Rosenegger et al., 2015). We found that hPMCA2w/b significantly reduced astrocyte basal Ca2+ levels (Figure S3A–C).
IP3R2 deletion reduced striatal astrocyte Ca2+ signaling (Jiang et al., 2016). We directly compared the degree of Ca2+ signaling attenuation produced by hPMCA2w/b (n = 14 cells, 6 mice) and IP3R2 deletion mice (n = 14 cells, 6 mice) in relation to controls (n = 20 cells, 7 mice) using GCaMP6f (Figure S3D). In regards to spontaneous Ca2+ signals, we found that IP3R2 deletion produced a statistically larger reduction of Ca2+ signal frequency than hPMCA2w/b across all astrocyte compartments. There was a trend for a larger reduction in Ca2+ signal amplitude in IP3R2 deletion mice, but this was only significant for astrocyte branches. However, hPMCA2w/b more strongly attenuated the duration of the Ca2+ signals. For PE-evoked responses, both hPMCA2w/b and IP3R2 deletion similarly (>80%) attenuated the responses, but the degree of reduction was statistically greater in IP3R2 deletion mice (Figure S3D). These data compare hPMCA2w/b expressed in the striatum of adult mice with global IP3R2 deletion mice.
Reducing striatal astrocyte calcium-dependent signaling leads to excessive self-grooming
We microinjected hPMCA2w/b (or control AAVs) bilaterally into the dorsolateral striatum to determine how attenuation of Ca2+-dependent signaling affected behavior. There are no past in vivo data available to indicate which striatal behaviors astrocyte Ca2+ signaling may regulate: we performed several tests and studied the phenotypes that we discovered.
Control and hPMCA2w/b mice were healthy, weighed similarly (26.8 ± 0.5 and 26.3 ± 0.5 g for control and hPMCA2w/b mice, respectively n = 8, P = 0.543; Student’s t test) and performed equally in a Rotarod test of motor function (latency to fall was 195.6 ± 19.3 and 162.5 ± 21.6 s for control and hPMCA2w/b mice, respectively n = 8 mice, P = 0.273; Student’s t test). However, in the open-field test, hPMCA2w/b mice displayed shorter travel distances and spent less time in the centre (Figure 2A–C; n = 8 mice, P = 0.001 and P = 0.003, Two-way ANOVA and Student’s t test). While avoidance of centre regions of the open field arena can be interpreted as reflecting phenotypes related to anxiety, other behavioural measures of anxiety produced no differences. For example, control and hPMCA2w/b mice were indiscernible in the elevated plus maze (Figure 2D–G). Furthermore, hyperlocomotion of hPMCA2w/b and control mice was indistinguishable following cocaine (Figure S4A–C), indicating that the dopaminergic pathway was intact and arguing against motor dysfunction. Notably, we found that hPMCA2w/b mice spent long periods in the corners of the open-field (Figure 2A) and closer examination showed that hPMCA2w/b mice were excessively self-grooming during such episodes (Figure 2H–K). The increased self-grooming was observed in the open-field and when the mice were placed in a clear chamber (Movie S2). We quantified the total duration of self-grooming by measuring dwell times in grooming and non-grooming states (Figure 2I), confirming that hPMCA2w/b mice self-groomed excessively and significantly by ~450% (Figure 2J; n = 8, P = 0.0034 Student’s t test). The number of self-grooming bouts was not significantly different for control and hPMCA2w/b mice (Figure 2K; P = 0.7008 Student’s t test), but the total bout length was significantly longer (Figure 2J). This was also significant for the duration of individual self-grooming bouts (16.6 ± 5.6 s and 60.3 ± 9.9 s for control and hPMCA2w/b, respectively; n = 7, P < 0.001 Student’s t test). We assessed stress-related grooming and found equally high levels of grooming in this spray test (Figure 2L–O). Self-grooming is an innate behavior for which syntax is controlled by the dorsolateral striatum, and excessive self-grooming is associated with obsessive-compulsive disorder phenotypes (Kalueff et al., 2016).
Figure 2: Reducing Ca2+-dependent signaling in striatal astrocytes increased self-grooming.
(A) Representative track traces of locomotor activity in open-field tests of control and hPMCA2w/b-expressing mice over 10 min. hPMCA2w/b mice spent extended periods in the corners (arrow). (B,C) Travel distance (B) and time spent in the center (C) were decreased in hPMCA2w/b mice. Graphs tracking open-field activity over time were assessed with two-way ANOVA tests. The comparisons between bar graphs were with Student’s t tests or Mann-Whitney tests. (D) Representative track traces of locomotor activity in the elevated plus maze for control and hPMCA2w/b mice. (E-G) Average data for zone entries (E), time in different zones (F) and the total travel distance (G) between control and hPMCA2w/b mice. (H) Images of non-grooming and self-grooming behaviors. (I) Behavioral traces of control and hPMCA2w/b-expressing mice for times in self-grooming and non-grooming episodes. (J-K) The number of self-grooming bouts (K) and duration in seconds per minute of self-grooming (J) in hPMCA2w/b and control mice (Student’s t tests). (L-O) Grooming evoked by spraying the mice with water for controls and hPMCA2w/b: dwell times (M), average duration (N) and number of bouts (O). The average data are shown as mean ± SEM. In some cases, the SEM bars are smaller than the symbol used to represent the mean.
For the remainder of this study, we focussed on self-grooming (Figure 2). For a specific set of experiments we expressed hPMCA2w/b in hippocampal astrocytes and found it significantly reduced both spontaneous and PE-evoked Ca2+ signals (Figure S5A–F; n = 27 and 24 cells, n = 4 and 6 mice for controls and hPMCA2w/b). However, we did not observe altered self-grooming relative to AAV controls following hippocampal expression (Figure S5G, H). These and other controls suggest that the self-grooming phenotype following hPMCA2w/b expression in the striatum is specific.
Reducing striatal astrocyte calcium-dependent signaling alters microcircuit function in vivo
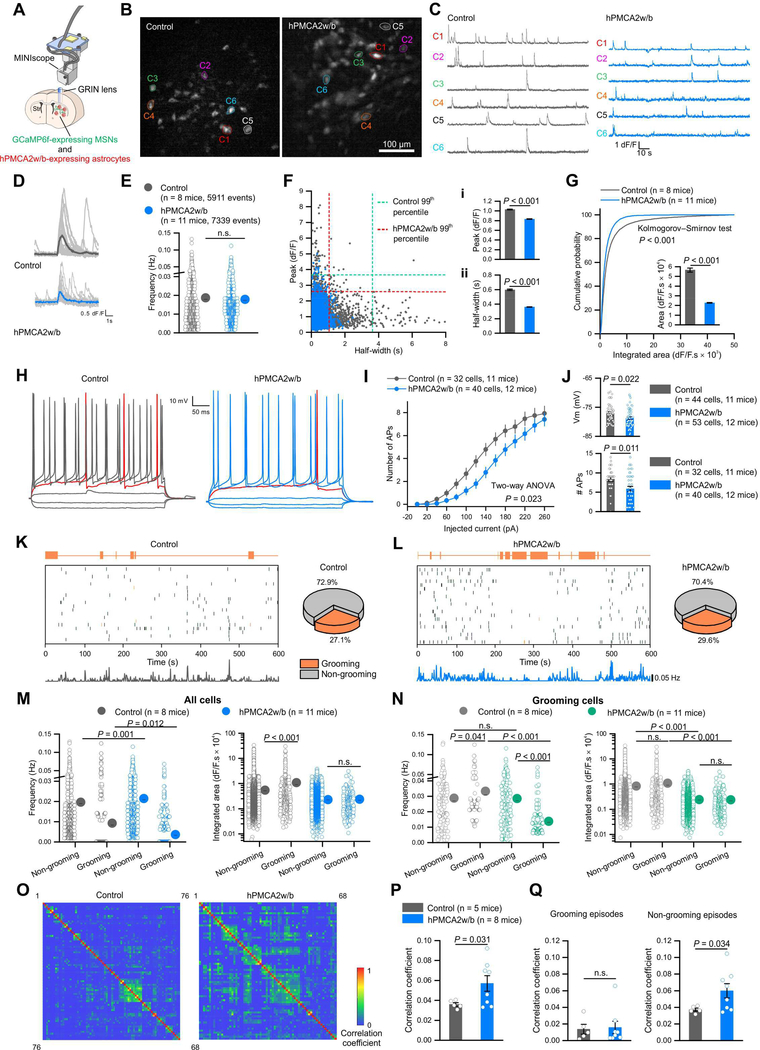
The dorsolateral striatum is involved in coordinating motor functions including self-grooming (Graybiel and Grafton, 2015; Kalueff et al., 2016). To explore how astrocyte hPMCA2w/b affected striatal microcircuits, we expressed GCaMP6f in MSNs (Figure S6A–D), and used Ca2+ imaging to measure MSN activity in freely behaving mice with head-mounted MINIscopes (Cai et al., 2016) (Figure 3A,B; Movie S3; Table S1). We detected activity in vivo from ~80 MSNs (Figure 3B–D; n = 8 & 11 mice). MSNs from hPMCA2w/b-expressing mice displayed the same frequency of activity as controls (Figure 3E), but with smaller amplitudes and shorter durations, reflected as smaller integrated areas (P < 0.001; Figure 3D,F,G). Furthermore, examination of the data showed that hPMCA2w/b caused a preferential loss of large amplitude MSN activity lasting several seconds (Figure 3F), suggesting that it may reduce MSN excitability associated with bursts of action potentials. To test this, we performed whole-cell patch-clamp recording from MSNs in acute brain slices. Relative to controls, MSNs from mice with astrocyte hPMCA2w/b displayed significantly hyperpolarized resting membrane potentials (~1.6 mV; Figure 3J), higher Rheobase and right-shifted input-output curves (Figure 3H–J), and decreased numbers of action potential bursts from Up state-like membrane potentials (Figure 3J; n = 32–40 cells from 11–12 mice). Thus, reducing astrocyte Ca2+ signaling produced significant effects on MSN activity in vivo, which we interpret to reflect reduced MSN excitability (Figure 3A-J).
Figure 3: Reducing striatal astrocyte Ca2+-dependent signaling altered MSN activity.
(A) The GRIN lens for MINIscopes was implanted above the dorsal striatum, near GCaMP6f-expressing MSNs and hPMCA2w/b-expressing astrocytes. (B) Z-projected Ca2+ images from MINIscope recordings in control and hPMCA2w/b mice. (C) Representative ΔF/F traces of individual MSNs recorded in B. (D) Average traces of thirty bouts of MSN activity measured as Ca2+ signals from control and hPMCA2w/b mice. (E) Frequency of MSN activity. (F) Scatter plot showing the relationship between peak amplitude and half-width of MSN Ca2+ signals. Peak amplitude (i) and half-width (ii) were significantly reduced in hPMCA2w/b mice. (G) Cumulative probability of integrated area of Ca2+ events in MSNs from control and hPMCA2w/b mice. Inset: integrated area plot. (H) Traces of MSN membrane responses to current injection in control and hPMCA2w/b mice. (I) Relationship of injected current to number of APs. (J) MSN resting membrane potentials (Vm) and the number of APs at Up state Vm values. The P values are from Mann-Whitney tests or a two-way ANOVA. (K) Left, raster plots of Ca2+ events with corresponding behaviors from 20 neurons in control mice. Bottom, event frequency of all neurons in single mice with 1 s time bins (76 neurons). Right, pie chart shows the percentage of imaged neurons active during grooming. (L) Left, raster plots of Ca2+ events with corresponding behaviors from 20 neurons in hPMCA2w/b mice. Bottom, event frequency of all recorded neurons in single mice with 1 s time bins (72 neurons). Right, pie chart shows the percentage of imaged neurons active during grooming. (M) Frequency and integrated area of Ca2+ events from all neurons during non-grooming and grooming episodes. (N) Frequency and integrated area of Ca2+ events during non-grooming and grooming episodes for the neurons active during grooming. (O) Correlation matrices showing the correlation coefficients between Ca2+ events of MSN pairs in control and hPMCA2w/b-expressing mice. (P) Average correlation coefficient of MSN pairs between all Ca2+ events was greater in hPMCA2w/b-expressing mice. (Q) Average correlation coefficients of Ca2+ events during non-grooming and grooming episodes in control and hPMCA2w/b-expressing mice. Data were assessed with Mann-Whitney tests or Student’s t tests as appropriate. The average data are shown as mean ± SEM. In some cases the SEM symbol is smaller than the symbol for the mean.
We also measured MSN activity in parallel with self-grooming (Figure 3K-Q; Table S1; n = 8 & 11 mice). We found that in control and hPMCA2w/b mice, a similar ~30% fraction of imaged MSNs defined an ensemble active during grooming (Figure 3K,L; P = 0.6574). In control mice, however, the frequency of MSN activity in these neurons was lower during grooming (Figure 3K,M; Table S1), but the event areas were larger (Figure 3M; Table S1). In hPMCA2w/b mice, the frequency of MSN activity was initially higher during non-grooming episodes than in control mice, and decreased significantly further during grooming episodes (Figure 3L,M; Table S1). Moreover, the event area did not increase during grooming episodes (Figure 3M; Table S1; P > 0.05), likely because large amplitude and long duration events were markedly reduced (Figure 3F; Table S1). These data establish that a pattern of MSN activity associated specifically with grooming episodes was altered significantly by reducing astrocyte Ca2+ signals. In particular, self-grooming episodes were associated with decreased frequency of MSN activity and this drop in frequency was greater when astrocyte Ca2+ signaling was attenuated (Figure 3K-M; Table S1). The activity of neurons that were active only during grooming is shown in Figure 3N. Next, we evaluated the degree of correlation between active MSNs and found a significantly greater correlation in hPMCA2w/b mice relative to controls irrespective of behavior (Figure 3O,P; Table S1; n = 5 and 8 mice). Greater correlated activity was also significant for non-grooming behaviors, but was equivalently low during grooming (Figure 3Q; Table S1). Thus, a predominant phenotype by reducing astrocyte Ca2+-dependent signaling in the striatum was greater correlated activity across MSNs during non-grooming episodes (Figure 3Q; Table S1). The resting correlation coefficient of ~0.04 observed between ~80 MSNs (400 × 400 μm; Figure 2A) is comparable with silicon probe recordings at ~0.01 (Bakhurin et al., 2016).
Astrocyte signaling controls self-grooming via tonic GABA-mediated neuromodulation
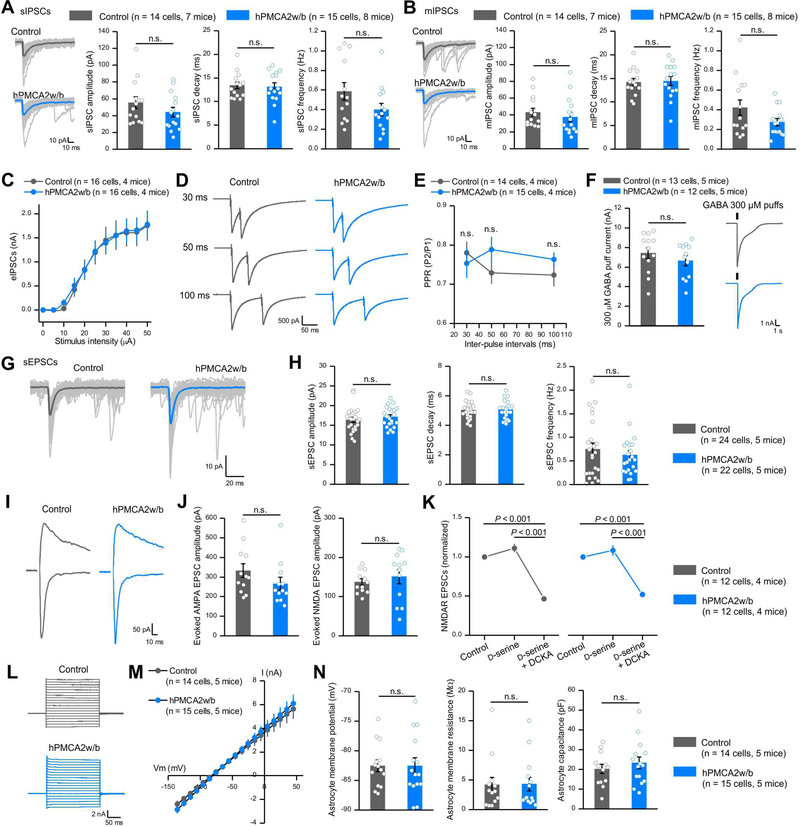
The greater coordination of MSNs covering an anatomically large area (~160,000 μm2) in hPMCA2w/b mice (Figure 3) prompted evaluation of mechanisms. Since MSNs are GABAergic, we examined fast phasic inhibition in control mice and those with hPMCA2w/b. We recorded phasic action potential-dependent sIPSCs (Figure 4A), action potential-independent mIPSCs (Figure 4B; in 500 nM TTX), evoked IPSCs at different stimuli (Figure 4C), paired-pulse responses (Figure 4D,E) and 300 μM GABA puff-evoked responses (Figure 4F). hPMCA2w/b had no significant effect on these metrics (Figure 4A-F, P > 0.05, un-paired Mann-Whitney or Student’s t test). We also evaluated fast excitatory synaptic transmission onto MSNs and found that AMPA receptor-mediated sEPSCs, AMPA receptor-mediated evoked EPSCs and NMDA receptor-mediated evoked EPSCs onto MSNs were not significantly different in hPMCA2w/b versus control (Figure 4G-J, n = 12 cells, 4 mice, un-paired Mann-Whitney or Student’s t test). Furthermore, by applying the glycine site agonists and antagonists, D-serine (10 μM) and DCKA (10 μM), respectively, we found that there were no differences between controls and hPMCA2w/b in D-serine mediated regulation of NMDA receptor evoked EPSCs (Figure 4K, n = 12 cells, 4 mice, un-paired Mann-Whitney or Student’s t test). Thus, significantly reducing Ca2+-dependent signaling in striatal astrocytes did not affect fast inhibitory or excitatory synaptic transmission. We do not imply that fast synaptic transmission is not regulated by astrocytes in brain areas that we have not examined.
Figure 4: Reducing striatal astrocyte Ca2+-dependent signaling in vivo did not affect fast synaptic transmission or astrocyte properties.
(A) Left, individual and average traces of thirty sIPSCs from control and hPMCA2w/b mice. Right, amplitude, decay time, and frequency of sIPSCs recorded from MSNs. (B) Left, individual and average traces of thirty mIPSCs from control and hPMCA2w/b-expressing mice. Right, mIPSC amplitude, decay time and frequency. (C) Evoked IPSCs from control and hPMCA2w/b mice. (D-E) Representative traces and average data for evoked IPSCs due to paired stimuli in control and hPMCA2w/b mice. (F) Traces and average data for currents evoked by 300 μM GABA puffs for control and hPMCA2w/b. Data in A-F were assessed with Student’s t or Mann-Whitney tests. (G,H) Traces (G) and average data (H) for sEPSCs in control and hPMCA2w/b mice. (I, J) Traces and average data for AMPA and NMDA receptor-mediated evoked EPSCs. AMPA EPSCs were measured at −70 mV and NMDA EPSCs were measured at +40 mV, 50 ms after the stimulus. (K) Normalized NMDA receptor-mediated evoked EPSCs. Responses to D-serine and DCKA were indiscernible for control and hPMCA2w/b- mice. (L) Current waveforms from astrocytes during 10 mV voltage jumps from - 140 mV to 40 mV. (M) Average current-voltage relations. (N) Average data for astrocyte membrane potential, membrane resistance and capacitance for control and hPMCA2w/b groups. Data in H, J and N were assessed by Student’s t or Mann-Whitney tests as appropriate; data in K were assessed by Two-way ANOVA tests. The average data are shown as mean ± SEM. In some cases, the bars used to represent SEM are smaller than the symbol for the mean.
We found that attenuation of Ca2+-dependent signaling did not alter astrocyte current waveforms in response to 10 mV voltage jumps from −140 to +40 mV (Figure 4L). Furthermore, membrane resistance (MOhms), capacitance (pF), macroscopic current-voltage relations and the resting membrane potential (mV) were all not significantly different between control and hPMCA2w/b groups (Figure 4L-N, n = 14 and 15 cells from 5 control and hPMCA2w/b mice). Insofar as these metrics frequently change during astrogliosis and disease (Tong et al., 2014), these data suggest that the astrocytes were healthy in hPMCA2w/b groups.
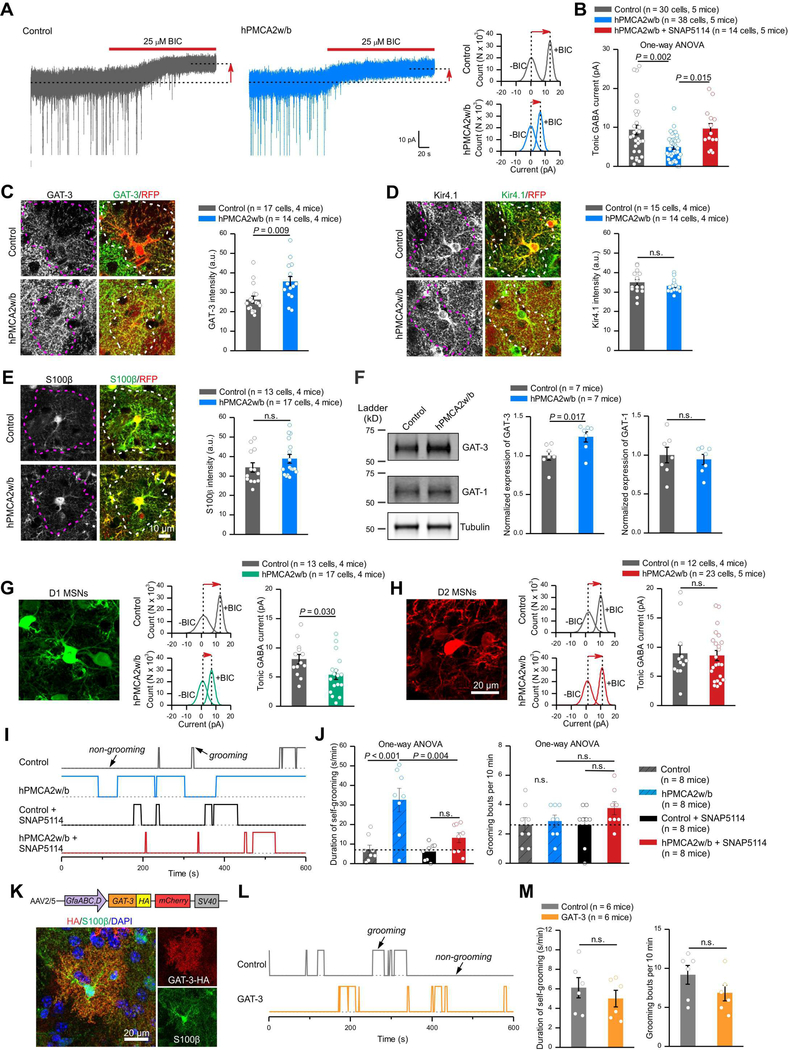
We next evaluated tonic inhibition of MSNs from brain slices expressing hPMCA2w/b in astrocytes (Figure 5A). GABA tonic inhibition was measured by applying bicuculline (BIC; 25 μM) and by recording the change in holding current at −60 mV (Figure 5A). Tonic inhibition was significantly smaller in MSNs from slices with reduced astrocyte Ca2+ signals (Figure 5A,B, P = 0.002, One-way ANOVA). This effect was not due to lower expression of GABAA receptors, because synaptic (Figure 4A-E) and extrasynaptic responses evoked by GABA puffs were unaffected by hPMCA2w/b (Figure 4F). Astrocytes are known to regulate ambient GABA levels and astrocytes preferentially express GAT-3 GABA transporters (Chai et al., 2017; Zhang et al., 2014). We explored if astrocyte hPMCA2w/b led to increased function of GAT-3, as more GABA uptake could result in reduced tonic inhibition (Figure 5A,B). We found that MSN tonic inhibition in hPMCA2w/b slices was rescued to control levels by 40 μM SNAP5114 (Figure 5B), a selective GAT-3 blocker. Moreover, immunohistochemistry (IHC) showed that hPMCA2w/b expressing astrocytes had significantly more GAT-3 (Figure 5C; n = 14–17 cells, n = 4 mice, P = 0.009, un-paired Mann-Whitney tests) and across whole fields of view (Figure S7A-C; n = 4 mice, P = 0.023), providing evidence for upregulation of GAT-3. In contrast, expression of Kir4.1 and S100β was unaffected by hPMCA2w/b (Figure 5D,E). To further explore GAT-3, we performed Western blots from whole striatum membrane protein preparations from control and hPMCA2w/b mice (Figure 5F; n = 7 mice). We found that attenuation of astrocyte Ca2+ signaling with hPMCA2w/b resulted in increased GAT-3 expression (relative to tubulin). However, the expression of GABA transporter GAT-1 was unaffected (Figure 5F).
Figure 5: Reducing striatal astrocyte Ca2+-dependent signaling in vivo altered MSN tonic inhibition, which contributed to excessive self-grooming.
(A) Current recordings in voltage-clamp (−60 mV) from MSNs in control and hPMCA2w/b-expressing mice. Dashed lines and arrows indicate the changes of baseline induced by bicuculline application (BIC; 25 μM). Histograms show the shift in baseline currents recorded during BIC application. (B) Tonic GABA currents were reduced in MSNs from hPMCA2w/b-expressing mice, and this effect was rescued by GAT-3 inhibitor SNAP5114 (40 μM). (C) Left, images of GAT-3 IHC in astrocytes expressing control protein or hPMCA2w/b. Right, quantification of GAT-3 fluorescence intensity in hPMCA2w/b-expressing astrocytes relative to those expressing tdTomato (Mann-Whitney test). (D-E) As in C, but for Kir4.1 and S100β immunostaining. (F) Western blot analysis of striatum membrane protein fractions showing for GAT-3 and GAT-1 levels in hPMCA2w/b groups relative to controls. The gel shown has been cropped to fit in the figure and the crop lines are shown as a solid border. The data were normalized to tubulin as a loading control. (G, H) Tonic GABA inhibition was decreased in D1-MSNs (G), but not D2-MSNs (H) in hPMCA2w/b-expressing mice (assessed with Student’s t or Mann-Whitney tests). The MSNs are colored, but were from Drd1-cre and Adora2a-cre mice with FLEX AAV tdTomato injections. (I) Behavioral traces of control and hPMCA2w/b-expressing mice under baseline and SNAP5114 treatment showing self-grooming and non-grooming episodes over 10 min. (J) Increased self-grooming behaviors in hPMCA2w/b-expressing mice were rescued by SNAP5114 (50 μmol/kg; data were assessed with One-way ANOVA followed by post hoc Bonferroni tests). (K) AAV2/5 overexpressing GAT-3 in astrocytes and GAT-3HA detection in S100β positive astrocytes. (L) Traces for self-grooming. (M) Average data for self-grooming duration and bouts for controls and GAT-3HA overexpressing mice. Data were assessed with Student’s t test. The average data are shown as mean ± SEM.
By recording from D1 and D2 MSNs from Drd1-cre and Adora2a-cre mouse lines injected with AAV-FLEX-tdTomato, we found that the hPMCA2w/b-mediated loss of tonic inhibition was significant for D1, but not D2 MSNs (Figure 5G,H). However, it is important to note that the genetic background of the Drd1- and Adora2a-cre mice is not identical to wild type mice. Hence, it is not possible to directly compare absolute amplitudes of responses between mice. Nonetheless, these data illustrate functional selectivity of reducing astrocyte Ca2+-dependent signaling in striatum.
The finding that astrocyte Ca2+ signaling regulates tonic inhibition in a GAT-3 dependent manner (Figure 5A-F) prompted us to explore if excessive self-grooming in hPMCA2w/b mice (Figure 2) was driven by decreased GABA-mediated tonic inhibition. To test this possibility, we administered SNAP5114 (50 μmol/kg i.p once per day for 1 week) using published methods (Wu et al., 2014) to hPMCA2w/b mice and found that the excessive self-grooming duration phenotype was rescued when assessed ~2 hours after the last dose (Figure 5I,J; n = 8 mice each, P > 0.05, One-way ANOVA). Importantly, the duration of self-grooming was not altered and there was no effect on the number of grooming bouts in either group (Figure 5I,J). Furthermore, the self-grooming phenotype observed in hPMCA2w/b mice was not affected by vehicle (Figure S7D,E, n = 8 mice).
Taken together, these data (Figures 1–5) provide strong evidence that hPMCA2w/b delivered selectively to the dorsolateral striatum leads to excessive self-grooming in a GAT-3 dependent manner, implying hPMCA2w/b has specific effects within the striatum. Furthermore, in control mice we found that GAT-3 inhibition did not alter tonic inhibition, which was 7.7 ± 0.8 pA in vehicle and 5.6 ± 0.9 pA in 40 μM SNAP5114 (Figure S7F,G; n = 11 & 13 cells, 5 mice; P > 0.05). However, inhibition of neuronal GAT-1 did, and tonic inhibition was significantly larger at 24.7 ± 3.4 pA (n = 8 cells, 4 mice; P < 0.0001) in the presence of the GAT-1 blocker NNC711 (10 μM) and was not further altered by SNAP5114 (Figure S7G). These data show that tonic inhibition in the striatal microcircuitry is controlled by GAT-1. However, when astrocyte Ca2+-dependent signaling was reduced, GAT-3 functional expression increased and also contributed (Figure 5). However, astrocyte overexpression of GAT-3 did not cause excessive self-grooming (Figure 5K-M), indicating that GAT-3 is necessary, but not sufficient for the phenotype.
We next explored which aspect(s) of the in vivo MSN activity altered in hPMCA2w/b mice was rescued by SNAP5114 by repeating all the in vivo experiments in Figure 3 following SNAP5114 treatment (Table S1). Relative to controls, hPMCA2w/b altered eight aspects of MSN activity in vivo (Figure 3). They were, (i) peak amplitude, (ii) half-width and (iii) correlation coefficient of MSN events analysed independently of behavior, (iv) frequency, (v) integrated area and (vi) correlation coefficient of MSN events during non-grooming episodes and (vii) frequency and (viii) integrated area of MSN events during grooming episodes (Figure 3, Table S1). Of these eight changes, five were fully rescued by SNAP5114 (Table S1; i, iii, iv, vi and vii, above). We suggest these five MSN properties drive GAT-3 dependent excessive self-grooming. Of particular interest, the heightened correlated activity across all events (iii) and during non-grooming episodes (vi) was rescued by SNAP5114, which recalls the loss of tonic inhibition at a cellular level and its rescue by SNAP5114 (Figure 5). Parenthetically, our data show three alterations of MSN activity in vivo that were not rescued by SNAP5114 (ii, v and viii, above), implying specificity of GAT-3 and additional circuit properties unrelated to self-grooming.
Astrocytic contributions to excessive self-grooming in Huntington’s disease model mice
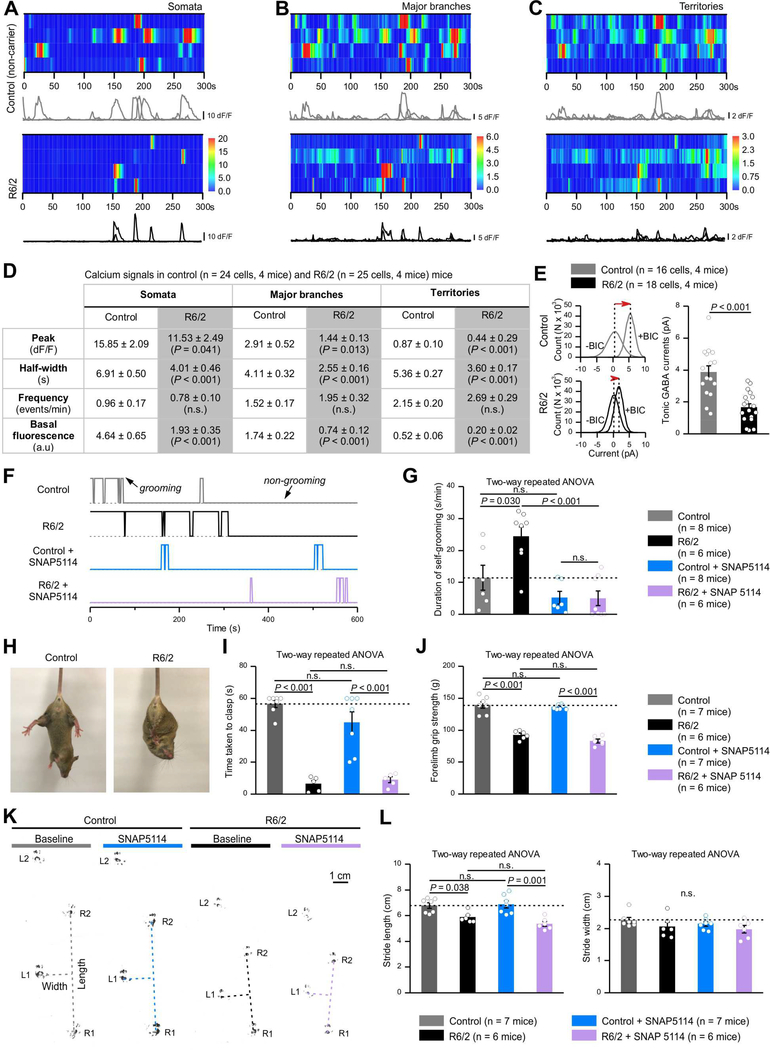
Huntington’s disease (HD) strongly affects the striatum and is associated with astrocyte dysfunction (Khakh et al., 2017). At early stages, HD patients display obsessive compulsive symptoms (Beglinger et al., 2008) that are seen in R6/2 HD model mice as excessive self-grooming (Steele et al., 2007). In light of our data (Figures 1–5) we explored if reduced astrocyte Ca2+ signaling in R6/2 mice (Jiang et al., 2016) was accompanied by loss of MSN tonic inhibition and if excessive self-grooming in R6/2 mice was GAT-3 dependent. We used the R6/2 mouse model, because most past studies on astrocytes have used this and AAV reagents have only been validated in R6/2 mice, which model early onset HD.
We found significantly reduced astrocyte Ca2+ signals in R6/2 mice relative to non-carrier controls (Figure 6A-D). We also found that tonic inhibition onto MSNs was significantly reduced in R6/2 mice relative to controls (Figure 6E). Moreover, we observed excessive self-grooming in R6/2 mice and this was relieved by SNAP5114 to control levels (Figure 6F,G). However, the excessive self-grooming phenotype in R6/2 mice was not affected by vehicle (Figure S7H,I, n = 8 mice). In contrast to self-grooming, later stage motor dysfunctions such as the time taken to clasp limbs when suspended by the tail, forelimb grip strength as well as walking stride that are proposed to be associated with striatal tissue loss were not affected by SNAP5114 (Figure 8H-L). Our data suggest that some HD phenotypes that occur during early stages such as repetitive self-grooming are correlated with reduced Ca2+ signaling and are corrected by GAT-3 blockers, recalling our data with wild type mice and hPMCA2w/b.
Figure 6: Early excessive self-grooming behaviors in Huntington’s disease model mice (7–8 weeks) were accompanied by reduced astrocyte Ca2+ signaling and were GAT-3 dependent. Later stage (10–11 weeks) motor dysfunctions were not.
(A-C) Kymographs and ΔF/F traces of Ca2+ signals in somata (A), major branches (B) and territories (C) of astrocytes in non-carrier control and R6/2 mice. (D) The inset table summarizes Ca2+ imaging data for control and R6/2 mice. (E) MSNs in R6/2 mice had significantly reduced tonic GABA currents (Student’s t tests). (F) Behavioral traces of non-carrier control and R6/2 mice under baseline and during SNAP5114 treatment (50 μmol/kg) showing self-grooming and non-grooming episodes over 10 min. (G) R6/2 mice exhibited excessive self-grooming, which were rescued by SNAP5114 treatment (Two-way ANOVA followed by a post hoc Bonferroni test). (H) Forelimb clasping of a R6/2 mouse and a control mouse. (I) Average time taken to clasp forelimbs was significantly less in R6/2 mice and SNAP5114 treatment did not improve this. (J) Forelimb grip strength of R6/2 mice was smaller than controls. SNAP5114 treatment did not change this. (K) Representative footprint tracks of control and R6/2 mice before and after SNAP5114 treatment. Dotted lines indicate stride lengths and widths. (L) Average footprint stride length and width of control and R6/2 mice before and after SNAP5114 treatment. The P values were from two-way ANOVA with a post hoc Bonferroni tests. The average data are shown as mean ± SEM.
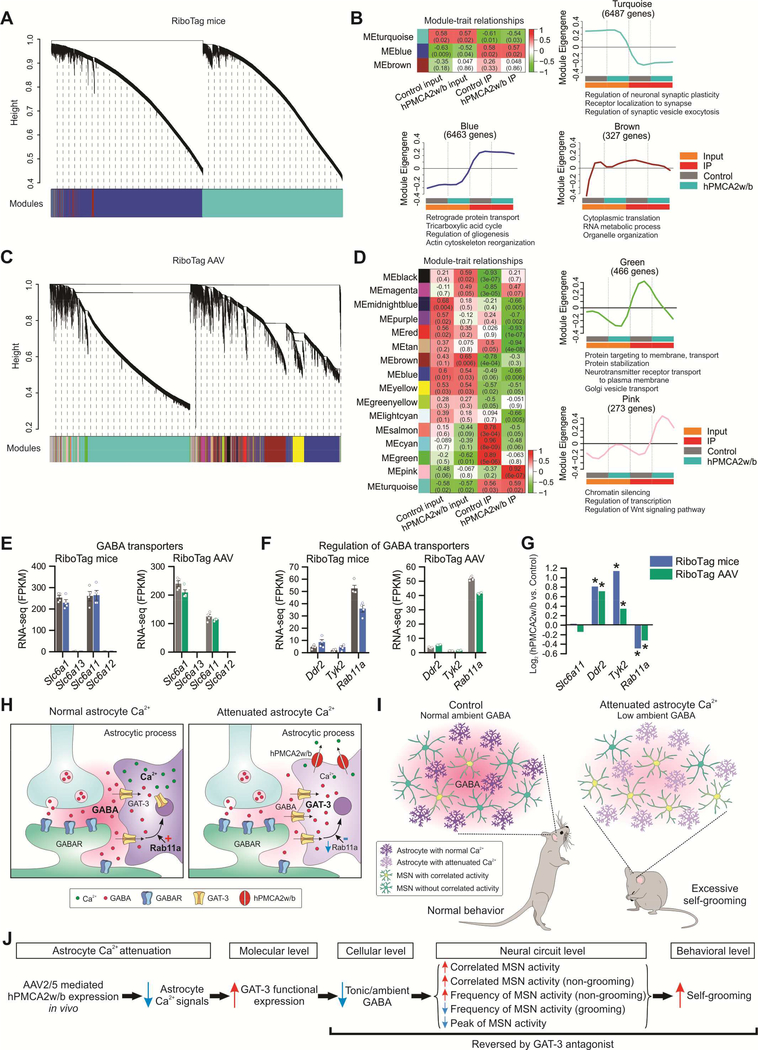
Figure 8: Weighted gene co-expression network analysis (WGCNA) identified distinct modules of co-expressed genes associated with attenuated astrocyte Ca2+ signaling.
(A) Hierarchical cluster dendrogram with colors underneath denoting three distinct modules of co-expressed transcripts in RiboTag mice RNA-seq data. (B) Heat map showing module-trait relationships, each row representing a module. Correlation coefficients between a Module Eigengene (ME) and trait (coded from −1 to 1) and corresponding P values are shown in each cell. The color indicates the level of correlation (positive correlation in red and negative correlation in green). Individual plots showing the expression of the module Eigengene across sample categories with significantly enriched terms for biological processes revealed by gene ontology enrichment analysis (FDR < 0.05). (C) Cluster dendrogram identified 16 co-expressed WGCNA modules for RiboTag AAV RNA-seq data. (D) Module-trait relationships revealed modules correlated with hPMCA2w/b expression in striatal astrocytes. Green module was composed of genes regulating protein transport, which were down-regulated in striatal astrocytes expressing hPMCA2w/b. Pink module was enriched in genes regulating chromatin silencing and transcription, which were upregulated in striatal astrocytes expressing hPMCA2w/b. (E) Expression levels of GABA transporters: Slc6a1 (GAT-1), Slc6a13 (GAT-2), Slc6a11 (GAT-3) and Slc6a12 (GAT-4) in RiboTag mice and RiboTag AAV RNA-seq data sets. (F) Expression levels of potential GABA transporter regulators suggested by previous work in RiboTag mice and RiboTag AAV RNA-seq data. (G) Changes in gene expression of striatal astrocytes expressing hPMCA2w/b, expressed as log2 (fold-change), in RiboTag mice and RiboTag AAV RNA-seq data. Asterisk (*) indicates differential expression between hPMCA2w/b IP and control IP using edgeR analysis on combined RNA-seq datasets (FDR < 0.1). (H-I) Cartoon summary of the main findings at synaptic (H) and in vivo levels (I). (J) Descriptive model based on our work (see discussion). Average data shown as mean ± SEM from 4 mice in each group.
Astrocyte-specific RNA-seq following Ca2+ signaling attenuation in vivo
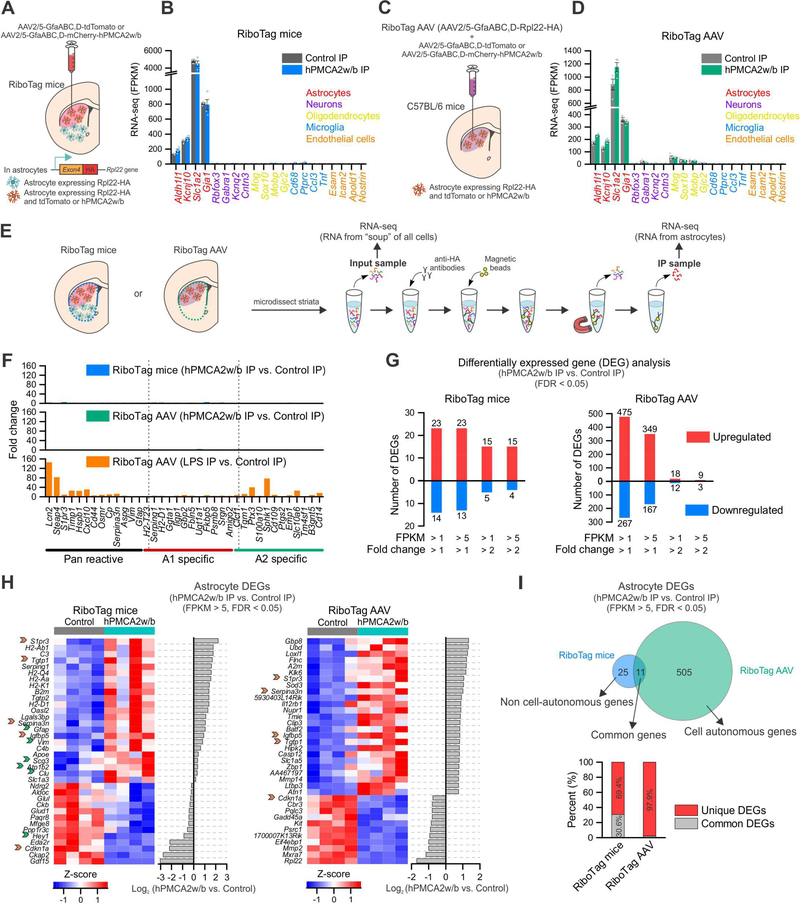
We performed RNA-seq to explore mechanisms that underlie GAT-3 dependent changes following attenuation of astrocyte Ca2+ signaling. First, we crossed astrocyte specific Aldh1l1-cre/ERT2 BAC transgenic mice with RiboTag mice and used offspring that express Rpl22HA in astrocytes to determine how astrocyte gene expression changed following Ca2+ attenuation with hPMCA2w/b in relation to controls (Figure 7A). In this approach we immunoprecipitated astrocyte-specific, actively translated RNA from the whole striatum following dorsolateral hPMCA2w/b microinjections (Figure 7A,E). In the second approach (Figure 7C,E), we used AAV2/5 GfaABC1D to deliver a Rpl22-HA cDNA (Sanz et al., 2015) to the same astrocytes that received hPMCA2w/b (or control AAV) in the dorsolateral striatum (Figure S8A-D). In this approach, because genes delivered by two AAVs are co-expressed at the ~98% level (Figure S2B), we immunoprecipitated astrocyte specific RNA mainly from astrocytes with attenuated Ca2+ signaling. By comparing RNA-seq data from RiboTag mice (i.e. astrocytes from whole striatum) with that from RiboTag AAV (i.e. astrocytes with attenuated Ca2+) we sought to identify non-cell autonomous and cell autonomous changes following attenuation of astrocyte Ca2+ signaling. RNA-seq samples were replete with astrocyte markers and depleted of other cell markers for both RiboTag mice and RiboTag AAV data in both control and hPMCA2w/b samples (Figure 7B,D; n = 4 mice; Table S2).
Figure 7: Ca2+ attenuation by hPMCA2w/b changed gene expression in astrocytes.
(A) RiboTag mice were crossed with Aldh1l1-cre/ERT2 mice to generate offspring expressing Rpl22HA in astrocytes (green cells in the cartoon). Control or hPMCA2w/b AAVs were injected into the dorsolateral striatum to target astrocytes (red cells). Astrocyte RNA from the whole striatal tissue was extracted; sequencing of IP and input material was performed (“RiboTag mice”). (B) Expression levels (in fragments per kilobase per million; FPKM) of cell-specific markers from RiboTag mice IP samples (n = 4 mice). (C) C57BL/6 mice received AAVs for Rpl22HA (“RiboTag AAV”) together with control or hPMCA2w/b AAVs in the dorsal lateral striatum (red cells). RNA from whole striatal tissue was extracted for IP of Rpl22HA: sequencing of IP and input material was performed (“RiboTag AAV”). (D) Expression levels of cell-specific markers in RiboTag AAV IP samples (n = 4 mice). (E) Schematic of the protocol used. (F) Fold-change of pan reactive, A1 and A2 astrocyte specific markers between hPMCA2w/b IP and control IP samples in RiboTag mice (top) and RiboTag AAV (middle). As a positive control, i.p. injection of LPS (5 mg/kg) was used. (G) Differentially expressed gene (DEG) analysis (limmaVoom, FDR < 0.05) showing the numbers of up- and down-regulated astrocyte genes between hPMCA2w/b IP and control IP in RiboTag mice and RiboTag AAV datasets. These analyses were restricted to genes with >2-fold enrichment in the IP compared with the input, and the plots show different threshold criteria. (H) Heatmaps of the top 36 astrocyte DEGs as ranked by differential expression log2 ratio between hPMCA2w/b IP and control IP (FPKM > 5). Orange arrows indicate genes that were common in the top 36 DEGs for both RiboTag mice and RiboTag AAV. Green arrows indicate additional DEGs in RiboTag mice that were also astrocyte DEGs in RiboTag AAV, but not in the top 36 for the latter. (I) Top, Venn diagram representing DEGs (FPKM > 5) in astrocytes from RiboTag mice and RiboTag AAV revealing putative cell autonomous genes and non cell-autonomous genes. Bottom, percent of unique astrocyte DEGs and common astrocyte DEGs between RiboTag mice and RiboTag AAV. Average data are shown as mean ± SEM. All RNA-seq data were for 4 separate replicates.
We evaluated if hPMCA2w/b caused astrogliosis by assessing the expression of pan reactive, A1 and A2 markers (Liddelow et al., 2017). None of these 37 markers were markedly elevated in hPMCA2w/b samples for either RiboTag mice or RiboTag AAV and the increase across the 37 markers was 1 ± 0.2 and 1 ± 0.1-fold for RiboTag mice and RiboTag AAV (Figure 7F; FDR > 0.05). However, 33/37 of these genes were increased when we used lipopolysaccharide (LPS) as a control to trigger neuroinflammation: the average increase across 37 markers was 16 ± 5-fold (Figure 7F; some > 40-fold). Furthermore, we analyzed the top 100 microglia markers and 23 activated microglia markers from the input RNA-seq (Figure S8E, F). For RiboTag mice and RiboTag AAV, 0/123 and 5/123 were altered respectively by hPMCA2w/b relative to control. The altered genes changed < 2-fold and were C3ar1, Lag3, Emr1, Lyz2 and Ccl3 (Figure S8E, F). In contrast, 52/123 genes were altered by LPS (range 1.5 to 16-fold). Our data provide strong evidence that reducing astrocyte Ca2+ signaling did not cause astrogliosis or overt microglial activation. The five microglial genes altered by astrocyte Ca2+ attenuation suggest microglial functions regulated by astrocytes.
Next, we measured astrocyte enriched (> 2-fold), differentially expressed genes (DEGs; Figure 7G, S8G). We parsed the data by FPKM and significant changes induced by hPMCA2w/b for both RiboTag mice and RiboTag AAV datasets (Figure S8G). By assigning a FPKM of 5 for the level of expression as the cut-off, we detected 36 and 516 significantly DEGs in the RiboTag mice and RiboTag AAV datasets (FDR < 0.05). This fairly low number of DEGs suggests specificity for the functions served by astrocyte Ca2+ signaling within striatal astrocytes and supports data showing attenuated astrocyte Ca2+ signaling did not alter astrocyte electrophysiological properties (Figure 4L-N), marker expression (Figure 5D,E) or cause reactivity (Figure 7F).
Figure 7H reports the top 36 up- and down-regulated genes for the RiboTag mice and RiboTag AAV datasets. Surprisingly, only five genes were shared. We interpret this to indicate that Ca2+ attenuation produced cell autonomous (RiboTag AAV) and non-cell autonomous effects on astrocyte gene expression (i.e. the difference between RiboTag AAV and RiboTag mice). This trend is summarized in Figure 7I for all DEGs (FPKM > 5, FDR < 0.05). Remarkably, these analyses show that of the 36 genes altered throughout the striatum, 25 are not altered within the astrocytes that received hPMCA2w/b. We propose that these genes define Ca2+-dependent functions that are regulated in astrocyte-astrocyte networks. This is an unexpected finding from our work. Data in Figure 7 provide a rich resource for exploring Ca2+-dependent astrocyte physiology in the cells that had attenuated Ca2+ signals and in those they impact in the striatum.
In order to shed light on the mechanisms that underlie the physiological changes in Figures 1–5, we used weighted gene co-expression network analysis (WGCNA) of RiboTag mice and RiboTag AAV RNA-seq data. WGCNA is a systems biology method for describing the correlation patterns among genes across data-rich samples, which can be used for identifying and interpreting clusters (modules) of highly co-expressed genes. WGCNA revealed 3 modules in the RiboTag mice data, but none of these differed between control and hPMCA2w/b groups (Figure 8A,B). In the RiboTag AAV data, we identified 16 modules (Figure 8C,D), some of which (e.g. green, salmon, cyan, pink) showed differences between control and hPMCA2w/b. Of note, the green module comprised a set of transcripts involved in membrane protein trafficking, Golgi vesicle transport and protein stabilization (Figure 8D; pink module shown for comparison). Upon closer examination, one down regulated gene belonging to the green module was Rab11a. This is notable because Rab11a regulates GAT-3 trafficking: a decrease in Rab11a increases cell surface GAT-3 expression by changing the balance between forward trafficking and removal (Zhang et al., 2017). These data suggest that GAT-3 functional upregulation that underlies cellular, neuronal and behavioral effects (Figures 1–5) involves trafficking. Consistent with this, we found no change in GAT-3 (Slc6a11) or GAT-1 (Slc6a1) FPKM levels (Figure 8E), whereas we detected significantly increased GAT-3 expression by immunostaining (Figure 5C, Figure S7A-C), Western blot of membrane protein fractions (Figure 5F) and by measurements of tonic GABA currents (Figure 5A,B). Furthermore, the FPKM values for Rab11a were lower in hPMCA2w/b groups relative to controls for both RiboTag mice and RiboTag AAV groups (Figure 8F). To assess this rigorously, Figure 8G shows analyses from the DEGs with FDR < 0.1 for Rab11a, Slc6a11 and two other exemplar kinase genes involved in posttranslational regulation of GABA transporters (Ddr2 and Tyk2). We found no significant change in Slc6a11 expression, increased Drd2 and Tyk2 expression and significantly decreased expression of Rab11a in hPMCA2w/b relative to controls in both RiboTag mice and RiboTag AAV data (Figure 8G). Table S2 reports expression data for other kinases, phosphatases and other proteins implicated in GABA transporter regulation. Taken together, our unbiased gene expression survey supports the notion that a posttranslational mechanism is involved in elevated GAT-3 functional expression following attenuation of astrocyte Ca2+ signaling and involves forward trafficking caused by the downregulation of Rab11a, without a change in Slc6a11 expression.
Discussion
There are four main findings from this work. First, hPMCA2w/b reduced Ca2+ signals in astrocyte somata, branches and processes, which resulted in GAT-3 dependent excessive self-grooming in mice. Second, reducing striatal astrocyte Ca2+ signals reduced tonic inhibition of striatal MSNs in a GAT-3 dependent manner and altered microcircuit activity in vivo. Third, at early stages in HD model mice, striatal astrocyte Ca2+ signaling was reduced and was associated with reduced MSN tonic inhibition and excessive self-grooming, which was also GAT-3 dependent. Fourth, two evaluations of astrocyte gene expression revealed molecular changes within astrocytes following attenuation of Ca2+ signaling, including of Rab11a – a known regulator of GAT-3 functional expression (Zhang et al., 2017). The main findings are schematized in Figure 8H-J.
The magnitude of striatal astrocyte Ca2+ signaling attenuation produced by hPMCA2w/b was smaller than that observed with IP3R2 deletion mice. However, using hPMCA2w/b we observed clear cellular and behavioural phenotypes. We interpret this to indicate that astrocyte functions within circuits are best explored by performing circuit specific astrocyte manipulations in the adult brain rather than across the whole brain and throughout development as with IP3R2 deletion mice. This is analogous to the use of optogenetics and pharmacogenetics aimed at exploring neuronal function within anatomically selected areas. By manipulating astrocytes in specific brain areas, it should be possible to determine if astrocyte function and/or dysfunction contributes to behaviours and disease-related phenotypes encoded by the local neurons. Once such behaviours are discovered, the underlying astrocyte-related molecular, cellular and circuit mechanisms could be teased apart. By performing such top-down studies for the dorsolateral striatum, our studies emphasize the importance of tonic GABA rather than direct effects of astrocytes on fast neurotransmitter release. Astrocytic regulation of GABA is considered a type of “gatekeeper” within circuits (Beenhakker and Huguenard, 2010; Muthukumar et al., 2014).
Although we focussed on the striatum and self-grooming, hPMCA2w/b could be used in other brain areas in order to explore the functions of astrocyte Ca2+ signaling. We do not imply that self-grooming is the only or the most important outcome of astrocyte Ca2+ signaling in the brain. Hence, systematic use of hPMCA2w/b constructs may reveal additional functions of astrocyte Ca2+ signaling in vivo. Such functions could include roles at quad-partite synapses (Schafer et al., 2013), release of trophic factors, phagocytosis, homeostasis, neurovascular coupling and responses to injury, disease, trauma and infection. hPMCA2w/b could also be improved: the current strategy uses AAV2/5-mediated expression that requires ~14–21 days. It may be possible to deliver the protein more expeditiously using mouse genetics and to engineer it so it can be activated by ligand.
In agreement with past work (Agulhon et al., 2010; Petravicz et al., 2008), we found that attenuation of astrocyte Ca2+ signaling did not affect striatal fast synaptic transmission. There are two interpretations of this. First, it is conceivable that the ~70–80% attenuation of Ca2+ signals produced by hPMCA2w/b was insufficient to reveal effects on fast synapses. Second, the result may indicate that astrocyte Ca2+ signaling does not in fact regulate fast excitation and inhibition in the striatum. On balance, we favor the latter view which is consistent with the paucity of astrocyte Ca2+-dependent glutamate release (Sloan and Barres, 2014), including from striatal astrocytes (Chai et al., 2017). Furthermore, the bulk of the evidence to suggest that astrocytes regulate fast synaptic transmission has been gathered with the use of Ca2+ chelators such as BAPTA, the validity of which has been questioned (Smith et al., 2018). We do not rule out astrocyte contributions to fast synaptic transmission in brain areas we have not studied.
By using in vivo imaging in freely behaving mice, we identified eight alterations of MSN activity that were caused by hPMCA2w/b expression within astrocytes in vivo. Of these eight, five were reversed by SNAP5114, which also rescued the self-grooming phenotype and the loss of tonic inhibition triggered by reducing astrocyte Ca2+ signals (Figure 8J). These data provide a strong correlation at different biological levels (from molecules to behavior) for a mechanism whereby reduced astrocyte Ca2+ signaling resulted in elevated functional activity of GAT-3, reduced ambient GABA, which regulated MSN activity in vivo and self-grooming (Figure 8J). Of the five GAT-3–dependent changes, two merit discussion. First, we recorded increased correlated activity between MSNs during non-grooming episodes following hPMCA2w/b expression in astrocytes: this effect was reversed by SNAP5114. We hypothesize that heightened correlated activity during non-grooming acts on downstream projection areas regulating self-grooming and results from reduced tonic inhibition. Second, we recorded a larger drop in the frequency of MSN activity during self-grooming in hPMCA2w/b mice, and this too was reversed by SNAP5114. These data suggest that specific GAT-3–dependent patterns of MSN activity are needed during self-grooming episodes in hPMCA2w/b mice. There is no circuit level model of self-grooming that can be used to model striatal astrocyte data. However, consistent with our data, there is good evidence that the dorsolateral striatum is involved in the execution of sequential patterns related to self-grooming (Kalueff et al., 2016). Furthermore, pharmacologically increasing or decreasing GABA decreases and increases grooming, respectively (Kalueff et al., 2016). Additionally, very precise lesions to the dorsolateral striatum result in the failure to complete self-grooming syntax (Cromwell and Berridge, 1996). This phenotype is separable from the prolonged self-grooming we observed, but consistent with our data showing little astrocyte reactivity that would have occurred in the event of an overt injury-related response. A satisfying microcircuit level model for astrocyte regulation of self-grooming is not yet possible, but our study provides an in vivo experimental platform and rational to explore an adult innate vertebrate behavior regulated by astrocytes located within an anatomically important circuitry providing syntax for sequential motor tasks. Our data also show three alterations of MSN activity in vivo that were not rescued by SNAP5114, implying specificity of the GAT-3 and astrocyte-related mechanisms as well as the existence of additional circuit properties unrelated to GAT-3.
Excessive self-grooming has also been observed in Hoxb8 null mutant mice that are proposed to alter neural circuits via microglia (Chen et al., 2010). We did not detect expression of Hoxb8 in astrocytes or the input samples in control or hPMCA2w/b data sets (Table S2). Hence, Hoxb8 related mechanisms are unrelated to our experiments. Excessive self-grooming has been observed in adolescent mice lacking the glutamate transporter GLT1 (Aida et al., 2015). These studies are tangential to our study as we did not delete GLT1, and GLT1 expression did not decrease following Ca2+ attenuation (Table S2).
At a molecular level, we found that attenuating astrocyte Ca2+ signaling increased GAT-3 functional expression and protein levels. However, RNA-seq showed no change in GAT-3 levels, suggesting that astrocyte Ca2+ signaling regulates GAT-3 in a posttranslational way. In accord, WGCNA and DEG analyses identified Rab11a as a significantly downregulated gene. Down regulation of Rab11a leads to functional upregulation of GAT-3 (Zhang et al., 2017), which supports our observations. In addition, the RNA-seq data provide a rich resource of gene expression changes and WGCNA modules that can be used in future, hypothesis-driven experiments aimed at evaluating astrocyte Ca2+ signaling. Pursuing such agnostically derived mechanisms complements strategies exploring other mechanisms such as gliotransmission. It should also be possible to make floxed Rab11a allele mice and image Rab11a regulation of GAT-3 in reduced preparations. Overall, RNA-seq data provide a valuable resource to design hypothesis-driven experiments to explore astrocyte Ca2+ signaling – a significant issue that has been difficult to address.
We have used several approaches to study the adult striatum as a model area to explore how astrocytes contribute to vertebrate microcircuit function (Chai et al., 2017; Jiang et al., 2016; Khakh et al., 2017; Octeau et al., 2018; Srinivasan et al., 2016; Tong et al., 2014). In the present study we found that striatal astrocyte Ca2+-dependent signaling affects microcircuit function in brain slices and in vivo, and that reducing such signaling triggers innate behavioral phenotypes in mice via altered neuromodulatory GABA signaling. Our data show that astrocytes regulate striatal MSNs and a specific behavior via ambient GABA mediated neuromodulation. Moreover, we show that both experimentally induced, and disease-model-associated reductions in striatal astrocyte Ca2+-dependent signaling in adult mice can produce abnormal repetitive behavioral phenotypes that are related to psychiatric disorders (Kalueff et al., 2016).
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Baljit S. Khakh (bkhakh@mednet.ucla.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mouse models
All animal experiments were conducted in accordance with the National Institute of Health Guide for the Care and Use of Laboratory Animals and were approved by the Chancellor’s Animal Research Committee at the University of California, Los Angeles. Male and female mice aged between 6- and 14-week old were used in this study. Mice were housed in the vivarium managed by the Division of Laboratory Animal Medicine at University of California, Los Angeles (UCLA) with a 12 h light/dark cycle and ad libitum access to food and water. Wild-type C57BL/6NTac mice were generated from in house breeding colonies or purchased from Taconic Biosciences. Transgenic mice expressing eGFP under an astrocyte specific promoter Aldh1l1 (Aldh1l1-eGFP mice) were maintained as a heterozygous line (Gong et al., 2003). D1-Cre (Strain B6.FVB(Cg)-Tg(Drd1-cre)EY262Gsat/Mmucd) and D2-Cre (Strain B6.FVB(Cg)-Tg(Adora2a-cre)KG139Gsat/Mmucd) BAC transgenic mouse lines were obtained from MMRRC at University of California Davis and maintained on a C57BL/6NTac background. R6/2 and non-carrier wild-type control mice were purchased from Jackson Laboratories (Strain B6CBA-Tg(HDexon1)62Gpb/3J). RiboTag mice were purchased from Jackson Laboratories (Strain B6N.129-Rpl22tm1.1Psam/J) and crossed with Aldh1l1-Cre/ERT2 mice (Srinivasan et al., 2016)to generate mice carrying both alleles for RNA-seq experiments. Ip3r2 knock-out (Itpr2tm1.1Chen) mice were available from previous work (Srinivasan et al., 2015) and were originally obtained from Dr. Ju Chen at University of California–San Diego and maintained as a heterozygous line.
Cell lines
HEK-293 cells (sex: female, RRID: CVCL_0045) were obtained from ATCC and maintained in 25 cm2 cell culture flasks in DMEM/F12 media with Glutamax (Invitrogen) supplemented with 10% fetal bovine serum and penicillin/streptomycin. Cells were grown in a humidified cell culture incubation with 95% air / 5% CO2 at 37 °C.
METHODS DETAILS
Molecular biology and generation of adeno-associated viruses
Adeno-associated viruses (AAVs) were generated as previously described (Shigetomi et al., 2013). In brief, the hPMCA2w/b and hPMCA2x/b sequences were removed by restriction digestion from Addgene plasmids 47586 and 47585, respectively. Together with a sequence encoding mCherry that was amplified by PCR from pCS2-mCherry, hPMCA2w/b and hPMCA2x/b were incorporated into pZac2.1 vector using In-Fusion Cloning Kits (Takara) to generate plasmid pZac2.1 GfaABC1D mCherry-hPMCA2w/b (Addgene, 111568) and pZac2.1 GfaABC1D mCherry-hPMCA2x/b (Addgene, 111569), which carried the minimal GfaABC1D astrocyte-specific promoter. After verifying the sequences, plasmids were used to make AAVs of the 5 serotype at the Penn Vector Core (AAV2/5 GfaABC1D mCherry-hPMCA2w/b; 9.2 × 1012 gc/ml and AAV5 GfaABC1D mCherry-hPMCA2x/b; 1.6 × 1013 gc/ml). For HEK-293 cell transfection, mCherry-hPMCA2w/b and mCherry-hPMCA2x/b sequences were cloned into pcDNA3.1 vectors between AflII and XbaI sites to generate pcDNA3.1-CMV-mCherry-hPMCA2w/b (Addgene, 111570) and pcDNA3.1-CMV-mCherry-hPMCA2x/b (Addgene, 111571). To generate RiboTag AAV, the cDNA of the ribosomal protein Rpl22 with three HA tags was amplified from pcDNA3.1-Rpl22–3xHA (Sanz et al., 2009) (a gift from Dr. McKnight) and cloned into pZac2.1 using In-Fusion Cloning Kits (Takara) to generate pZac2.1- GfaABC1D-Rpl22-HA (Addgene, 111811). The plasmid was then sent to the Penn Vector Core for AAV2/5 production (AAV2/5 GfaABC1D-Rpl22-HA; 2.1 × 1013 gc/ml). All the new plasmids generated in this study that are listed in Key Resources Table have been deposited at Addgene in the Khakh laboratory repository (http://www.addgene.org/Baljit_Khakh).
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| rabbit anti-S100β | Abcam | Cat#ab41548; RRID: AB_956280 |
| mouse anti-NeuN (clone A60) | Millipore | Cat#MAB377; RRID: AB_2298772 |
| chicken anti-GFP | Abcam | Cat#ab13970; RRID: AB_300798 |
| rabbit anti-RFP | Rochland | Cat#600–401-379; RRID:AB_2209751 |
| chicken anti-RFP | Brecha Lab, University of California at Los Angeles | N/A |
| Mouse anti-HA 11 antibody (clone 16B12) | Covance | Cat#MMS-101R; RRID AB_291263 |
| rabbit anti-GAT-3 | Brecha Lab, University of California at Los Angeles | Cat# 374D; RRID:AB_2313749 |
| rabbit anti-GAT1 | Abcam | Cat# ab426, RRID:AB_2189971 |
| rabbit anti-Kir4.1 | Alomone | Cat#APC-035; RRID: AB_2040120 |
| rabbit anti-DARPP-32 | Abcam | Cat#ab40801; RRID: AB_731843 |
| Mouse anti-β-tubulin | Millipore | Cat# 05–559, RRID:AB_309804 |
| Alexa Fluor 488 goat anti-chicken | Molecular Probes | Cat#A11039; RRID: AB_2534096 |
| Alexa Fluor 488 goat anti-rabbit | Molecular Probes | Cat#A11008; RRID: AB_143165 |
| Alexa Fluor 546 goat anti-mouse | Molecular Probes | Cat#A11003; RRID: AB_2534071 |
| Alexa Fluor 546 goat anti-chicken | Molecular Probes | Cat#A11040; RRID: AB_2534097 |
| Alexa Fluor 594 goat anti-rabbit | Molecular Probes | Cat# R37117; RRID: AB_2556545 |
| IRDye 800CW anti-rabbit | Li-Cor | Cat#827–08365 RRID: AB_10796098 |
| IRDye 680RD anti-mouse | Li-Cor | Cat#926–68170 RRID: AB_10956589 |
| Bacterial and Virus Strains | ||
| pcDNA3.1 mCherry-hPMCA2w/b | This study: Addgene | 111570 |
| pcDNA3.1 mCherry-hPMCA2x/b | This study: Addgene | 111571 |
| pZac2.1 GfaABC1D mCherry-hPMCA2w/b | This study: Addgene | 111568 |
| pZac2.1 GfaABC1D mCherry-hPMCA2x/b | This study: Addgene | 111569 |
| pZac2.1 GfaABC1D Rpl22HA | This study: Addgene | 111811 |
| AAV5 GfaABC1D cyto-GCaMP6f | Haustein et al., 2014; UPenn Vector Core Addgene Vectors | Cat#AV-5–52925 #52925-AAV5 |
| AAV5 GfaABC1D tdTomato | Tong et al., 2014 UPenn Vector Core Addgene Vectors | Cat#AV-5-PV3106 44332-AAV5 |
| AAV2/5 GfaABC1D Rpl22HA | This study: UPenn Vector Core | In process |
| AAV2/5 GfaABC1D mCherry-hPMCA2w/b | This study: UPenn Vector Core | In process |
| AAV2/5 GfaABC1D mCherry-hPMCA2x/b | This study: UPenn Vector Core | In process |
| AAV1 Synapsin GCaMP6f | UPenn Vector Core | Cat#AV-1-PV2822 |
| AAV1 CAG-Flex tdTomato | UPenn Vector Core | Cat#AV-1-PV2816 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Paraformaldehyde, EM grade | EMS | Cat#19202 |
| TTX | Cayman Chemical Company | Cat#14964 |
| Bicuculline | Sigma-Aldrich | Cat#14340 |
| CNQX disodium salt | Abcam | Cat#ab120044 |
| Cyclopiazonic acid (CPA) | Tocris | Cat#1235 |
| Clozapine N-oxide (CNO) | Tocris | Cat#4936 |
| Phenylephrine | Tocris | Cat#2838 |
| Papain | Worthington | Cat#LS003126 |
| Ovomucoid trypsin inhibitor | Worthington | Cat#LS003086 |
| EBSS | Sigma-Aldrich | Cat#E7510 |
| Halt protease inhibitor cocktail | Thermo | Cat#1861278 |
| Critical Commercial Assays | ||
| Ovation PicoSL WTA System V2 | Nugen | Cat#3312 |
| QIAquick PCR Purification Kit | Qiagen | Cat#28104 |
| Qiagen RNeasy Plus Micro Kit | Qiagen | Cat#74034 |
| Pierce BCA protein assay kit | Thermo Scientific | Cat#23225 |
| Experimental Models: Cell lines | ||
| Human: HEK-293 cell line; sex: female | ATCC | Cat# PTA-4488; RRID: CVCL_0045 |
| Experimental Models: Organisms/Strains | ||
| Mouse: Aldh1l1-eGFP | MMRRC | Stock#011015-UCD; RRID: MMRRC_011015-UCD |
| Mouse: B6N.129-Rpl22tm1.1Psam/J | Jackson Laboratories | Stock#011029 RRID: IMSR_JAX:011029 |
| Mouse: Aldh1l1-cre/ERT2 | Jackson Laboratories | Stock#029655 RRID: IMSR_JAX:029655 |
| Mouse: C57Bl/6NTac inbred mice | Taconic | Stock#B6; RRID:IMSR_TAC:b6 |
| Mouse: B6.FVB(Cg)-Tg(Drd1-cre)EY262Gsat/Mmucd | MMRRC | Stock# 030989-UCD; RRID: MMRRC_030989-UCD |
| Mouse: B6.FVB(Cg)-Tg(Adora2a-cre)KG139Gsat/Mmucd | MMRRC | Stock# 036158-UCD; RRID:MMRRC_036158-UCD |
| Mouse: B6CBA-Tg(HDexon1)62Gpb/3J | Jackson Laboratory | Stock#006494; RRID:IMSR_JAX:006494 |
| Ip3r2 knock-out (Itpr2tm1.1Chen) | MGI | Cat# 3641042, RRID:MGI:3641042 |
| Oligonucleotides | ||
| qPCR Atp2b1 forward primer
sequence: 5' TTCCGGAACCAGAAGCCATC 3' |
This paper | N/A |
| qPCR Atp2b1 reverse primer
sequence: 5' AGAAACCCCAACAAGGCACA 3' |
This paper | N/A |
| qPCR Atp2b2 forward primer
sequence: 5' TCGAAGACCCAGTACGACCT 3' |
This paper | N/A |
| qPCR Atp2b2 reverse primer
sequence: 5' CCCTTCTCGTTGCGAATCCT 3' |
This paper | N/A |
| qPCR Atp2b3 forward primer
sequence: 5' TTCTGGTTCACGCCATCTCC 3' |
This paper | N/A |
| qPCR Atp2b3 reverse primer
sequence: 5' TATCTGCTCCCGTACTGGCT 3' |
This paper | N/A |
| qPCR Atp2b4 forward primer
sequence: 5' TACATCGGGGGCACTCACTA 3' |
This paper | N/A |
| qPCR Atp2b4 reverse primer
sequence: 5' CAGCCCACACTCTGTCTTGT 3' |
This paper | N/A |
| qPCR Arbp forward primer
sequence: 5' TCCAGGCTTTGGGCATCA 3' |
Jiang et al., 2016 | N/A |
| qPCR Arbp reverse primer
sequence: 5' AGTCTTTATCAGCTGCACATCAC 3' |
Jiang et al., 2016 | N/A |
| Software and Algorithms | ||
| OriginPro 8 | Origin Lab Corporation | RRID:SCR_015636 |
| SciScan 2014 | Scientifica | http://www.scientifica.uk.com/products/scientifica-sciscan |
| pCLAMP10.4 | Molecular Devices | RRID:SCR_011323 |
| ClampFit10.4 | Molecular Devices | N/A |
| Fluoview FV3000 | Olympus | N/A |
| ImageJ v1.51h | NIH | RRID:SCR_003070 |
| MiniAnalysis 6.0.7 | Synaptosoft Inc. | RRID:SCR_002184 |
| MATLAB | MathWorks | RRID:SCR_001622 |
| R v3.3.2 | RCoreTeam, 2016 | https://cran.r-project.org/bin/windows/base/old/3.2.2/ |
| Ethovision XT | Noldus Information Technology | RRID:SCR_000441 |
| CorelDraw12 | Corel Corporation | RRID:SCR_014235 |
| Bioconductor | Law et al., 2015 | |
| Htseq-count | Anders et al., 2015 | N/A |
HEK-293 cell imaging
HEK-293 cell imaging methods have been described (Richler et al., 2008). HEK-293 cells were prepared for transfection by plating onto six-well plates and transfection was performed when cells reached 80–90% confluence. For transient expression in HEK-293 cells, 0.5–1 μg plasmid DNA were transfected using Effectene transfection reagent (Qiagen). Cells were imaged 24–72 hours after transfection in HEK cell recording buffer (150 mM NaCl, 1 mM MgCl2, 1 mM CaCl2, 10 mM D-glucose, 10 mM HEPES, with pH adjusted to 7.4). Ca2+ signals were recorded under a laser-scanning confocal microscope (Olympus) with a 40× water-immersion objective lens (NA 0.8). To activate P2Y1 receptors, ATP (50 μM) was bath-applied in the recording solution.
Astrocyte purification and quantitative PCR
Fluorescence-activated cell sorting (FACS) was performed to purify striatal astrocytes from Aldh1l1-eGFP mice at P30, as previously described (Jiang et al., 2016). Briefly, the striata from 4 mice were dissected and digested at 36°C for 45 min with papain solution (1 × EBSS, 0.46% D-glucose, 26 mM NaHCO3, 50 mM EDTA, 75 U/ml DNase 1, 300 units of papain, and 2 mM L-cysteine). The tissue was then rinsed with ovomucoid solution (1× EBSS, 0.46% Dglucose, 26 mM NaHCO3, 1 mg/ml ovomucoid, 1 mg/ml BSA, and 60 U/ml DNase 1) and mechanically dissociated with borosilicate glass pipettes. A concentrated ovomucoid solution (1× EBSS, 0.46% D-glucose, 26 mM NaHCO3, 5.5 mg/ml ovomucoid, 5.5 mg/ml BSA, and 25 U/ml DNase 1) was layered at the bottom of the cell suspension. Cells were precipitated by centrifuging and the pellet was resuspended and filtered. FACS was performed using a FACSAria II (BD Biosciences) with a 70 μm nozzle at UCLA Cell Sorting Core. Total RNA was extracted from sorted GFP+ cells (RNeasy Plus Micro Kit; Qiagen) and cDNA was amplified (Ovation PicoSL WTA System V2; Nugen) and purified (QIAquick PCR Purification Kit; Qiagen). Quantitative PCR was performed in a LightCycler 96 Real-Time PCR System (Roche). Ten nanograms of cDNA were loaded per well and the expression of Atp2b1, Atp2b2, Atp2b3, Atp2b4 and Arbp was analyzed using the primers listed in Key Resources Table. Arbp was used as an internal control to normalize RNA content. To calculate the expression of gene of interest, the following formula was used: 2−ΔCt (Gene of interest-Arbp).
Surgical procedure of in vivo microinjection
Surgical procedure of viral microinjection has been described previously (Haustein et al., 2014; Jiang et al., 2016; Jiang et al., 2014; Shigetomi et al., 2013). In brief, mice aged 6- to 8week-old were anesthetized and placed onto a stereotaxic frame (David Kopf Instruments, Tujunga CA). Continuous anesthesia using isoflurane was carefully monitored and adjusted throughout the surgery. Mice were injected with buprenorphine (Buprenex; 0.1 mg/kg) subcutaneously before surgery. Scalp incisions were made and craniotomies (1–2 mm in diameter) above the left parietal cortex were created using a high-speed drill (K.1070; Foredom) for unilateral viral injections while two craniotomies were made above both parietal cortices for bilateral viral injections. Beveled glass pipettes (1B100–4; World Precision Instruments) filled with viruses were placed into the striatum (0.8 mm anterior to the bregma, 2.0 mm lateral to the midline, and 2.4 mm from the pial surface) or the hippocampus CA1 (2 mm posterior to the bregma, 1.5 mm lateral to the midline, and 1.6 mm from the pial surface). For slice Ca2+ imaging, 1 μl AAV2/5 GfaABC1D GCaMP6f (2.4 × 1013 gc/ml) with either 1 μl of AAV2/5 GfaABC1D mCherry-hPMCA2w/b (9.2 × 1012 gc/ml) or 0.5 μl AAV5 GfaABC1D tdTomato (5.0 × 1013 gc/ml) were injected at 200 nl/min using a syringe pump (Pump11 PicoPlus Elite; Harvard Apparatus). For MINIscope Ca2+ imaging, 0.5 μl AAV1 Synapsin GCaMP6f (2.0 × 1013 gc/ml) with either 1 μl of AAV5 GfaABC1D mCherry-hPMCA2w/b or 0.5 μl AAV5 GfaABC1D tdTomato were injected. For D1- and D2-MSN physiological recording, 0.5 μl AAV1 CAG Flex tdTomato (1.1 × 1013 gc/ml) with either 1 μl of AAV5 GfaABC1D mCherry-hPMCA2w/b or 0.5 μl AAV5 GfaABC1D tdTomato were injected. For Ca2+ imaging in R6/2 and non-carrier control mice, 1.5 μl AAV5 GfaABC1D GCaMP6f were injected. For RiboTag mice RNA-seq experiments, 1 μl of AAV2/5 GfaABC1D-mCherry-hPMCA2w/b or 0.5 μl AAV2/5 GfaABC1D-tdTomato were injected into the striatum of RiboTag mice. For RiboTag AAV RNA-seq experiments, 0.7 μl AAV2/5 GfaABC1D-Rpl22-HA with either 1 μl of AAV2/5 GfaABC1D-mCherry-hPMCA2w/b or 0.5 μl AAV2/5 GfaABC1D-tdTomato were injected into the striatum of C57BL/6NTac mice. Glass pipettes were withdrawn after 10 min and scalps were cleaned and sutured with sterile surgical sutures. Mice were allowed to recover in clean cages with food containing Trimethoprim/Sulfamethoxazole and water for 7 days. Imaging and behavioral experiments were performed at least three weeks after surgeries.
Brain slice preparation
For imaging and electrophysiological recordings, mice were transferred from the vivarium to the laboratory during the light cycle between 8 am and 10 am. Brain slices were prepared 30 min to 1 hour afterwards and were used for experiments within 8 h of slicing (mostly within 6 hours). Both male and female mice (9–11 weeks old) with AAV injection were anesthetized and decapitated ~2–4 weeks. Coronal slices (300 μm thick) of the striatum or hippocampus were prepared in ice-cold sucrose cutting solution (30 mM NaCl, 4.5 mM KCl, 1.2 mM NaH2PO4, 26 mM NaHCO3, 10 mM D-glucose, 194 mM sucrose and 1 mM MgCl2) using a vibratome (DSK Microslicer; Ted Pella, Inc.). Slices were then incubated in artificial cerebrospinal fluid (ACSF) (124 mM NaCl, 4.5 mM KCl, 1 mM MgCl2, 1.2 mM NaH2PO4, 26 mM NaHCO3, 10 mM D-glucose, and 2.0 mM CaCl2) for 30 min at 32°C and 1 h at room temperature before recording. All the solutions were oxygenated with 95% O2/5% CO2. Brain slices were used for experiments within 8 h of slicing. Procedures have been described in detail (Jiang et al., 2016; Tong et al., 2014).
Intracellular Ca2+ imaging of striatal astrocytes
Ca2+ imaging in striatal slices was performed using a Scientifica two-photon laser-scanning microscope (2PLSM) equipped with a MaiTai laser (Spectra-physics). To image GCaMP6f signals, laser was tuned at 920 nm wavelength. The laser power measured at the sample was less than 30 mW with a 40× water-immersion objective lens (Olympus). Astrocytes located in the dorsolateral striatum and at least 40 μm from the slice surface were selected for imaging. Images were acquired at 1 frame per second using SciScan software (Scientifica). Striatal slices were maintained in ACSF (124 mM NaCl, 4.5 mM KCl, 1 mM MgCl2, 1.2 mM NaH2PO4, 26 mM NaHCO3, 10 mM D-glucose, and 2.0 mM CaCl2) through a perfusion system. Phenylephrine (10 μM) was bath-applied to activate GPCR-mediated Ca2+ signals. Cyclopiazonic acid (20 μM) was applied to deplete intracellular calcium stores. Slices were pre-incubated with SR101 (1 μM) for 10 min for some imaging experiments. Ca2+ signals were processed in ImageJ (NIH) and presented as the relative change in fluorescence (ΔF/F). Peak amplitude, half-width, and frequency of Ca2+ signals were analyzed in ClampFit10.4, while integrated areas were calculated in OriginPro 8.
Intracellular Ca2+ imaging of hippocampal astrocytes
Ca2+ imaging in hippocampal slices was performed using a confocal microscope (Fluoview 1000; Olympus) with a 40X water-immersion objective lens (NA = 0.8). To image GCaMP6f signals, 488 nm line of an Argon laser was used with the intensity adjusted to 10–14% of the maximum output of 10 mW. The emitted light pathway consisted of an emission high pass filter (505–525 nm) before the photomultiplier tube. Astrocytes located in the hippocampal CA1 and at least 40 μm from the slice surface were selected for imaging. Images were acquired at 1 frame per second. hippocampal slices were maintained in ACSF (124 mM NaCl, 4.5 mM KCl, 1 mM MgCl2, 1.2 mM NaH2PO4, 26 mM NaHCO3, 10 mM D-glucose, and 2.0 mM CaCl2) through a perfusion system. Phenylephrine (10 μM) was bath-applied to activate GPCR-mediated Ca2+ signals. Ca2+ signals were processed in ImageJ (NIH) and presented as the relative change in fluorescence (ΔF/F). Peak amplitude, half-width, and frequency of Ca2+ signals were analyzed in ClampFit10.4, while integrated areas were calculated in OriginPro 8.
MINIscope implantation and imaging in the striatum
Both male and female mice (9 weeks old) were anesthetized and placed onto a stereotaxic frame (David Kopf Instruments, Tujunga, CA) one week after viral injection in the striatum. Dexamethasone (0.2 mg/kg) and carprofen (5 mg/kg) were administered subcutaneously during the surgery. After hair removal and scalp incision, the skull was exposed and the connective tissue underneath was removed. A 2-mm diameter circular craniotomy was made with the center located 0.5 mm from the viral injection site. The cortex underneath the craniotomy was aspirated using a blunt syringe needle connected to a vacuum pump until a layer of corpus callosum was exposed. ACSF was constantly applied to the surgery site to prevent tissue from drying. Sterilized GRIN lens (0.25 pitch, 2 mm in diameter and 4.79 mm in length; Grintech Gmbh) attached to a stereotaxic arm was slowly lowered into the aspirated space until the bottom of the lens was 2 mm below the surface of the skull. Implanted GRIN lens was then anchored to the skull as well as a custom-made head bar that was glued to the back of the occipital bone with cyanoacrylate (Krazy glue) and dental cement (Ortho-Jet, Lang Dental). After surgery, mice were given carprofen (5 mg/kg) and amoxicillin (BIOMOX, Virbac Animal Health) for at least 7 days. At least 3 days before imaging, a light-weight aluminum baseplate was glued on top of the dental cement to secure the miniature microscope (MINIscope), a procedure called “baseplating”. During baseplating, the mouse was head-fixed without anesthesia so that the appropriate field of view could be determined based on neuronal activities. Mice were accustomed to head-fixation after GRIN lens implantation and before baseplating. SNAP5114 (50 μmol/kg) was intraperitoneally injected for 7 days before imaging. On the day of imaging, MINIscopes were placed on top of the baseplates and secured while the mice were awake (see www.miniscope.org for further details). Both MINIscope and behavior camera (Logitech) were controlled by a custom data acquisition software written in C++. Images were acquired at 30 frames per second and time stamped by the internal CPU clock for subsequent analysis.
Electrophysiological recordings in the striatal slices
Electrophysiological recordings were performed as described previously by us for striatal slices (Chai et al., 2017; Jiang et al., 2016; Octeau et al., 2018; Srinivasan et al., 2015; Srinivasan et al., 2016; Tong et al., 2014), which uses methods reported earlier in landmark studies of striatal astrocytes (Adermark and Lovinger, 2006). Of note, the brain slice recordings were performed at room temperature (21–23°C). This is because in our past work over a period of 5 years we have found that incubating striatal slices from adult mice (>P60) at ~33°C closer to mouse body temperature caused swelling, appearance of sick cells and the rapid loss of whole-cell seal integrity (particularly for astrocytes – the focus of our work). In order to minimize such effects, we had to balance the requirement for healthy tissue with that of recording at warmer temperatures. By opting to record at room temperature we prioritized the quality of the recordings over temperature. However, the in vivo imaging, behavior, immunohistochemistry and RNA-seq work described all reflect measurements at mouse body temperature. Hence, our studies need to be interpreted with the knowledge that the kinetics of the synaptic responses we report are likely slowed. Our use of room temperature is identical to that used in past striatal astrocyte studies by other labs (Adermark and Lovinger, 2006; D’Ascenzo et al., 2007; Martín et al., 2015).
Whole-cell patch-clamp recordings were made from medium spiny neurons (MSNs) or astrocytes morphologically identified under infrared optics from the dorsolateral striatum. D1-and D2-MSNs were selected based on tdTomato fluorescence. The intracellular solution for MSN IPSCs recordings comprised the following: 138 mM KCl, 10 mM HEPES, 1 mM EGTA, 0.3 mM Na-ATP, 4 mM Mg-ATP, 0.1 mM CaCl2, 8 mM Na2-phosphocreatine, with pH adjusted to 7.3. The intracellular solution for MSN EPSCs recordings comprised the following: 120 mM CsMeSO3, 15 mM CsCl, 8 mM NaCl, 10 mM HEPES, 0.2 mM EGTA, 0.3 mM Na-GTP, 2 mM Mg-ATP, 10 mM TEA-Cl, with pH adjusted to 7.3 with CsOH. The intracellular solution for astrocyte recordings comprised the following: 135 mM potassium gluconate, 3 mM KCl, 10 mM HEPES, 1 mM EGTA, 0.3 mM Na-ATP, 4 mM Mg-ATP, 0.1 mM CaCl2, 8 mM Na2-phosphocreatine, with pH adjusted to 7.3. To isolate sIPSCs, MSNs were voltage-clamped at −60 mV and pre-incubated with ACSF (124 mM NaCl, 4.5 mM KCl, 1 mM MgCl2, 1.2 mM NaH2PO4, 26 mM NaHCO3, 10 mM D-glucose, and 2.0 mM CaCl2) in the presence of 10 μM CNQX for 10 min before recording. Note, that all our recordings used whole-cell patch clamp and not perforated patch clamp. Brain slice neuronal perforated patch recordings most successfully employ antibiotics such as nystatin and amphotericin that rapidly form pores in the membrane patch. However, these pores are permeable to monovalent anions and cations, including Cl- (Lippiat, 2008; Sherman-Gold, 2006). Hence, perforated patch recordings would not allow us to maintain intracellular Cl- at its endogenous value. The pores in perforated patches do discriminate between monovalent and multivalent cations and larger organic species, thus allowing researchers to preserve intracellular metabolites such at ATP. However, such recordings do not prevent equilibration of Cl- (Lippiat, 2008; Sherman-Gold, 2006). Gramicidin may more favorably discriminate between monovalent anions and cations, but the procedure is very slow, leads to high access resistances and also creates a Donnan equilibrium potential that in turn causes a voltage offset and change in cell volume. These factors would impair our ability to record tonic currents (which are far smaller than phasic GABA IPSCs and exogenous GABA puff currents). For all these reasons, we chose whole-cell patch clamp, which is the same as that used in past studies on tonic inhibition in MSNs (Kirmse et al., 2009; Wójtowicz et al., 2013). mIPSCs were recorded after incubation with 10 μM CNQX and 0.5 μM TTX for 10 min. To assess extrasynaptic GABAA receptor-mediated responses, GABA (300 μM) was puffed onto the cell bodies of MSNs from a micropipette connected to Picospritzer III (Parker) for 100 ms to evoke GABA currents. To evoke IPSCs in MSNs, a bipolar stimulating electrode was placed approximately 100–150 μm from the recorded neuron in the dorsal striatum. Glutamate receptors were blocked by incubation with 10 μM CNQX and 25 μM APV for 10 min. To assess input-output function, test-stimuli were applied at increasing intensities ranging from 0 to 50 μA. Stimulation intensities were set to evoke responses at 50% maximal amplitude to induce paired pulse responses. Paired pulses were delivered at three different inter-pulse intervals: 30, 50 and 100 ms apart. To access tonic GABA currents, 25 μM bicuculline was bath-applied in the presence of 10 μM CNQX, 0.5 μM TTX and 25 μM APV. To investigate the contributions of GABA transporters to tonic GABA currents, striatal slices were pre-incubated with GAT-1 inhibitor NNC711 (10 μM; Tocris Bioscience) or GAT-3 inhibitor SNAP5114 (40 μM; Tocris Bioscience) or both for 10 min before recording. To isolate spontaneous EPSCs, MSNs were voltage-clamped at −60 mV and pre-incubated with ACSF in the presence of 25 μM bicuculline for 10 min before recording. To assess evoked EPSCs, electrical field stimulation (EFS) were achieved using a bipolar matrix electrode (FHC) that was placed on the dorsolateral corpus callosum to evoke glutamate release from cortico-striatal pathway (Jiang et al., 2016). The MSNs to be assessed were typically located ~250–300 μm away from the stimulation site. To isolate the AMPAR- and NMDAR-mediated evoked EPSCs, MSNs were voltage-clamped at −70 mV or +40 mV in the presence of 10 μM bicuculline. The AMPAR-mediated EPSC was measured at the peak amplitude of the EPSC at −70 mV, while the amplitude of the EPSC 50 ms after stimulation at +40 mV was used to estimate the NMDAR-mediated component. All recordings were performed at room temperature, using pCLAMP10 (Axon Instruments, Molecular Devices) and a MultiClamp 700B amplifier (Axon Instruments, Molecular Devices). Cells with Ra exceeded 20 MΩ were excluded from analysis. Analysis was performed using ClampFit 10.4 software.
Immunohistochemistry (IHC)
Both male and female mice (9–14 weeks old) were transcardially perfused with 0.1 M phosphate buffered saline (PBS) followed by 10% formalin. Brains were dissected out and post-fixed in 10% formalin for several hours and then cryoprotected in 30% sucrose (0.1 M PBS). Serial 40 μm sections were collected and incubated with the following primary antibodies overnight at 4°C: rabbit anti-S100β (1:1,000; Abcam, ab41548), mouse anti-NeuN (1:1,000; Millipore, MAB377), chicken anti-GFP (1:1,000; Abcam, ab13970), rabbit anti-RFP (1:1,000; Rockland, 600–401-379), chicken anti-RFP (1:1,000; a gift from Dr. Brecha’s laboratory at UCLA), rabbit anti-GAT-3 (1:500; a gift from Dr. Brecha’s laboratory at UCLA), rabbit antiKir4.1 (1:1,500; Alomone, APC-035), rabbit anti-DARPP-32 (1:200; Abcam, ab40801) or mouse anti-HA (1:1,000; Covance, MMS-101R). Sections were then incubated with the following secondary antibodies (1:1,000; Molecular Probes): Alexa Fluor 488 goat anti-chicken (A11039), Alexa Fluor 488 goat anti-rabbit (A11008), Alexa Fluor 546 goat anti-mouse (A11003), Alexa Fluor 546 goat anti-chicken (A11040) and Alexa Fluor 594 goat anti-rabbit (R37007) at room temperature for 2 h. Fluorescence imaging was acquired with a 40× oil-immersion objective lens (NA 1.3) under a laser-scanning confocal microscope (Olympus).
Western blot analyses
To extract membrane fraction, mice were decapitated and striata were dissected and homogenized in ice-cold buffer (250 mM sucrose, 1 mM EDTA, 10 mM Tris-HCl and protease inhibitor cocktail, adjusted to pH 7.2). The homogenate was centrifuged at 500 × g for 15 min at 4 °C. The pellet was discarded and the supernatant was centrifuged twice at 75,000 × g for 30 min at 4 °C. The pellet containing the membrane fraction was dissociated in RIPA buffer with protease inhibitors by sonication for 30 min. Protein concentrations were measured using BCA protein assay (Thermo Scientific). Membrane fraction was mixed with 2× Laemmli buffer containing 8 M urea. The samples were then electrophoretically separated by 12% SDS-PAGE (5 μg protein per lane) and transferred onto nitrocellulose membrane (0.45 μm). The membrane was probed at 4 °C overnight with the following primary antibodies: rabbit anti-GAT-3 (1:1,000; a gift from Dr. Brecha’s laboratory at UCLA), rabbit anti-GAT-1 (1:500; Abcam) and mouse anti-β-tubulin (1:5,000; Millipore). IRDye 680RD anti-mouse (1:10,000; Li-Cor) and IRDye 800CW anti-rabbit (1:10,000; Li-Cor) secondary antibodies were used and images were acquired on a Li-Cor Odyssey infrared imager. Signal intensities at expected molecular weight were quantified using ImageJ (NIH). GAT-1 and GAT-3 levels were normalized to β-tubulin.
Behavioral tests
Behavioral tests were performed during the light cycle between 12:00 pm and 6:00 pm. Only male mice (9–13 weeks old) were used in behavioral tests because of sex-dependent differences known for striatal physiology. All the experimental mice were transferred to the behavior testing room at least 30 min before the tests to acclimatize to the environment and to reduce stress.
Open field test:
The open field chamber consisted of a square arena (28 cm × 28 cm) enclosed by walls made of Plexiglass (19 cm tall). The periphery of the arena was defined as the area within 2.5 cm adjacent to the walls of the chamber and the center of the arena was defined as the area 2.5 cm away from the chamber walls. Each mouse was placed into an open field box to habituate for 20 min. Locomotor activity was then recorded for 1 h using an infrared camera located underneath the open field chamber. Recording camera was connected to a computer operating an automated video tracking software Ethovision XT (Noldus Information Technology). Parameters analyzed included distance traveled and time spent in the center, with 5 min and 1 h time bins. After recordings finished, mice were returned to their home cages.
Acute locomotor effects of cocaine:
Each mouse was weighed and received single intraperitoneal injection of saline or cocaine (10 mg/kg) and then placed into the open field chamber. Locomotor activities was recorded for 15 min and distance travelled was assessed using Ethovision XT with 3 min time bins.
Locomotor sensitization induced by cocaine:
Each mouse was weighed and received intraperitoneal injection of saline or cocaine (10 mg/kg) for 5 consecutive days. After 3 weeks of withdrawal from repeated cocaine injection, cocaine challenge was performed with mice injected with either saline or cocaine (10 mg/kg) to test the sensitization. Locomotor activity was recorded for 15 min in the open field chamber and distance travelled was assessed using Ethovision XT with 15 min time bins.
Self-grooming behavior:
The procedure of self-grooming behavior measurement was adapted from previously published study (Kalueff et al., 2007). Mice were placed individually into the open field chambers or plastic cylinders (15 cm in diameter and 35 cm tall), and allowed to habituate for 20 min. Self-grooming behavior was recorded for 10 min. SNAP5114 (50 μmol/kg) was intraperitoneally injected for 7 days and the experiments were performed again. A timer was used to assess the cumulative time spent in self-grooming behavior, which included paw licking, unilateral and bilateral strokes around the nose, mouth and face, paw movement over the head and behind ears, body fur licking, body scratching with hind paws, tail licking and genital cleaning. The number of self-grooming bouts was also counted. Separate grooming bouts were considered when the pause was more than 5 s or behaviors other than self-grooming occurred. Self-grooming microstructure was not assessed.
Spray test:
A standard spray bottle was filled with distilled water and the nozzle was adjusted to the “misting” mode. Mice were held by the tails on the bench and sprayed three times from 30 cm away to be adequately covered with mist. Mice were placed individually into the plastic cylinders and grooming behavior was recorded for 10 min following the spray and then analyses as described in the preceding section.
Rotarod test:
Mice were held by the tails and placed on the rod (3 cm diameter) of a single lane rotarod apparatus (ENV-577M, Med Associates Inc.), facing away from the direction of rotation. The rotarod was set with a start speed of 4 rpm. Acceleration started 10 seconds later and was set to 20 rpm per minute with a maximum speed 40 rpm. Each mouse received two trials 30 min apart and the latency to fall was recorded for each trial. The average latency to fall was used as a measurement for motor coordination (Deacon, 2013).
Elevated plus maze:
Modified from the protocol described previously (Komada et al., 2008), the elevated plus maze apparatus was a cross-shaped platform that comprised two open arms and two closed arms (29 cm × 9 cm) with a center platform (9 cm × 9 cm). Open arms and closed arms were perpendicular to each other and elevated about 50 cm from the floor. Closed arms had walls (15 cm) to enclose the arms. Each mouse was placed in the center platform with its nose directed toward the same closed arm and allowed to explore the entire apparatus for 10 min. Anxiety-like behavior was evaluated with the number of entries into each compartment and the time spent in each compartment. Total travel distance was also determined. All the parameters were analyzed in Ethovision XT.
Clasping test:
R6/2 mice and non-carrier controls at 10–11 weeks old were used in this test. As previously described (Tong et al., 2014), each mouse was suspended by holding the tail. The experiments were conducted for 1 min. Time taken for the mouse to move both forelimbs close to the body and both paws clasp was recorded. Mice did not clasp in this time period were given a data value of 60 s. SNAP5114 (50 μmol/kg) was intraperitoneally injected for 7 days and the test was performed again.
Grip strength test:
R6/2 mice and non-carrier controls at 10–11 weeks old were used in this test. Forelimb strength was measured using a customized grip strength meter (Chatillon Digital force gauge) with an adjustable angel wire mesh grip (San Diego Instruments). Both the grip strength meter and the mesh grip were positioned horizontally. Each mouse was held by the base of the tail and lowered to grasp the mesh with both forelimbs. A steady horizontal force was then applied to extend the mouse away from the meter until it relinquished its grip on the mesh. Maximum pulling force required to separate the mouse from the mesh grip was recorded by the meter. Each mouse was tested in five trials with a 1 min intertrial interval and average maximum pulling force was used to assess the muscle strength. SNAP5114 (50 μmol/kg) was intraperitoneally injected for 7 days and the test was performed again.
Footprint test:
R6/2 mice and non-carrier controls at 10–11 weeks old were used in this test. A one meter long runway (8 cm wide) was lined with paper. Each mouse with hind paws painted with non-toxic ink was placed at an open end of the runway and allowed to walk to the other end with a darkened box. For the gait analysis, stride length and width were measured and averaged for both left and right hindlimbs over 5 steps. SNAP5114 (50 μmol/kg) was intraperitoneally injected for 7 days and the test was performed again.
RNA-seq analysis of striatal astrocyte transcriptomes
Total RNA was extracted from the striatum of male and female mice (aged 9–10 weeks) as previously described (Chai et al., 2017). For RiboTag mice RNA-seq experiments, tamoxifen (Sigma, 20 mg/ml) was administered intraperitoneally for five consecutive days at 100 mg/kg body weight to Aldh1l1-Cre/ERT2 × RiboTag mice. One day after the last tamoxifen injection, 1 μl of AAV2/5 GfaABC1D-mCherry-hPMCA2w/b or 0.5 μl AAV2/5 GfaABC1D-tdTomato were injected into the striatum of RiboTag mice. For RiboTag AAV RNA-seq experiments, 0.7 μl AAV2/5 GfaABC1D-Rpl22-HA with either 1 μl of AAV2/5 GfaABC1D-mCherryhPMCA2w/b or 0.5 μl AAV2/5 GfaABC1D-tdTomato were injected into the striatum of C57BL/6NTac mice. Three weeks after the AAV injection, RNA extraction was performed as previously described (Chai et al., 2017). Briefly, striatal tissues were dissected and homogenized. RNA was extracted from 10–20% of homogenate after centrifugation as input sample, which contained RNA from all cell types in the striatum (Qiagen Rneasy Plus Micro #74034). The remaining homogenate was incubated with mouse anti-HA antibody (1:250; Covance, #MMS101R) for 4 hours followed by the addition of magnetic beads (Invitrogen, Dynabeads #110.04D) for overnight incubation at 4°C. RNA was purified from the immunoprecipitation (IP) sample, which contained astrocyte-enriched RNA (Qiagen Rneasy Plus Micro #74034). RNA concentration and quality were assessed with Agilent 2100 Bioanalyzer. RNA samples with RNA integrity number (RIN) greater than 7 were used for multiplexed library prep with Nugen Ovation RNA-Seq System V2. For each experiment, all samples were multiplexed into a single pool in order to avoid batch effects (Auer and Doerge, 2010), and sequencing was performed on Illumina NextSeq 500 for 2 × 75 yielding between 58 and 83 million reads per sample. Demultiplexing was performed with Illumina Bcl2fastq2 v 2.17 program. Reads were aligned to the mouse mm10 reference genome using the STAR spliced read aligner (Dobin et al., 2013) with default parameters and fragment counts were derived using HTS-seq program. Between 66 and 91% of the reads mapped uniquely to the mouse genome and were used for subsequent analyses. Differential gene expression analysis was performed with genes with CPM > 3 in at least 4 samples, using Bioconductor packages edgeR (Robinson et al., 2010) and limmaVoom (Law et al., 2014) with false discovery rate (FDR) threshold < 0.1 or 0.05 (http://www.bioconductor.org). Weighted gene co-expression network analysis (WGCNA) was performed using an R package of WGCNA for finding modules of highly correlated genes. The gene ontology (GO) enrichment analysis was performed using PANTHER overrepresentation test (GO ontology database released 2018–04-04) with FDR < 0.05. RNAseq data have been deposited within the Gene Expression Omnibus (GEO) repository (www.ncbi.nlm.nih.gov/geo) with accession numbers GSE114757 and GSE114756.
QUANTIFICATION AND STATISTICAL ANALYSIS
Data from every experiment represent at least four replicates. Sample sizes were not calculated a priori. Mice were randomly assigned to different experimental groups. For 2PLSM Ca2+ imaging experiments, Ca2+ transients recorded in a single optical plane and were measured by plotting the intensity of regions of interest over time. Slow drifts in astrocyte position (~25 μm) were corrected with a custom Plugin in NIH ImageJ. A signal was declared as a Ca2+ transient if it exceeded the baseline by greater than twice the baseline noise (SD). Ca2+ signals were processed in ImageJ and presented as the relative change in fluorescence (ΔF/F). Peak amplitude, half-width, and frequency of Ca2+ signals were analyzed in ClampFit10.4 (Molecular Devices), while integrated areas were calculated in OriginPro 8. To evaluate the resting astrocyte Ca2+ level, basal GCaMP6f fluorescence intensity was normalized to SR101 fluorescence intensity. For MINIscope experiments, a custom script written in MATLAB (MathWorks, Natick, MA) was used to correct column-wise ACD variation and small movement artifacts. Active neurons (ROIs) were segmented manually and Ca2+ traces were extracted in ImageJ. Peak amplitude, half-width, integrated area and frequency of individual Ca2+ events were analyzed in MiniAnalysis Program 6.0.7 (Synaptosoft, Decatur, GA). Ca2+ traces were aligned with behavioral analysis using custom-written scripts in R (version 3.3.2). To assess correlation coefficient between neuronal pairs, Ca2+ traces were binned every 1 s and given binary variables without deconvolution. A matrix with columns representing discrete time intervals of 1 s each and rows representing each ROI was created and the Pearson correlation matrix was calculated using custom-written scripts in R. Since the correlation matrix is a symmetric square matrix with 1s along the main diagonal, the absolute mean correlation of the ROIs was calculated after excluding the values along and below the main diagonal. Results were also confirmed with deconvolved data (Friedrich et al., 2017). For electrophysiological recordings, synaptic currents and tonic GABA currents were analyzed using MiniAnalysis Program 6.0.7 (Synaptosoft, Decatur, GA) or Clampfit10.4 (Molecular Devices).
All statistical tests were performed in OriginPro 8. The graphs were created in OriginPro 8 and assembled in CorelDraw12. Data are presented as mean ± SEM. Note that in some of the graphs, the SEM. bars were smaller than the symbols used to represent the mean. For each set of data to be compared, OriginPro 8 was used to determine whether the data were normally distributed or not. Parametric tests were used for normally distributed data sets while nonparametric tests were applied to data not normally distributed. Paired and unpaired Student’s two tailed t tests (as appropriate) and two tailed Mann–Whitney tests were used for most statistical analyses with significance declared at P < 0.05, but stated in each case with a precise P value. Comparisons of data sets with more than two groups or conditions were calculated with one-way or two-way ANOVA tests followed by Bonferroni post hoc tests. When the P value was less than 0.001, it is stated as P < 0.001 to save space on the figure panels and tables. When the P values was greater than 0.05, it is stated as non-significant (n.s.). Throughout the manuscript, the results of statistical tests (P values and n numbers) are reported in the figure panels. N numbers are defined as the numbers of cells/slices or mice throughout.
Supplementary Material
Highlights.
Multiple approaches were used to assess striatal astrocyte Ca2+ signaling in vivo
Ambient GABA was altered when astrocyte signaling was reduced
Reduced astrocyte signaling resulted in excessive self-grooming in mice
Astrocytes contribute to neural circuits in vivo and OCD-like phenotypes in mice
Acknowledgements:
Supported by NIH grants NS060677 and MH104069 (BSK). MINIscopes supported by U01NS094286 (BSK and PG). AMWT and CJE supported by K99DA004016 (AMWT), DA005010 (CJE), and the Shirley and Stefan Hatos Foundation. We acknowledge the NINDS Informatics Center for Neurogenetics and Neurogenomics (P30 NS062691 to GC) and the Genetics, Genomics and Informatics Core of the Semel Institute of Neuroscience at UCLA (supported by grant U54HD087101–01 from the Eunice Kennedy Shriver National Institute of Child Health and Human Development). XY was supported by American Heart Association (16POST27260256). JN supported by a JSPS Overseas Research Fellowship (H28–729). Thanks to Daniel Aharoni for help with MINIscopes, to Blanca Diaz-Castro for help with qPCR, Fuying Gao for help with RNA-seq data processing, and the UCLA Neuroscience Genomics Core for sequencing data. J Christopher Octeau and Blanca Diaz-Castro helped with AAV2/5 Rpl22HA testing. Thanks to Michael V. Sofroniew and Wendy Walwyn for equipment, and Nicholas Brecha for GAT-3 antibody. Thanks to Jonathan Flint, Vahri Beaumont, Ignacio Munoz-Sanjuan, Michael V. Sofroniew and Daniel Geschwind for comments. Thanks to Emanuel E. Strehler for sharing tips. Thanks to Brandon Brown for R scripts.
Footnotes
Declaration of interests: none
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adermark L, and Lovinger DM (2006). Ethanol effects on electrophysiological properties of astrocytes in striatal brain slices. Neuropharmacology 51, 1099–1108. [DOI] [PubMed] [Google Scholar]
- Agulhon C, Fiacco TA, and McCarthy KD (2010). Hippocampal short- and long-term plasticity are not modulated by astrocyte Ca2+ signaling. Science 327, 1250–1254. [DOI] [PubMed] [Google Scholar]
- Aida T, Yoshida J, Nomura M, Tanimura A, Iino Y, Soma M, Bai N, Ito Y, Cui W, Aizawa H, et al. (2015). Astroglial glutamate transporter deficiency increases synaptic excitability and leads to pathological repetitive behaviors in mice. Neuropsychopharmacology 40, 1569–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auer PL, and Doerge RW (2010). Statistical design and analysis of RNA sequencing data. Genetics 185, 405–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhurin KI, Mac V, Golshani P, and Masmanidis SC (2016). Temporal correlations among functionally specialized striatal neural ensembles in reward-conditioned mice. J Neurophysiol 115, 1521–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazargani N, and Attwell D (2016). Astrocyte calcium signalling: the third wave. Nat Neurosci 19, 182–189. [DOI] [PubMed] [Google Scholar]
- Beenhakker MP, and Huguenard JR (2010). Astrocytes as gatekeepers of GABAB receptor function. J Neurosci 30, 15262–15276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beglinger LJ, Paulsen JS, Watson DB, Wang C, Duff K, Langbehn DR, Moser DJ, Paulson HL, Aylward EH, Carlozzi NE, et al. (2008). Obsessive and compulsive symptoms in prediagnosed Huntington’s disease. J Clin Psychiatry 69, 1758–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai DJ, Aharoni D, Shuman T, Shobe J, Biane J, Song W, Wei B, Veshkini M, La-Vu M, Lou J, et al. (2016). A shared neural ensemble links distinct contextual memories encoded close in time. Nature 534, 115–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai H, Diaz-Castro B, Shigetomi E, Monte E, Octeau JC, Yu X, Cohn W, Rajendran PS, Vondriska TM, Whittelegge JP, et al. (2017). Neural circuit-specialized astrocytes: transcriptomic, proteomic, morphological and functional evidence. Neuron 95, 531–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SK, Tvrdik P, Peden E, Cho S, Wu S, Spangrude G, and Capecchi MR (2010). Hematopoietic origin of pathological grooming in Hoxb8 mutant mice. Cell 141, 775–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromwell HC, and Berridge KC (1996). Implementation of action sequences by a neostriatal site: a lesion mapping study of grooming syntax. J Neurosci 16, 3444–3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Ascenzo M, Fellin T, Terunuma M, Revilla-Sanchez R, Meaney DF, Auberson YP, Moss SJ, and Haydon PG (2007). mGluR5 stimulates gliotransmission in the nucleus accumbens. Proc Natl Acad Sci U S A 104, 1995–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deacon RM (2013). Measuring motor coordination in mice. J Vis Exp May 29;(75):e2609. doi: 10.3791/2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich J, Zhou P, and Paninski L (2017). Fast online deconvolution of calcium imaging data. PLoS Comput Biol Mar 14;13(3):e1005423. doi: 10.1371/journal.pcbi.1005423 eCollection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, Nowak NJ, Joyner A, Leblanc G, Hatten ME, et al. (2003). A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature 425, 917–925. [DOI] [PubMed] [Google Scholar]
- Graybiel AM, and Grafton ST (2015). The striatum: where skills and habits meet. Cold Spring Harb Perspect Biol Aug 3;7(8):a021691. doi: 10.1101/cshperspect.a021691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haustein MD, Kracun S, Lu XH, Shih T, Jackson-Weaver O, Tong X, Xu J, Yang XW, O’Dell TJ, Marvin JS, et al. (2014). Conditions and constraints for astrocyte calcium signaling in the hippocampal mossy fiber pathway. Neuron 82, 413–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R, Diaz-Castro B, Tong X, Looger LL, and Khakh BS (2016). Dysfunctional calcium and glutamate signaling in striatal astrocytes from Huntington’s disease model mice. J Neurosci 36, 3453–3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R, Haustein MD, Sofroniew MV, and Khakh BS (2014). Imaging intracellular Ca2+ signals in striatal astrocytes from adult mice using genetically-encoded calcium indicators. J Vis Exp Nov 19;(93):e51972. doi: 10.3791/51972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalueff AV, Aldridge JW, LaPorte JL, Murphy DL, and Tuohimaa P (2007). Analyzing grooming microstructure in neurobehavioral experiments. Nat Protoc 2, 2538–2544. [DOI] [PubMed] [Google Scholar]
- Kalueff AV, Stewart AM, Song C, Berridge KC, Graybiel AM, and Fentress JC (2016). Neurobiology of rodent self-grooming and its value for translational neuroscience. Nat Rev Neurosci 17, 45–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khakh BS, Beaumont V, Cachope R, Munoz-Sanjuan I, Goldman SA, and Grantyn R (2017). Unravelling and exploiting astrocyte dysfunction in Huntington’s disease. Trends Neurosci 40, 422–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khakh BS, and McCarthy KD (2015). Astrocyte calcium signaling: from observations to functions and the challenges therein. Cold Spring Harb Perspect Biol January 20;7(4):a020404. doi: 10.1101/cshperspect.a020404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khakh BS, and Sofroniew MV (2015). Diversity of astrocyte functions and phenotypes in neural circuits. Nat Neurosci 18, 942–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirmse K, Kirischuk S, and Grantyn R (2009). Role of GABA transporter 3 in GABAergic synaptic transmission at striatal output neurons. Synapse 63, 921–929. [DOI] [PubMed] [Google Scholar]
- Komada M, Takao K, and Miyakawa T (2008). Elevated plus maze for mice. J Vis Exp December 22;(22). pii: 1088. doi: 10.3791/1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law CW, Chen Y, Shi W, and Smyth GK (2014). voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol February 3;15(2):R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liddelow SA, Guttenplan KA, Clarke LE, Bennett FC, Bohlen CJ, Schirmer L, Bennett ML, Münch AE, Chung WS, Peterson TC, et al. (2017). Neurotoxic reactive astrocytes are induced by activated microglia. Nature 541, 481–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippiat JD (2008). Whole-cell recording using the perforated patch clamp technique. Methods Mol Biol 491, 141–149. [DOI] [PubMed] [Google Scholar]
- Martín R, Bajo-Grañeras R, Moratalla R, Perea G, and Araque A (2015). Circuit-specific signaling in astrocyte-neuron networks in basal ganglia pathways. Science 349, 730–734. [DOI] [PubMed] [Google Scholar]
- Muthukumar AK, Stork T, and Freeman MR (2014). Activity-dependent regulation of astrocyte GAT levels during synaptogenesis. Nat Neurosci 17, 1340–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmerjahn A, and Bergles DE (2015). Large-scale recording of astrocyte activity. Curr Opin Neurobiol 32, 95–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Octeau JC, Chai H, Jiang R, Bonanno SL, Martin KC, and Khakh BS (2018). An Optical Neuron-Astrocyte Proximity Assay at Synaptic Distance Scales. Neuron 98, 49–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petravicz J, Boyt KM, and McCarthy KD (2014). Astrocyte IP3R2-dependent Ca(2+) signaling is not a major modulator of neuronal pathways governing behavior. Front Behav Neurosci November 12;8:384. doi: 10.3389/fnbeh.2014.00384. eCollection 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petravicz J, Fiacco TA, and McCarthy KD (2008). Loss of IP3 receptor-dependent Ca2+ increases in hippocampal astrocytes does not affect baseline CA1 pyramidal neuron synaptic activity. J Neurosci 28, 4967–4973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richler E, Chaumont S, Shigetomi E, Sagasti A, and Khakh BS (2008). Tracking transmitter-gated P2X cation channel activation in vitro and in vivo. Nat Methods 5, 87–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, and Smyth GK (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenegger DG, Tran CH, J.I. WC, and Gordon GR (2015). Tonic Local Brain Blood Flow Control by Astrocytes Independent of Phasic Neurovascular Coupling. J Neurosci 35, 13463–13474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rungta R, Bernier L-P, Dissing-Olesen L, Groten C, LeDue J, Drissler S, and MacVicar B (2016). Ca2+ transients in astrocyte fine processes occur via Ca2+ influx in adult mouse hippocampus. Glia August 1. doi: 10.1002/glia.23042 [Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- Sanz E, Quintana A, Deem JD, Steiner RA, Palmiter RD, and McKnight GS (2015). Fertility-regulating Kiss1 neurons arise from hypothalamic POMC-expressing progenitors. J Neurosci 35, 5549–5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz E, Yang L, Su T, Morris DR, McKnight GS, and Amieux PS (2009). Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proc Natl Acad Sci U S A 106, 13939–13944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer DP, Lehrman EK, and Stevens B (2013). The “quad-partite” synapse: microglia-synapse interactions in the developing and mature CNS. Glia 61, 24–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd GM, and Grillner S (2010). Handbook of brain microcircuits. Oxford University Press, p xvii. [Google Scholar]
- Sherman-Gold R (2006). The axon CNS guide guide to electrophysiology and biophysics laboratory techniques (2nd edition). Molecular Devices Corporation. [Google Scholar]
- Shigetomi E, Bushong EA, Haustein MD, Tong X, Jackson-Weaver O, Kracun S, Xu J, Sofroniew MV, Ellisman MH, and Khakh BS (2013). Imaging calcium microdomains within entire astrocyte territories and endfeet with GCaMPs expressed using adeno-associated viruses. J Gen Physiol 141, 633–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigetomi E, Patel S, and Khakh BS (2016). Probing the Complexities of Astrocyte Calcium Signaling. Trends Cell Biol 26, 300–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan SA, and Barres BA (2014). Looks can be deceiving: reconsidering the evidence for gliotransmission. Neuron 84, 1112–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith NA, Kress BT, Lu Y, Chandler-Militello D, Benraiss A, and Nedergaard M (2018). Fluorescent Ca2+ indicators directly inhibit the Na,K-ATPase and disrupt cellular functions. Sci Signal January 30;11(515). pii: eaal2039. doi: 10.1126/scisignal.aal2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan R, Huang BS, Venugopal S, Johnston AD, Chai H, Zeng H, Golshani P, and Khakh BS (2015). Ca(2+) signaling in astrocytes from Ip3r2(−/−) mice in brain slices and during startle responses in vivo. Nat Neurosci 18, 708–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan R, Lu T-Y, Chai H, Xu J, Huang BS, Golshani P, Coppola G, and Khakh BS (2016). New Transgenic Mouse Lines for Selectively Targeting Astrocytes and Studying Calcium Signals in Astrocyte Processes In Situ and In Vivo. Neuron 92, 1181–1195: PMID: 27939582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele AD, Jackson WS, King OD, and Lindquist S (2007). The power of automated high-resolution behavior analysis revealed by its application to mouse models of Huntington’s and prion diseases. Proc Natl Acad Sci U S A 104, 1983–1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stobart JL, Ferrari KD, Barrett MJP, Stobart MJ, Looser ZJ, Saab AS, and Weber B (2016). Long-term In Vivo Calcium Imaging of Astrocytes Reveals Distinct Cellular Compartment Responses to Sensory Stimulation. Cereb Cortex Nov 19:1–15. doi: 10.1093/cercor/bhw366 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Strehler EE (2015). Plasma membrane calcium ATPases: From generic Ca(2+) sump pumps to versatile systems for fine-tuning cellular Ca(2+). Biochem Biophys Res Commun 460, 26–33. [DOI] [PubMed] [Google Scholar]
- Tong X, Ao Y, Faas GC, Nwaobi SE, Xu J, Haustein MD, Anderson MA, Mody I, Olsen ML, Sofroniew MV, et al. (2014). Astrocyte Kir4.1 ion channel deficits contribute to neuronal dysfunction in Huntington’s disease model mice. Nat Neurosci 17, 694–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volterra A, Liaudet N, and Savtchouk I (2014). Astrocyte Ca(2)(+) signalling: an unexpected complexity. Nat Rev Neurosci 15, 327–335. [DOI] [PubMed] [Google Scholar]
- Wójtowicz AM, Dvorzhak A, Semtner M, and Grantyn R (2013). Reduced tonic inhibition in striatal output neurons from Huntington mice due to loss of astrocytic GABA release through GAT-3. Front Neural Circuits November 26;7:188. doi: 10.3389/fncir.2013.00188. eCollection 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Guo Z, Gearing M, and Chen GG (2014). Tonic inhibition in dentate gyrus impairs long-term potentiation and memory in an Alzheimer’s [corrected] disease model. Nat Commun Jun 13;5:4159. doi: 10.1038/ncomms5159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Wang T, Sun GY, and Ding S (2010). Specific disruption of astrocytic Ca2+ signaling pathway in vivo by adeno-associated viral transduction. Neuroscience 170, 992–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O’Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N, et al. (2014). An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci 34, 11929–11947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YV, Ormerod KG, and Littleton JT (2017). Astrocyte Ca2+ Influx Negatively Regulates Neuronal Activity. eNeuro March 10;4(2). pii: ENEURO.0340–16.2017. doi: 10.1523/ENEURO.0340-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.